Abstract
Background
Occurrences of pathogens in environmental and irrigation waters, as well as the use of inadequately treated sewage for fresh produce constitute potential public health threats worldwide.
Objective
To investigate the treated wastewater used in fresh produce irrigation in Nsuskka, Southeastern Nigeria, as a reservoir enterotoxigenic and multidrug-resistant Escherichia coli.
Methods
Treated wastewater (from the sewage treatment facility at Nsukka, Southeast Nigeria), soil and irrigated vegetable samples were collected and analyzed using standard procedures. Escherichia coli isolated from the samples were screened for the presence of enterotoxigenic E. coli strain encoding lt gene and profiled for antibiotic resistance using the conventional PCR and standardized agar disk diffusion assays respectively.
Results
Of the total presumptive 103 isolates, PCR detected uidA gene in 87 (84 %), of which 23 (26 %) harboured the lt encoding ETEC gene. Generally, imipenem, cefuroxime and norfloxacin proved to be most effective of all the antibiotics employed. Wastewater isolates were variously susceptible to ciprofloxacin (95 %), norfloxacin (95 %), cefuroxime (93 %), chloramphenicol (93 %), trimethoprim and tetracycline (88 %), soil isolates to streptomycin (75 %) and vegetable isolates to cefuroxime (90 %), norfloxacin (86 %), ciprofloxacin (81 %) and chloramphenicol. Contrariwise, high resistances observed to other antibiotics were in the order; ampicillin (95 %), penicillin (93 %), erythromycin (90 %) and clarithromycin (83 %) among wastewater isolates, ciprofloxacin and norfloxacin (75 %) in soil isolates; penicillin, vancomycin and erythromycin (98 %), rifampicin and clarithromycin (93 %), sulphamethoxazole (83 %), ampicillin (81 %), tetracycline and imipenem (76 %), trimethoprim (72 %) and amoxicillin (71 %) among vegetable isolates, with multidrug resistance patterns ranging from three to seventeen.
Conclusions
Our results reveal the treated wastewater as a reservoir of enterotoxigenic E. coli as well as multidrug resistance that may pose a health hazard for humans and animals when released to the natural environment. Hence, there is need to develop management strategies and ensure compliance in order to prevent water-borne diarrhoea caused by ETEC and reduce the menace of antibiotic resistance in the environment.
Keywords: Microbiology, Environmental science, Enterotoxigenic E. coli, Multidrug resistance, Irrigation, Wastewater, Public health
Microbiology; Environmental science; Enterotoxigenic E. coli; Multidrug resistance; Irrigation; Wastewater; Public health
1. Introduction
Treated wastewater serves as an alternative source to scarce quality water, thus reducing excessive application of fertilizer drastically (Martinez et al., 2013). However, poorly operated plants and grossly inadequate disinfection processes can enable various pathogenic bacteria survive or even multiply in treated wastewater effluents discharged into aquatic and soil environments (Abbas et al., 2017; Christou et al., 2017; Mukatea et al., 2018). The application of such effluents or contaminated water bodies for fresh produce irrigation constitutes potential public health risks worldwide and the potential for producing contamination using untreated wastewater for irrigation of crops increases, especially in the developing world (United Nations Educational, Scientific and Cultural Organization, 2003; Thebo et al., 2017). Vegetables form an indispensable constituent of human diet and the consumption of contaminated vegetables has been attributed to disease outbreaks (Lynch et al., 2009; Berger et al., 2010), with concomitant heightening in the contamination of farm yield resulting in subsequent outbursts of food-borne illnesses (Chigor et al., 2010; Allende and Monaghan, 2015).
Enterobacteriaceae constitute integral part of the normal microflora of animals and humans. Escherichia coli, as one of the famous member species of this family, can disperse widely through faecal materials and wastewaters in diverse environments including soil, vegetables, among others (Bain et al., 2014). Globally, E. coli is a significant agent of food and water-borne illnesses in humans (Duffy, 2003). Diarrhoeagenic E. coli (DEC) strains are essential causes of diarrhoea among children in many developing nations, and have been established as emerging enteropathogens (Okeke et al., 2003; Nweze, 2009; Bielaszewska et al., 2011). They have been grouped into six which include enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enterohaemorrhagic E. coli (EHEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC). Others including cell-detaching E. coli (CDEC) and necrotoxic E. coli (NTEC) have been also documented (Hamelin et al., 2007; Guion et al., 2008). Of all, ETEC is the most frequently detected enteric bacterial pathogens among infants usually below 5 years (Ahmed et al., 2005; Walk et al., 2007; Nweze, 2009). Often times, it is the first enteric infection experienced by infants in low income economies, and in endemic regions almost all children will have had one ETEC diarrhoea episode in their first year of life, and one out of every six travellers to these areas has been observed to be infected with ETEC (Centres for Disease Control, 2004; Steffen et al., 2005).
The excessive and inappropriate usage of antimicrobials in preventing or treating human and veterinary bacterial infectious diseases has led to increased antimicrobial and multidrug resistance, yet no concomitant intervention (Freire-Moran et al., 2011; Garbisu et al., 2018). The extensive use of antimicrobials in animal and human therapy, subsequent release of treated or untreated wastewater into the environment, continuous application of recycled water in agricultural practices and agricultural runoff, may pose direct consequences for indigenous microbiomes, especially in freshwater milieus (Vaz-Moreira et al., 2014; Manaia et al., 2016). Prior investigations have reported the injurious effects of antibiotics on the environment, bacterial inhabitants, biogeochemical procedures and organic pollutants’ degradation (Proia et al., 2013; Roose-Amsaleg and Laverman, 2016; Zhang et al., 2019). Antimicrobial residues present in the environment have been found attributable to hospital use and subsequent release into effluents, then treated in WWTPs (Amos et al., 2014; Ben et al., 2019). Treated effluents, alongside any antimicrobial, is discharged into the environment and may encourage development of resistant bacterial strains (Korzeniewska et al., 2013; Pan and Chu, 2018; Ben et al., 2019).
In many developing nations, both low and middle income, including Nigeria, farmers in urban and semi-urban settlements usually obtain waters for irrigating their crops via different wastewater sources, and the associated dangers in agricultural practices is heightened by the proliferation and persistence of DEC in those waters. Its spread to vegetables and subsequent incorporation into the tissues of plants that shield the pathogens from the effects of sanitizers and disinfectants have been documented (Chigor et al., 2010; Titilawo et al., 2015a). For over 3 decades, the Nsukka wastewater treatment plant (WWTP) has been reliable and consistent in providing waters for irrigation purpose mainly for cultivation of vegetables. Recently, Echiegu et al. (2016) evaluated the physicochemical statuses of effluents from the wastewater treatment plant for possible irrigation suitability but excluded the microbial quality status of the treated sewage. Therefore, in this study for the first time, final effluent, irrigated soil and vegetable samples from Nsukka WWTP were assessed for the likely detection of enterotoxigenic and multidrug-resistant Escherichia coli.
2. Methods
2.1. Description of study area
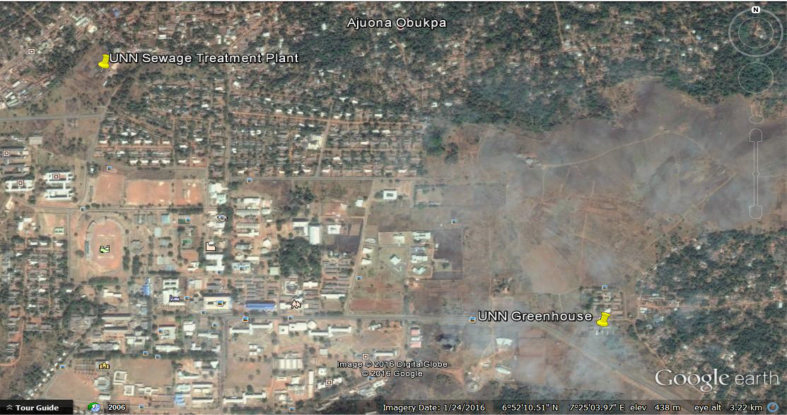
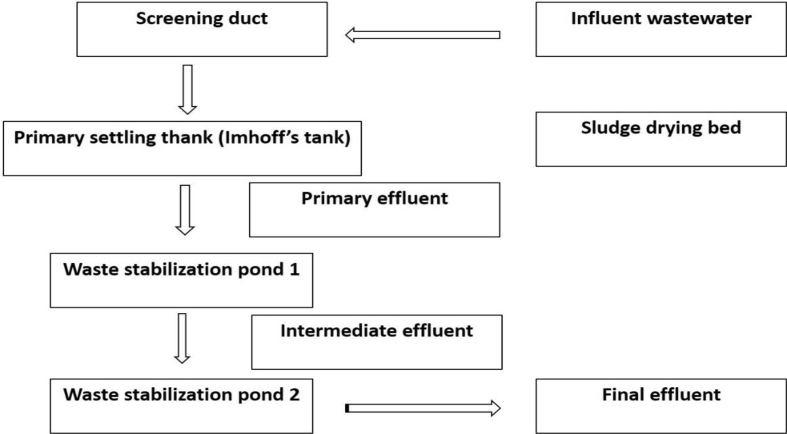
The sewage treatment facility in Nsukka is situated at the northwest end, about 500m away from the Junior Staff residences, of the University of Nigeria, Nsukka (Figure 1). The WWTP consists of a screen, primary settling (Imhoff) tank, sludge drying beds and two oxidation ponds (Figure 2). The treatment procedures invlove channeling domestic sewage through pipes and screens directly into the primary treatment tank and from there enters the first oxidation pond for the production of intermediate effluents. These effluents undergo further treatment by allowing them pass through the second oxidation pond to yield the final effluents utilized for irrigation purpose by some members of the community (Echiegu et al., 2016). The major crops cultivated during the dry season include the green vegetable (Amaranthus spp) and fluted pumpkin (Telfaria occidentalis). The corrugated system of furrow irrigation is widely practised whereby wastewater is transported from the treatment sites to the corrugation by gravity through diversions and by direct lifting of effluents from the source using metal and plastic buckets or pails (Echiegu et al., 2016).
Figure 1.
Aerial view of University of Nigeria, Nsukka campus and its environment.
Figure 2.
Schematic illustration of the Nsukka wastewater treatment plant.
2.2. Collection of wastewater effluent, soil and irrigated vegetable samples
Wastewater samples were collected using sterile wide-mouthed, screw-capped 250-ml bottles. In the same vein, composite surface soils (0–20 cm) were collected from each of the earthen pots on which the test crops were grown individually into sterile sampling bottles.
Amaranths grown in earthen pots in the Soil Science Departmental greenhouse and the vegetables were irrigated twice daily, using the sprinkler method. After six weeks of cultivation, vegetables were collected from all earthen pots into sterile beakers. All samples were conveyed on ice to the Microbiology Postgraduate laboratory of the institution and processed within 6 h of collection. All trials were performed in triplicates.
2.3. Isolation and presumptive identification of E. coli from wastewater, soil and irrigated vegetable samples
Water samples were analysed following Standard Methods (American Public Health Association, 2012). Exactly 100 ml aliquots of the water samples were filtered through a 90-mm diameter, 0.45-μm pore-sized membrane filters (Millipore, Ireland). The filters were incubated overnight at 44.5 °C on eosin methylene blue (EMB) agar (Oxoid, UK) plates. Similarly, soil samples were serially diluted (1/10) in a normal physiological saline (0.9 M NaCl) and 0.1 ml of the diluted suspension spread on to EMB agar plates using sterile glass rods. Also, 2.5 g each of vegetable samples was weighed, washed and homogenized in 45 ml of sterile distilled water using an electric blender. Aliquots of the extract were then streak-plated on EMB agar plates and incubated at 44 °C for 24 h. Characteristic metallic-sheen colonies were picked and purified on E. coli chromogenic agar (Conda Pronadisa, Spain) plates before preserving on 50 % glycerol for further studies.
2.4. DNA extraction and molecular identification of E. coli isolates and enterotoxigenic E. coli strain
Genomic DNA was isolated from all presumptive E. coli using boiling method as previously described (Chapman et al., 2006). Briefly, a single colony was inoculated onto 2 ml LB broth and incubated at 37 °C with moderate shaking (100 rpm) overnight. The culture was then centrifuged at 13,000 rpm, the supernatant was removed, and the pellet re-suspended in 200 μl of sterile distilled water, followed by heating at 100 °C for 10 min. After centrifugation, the supernatant which is the DNA was carefully transferred into a sterile 1.5 ml Eppendorf tube and stored at -20 °C for further analysis. The PCR reaction mixtures consisted of 25 μl of PCR Master Mix (Thermo Scientific, (EU) Lithuania), 0.5 μl each of oligonucleotide primers (Inqaba Biotech, Pretoria, South Africa), 10 μl of template DNA and 14 μl of nuclease free water constituting a total reaction volume of 50 μl. The PCR cycling conditions were in conformity with the protocols prescribed elsewhere (Bej et al., 1991; Lopez-Saucedo et al., 2003), with slight modifications though. The oligonucleotide primers used, target genes and expected amplification products are listed in Table 1.
Table 1.
Primers used for the detection of E. coli and ETEC.
| Genus/strain | Target gene | Oligonucleotide sequence (5′→3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| E. coli | uidA | F: AAAACGGCAAGAAAAAGCAG R: ACGCGTGGTTAACAGTCTTGCG |
147 | Bej et al. (1991) |
| ETEC | lt | F: GGCGACAGATTATACCGTGC R: CGGTCTCTATATTCCCTGTT |
450 | Lopez-Saucedo et al. (2003) |
2.5. Controls and product visualization
Both positive and negative controls were included in each PCR reaction. Reference strains ATCC 25922 (ATCC, USA) and DSM 10973 (DSMZ, Germany) were used for E. coli genus identification and enterotoxigenic E. coli strain detection respectively. Samples were labelled positive for a specific gene when the visible band had the same size of the positive control DNA. Exactly 5 μl aliquots of the amplicons were analyzed on a 1 % horizontal agarose gel (Merck, SA) for 45 min at 100 V in 0.5X TBE buffer and stained with ethidium bromide (Sigma-Aldrich, USA), using the gel documentation system (Alliance 4.7, France). Identification of the bands was established by comparing the band sizes with 100-bp molecular-weight size marker (Thermo Scientific, (EU) Lithuania).
2.6. Antibiotic susceptibility testing of E. coli isolates
Susceptibility of E. coli isolates to selected antimicrobials was carried out by the standardized disk diffusion assay of Kirby-Bauer et al. (1966). Briefly, isolates grown on nutrient agar were suspended into normal saline with the aid of sterile wire loop until the turbidity equalled 0.5 MacFarland standard. Sterile non-toxic cotton swabs were used to streak the entire surface of Mueller-Hinton agar plates and antibiotic discs were applied aseptically on Muller-Hinton incubated for 18 h at 37 °C using antibiotic dispenser (Mast Diagnostics, UK). Selection of antibiotic discs was performed according to the guidelines previously recommended (Clinical and Laboratory Standards Institute, 2015). Exactly 18 different antibiotics belonging to 11 antimicrobial families together with their concentrations used are as follows: [Beta-lactams: amoxicillin (10 μg), ampicillin (5 μg), penicillin (10 U) and cloxacillin (5 μg)]; [Cephems: cefuroxime (30 μg)]; [Aminoglycosides: streptomycin (10 μg)]; [Ansamycins: rifampicin (5 μg)]; [Folate pathway inhibitors: metronidazole (50 μg), sulphamethoxazole (25 μg) and trimethoprim (5 μg)]; [Glycopeptides: vancomycin (30 μg) and erythromycin (15 μg)]; [Macrolides: clarithromycin (15 μg)]; [Phenicols: chloramphenicol (30 μg)]; [Fluoroquinolones: ciprofloxacin (5 μg) and norfloxacin (10 μg)]; [Tetracyclines: tetracycline (30 μg)]; [Carbapenems: imipenem (10 μg)] (Mast Diagnostics, UK). All plates were incubated at 35 °C for 18 h. Zones showing complete inhibitions around the discs were observed, recorded and interpreted as resistant (R), intermediate (I) and susceptible (S) accordingly (Clinical and Laboratory Standards Institute, 2015). E. coli ATCC 25922 (ATCC USA) was used as negative control.
2.7. Multi-antibiotic resistance phenotypes and indexing of E. coli isolates
Multi-antibiotic resistance phenotypes of E. coli isolates were evaluated and mapped out accordingly. The multiple antibiotic resistance index (MARI) of isolated was mathematically expressed as:
| MARindex = a/b; |
“a” represents the number of antibiotics to which the isolates are resistant whereas “b” is the total number of antibiotics exposed (Titilawo et al., 2015a).
2.8. Data analysis
Data analysis was performed following the statistical package for the social sciences [IBM version 24]. One way ANOVA was performed to determine the variation in resistances and susceptibilities among the isolates with respect to sampling locations and a P value of 0.05 was used to declare significance.
3. Results
3.1. Presumptive identification, PCR confirmation of E. coli isolates and detection of ETEC strain
A total of 103 E. coli isolates, involving 55, 4 and 44 from wastewater, soil and irrigated vegetables respectively were obtained, of which 87 of them harboured the housekeeping uidA gene, ascertaining their identity. While 41 and 42 isolates were uidA positive for wastewaters and irrigated vegetables respectively, all 4 soil isolates had the gene (Table 2). Figure 3 shows the PCR products of E. coli confirmation by the uidA gene amplification. Likewise, the PCR-identified 87 E. coli isolates were further screened for probable detection of lt gene encoding ETEC, only 23 (26 %) isolates consisting of 12 (52 %) from wastewater, 1 (4 %) from soil and 10 (44 %) from irrigated vegetables harboured the gene (Table 2). Figure 4 shows the PCR products of the lt gene amplification.
Table 2.
Presumptive identification, PCR confirmation of E. coli and detection of ETEC isolates.
| Sample type | No. of presumptive E. coli isolates | No. of isolates positive for uidA gene | No. of isolates positive for lt gene |
|---|---|---|---|
| Wastewater | 55 | 41 | 12 |
| Soil | 4 | 4 | 1 |
| Irrigated vegetable | 44 | 42 | 10 |
| Total | 103 | 87 | 23 |
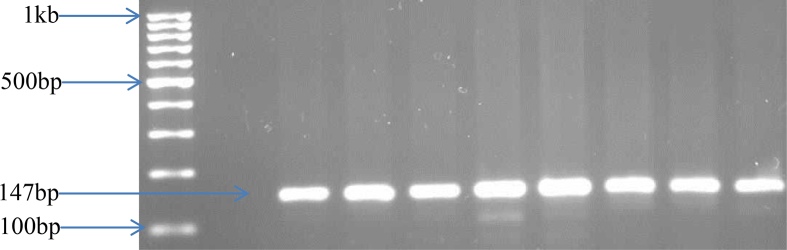
Figure 3.
PCR products for E. coli confirmation by uidA gene amplification. Legend: MWM: Molecular-weight marker; NC: Negative control; PC: Positive control; Lanes 1–7: Positive isolates. See Figure S1 for full blot.
Figure 4.
PCR products for ETEC strain confirmation by lt gene amplification. Legend: MWM: Molecular weight marker; PC: Positive control; NC: Negative control; Lanes 1–7: Positive isolates. See Figure S2 for full blot.
3.2. Antimicrobial susceptibility profiles of E. coli isolates
All 87 E. coli isolates were elucidated for susceptibility to 18 antimicrobial agents with noticeable varying degrees of susceptibility towards antimicrobials tested. While all were susceptible to imipenem, only soil isolates were susceptible to cefuroxime and chloramphenicol (Figures 5 and 6). Furthermore, wastewater isolates were variously susceptible to ciprofloxacin and norfloxacin (95 %), cefuroxime and chloramphenicol (93 %), trimethoprim and tetracycline (88 %); soil isolates to streptomycin (75 %) and irrigated vegetable isolates to cefuroxime (90 %), norfloxacin (86 %), ciprofloxacin (81 %) and chloramphenicol (79 %) (Figures 5, 6, and 7).
Figure 5.
Percentage frequency of antibiotic susceptibility patterns of wastewater isolates (n = 41).
Figure 6.
Percentage frequency of antibiotic susceptibility patterns of soil isolates (n = 4).
Figure 7.
Percentage frequency of antibiotic susceptibility patterns of irrigated vegetable isolates (n = 42).
On the contrary, all isolates from the three sources were resistant to cloxacillin and metronidazole. While all isolates from wastewater and vegetables were resistant to rifampicin and vancomycin treatments (Figures 5 and 7), all soil isolates displayed resistance to amoxicillin, ampicillin, penicillin, sulphamethoxazole, trimethoprim, erythromycin, clarithromycin and tetracycline (Figure 6). High resistances observed against other antibiotics were in the order; ampicillin (95 %), penicillin (93 %), erythromycin (90 %) and clarithromycin (83 %) among wastewater isolates; ciprofloxacin and norfloxacin (75 %) in soil isolates (Figure 4); penicillin, vancomycin and erythromycin 98 %, rifampicin and clarithromycin (93 %), sulphamethoxazole (83 %), ampicillin (81 %), tetracycline and imipenem (76 %), trimethoprim (72 %) and amoxicillin (71 %) among irrigated vegetable isolates (Figure 7).
3.3. Multi-antimicrobial resistance phenotypes of E. coli isolates
The percentage of resistant-E. coli isolates to five or more antimicrobials was noticed to be highest in soil (100 %), afterwards irrigated vegetables (90 %) and wastewater (83 %), with an overall MAR index of above 0.2. Table 3 depicts the resistance pattern to ≥ three antimicrobials, number of isolates and multiple antibiotic resistance indices with respect to sampling sites.
Table 3.
Multidrug resistance phenotypes of E. coli isolates from wastewater treatment plant, soils and vegetables.
| No. of antimicrobials | Resistance pattern∗ | No. of isolates | MARI |
|---|---|---|---|
| Wastewater isolates (n = 21) | |||
| 4 | COX, RD, MTZ, VA | 5 | 0.3 |
| 5 | P, COX, MTZ, VA, E | 3 | 0.3 |
| 7 | AMP, P, COX, RD, MTZ, VA, CLA | 1 | 0.4 |
| 8 | P, COX, RD, MTZ, SMZ, VA, E, CLA | 1 | 0.4 |
| 10 | AMX, AMP, P, COX, RD, MTZ, SMZ, VA, E, CLA | 4 | 0.5 |
| 11 | AMP, P, COX, RD, MTZ, SMZ, TMP, VA, E, CLA, TE | 1 | 0.6 |
| 13 | AMX, AMP, P, COX, S, RD, MTZ, SMZ, TMP, VA, E, CLA, TE | 2 | 0.7 |
| 14 | AMX, AMP, P, COX, RD, MTZ, SMZ, TMP, VA, E, CLA, CIP, NOR, TE | 1 | 0.7 |
| 16 | AMX, AMP, P, COX, CXM, S, RD, MTZ, SMZ, TMP, VA, E, CLA, CIP, NOR, TE | 2 | 0.9 |
| 17 | AMX, AMP, P, COX, CXM, S, RD, MTZ, SMZ, TMP, VA, C, E, CLA, CIP, NOR, TE | 1 | 0.9 |
| Soil isolates (n=4) | |||
| 12 | AMX, AMP, P, COX, RD, MTZ, SMZ, TMP, VA, E, CLA, TE | 2 | 0.7 |
| 15 | AMX, AMP, P, COX, S, RD, MTZ, SMZ, TMP, VA, E, CLA, CIP, NOR, TE | 2 | 0.8 |
| Vegetable isolates (n=29) | |||
| 3 | AMP, COX, MTZ | 6 | 0.2 |
| 3 | COX, MTZ, E | 2 | 0.2 |
| 4 | COX, MTZ, CIP, NOR | 3 | 0.2 |
| 4 | COX, CXM, MTZ, C | 2 | 0.2 |
| 5 | AMX, COX, CXM, MTZ, C | 3 | 0.3 |
| 6 | COX, CXM, S, RD, MTZ, TE | 1 | 0.4 |
| 6 | COX, CXM, MTZ, C, CIP, NOR | 2 | 0.4 |
| 7 | AMP, P, COX, RD, MTZ, VA, E | 1 | 0.4 |
| 8 | AMP, P, COX, S, RD, MTZ, VA, E | 1 | 0.4 |
| 9 | AMP, P, COX, MTZ, SMZ, VA, E, CLA, TE | 2 | 0.5 |
| 10 | P, COX, CXM, RD, MTZ, SMZ, VA, C, E, CLA | 1 | 0.5 |
| 12 | AMP, AMX, P, COX, RD, MTZ, SMZ, TMP, VA, E, CLA,TE | 1 | 0.6 |
| 14 | AMP, AMX, P, COX, S, RD, MTZ, SMZ, TMP, VA, C, E, CLA, TE | 2 | 0.7 |
| 16 | AMP, AMX, P, COX, S, RD, MTZ, SMZ, TMP, VA, C, E, CLA, CIP, NOR, TE | 1 | 0.8 |
| 17 | AMP, AMX, P, COX, S, RD, CXM, MTZ, SMZ, TMP, VA, C, E, CLA, CIP, NOR, TE | 1 | 0.9 |
Symbols: AMX: Amoxicillin; AMP: Ampicillin, P: Penicillin, COX Cloxacillin, CXM: Cefuroxime, S: Streptomycin, RD: Rifampicin, MTZ: Metronidazole, SMZ: Sulphamethoxazole, TMP: Trimethoprim, C: Chloramphenicol, E: Erythromycin, VA: Vancomycin, NOR: Norfloxacin, CIP: Ciprofloxacin, IPM: Imipenem, TE: Tetracycline, CLA: Clarithromycin.
3.4. Statistical analysis
The one way ANOVA revealed that susceptibilities of cefuroxime, ciprofloxacin, norfloxacin and imipenem were significantly associated with ampicillin, penicillin, rifampicin, metronidazole, sulphamethoxazole, vancomycin, erythromycin and clarithromycin (P < 0.05), whereas amoxicillin, streptomycin, trimethoprim and tetracycline revealed no significant susceptibilities to other antimicrobials tested (P > 0.05). Likewise, it was observed that the resistances of amoxicillin, ampicillin, streptomycin, sulphamethoxazole, trimethoprim, clarithromycin, chloramphenicol, ciprofloxacin, norfloxacin and tetracycline did not significantly differ (P > 0.05), whereas cefuroxime and imipenem varied significantly in their resistances to other antimicrobials screened (P < 0.05).
4. Discussion
The occurrence of pathogens in environmental and irrigation waters has become an ongoing global concern (Anastasi et al., 2012; Titilawo et al., 2015a), and the incidence of gastroenteritis outbreaks caused by food-borne pathogens after ingestion of uncooked vegetables has greatly increased (Lynch et al., 2009; Berger et al., 2010). This study sought to profile enterotoxigenic E. coli strains and multidrug E. coli isolates from wastewater, soil and irrigated vegetables. The E. coli isolates were PCR-confirmed using the housekeeping uidA gene. Recovery of E. coli in this study suggests that the samples have been subjected to faecal pollution due to poor sanitation, improper disposal of sewage, surface runoff and leakage from nearby defective sewage, contaminated ground and waste waters possibly (Igbinosa and Okoh, 2009; Odjadjare and Okoh, 2010; Titilawo et al., 2015a). The existence of E. coli in waters is commonly employed to signal faecal contamination and microbial water quality evaluation (World Health Organization, 2006; Sazakli et al., 2007).
Likewise, heat-labile (lt) toxin encoding ETEC pathogroup was variously detected among the samples. Of the 23 ETEC isolates harbouring the gene, 12 (52 %), 1 (4 %) and 10 (44 %) were from wastewater, soil and vegetables respectively (Table 2). It has been noted earlier that leakages from sewage lines and discharge of animal and human excreta discharged into open drains are responsible for the contamination sources of pathogenic E. coli in defective drinking water supplies (Ram et al., 2008b; Titilawo et al., 2015c). ETEC is the most important but under-recognized bacterial cause of diarrhoea or cholera-like disease in all age groups especially in areas with population pressure, poor sanitation and inadequate drinking water (Kaper et al., 2004). It induces watery diarrhoea in humans, distressing mostly infants and travelers (Turner et al., 2006). The strains, aside being extensively responsible for millions of infection cases, are significant pathogens linked to mortality from severe infantile diarrhea worldwide (Luo et al., 2014). The lt gene in ETEC strains associated has been frequently detected among those recovered from surface waters of India and other South Asian countries polluted by human faeces (Begum et al., 2005; Ram et al., 2008c). Similarly, our findings on ETEC detection variously corroborates the findings of some previous studies on strains from sewage treatment plant's final effluents (Anastasi et al., 2012) and untreated surface waters used in potable water production (Begum et al., 2005; Ram et al., 2007; Lothigius et al., 2008; Singh et al., 2010).
The presence of pathogenic E. coli in environmental waters poses impending health dangers in animals and humans especially since water is used for drinking, irrigation and recreations (Koczura et al., 2012). Several studies on incidences of virulence gene properties of E. coli have dealt extensively with isolates of clinical and surface water origin, thereby making it difficult to compare and contrast between our findings and previous investigations, especially soils and vegetables. Frequent detections of potential E. coli pathotypes from clinical settings in Mexico (Estrada-Garcia et al., 2005), diarrhoeal stool isolates from Southeast Nigeria (Nweze, 2009), river water E. coli isolates of Osun State, Southwest Nigeria (Titilawo et al., 2015c), Kat river and Fort Beaufort abstraction water in South Africa (Nontongana et al., 2014), marine recreational water in the USA (Hamilton et al., 2010), Minjiang river in China (Chen et al., 2011), surface water in Brisbane, Australia (Sidhu et al., 2013) and Warta river in Poland (Koczura et al., 2013) have been properly documented.
The spread of antibiotic resistance phenomena has drawn increasing attention in recent years, because various infectious diseases that were once considered susceptible have steadily begun to be resistant to antimicrobial therapy, thus dashing man's hope in recovering from ailments (Titilawo et al., 2015b). In the present survey, antimicrobial resistance analysis reveals that all the strains were resistant to sulphamethoxazole and displayed a significantly high resistance to ampicillin, amoxycillin, gentamycin, cefuroxime, tetracycline and chloramphenicol (Figure 5). Our findings concur with previous reports on the high level of resistance among isolates from waters (Chigor et al., 2010; Titilawo et al., 2015a). This statement is premised on the definition of MDR that a strain can demonstrate resistance to three or more antimicrobials (Doyle et al., 2013). The occurrence of antimicrobial-resistant E. coli was equally noticed in some reports on animal and human faeces, wastewater treatment plants and surface waters (Mokracka et al., 2011; Sun et al., 2012).
Similarly, high resistances against β-lactams and cephalosporins were observed, though somewhat higher than those reported elsewhere (Schroeder et al., 2002; Mohanta and Goel, 2014). Recently, trimethoprim-sulfamethoxazole and quinolones have been alternatively prescribed in place of tetracycline as empirical therapy for community acquired UTIs, owing to the emergence of E. coli resistant strains (Karlowsky et al., 2002). Similarly, high resistances against ampicillin, trimethoprim-sulfamethoxazole, chloramphenicol, gentamicin and tetracycline have been observed in E. coli strains from drinking waters in Jordan (Shehabi et al., 2006). The current study reveals multidrug resistance against several antimicrobials ranging from three to seventeen. Overall, the MARPs of all the E. coli isolates include three drugs (8), four drugs (12), five drugs (5), six drugs (3), seven drugs (2), eight drugs (2) and nine drugs (2), ten drugs (5), eleven drugs (1), twelve drugs (3), thirteen drugs (2), fourteen drugs (3), fifteen drugs (2), sixteen drugs (3) and seventeen drugs (2) resistances (Table 3). The differences in antimicrobial agent resistance patterns might have been resulted from exposure to different agents which can be used to differentiate water contamination sources (Graves et al., 2002; Guan et al., 2002; Titilawo et al., 2015a). Also, multidrug resistant E. coli strains have been frequently recovered from surface and ground waters in KwaZulu-Natal and North-West Provinces of South Africa respectively (Olaniran et al., 2009; Wose et al., 2010).
A MAR index of 0.2 is used to distinguish between low and high-risk contamination sources (Krumperman, 1983). The calculated MARI values obtained in this study reflect high exposure to antibiotics as they exceeded the threshold value of 0.2, suggesting contamination from high-risk origin (Table 3). MAR indices arbitrarily estimate the relative abundance of resistant E. coli stains in the environment. The major concern in our findings was that the isolates showed high resistance to ≥3 antibiotics with an overall MAR index >0.2. Multiple antibiotic resistance in bacteria is generally connected to plasmids containing one or more resistance determinants, each encoding a single phenotype of antibiotic resistance, and multiple bacterial resistance to drugs had earlier been reported (Daini et al., 2005; Baker et al., 2018). The relatively high antimicrobial-resistant E. coli isolates in our study corresponds with reports previously documented (Rakic-Martinez et al., 2011; Puah et al., 2013). Multi-antimicrobial resistance has the tendency to breed newly emerging resistant bacteria, which may be spread to consumers, triggering infections that are very hard to cure. The observed high resistance frequency may not only result in the treatment letdown, but equally poses risk of infection with pathogens to all and sundry using the waters (Schmidt et al., 2001). Acquisition of resistance can also occur via uptake of resistance determinants through conjugation, transduction and transformation transference mechanisms of gene material (Zhu et al., 2017; Chen et al., 2018; Zhang et al., 2019). Anthropogenic-motivated selective burdens may be contributing to the distribution of antimicrobial resistant bacteria, and resistance genes in hospital settings (Titilawo et al., 2015b; Xu et al., 2015).
5. Conclusions
Antimicrobial resistance in E. coli is a key indicator of the emerging resistant microbes in diverse milieus. This study, the first of its kind, investigated the prevalence of ETEC strains and elucidated the multidrug resistance patterns among E. coli isolates from sewage treatment facility in Nsukka, Nigeria. The study concludes that the treated sewage, irrigated soils and vegetables harbour ETEC strains as detected by the conventional PCR assays, signifying that the waters are unsuitable for domestic and irrigation. Our demonstration clearly indicates that the percentage of resistance to antibiotics including cloxacillin, metronidazole, rifampicin, vancomycin, amoxicillin, ampicillin, penicillin sulfamethoxazole and clarithromycin was significantly high, suggesting wastewater treatment plant as a chief reservoir of multi-antimicrobial resistant E. coli strains, which could consequently pose a significant public health threat. Hence, there is need for regular monitoring of the sewage treatment plant for compliance in order to prevent water-borne diarrhoea caused by ETEC, effective intervention strategies, wide-ranging and confined antimicrobial resistance surveillance to lessen multidrug resistance in the environments.
Declarations
Author contribution statement
Vincent Chigor: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ini-Abasi Ibangha: Performed the experiments; Wrote the paper.
Chinyere Chigor: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yinka Titilawo: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors appreciate the University of Nigeria, Nsukka, Enugu State, Nigeria for supporting the study.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abbas Q., Yousaf B., Liu G., Ali M.U., Munir M.A.M., Zia-ur-Rehman M., Husain S.A. Evaluating the health risks of potentially toxic elements through wheat consumption in multi-industrial metropolis of Faisalabad, Pakistan. Environ. Sci. Pollut. Res. 2017;24:26646–26657. doi: 10.1007/s11356-017-0311-9. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Neller R., Katouli M. Host species-specific metabolic fingerprint database for enterococci and Escherichia coli and its application to identify sources of faecal contamination in surface waters. Appl. Environ. Microbiol. 2005;71:4461–4468. doi: 10.1128/AEM.71.8.4461-4468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende A., Monaghan J. Irrigation water quality for leafy crops: a perspective of risks and potential solutions. Int. J. Environ. Res. Publ. Health. 2015;12:7457–7477. doi: 10.3390/ijerph120707457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos G.C.A., Zhang L., Hawkey P.M., Gaze W.H., Wellington E.M. Functional metagenomic analysis reveals rivers are a reservoir for diverse antibiotic resistance genes. Vet. Microbiol. 2014;171:441–447. doi: 10.1016/j.vetmic.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Anastasi E.M., Matthews B., Stratton H.M., Katouli M. Pathogenic Escherichia coli found in sewage treatment plants and environmental waters. Appl. Environ. Microbiol. 2012;78:5536–5541. doi: 10.1128/AEM.00657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Public Health Association . twenty-second ed. APHA; Washington, DC: 2012. Standard Methods for the Examinations of Water and Wastewater. [Google Scholar]
- Bain R., Cronk R., Hossain R., Bonjour S., Onda K., Wright J. Global assessment of exposure to faecal contamination through drinking water based on a systematic review. Trop. Med. Int. Health. 2014;19:917–927. doi: 10.1111/tmi.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S., Thomson N., Weill F.X., Holt K.E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018;360:733–738. doi: 10.1126/science.aar3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A.K., DiCesare J.L., Haff L., Atlas R.M. Detection of Escherichia coli and Shigella spp. in water by using the polymerase chain reaction and gene probes for uid. Appl. Environ. Microbiol. 1991;57:1013–1017. doi: 10.1128/aem.57.4.1013-1017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum Y.A., Talukder K.A., Nair G.B., Qadri F., Sack R.B., Svennerholm A.M. Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. J. Clin. Microbiol. 2005;43:3582–3583. doi: 10.1128/JCM.43.7.3582-3583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Y., Fu C., Hu M., Liu L., Wong M.H., Zheng C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: a review. Environ. Res. 2019;169:483–493. doi: 10.1016/j.envres.2018.11.040. [DOI] [PubMed] [Google Scholar]
- Berger C.N., Sodha S.V., Shaw R.K. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- Bielaszewska M., Mellmann A., Zhang W., Kock R., Fruth A., Bauwens A. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- Centres for Disease Control . Centers for Disease Control and Prevention; United States: 2004. Health. [Google Scholar]
- Chapman T.A., Wu X.Y., Barchia I., Bettelheim K.A., Driesen S., Trott D. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrhoeaic swine. Appl. Environ. Microbiol. 2006;72:4782–4795. doi: 10.1128/AEM.02885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Zheng W., Yu Y., Huang W., Zheng S., Zhang Y. Class 1 integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the Minjiang River Fujian Province, China. Appl. Environ. Microbiol. 2011;77:148–155. doi: 10.1128/AEM.01676-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Quiles-Puchalt N., Chiang Y.N., Bacigalupe R., Fillol-Salom A., Chee M.S.J., Fitzgerald J.R., Penadés J.R. Genome hypermobility by lateral transduction. Science. 2018;362:207–212. doi: 10.1126/science.aat5867. [DOI] [PubMed] [Google Scholar]
- Chigor V.N., Umoh V.J., Smith S.I. Occurrence of Escherichia coli O157 in a river used for fresh produce irrigation in Nigeria. Afr. J. Biotechnol. 2010;9:178–182. [Google Scholar]
- Christou A., Ag¨uera A., Bayona J.M., Cytryn E., Fotopoulos V., Lambropoulou D. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: the knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes—a review. Water Res. 2017;123:448–467. doi: 10.1016/j.watres.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . 2015. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria. M45, 3rd edition, CLSI Informational Supplement, Wayne, PA, 19087, USA. [DOI] [PubMed] [Google Scholar]
- Daini O.A., Ogbolu O.D., Ogunledun A. Quinolone resistance and R-plasmids of some Gram negative enteric bacilli. Afr. J. Clin. Exp. Microbiol. 2005;6(1):14–20. [Google Scholar]
- Doyle M.P., Loneragan G.H., Scott H.M., Singer R.S. Antimicrobial resistance: challenges and perspectives. Compr. Rev. Food Sci. Food Saf. 2013;12:234–248. [Google Scholar]
- Duffy G. Verocytotoxigenic Escherichia coli in animal faeces, manures and slurries. J. Appl. Microbiol. 2003;94:94–103. doi: 10.1046/j.1365-2672.94.s1.11.x. [DOI] [PubMed] [Google Scholar]
- Echiegu E.A., Eya O.R., Ugwu S.N., Ugwuishiwu B. Evaluation of effluent from the University of Nigeria, Nsukka wastewater treatment plant for irrigation purposes. Am. J. Environ. Sci. 2016;12(2):122–130. [Google Scholar]
- Estrada-Garcia T., Cerna J.F., Paheco–Gil L., Velazquez R.F., Ochoa T.J., Torres J. Drug-resistant diarrhoeagenic Escherichia coli, Mexico. Emerg. Infect. Dis. 2005;11:1306–1308. doi: 10.3201/eid1108.050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Moran L., Aronsson B., Manz C., Gyssens I.C., So A.D., Monnet D.L. Critical shortage of new antibiotics in development against multidrug-resistant bacteria—time to react is now. Drug Resist. Updates. 2011;14:118–124. doi: 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Garbisu C., Garaiyurrebaso O., Lanz´en A., Alvarez-Rodr´ıguez I., Arana L., Blanco F., Smalla K., Grohmann E., Alkorta I. Mobile genetic elements and antibiotic resistance in mine soil amended with organic wastes. Sci. Total Environ. 2018;621:725–733. doi: 10.1016/j.scitotenv.2017.11.221. [DOI] [PubMed] [Google Scholar]
- Graves A.K., Hagedorn C., Teetor A., Mahal M., Booth A.M., Reneau R.B. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 2002;31:1300–1308. doi: 10.2134/jeq2002.1300. [DOI] [PubMed] [Google Scholar]
- Guan S., Xu R., Chen S., Odumeru J., Gyles C. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Technol. 2002;68:2690–2698. doi: 10.1128/AEM.68.6.2690-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guion C.E., Ochoa T.J., Walker C.M. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microbiol. 2008;46:1752–1757. doi: 10.1128/JCM.02341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin K., Bruant G., El-Shaarawi A., Hill S., Edge T., Fairbrother J. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair river and Detroit river areas. Appl. Environ. Microbiol. 2007;73:477–484. doi: 10.1128/AEM.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M.J., Hadi A.Z., Griffith J.F., Ishii S., Sadowsky M.J. Large-scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 2010;44:5463–5473. doi: 10.1016/j.watres.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa E.O., Okoh A.I. Impact of discharge wastewater effluents on the physicochemical qualities of a receiving watershed in a typical rural community. Int. J. Environ. Sci. Technol. 2009;6(2):175–182. [Google Scholar]
- Kaper J.B., Nataro J.P., Mobley H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Karlowsky J.A., Kelly L.J., Thornsberry C., Jones M.E., Sahm D.F. Trends in antimicrobial resistance among urinary tract infection isolates of Escherichia coli from female outpatients in the United States. Antimicrob. Agents Chemother. 2002;46:2540–2545. doi: 10.1128/AAC.46.8.2540-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby-Bauer W.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by single disc method. Am. J. Clin. Pathol. 1966;45:4. [PubMed] [Google Scholar]
- Koczura R., Mokracka J., Barczak A., Krysiak N., Kaznowski A. Association between the presence of class 1 integrons, virulence genes, and phylogenetic groups of Escherichia coli isolates from river water. Microb. Ecol. 2013;65:84–90. doi: 10.1007/s00248-012-0101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczura R., Mokracka J., Jabłonska L., Gozdecka E., Kubek M., Kazonowksi A. Antimicrobial resistance of integron-harbouring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water. Sci. Total Environ. 2012;414:680–685. doi: 10.1016/j.scitotenv.2011.10.036. [DOI] [PubMed] [Google Scholar]
- Korzeniewska E., Korzeniewska A., Harnisz M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013;91:96–102. doi: 10.1016/j.ecoenv.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Saucedo C., Cerna J.F., Villegas-Sepulveda N., Thompson R., Velazquez F.R., Torres J. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrhoeagenic Escherichia coli. Emerg. Infect. Dis. 2003;9:127–131. doi: 10.3201/eid0901.01-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lothigius A., Janzon A., Begum Y., Sjoling A., Qadri F., Svennerholm A.M. Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area. J. Appl. Microbiol. 2008;104:1128–1136. doi: 10.1111/j.1365-2672.2007.03628.x. [DOI] [PubMed] [Google Scholar]
- Luo Q., Kumar P., Vickers T.J., Sheikh A., Lewis W.G., Rasko D.A. Enterotoxigenic Escherichia coli secretes a highly conserved Mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect. Immun. 2014;82(2):509–521. doi: 10.1128/IAI.01106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M.F., Tauxe R.V., Hedberg C.W. The growing burden of food-borne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol. Infect. 2009;137:307–315. doi: 10.1017/S0950268808001969. [DOI] [PubMed] [Google Scholar]
- Manaia C.M., Macedo G., Fatta-Kassinos D., Nunes O.C. Antibiotic resistance in urban aquatic environments: can it be controlled? Appl. Microbiol. Biotechnol. 2016;100:1543–1557. doi: 10.1007/s00253-015-7202-0. [DOI] [PubMed] [Google Scholar]
- Martinez S., Suay R., Moreno J., Segura M.L. Reuse of tertiary municipal wastewater effluent for irrigation of Cucumismelo L. Irrigation Sci. 2013;31:661–672. [Google Scholar]
- Mohanta T., Goel S. Prevalence of antibiotic-resistant bacteria in three different aquatic environments over three seasons. Environ. Monit. Assess. 2014;186:5089–5100. doi: 10.1007/s10661-014-3762-1. [DOI] [PubMed] [Google Scholar]
- Mokracka J., Koczura R., Jablonska L., Kaznowski A. Phylogenetic groups, virulence genes and quinolone resistance of integron-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Van Leeuwenhoek. 2011;99:817–824. doi: 10.1007/s10482-011-9555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukatea S., Panaskara D., Wagha V., Muleyb A., Jangama C., Pawarc R. Impact of anthropogenic inputs on water quality in Chincholi industrial area of Solapur, Maharashtra, India. Groundwater Sust. Dev. 2018;7:359–371. [Google Scholar]
- Nontongana N., Sibanda T., Ngwenya E., Okoh A.I. Prevalence and antibiogram profiling of Escherichia coli pathotypes isolated from the Kat River and the Fort Beaufort abstraction water. Int. J. Environ. Res. Publ. Health. 2014;11:8213–8227. doi: 10.3390/ijerph110808213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nweze E.I. Virulence properties of diarrheagenic E. coli and etiology of diarrhea in infants, young children and other age groups in Southeast, Nigeria. Am.-Eurasian J. Sci. Res. 2009;4(3):173–179. [Google Scholar]
- Odjadjare E.E.O., Okoh A.I. Prevalence and distribution of Listeria pathogens in the final effluents of a rural wastewater treatment facility in the Eastern Cape Province of South Africa. World J. Microbiol. Biotechnol. 2010;26:297–307. [Google Scholar]
- Okeke I.N., Ojo O., Lamikanra A., Kaper J.B. Etiology of acute diarrhea in adults in Southwestern Nigeria. J. Clin. Microbiol. 2003;41:4525–4530. doi: 10.1128/JCM.41.10.4525-4530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaniran A.O., Naicker K., Pillay B. Antibiotic resistance profiles of Escherichia coli isolates from river sources in Durban, South Africa. World J. Microbiol. Biotechnol. 2009;25:1743–1749. [Google Scholar]
- Pan M., Chu L.M. Occurrence of antibiotics and antibiotic resistance genes in soils from wastewater irrigation areas in the Pearl River Delta region, southern China. Sci. Total Environ. 2018;624:145–152. doi: 10.1016/j.scitotenv.2017.12.008. [DOI] [PubMed] [Google Scholar]
- Proia L., Osorio V., Soley S., Kock-Schulmeyer M., Perez S., Barcelo D., Sabater S. Effects of pesticides and pharmaceuticals on biofilms in a highly impacted river. Environ. Pollut. 2013;178:220–228. doi: 10.1016/j.envpol.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Puah S., Puthucheary S.D., Liew F., Chua K. Aeromonas aquariorum clinical isolates: antimicrobial profiles, plasmids and genetic determinants. Int. J. Antimicrob. Agents. 2013;41:281–284. doi: 10.1016/j.ijantimicag.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Ram S., Vajpayee P., Shanker R. Prevalence of multi-antimicrobial-agent resistant, Shiga toxin and enterotoxin producing Escherichia coli in surface waters of River Ganga. Environ. Sci. Technol. 2007;41:7383–7388. doi: 10.1021/es0712266. [DOI] [PubMed] [Google Scholar]
- Ram S., Vajpayee P., Shanker R. Contamination of potable water distribution systems by multiantimicrobial-resistant enterohaemorrhagic Escherichia coli. Environ. Health Perspect. 2008;116:448–452. doi: 10.1289/ehp.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram S., Vajpayee P., Tripathi U., Singh R.L., Seth P.K., Shanker R. Determination of antimicrobial resistance and virulence gene signatures in surface water isolates of Escherichia coli. J. Appl. Microbiol. 2008;105:1899–1908. doi: 10.1111/j.1365-2672.2008.03879.x. [DOI] [PubMed] [Google Scholar]
- Rakic-Martinez M., Drevets D.A., Dutta V., Katic V., Kathariou S. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 2011;77:8714–8721. doi: 10.1128/AEM.05941-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose-Amsaleg C., Laverman A.M. Do antibiotics have environmental side effects? Impact of synthetic antibiotics on biogeochemical processes. Environ. Sci. Pollut. Res. 2016;23(5):4000–4012. doi: 10.1007/s11356-015-4943-3. [DOI] [PubMed] [Google Scholar]
- Sazakli E., Alexopoulos A., Leotsinidis M. Rainwater harvesting, quality assessment and utilization in Kefalonia Island, Greece. Water Res. 2007;41(9):2039–2047. doi: 10.1016/j.watres.2007.01.037. [DOI] [PubMed] [Google Scholar]
- Schmidt A.S., Bruun M.S., Dalsgaard I., Larsen J.L. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 2001;67:5675–5682. doi: 10.1128/AEM.67.12.5675-5682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C.M., Zhao C., DebRoy C., Torcolini J., Zhao S., White D.G. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine and food. Appl. Environ. Microbiol. 2002;68:576–581. doi: 10.1128/AEM.68.2.576-581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehabi A.A., Odeh J.F., Fayyad M. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from human stools and drinking water sources in Jordan. J. Chemother. 2006;18(5):468–472. doi: 10.1179/joc.2006.18.5.468. [DOI] [PubMed] [Google Scholar]
- Sidhu J.P., Ahmed W., Hodgers L., Toze S. Occurrence of virulence genes associated with diarrheagenic pathotypes in Escherichia coli isolates from surface water. Appl. Environ. Microbiol. 2013;79:328–335. doi: 10.1128/AEM.02888-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Vajpayee P., Ram S., Shanker R. Environmental reservoirs for enterotoxigenic Escherichia coli in South asian gangetic riverine system. Environ. Sci. Technol. 2010;44:6475–6480. doi: 10.1021/es1004208. [DOI] [PubMed] [Google Scholar]
- Steffen R., Castelli F., Dieter Nothdurft H., Rombo L., Jane Zuckerman N. Vaccination against enterotoxigenic Escherichia coli, a cause of travelers-diarrhoea. J. Trav. Med. 2005;12(2):102–107. doi: 10.2310/7060.2005.12207. [DOI] [PubMed] [Google Scholar]
- Sun J., Hu J., Peng H., Shi J., Dong Z. Molecular and physiological characterization of fluoroquinolone resistance in relation to uropathogenicity among Escherichia coli isolates isolated from Wenyu River, China. Chemosphere. 2012;87:37–42. doi: 10.1016/j.chemosphere.2011.11.050. [DOI] [PubMed] [Google Scholar]
- Thebo A.L., Drechsel P., Lambin E.F., Nelson K.L. A global, spatially-explicit assessment of irrigated croplands influenced by urban wastewater flows. Environ. Res. Lett. 2017;12 [Google Scholar]
- Turner S.M., Scott-Tucker A., Cooper L.M., Henderson I.R. Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 2006;263:10–20. doi: 10.1111/j.1574-6968.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- Titilawo O.Y., Obi C.L., Okoh A.I. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015;15:204. doi: 10.1186/s12866-015-0540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titilawo Y., Obi L., Okoh L. Antimicrobial resistance determinants of Escherichia coli isolates recovered from some rivers in Osun State, Southwestern Nigeria: implications for public health. Sci. Total Environ. 2015;523:82–94. doi: 10.1016/j.scitotenv.2015.03.095. [DOI] [PubMed] [Google Scholar]
- Titilawo Y., Sibanda T., Obi L., Okoh A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 2015;22(14):10969–10989. doi: 10.1007/s11356-014-3887-3. [DOI] [PubMed] [Google Scholar]
- United Nations Educational, Scientific and Cultural Organization . United Nations World Water Development Report 2003. UNESCO Publication; Paris, France: 2003. Water for People, Water for Life: Executive Summary.http://unesdoc.unesco.org/images/0012/001295/129556e.pdf Available at: [Google Scholar]
- Vaz-Moreira I., Nunes O.C., Manaia C.M. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol. Rev. 2014;38:761–778. doi: 10.1111/1574-6976.12062. [DOI] [PubMed] [Google Scholar]
- Walk S.T., Alm E.W., Clahoun L.M., Mladonicky J.M., Whittam T. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 2007;9:2274–2288. doi: 10.1111/j.1462-2920.2007.01341.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . WHO; Geneva: 2006. Burden of Disease and Cost Effectiveness Estimates. [Google Scholar]
- Wose K.C.N., Ateba N., Kawadza T.D. Antibiotic resistance profiles of Escherichia coli isolated from different water sources in the Mmabatho locality, North-West Province, South Africa. Res. Lett. 2010;106(1–2):44–49. [Google Scholar]
- Xu J., Xu Y., Wang H., Guo C., Qiu H., He Y., Zhang Y., Li X., Meng W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere. 2015;119:1379–1385. doi: 10.1016/j.chemosphere.2014.02.040. [DOI] [PubMed] [Google Scholar]
- Zhang C.M., Xu L.M., Mou X., Xu H., Liu J., Miao Y.H., Wang X.C., Li X. Characterization and evolution of antibiotic resistance of Salmonella in municipal wastewater treatment plants. J. Environ. Manag. 2019;251:109547. doi: 10.1016/j.jenvman.2019.109547. [DOI] [PubMed] [Google Scholar]
- Zhu Y.G., Zhao Y., Li B., Huang C.L., Zhang S.Y., Yu S., Chen Y.S., Zhang T., Gillings M.R., Su J.Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017;2:1–7. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.