Abstract
Mitochondrial reactive oxygen species (ROS) production, specifically at complex I (Cx I), has been widely suggested to be one of the determinants of species longevity. The present study follows a comparative approach to analyse complex I in heart tissue from 8 mammalian species with a longevity ranging from 3.5 to 46 years. Gene expression and protein content of selected Cx I subunits were analysed using droplet digital PCR (ddPCR) and western blot, respectively. Our results demonstrate: 1) the existence of species-specific differences in gene expression and protein content of Cx I in relation to longevity; 2) the achievement of a longevity phenotype is associated with low protein abundance of subunits NDUFV2 and NDUFS4 from the matrix hydrophilic domain of Cx I; and 3) long-lived mammals show also lower levels of VDAC (voltage-dependent anion channel) amount. These differences could be associated with the lower mitochondrial ROS production and slower aging rate of long-lived animals and, unexpectedly, with a low content of the mitochondrial permeability transition pore in these species.
Keywords: Complex I, Droplet digital PCR, Longevity, Mammals, Mitochondria, NDUFV2 subunit, NDUFS4 subunit, VDAC, Western blot
Graphical abstract
Highlights
-
•
There are species-specific differences in gene expression and protein content of Cx I.
-
•
The achievement of a longevity phenotype is associated with low protein abundance of subunits NDUFV2 and NDUFS4 from the matrix hydrophilic domain of Cx I.
-
•
Long-lived mammals show also lower levels of VDAC (voltage-dependent anion channel) amount.
-
•
These differences can be causally associated with the aging rate of long-lived animals.
1. Introduction
Complex I (Cx I) (NADH-ubiquinone oxidoreductase; EC 1.6.5.3) is an electron entry point in the mitochondrial respiratory electron transport chain (ETC). Cx I catalyses NADH oxidation reducing ubiquinone to ubiquinol, importantly contributing to the proton motive force used to synthesize ATP by the oxidative phosphorylation [1]. Cx I also produce reactive oxygen species (ROS), initially superoxide radicals, which can damage all cellular components. Although at least 11 sites producing ROS have been identified, Cx I and complex III (Cx III) are conventionally recognized as the major sources of ROS at the ETC [2]. Mitochondrial ROS production (mitROSp) has been considered one of the main effectors responsible for aging and longevity [3,4].
Low rates of mitROSp have been described in many long‐lived mammalian and bird species [[3], [4], [5]]. These studies generally demonstrated the existence of a negative correlation between mitROSp and longevity. Among the two main ROS generating ETC complexes, the low ROS production of various long-lived species has been localized at Cx I [[6], [7], [8]]. Interestingly, different pro-longevity nutritional and pharmacological interventions like dietary (DR) and methionine restriction, and rapamycin treatment have been also associated with decreased mitROSp at Cx I [3,9]. Although is a matter of controversy, within complex I longevity-related ROS production has been observed in the hydrophilic domain [10], which could be due either to the flavin semiquinone, to some of the eight different FeS clusters of this domain [[11], [12], [13]] or both to flavin and FeS centers.
The underlying mechanism responsible, at least in part, for the low mitROSp of some long‐lived species has been attributed to a smaller degree of electronic reduction of Cx I under basal conditions [4,8]. Additional studies demonstrated that a lower Cx I content could also explain the low ROS production of those long-lived species [14,15]. Pro-longevity nutritional interventions also induced decreased Cx I content concomitantly with a lower mitROSp [10,[16], [17], [18], [19]].
Mammalian Cx I is the largest component of the ETC built of 45 different subunits in mammals [1]. The conserved L-shaped core, formed by 14 subunits sufficient for catalysis, is surrounded by 31 accessory subunits forming an interlinked shell with unclear function [20,21]. Among the 14 core subunits, the 7 mitochondrial-encoded ND subunits are present in the hydrophobic membrane domain, and the other 7 nuclear-encoded (NDUF) subunits are present in the hydrophilic matrix domain [22]. The 31 supernumerary NDUF accessory subunits are also nuclear coded [20]. However, it is totally unknown if some particular Cx I subunits, especially some NDUF subunits of the Cx I hydrophilic domain, could be involved in the determination of the longevity-related low complex I ROS production of long-lived animal species.
On the other hand, the mitochondrial permeability transition pore (mPTP) is a protein complex that permits diffusion of molecules of molecular mass up to 1500 Da across the mitochondrial inner membrane [23,24]. Many substrates can pass from mitochondrial matrix to cytosol through this pore. The mPTP opening is promoted through oxidation of lipids and proteins of mitochondrial membrane by ROS [23] and is related to pathological stages. However, some studies suggest that mPTP opening by mitROS can also drive the progression of aging [23,25]. It has been hypothesized that mPTP comprises inner and outer mitochondrial membrane proteins such as the ATP synthase, key in the channel formation, and a diversity of regulatory components including the voltage‐dependent anion channel (VDAC), the pro-death Bcl-2 family member proteins Bax and Bak, the translocator protein (TSPO), the hexokinase (HK), the adenine nucleotide translocase (ANT), the phosphate carrier (PiC), and the cyclophilin-D [[26], [27], [28]], all of which have been studied as candidates for exact molecular mPTP identity which remains uncertain. Recent studies have shown that decreased VDAC promotes longevity by decreasing mitochondrial permeability in C. elegans [29].
The purpose of this study was to investigate the abundance of Cx I hydrophilic domain selected subunits and VDAC as possibly related to the low mitROSp of long‐lived mammalian species. We used droplet digital PCR (ddPCR) and western blot methods to define the steady-state levels of gene expression and protein content of mitochondrial electron transport chain Cx I subunits and VDAC in heart tissue of eight mammalian species showing more than one order of magnitude of difference in longevity, from 3.5 years in mice to 46 years in horses. Cx I hydrophilic subunits were selected based on: i) previous partial proteomic studies of the ETC revealing the subunits more acutely decreasing in dietary restriction or varying in aged or longevity mutant mice [10]; ii) their core (NDUFV2 and NDUFS3) or supernumerary (NDUFS4 and NDUFS5) character; iii) their especial situation in the electron path of the hydrophilic domain (NDUFV2); or iv) their lack of variation in those longevity models to be potentially used as a reference (NDUFA9). Abundance of the VDAC component of the mPTP was also measured due to its possible relationship with mitROSp and aging.
2. Results
2.1. Multivariate statistics reveals a species-specific Cx I profile
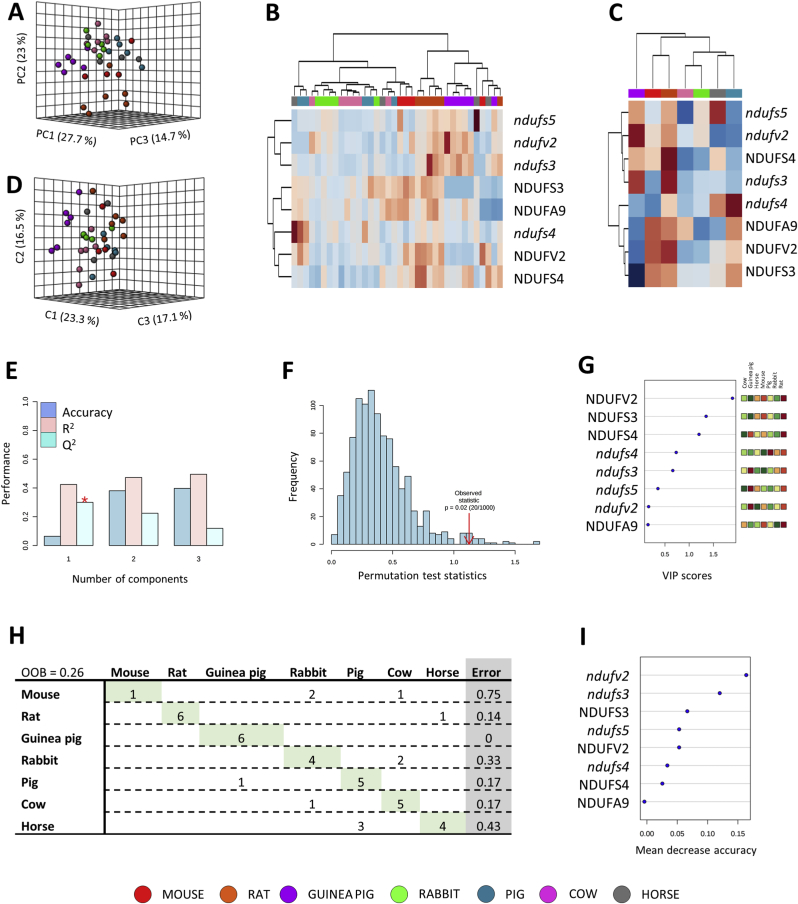
Multivariate statistics was applied to determine whether specific Cx I subunits gene expression (ndufv2, ndufs3, ndufs4 and ndufs5) and protein content (NDUFV2, NDUFS3, NDUFS4 and NDUFA9) differ among mammals. Non-supervised principal component analysis (PCA) revealed the existence of a species-specific gene and protein profile of the Cx I, capable to explain up to 65.4% samples variability (Fig. 1A). A hierarchical clustering of the samples represented by a heat map revealed specific Cx I patterns for rodents (mouse, rat and guinea pig) and non-rodents (rabbit, pig, cow and horse) (Fig. 1B and C). These results were confirmed by performing a supervised analysis, such as partial least squares discriminant analysis (PLS-DA) (Fig. 1D). Cross-validation values of PLS-DA model scored a maximum Q2 of 0.3 and R2 of 0.4 when using only one component (Fig. 1E). Permutation tests (1000 repeats) yielded a low p = 0.02, indicating that none of the distributions formed by the permuted data was better than the observed statistic based on the original data (Fig. 1F). The inter-group discriminating power of the different measured features is evaluated by a variable importance projection (VIP) score > 1.5, which was obtained only for NDUFV2 (Fig. 1G). These results were supported by applying a Random Forest (RF) classification algorithm which revealed a species overall classification error of 0.26, being ndufv2 the variable with the highest contribution to classification accuracy (Fig. 1H and I).
Fig. 1.
Multivariate statistics reveals a species-specific gene expression and protein content of Cx I. A) Principal component analyses (PCA) representation of gene expression and protein content of CI subunits. X: Principal component 1 (PC1); Y: Principal component 2 (PC2); Z: Principal component 3 (PC3). B) Hierarchical clustering of individual animal samples according to gene expression and protein content of CI. C) Hierarchical clustering of animal species according to average sample values of gene expression and protein content of CI. D) Partial least squares discriminant analysis (PLS-DA) representation of gene expression and protein content of CI. X: Component 1 (C1); Y: Component 2 (C2); Z: Component 3 (C3). E) Cross validation (CV) analyses (10-fold CV method) of the PLS-DA model. F) Permutation test (1000 repeats) using separation distance. G) Variable importance projection (VIP) scores indicating the elements which contribute the most to define the first component of a PLS-DA. H) Random Forest (RF) classification algorithm. I) VIP scores for RF.
2.2. The species-specific Cx I profile is associated with animal longevity
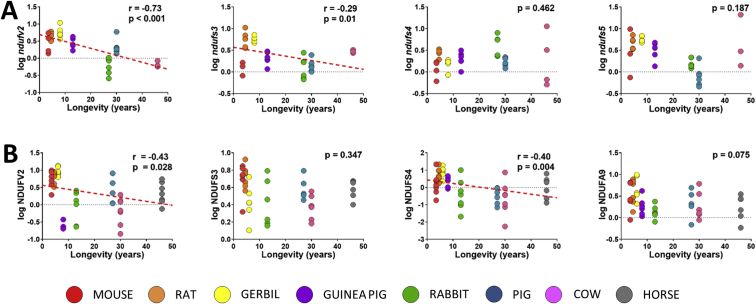
Gene expression and protein content of Cx I were also correlated with mammalian longevity. Specifically, long-lived animals showed decreased gene expression of ndufv2 and ndufs3 (Fig. 2A). Moreover, protein content of NDUFV2 and NDUFS4 were also decreased in long-lived mammals (Fig. 2B). Yet no correlation between protein content and gene expression was found (Supplementary Fig. 1).
Fig. 2.
Cx I gene expression (A) and protein content (B) are linearly correlated with mammalian longevity. Pearson correlation was performed between maximum longevity (ML) and gene expression or protein content. Linear regression was applied when significant relationships were found. R2(ndufv2) = 0.48; R2(ndufs3) = 0.20; R2(NDUFV2) = 0.10; R2(NDUFS4) = 0.15. NDUFA5 protein results aren't reported due to technical issues. Expression of ndufs9 is not reported since is used as a housekeeping to normalize the data. Minimum signification level was set at p<0.05. Gene expression and protein content were log-transformed to accomplish the assumptions of normality.
2.3. The VDAC content is associated with animal longevity
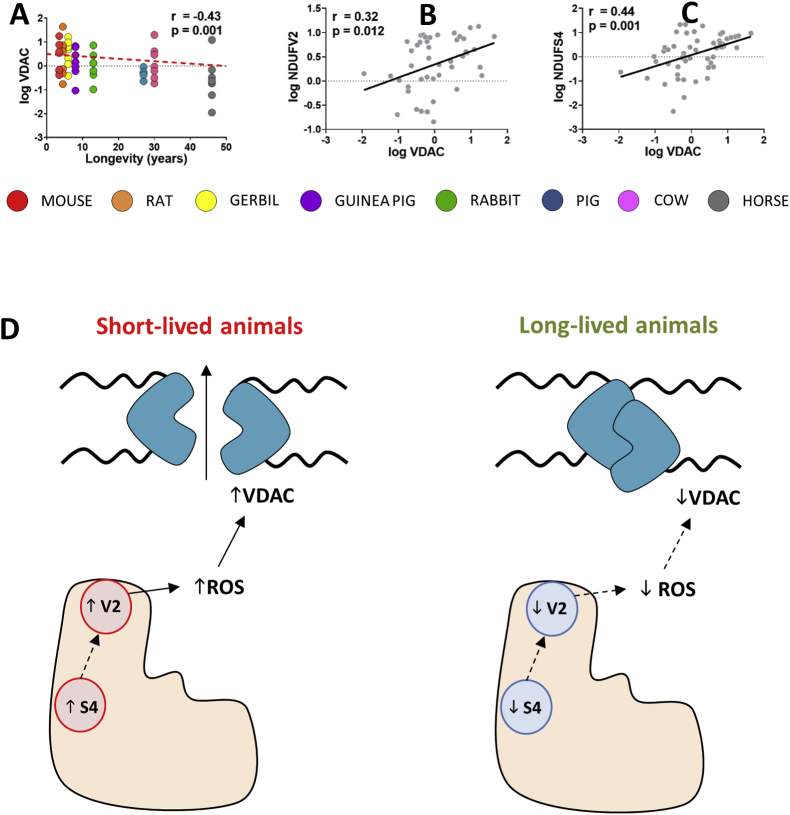
VDAC is a component of the mitochondrial permeability transition pore (mPTP) and is commonly used as a loading control for mitochondrial proteins. In order to check its suitability when comparing different animals, we've compared total VDAC content by performing an ANOVA analysis. Unexpectedly, we've found that total VDAC content changes across the different animal species and is negatively correlated with animal longevity (Fig. 3A). Furthermore, VDAC total content was positively correlated with NDUFV2 and NDUFS4 protein content (Fig. 3B and C, Supplementary Fig. 1).
Fig. 3.
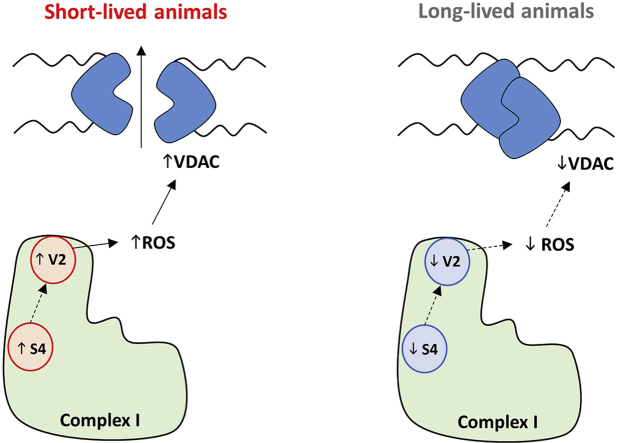
VDAC-1 changes across animal longevity and its association with Cx I subunits and animal longevity. A) Pearson correlation between VDAC-1 protein content and animal longevity. R2(VDAC) = 0.18. B) PGLS regression between VDAC-1 protein content and animal longevity according to phylogenetic tree reported in Fig. 3A. C) Pearson correlation between VDAC-1 and complex 1 subunits. Linear regression (LR) model was performed when significant relationships were found. R2(VDAC vs. NDUFV2) = 0.13; R2(VDAC vs. NDUFS4) = 0.19. Minimum signification level was set at p<0.05. Protein content was log-transformed to accomplish the assumptions of normality. D) Longevity model of longevity modulation via Cx I, ROS and VDAC.
2.4. ndufv2 and VDAC correlate with longevity after controlling for phylogenetic relationships
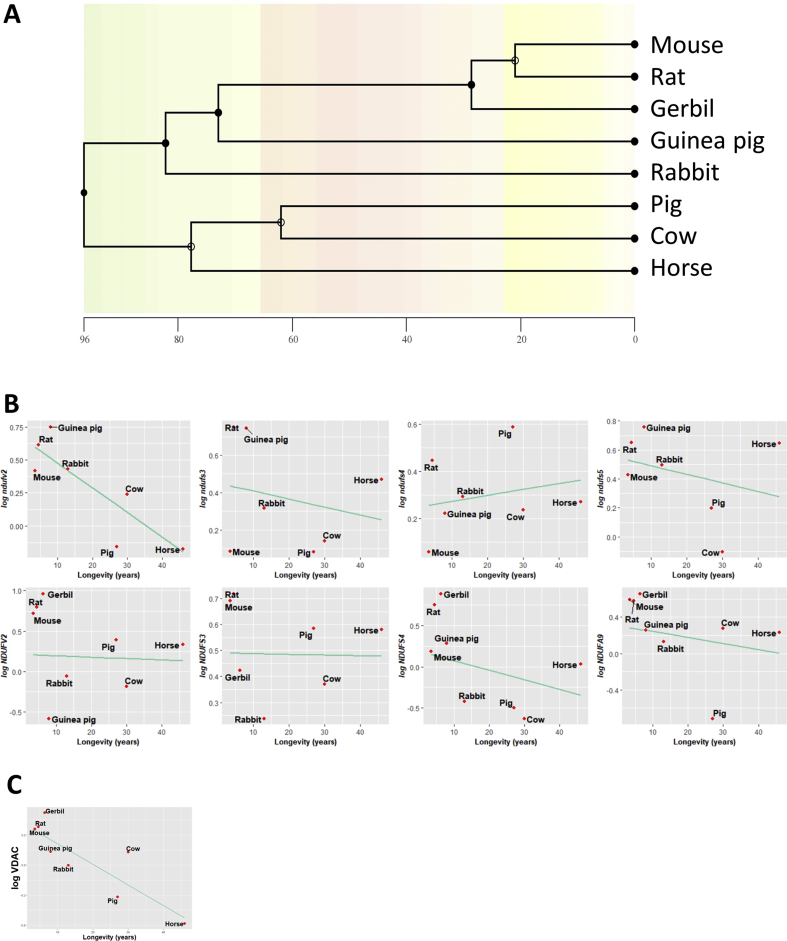
Animal species are evolutionarily related, and closely related species often have similar traits due to inheritance from a common ancestor. Most of statistical analysis, such as linear regression, assume the independence of the data, which might not be accomplished from data obtained from these close-related species. In order to find associations between longevity and gene expression and protein content of selected Cx I subunits and VDAC protein content, we have applied a phylogenetic comparative method, such as phylogenetically generalised least squares (PGLS) regression. A phylogenetic tree allowing to evolutionary relate the species in our study was inferred and is constructed in Supplementary Fig. 2A.
First of all, under the assumption of a Brownian motion model of evolution (a branching, random walk of trait values from an ancestral value at the root to the tips of the tree [30]), we have estimated the Pagels λ. It allows to measure phylogenetic signal and indicates the relative extent to which a traits’ correlation among close relatives match a Brownian motion model of trait evolution. Pagels λ range from 0 to 1, where λ = 1 indicate that trait similarities between species are influenced by phylogenetic relationships; λ = 0 indicate that trait similarities between species are independent of phylogenetic relationships; and 0<λ<1 indicate different levels of phylogenetic signal. According to the estimated λ value, we have classified the measured traits according to its association degree with phylogenetic relationships: i) independent (λ = 0): ndufs5 and NDUFS3; ii) low dependence: ndufs3 and NDUFA9; iii) mild dependence: ndufv2 (λ = 0.65), NDUFV2 (λ = 0.81) and VDAC (λ = 0.53); and strong dependence (λ = 1): ndufs4 and NDUFS4. Finally, we have applied a PGLS regression, which revealed that only gene expression of ndufv2 (p = 0.041, r = −0.80) (Supplementary Fig. 2B) and protein content of VDAC (p = 0.015, r = −0.79) (Supplementary Fig. 2C) were positively correlated with animal longevity after controlling for phylogenetic relationships.
3. Discussion
3.1. Longevity-associated Cx I profile
In this investigation we have found that the mitochondrial Cx I profile is associated with mammalian longevity. Our model revealed that Cx I accounts for 40% of inter-species variation, being ndufv2 gene expression and protein content, followed by NDUFS4 protein, the highest longevity predictor. To the best of our knowledge, ours is the first study elucidating the genetic changes in particular Cx I subunits across mammals of different orders revealing new insights on animal longevity.
3.2. Low Cx I content in long-lived mammalian species
Mitochondrial-related genes have been positively selected in long-living invertebrates [31] and mammals, including rodents [32], bats [33] and primates [34]. Our results showed a decreased matrix Cx I subunits gene expression in the same direction that protein content. These findings agree with previous studies that show a positive selection during evolution of gene expression of Cx I subunits, as well as genes that regulate its assembly [35]. Our study design revealed decreased gene expression and/or amount of particular Cx I subunits, suggesting that longevity is associated with a genetic modulation targeted to maintain a lower content of those hydrophilic Cx I matrix domain subunits.
The core Cx I structure is highly conserved across animal species [36] and maintains a fixed stoichiometric relationship of 1:1 in their forming subunits [37]. Since we have found different protein content of the core subunits NDUFV2 and NDUFS4 in different animal species, we suggest that the lowest amount of core subunits determines the total amount of fully assembled Cx I. Accordingly, several studies extrapolated changes in specific Cx I subunits to total Cx I content variations [14,18,38,39]. Furthermore, we are measuring the total content of these proteins, but not its intracellular location. Therefore, we cannot discard that these stoichiometric differences are due to the presence of free subunits at the mitochondrial matrix. Supporting the latter, it has been described different protein turnover rates for each Cx I subunit (up to 4.6-fold difference), which is higher in matrix subunits compared to those located in the membrane [40].
Although ROS can be produced at many different mitochondrial sites, studies including those with endogenous reporters in the absence of inhibitors have localized the main ROS generator sites at the hydrophilic Cx I domain and at Cx III [2,41]. Among these two, the low ROS production of long-lived species and dietary restricted animals has been localized at Cx I [3,[6], [7], [8], [9]], particularly to its matrix hydrophilic domain [10] which catalyses oxidation-reduction reactions, while the hydrophobic membrane domain catalyses proton pumping (Fig. 4). ROS production at the hydrophilic Cx I domain can occur either at the flavin (IF site) or at some of its FeS clusters [[11], [12], [13]], which can also further contribute to secondary ROS formation due to Fe liberation under oxidative stress [1]. Adscription of ROS production to the IF site has been based on experiments increasing or decreasing flavin reduction concomitant with increases and decreases in ROS production respectively. However, these experiments can't rule out Cx I FeS clusters as potential ROS generators because they are located downstream of the flavin, and then increases and decreases in FeS reduction will also occur in those experiments. Studying the amounts of particular Cx I subunits of the hydrophilic Cx I domain can further localize the sites, subunits and genes responsible for the longevity related decrease in mitROSp.
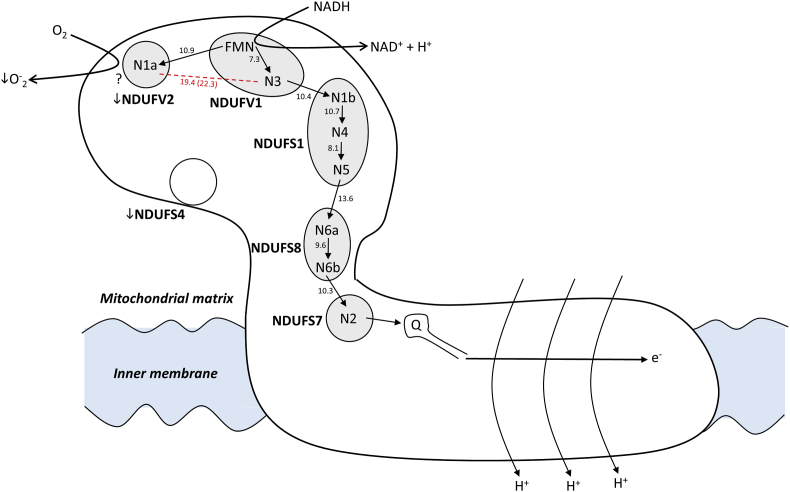
Fig. 4.
Low NDUFV2 and NDUFS4 in long-lived species can decrease mitROSp from the hydrophilic domain of Cx I. The figure shows the pathway of electron transfer. The human nomenclature for Cx I subunits (NDUF) is used. Electron transfer is indicated by solid arrows from FMN to Fe S clusters N1a, N3, N1b, N4, N5, N6a, N6b and N2 which reduces the quinone. The conserved off-pathway bifurcation from the flavin to FeS cluster N1a (in NDUFV2) is shown as alternative to main electron transfer to N3. Edge-to-edge distances between cofactors indicated in Å correspond to those described for the mammalian ovine enzyme studied by cryo-Electron Microscopy [20]. The distance between iron-sulphur clusters N1a and N3 (T. thermophilus, Sazanov 2015) is too long for physiological electron transfer to occur either edge-to-edge (19,4 Å) or centre-to-centre (22.3 Å, shown inside brackets). Therefore, the electron transferred from FMN to 2Fe2s cluster N1a could then reduce oxygen to superoxide radical at NDUFV2. A low amount of NDUFV2 in long-lived species would divert less electrons from the main pathway and would thus lead to a lower rate of complex I ROS production. A lower amount of the accessory subunit NDUFS4 in long-lived species can also downregulate ROS generation at N1a through its known interaction with NDUFV2.
3.3. Low abundance of NDUFV2 and NDUFS4 subunits is associated with mammalian longevity
The functional role of the Cx I accessory subunits is fragmentary and not clear [21,22] although it has been proposed that they have regulatory roles participating in Cx I stabilization, assembly, subunit interactions or limitation of ROS production [21].
Among the CxI subunits selected, NDUFVS3 and NDUFS5 did not show correlation with species longevity in spite of their decrease in DR and in the long-lived ICRFa mouse strain (moderate decreases), and the increase in NDUFS3 with age in C57Bl/6 mice [10]. The lack of correlation with longevity observed for NDUFS5 is not strange however, since recent description of the near atomic structure of bovine and ovine Cx I at around 4 Å resolution by cryoEM localized NDUFS5 at the bottom of the hydrophobic domain near the intermembrane space [20,21], far away from the Cx I hydrophilic domain sites of ROS production. Concerning NDUFS3, it is an accessory subunit without cofactor that does not take part directly on the main electron transfer but rather provides the platform required for complex assembly [22]. The lack of correlation of the NDUFA9 subunit with species longevity was expected since it did not show correlation in other longevity models [10], it is a subunit essential for activity that binds NADPH to indirectly regulate the terminal N2 cluster [22], and is present at the bottom of the hydrophilic domain near the hinge connecting the two Cx I domains. This protein also has a structural function because it stabilizes the junction between the membrane and matrix domains of Cx I [42].
The remaining two subunits studied, the NDUFV2 and NDUFS4 proteins, showed negative correlation with species longevity. In principle, regulation of ROS production should be left for accessory subunits because the central core is needed for the most fundamental function, electron transfer coupled to H+ pumping and ATP production. However, NDUFV2, which was lower in long-lived species both at mRNA and protein levels, is a special case because it does not take part in the main electron path of the hydrophilic domain of Cx I. Instead, the highly conserved off-pathway N1a FeS cofactor of NDUFV2 can take one electron from FMN but cannot give it to N3 because the distance from N1a (in subunit NDUFV2) to N3 (19.4 to 22.3 Å, Fig. 4) is too long compared to the maximum distance for physiological electron transfer between redox centers, 14 Å [43]. It has been proposed that N1a can function to prevent excessive ROS production [44,45]. The very low one electron potential of N1a (around −370mV) compared to all the other isopotential FeS clusters (N3 to N6b at around −250 mV) makes it highly susceptible to one electron reduction of oxygen to superoxide radical (Fig. 4). The low NUDFV2 (and N1a) amount present in long-lived species agrees with the hypothesis that it can serve to limit ROS production, since the smaller the NDUFV2 amount, the less the number of electrons transferred from FMN to N1a, and thus less would reduce O2 to O2.-, while more electrons would follow the central path from the flavin to N3 and then the rest of the FeS until N2 (Fig. 4). This could help to explain the smaller amount of ROS produced per unit electron flow in long-lived species. This mechanism is simple and avoids the need for electrons to travel bidirectionally between the flavin and N1a [44]. Therefore, we postulate that the amount of NDUFV2 subunit could serve to modulate ROS production in each species in accordance with its longevity and would help to explain the functional significance of the off-path situation of cluster N1a. Interestingly, it has been found that phosphorylation of NDUFV2 tyrosine 118 decreases ROS production leading to cardioprotection after ischemia-reperfusion [46].
The supernumerary non cofactor containing NDUFS4 was the other subunit showing less protein level amount in long-lived species, although its mRNA levels did not show significant differences. Such low level agrees with its strongly decreased levels in long-lived DR mice [10]. The accessory NDUFS4 subunit is located where the NADH dehydrogenase domain meets the rest of the hydrophilic arm [21] and is not likely to play a catalytic role but it is important for assembly of the complex [47]. Various NDUFS4 mutations lead to total loss of Cx I activity [48].
It is known that mammalian NDUFSV1, V3, S1, S4 and S6 interact with NDUFV2 [20,21]. Among all those, only NDUFS4 shows a strong decrease in DR [10] the rest being invariant or showing only modest decreases. Therefore, although NDUFS4 does not have an electron transport cofactor, it could modulate ROS production through its interaction with NDUFV2, which would be affected by the lower NDUFS4 levels of long-lived species. In agreement with this possibility, it is known that cAMP associated serine phosphorylation of NDUFS4 by PKA lowers ROS production and increases Cx I activity [49], and heart-specific NDUFS4 knockout mice have normal heart energetics and function together with less ROS generation and better functional recovery after ischemia-reperfusion while reintroduction of NDUFS4 abolishes this protection [50].
Although our finding of smaller amounts of NDUFV2 and NDUFS4 can contribute to decrease Cx I ROS production in long-lived species, association of complex I within supercomplexes could perhaps also contribute to ROS regulation. Possible functional roles of respiratory supercomplexes remain controversial, proposals including roles in stability of individual complexes, CoQ substrate channeling between Cx I and Cx III2, prevention of protein aggregation, and reduction in ROS production. Various authors have suggested that assembly of Cx I into supercomplexes prevents excessive ROS production during oxidation of NADH-linked substrates because more efficient CoQ channeling helps in maintaining the chain in a more oxidized state [51,52]. However, despite progress in the determination of the structures of respiratory supercomplexes their physiological role is not yet understood [22] and more research is needed to clarify if they could also contribute to modulate mitROSp in relation to animal longevity.
3.4. There is an association between mitROSp and VDAC content in animal longevity
ROS production promotes the opening of the mitochondrial permeability transition pore mPTP [25] which is related to mitochondrial swelling and fragmentation, ischemia-reperfusion injury, and cell death [1,50,53]. Specifically, ROS generation and reverse electron transport through Cx I increase the sensitivity of mPTP opening [54]. Previous studies have shown that dietary restriction decreases both mPTP [55,56] and mitROSp at Cx I [3,9]. Since mPTP opening is promoted through oxidation of mitochondrial membrane lipids and proteins by ROS [23], the decrease in mitochondrial permeability in DR could be due to the decrease in mitROSp that occurs in this dietary manipulation.
VDAC is a component of mPTP [[26], [27], [28],57]. Some studies indicate that VDAC content regulates the opening of mPTP in response to modifications in mitROSp [23,25]. Since decreases in Cx I content correlate with reduction in mitROSp [10,[16], [17], [18], [19]], Cx I content could regulate the mPTP opening mediated by mitROSp modifications.
On the other hand, our results show that a lower VDAC content is associated with mammalian longevity. Furthermore, VDAC is positively correlated with NDUFV2 and NDUFS4 Cx I subunits which are present at higher levels in short-lived animals. Thus, we hypothesize that a lower level of NDUFV2 and NDUFS4 would lead to lower complex I ROSp, lower ROS-mediated VDAC modification and lower mPTP in long-lived animal species (Fig. 3). This agrees with the recent description that high VDAC levels lead to mPTP and to a shortened lifespan [29], as well as with the decrease in mPTP observed in dietary restriction studies [55,56].
3.5. Dealing with Cx I and VDAC variations in evolutionary-related species
Comparative studies across species with different lifespan are a powerful source of information to identify mechanisms linked to extended longevity [58,59]. However, those studies come up with several technical limitations. Specifically, in terms of protein recognition, the presence of SNPs inducing little sequence or amino acid changes might alter protein structure, even in highly conserved structure, possibly affecting antibody recognition. Since we lack of methods to improve inter-species antibody reactivity, we have increased the number of repeated measurements up to 7, in order to capture higher inter-individual variability in each specie, and measured gene expression using a very sensible technique in order to confirm those results. However, we didn't find significative correlations between gene expression and protein content. Although we cannot omit technical issues, the lack of correlation doesn't necessarily invalidate the presented results. Accordingly, it had been reported that mRNA and protein content doesn't need to be necessarily correlated [60], even in mammals [61]. Furthermore, a comparative study has demonstrated an increasing protein divergence with higher evolutionary distance, since expression divergence seems to be more stable [62], which agrees with the obtained data.
Even after dealing with technical issues, inter-species studies have to face that evolutionary relationships don't allow for independence of the data [63]. Therefore, it's important to elucidate whether a specific trait correlates with longevity differences, or alternatively, these differences arise because of the data similarity. To overcome this limitation, several longevity cross-species studies in birds [64], bats [65], primates [34], mammals [66,67] and vertebrates [68] have applied phylogenetic comparative methods, such as PGLS. In our data, PGLS revealed that ndufv2 gene expression and VDAC protein content changes associated with longevity weren't due interspecies relationships. Although robust, the main limitation of the methods for phylogenetic analysis is that they rely on the construction of a phylogenetic tree, which is unknown. Phylogenies are estimated, mainly by aligning homologue gene sequences and using models of mutation to infer most-likely evolutionary histories. However, depending on the genes chosen, how the sequences are aligned and which method is used to infer evolutionary histories we get different phylogenies. And errors in phylogenetic inferences propagate to errors in phylogenetic analyses [30]. These limitations arise the need of the development of more robust bioinformatic methodologies for phylogenetic tree inferences.
4. Conclusions
Our study indicates that long-lived mammalian species have low levels of NDUFV2 and NDUFS4 subunits of the hydrophilic domain of complex I which could be responsible in part for their lower mitROSp and superior longevity. The lower VDAC content of these animals could also contribute to their high longevity due to lower ROS-mediated opening of the mitochondrial permeability transition pore.
5. Methods
5.1. Animals
Mammalian species included in the study were male adult specimens with an age representing their 15–30% of their longevity. The recorded values for longevity (in years) were: mouse (Mus musculus), 3.5; rat (Rattus norvegicus), 4.5; gerbil (Meriones unguiculatus), 6.3 guinea pig (Cavia porcellus), 8; rabbit (Oryctolagus cuniculus), 13; pig (Sus scrofa), 27; cow (Bos taurus), 30; and horse (Equus caballus), 46. Rodents and rabbits were obtained from rodent husbandries and sacrificed by decapitation, whereas sheep, pigs, cow, and horses were obtained from abattoirs. The animal care protocols were approved by the Animal Experimentation Ethics committee of the University of Lleida. Heart ventricles from 5-7 animals were removed and immediately frozen in liquid nitrogen and transferred to −80 °C until analyses.
5.2. Sample homogenization and quantification
Heart tissue (≈50 mg of whole tissue) was homogenized in a buffer containing 180 mM KCl, 5 mM MOPS, 2 mM EDTA, 1 mM DTPAC adjusted to pH = 7.4. Prior homogenization, 1 μM BHT and a mix of proteases inhibitors (GE80-6501-23, Sigma, Madrid, Spain) and phosphatase inhibitors (1 mM Na3VO4, 1 M NaF) were added. After a brief centrifugation (1000 rpm for 3 min at 4 °C), supernatants protein concentration was measured using the Bradford method (500-0006, Bio-Rad Laboratories, Barcelona, Spain).
5.3. Protein content determination
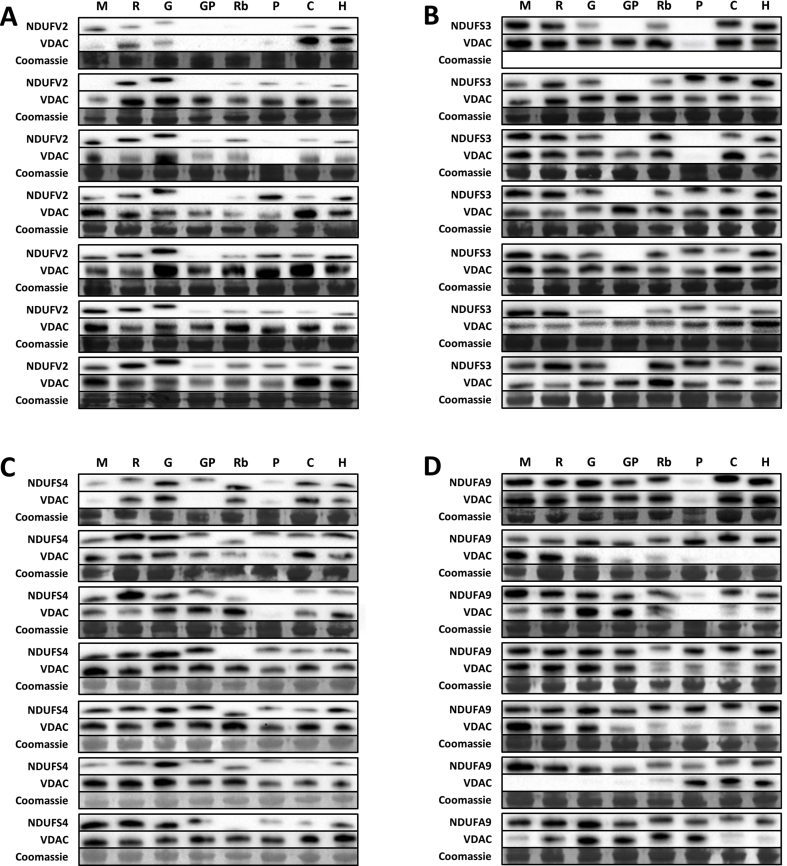
The amount of the specific Cx I subunits NDUFV2, NDUFS3, NDUFS4, NDUFS5 and NDUFA9, as well as VDAC-1/Porin, were estimated using western blot analyses as previously described [39]. Anticipating potential technical limitations mainly due to antibody reactivity, and before performing the comparative approach, we have evaluated the similarity of the protein sequences using a BLAST algorithm (Supplementary Table 1). Although the exact target sequence of the commercial antibodies is unknown, we have found a high similarity between sequences, and have chosen the most appropriate antibodies available according to their proved or predicted reactivity against the species of our study.
Briefly, heart homogenates were mixed with a buffer containing 62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 20% β-mercaptoethanol and 0.02% bromophenol blue, and heated for 3 min at 95 °C. Then, proteins were subjected to one-dimensional electrophoresis on SDS and transferred to PVDF membranes. Membranes were maintained in blocking solution containing Tris 2 M, NaCl 2.5 M, 5% BSA and 0.01% Tween for 1 h at room temperature. Immunodetection was performed using antibodies against NDUFV2 (SAB2107279, Sigma, Madrid, Spain), NDUFS3 (459130, ThermoFisher, Barcelona, Spain), NDUFS4 (ab96549, Abcam, Cambridge, UK), NDUFS5 (ab188510, Abcam, Cambridge, UK), NDUFA9 (459100, ThermoFisher, Barcelona, Spain), and VDAC-1/Porin (ab15898, Abcam, Cambridge, UK).
Secondary antibodies were anti-mouse (GENA931, Sigma, Madrid, Spain) and anti-rabbit (31460, ThermoFisher, Barcelona, Spain). Bands were visualized using an enhanced chemiluminescence HRP substrate (Millipore, MA, USA). Signal quantification and recording was performed with ChemiDoc equipment (Bio-Rad Laboratories, Barcelona, Spain). The protein amount was calculated from the ratio of their densitometry values referred to its respective Coomassie staining (1610436, Bio-Rad Laboratories, Barcelona, Spain) (Supplementary Fig. 3). Due to technical issues (antibody recognition), NDUFA5 amount is not reported. Lack of protein recognition might be due to technical issues, not to missing proteins, such as for NDUFS3 in guinea pig.
5.4. Primers design
Gene cDNA sequences coding for the Cx I subunits NDUFV2 (ndufv2), NDUFS3 (ndufs3), NDUFS4 (ndufs4), NDUFS5 (ndufs5) and NDUFA9 (ndufa9) were obtained from Ensmbl (http://www.ensmbl.org). Due to gene cDNA sequences limitations for gerbil, that specie wasn't included in the gene expression analyses. Degenerate primers were designed to amplify conserved regions among mammalian sequences using the software PriFi [69], and are listed in Supplementary Table 2. Primers were purchased from Isogen (LifeSciences, Utretch, Netherlands).
5.5. Gene expression: Droplet digital PCR
Prior DNA amplification, RNA from 15 mg whole heart tissue was extracted using RNeasy Fibrous Tissue Mini Kit (Qiagen, Hilden, Germany), and retro-transcribed to cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, CA, USA).
For DNA absolute quantification (copies/μl), reaction mixture contained 1x of EvaGreen ddPCR Supermix, 200 nM primers and 0.01–16 ng of template cDNA. For droplet generation, 20 μL of reaction mixture and 70 μL of Droplet Generation Oil for EvaGreen were loaded in the Droplet Generation Cartridge, which was placed into the Droplet generator. From each PCR reaction mixture, 20 μL were transferred to a 96-well PCR plate, which was sealed with a foil heat using PX1 PCR plate sealer. Amplification was performed in a C1000 Touch Thermal Cycler following an initial DNA Polymerase activation (95 °C, 5 min), and 40 cycles consisting of a DNA denaturation (95 °C, 30 s), primer annealing (58 °C, 1 min) and extension (60 °C, 1 min). A final dye-stabilization step was included (4 °C 5 min, and 90 °C 5 min). Droplets were read with a QX200 Droplet Reader and analysed using QuantaSoft software (Bio-Rad). The results from more than 12,000 droplets were accepted and normalised to an appropriate housekeeping gene (ndufa9) as suggested by previous work [10]. Values are reported as cDNA gene units per cDNA housekeeping units. All equipment and reagents were purchased from Bio-Rad (Bio-Rad Laboratories, Barcelona, Spain).
5.6. Statistics
Multivariate statistics was performed using Metaboanalyst software [70]. Pearson correlation and Pearson partial correlation were performed using RStudio (v1.1.453). Linear models and phylogenetic generalised least squares regression (PGLS) were constructed using RStudio (v1.1.453). The phylogenetic tree was constructed using taxa names as described previously [71]. Functions used were included in the package caper. Data was log-transformed and mean-centred prior statistical analyses in order to accomplish the assumptions of linearity.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness, Institute of Health Carlos III (grant number PI14/00328), the Spanish Ministry of Science, Innovation and Universities (RTI2018-099200-B-I00), and the Generalitat of Catalonia, Agency for Management of University and Research Grants (2017SGR696) and Department of Health (SLT002/16/00250) to R.P, and PR [19] BIO MET 0155 to GB.This study has been co-financed by FEDER funds from the European Union (“A way to build Europe”). IRBLleida is a CERCA Programme/Generalitat of Catalonia.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
M.J. is a ‘Serra Hunter’ Fellow. N.M-M received a predoctoral fellowship from the Generalitat of Catalonia (AGAUR, ref 2018FI_B2_00104). We thank Dr. Eloi Gari, from the Basic Medical Sciences Department (UdL-IRBLleida) for technical support in gene expression analysis. We thank Salvador Batolome, from the Laboratory of Luminescense and Biomolecular Spectroscopy (Autonomous University of Barcelona, Barcelona, Catalonia, Spain), for ddPCR technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101539.
Contributor Information
Natalia Mota-Martorell, Email: Natalia.mota@mex.udl.cat.
Mariona Jove, Email: mariona.jove@udl.cat.
Irene Pradas, Email: ipradas@mex.udl.cat.
Isabel Sanchez, Email: Isabel.sanchez@udl.cat.
José Gómez, Email: jose.gomezs@urjc.es.
Alba Naudi, Email: alba.naudi@mex.udl.cat.
Gustavo Barja, Email: gbarja@bio.ucm.es.
Reinald Pamplona, Email: reinald.pamplona@mex.udl.cat.
Author contributions
G.B and R.P designed the study. N.M.M., M.J., I.P., I.S., J.G., and A.N. performed experimental work. N.M.M and R.P. analysed the data. R.P. supervised the design and data interpretation. The manuscript was written by N.M.M, J.G, G.B. and R.P. and edited by R.P. All authors discussed the results and commented on the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
References
- 1.Koopman W.J.H., Nijtmans L.G.J., Dieteren C.E.J., Roestenberg P., Valsecchi F., Smeitink J.A.M., Willems P.H.G.M. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxidants Redox Signal. 2010;12:1431–1470. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- 2.Wong H.-S., Dighe P.A., Mezera V., Monternier P.-A., Brand M.D. Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J. Biol. Chem. 2017;292:16804–16809. doi: 10.1074/jbc.R117.789271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pamplona R., Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Res. Rev. 2007;6:189–210. doi: 10.1016/j.arr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Barja G. Updating the mitochondrial free radical theory of aging: an integrated view, key aspects, and confounding concepts. Antioxidants Redox Signal. 2013;19:1420–1445. doi: 10.1089/ars.2012.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku H.H., Brunk U.T., Sohal R.S. Relationship between mitochondrial superoxide and hydrogen peroxide production and longevity of mammalian species. Free Radic. Biol. Med. 1993;15:621–627. doi: 10.1016/0891-5849(93)90165-q. [DOI] [PubMed] [Google Scholar]
- 6.Herrero A., Barja G. Sites and mechanisms responsible for the low rate of free radical production of heart mitochondria in the long-lived pigeon. Mech. Ageing Dev. 1997;98:95–111. doi: 10.1016/S0047-6374(97)00076-6. [DOI] [PubMed] [Google Scholar]
- 7.Herrero A., Barja G. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: sites of free radical generation and mechanisms involved. Mech. Ageing Dev. 1998;103:133–146. doi: 10.1016/S0047-6374(98)00035-9. [DOI] [PubMed] [Google Scholar]
- 8.Barja G., Herrero A. Localization at complex I and mechanism of the higher free radical production of brain nonsynaptic mitochondria in the short-lived rat than in the longevous pigeon. J. Bioenerg. Biomembr. 1998;30:235–243. doi: 10.1023/a:1020592719405. [DOI] [PubMed] [Google Scholar]
- 9.Pamplona R., Barja G. Mitochondrial oxidative stress, aging and caloric restriction: the protein and methionine connection. Biochim. Biophys. Acta Bioenerg. 2006;1757:496–508. doi: 10.1016/j.bbabio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Miwa S., Jow H., Baty K., Johnson A., Czapiewski R., Saretzki G., Treumann A., Von Zglinicki T. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero A., Barja G. Localization of the site of oxygen radical generation inside the complex I of heart and nonsynaptic brain mammalian mitochondria. J. Bioenerg. Biomembr. 2000;32:609–615. doi: 10.1023/a:1005626712319. [DOI] [PubMed] [Google Scholar]
- 12.Genova M.L., Ventura B., Giuliano G., Bovina C., Formiggini G., Parenti Castelli G., Lenaz G. The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett. 2001;505:364–368. doi: 10.1016/S0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 13.Kushnareva Y., Murphy A.N., Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation–reduction state. Biochem. J. 2002;368:545–553. doi: 10.1042/bj20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Pamplona, M. Portero-Otin, A. Sanz, V. Ayala, E. Vasileva, G. Barja, Protein and lipid oxidative damage and complex I content are lower in the brain of budgerigar and canaries than in mice. Relation to aging rate, Age (Omaha). 27 (2005) 267-280. 10.1007/s11357-005-4562-x. [DOI] [PMC free article] [PubMed]
- 15.Lambert A.J., Buckingham J.A., Boysen H.M., Brand M.D. Low complex I content explains the low hydrogen peroxide production rate of heart mitochondria from the long-lived pigeon, Columba livia. Aging Cell. 2010;9:78–91. doi: 10.1111/j.1474-9726.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 16.Sanz A., Caro P., Ayala V., Portero-Otin M., Pamplona R., Barja G. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. Faseb. J. 2006;20:1064–1073. doi: 10.1096/fj.05-5568com. [DOI] [PubMed] [Google Scholar]
- 17.Gómez J., Caro P., Naudí A., Portero-Otin M., Pamplona R., Barja G. Effect of 8.5% and 25% caloric restriction on mitochondrial free radical production and oxidative stress in rat liver. Biogerontology. 2007;8:555–566. doi: 10.1007/s10522-007-9099-1. [DOI] [PubMed] [Google Scholar]
- 18.Ayala V., Naudi A., Sanz A., Caro P., Portero-Otin M., Barja G., Pamplona R. Dietary protein restriction decreases oxidative protein damage, peroxidizability index, and mitochondrial complex I content in rat liver. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2007;62:352–360. doi: 10.1093/gerona/62.4.352. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Roman I., Gomez A., Gomez J., Suarez H., Sanchez C., Naudi A., Ayala V., Portero-Otin M., Lopez-Torres M., Pamplona R., Barja G. Forty percent methionine restriction lowers DNA methylation, complex I ROS generation, and oxidative damage to mtDNA and mitochondrial proteins in rat heart. J. Bioenerg. Biomembr. 2011;43:699–708. doi: 10.1007/s10863-011-9389-9. [DOI] [PubMed] [Google Scholar]
- 20.Fiedorczuk K., Letts J.A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Vinothkumar K.R., Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiedorczuk K., Sazanov L.A. Mammalian mitochondrial complex I structure and disease-causing mutations. Trends Cell Biol. 2018;28:835–867. doi: 10.1016/j.tcb.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Panel M., Ghaleh B., Morin D. Mitochondria and aging: a role for the mitochondrial transition pore? Aging Cell. 2018;17 doi: 10.1111/acel.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haworth R.A., Hunter D.R. The Ca2+-induced membrane transition in mitochondria. Arch. Biochem. Biophys. 1979;195:460–467. doi: 10.1016/0003-9861(79)90372-2. [DOI] [PubMed] [Google Scholar]
- 25.Rottenberg H., Hoek J.B. The path from mitochondrial ROS to aging runs through the mitochondrial permeability transition pore. Aging Cell. 2017;16:943–955. doi: 10.1111/acel.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halestrap A.P. What is the mitochondrial permeability transition pore? J. Mol. Cell. Cardiol. 2009;46:821–831. doi: 10.1016/j.yjmcc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Mnatsakanyan N., Beutner G., Porter G.A., Alavian K.N., Jonas E.A. Physiological roles of the mitochondrial permeability transition pore. J. Bioenerg. Biomembr. 2017;49:13–25. doi: 10.1007/s10863-016-9652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mnatsakanyan N., Llaguno M.C., Yang Y., Yan Y., Weber J., Sigworth F.J., Jonas E.A. A mitochondrial megachannel resides in monomeric F1FO ATP synthase. Nat. Commun. 2019;10:5823. doi: 10.1038/s41467-019-13766-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou B., Kreuzer J., Kumsta C., Wu L., Kamer K.J., Cedillo L., Zhang Y., Li S., Kacergis M.C., Webster C.M., Fejes-Toth G., Naray-Fejes-Toth A., Das S., Hansen M., Haas W., Soukas A.A. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell. 2019;177:299–314. doi: 10.1016/j.cell.2019.02.013. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washburne A.D., Morton J.T., Sanders J., McDonald D., Zhu Q., Oliverio A.M., Knight R. Methods for phylogenetic analysis of microbiome data. Nat. Microbiol. 2018;3:652–661. doi: 10.1038/s41564-018-0156-0. [DOI] [PubMed] [Google Scholar]
- 31.Roux J., Privman E., Moretti S., Daub J.T., Robinson-Rechavi M., Keller L. Patterns of positive selection in seven ant genomes. Mol. Biol. Evol. 2014;31:1661–1685. doi: 10.1093/molbev/msu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahm A., Bens M., Szafranski K., Holtze S., Groth M., Görlach M., Calkhoven C., Müller C., Schwab M., Kraus J., Kestler H.A., Cellerino A., Burda H., Hildebrandt T., Dammann P., Platzer M. Long-lived rodents reveal signatures of positive selection in genes associated with lifespan. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y.-Y., Liang L., Zhu Z.-H., Zhou W.-P., Irwin D.M., Zhang Y.-P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107:8666–8671. doi: 10.1073/pnas.0912613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muntané G., Farré X., Rodríguez J.A., Pegueroles C., Hughes D.A., de Magalhães J.P., Gabaldón T., Navarro A. Biological processes modulating longevity across primates: a phylogenetic genome-phenome analysis. Mol. Biol. Evol. 2018;35:1990–2004. doi: 10.1093/molbev/msy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahm A., Bens M., Platzer M., Cellerino A. Parallel evolution of genes controlling mitonuclear balance in short-lived annual fishes. Aging Cell. 2017;16:488–496. doi: 10.1111/acel.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinothkumar K.R., Zhu J., Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515:80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth C., Brandt U., Hunte C., Zickermann V. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta Bioenerg. 2016;1857:902–914. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-Cisuelo V., Gómez J., García-Junceda I., Naudí A., Cabré R., Mota-Martorell N., López-Torres M., González-Sánchez M., Pamplona R., Barja G. Rapamycin reverses age-related increases in mitochondrial ROS production at complex I, oxidative stress, accumulation of mtDNA fragments inside nuclear DNA, and lipofuscin level, and increases autophagy, in the liver of middle-aged mice. Exp. Gerontol. 2016;83 doi: 10.1016/j.exger.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Gomez A., Gomez J., Torres M.L., Naudi A., Mota-Martorell N., Pamplona R., Barja G. Cysteine dietary supplementation reverses the decrease in mitochondrial ROS production at complex I induced by methionine restriction. J. Bioenerg. Biomembr. 2015;47 doi: 10.1007/s10863-015-9608-x. [DOI] [PubMed] [Google Scholar]
- 40.Karunadharma P.P., Basisty N., Chiao Y.A., Dai D.-F., Drake R., Levy N., Koh W.J., Emond M.J., Kruse S., Marcinek D., Maccoss M.J., Rabinovitch P.S. Respiratory chain protein turnover rates in mice are highly heterogeneous but strikingly conserved across tissues, ages, and treatments. Faseb. J. 2015;29:3582–3592. doi: 10.1096/fj.15-272666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlan C.L., Treberg J.R., Perevoshchikova I.V., Orr A.L., Brand M.D. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radic. Biol. Med. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroud D.A., Formosa L.E., Wijeyeratne X.W., Nguyen T.N., Ryan M.T. Gene knockout using transcription activator-like effector nucleases (TALENs) reveals that human NDUFA9 protein is essential for stabilizing the junction between membrane and matrix arms of complex I. J. Biol. Chem. 2013;288:1685–1690. doi: 10.1074/jbc.C112.436766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page C.C., Moser C.C., Chen X., Dutton P.L. Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 44.Sazanov L.A. Structure of the hydrophilic domain of respiratory complex I from thermus thermophilus. Science. 2006;311:1430–1436. doi: 10.1126/science.1123809. (80-. ) [DOI] [PubMed] [Google Scholar]
- 45.Sazanov L.A. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 46.Xu J., Bian X., Liu Y., Hong L., Teng T., Sun Y., Xu Z. Adenosine A 2 receptor activation ameliorates mitochondrial oxidative stress upon reperfusion through the posttranslational modification of NDUFV2 subunit of complex I in the heart. Free Radic. Biol. Med. 2017;106:208–218. doi: 10.1016/j.freeradbiomed.2017.02.036. [DOI] [PubMed] [Google Scholar]
- 47.Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., Salim A., Ryan M.T. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 48.Budde S.M.S., van den Heuvel L.P.W.J., Janssen A.J., Smeets R.J.P., Buskens C.A.F., DeMeirleir L., Van Coster R., Baethmann M., Voit T., Trijbels J.M.F., Smeitink J.A.M. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 2000;275:63–68. doi: 10.1006/bbrc.2000.3257. [DOI] [PubMed] [Google Scholar]
- 49.Papa S., De Rasmo D., Scacco S., Signorile A., Technikova-Dobrova Z., Palmisano G., Sardanelli A.M., Papa F., Panelli D., Scaringi R., Santeramo A. Mammalian complex I: a regulable and vulnerable pacemaker in mitochondrial respiratory function. Biochim. Biophys. Acta Bioenerg. 2008;1777:719–728. doi: 10.1016/j.bbabio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Gong G., Wang P., Zhang Z., Kolwicz S.C., Rabinovitch P.S., Tian R., Wang W. Heart specific knockout of Ndufs4 ameliorates ischemia reperfusion injury. J. Mol. Cell. Cardiol. 2018;123:38–45. doi: 10.1016/j.yjmcc.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maranzana E., Barbero G., Falasca A.I., Lenaz G., Genova M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxidants Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Fabuel I., Le Douce J., Logan A., James A.M., Bonvento G., Murphy M.P., Almeida A., Bolaños J.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy E., Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol. Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Briston T., Roberts M., Lewis S., Powney B., Staddon J.M., Szabadkai G., Duchen M.R. Mitochondrial permeability transition pore: sensitivity to opening and mechanistic dependence on substrate availability. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-10673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrosillo G., Moro N., Paradies V., Ruggiero F.M., Paradies G. Increased susceptibility to Ca 2+ -induced permeability transition and to cytochrome c release in rat heart mitochondria with aging: effect of melatonin. J. Pineal Res. 2010;48:340–346. doi: 10.1111/j.1600-079X.2010.00758.x. [DOI] [PubMed] [Google Scholar]
- 56.Amigo I., Menezes-Filho S.L., Luévano-Martínez L.A., Chausse B., Kowaltowski A.J. Caloric restriction increases brain mitochondrial calcium retention capacity and protects against excitotoxicity. Aging Cell. 2017;16:73–81. doi: 10.1111/acel.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leanza L., Checchetto V., Biasutto L., Rossa A., Costa R., Bachmann M., Zoratti M., Szabo I. Pharmacological modulation of mitochondrial ion channels. Br. J. Pharmacol. 2019;176:4258–4283. doi: 10.1111/bph.14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma S., Yim S.H., Lee S.-G., Kim E.B., Lee S.-R., Chang K.-T., Buffenstein R., Lewis K.N., Park T.J., Miller R.A., Clish C.B., Gladyshev V.N. Organization of the mammalian metabolome according to organ function, lineage specialization and longevity. Cell Metabol. 2015;22:332–343. doi: 10.1016/j.cmet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bozek K., Khrameeva E.E., Reznick J., Omerbašić D., Bennett N.C., Lewin G.R., Azpurua J., Gorbunova V., Seluanov A., Regnard P., Wanert F., Marchal J., Pifferi F., Aujard F., Liu Z., Shi P., Pääbo S., Schroeder F., Willmitzer L., Giavalisco P., Khaitovich P. Lipidome determinants of maximal lifespan in mammals. Sci. Rep. 2017;7:1–5. doi: 10.1038/s41598-017-00037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier T., Güell M., Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 61.Tian Q., Stepaniants S.B., Mao M., Weng L., Feetham M.C., Doyle M.J., Yi E.C., Dai H., Thorsson V., Eng J., Goodlett D., Berger J.P., Gunter B., Linseley P.S., Stoughton R.B., Aebersold R., Collins S.J., Hanlon W.A., Hood L.E. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Warnefors M., Kaessmann H. Evolution of the correlation between expression divergence and protein divergence in mammals. Genome Biol. Evol. 2013;5:1324–1335. doi: 10.1093/gbe/evt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper N., Thomas G.H., FitzJohn R.G. Shedding light on the ‘dark side’ of phylogenetic comparative methods. Methods Ecol. Evol. 2016;7:693–699. doi: 10.1111/2041-210X.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minias P., Podlaszczuk P. Longevity is associated with relative brain size in birds. Ecol. Evol. 2017;7:3558–3566. doi: 10.1002/ece3.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkinson G.S., Adams D.M. Recurrent evolution of extreme longevity in bats. Biol. Lett. 2019;15 doi: 10.1098/rsbl.2018.0860. 20180860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindborg C.M., Propert K.J., Pignolo R.J. Conservation of pro-longevity genes among mammals. Mech. Ageing Dev. 2015;146–148:23–27. doi: 10.1016/j.mad.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jové M., Naudí A., Aledo J.C., Cabré R., Ayala V., Portero-Otin M., Barja G., Pamplona R. Plasma long-chain free fatty acids predict mammalian longevity. Sci. Rep. 2013;3:3346. doi: 10.1038/srep03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayne B., Berry O., Davies C., Farley J., Jarman S. A genomic predictor of lifespan in vertebrates. Sci. Rep. 2019;9:17866. doi: 10.1038/s41598-019-54447-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fredslund J., Schauser L., Madsen L.H., Sandal N., Stougaard J. PriFi: using a multiple alignment of related sequences to find primers for amplification of homologs. Nucleic Acids Res. 2005;33:516–520. doi: 10.1093/nar/gki425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinforma. 2019;68 doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.