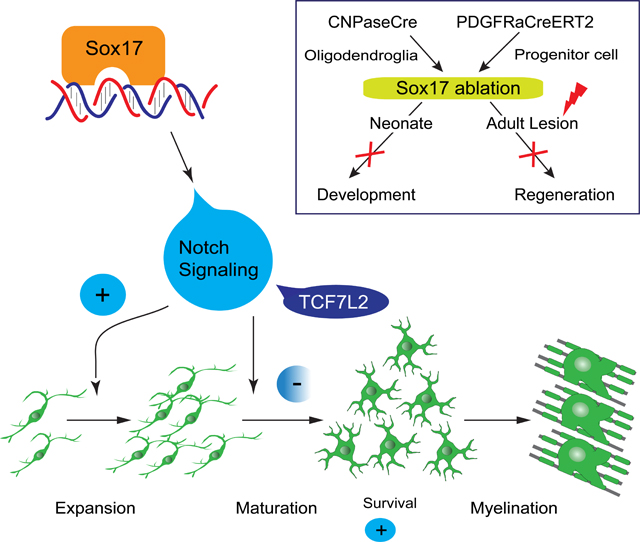
Summary
Sox17, a SoxF family member transiently upregulated during postnatal oligodendrocyte development, promotes oligodendrocyte cell differentiation, but its function in white matter development and pathology in vivo is unknown. Our analysis of oligodendroglial- and oligodendrocyte progenitor cell-targeted ablation in vivo using a floxed Sox17 mouse establishes a dependence of postnatal oligodendrogenesis on Sox17, and reveals Notch signaling as a mediator of Sox17 function. Following Sox17 ablation, reduced numbers of Olig2-expressing cells and mature oligodendrocytes led to developmental hypomyelination and motor dysfunction. After demyelination, Sox17 deficiency inhibited oligodendrocyte regeneration. Oligodendrocyte decline was unexpectedly preceded by transiently increased differentiation, and a reduction of oligodendrocyte progenitor cells. Evidence of a dual role for Sox17 in progenitor cell expansion via Notch, and differentiation involving TCF7L2 expression was found. A program of progenitor expansion and differentiation promoted by Sox17 through Notch thus contributes to oligodendrocyte production and determines the outcome of white matter repair.
GRAPHICAL ABSTRACT
Introduction
SRY-Box (Sox) containing transcription factors are evolutionarily conserved proteins (Gubbay et al., 1990) that are essential for the differentiation and maturation of a variety of tissue systems, including the developing nervous system (Chew and Gallo, 2009; Stolt and Wegner, 2010). Unlike the Sox D and E families, studies showing the physiological role of Sox F family members in the CNS in vivo are lacking, and Sox17 remains as the only member of the Sox F with established involvement in CNS glia development (Sohn et al., 2006). Sox17 was originally identified as an obligate endodermal determinant (Kanai-Azuma et al., 2002), while Sox7, 17 and 18 regulate the vasculature (Matsui et al., 2006; Wat and Wat, 2014). In the postnatal mouse white matter (WM), Sox17 expression is developmentally associated with that of multiple myelin genes, and its peak of expression in pre-myelinating oligodendrocytes is consistent with a role in regulating the transition to immature oligodendrocytes (Sohn et al., 2006). In the oligodendrocyte lineage, Sox17 regulates the Wnt/beta catenin signaling pathway and progenitor cell differentiation (Chew et al., 2011). Consistent with a role in oligodendrocyte regeneration, recent studies have shown that Sox17 expression in multiple sclerosis and experimental demyelinated lesions is localized in newly generated oligodendrocyte cells of actively remyelinating WM (Moll et al., 2013). However, functional involvement of endogenous Sox17 in postnatal oligodendrocyte development and regeneration in WM in vivo has not been investigated.
We have generated a conditional mouse allele to study Sox17 function in the oligodendroglia lineage in vivo by breeding this floxed strain with the CNP-Cre strain (Lappe-Seifke et al., 2003). Our characterization shows that Sox17 ablation disrupts oligodendrocyte differentiation in the postnatal subcortical WM. In contrast to previous in vitro studies of Sox17, the evidence indicates that oligodendrocyte loss arises initially from a reduction in OPCs. The eventual decrease in oligodendrocyte lineage cells was accompanied by reduced myelin protein expression, thin myelin sheaths and motor deficits. Sox17 ablation using PDGFRαCreERT2 produced similar changes in progenitor cells and oligodendrocytes, indicating a progenitor-specific role for Sox17. In addition, Sox17 ablation resulted in a significantly reduced capacity for oligodendrocyte regeneration following LPC-induced demyelination. Similar to postnatal development, OPC induction was reduced and fewer mature oligodendrocytes were observed in Sox17-deficient WM lesions. In addition to Sox17 regulation of Notch1 receptor and Hes effectors, we identified TCF7L2 expression to be regulated by both Sox17 and Notch. These studies demonstrate that Sox17 plays an important role in promoting a program of oligodendrocyte development via progenitor expansion and maturation, and which operates to regulate the regenerative potential of the adult WM.
Results
Targeted ablation of Sox17 in the oligodendroglial lineage decreases Sox17 expression
To determine the physiological role of Sox17 in developing oligodendrocytes, a conditional mouse Sox17 allele was created. This Sox17 allele has exons 4 and 5 flanked by loxP sites, allowing for Cre-mediated excision of the two largest exons (Figure 1A). Genomic deletion of Sox17 exons 4 and 5 was validated by PCR was performed on DNA isolated from SCWM of CNP-Cre/+; Sox17f/f (CKO) and Cre-negative (Ctrl) littermates (Figure S1A). Decreased Sox17 expression in vivo was previously shown in FACS-purified oligodendrocyte lineage cells of this conditional mutant at P10 (Chew et al., 2011). Quantitative PCR using P18 WM tissue showed reduced Sox17 and myelin protein RNA (Figure S1B).
Figure 1.
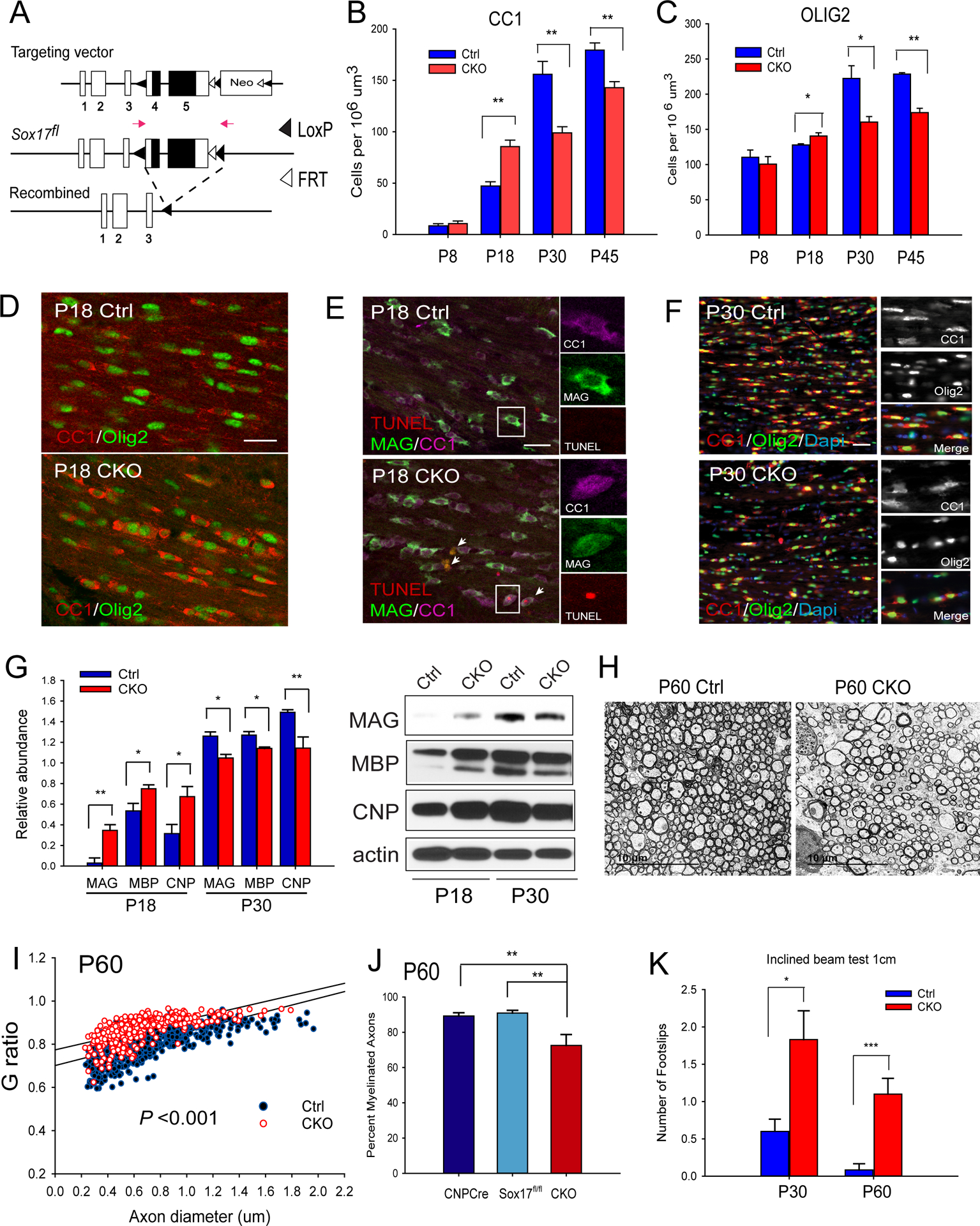
Targeted Sox17 ablation impairs oligodendrocyte development and myelination. (A). Sox17 conditional allele with LoxP sites (black triangles) flanking Exons 4 and 5. CNP-Cre-mediated excision produces the recombined allele lacking the protein-coding region (black boxes). See also Figure S1. (B–C) Immunohistochemical analysis of postnatal oligodendrocyte development, with quantification of CC1 (B) and Olig2 (C) cells. Note increased CC1 and Olig2 cells at P18 (D). N=3–6. *p < 0.05, and **p < 0.01, Student’s unpaired T test, mean ± SEM. (E) TUNEL analysis in P18 CKO shows colocalized TUNEL (red) with CC1 (magenta) and MAG(green). (F) Decreased oligodendrocytes CC1 (red) and Olig2 (green) in P30 CKO indicates oligodendrocyte deficit. Color information was removed for CC1 and Olig2 insets. (G) Western blotting indicates biphasic change in myelin proteins between P18 and P30. N=3. *p < 0.05, and **p < 0.01, Student’s unpaired T test, mean ± SEM. (H) Transmission electron microscopy indicates hypomyelination in P60 CKO. N=3–11. (I) G-ratio analysis shows significantly increased values for CKO at P60 arising from thinner myelin, as the average diameter of myelinated axons remained unchanged. (J) Quantitative analysis revealed decreased percentage of myelinated axons in the P60 CKO compared with CNP-Cre and Sox17f/f groups as separate control groups. N=6–12 (K) Behavior analysis using the 1cm inclined beam test distinguishes CKO from Ctrl via increased footslips at P30 and P60, supporting abnormal WM function resulting from Sox17 deficiency. n=6. ***P< 0.001,**P<0.005, *P<0.05 Student’s T-test vs Ctrl. Scale bars = 50 um, except in H = 10 um.
Sox17 ablation decreases the number of WM oligodendrocytes
Previous characterization of Sox17 expression in WM oligodendrocytes, as well as loss- and gain-of function studies in cultured OPCs indicate a role for Sox17 in oligodendrocyte development (Sohn et al., 2006). We therefore hypothesized that Sox17 ablation in the oligodendrocyte lineage would impair progenitor cell differentiation and lead to a reduction of mature oligodendrocytes, with functional deficits resulting from developmental hypomyelination. Immunohistochemical characterization of subcortical WM in CNP-Cre/+; Sox17+/+ (Ctrl) and CNP-Cre/+; Sox17f/f (CKO) mice over the course of postnatal development revealed age-specific changes in the numbers of oligodendroglial lineage cells. Myelinating oligodendrocytes (OL) labeled with CC1 were significantly reduced in the Sox17 CKO animals at P30 and P45 (Figure 1B). A reduction in mature MAG+ oligodendrocytes was observed at P30 and P45 (Figure S1C). The number Olig2 cells was also decreased at P30 and P45 (Figure 1C). Interestingly, there was a significant increase in oligodendrocytes at P18 (Figure 1B–D). This transient rise in differentiation was accompanied by reduced cell survival, as total TUNEL staining showed a modest, but significant increase in CKO WM (1.6714 ± 0.922 vs 5.987 ± 1.065 per 106 um3, P=0.01085 Student’s unpaired T-test). TUNEL+ cells included CC1+ and MAG+ oligodendrocytes (Figure 1E), likely contributing to oligodendrocyte loss by P30 (Figure 1F). The protein levels of myelin proteins were also analyzed across development using micro-dissected corpus callosum from CKO and WT siblings. Accordingly, a transient increase in MBP, CNP, MAG protein levels at P18 in Sox17 mutants was followed by significant decrease in these proteins at P30 compared with littermate controls (Figure 1G).
Sox17 ablation causes myelin thinning and impairs motor coordination
To determine whether the decline in oligodendrocytes affected myelination, we analyzed axonal ultrastructure by transmission electron microscopy. Figure 1H shows that, although the average diameter of myelinated axons and axonal integrity remained unchanged, myelin thickness, as quantified in Figure 1I by G ratio, was significantly reduced, together with a decrease in myelinated axons (Figure 1J). The size of the corpus callosum was also found to be reduced in P30 CKO (Figure S1D–E). To determine whether these changes led to functional impairment in behavioral tasks, control and Sox17 conditional knockout animals were tested on the inclined beam task at both P30 and P60. While the 2cm beam could not distinguish between controls and CKO, the more challenging 1cm beam revealed significant functional deficit of the Sox17 CKO at both P30 and P60 (Figure 1K; 1 cm control 0.13 ± 0.09 foot slips/trial, CKO 1.10 ± 0.23 foot slips/trial, p=0.0002; 2cm control 0.20 ± 0.14, CKO 0.60 ± 0.22 foot slips/trial, p=0.13).
Sox17 regulates OPC expansion and sustains differentiation
Since the decrease in oligodendrocytes occurs during active postnatal oligodendrogenesis and myelination, it is possible that Sox17 deficiency disrupted OPC differentiation and/or OPC production. NG2+ cells were found significantly decreased in the P18 CKO (Figure 2A–B). This is due to reduced proliferation, as evidenced by reduced Ki67+ and NG2+BrdU+ cells (Figure 2C). To determine whether this change arose from the cell-autonomous loss of Sox17, analysis of NG2 cell proliferation in P18 CNP-Cre/+;Sox17f/f;Rosa26YFP mice was performed. As shown in Figure 2D–E, compared with CNP-Cre/+;Rosa26YFP, fewer NG2+YFP+ cells were present in the P18 CKO WM that were BrdU+. CNP-Cre-targeted recombination rate within the NG2 cell population was estimated at about 25% (Figure S2A,B). Within the YFP+ population, Sox17 ablation produced a significant decrease in proliferating NG2+ cells. Among CNP-Cre/+;Rosa26YFP-expressing cells, the loss of Sox17 caused a decrease in the percentage of Sox2-expressing progenitor cells at P18 (Figure 2F). Sox2+BrdU+YFP+ OPCs are detectable at P18 in less intense YFP+ cells in this mouse strain (Figure S2C). When the total Sox2 population was analyzed, there was a significant decrease over postnatal WM development in the CKO (Figure 2 G–H). This was due to reduced cell proliferation (Figure S2D–F).
Figure 2.
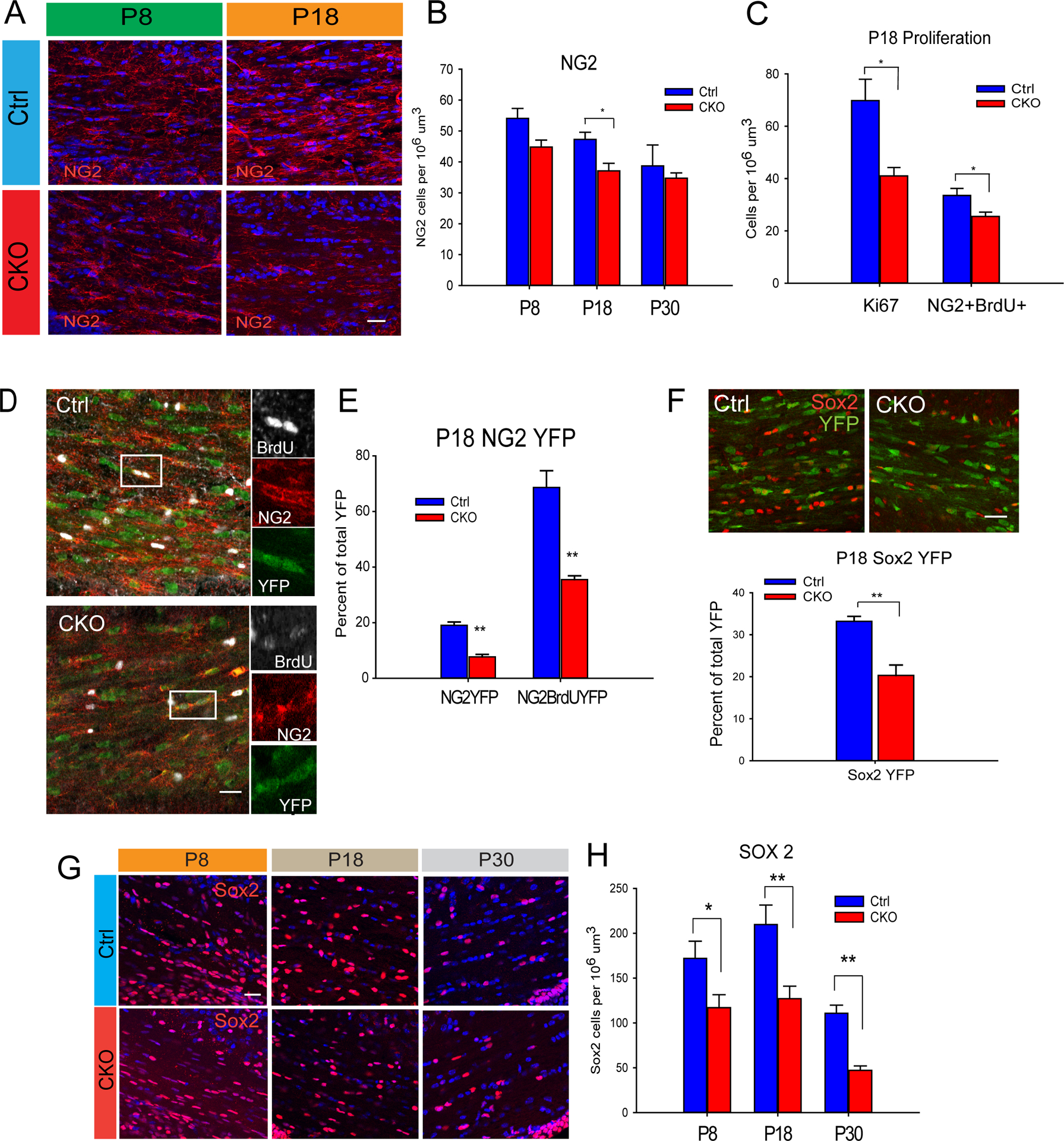
Sox17 regulates OPC expansion. (A–B) Immunohistochemical analysis of NG2-expressing OPCs in P8 and P18 WM shows little change at P8, but significant decrease at P18. (C) Analysis of proliferation using Ki67 and BrdU reveals decreased Ki67 and BrdU labeling in WM at P18. N=4. *p < 0.05, and **p < 0.005, Student’s unpaired T test, mean ± SEM. 50mg/kg BrdU was injected i.p. 24 hr prior to sacrifice. (D) Decreased total NG2 and BrdU-labeled OPCs within the CNP-Cre;RosaYFP population at P18 indicates an underlying deficit in proliferation (E). N=3. *p < 0.05, and **p < 0.01, Student’s unpaired T test, mean ± SEM. (F) The number of Sox2-expressing progenitor cells is decreased in P18 CKO WM. N=3. *p < 0.05, and **p < 0.005, Student’s unpaired T test, mean ± SEM (G) The total number of Sox2-expressing cells is decreased in developing WM of the postnatal CKO. N=4 **P<0.005, *P<0.05 Student’s T-test vs Ctrl. Scale bars: A,D,F 20 um, G,50 um.
Sox17 ablation in OPCs impairs oligodendrocyte production
To determine whether Sox17 loss in OPCs was sufficient to produce the observed biphasic change in oligodendrocytes during postnatal development, we generated CNS-progenitor-targeted Sox17 mutants by breeding PDGFRaCreERT2 with Sox17f/f mice. Analysis of PDGFRaCreERT2/+;Sox17f/f (PCKO) mice over postnatal development revealed a pattern of change in the oligodendroglial lineage similar to that observed in CKO. Figure 3A shows that early postnatal ablation with Tam at P4–P6 reduces NG2 progenitor cells at P8 without altering CC1 oligodendrocytes. Subsequently, analysis at P18 shows a smaller reduction in NG2 progenitors, but this change is accompanied by an increase in CC1 oligodendrocytes (Figure 3B). Tamoxifen injections at P7 were estimated by in situ hybridization to lead to Cre-based recombination in about 70% of PDGFRa-expressing OPCs (Figure S3A–B), which was sufficient to induce a detectable change in overall Sox17 expression (Figure S3C–E). By P28, CC1 oligodendrocytes are significantly reduced (Figure 3C). At this time point, the numbers of progenitor cells are no longer significantly different, based on NG2 and Sox2 cell analysis (Figure 3C). The decrease in Olig2- and MAG-expressing cells (Figure 3C) indicates a sustained disruption of differentiation despite progenitor recovery.
Figure 3.
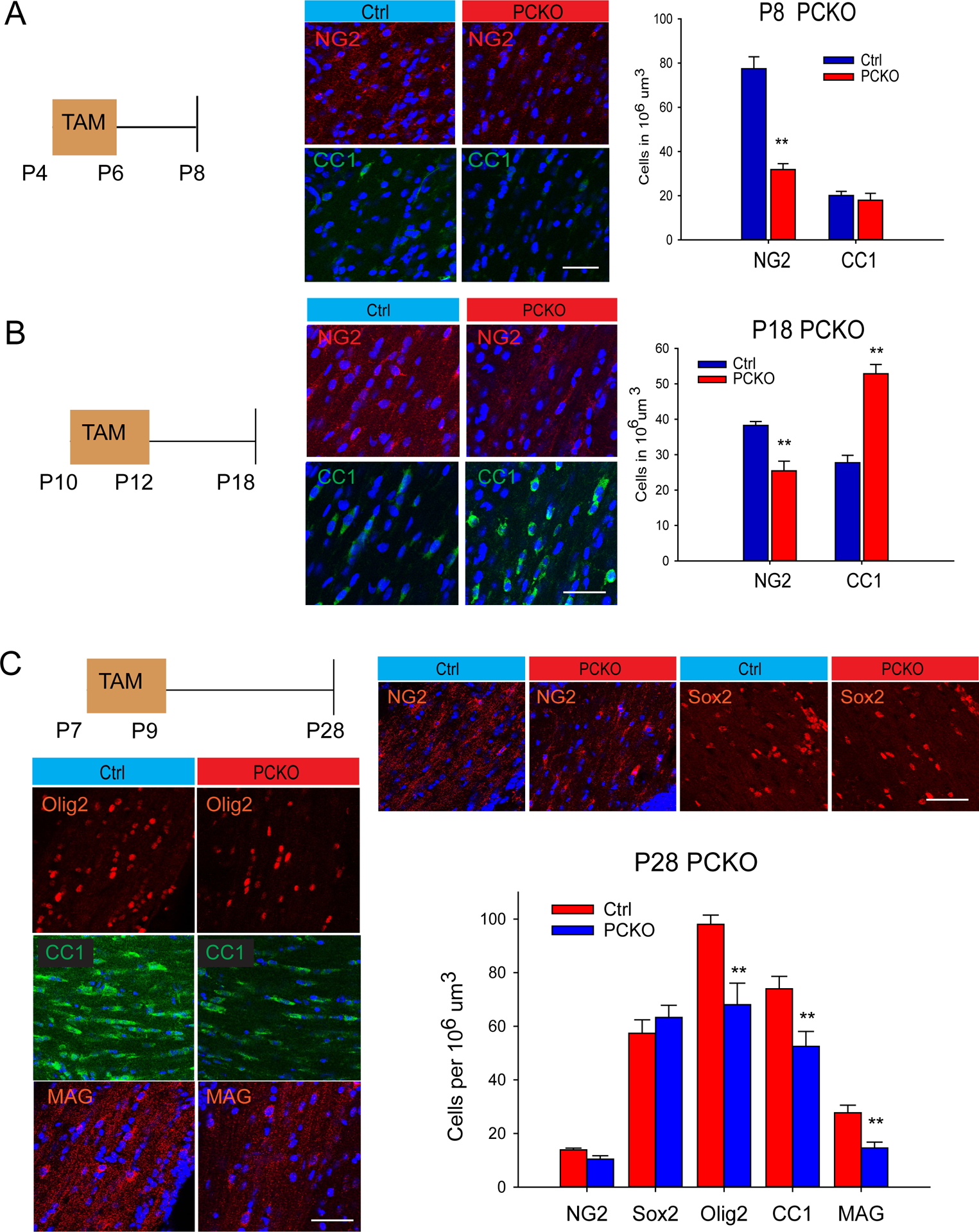
Sox17 ablation in progenitor cells regulates oligodendrocytes in biphasic manner. (A) Tamoxifen-induced ablation (TAM) in PDGFRaCreERT2 (PCKO) between P4 and P6 reduces NG2 (red) but not CC1 (green) in P8 WM. N=3. (B) TAM-induced Sox17 ablation between P10 and P12 reduces NG2 (red) and increases CC1 (green) in P18 WM. N=3. (C) TAM-induced Sox17 ablation between P7 and P9 reduces oligodendrocytes in P28 PCKO WM. Significant decreases in Olig2(red), CC1(green) and MAG(red) oligodendrocytes are observed without changes in NG2(red) or Sox2(red)-expressing progenitor cells. N=3. **p<0.005, Student’s T test, mean ± SEM. Scale bars = 50 um.
Sox17 ablation impairs remyelination
Given the importance of Sox2-expressing progenitor cells in both OPC proliferation and differentiation (Zhang et al., 2018), it is likely that Sox17 control of Sox2 and NG2 cell populations impacts progenitor-dependent cell regeneration following injury in the adult WM. To determine whether Sox17 ablation impedes oligodendrocyte regeneration in demyelinating lesions, lyso-phosphatidylcholine (LPC)-induced focal demyelination lesions in the P60 cingulate WM were analyzed. The density of MAG+ cells labeled by cytoplasmic staining was significantly lower in the CKO at 14 days post lesion (DPL) (Figure 4A and B). At 10 DPL, MAG cells were reduced in CKO lesions (Ctrl 22.36 ± 2.41 vs CKO 15.52 ± 1.55 per 106 um3, P<0.05, Student’s unpaired T-test), and BrdU pulse labeling (Figure 4C) revealed fewer newly formed MAG+ cells in CKO lesions (Figure 4D). The diminished response of Olig2 cells in CKO lesions (Figure 4E) suggests altered induction or activation of OPCs. Indeed, the production of NG2 cells in response to demyelination at 7 DPL was deficient in the CKO (Figure 4F–G). Similar to development, Sox17 ablation diminished the demyelination-induced increase in BrdU+ cells, including regenerative Sox2-expressing cells in WM at 7 DPL (Figures 4H–J).
Figure 4.
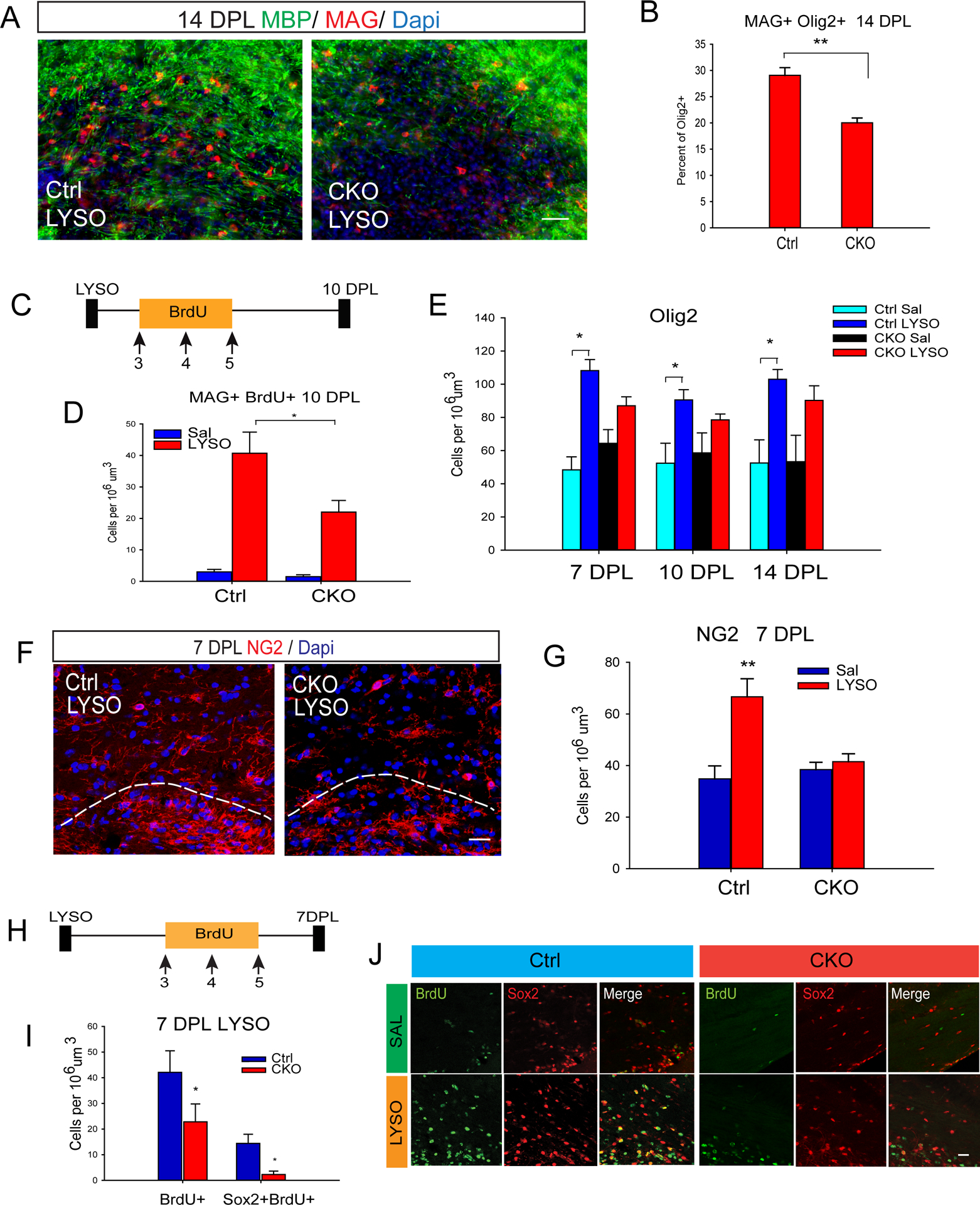
Sox17 promotes oligodendrocyte regeneration after LPC demyelination (LYSO). (A) Immunohistochemical analysis showing WM lesion area in CKO at 14 DPL that remains largely devoid of MBP (green) and MAG (red) immunostaining which indicates regenerating oligodendrocytes. (B) Analysis of MAG+Olig2+ cells observed in the lesion area shows decreased numbers in the CKO at 14 DPL. N=4. (C) Schematic illustration of the labeling paradigm to determine regenerative oligodendrogenesis. 50mg/kg BrdU was injected i.p., daily from 3–5 DPL, followed by sacrifice at 10 DPL. (D) The number of BrdU-labeled MAG cells is reduced in CKO lesions relative to control at 10 DPL. N=4 (E) Analysis of Olig2-expressing cells from 7–14 DPL reveals a defective oligodendroglial response in CKO lesions. N=3 (F–G) Confocal images of WM lesions at 7DPL showing reduced numbers of NG2 cells (red) populating the subcortical WM region adjacent to the subventricular area delineated by the white dashed line. Quantification reveals the absence of a significant progenitor cell response to demyelination in CKO. N=3. **P<0.005, ANOVA, Holm-Sidak post-hoc test. (H) Schematic representation of BrdU labeling paradigm followed by sacrifice at 7 DPL. (I-J) Immunohistochemical analysis of subcortical WM lesions showing decreased numbers of BrdU-labeled (green) Sox2-expressing cells (red) in the CKO. N=3. Scale bars in A= 50 um; F,J = 20um **P<0.005, *P<0.05 Student’s T-test vs Ctrl.
Sox17 regulates the Notch signaling pathway that controls oligodendrocyte progenitor expansion
To understand mechanisms that underlie Sox17 regulation of oligodendrocyte formation in the WM, we sought to determine the expression of factors that control progenitor cell expansion and maturation, such as Sox2 (Zhang et al., 2018) and TCF7L2 (Hammond et al., 2015; Zhao et al., 2016). Figure 5A shows that Sox17 knockdown in cultured OPCs revealed a surprisingly selective effect on TCF7L2 protein, rather than Sox2. This selectivity was confirmed at the RNA level by quantitative PCR (Figure 5B). We hypothesized that, since Sox17 did not regulate Sox2 expression in OPCs, its control of the Sox2 cells could be mediated through signaling which regulates the progenitor population (Zhang et al., 2009), such as Notch. Indeed, the protein levels of cleaved or activated Notch1 (Act N1, Figure 5A) and RNA levels of Notch1 receptor and signaling effectors Hes1 and Hes5 were significantly reduced by Sox17 siRNA (Figure 5B). To determine whether Notch signaling regulated TCF7L2 or Sox2 gene expression in cultured OPCs, we applied the gamma secretase inhibitor DAPT for 2 – 3 days and found that TCF7L2 RNA was reduced, while Sox2 remained unaffected (Figure 5C). Notch1 was found to mediate this change in TCF7L2 expression, since Notch1 siRNA transfection of cultured OPCs also decreased TCF7L2 without affecting Sox2 RNA (Figure 5D). DAPT decreased the percentage of proliferating Sox2 cells in culture (Figure 5E–F), ultimately decreasing the total number of Sox2 cells (Figure S4A) indicating that Notch regulates the expansion of Sox2-expressing OPCs, rather than its expression. Although 1uM and 2uM DAPT were found to reduce cell viability to a similar extent (Figure S4B), 1uM was selected for subsequent assays analyzing differentiation (below).
Figure 5.
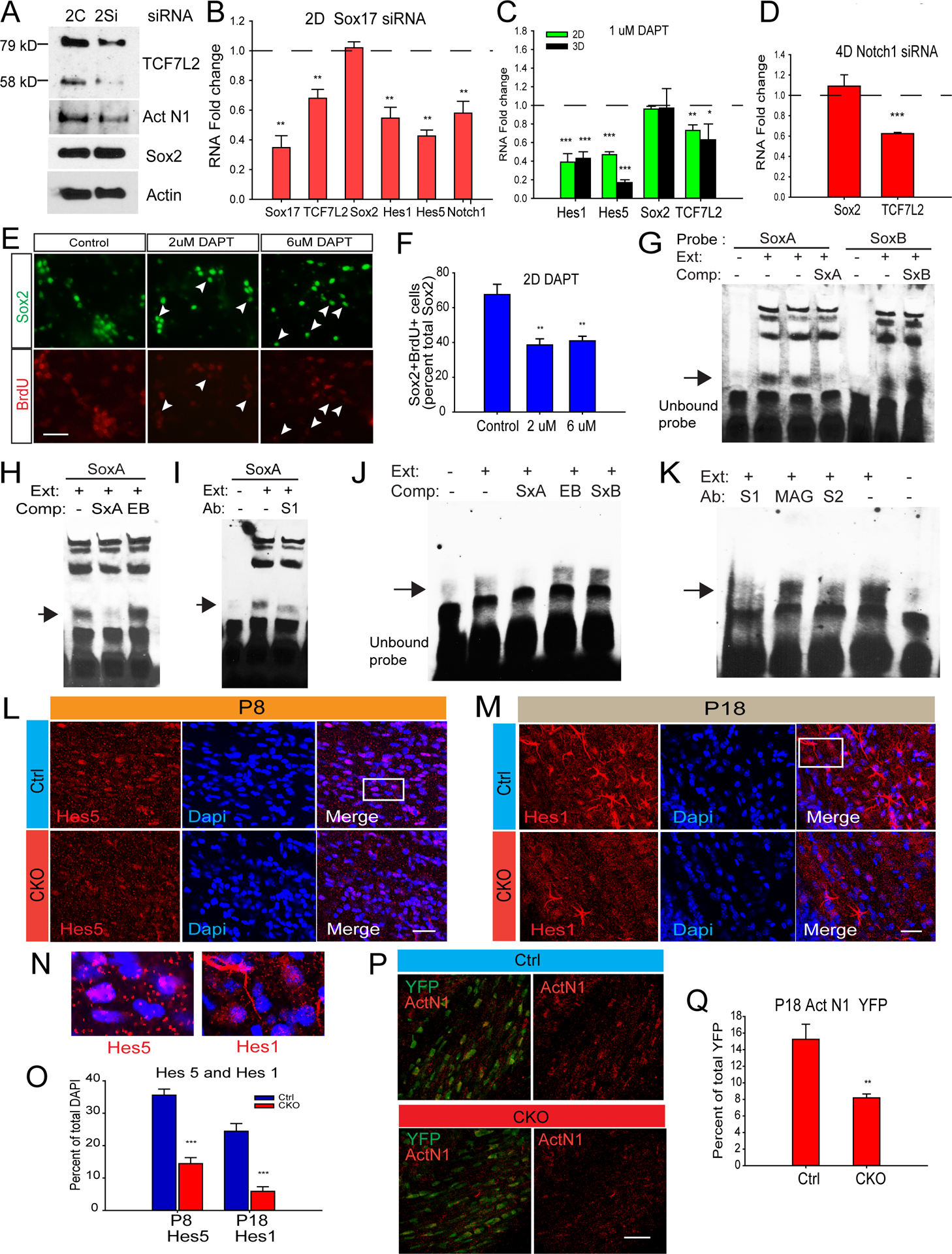
Sox17 regulates oligodendroglial Notch signaling. (A) Western blots of cultured OPCs transfected with siRNA and analyzed 2 days after transfection. Sox17 siRNA (2S) reduces activated Notch1 (Act N1) and both bands of TCF7L2 protein, not Sox2, when compared with control siRNA (2C). N=2 (B) Quantitative/real-time PCR analysis showing OPC RNA changes in TCF7L2, Notch1 and Hes genes after 2 days of Sox17 siRNA transfection. N=4 (C) Quantitative PCR analysis showing RNA changes in Hes genes and TCF7L2 following 2 and 3 days treatment of OPCs with 1uM DAPT. N=4. (D) Quantitative PCR analysis showing decreased TCF7L2 and not Sox2RNA after 2 days of Notch1 siRNA transfection. (E–F) The percentage of Sox2 cells (green) labeled with BrdU (red) was decreased after 2D co-treatment with PDGF and 2uM or 6uM DAPT. 50 uM BrdU was added 12 hr before analysis. Arrowheads indicate Sox2 cells with weak or no BrdU labeling. N=3. Scale bar= 50 um. *P <0.05, **P<0.01, ***P<0.005 Student’s unpaired T-test, mean ± SEM. (G–I) EMSA analysis with P12 WM extract (Ext) shows sequence-specific complex formation on a Sox binding sequence (SoxA) probe derived from the Notch1 intron. (G) Sequence-specific complexes do not form on SoxB probe. SxA is the unlabeled competitor oligo for SoxA, SxB for SoxB. (H) Using SoxA as probe, unrelated oligonucleotide sequence EBNA (EB) fails to competitively disrupt the complex on SoxA. (I) With SoxA as probe, Sox17-specific antibodies (S1) disrupt the SoxA complex. All arrowheads indicate position of Sox17-specific DNA-protein complex. (J-K) EMSA analysis using OPC nuclear extracts with SoxA probe. (J) SxA competitor (Comp) successfully abolishes complex formation by OPC nuclear extract (Ext), but not EB or SxB. (K) Antibodies (Ab) against Sox17 (S1 and S2) reduce complex formation on SoxA probe while MAG antibody is without effect. (L-M) The percentage of WM cells expressing Hes5 and Hes1 (red) at P8 and P18 respectively are decreased in CKO. Note the numbers of DAPI nuclei are comparable in Ctrl and CKO. (N) Magnified views of boxed areas in A and B showing cellular distribution of Hes proteins. Hes1 is less nuclear. (P) Quantitation shows that total Hes5 and Hes1 cells are decreased by Sox17 ablation. (Q–R) Act N1(red) levels are decreased in YFP reporter cells (green) of CKO at P18. N=3. Scale bars=50um except in D where scale bar=20um **P<0.01, ***P<0.005. ANOVA Holm-Sidak posthoc (D) or Student’s unpaired T-test (F).
Based on Sox17-induced changes in Notch1 expression, we investigated the possibility that Sox17 might interact with a Notch1 enhancer region in developing white matter. A recent study of SoxF factors by Chiang et al in arterial development identified two Sox consensus-containing intronic enhancers within the Notch1 gene (Chiang et al., 2017) which bound recombinant Sox7 and Sox18. Using these sequences as EMSA probes, we found that the sequence corresponding to HmSox-a (Chiang et al., 2017), which we call SoxA in this study, formed a sequence-specific complex with nuclear proteins from P12 WM (Figure 5G–H), whereas HmSox-b or SoxB did not. Figure 5I shows that the complex is vulnerable to disruption by anti-Sox17 antibody, indicating the presence of Sox17 in this complex from WM. To determine whether Sox17 in proliferating OPCs bound the SoxA sequence, we performed EMSA analysis using nuclear extract isolated from rat OPCs cultured for 2 days in PDGF. Figures 5J shows that a SoxA-specific complex was detected which was disrupted by Sox17 antibodies from two vendor sources (Figure 5K, Antibody S1 and S2). These data is consistent with the notion of direct control of Notch1 expression by Sox17 that could underlie progenitor cell expansion.
To determine whether Notch signaling in vivo was altered by Sox17 loss, the number of WM cells expressing Notch mediators Hes1 and Hes5 was analyzed. These were decreased in CKO WM at P8 and P18 (Figure 5L–M). Hes1 distribution at P18 was not exclusively nuclear, unlike Hes5 at P8 (Figure 5N), but total Hes1 and Hes5-expressing cells were decreased in number in CKO (Figure 5P). We then determined whether Notch1 activation was regulated in CNPCre-targeted cells. Figure 5Q shows that in P18 Controls, CNPCre-targeted YFP reporter expression (green) was colocalized with activated Notch1(ActN1,red). The percentage of YFP cells that colocalized with ActN1 was significantly decreased (Figure 5R), indicating that Sox17 ablation cell-autonomously downregulated Notch1 activation in P18 CKO WM.
TCF7L2 is regulated by Sox17 and Notch signaling
Given the function of Notch in progenitor maintenance, the biphasic change in TCF7L2 cells of the developing CKO WM (Figure 6A–B) suggests bona fide changes in differentiation events - disrupted progenitor expansion and enhanced precocious differentiation, followed by unsustained differentiation and eventual oligodendrocyte reduction. Progenitor-targeted Sox17 ablation reproduced the biphasic change in TCF7L2-expressing cells between P18 and P28 (Figure 6C, and Figure S5). We wanted to determine whether Sox17 also similarly regulated TCF7L2 in oligodendrocyte regeneration. In the intact adult WM, TCF7L2 is undetectable (Zhao et al., 2016), but its upregulation upon demyelination is dependent on Sox17 (Figure 6D–E). As in CKO development, the change in total TCF7L2-expressing cells undergoes an initial increase at 7 DPL, followed by decrease at 10 DPL (Figure 6F). BrdU pulse-labeling showed fewer newly formed TCF7L2 cells in the CKO by 10 DPL (Figure 6G). It is possible that the initial increase in TCF7L2 lies downstream from Notch inhibition, as cultured OPCs treated with DAPT also show enhanced differentiation based on increased O4 and TCF7L2-expressing cells (Figure 6H–I). Together, the observations are consistent with the interpretation that TCF7L2 may contribute to Sox17 functions in differentiation, and thus Sox17 regulates the number of oligodendrocytes in the adult WM through progenitor expansion and subsequent differentiation.
Figure 6.
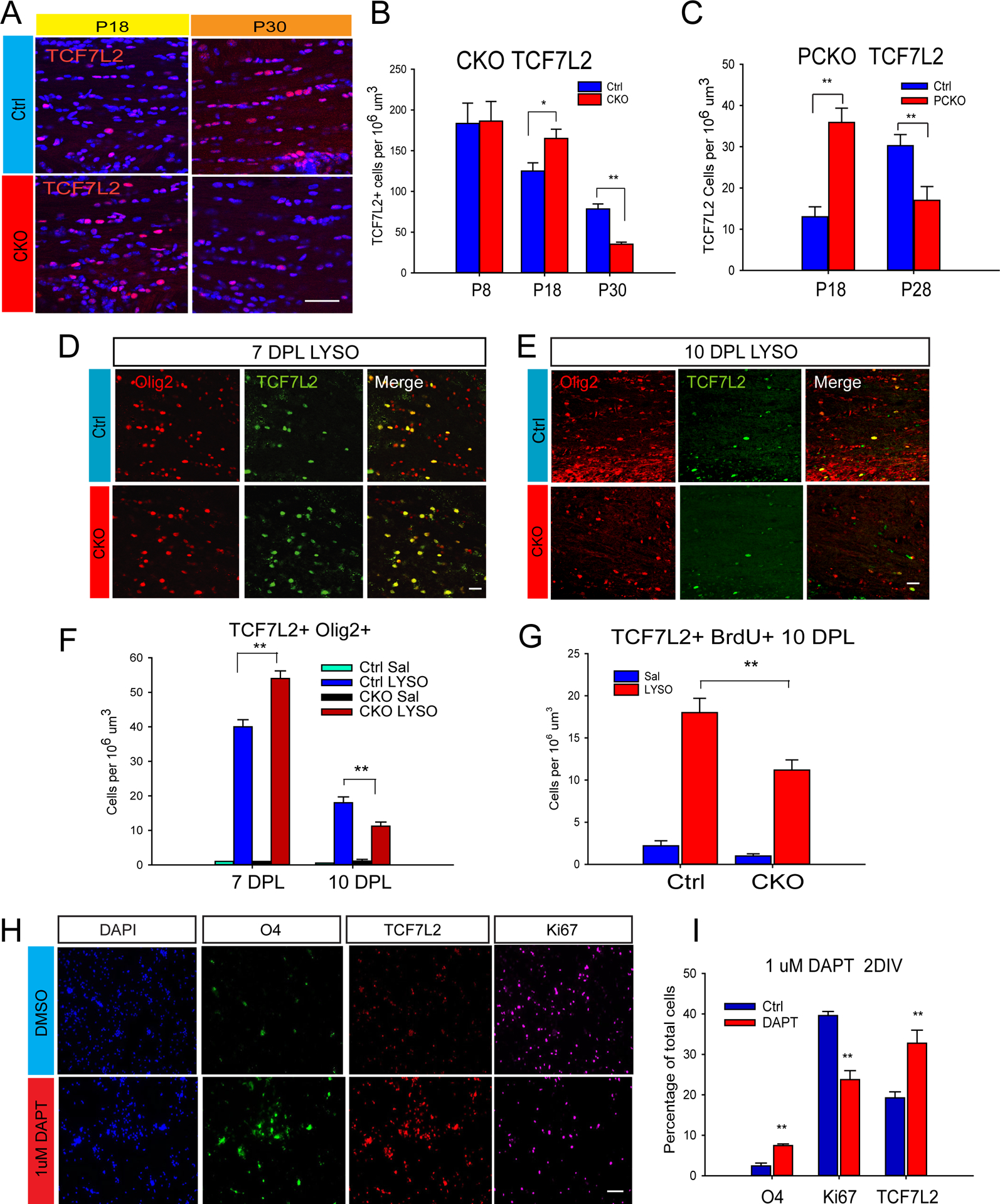
Biphasic regulation of TCF7L2-expressing cells by Sox17. (A,B) Biphasic change in TCF7L2- expressing cells (red) in developing CKO WM at P18 and P30, consistent with premature and transiently increased differentiation. Scale bar=50um. (C) Sox17 ablation in PCKO causes biphasic changes in TCF7L2-expressing cells during WM development. TAM was administered as in Figure 3B and 3C. (D–E) Confocal microscope images showing biphasic changes in Olig2 (red) and TCF7L2 (green) cells in Sox17-deficient WM following LPC demyelination (LYSO). Scale bars=20um. (F) Cell quantification showing increased TCF7L2+Olig2+ cells in CKO lesions at 7DPL while decreased numbers are observed at 10 DPL. N=3 (G) Reduced BrdU pulse labeling in CKO at 3–5 DPL indicates that Sox17 promotes the generation of TCF7L2-expressing cells at 10 DPL. (H–I) Exposure of proliferating OPC cultures to 1uM DAPT for 2 days decreases the percentage of cells that are proliferating (Ki67,red), and increases the proportion of differentiating cells expressing O4 (green) and TCF7L2 (red). Scale bar= 50 um. N=4. *P,0.05, **P<0.01, Student’s unpaired T-test.
Sox17 function in adult OPCs regulates oligodendroglial response to demyelination
To determine whether Sox17 ablation in adult OPCs regulated oligodendroglial regeneration, we performed LPC lesions in P60 PCKO mice, which received Tam injections 3 days prior to demyelination (Figure 7A). At 7 DPL, the number of NG2 progenitor cells was attenuated in PCKO (Figure 7B–C), accompanied by decreased ActN1 (Figure 7D–E). Lesion-induced Sox2-expressing oligodendroglial lineage cells, which co-express YFP reporter in controls (Figure S6A), were found to be significantly reduced in PCKO lesions along with Hes1 (Figure S6B-C). Olig2 cells were decreased in PCKO lesions, indicating an impaired oligodendroglial cell response (Figure 7F–G). The increased numbers of lightly stained Olig2 cells in intact PCKO may be due to progenitor maturation (Figure 7G, Sal). These observations support the interpretation that Sox17 in OPCs controls Notch1 activation, which enhances progenitor cell expansion in response to demyelination.
Figure 7.
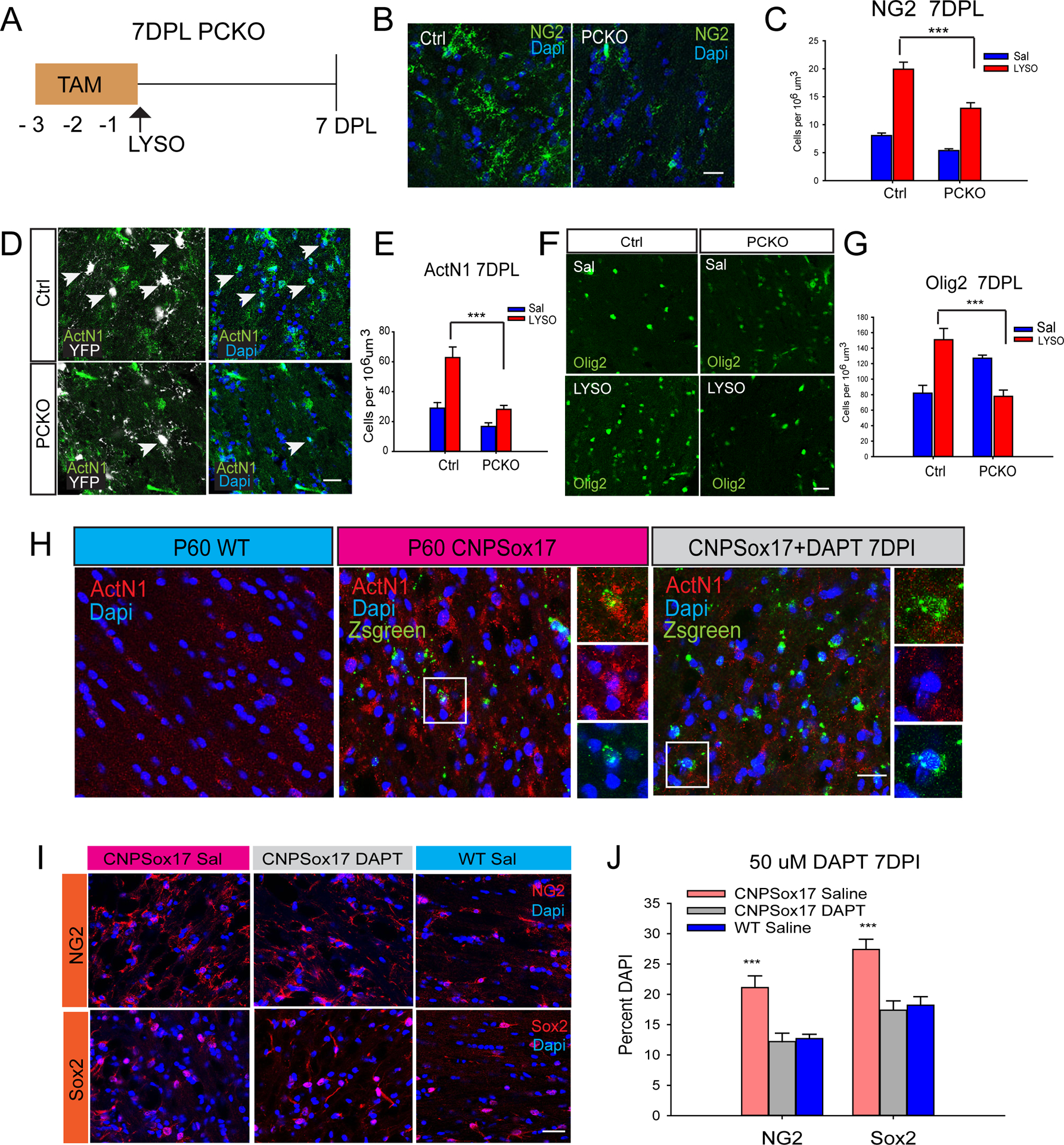
Adult OPC regulation by Sox17 involves Notch. (A) Illustration of daily TAM injections and demyelination paradigm (LYSO) in PCKO. Analysis was performed at 7 DPL. (B-C) In LYSO lesions, NG2 (green) progenitor cell response is attenuated by Sox17 ablation in PCKO. (D-E) In LYSO lesions, activated Notch1 (ActN1, green) is decreased in PCKO. Arrowheads indicate nuclear ActN1(green) colocalized with Rosa26YFP reporter (white). (F-G) Sox17 ablation decreases Olig2 (green) cells at 7 DPL. (H) Activated Notch1 (ActN1, red) is increased in intact WM of P60 CNPSox17 transgenic mice relative to WT. Inset shows a Zsgreen reporter (green)-expressing cell colocalized with ActN1(red). Notch inhibition in WM of P60 CNPSox17 mice with 50uM DAPT decreases Notch activation (ActN1, red) at 7 days post injection (7DPI). (I-J) DAPT decreases mean percentages of NG2 and Sox2 progenitor cells (red) at 7 days post injection (7DPI). N=3. Scale bars= 20um. *P <0.05, **P<0.01, ***P<0.005, ANOVA Holm-Sidak posthoc (C,E,G,J).
Notch inhibition in adult Sox17 overexpressing WM decreases progenitor cells
To determine whether Sox17 gain-of-function in vivo increases the progenitor cell population through Notch, we analyzed adult CNP-Sox17 transgenic mice at P60 when Ctrl progenitor proliferation had declined. This transgenic strain, which overexpresses recombinant, full length mouse Sox17 in NG2, O4 and CC1 cells of subcortical WM, displays increased numbers of oligodendrocytes in adult WM (Ming et al., 2013). Figures S7 A-B show that Sox2-expressing cells in the intact CNP-Sox17 WM were significantly increased over Ctrl (Saline), consistent with previous findings of greater numbers of NG2-expressing cells (Ming et al., 2013). Following LPC demyelination, at 3DPL - before the well-defined peak of progenitor proliferation in Ctrl mice reported to be at 7 days (Aguirre et al., 2007; Nait-Oumesmar et al., 1999; Watanabe et al., 2002; Woodruff and Franklin, 1999)- the CNP-Sox17 mouse showed an additional Sox2 cell response that was absent in Ctrl (Figure S7B). Sox17 overexpression was also previously shown to elevate the numbers of TCF7L2 cells (Ming et al., 2013). Our observation that WM lesions selectively stimulated an increase in TCF7L2 cells only in WT mice (Figure S7C) suggests that signaling mechanisms upregulating TCF7L2 and Sox2 cells in the CNP-Sox17 mouse were already intrinsically stimulated. Figure 7H shows that ActN1 is elevated in intact P60 CNPSox17 WM over wild type (WT) controls. A single stereotaxic injection of 50uM DAPT significantly reduced ActN1 levels in CNPSox17 (Figure 7H). To determine whether the increased population of NG2 and Sox2-expressing cells in the intact adult CNP-Sox17 transgenic WM was produced by enhanced Notch signaling, the numbers of NG2 and Sox2 cells were analyzed after stereotaxic injection of 50 uM DAPT (Figure 7 I- J). These experiments showing a reduction of NG2 and Sox2 cells in the CNP-Sox17 WM by DAPT implicate Notch signaling in progenitor maintenance. In Lyso lesions of the CNPSox17, this dose of DAPT was effective in decreasing lesion-induced ActN1, NG2 and Sox2 cells and increasing OPC differentiation, indicated by Olig2 and CC1 cells at 3DPL (Figure S7 D-E). Taken together, our studies provide evidence of a programmatic role for Sox17 in the control of Notch expression and in oligodendrocyte production through progenitor expansion and subsequent differentiation.
Discussion
Sox factors control many aspects of development in oligodendrocytes, ranging from lineage specification to effects on proliferation and survival, with the majority of the literature focused on the SoxD and SoxE families. SoxF factors are involved in hematopoietic progenitor regulation (He et al., 2011), tumor angiogenesis (Yang et al., 2013), and arterial development (Chiang et al., 2017). To date, Sox17 is the only SoxF family member whose expression has been shown to be developmentally regulated in postnatal WM, and the physiological function of endogenous Sox17 within oligodendrocytes in vivo is poorly understood. This study demonstrates that Sox17 contributes to the size of the WM oligodendrocyte population by promoting a program of OPC expansion and maturation that impacts both myelination and functional behavior.
Our laboratory has previously reported pro-differentiation functions of Sox17 in cultured OPCs (Chew et al., 2011; Sohn et al., 2006). Sox17 expression is lower in progenitor cells and mature oligodendrocytes, and peaks at the intermediate O4 stage (Sohn et al., 2006). While the significant decline in oligodendrocyte number was not surprising in CKO WM, the detection of biphasic Olig2 cell changes and precocious differentiation was unanticipated. The observations reflect the temporal and spatial pattern of Sox17 expression in the lineage, with greater and more persistent loss of oligodendrocytes than OPCs. In regulating multiple stages of the oligodendroglial lineage, Sox17 appears to stand apart from the better established SoxE members. Sox9 critically specifies OPCs, but its loss does not affect Olig2 cells in the spinal cord (Stolt et al., 2003). Although both Sox17 and Sox10 regulate myelin genes (Sohn et al., 2006), Sox17 ablation in vivo leads to a deficit in OPCs in CKO and PCKO, an effect not reported with Sox10 loss (Stolt et al., 2002). The decrease in OPCs labeled with ROSA26YFP following Sox17 loss in CKO indicates that cell-autonomous Sox17 activity contributes to the proliferation and maintenance of the OPC population. Indeed, the change in Sox2+;Rosa26YFP+ cells during postnatal development supports this interpretation. Our findings using a time-course series of Tam-induced ablation of Sox17 in developing PCKO (Figure 3) indicate that the biphasic pattern of oligodendrocyte changes is initiated at the level of the OPC. This suggests that the changes in CKO could also have been initiated at the level of the OPC, despite broader expression of the CNP-Cre. While Sox2 also labels adult GFAP-expressing astrocytes (Zhao et al., 2015), we detect Sox2 in cells which co-express Rosa26YFP reporter in LYSO lesions of adult PDGFRaCreERT2/+;Rosa26YFP mice, arguing that a population of adult Sox2-expressing OPCs (Zhang et al., 2018) may be responsible for the oligodendroglial responses observed in the absence of Sox17.
Although our current findings of Sox17 in progenitor expansion appear to be at odds with our previous reports of Sox17 in OPC differentiation (Chew et al., 2011; Sohn et al., 2006), it should be noted that these studies were primarily performed in cultured cells maintained under highly proliferating conditions. It is possible that transient effects on OPC expansion in this system would be less readily detectable than lasting effects on differentiation. Emerging evidence from studies of Sox proteins indicate that changes in both progenitor cells and mature oligodendrocytes can result from the loss of a single factor. Sox2 not only possesses “stem-ness” properties (Zhao et al., 2015), but also promotes progenitor proliferation and differentiation (Hoffmann et al., 2014; Zhang et al., 2018; Zhao et al., 2015). It is presently not clear how TCF7L2-expressing cells are regulated by Sox17 ablation. As a differentiation factor, Sox2 is known to be upregulated in TCF7L2-expressing newly formed oligodendrocytes (Hammond et al., 2015; Zhang et al., 2018). We do observe an increase in Sox2 intensity despite reduced Sox2 cell number in Sox17 mutants; suggesting Sox2 involvement in early differentiation events in parallel with TCF7L2 (Zhao et al., 2016). It is possible that the subsequent accelerated death of CKO CC1 cells arises from the lack of secondary or indirect targets of Sox17 that promote progenitor differentiation. Although progenitor depletion can contribute to the mechanism of oligodendrocyte loss, our observation of substantial NG2 cell recovery in Sox17 mutants argues that other processes initiated in OPCs, such as differentiation and survival, are likely to be involved in the functions of Sox17 in this lineage. Our studies showing regulation of TCF7L2 support the notion of a differentiation target of Sox17 and of Notch, although future studies will determine its underlying molecular mechanisms.
Both Sox2 and TCF7L2 have been reported to be regulated by canonical Notch signaling in neural stem cells (Li et al., 2012); however less is known about their functional relationships in OPCs. Sox17 overexpression increases the number of WM cells expressing TCF7L2/TCF4 (Ming et al., 2013). Our evidence shows that Sox17 regulates TCF7L2 gene expression in OPCs along with Notch1 receptor, and that DAPT and Notch1 siRNA also decrease TCF7L2 RNA. These findings suggest a requirement for Notch1 signaling in TCF7L2 control by Sox17. Together with the previous observation that Notch1 signaling supports OPC proliferation (Zhang et al., 2009), our results are consistent with the notion that Sox17 contributes to developmental OPC expansion by regulating Notch signaling. Indeed, we have identified a Notch1 enhancer as a binding target of Sox17 in developing WM and in proliferating OPCs, which provides support for direct control by Sox17. We also found Notch-related genes among differentially expressed genes from microarray analysis after Sox17 siRNA knockdown in cultured OPCs, a paradigm which dysregulates Wnt and differentiation ((Chew et al., 2011) and data not shown). Interestingly, Notch targets were not identified by RNAseq screening using a gain-of-function approach by inducible Sox17 overexpression in oligodendroglial cells (Fauveau et al., 2018), a paradigm which led to severe developmental hypomyelination. Nevertheless, loss-of-function studies suggest that Notch-dependent mechanism may be shared among SoxF factors in different tissue systems, as SoxF control Notch receptor expression in arterial specification (Chiang et al., 2017). Although Sox2 has been identified as a bona fide binding target for the Notch DNA-binding cofactor, RBPJ in neural stem cells (Li et al., 2012), we have failed to observed acute regulation of Sox2 RNA and protein in cultured OPCs by Sox17 knockdown or Notch inhibition. This suggests a context-specific relationship between Notch1 and Sox2, whose effects on progenitor development are mediated by distinct mechanisms (Holmberg et al., 2008). These results nonetheless support the interpretation that Sox17 regulates the progenitor cell population at least partly through Notch signaling in WM tissue. Given the complexity of Notch functions, it is tempting to speculate that its many lineage-relevant signaling activities (Stasiulewicz et al., 2015; Yabut et al., 2015) may underlie downstream oligodendrocyte changes resulting from Sox17 loss.
Our studies provide in vivo evidence that Sox17 controls the size of the oligodendrocyte cell population through progenitor expansion and maturation in both WM development and regeneration. Although its regulatory functions in relation to other SoxE and SoxF family members in this cell lineage await further analysis, our findings demonstrate that, by regulating the expression of a Notch receptor and its targets, Sox17 functions in a complex transcription factor network to coordinate a program of cell production and tissue homeostasis.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
The mouse strain containing the Sox17 floxed allele was generated in this study. As this has not been deposited with an external centralized repository for its distribution, the Sox17 floxed allele generated in this study is available for sharing from the Lead Contact with a completed Materials Transfer Agreement. The CNPSox17 mouse line is available for sharing from the Lead Contact with a completed Materials Transfer Agreement. Other resources and reagents are available without restriction from the Lead Contact, Vittorio Gallo (VGallo@childrensnational.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Generation of Sox17 conditional knockout animals
A targeting construct containing all 5 exons of the mouse Sox17 gene was generated by Ingenious Targeting Laboratory (Stony Brook, NY). In this construct, exons 4 and 5, which encode the entire Sox17 coding region, are flanked by loxP sites, and the pGK-gb2 loxP/FRT Neo cassette lies 3’ to exon 5. The construct was linearized and electroporated into ES cells, and positive clones were isolated. Resultant chimeric mice with the targeted Sox17flox conditional allele were bred with C57BL/6J mice (Stock 000664, Jackson Laboratory) to obtain heterozygous Sox17 flox offspring. The Neo cassette, which is flanked by two FRT sites, was excised by breeding with transgenic “flipper” mice ubiquitously expressing FLP recombinase. These mice were crossed with C57BL/6J mice, resulting in Neomycin-deleted Sox17 flox animals. Conditional Sox17 mutants were subsequently generated by crossing female Sox17 flox/flox mice with male CNP-crecre/+; Sox17 flox/flox mice. The CNP-Cre knock-in mouse strain (Lappe-Seifke et al., 2003) is a kind gift from Dr Klaus-Armin Nave (Gottingen, Germany). The R26R-EYFP strain was purchased from the Jackson Laboratory (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J, Stock 006148). The sequences of genotyping primers used to identify the floxed Sox17 allele, and CNP-Cre mice are as follows: lox1 F- CAG CCT TCC TAT TTC CCC AAG AGG; lox3 R- CTG GTC GTC ACT GGC GTA TCC; cre F- GCG GTC TGG CAG TAA AAA CTA TC; cre R - GTG AAA CAG CAT TGC TGT CAC TT. The deleted gene is identified with a 1000 bp product using lox1- CAG CCT TCC TAT TTC CCC AAG AGG; KO R CTA GTG TCA GGG ACT AGG AGG GAG (arrows Figure 1A and Figure S1A). Although female CKO mice are sterile, the mutant mice are viable albeit producing small litters, with an apparently normal development and life span. OPC-specific Sox17 ablated mice were generated by breeding Sox17flox with PDGFRacreERT2 (018280, Jackson laboratory). Primers used genotyping are listed in Table S1. Tamoxifen (Sigma, 90 mg/ml) was dissolved in 100% ethanol and then diluted in autoclaved sunflower oil (Sigma) to a final concentration of 10 mg/ml. Sox17flox/flox; PDGFRacreERT2/+were injected with 1 mg (neonates) or 75 mg/kg (P60) Tam once per day from −3 to - 1 DPL. Control mice received 10% ethanol in sunflower oil. Due to the low frequency of mutant generation, results from male and female mutant mice are combined and averaged.
Mouse studies, Lysolecithin (LPC) demyelination and DAPT injections
All mouse experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at Children’s National Hospital, Children’s National Research Institute (Protocols 00030331, 00030337). Male and female mice were not analyzed as separate groups for the following reasons: neonates were not sexed, and genetic mutants are produced at very low frequency e.g. 1 in 10, and no more than 3 litters per breeder pair. For focal demyelination, adult mice were deeply anaesthetized using 100 mg/kg Ketamine and 10mg/kg Xylazine. Lysolecithin (1% L-α-lysophosphatidylcholine, 2μL, EMD chemicals) was injected unilaterally into the external capsule of 9–10 week old mice using a Hamilton syringe. On the contralateral side, 2μL of 0.9% NaCl was injected for control purposes. Injections were made using a stereotaxic apparatus at the following coordinates: 1.0 mm anterior to Bregma, 1.5 mm lateral, 2.0 mm deep. The date of injection was denoted as 0 days post lesion (DPL). Mice were allowed to recover for 7, 10, or 14 DPL and then perfusion-fixed with 4% paraformaldehyde for immunohistochemical analysis. DAPT (gamma secretase inhibitor N-[(3,5-Difluorophenyl)acetyl]-L-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester. Tocris Bioscience, UK) was prepared in a final concentration of 50 uM in 2% DMSO in saline and injected using the same stereotaxic coordinates for cingulate WM as for LPC. Perfusion fixation was performed at 7 days post injection.
Oligodendrocyte Progenitor cell culture, loss-of-function and DAPT studies
Purified rat cortical OPC cultures were prepared as previously described from embryonic day 20 SAS Sprague Dawley rats (Charles River Laboratories, Wilmington, MA), using a modified protocol for mixed glia culture (Ghiani et al., 1999a; Ghiani et al., 1999b; McCarthy and de Vellis, 1980). The E20 embryos isolated from uteri of timed pregnant dams are not sexed, and all embryonic cortical hemisphere tissue was combined. OPCs were isolated by differential plating following overnight shaking of flasks of mixed glia. OPCs were plated at a density of 500,000 cells per dish on poly-ornithine-coated 60 mm plastic Petri dishes or at 125,000 per well on poly-ornithine-coated, acid-cleaned 18 mm glass coverslips, and maintained in Dulbecco’s modified Eagle’s DME–N1 supplemented with 10 ng/ml platelet-derived growth factor (PDGF) (human AB, heterodimer form; Upstate Biotechnology, Lake Placid, NY) in biotin-containing DME-N1. Double-stranded Sox17 small interfering RNAs (siRNA) were purchased from Applied Biosystems. A combination of three Sox17 siRNAs containing ID101960, ID 69339 and ID 101869 was used (Sohn et al., 2006). OPCs were plated at 5 × 105 cells per 60 mm dish and treated with PDGF for 36 hr, then transfected in antibiotic-free DME-N1 with a 1:1:1 mixture of Sox17 siRNAs at a final concentration of 45 nM. using Lipofectamine 2000 (Invitrogen) at a siRNA:lipofectamine ratio of 20pmol:1ul for 24 or 48 h. Controls were mock-transfected with Lipofectamine reagent. Cells were allowed to recover for 48h before analysis by qPCR. Notch1 siRNA (ID s129953) was treated similarly. DAPT was added to PDGF-containing DME-N1 with biotin 6–24h after plating at the indicated final concentrations. 5-Bromo-2’-deoxyuridine (BrdU, MilliporeSigma) was added to a final concentration of 50 uM 12 h before fixation with 4% paraformaldehyde and analysis by immunocytochemistry for Sox2 and BrdU.
METHOD DETAILS
Immunohistochemistry and TUNEL assay
Mice were anesthetized with isoflurane and perfused with 0.1M PBS, pH 7.4, followed by 4% paraformaldehyde (PFA). The brains were removed and fixed in 4% PFA overnight at 4 °C. 45 μm coronal sections cut using a vibratome (Leica) or freezing microtome (Leica) following cryoprotection in 20% and 10% glycerol. Immunohistochemistry was performed on floating sections using methods previously described in Yuan et al., 2004. The following antibodies were used: rabbit anti-NG2 (1:500,Millipore), rabbit anti-Olig2 (1:500; Millipore), mouse anti-CC1 (1:1000; Calbiochem), rabbit MAG (1:300, Santa Cruz Biotechnology), rabbit anti-caspase3 (1:250; Cell Signaling), rabbit anti-ki67 (1:200; Abcam), rat anti-BrdU (1:250; Abcam), goat anti-TCF7L2 (1:150, Santa Cruz Biotechnology), rabbit anti-TCF7L2 (1:500, Abcam), Rabbit anti-Sox2 (1:500, Abcam), rabbit anti-Hes1 and Hes5 (1:300, Millipore), rabbit anti-Activated Notch1 (1:400, Abcam), and chicken anti-GFP/YFP (1:1000, Aves labs, Inc). Corresponding secondary antibodies were used at a 1:600 dilution (Jackson Immunoresearch Laboratories). Sections were mounted on slides and covered in Fluoromount G + DAPI (Southern Biotechnology). TUNEL assays were performed with tissue sections using the In Situ Cell Death detection kit, TMR red (Roche Life Science) according to manufacturer’s instructions. TUNEL procedures preceded immunohistochemistry for CC1 and MAG.
Confocal Microscopy and image acquisition
Stained tissue sections were imaged on either a Zeiss LSM 510, Olympus FV1000 or Leica TCS SP8 laser scanning confocal microscope, acquired with Zen or LasX under 40X or 63X oil objective respectively, using a Z step of 0.75 um. Demyelinated lesions were confirmed by a decrease or absence of CC1 and MBP immunoreactivity, and disruption of WM cytoarchitecture. For saline-injected tissue, the injection site was located based on identification of the needle track, and injection site on the skull surface.
Western Blot analyses
Coronal brain sections 200 um thick were made on a McIlwain tissue chopper and placed in ice-cold DMEM. The corpus callosum was carefully micro-dissected from tissues between bregma 1.32mm and −2.12mm and homogenized in RIPA-lysis buffer (Santa Cruz Biotechnology, Inc). Cultured oligodendrocyte progenitor cells in dishes were harvested gently by rubber cell scraper into cold PBS containing Halt protease inhibitors(ThermoFisher) and pelleted by centrifugation. Total protein extracts and Western blot analysis were prepared as previous described (Chew et al., 2011). Briefly, tissue was extracted by homogenization in RIPA buffer containing Halt protease inhibitors (ThermoFisher), cleared by centrifugation at 13,000 rpm at 4C and the supernatant analyzed for protein concentration. 15–30 ug protein per well was resolved on 4–20% Tris-Glycine-SDS polyacrylamide gels (Biorad) and transferred onto PVDF membranes. After transfer, the membranes were blocked in either 4% milk or 5% BSA for 1 hr at room temperature, and then incubated overnight with primary antibodies diluted in either 4% milk or 5% BSA. The following primary antibodies were used: mouse anti-MBP (1:1000, Covance), anti-actin (1:5000, Chemicon), mouse anti-CNP (1:500; Covance), rabbit anti-MAG (1:500; Santa Cruz), rabbit anti-Sox2 and TCF7L2 (1:1000; Abcam). Activated Notch1 (Cell Signaling) was used at 1:1200. After washing with TBS-containing Tween20 (0.1%v/v), membranes were incubated with HRP-conjugated secondary antibodies (Cell Signaling Technology) diluted at a concentration of 1:5000 in blocking buffer for 1hr at room temperature. After washing, signals were developed using either Lightning Plus ECL (Perkin Elmer) or Supersignal West Dura Extended Duration ECL substrates (Thermo Fisher).
Quantitative PCR
The corpus callosum was micro-dissected as described above and RNA was extracted with RNeasy Micro Kit (Qiagen). Primary OPCs were collected in 350ul RLT buffer per 60mm dish and extracted in a manner similar to tissue. Total RNA (0.5–1 ug) was used to synthesize cDNA using SuperScript III First strand synthesis system (Life Technologies) or iScript synthesis kit (BioRad) in a 20ul reaction. Primer sequences are provided in Table S1. qPCR was carried out using diluted cDNA with SYBR Green qPCR mix (Sigma-Aldrich or GeneCopoiea) in 20–25 ul reactions a 96 well spectrofluorometric thermal cycler (ABI Prism 7900 HT Sequence Detector System; Applied Biosystems). Cycling conditions were as follows: Stage 1 95°C 10 min, Stage 2 40 cycles 95°C 45 s, 56–60°C 45 s, 72°C 45 s. Baseline values lie between cycles 8 and 15. Mean fold gene expression normalized against beta-actin was calculated with ABI software with the 2 (Delta Delta CT) Method. Data are expressed as mean fold change ± SEM with respect to reference control group.
Behavioral analysis
Mice were tested on the inclined, elevated beam-walking task at ages P30 and P60 as previously described (Brooks and Dunnett, 2009; Carter et al., 2001; Scafidi et al., 2014). Briefly, a 80 cm long wooden beam of 2cm or 1cm width was placed at a 30-degree angle, raised about 45 cm from the bench surface. A dark box with bedding was positioned at the top of the incline and served as the destination. Mice were placed at the lower end of the 2cm beam, and trained to walk across it into the dark box. After training, the width of the beam was reduced to 1cm to assess motor coordination. Four consecutive trials were recorded, and between each trial the animal was returned to its home cage for at least 15 minutes. A blinded experimenter observing from above, assessed mouse performance by documenting the number of foot slips (either hind legs or front legs) and the time to traverse the beam.
Electron Microscopy
Mice were prepared for electron microscopic analysis as previously described (Aguirre et al., 2007; Marcus et al., 2006). Following transcardial perfusion with 0.1M Millonig’s buffer containing 4% paraformaldehyde and 2.5% glutaraldehyde and brain removal, a coronal brain matrix (Ted Pella, Redding, CA) was used to cut the brain in serial 1 mm slices. Comparable slices for each brain (~0.5mm anterior to bregma were rinsed 3X in 0.1M cacodylate buffer, post fixed in 1% osmium tetroxide (in cacodylate buffer), dehydrated in increasing dilutions of ethanol and embedded in PolyBed resin. Thick (1μm) and thin (70nm) sections were stained with toluidine blue or with a combination of uranyl acetate and lead citrate, respectively. A minimum of 10 electron micrographs within the corpus callosum region were captured at both low (5,000X) and high (20,000X) magnifications per mouse using a JEOL JEM 1230 transmission electron microscope equipped with a Gatan 4K × 4K Ultrascan digital camera.
Electrophoretic Mobility Shift Assays
Nuclear lysates from P12 white matter tissue and OPC cells cultured for 2 days in 10 ng/ml PDGF were prepared by using the Thermo Scientific NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer’s instructions. The SoxA(SxA) probe was derived from hmSox-a, (Chiang et al., 2017) and SoxB(SxB) probe was derived from hmSox-b,(Chiang et al., 2017) (Table S1). Double-stranded probes were labeled with biotin on both ends of one strand (IDTDNA, Coralville, IA). Binding assays were performed using the LightShift™ Chemiluminescent EMSA kit according to manufacturer’s directions. Briefly, approximately 7–10 ug of nuclear protein was incubated with 20 fmol of biotinylated probe in 20ul reactions containing 1x Binding buffer, 2.5% glycerol, 50 ng/ml poly (dI.dC) and 5 mM MgCl2. 4 pmol unlabeled oligonucleotides (EBNA Control 5’-TAGCATATGCTA-3’ from LightShift™ kit, or SoxA(SxA) or SoxB(SxB)) were added in competition experiments, and 1–2ul of Sox17 antibody (Abcam ab224637 (Antibody S1,Figure 5)) or Thermo PA5 72815 (S2,Figure 5)) was added to the reactions 30 mins prior to loading the gel. After adding 5 ul of 5X loading buffer, the reactions were resolved on 5% nondenaturing polyacrylamide gels and subsequently transferred onto Biodyne™ Nylon membrane at 100V in 0.5X TBE. Blocking, incubation with Streptavidin-HRP, washing and visualization procedures were performed using reagents in the Chemiluminescent Nucleic Acid Detection Module (ThermoFisher Scientific). The membranes were exposed to X-ray film for 2–5 mins.
In situ hybridization
Following perfusion fixation with 4% paraformaldehyde, whole brain specimens were post-fixed in 4% paraformaldehyde for 24 h, then cryoprotected through 10%, 20% and 30% sucrose solutions in PBS, each for 24h at 4°C. After freezing in O.C.T., frozen coronal sections 14 um thick were used for in situ hybridization experiments with RNAscope reagents (Advanced Cell Diagnostics), and according to the vendor’s protocols for fluorescent multiplex assays. Only minor modifications were taken during pretreatment procedure: Tissue sections were pretreated first with PBS, then 4% paraformaldehyde and progressively dehydrated through a 50, 70, 100 % ethanol series of 2 mins each and bake-dried for 10 mins at 60°C before incubation with hydrogen peroxide from the ACD RNAscope Pretreatment kit. After rinsing with water twice and allowing the slides to dry, a hydrophobic barrier was traced around the sections and dried for 20 mins before proceeding with Protease III incubation for 30 minutes in the Hyb-EZ oven (ACD). After rinsing in water, probe hybridization was performed in a volume of 140 ul. The remainder of the procedure- probe hybridization, amplification and TSA fluorophore tagging (diluted to 1:1500) - were performed according to the ACD protocol for RNAscope Multiplex Fluorescent Reagent Kit v2 Assay, Document number 323100-USM without modification. Images were captured on the Leica TCS SP8 laser scanning confocal microscope with the 63X oil objective with Z step of 0.5 um. Higher resolution images were captured with the same objective using Zoom scaling of 1.5x.
QUANTIFICATION AND STATISTICAL ANALYSIS
Confocal image analysis
Images were analyzed using ImageJ software for the parameter outlined for each experiment. At least 3 sections per lesion were imaged per animal, at evenly spaced intervals (e.g. one in every 4 sections). The lesion area was imaged using tiling. For cell density calculations, the number of cells per parameter was divided by the total volume of the z-stacked image (length × width × stacked depth) to give cells/um3. Measurements were then multiplied by 106 to give cells/106 um3. Images of cingulate white matter and corpus callosum white matter up to the dorsal edge of, but not including, the subventricular zone, were taken at the same laser intensity and microscope acquisition settings for 3 sections per animal. ImageJ was used to image the WM area, form z-stacks and adjust the threshold. Images were minimally adjusted for clarity and size using Adobe Photoshop software.
Electron microscope image analysis
Low magnification images were used to qualitatively assess the overall extent of myelination; high magnification images were used for quantitative analyses. The percent myelinated and unmyelinated axons was determined and G ratio analysis was conducted. Any axon with a single wrap of an oligodendrocyte process was classified as “myelinated”; G ratio analyses were limited to only myelinated axons and were calculated by dividing the average axon diameter determined by measuring the longest and shortest diameter by the average axon diameter plus the width of the thickest and narrowest span of the myelin sheath in cross section. For myelin thickness measurements, regions with expanded myelin due to structural artifact were not used. Analyses were limited to axons with a caliber of >0.3 μm since previous work has shown that CNS axons ≤ 0.3μm are not myelinated (Mason et al., 2001).
Corpus Callosum thickness
Measurement of corpus callosum thickness in coronal sections was performed on images captured on the Olympus BX63 fluorescence microscope using the 10X objective and by using the line drawing tool of Image J. Six lines, three on either side of the midline, distributed evenly between the midline and cingulate white matter, were drawn perpendicular to opposite edges of the corpus callosum. Their lengths in um were recorded. At least 2 lines were drawn and their average recorded for each location. Two consecutive sections were measured per brain sample, and averages calculated for each location. 4 brain samples (n=4) were analyzed per group.
In situ hybridization signal analysis
PDGFRa-expressing cells were identified based on the presence of dot clustering in its channel. Only PDGFRa cells with 10 or more dots were considered positive. Olig2-expressing cells were treated similarly, whereas YFP-C3 criterion was a minimum of 5 dots. These criteria were applied in the estimation of Cre recombination efficiency by YFP expression. For quantification of fluorescent RNA signal, the number of dots or grains per cell with visible DAPI-stained nucleus was estimated by manual counting in ImageJ. The absolute number of Sox17-C2 dots was counted only in PDGFRa-positive cells. Sox17-C dots were counted in a total of about 60 PDGFRa cells in each group i.e. 2 higher magnification images from each of 3 brains. The average number of dots per PDGFRa-expressing cell was compared between groups.
Statistical analysis
All statistical analyses were performed with Sigmaplot 13, and all values are expressed as mean ± SEM, and illustrations were created using SigmaPlot 13. Neonatal mice sometimes died from early tamoxifen injections, so these samples could not be analyzed. The specific numbers of animals or cultures are denoted in each figure legend. Significance was determined using SigmaPlot13 software, with Student’s unpaired, two-tailed T-tests for comparisons between two groups and one-way ANOVA with Holm-Sidak post hoc tests for multiple comparisons. N values denote independent experiments. The degree of statistical significance was indicated with asterisks as defined in figure legends.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal NG2 | MilliporeSigma | Cat#AB5320; RRID:AB 91789 |
| Rabbit polyclonal Olig2 | EMDMillipore | Cat# AB9610 RRID:AB 570666 |
| Mouse monoclonal CC1 | Calbiochem | Cat # OP-80 RRID:AB 2057371 |
| Rabbit polyclonal MAG (H-300) | Santa Cruz Biotechnology | Cat # Sc-15324 RRID:AB 670104 |
| Rabbit cleaved caspase 3 | Cell Signaling Technologies | Cat #9661 RRID:AB 2341188 |
| Rabbit Ki67 | Abcam | Cat # Ab15580 RRID:AB 443209 |
| Rat BrdU | Abcam | Cat# Ab6326 RRID:AB 305426 |
| Rabbit TCF7L2 | Abcam | Cat # Ab76151 RRID:AB 1310728 |
| Goat TCF7L2 | Santa Cruz Biotechnology | Cat # sc-8631 RRID:AB 2199826 |
| Rabbit Sox2 | Abcam | Cat# Ab97959 RRID:AB 306863 |
| Rabbit Hes1 | Abcam | Cat# Ab71559 RRID:AB 1209570 |
| Rabbit Hes5 | EMDMillipore | Cat# Ab5708 RRID:AB 91988 |
| Rabbit Activated Notch1 | Abcam | Cat# Ab8925 RRID:AB 306863 |
| Rabbit Activated Notch1(Cl Val1744) | Cell Signaling Technologies | Cat #4147 RRID:AB 2153348 |
| Chicken GFP/YFP | Aves | Cat # GFP-1020 RRID:AB 1000024 0 |
| Mouse monoclonal MBP | Biolegend | Cat# 808402 RRID:AB 2564742 |
| Mouse monoclonal CNPase | Biolegend | Cat# 836404 RRID:AB 2566639 |
| Mouse monoclonal Actin | EMDMillipore | Cat# MAB1501R RRID:AB 2223041 |
| Rabbit polyclonal Sox17 | ThermoFisher | Cat# PA5–72815 RRID:AB_2718669 |
| Rabbit polyclonal Sox17 | Abcam | Cat# Ab224637 RRID:AB 2801385 |
| Alexa Fluor 647-Donkey Anti-Mouse IgG (H+L) | Jackson Immunoresearch Labs | Cat# 715–605-151 RRID:AB_2340863 |
| Alexa Fluor 647-Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch Labs | Cat# 712–605-150 RRID:AB_2340693 |
| Alexa Fluor 647-Donkey Anti-Chicken IgG (H+L) | Jackson Immunoresearch Labs | Cat# 703–605-155 RRID:AB_2340379 |
| Alexa Fluor 594-Donkey Anti-Rabbit IgG (H+L) | Jackson Immunoresearch Labs | Cat# 711–585-152 RRID:AB_2340621 |
| Alexa Fluor 594-Donkey Anti-Mouse IgG (H+L) | Jackson Immunoresearch Labs | Cat# 715–585-151 RRID: AB_2340855 |
| Alexa Fluor 594 F(ab’)2 fragment Donkey Anti-Goat IgG (H+L) | Jackson Immunoresearch Labs | Cat #705–586-147 RRID:AB_2340434 |
| Alexa Fluor 594- F(ab’)2 fragment Donkey Anti-Rat IgG(H+L) | Jackson Immunoresearch Labs | Cat# 711–586-153 RRID:AB_2340434 |
| Alexa Fluor 488-Donkey Anti-Rabbit IgG (H+L) | Jackson Immunoresearch Labs | Cat# 711–545-152 RRID:AB_2340621 |
| Alexa Fluor 488-Donkey Anti-Mouse IgG (H+L) | Jackson Immunoresearch Labs | Cat# 715–545-151 RRID:AB_2341099 |
| Alexa Fluor 488-Donkey Anti-Mouse IgM , u chain specific | Jackson Immunoresearch Labs | Cat# 715–545-020 RRID:AB_2340844 |
| Alexa Fluor 488-Donkey Anti-Rat IgG (H+L) | Jackson Immunoresearch Labs | Cat# 712–545-150 RRID:AB_2340683 |
| Alexa Fluor 488-Donkey Anti-Goat IgG (H+L) | Jackson Immunoresearch Labs | Cat# 705–545-147 RRID:AB_2336933 |
| Alexa Fluor 488-Donkey Anti-Chicken IgG (H+L) | Jackson Immunoresearch Labs | Cat# 703–545-155 RRID:AB_2340375 |
| Goat anti-rabbit IgG-HRP | Cell Signaling Technology | Cat# 7074 RRID:AB_2099233 |
| Goat anti-mouse IgG-HRP | Cell Signaling Technology | Cat# 7076 RRID:AB_330924 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| PDGF-AB, recombinant human | MilliporeSigma | Cat# GF106 |
| N2 supplement | Life Technologies | Cat# 17502048 |
| D-Biotin | Sigma Aldrich | Cat# B4639 |
| 5-Bromo-2’-deoxyuridine (BrdU) | Sigma Aldrich | Cat# 203806 CAS number 5914–3 |
| 4’,6’-diamidino-2-phenylindole (DAPI) | Sigma Aldrich | Cat# D9542 CAS:28718–90-3 |
| Fluoromount-G | Fisher Scientific | Cat# OB100–01 |
| DMEM high glucose | Life Technologies | Cat# 11995065 |
| Hank’s Balanced Salt Solution | Life Technologies | Cat# 14170112 |
| OPTIMEM Reduced serum medium | Life Technologies | Cat# 31985070 |
| Penicillin-Streptomycin 100X | Life Technologies | Cat# 15070063 |
| Fetal Bovine Serum, Premium | Atlanta Biologicals | Cat# S11150 |
| Lipofectamine 2000 | Life Technologies | Cat# 11668–030 |
| Poly L Lysine 30000–70000 MW | Sigma Aldrich | Cat# P9155 CAS:25988–63-0 |
| Poly L ornithine 30000–70000 MW | Sigma Aldrich | Cat# P3655 CAS:27378–49-0 |
| Paraformaldehyde | Sigma Aldrich | Cat# P6148 CAS:30525–89-4 |
| Methanol | BDH | Cat# BDH1135– 4LP CAS: 67–56-1 |
| Ethanol | KOPTEC | Cat# DSP-MD-43 CAS: 6417–5 |
| Hydrochloric Acid | Fisher Scientific | Cat# A144SI-212 CAS:7646–01-0 |
| RIPA Buffer | Thermo Scientific | Cat# 89900 |
| Halt Protease Inhibitor Cocktail | Thermo Scientific | Cat# 87786 |
| DAPT N-[(3,5-Difluorophenyl)acetyl]-L-alanyl-2- phenyl]glycine-1,1 -dimethylethyl ester | Millipore Sigma | Cat# 565770 CAS# 208255–805 |
| T amoxifen | Sigma Aldrich | Cat# T5648 |
| Sunflower seed oil | Sigma Aldrich | Cat# 47123 |
| L-a-lyso-lecithin | CalBiochem | Cat# 440154100MG |
| Ketamine | Henry SCHEIN | ANADA 200–055 NDC 11695–68351 |
| Xylazine | AKORN | NADA 139–236 NDC 59399–11020 |
| Bovin Serum Albumin | Sigma Aldrich | Cat# A7906 CAS:9048–46-8 |
| Donkey Serum | Jackson ImmunoresearchLab | Cat# 017–000-121 |
| Laemmli Sample buffer | Bio-Rad | Cat# 161–0737 |
| 10X TAE | K-D Medical | Cat# RGF-3310 |
| 10X PBS | K-D Medical | Cat# RGF-3210 |
| 10X Tris/Glycine/SDS buffer | K-D Medical | Cat# RGF-3390 |
| 10X Transfer Buffer | K-D Medical | Cat# RGF-3395 |
| 20X TBS | K-D Medical | Cat# RGF-3346 |
| 10X TBE | K-D Medical | Cat# RGE-3330 |
| 20X SSC | K-D Medical | Cat# RGF-3240 |
| Triton X-100 | Sigma Aldrich | Cat# T8787 CAS:9002–93-1 |
| Tween-20 | Sigma Aldrich | Cat# P9416 CAS:9005–64-5 |
| Critical Commercial Assays | ||
| In situ Cell Death detection kit, TMR red | Sigma Aldrich | Cat# 12156792910 |
| Pierce BCA Protein Assay kit | ThermoFisher | Cat# 23227 |
| Lightning Plus ECL | Perkin Elmer | Cat# NEL 103001EA |
| Supersignal West Dura Extended Duration ECL substrate | Thermo Fisher | Cat# 34075 |
| RNeasy ® Micro kit | Qiagen | Cat# 74004 |
| iScript™ first strand cDNA synthesis kit | BioRad | Cat# 1708890 |
| SYBR green qPCR mix with ROX | GeneCopoiea | Cat# QP031 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagents | ThermoFisher | Cat# 78833 |
| LightShift™ Chemiluminescent EMSA kit | ThermoFisher | Cat# 20148 |
| RNAScope - ms Sox17 probe | Advanced Cell Diagnostics | Cat# 493151-C2 |
| RNAScope - ms Olig2 probe-C1 | Advanced Cell Diagnostics | Cat# 447091 |
| RNAScope- ms PDGFRa probe-C1 | Advanced Cell Diagnostics | Cat# 480661 |
| RNAScope- EYFP-C3 | Advanced Cell Diagnostics | Cat# 312131-C3 |
| RNAScope pretreatment reagents | Advanced Cell Diagnostics | Cat# 322380 |
| RNAScope Wash buffer | Advanced Cell Diagnostics | Cat# 310091 |
| RNAScope Multiplex Fluorescent Reagent kit V2 | Advanced Cell Diagnostics | Cat# 323100 |
| TSA Plus Cyanine3/Cyanine5 System | Perkin Elmer | Cat# NEL 752001KT |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| CNP-Cre knock-in mouse strain | Klaus Nave lab, Max Planck Inst | (Lappe-Seifke et al., 2003) |
| Sox17 floxed allele mouse strain | This study | N/A |
| R26R-EYFP mouse strain | Jackson Labs | Cat# 006148 |
| PDGFRaCreERT2 mouse strain | Jackson Labs | Cat# 018280 |
| CNPSox17ZsGreen transgenic mouse strain | Gallo lab, Children’s National | (Ming et al., 2013) |
| Rat CRL SAS Sprague Dawley Timed pregnant E18 | Charles River | Cat#400 |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| LAS × Leica confocal software | Leica Microsystems | http://softadvice.informer.com/Leica_Confocal_Software.html |
| Olympus BX63 camera CellSens Standard 1.16 imaging software | Olympus Life Science | https://www.olympuslifescience.com/en/software/cellsens/ |
| SigmaPlot 13 | Systat software, Inc | https://systatsoftware.com/products/sigmaplot/ |
| Fiji ImageJ | Image J public freeware | https://imagej.net/Fiji/Downloads |
| Adobe Photoshop CC | Adobe Inc. Systems | https://www.adobe.com/creativecloud/plans |
| Adobe Illustrator CC | Adobe Inc. Systems | https://www.adobe.com/creativecloud/plans |
| Other | ||
| Ms Sox17 Silencer Pre-designed siRNA | Ambion | Cat #AM16708A ID#69339,101960, 101869 |
| Rat Notch1 Select Pre-designed siRNA | Ambion | Cat #4390771 ID#s135114 |
| 4–20% polyacrylamide tris glycine SDS gel | BioRad | Cat#4561095 |
ACKNOWLEDGEMENTS
This work was supported by the National Multiple Sclerosis Society RG4706A4/2 and RR-1512–07066 (V.G and BNO), and partially supported by FISM 2015/R/13 (V.G) and VA I01BX002565–05 (J.D). Confocal and fluorescence microscopy, and neurobehavioral analysis portions of this project were performed in the Neuroimaging and Neurobehavior Cores, supported by Award number 1U54HD090257 from the NIH, District of Columbia Intellectual and Developmental Disabilities Research Center Award (DC-IDDRDC, VG). Its contents are solely the responsibility of the authors and do not represent the official views of the DC-IDDRC or NIH. Further support was provided through the Fondation pour l’Aide à la Recherche sur la Sclérose en Plaques (ARSEP, BNO) and the “Investissements d’avenir” ANR-10-IAIHU-06 (BNO).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL ITEMS
PDF with 7 Supplemental Figures and 1 Table
REFERENCES
- Aguirre AA, Dupree JL, Mangin JM, and Gallo V (2007). A functional role for EGFR signalling in myelination and remyelination. Nat Neurosci 10, 990–1002. [DOI] [PubMed] [Google Scholar]
- Brooks SP, and Dunnett SB (2009). Tests to assess motor phenotype in mice: a user’s guide. Nat Rev Neurosci 10, 519–529. [DOI] [PubMed] [Google Scholar]
- Carter RJ, Morton J, and Dunnett SB (2001). Motor coordination and balance in rodents. Curr Protoc Neurosci Chapter 8. [DOI] [PubMed] [Google Scholar]
- Chew LJ, and Gallo V (2009). The Yin and Yang of Sox proteins: activation and repression in development and disease. J Neurosci Res 87, 3277–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew LJ, Shen W, Ming X, Senatorov VVJ, Chen H-L, Cheng Y, Hong E, Knoblach S, and Gallo V (2011). SRY-box containing gene 17 regulates the Wnt/beta-catenin signaling pathway in oligodendrocyte progenitor cells. J Neurosci 31, 13921–13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K-N, Fritzsche M, Pichol-Thievend C, Neal A, Holmes K, Lagendijk A, Overman J, D’Angelo D, Omini A, Hermkens D, et al. (2017). SoxF factors induce Notch1 expression via direct transcriptional regulation during early arterial development. Development 144, 2629–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauveau M, Wilmet B, Deboux C, Benardais K, Bachelin C, Temporao C, Kerninon C, and Nait Oumesmar B (2018). Sox17 transcription factor negatively regulates oligodendrocyte precursor cell differentiaiton. GLIA 66, 2221–2232. [DOI] [PubMed] [Google Scholar]
- Ghiani CA, Eisen AM, Yuan X, DePinho RA, McBain CJ, and Gallo V (1999a). Neurotransmitter receptor activation triggers p27(Kip1) and p21(CIP1) accumulation and G1 cell cycle arrest in oligodendrocyte progenitors. Development 126, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Ghiani CA, Yuan X, Eisen AM, Knutson PL, DePinho RA, McBain CJ, and Gallo V (1999b). Voltage-activated K+ channels and membrane depolarization regulate accumulation of the cyclin-dependent kinase inhibitors p27(Kip1) and p21(CIP)1 in glial progenitor cells. J Neurosci 19, 5380–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, and Lovell-Badge R (1990). A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346, 245–250. [DOI] [PubMed] [Google Scholar]
- Hammond E, Lang J, Maeda Y, Pleasure D, Angus-Hill M, Xu J, Horiuchi M, Deng W, and Guo F (2015). The Wnt effector transcription factor 7-like 2 positively regulates oligodendrocyte differentiation in a manner independent of Wnt/beta-catenin signaling. J Neurosci 35, 5007–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Kim I, Lim MS, and Morrison SJ (2011). Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev 25, 1613–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann SA, Hos D, Kuspert M, Lang RA, Lovell-Badge R, Wegner M, and Reiprich S (2014). Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development 141, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Hansson E, Malewicz M, Sandberg M, Perlmann T, Lendahl U, and Muhr J (2008). SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development 135, 1843–1851. [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PPL, et al. (2002). Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367–2379. [DOI] [PubMed] [Google Scholar]
- Lappe-Seifke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, and Nave KA (2003). Disruption of CNP1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet 33, 366–374. [DOI] [PubMed] [Google Scholar]
- Li Y, Hibbs MA, Gard AL, Shylo NA, and Yun K (2012). Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and Hippo pathway effectors by Notch1. Stem Cells 30, 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J, Honigbaum S, Shroff S, Honke K, Rosenbluth J, and Dupree JL (2006). Sulfatide is essential for the maintenance of CNS myelin and axon structure. GLIA 53, 372–381. [DOI] [PubMed] [Google Scholar]
- Mason JL, Langaman C, Morell P, Suzuki K, and Matsushima GK (2001). Episodic demyelination and subsequent remyelination within the murine central nervous system: changes in axonal calibre. Neuropathol Appl Neurobiol 27, 50–58. [DOI] [PubMed] [Google Scholar]
- Matsui T, Kanai-Azuma M, Hara K, Matoba S, Hiramatsu R, Kawakami H, Kurohmaru M, Koopman P, and Kanai Y (2006). Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci 119, 3513–3526. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, and de Vellis J (1980). Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 85, 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming X, Chew LJ, and Gallo V (2013). Transgenic overexpression of Sox17 promotes oligodendrocyte development and attenuates demyelination. J Neurosci 33, 12528–12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll NM, Hong E, Fauveau M, Naruse M, Kerninon C, Tepavcevic V, Klopstein A, Seilhean D, Chew LJ, Gallo V, et al. (2013). Sox17 is expressed in regenerating oligodendrocytes in experimental models of demyelination and in multiple sclerosis. GLIA 61, 1659–1672. [DOI] [PubMed] [Google Scholar]
- Nait-Oumesmar B, Decker L, Lachapelle F, Avellana-Adalid V, Bachelin C, and Baron-Van Evercooren A (1999). Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. European Journal of Neuroscience 11, 4537–4366. [DOI] [PubMed] [Google Scholar]
- Scafidi J, Hammond TR, Scafidi S, Ritter J, Jablonska B, Roncal M, Szigeti-Buck K, Coman D, Huang Y, McCarter RJ Jr., et al. (2014). Intranasal epidermal growth factor treatment rescues neonatal brain injury. Nature 506, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Natale JE, Chew LJ, Belachew S, Cheng Y, Aguirre AA, Lytle J, Nait-Oumesmar B, Kerninon C, Kanai-Azuma M, et al. (2006). Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci 26, 9722–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiulewicz M, Gray SD, Mastromina I, Silva JC, Bjorklund M, Seymour PA, Booth D, Thompson CB, Green RJ, Hall EA, et al. (2015). A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development 142, 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, and Wegner M (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17, 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, and Wegner M (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, and Wegner M (2010). SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol 42, 437–440. [DOI] [PubMed] [Google Scholar]
- Wat JJ, and Wat MJ (2014). Sox7 in vascular development: review, insights and potential mechanisms. Int J Dev Biol 58, 1–8. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Toyama Y, and Nishiyama A (2002). Differentiation of proliferated NG2-positive glial progenitor cells in a remyelinating lesion. J Neurosci Res 69, 826–836. [DOI] [PubMed] [Google Scholar]
- Woodruff RH, and Franklin RJM (1999). Demyelination and remyelination of the caudal cerebellar peduncle of adult rats following stereotaxic injections of lysolecithin, ethidium bromide and complement/anti-galactocerebroside: a comparative study. GLIA 25, 216–228. [DOI] [PubMed] [Google Scholar]
- Yabut O, Pleasure SJ, and Yoon K (2015). A Notch above Sonic Hedgehog. Dev Cell 33, 371–372. [DOI] [PubMed] [Google Scholar]
- Yang H, Lee S, Lee S, Kim K, Yang Y, Kim JH, Adams RH, Wells JM, Morrison SJ, Koh GY, et al. (2013). Sox17 promotes tumor angiogenesis and destabilizes tumor vessels in mice. J Clin Invest 123, 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhu X, Gui X, Croteau C, Song L, Xu J, Wang A, Bannerman P, and Guo F (2018). Sox2 is essential for Oligodendroglial proliferation and differentiation during postnatal brain myelination and CNS remyelination. J Neurosci 38, 1802–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Argaw AT, Gurfein BT, Zameer A, Snyder BJ, Ge C, Lu RQ, Rowitch D, Raine CS, Brosnan CF, et al. (2009). Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci (USA) 106, 19162–19167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng Y, Liu L, Yu K, Zhang L, Wang H, He X, Wang J, Lu C, Wu LN, et al. (2016). Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun 7, 10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ma D, Zawadzka M, Fancy SPJ, Elis-Williams L, Bouvier G, Stockley JH, Monteiro de Castro G, Wang B, Jacobs S, et al. (2015). Sox2 sustains recruitment of oligodendrocyte progenitor cells following CNS demyelination and primes them for differentiation during remyelination. J Neurosci 35, 11482–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.


