Abstract
During infection, the host restrains Mycobacterium tuberculosis (Mtb) from proliferating by imposing an arsenal of stresses. Despite this onslaught of attacks, Mtb is able to persist for the lifetime of the host, indicating that this pathogen has substantial molecular mechanisms to resist host-inflicted damage. The stringent response is a conserved global stress response in bacteria that involves the production of the hyperphosphorylated guanine nucleotides ppGpp and pppGpp (collectively called (p)ppGpp). (p)ppGpp then regulates a number of cellular processes to adjust the physiology of the bacteria to promote survival in different environments. Survival in the presence of host-generated stresses is an essential quality of successful pathogens, and the stringent response is critical for the intracellular survival of a number of pathogenic bacteria. In addition, the stringent response has been linked to virulence gene expression, persistence, latency and drug tolerance. In Mtb, (p)ppGpp synthesis is required for survival in low nutrient conditions, long term culture and during chronic infection in animal models, all indicative of a strict requirement for (p)ppGpp during exposure to stresses associated with infection. In this review we discuss (p)ppGpp metabolism and how this functions as a critical regulator of Mtb virulence.
Keywords: tuberculosis, stringent response, mycobacteria, starvation, polyphosphate, Rel
In this review we discuss (p)ppGpp metabolism and how this functions as a critical regulator of Mycobacterium tuberculosis pathogenesis.
(p)ppGpp SYNTHESIS IN BACTERIA
(p)ppGpp (also referred to as magic spot due to initial identification as a spot in chromotagraphy experiments (Cashel 1969; Cashel and Gallant 1969)) is the effector molecule of the stringent response. (p)ppGpp metabolism in bacteria is controlled by Rel/SpoT homolog proteins (RSHs), named for their sequence similarity to RelA and SpoT enzymes in Escherichia coli (Atkinson, Tenson and Hauryliuk 2011). RelA is a monofunctional (p)ppGpp synthetase while SpoT is a bifunctional enzyme with (p)ppGpp hydrolysis activity and weak (p)ppGpp synthetase activity. Despite being the basis for the nomenclature of RSH proteins, phylogenetic analyses indicate that the presence of two functionally divergent RSH homologs (RelA and SpoT) is unique to the γ- and β- proteobacteria lineages (Atkinson, Tenson and Hauryliuk 2011). Most other bacteria encode a single bifunctional RSH enzyme that contains both a (p)ppGpp synthetase and hydrolase domain as well as several regulatory domains. The common nomenclature for multi-domain RSH proteins is long RSHs. Orthologs of long RSHs are widely-distributed throughout bacteria and are believed to originate from a common ancestral long RSH (Atkinson, Tenson and Hauryliuk 2011). The E. coli homologs RelA and SpoT likely arose from a lineage-specific duplication.
Mtb encodes a single bifunctional long RSH named RelMtb (Avarbock et al.1999) (Fig. 1) that is conserved in all Mycobacterium species and has been shown to complement an E. coli relA mutant for growth on minimal media (Ojha, Mukherjee and Chatterji 2000), confirming its ability to promote the stringent response. The relMtb locus is syntenous with the rel locus in Bacillus subtilis, encoding apt (adenine phosphoribosyltransferase) upstream and cypH (cyclophilin) downstream (Avarbock et al.1999). Deletion of relMtb(ΔrelMtb) results in a (p)ppGpp null mutant ((p)ppGpp0) (Primm et al.2000), suggesting that RelMtb is the only functional (p)ppGpp synthetase in Mtb. (p)ppGpp synthesis by RelMtb is necessary for chronic Mtb infection in both mice and guinea pigs (Dahl et al.2003; Karakousis et al.2004; Klinkenberg et al.2010; Weiss and Stallings 2013), which is when the immune system is activated to impose growth restrictive stresses on the bacteria. In contrast, the ΔrelMtb mutant is not attenuated for growth in THP-1 macrophages in cell culture (Primm et al.2000), indicating that the stringent response is essential for Mtb survival during stress conditions specific to the chronic phase of in vivo infection.
Figure 1.
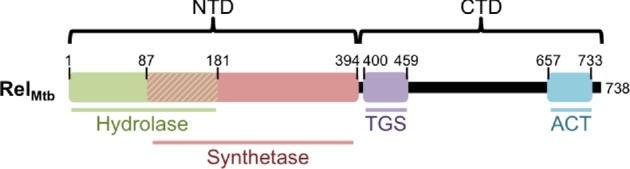
Diagram of the full-length RelMtb protein illustrating functional domains. The N-terminal domain (NTD) contains the catalytic domains and the C-terminal domain (CTD) contains the regulatory domains. The CTD contains two structural domains: the TGS domain (named for its initial identification in ThrRS, GTPase and SpoT) and the ACT domain (named for its initial identification in aspartate kinase, chorismite mutase and TyrA). The numbers designate the amino acid positions that form the borders of each domain.
Streptomyces, Frankia and Salinispora species encode a second long RSH homolog named RshA, which is believed to have arisen from an Actinobacteria-specific duplication but has subsequently been lost in Mycobacterium species (Sun, Hesketh and Bibb 2001; Atkinson, Tenson and Hauryliuk 2011). In addition to long RSHs, two other classes of RSH proteins have been discovered, the Small Alarmone Synthetases (SASs) and Small Alarmone Hydrolases (SAHs). In contrast to the long RSHs, SASs and SAHs are monofunctional, single-domain enzymes. Actinobacteria SASs belong to their own monophyletic clade (actRel) and are orthologous to the well-studied B. subtilis SAS proteins RelP (YjbM) and RelQ (YwaC) (Atkinson, Tenson and Hauryliuk 2011). Mtb encodes a single SAS (Rv1366), but the enzyme is believed to be non-functional (Bag et al.2014) and deletion of rv1366 in Mtb has no phenotypes in culture or during infection of mice (Weiss and Stallings 2013). In other bacteria, SAS activity is toxic in the absence of a functional (p)ppGpp hydrolase (Lemos et al.2007; Nanamiya et al.2008; Abranches et al.2009; Das et al.2009; Natori et al.2009), which is not observed in Mtb, further supporting that Rv1366 is not a functional (p)ppGpp synthetase (Weiss and Stallings 2013). The only other SAS that has been studied in mycobacteria is MS_RHII-RSD in Mycobacterium smegmatis (Msm), a non-pathogenic model organism for Mtb. Unlike Rv1366, MS_RHII-RSD has been shown to be a functional pppGpp synthetase and encodes an amino-terminal RNase HII domain (Murdeshwar and Chatterji 2012) that can hydrolyze R-loops (Krishnan et al.2016) that may form as a result of collisions between the replication and transcription machinery, indicating that MS_RHII-RSD has evolved functions distinct from Rv1366.
(p)ppGpp METABOLISM BY RelMtb
RelMtb is comprised of two catalytic domains, a (p)ppGpp hydrolysis domain (1-181 amino acids (aa)) and a (p)ppGpp synthetase domain (87-394 aa), that include an overlapping three helix bundle (87-181), as well as a regulatory C-terminal domain (CTD, 395-738 aa) (Fig. 1) (Avarbock et al.2005; Singal et al.2017). The (p)ppGpp synthetase domain transfers the 5΄-β,γ-pyrophosphate from ATP to the 3΄-OH of GDP or GTP to synthesize ppGpp and pppGpp, respectively (Avarbock et al.1999). Crystal structures of the N-terminus of RelMtb (1-394 aa) and the N-terminus of a homolog from Streptococcus dysgalactiae subspecies equisimilis (RelSeq, 1-385 aa included in the crystal structure) have provided insight to the functional features of Rel catalytic domains (Hogg et al.2004; Singal et al.2017). The (p)ppGpp synthetase domain is comprised of five β-sheets surrounded by five α-helices (Singal et al.2017). The (p)ppGpp synthetase domain was determined to be structurally homologous to DNA polymerase β (pol β) and the D190 and D256 residues in pol β that are essential for coordinating the Mg2+ cofactor correspond to D265 and E325 in RelMtb (Hogg et al.2004). Both D265 and E325 of RelMtb are required for (p)ppGpp synthesis in vitro and RelMtb rescue of an E. coli ΔrelAΔspoT ((p)ppGpp0) strain growth on minimal media (Hogg et al.2004; Bag et al.2014). RelSeq co-crystallized with GDP in a ligand-binding pocket partially formed by 176-185 aa within the synthetase domain. The conservation of the majority of the residues comprising the GDP binding pockets between RelSeq and RelMtb suggests that RelMtb and RelSeq recognize and bind to GDP in a similar fashion. The GDP pocket also includes RelSeq Y308 (RelMtb Y309), which face-to-face stacks with GDP and is required for Relseq to rescue growth of an E. coli (p)ppGpp0 strain on minimal media (Hogg et al.2004; Bag et al.2014). The RelMtb ATP binding pocket likely involves the positively charged residues R242, K244 and K252 coordinating the β and γ phosphates of ATP at a site proximal to the GDP binding pocket and the Mg2+ binding site (Hogg et al.2004; Singal et al.2017). The importance of the basic patch created by R242, K244 and K252 is supported by experiments showing that RelMtb R242H is unable to rescue growth of an E. coli (p)ppGpp0 strain on minimal media (Bag et al.2014). In addition, RelMtb G241 and H344, which were initially identified in E. coli RelA as being important for ATP binding (Gropp et al.2001), are required for pppGpp synthesis in vitro (Avarbock et al.2005). Additionally, H344 is required for RelMtb to rescue growth of an E. coli (p)ppGpp0 strain on minimal media (Bag et al.2014) and mutation of RelMtb H344 attenuates Mtb in mice (Weiss and Stallings 2013).
In addition to being able to synthesize (p)ppGpp, RelMtb also hydrolyzes (p)ppGpp into pyrophosphate (PPi) and GDP or GTP via an N-terminal catalytic HD superfamily domain (Avarbock et al.2005) (Fig. 1). HD superfamily members are phosphohydrolases with conserved histidine and aspartate residues that are involved in the coordination of divalent cations, which is essential for their activity (Aravind and Koonin 1998). The RelMtb hydrolase domain is comprised of 11 α-helices, which includes a (p)ppGpp binding pocket spanning residues 41-53 located in the region connecting the second and third α-helices (α2 and α3). The HD motif comprised of the conserved histidine (H80) and aspartate (D81) residues is located in a β-turn that connects α4 and α5 (Singal et al.2017). Alanine substitution of H80 or D81 in RelMtb abolishes hydrolase activity in vitro without affecting pppGpp synthesis (Avarbock et al.2005).
The RelMtb catalytic domains have strict cation co-factor requirements where (p)ppGpp synthesis requires Mg2+ or Mn2+ and RelMtb (p)ppGpp hydrolysis requires Mn2+. The Mg2+ and Mn2+ concentrations required for optimal (p)ppGpp synthesis is equal to the combined concentration of the substrates, ATP and GTP. RelMtb synthetase activity is inhibited by concentrations of Mg2+ (or Mn2+) that exceed the GTP and ATP substrate concentrations due to an RXKD motif within the synthetase domain that is highly conserved among bifunctional Rel enzymes (Avarbock, Avarbock and Rubin 2000; Sajish et al., 2007, 2009). The model for how the RXKD motif sensitizes the RelMtb synthetase catalytic activity to Mg2+ concentrations that exceed ATP and GTP substrate concentrations is that the negatively charged phosphates of GTP can bind to either the positive charges of RelMtb RXKD motif (R and D) or positively charged Mg2+ ions. Mg2+ bound GTP is unable to bind the positive charges of RXKD in a way that is required for (p)ppGpp synthesis. Thus, Mg2+ ions compete with the RelMtb RXKD motif for GTP binding. In contrast, E. coli RelA has an EXDD motif instead of the RXKD motif and is not sensitive to high concentrations of Mg2+. Instead, due to the presence of two negatively charged residues (E and D), the E. coli EXDD motif requires Mg2+ to bind GTP, allowing the E. coli RelA synthetase domain to function when Mg2+ concentrations are higher than the combined ATP and GTP concentration (Sajish et al., 2007, 2009).
REGULATION OF (p)ppGpp SYNTHESIS BY RelMtb
RelMtb is constitutively expressed at basal levels, likely via an almost consensus -10 promoter element upstream of the relMtb gene that would be recognized by the housekeeping σ-factor σA (Jain et al.2005). However, in the absence of stress or stimuli, the RelMtb synthetase activity is repressed by the presence of the CTD (Avarbock et al.1999; Jain et al.2006) and only a low level of (p)ppGpp is produced (Stallings et al.2009; Weiss and Stallings 2013). Although the level of (p)ppGpp produced in unstressed conditions is low, this low level production is still required for WT growth rates and a ΔrelMtb mutant is attenuated for growth in unstressed, nutrient-rich media under aerobic conditions (Primm et al.2000; Weiss and Stallings 2013), suggesting that the low levels of (p)ppGpp produced in unstressed conditions still provide a growth advantage to Mtb. In addition, (p)ppGpp hydrolysis is essential for preventing toxic (p)ppGpp accumulation in Mtb in unstressed culture conditions, as well as during both acute and chronic stages of infection (Weiss and Stallings 2013). These observations demonstrate that RelMtb constitutively produces (p)ppGpp and may act to maintain homeostasis in all growth conditions. This is also similar to observations in other bacteria where (p)ppGpp hydrolysis is essential for viability in unstressed conditions due to the presence of a constitutive level of (p)ppGpp synthesis (Lemos et al.2007; Nanamiya et al.2008; Das et al.2009).
The induction of (p)ppGpp synthesis by Rel homologs is best characterized during amino acid starvation where Rel is associated with ribosomes when an uncharged tRNA enters the A site of the ribosome and stimulates Rel-mediated (p)ppGpp production (Haseltine and Block 1973; Avarbock, Avarbock and Rubin 2000). Accordingly, Mtb accumulates (p)ppGpp in a RelMtb-dependent manner during nutrient starvation in liquid culture (Primm et al.2000; Stallings et al.2009). In agreement with stringent response mutants in other bacteria, deletion of relMtb reduces the viability of Mtb in nutrient-poor conditions (Primm et al.2000), supporting a role for the Mtb stringent response in responding to starvation. In addition to during starvation, (p)ppGpp accumulates in Mtb in response to hypoxia and oxidative stress (Primm et al.2000; Stallings et al.2009), and it is not known whether this is because these stresses cause amino acid starvation that triggers RelMtb (p)ppGpp synthetase activity or if there are other ways to induce RelMtb-mediated (p)ppGpp synthesis.
The induction of (p)ppGpp synthesis during amino acid starvation requires the RelMtb CTD and can be recapitulated in vitro by adding the Rel activating complex (RAC) consisting of ribosomes, uncharged tRNAs, and mRNA (Avarbock, Avarbock and Rubin 2000; Avarbock et al.2005; Jain et al.2006). The RelMtb C-terminal domain (CTD) contains two structural domains: the TGS domain (400-459 aa, named for its initial identification in ThrRS (threonyl tRNA synthetase), GTPase (Obg family of GTPases) and SpoT) and the ACT domain (657-733 aa, named for its initial identification in aspartate kinase, chorismite mutase and TyrA) (Wolf et al.1999; Chipman and Shaanan 2001) (Fig. 1). Although a crystal structure for the RelMtb CTD has not yet been solved, multiple cryo-EM structures of E. coli RelA in complex with a ribosome with an uncharged tRNA (Phe) occupying the A site have provided a structure-based model for the function of TGS and ACT domains (Arenz et al.2016; Brown et al.2016; Loveland et al.2016). These structures suggest that the TGS domain acts as a sensor to distinguish charged versus uncharged tRNAs that enter the A-site of the ribosome. The sensor function of the TGS domain is accomplished by the ability of the TGS domain to interact with the 3’CCA of the A-site tRNA. If an aminoacylated tRNA occupies the A-site of the ribosome, the amino acid attached to the 3’CCA will sterically occlude the interaction of the 3’CCA with the TGS domain. However, when a deacylated tRNA occupies the A-site of the ribosome, the TGS domain is able to interact with the 3’CCA, resulting in structural changes within the RelA synthetase domain that activates (p)ppGpp synthesis. The ACT domain serves an anchor function, allowing RelA to bind the ribosome regardless of whether the A-site is occupied by a charged or uncharged tRNA. The ACT domain occupies a cavity between the A and P sites of the ribosome and is held in place through several interactions with the A-site finger (helix 38) of the 23S ribosomal RNA (rRNA), the L16 ribosomal protein and the P-site tRNA. The cryo-EM structures also revealed a domain between the TGS and ACT domains of RelA that resembles a zinc-finger domain. This previously uncharacterized domain provides a second binding interface between RelA and the ribosome, and is expected to be conserved in mycobacteria. Loveland et al. (2016) termed this new domain in RelA the Ribosomal-InterSubunit (RIS) domain because it bridges the 30S and 50S ribosomal subunits through interactions with the A-site finger and the hydrophobic patch on the 30S S19 ribosomal protein.
Induction of (p)ppGpp synthesis is accompanied by a repression of (p)ppGpp hydrolysis. The RelSeq crystal structure supports a model where Rel does not simultaneously catalyze (p)ppGpp synthesis and (p)ppGpp hydrolysis (Hogg et al.2004). In this study, two conformations of the RelSeq monomer (1-385) were observed, a hydrolase-OFF/synthetase-ON conformation and a hydrolase-ON/synthetase-OFF conformation. Transition between the two conformations is dependent on ligand binding to the respective active site and indicates that at any given time only one enzymatic function is active, thus preventing simultaneous synthesis and hydrolysis of (p)ppGpp (Hogg et al.2004). In support of this model, Avarbock et al. have shown that RelMtb hydrolase activity is inhibited in the presence of synthetase substrates ATP and GTP (Avarbock, Avarbock and Rubin 2000).
In what appears to be a negative feedback loop, pppGpp has been shown to bind to the CTD in the region that separates the TGS and ACT of the Rel protein from Msm (Syal et al.2015). Binding of pppGpp to the RelMsm CTD represses pppGpp synthesis while increasing pppGpp hydrolysis. As such, RelMsm-mediated pppGpp synthesis is decreased at saturating pppGpp concentrations compared to synthesis at non-saturating pppGpp concentrations. RelMsm point mutants with lower affinity for pppGpp (F533A, R615A, F620A and R633A) lose this effect. pppGpp binding to RelMsm decreases protein compaction and causes RelMsm to become less structured, suggesting that conformational changes within RelMsm may lead to repression of the synthetase activity (Jain, Saleem-Batcha and Chatterji 2007).
Studies in vitro have suggested that RelMtb could also be regulated through oligomerization (Avarbock et al.2005; Jain et al.2006; Singal et al.2017). The full-length RelMtb has been shown to form trimers in gel filtration experiments, which is facilitated by the CTD, although this domain is not required for oligomerization (Avarbock et al.2005). The RelMtb NTD (amino acids 1-394) alone formed dimers in small angle X-ray scattering analysis and X-ray crystallography experiments (Singal et al.2017). In both cases, oligomerization was shown to inhibit pppGpp synthesis (Avarbock et al.2005; Jain et al.2006). These in vitro observations have led to some speculation that this could be another way that RelMtb activity is regulated, however, it is still unknown if oligomerization occurs in vivo.
REGULATION OF TRANSCRIPTION BY (p)ppGpp
Once (p)ppGpp is synthesized, it has been shown to bind multiple targets in bacteria to modify their function and promote changes in physiology that allow for survival during stress. The most well-characterized outcome of the stringent response in bacteria is a downregulation of rRNA and ribosomal proteins accompanied by an upregulation of amino acid biosynthesis genes to restore amino acid levels during starvation (Eymann et al.2002; Traxler et al.2008). Hallmarks of this canonical stringent response appear to be conserved in Mtb. In a microarray experiment comparing expression profiles of a ΔrelMtb strain to WT Mtb starved in Tris-buffered saline with Tween-80 (TBS-T), expression of 54 of the 58 ribosomal proteins was downregulated during starvation in WT Mtb but not in the ΔrelMtb mutant (Dahl et al.2003). In addition, quantitative reverse transcription-PCR (qRT-PCR) experiments showed that the ΔrelMtb strain was unable to downregulate rRNA levels during starvation (Stallings et al.2009). These data support the conservation of the canonical stringent response involving a Rel-dependent repression of genes encoding the translation machinery. However, WT Mtb starved in TBS-T does not induce expression of amino acid biosynthesis genes (Betts et al.2002; Dahl et al.2003). One potential explanation for Mtb failing to induce expression of amino acid biosynthesis genes is that the studies with Mtb used a general carbon starvation condition by incubating Mtb in TBS-T, whereas studies in E. coli and B. subtilis have used amino acid analogs to directly interfere with tRNA charging. Therefore, it is possible that Mtb would upregulate amino acid biosynthesis genes in response to specifically interfering with tRNA charging, but this has yet to be tested. Alternatively, Zhang and Rubin (2013) attribute this observation to the relatively unique amino acid prototrophy of Mtb where the bacteria constitutively synthesizes its own amino acids and these pathways may have evolved to become independent of stress signaling.
The mechanisms by which (p)ppGpp alters gene expression during starvation have been most well-characterized in the gram-negative γ-proteobacteria E. coli and the gram-positive firmicute B. subtilis. Numerous differences in the mechanisms used by (p)ppGpp in these two organisms has established two divergent paradigms of the stringent response: the E. coli paradigm, which is utilized by most proteobacteria, and the B. subtilis paradigm, which occurs in most other bacteria species (Boutte and Crosson 2013; Hauryliuk et al.2015; Liu et al.2015). One of the primary differences between the two stringent response paradigms is the direct binding target of (p)ppGpp. In E. coli, (p)ppGpp directly binds to RNA polymerase (RNAP) and changes gene expression by destabilizing RNAP-promoter open complexes (RPo) that form during transcription initiation (Barker, Gaal and Gourse 2001; Ross et al.2013; Zuo, Wang and Steitz 2013). Two distinct ppGpp binding sites have been identified on the E. coli RNAP. The first ppGpp binding site (Site 1) is a pocket in between the β’ and ω subunits of RNAP (Ross et al.2013; Zuo, Wang and Steitz 2013). ppGpp binding to Site 1 is thought to allosterically prevent the RNAP from making stable contacts with the promoter DNA, thus destabilizing RPo (Ross et al.2013; Zuo, Wang and Steitz 2013). A second ppGpp binding site (Site 2) is introduced by the binding of the DksA protein to RNAP (Ross et al.2016). DksA binds to the β’ subunit of RNAP and inserts its coiled-coil domain into the RNAP secondary channel, creating (p)ppGpp binding Site 2, located at the interface between the RNAP β’ subunit and DksA. Recent crystal structures revealed that ppGpp binding stabilizes the DksA-RNAP interaction by relieving the mechanical stress in RNAP caused by DksA binding (Molodtsov et al.2018). In addition, ppGpp binding induces a conformational change in DksA, allowing it to insert its coiled-coil domain further into the RNAP secondary channel to potentially interfere with RNAP catalytic activity (Molodtsov et al.2018). Binding of DksA is necessary for the full effects of ppGpp on RPo stability in vitro (Paul et al.2004; Paul, Berkmen and Gourse 2005; Lennon et al.2012; Molodtsov et al.2018). (p)ppGpp and DksA are thought to regulate transcription initiation by destabilizing the transition states between the RNAP-promoter closed complex (RPc) and RPo complex, thus decreasing the activation energy of this transition (Paul, Berkmen and Gourse 2005; Rutherford et al.2009). At promoters that form intrinsically unstable RPo, such as E. coli rRNA promoters, this effect of ppGpp and DksA negatively regulates transcription by facilitating RPo collapse back to RPc (Paul et al.2004). In contrast, at promoters where transcription is not rate-limited by RPo stability, such as promoters of the E. coli amino acid biosynthesis genes, this effect of ppGpp and DksA positively regulates transcription by allowing the isomerization of RPc to RPo to occur more readily (Paul, Berkmen and Gourse 2005). Thus, (p)ppGpp is able to exert differential effects on transcription initiation based on the kinetics of the RNAP complex at a given promoter.
In B. subtilis, (p)ppGpp exerts its effects on transcription indirectly through changes in intracellular guanosine nucleobase metabolism (Kriel et al.2012). (p)ppGpp directly binds and inhibits enzymes involved in both de novo and salvage pathways of GTP synthesis. Specifically, (p)ppGpp potently inhibits the activity of guanylate kinase (GMK), which phosphorylates GMP to GDP (Kriel et al.2012; Liu et al.2015), and HprT, which converts hypoxanthine to inosine monophosphate (IMP) in the purine salvage pathway (Kriel et al.2012), collectively inhibiting GTP biosynthesis. At high (millimolar) concentrations, (p)ppGpp also inhibits the enzymatic activity of IMP dehydrogenase (IMPDH or GuaB), which converts IMP to xanthosine monophosphate (XMP) and controls a branch point between ATP and GTP synthesis in the purine salvage pathway, in both B. subtilis (Kriel et al.2012) and E. coli (Pao and Dyess 1981). Inhibition of the GTP biosynthesis pathways upon nutrient starvation alters the metabolite landscape, most notably causing depletion of GTP and concordant accumulation of ATP (Kriel et al.2012; Liu et al.2015). In addition, pppGpp synthesis depletes free GTP by using it as a substrate for pppGpp synthesis, further decreasing intracellular GTP levels and increasing the intracellular ATP:GTP ratio. ATP and GTP are the most common initiating nucleotide triphosphates (iNTP) used in B. subtilis and since binding of the iNTP to the transcription initiation complex stabilizes RPo (Kuzmine, Gottlieb and Martin 2003), changes in the ATP:GTP ratio impacts gene expression. Consequently, B. subtilis genes that initiate with ATP, such as amino acid biosynthesis genes, are up-regulated while genes that initiate with GTP, such as rRNA genes, are down-regulated (Krásný et al.2008; Tojo et al.2010; Kriel et al.2012). Depletion of GTP also activates a starvation response through the transcription factor CodY (Handke, Shivers and Sonenshein 2008; Kriel et al.2012). In nutrient replete conditions, GTP binds CodY and blocks transcription of stress adaptive genes such as those involved in sporulation and branched chain amino acid biosynthesis (Ratnayake-Lecamwasam et al.2001; Belitsky and Sonenshein 2008). When GTP levels decrease during the stringent response, CodY-mediated repression is relieved and transcription of these genes is activated. Together, the ATP:GTP ratio and CodY contribute to the gene expression profile that results from the B. subtilis stringent response. However, the role of guanosine metabolites in the stringent response is likely more complex because GTP depletion alone is not sufficient for starvation survival (Liu et al.2015).
Intriguingly, Mtb does not appear to strictly follow either stringent response paradigm, and the mechanisms underpinning the Mtb stringent response remain mostly unknown. In terms of the E. coli paradigm of the stringent response, the sites known to be targeted by ppGpp on the E. coli RNAP are largely not conserved in the Mtb RNAP. Site 1 where ppGpp binds the E. coli RNAP involves numerous residues at the interface of the β’ and ω subunits, including β’ K615, β’ I619, β’ D622, β’ Y626, β’ R632, β’ R417, ω A2, ω R3, ω V4 and ω T5 (Ross et al.2013; Zuo, Wang and Steitz 2013). The Mtb RNAP shares identity or similarity with only two of these residues, E. coli β’ D622 (Mtb β’ D714) and E. coli β’ R362 (Mtb β’ K437). Site 2 on the E. coli RNAP that is targeted by ppGpp is DksA-dependent (Ross et al.2016) and Mtb does not encode DksA. In addition, the Site 2 residues located in the E. coli β’ subunit are not conserved in the Mtb RNAP.
Nonetheless, there is evidence that pppGpp directly binds the Mtb RNAP, raising the possibility that although the features of the (p)ppGpp-RNAP interaction in Mtb may be different than described in E. coli, this interaction could still occur. Addition of pppGpp to in vitro transcription reactions inhibited RPo formation by Mtb RNAP at the Mtb rRNA rrnAP3 promoter (China et al.2012) and transcript formation by Mtb RNAP at the Mtb rrnAP3 and gyrB promoters (Tare, China and Nagaraja 2012; Tare, Mallick and Nagaraja 2013). Addition of Rv3788, a protein that binds near the secondary channel of the Mtb RNAP, blocks the effects of pppGpp on RPo formation at the Mtb rrnAP3 promoter (China et al.2012), suggesting that the effects of pppGpp require its interaction with the RNAP. The effect of pppGpp on transcript formation by Mtb RNAP at the Mtb rrnAP3 promoter was also dependent on a thymine positioned three nucleotides downstream of the -10 element and L232 in σA, both of which affected RPo stability, thus supporting a link between pppGpp binding to the RNAP and RPo stability (Tare, Mallick and Nagaraja 2013). However, in all of these cases, effects of pppGpp were only observed at concentrations ≥100 μM, and often only at ≥500 μM, whereas the IC50 of ppGpp on the E. coli RNAP at the rrnBP1 promoter is 12 μM (Ross et al.2013). Therefore, differences exist in how (p)ppGpp impacts RNAP activity in E. coli and Mtb and future studies will be necessary to determine whether the high concentrations of pppGpp used in vitro are physiologically relevant in vivo in Mtb.
Mycobacteria also share some similarities with the B. subtilis stringent response paradigm. For example, when M. smegmatis was engineered to accumulate higher levels of (p)ppGpp through mutation of the RelMsm hydrolase domain, ATP levels were increased and GTP levels were decreased (Weiss and Stallings 2013), indicating that (p)ppGpp synthesis affected purine metabolism. Accordingly, rRNA production is downregulated in mycobacteria during the stringent response and the two promoters (AP1 and AP3) upstream of the Mtb rRNA locus both initiate with GTP, which mirrors the pattern for transcriptional start sites at rRNA operons that has been reported in B. subtilis (Krásný et al.2008). Nonetheless, it has yet to be directly tested whether the identity of the iNTP for Mtb promoters is sufficient to determine the direction of gene expression during the stringent response. Unlike B. subtilis, pppGpp is not able to inhibit Mtb GMK (Rv1389) in vitro, even though GMK homologs from other Actinobacteria such as Streptomyces coelicolor were inhibited by pppGpp (Liu et al.2015). This is possibly explained by the lack of conservation in multiple residues in the Rv1389 active site that are required for the interaction of GMK with pppGpp in B. subtilis and S. coelicolor (Fig. 2). These polymorphisms are exclusive to Mtb, as even the (p)ppGpp-insensitive proteobacteria GMK homologs are conserved at these residues and their (p)ppGpp insensitivity is due to polymorphisms elsewhere in the protein (Liu et al.2015). Another difference between the B. subtilis paradigm and Mtb is that mycobacteria do not encode a CodY homolog. The effect of (p)ppGpp on the Mtb homologs of other B. subtilis (p)ppGpp targets in the purine biosynthesis pathway (HprT and IMP dehydrogenase) has not yet been tested and, therefore, could be (p)ppGpp targets in Mtb and explain the decreased GTP levels during (p)ppGpp synthesis. Alternatively, (p)ppGpp synthesis could be sufficient to deplete GTP by using it as a substrate. Together these data indicate that the classic model of (p)ppGpp impacting RPo to alter gene expression during the stringent response could be conserved in Mtb, but the mechanistic features of this response are unique from model organisms.
Figure 2.
Sequence alignment of GMK homologs from Mycobacterium tuberculosis (Mtb), Streptomyces coelicolor and Bacillus subtilis. Yellow boxes indicate residues that interact with pppGpp and are conserved in all three species. Red boxes indicate residues that interact with pppGpp and are polymorphic in Mtb.
(p)ppGpp, POLYPHOSPHATE AND PERSISTENCE
In addition to RNAP and enzymes involved in GTP metabolism, polyphosphatase, a key regulator of polyphosphate (polyP) metabolism, has been investigated as a target of (p)ppGpp in Mtb. PolyP chains are linear polymers of phosphates that can reach hundreds to thousands of phosphates in length (Rao, Roberts and Torriani 1985). Mtb accumulates polyP chains when exposed to nutritional, nitrosative, oxidative, acidic and antibiotic stresses as well as during its transition to stationary phase (Thayil et al.2011; Singh et al.2013; Chuang et al.2015), overlapping with conditions that induce (p)ppGpp accumulation (Primm et al.2000; Stallings et al.2009). PolyP accumulation results in an overhaul of Mtb metabolism that arrests growth and facilitates Mtb persistence. In Mtb, polyP is synthesized primarily by the polyphosphate kinase PPK1 (Rv2984). Mtb encodes another polyphosphate kinase PPK2 (Rv1026) that can synthesize polyP, but PPK2 more efficiently carries out the reverse reaction as a polyphosphate dependent nucleoside diphosphate kinase that utilizes polyP to produce ATP and GTP (Sureka et al.2009; Shum et al.2011; Chuang, Belchis and Karakousis 2013). In addition, Δppk1 mutants produce only negligible amounts of polyP (Singh et al.2013), supporting that PPK1 serves as the primary polyP synthase in Mtb. Mtb encodes two polyphosphatases, PPX1 (Rv0496) and PPX2 (Rv1026), that hydrolyze polyP by cleaving phosphates from the termini of polyP chains. PPX1 and PPX2 differ in their substrate preference where PPX1 preferentially catabolizes short polyP chains and PPX2 specializes in breaking down long polyP chains (Choi et al.2012; Chuang et al.2015). (p)ppGpp directly inhibits the hydrolytic activity of Mtb PPX1 and PPX2 during in vitro exopolyphosphatase assays (Choi et al.2012; Chuang et al.2015). Inhibition of PPX1 or PPX2 would lead to the accumulation of polyP in Mtb and this has been proposed to contribute to the role of the stringent response in persistence (Thayil et al.2011; Chuang et al.2015). In support of the physiological relevance of this mechanism, deleting relMtbcauses mid- and late- log phase cultures of Mtb to accumulate less polyP (Singh et al.2013).
How changes in polyP levels affect Mtb has been investigated using Mtb mutants that produce lower levels of polyP (Δppk1) or over-accumulate polyP (ppx1:Tn, ppx2 knockdown, and ppk2:Tn) (Table 1). The Δppk1 and ppx1:Tn strains grow slower in liquid culture and the Δppk1, ppx1:Tn, ppx2 knockdown and ppk2:Tn mutants all reach a lower maximum optical density at stationary phase (Thayil et al.2011; Chuang, Belchis and Karakousis 2013; Singh et al.2013; Chuang et al.2015). Δppk1, ppx1:Tn, and ppk2:Tn mutant strains are also attenuated in vivo (Thayil et al.2011; Chuang, Belchis and Karakousis 2013; Singh et al.2013). The Δppk1 and ppx1:Tn strains both showed decreased lung CFU starting at 14 days post-infection in the guinea pig infection model (Thayil et al.2011; Singh et al.2013). The ppk2:Tn strain showed decreased lung CFU at 14 days but grew back to levels comparable to WT Mtb in a mouse infection model (Chuang, Belchis and Karakousis 2013). Unlike the ΔrelMtb mutant, the Δppk1, ppx1:Tn, and ppk2:Tn strains were impaired for survival in macrophages (Thayil et al.2011; Chuang, Belchis and Karakousis 2013; Singh et al.2013), suggesting that the virulence mechanisms mediated by polyP and the stringent response are not completely overlapping. Unexpectedly, the ppx2 knockdown strain grew better in macrophages (Chuang et al.2015), suggesting that the role of polyP metabolism in virulence requires further study to reconcile these seemingly contradictory findings. Nonetheless, these data demonstrated that proper regulation of polyP levels is important for Mtb virulence and the ability of (p)ppGpp to modulate polyP levels by direct inhibition of polyphosphatases could be one mechanism by which the stringent response impacts pathogenesis.
Table 1.
Summary of the phenotypes of polyP accumulating and polyP deficient Mtb strains. References are provided within the main text.
| Phenotype | Δppk1 | ppx1:Tn | ppx2 knockdown | ppk2:Tn |
|---|---|---|---|---|
| polyP levels | Decreased | Increased | Increased | Increased |
| Growth in liquid culture | Decreased growth rate in late log and stationary phase | Decreased growth rate | Decreased cell density during stationary phase | Decreased cell density during stationary phase |
| Growth in macrophages | Decreased survival in macrophages | Decreased survival in macrophages | Increased survival in macrophages | Decreased survival in macrophages |
| Virulence | Decreased virulence in guinea pigs | Decreased virulence in guinea pigs | Unknown | Decreased virulence in mice |
| Biofilm formation | Unknown | Decreased biofilm formation | Decreased biofilm formation | Decreased biofilm formation |
| Antibiotic and Stress tolerance | Increased sensitivity to isoniazid, levofloxacin, rifampicin and nitrosative stress | Decreased sensitivity to isoniazid. Increased sensitivity to clofazimine and hypoxia | Decreased sensitivity to isoniazid, heat, cell surface and acid stress | Decreased sensitivity to isoniazid. Increased sensitivity to naphthoquinone plumbagin and meropenem |
| Metabolism | Unknown | Decreased levels of peptidoglycan and phospholipid metabolism intermediates. Increased levels of TCA cycle and arginine metabolism intermediates | Decreased levels of glycolysis, fatty acid metabolism, phospholipid metabolism and pentose phosphate pathway intermediates | Decreased levels of peptidoglycan and phospholipid metabolism intermediates. Increased levels of arginine metabolism intermediates |
Metabolomics studies revealed that the ppx1:Tn, ppk2:Tn and ppx2 knockdown mutants that accumulate polyP contain significantly less glycerol-3-phosphate (G3P), a major scaffold for phospholipids, accompanied by differences in the expression of enzymes involved in G3P metabolism. The enzymes responsible for G3P synthesis from dihydroxyacetone phosphate and glycerol, Rv0564c and GlpK, respectively, were significantly downregulated in the ppx1:Tn mutant and the enzyme responsible for the reverse reaction, Rv1692, which hydrolyses G3P to glycerol, was significantly upregulated in the ppk2:Tn strain. In addition, an enzyme involved in the recycling of glycerophospholipids for G3P synthesis, Rv2182c, was significantly downregulated in both the ppx1:Tn and ppk2:Tn mutants (Chuang et al., 2015, 2016). These data suggest that polyP accumulation signals for changes in gene expression that are linked to a reduction in de novo G3P synthesis, a reduction in recycling pathways for G3P synthesis, and increased G3P turnover. The ppx1:Tn, ppk2:Tn, and ppx2 knockdown mutants also exhibit decreased levels of metabolites involved in peptidoglycan synthesis and both ppx1:Tn and ppk2:Tn mutants express lower transcript levels for the peptidoglycan L,D-transpeptidases ldtA and ldtB (Chuang et al., 2015, 2016). Several metabolites involved in NADH metabolism, the TCA cycle and arginine metabolism also accumulate in the ppx1:Tn and ppk2:Tn mutants (Chuang et al.2016), while fatty acid biosynthesis and nucleotide biogenesis metabolites are reduced in the ppx2 knockdown strain (Chuang et al.2015). The accumulation of metabolites involved in the TCA cycle also occurs during exposure to hypoxia, a condition that induces the formation of drug and stress tolerant persisters (Wayne and Hayes 1996; Watanabe et al.2011; Eoh and Rhee 2013). Overall, the decrease in central metabolism and anabolic pathways during polyP accumulation is indicative of a shift towards metabolic quiescence.
PolyP accumulation in Mtb also affects antibiotic tolerance. The Δppk1 mutant that does not produce polyP showed increased susceptibility to rifampicin, levofloxacin and isoniazid (Singh et al.2013). In contrast, the polyP accumulating ppx1:Tn, ppk2:Tn and ppx2 knockdown mutants displayed decreased sensitivity to isoniazid (Thayil et al.2011; Chuang, Belchis and Karakousis 2013; Chuang et al.2015). However, the ppx1:Tn strain displayed increased sensitivity to clofazimine, which is thought to kill Mtb through membrane destabilization and altered redox cycling (Cholo et al.2017), while the ppk2:Tn strain displayed increased sensitivity to meropenem, which inhibits peptidoglycan L,D-transpeptidases (Chuang et al.2016). Therefore, although polyP production is important for antibiotic tolerance, over-accumulation of polyP results in changes in physiology that compromise antibiotic tolerance, indicating the polyP levels must be tightly regulated. Together, the changes that result from polyP accumulation coordinate a program that favors persistence, stress resistance and antibiotic tolerance, which would be induced when (p)ppGpp inhibits PPX1 and PPX2 and polyP accumulates.
In addition to (p)ppGpp promoting polyP accumulation, polyP also promotes RelMtb expression and the production of (p)ppGpp through a signaling cascade involving the two component system MprAB and the alternative σ-factor σE (Sureka et al.2007) (Fig. 3). PolyP serves as a phosphodonor to the histidine kinase MprB, which then phosphorylates the response regulator MprA. Phosphorylated MprA activates expression of σE, which binds to a promoter sequence upstream of the relMtb gene to activate transcription of relMtb (He et al.2006; Sureka et al.2007). Accordingly, increasing intracellular polyP levels via mutation of ppx1 also increased transcript levels of relMtb (Thayil et al.2011). However, the Δppk1 mutation does not affect the expression of relMtb, suggesting that polyP is not necessary for RelMtb expression but can induce higher expression (Singh et al.2013). This signaling cascade is amplified by several layers of positive feedback loops that rest on three molecular fulcrums: σE, MprA and (p)ppGpp. MprAB activates its own expression in addition to sigE (He and Zahrt 2005; He et al.2006) and σE activates expression of ppk1, the mprAB operon, and itself (Manganelli et al.2001; Sanyal et al.2013), thereby positively regulating this signaling cascade. (p)ppGpp also positively regulates its own production by inhibiting polyphosphatases to promote polyP accumulation. Thus, crosstalk between the polyP metabolism and stringent response activation pathways results in synchronized promotion of polyP and (p)ppGpp production.
Figure 3.
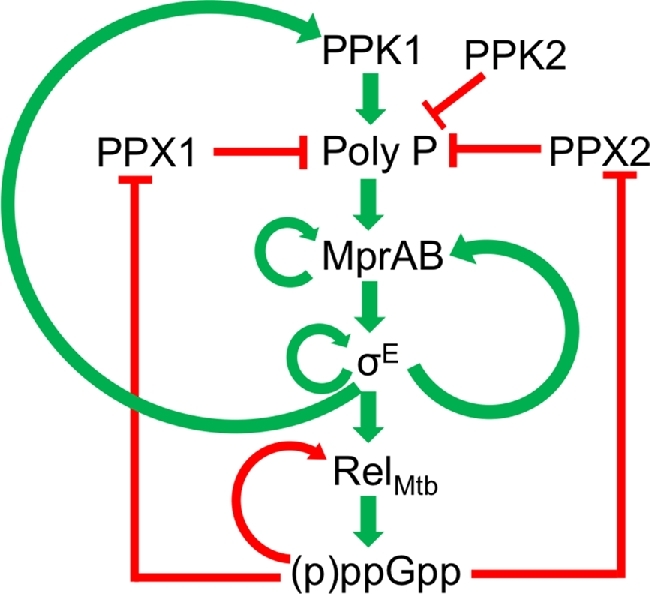
Signaling network involving RelMtb, (p)ppGpp and polyP in Mtb. The network shows the proteins and molecules comprising the signaling cascades that positively and negatively regulate (p)ppGpp and polyP accumulation. Polyphosphate kinase 1 (PPK1), Polyphosphate kinase 2 (PPK2), Polyphosphatase 1 (PPX1) and Polyphosphatase 2 (PPX2) are shown. Green arrows indicate positive regulation. Red lines indicate negative regulation.
PolyP accumulation and the stringent response are both linked to Mtb persistence. The ability of Mtb to persist in the host is attributed to the formation of persister cells that exhibit decreased replication, altered metabolism, increased antibiotic tolerance and increased stress resistance (Gomez and McKinney 2004; Keren et al.2011). Persister cell formation occurs through bacterial bistability, a strategy used by bacterial populations to maintain phenotypically heterogeneous subpopulations that protect the population from environmental insults (Dubnau and Losick 2006; Smits, Kuipers and Veening 2006). Cells transition between phenotypic states through stochastic changes in gene expression that are reinforced by feedback loops. In E. coli, a subpopulation of persister cells is maintained through phenotypic heterogeneity in relA expression (Balaban et al.2004). Similarly, the mprAB-sigE-rel feedback loops underpin the maintenance of ‘bistable’ rel expression in mycobacteria (Sureka et al.2008; Ghosh et al.2011). When Sureka et al. sorted a population of M. smegmatis cells based on GFP expression from the relMsm promoter, they observed a bi-modal distribution of GFP expression with ‘high’ and ‘low’ subpopulations (Sureka et al.2008). As the cultures of M. smegmatis approach stationary phase, the ‘low’ relMsm-expressing subpopulation declines and the ‘high’ population increases. When the mprAB-sigE-rel feedback loop is disrupted by mutations to the σE binding site in the relMsm promoter, the ‘high’ subpopulation does not form (Sureka et al.2008), suggesting that this loop is necessary for maintaining high relMsmexpression. Although the existence of bistable relMtb expression has not yet been examined in Mtb, the mprAB-sigE-rel feedback loop presents a model that connects the stringent response, polyP and mycobacterial persistence.
THE STRINGENT RESPONSE AND Mtb VIRULENCE GENE EXPRESSION
(p)ppGpp accumulates in Mtb during exposure to multiple stresses, including starvation, hypoxia and oxidative stress (Primm et al.2000; Stallings et al.2009), which are all conditions Mtb is exposed to during infection (Stallings and Glickman 2010). As a result, stress survival strategies and virulence mechanisms have converged such that activating the stringent response leads to expression of multiple virulence factors (Dahl et al.2003; Dalebroux et al.2010). A microarray study examining the effect of nutrient starvation on gene expression in WT Mtb versus the ΔrelMtb mutant identified a RelMtb regulon as the set of 159 genes whose expression changed significantly during starvation in WT bacteria but not in the ΔrelMtb mutant (Dahl et al.2003). Genes involved in lipid metabolism comprised a significant proportion of the RelMtb regulon indicating that RelMtb may coordinate changes in the production of Mtb lipids and lipoproteins that can play important protective and immunomodulatory roles during infection (Dahl et al.2003; Cambier et al.2014). With a few exceptions, the trend of gene expression suggests that RelMtb may coordinate a shift from lipid anabolism to catabolism during starvation, which could represent the bacteria liberating nutrients from lipids but may also affect lipid modulators of virulence. Indeed, the RelMtb regulon also included several polyketide synthases (PKS) and polyketide associated protein (Pap) genes that are required to synthesize complex lipids (Dahl et al.2003; Gokhale et al.2007). Notably, during starvation, RelMtb upregulates pks2 and papA1, which are responsible for synthesizing sulfolipid-1 (SL-1) (Dahl et al.2003; Bhatt et al.2007). SL-1 is the most abundant sulfolipid in the Mtb cell wall and purified SL-1 has been reported to have immunomodulatory effects (Pabst et al.1988). PKS-synthesized surface glycolipids are also critical for biofilm formation in vitro (Pang et al.2012; Cambier et al.2014). Accordingly, loss of RelMtb-mediated (p)ppGpp synthesis resulted in delayed formation of biofilms and pellicles in static liquid cultures (Weiss and Stallings 2013). The relevance of biofilm formation for Mtb pathogenesis is currently uncertain, but Mtb during chronic infection shares many characteristics with Mtb within biofilms, including decreased replication rates, and increased stress tolerance (Ojha et al.2008; Richards and Ojha 2014). In addition, multiple groups have also reported the observation of extracellular communities of Mtb that resemble biofilms during infection (Lenaerts et al.2007; Wong and Jacobs 2016).
The ΔrelMtb mutant also exhibited lower expression of the mce1, mce3 and mce4 (mammalian cell entry) operons that have been associated with macrophage entry and are critical for Mtb infection (Zhang and Xie 2011) as well as lower expression of the major secreted antigen ESAT-6 (Dahl et al.2003; Ganguly et al.2007; Wang et al.2009; Sreejit et al.2014). Additionally, multiple genes belonging to the PE-PPE (Pro-Glu-rich) family of Mtb proteins were up- or downregulated in the ΔrelMtb mutant (Dahl et al.2003). PE-PPE proteins are either secreted or are cell surface proteins that are thought to contribute to Mtb virulence (Singh et al.2016). Collectively, this data suggests that the Mtb stringent response has crosstalk with numerous virulence pathways. The connection between the stringent response and virulence suggests that Mtb may have coopted sensing changes in the host environment to activate virulence mechanisms and changes in metabolism to promote adaptation and persistence.
TARGETING RelMtb AS AN ANTI-VIRULENCE STRATEGY TO TREAT TUBERCULOSIS
There has been significant interest in identifying compounds that target (p)ppGpp synthesis as an anti-virulence and anti-persistence strategy in many pathogenic bacteria, including Mtb (Zhang, Yew and Barer 2012). Relacin, a 2΄-deoxyguanosine-based analogue of ppGpp with the original pyrophosphate moieties at positions 5΄ and 3΄ replaced by glycyl-glycine dipeptides, was designed based on the RelSeq crystal structure and shown to inhibit E. coli RelA in vitro and (p)ppGpp synthesis in B. subtilis (Wexselblatt et al., 2010, 2012). However, Relacin is unable to penetrate the mycobacterial cell envelope to access its cytoplasmic target RelMtb. More recently, Syal et al. designed and synthesized analogs of Relacin that had activity in mycobacteria by functionalization of the amine group at C-2 position of the guanine base in Relacin (Syal et al.2017). These next generation Rel inhibitors were able to inhibit long-term survival and biofilm formation of mycobacteria. However, all compounds that have been designed to target the stringent response so far are analogs of ppGpp, which has limits in terms of improving potency and cell permeability. Therefore, the identification of new chemical matter will likely be important to translate (p)ppGpp synthesis inhibitors to clinical use.
SUMMARY AND OPEN QUESTIONS
A model has emerged where RelMtb-synthesized (p)ppGpp alters gene expression profiles, purine metabolism and polyP metabolism to enhance Mtb stress responses, virulence and persistence (Fig. 4 and Table 2). In addition to the RNAP, GTP synthesis enzymes, and polyphosphatases, other molecular targets of (p)ppGpp have been identified in non-mycobacterial species, but have yet to be explored in mycobacteria (Table 3) (Kanjee, Ogata and Houry 2012; Hauryliuk et al.2015; Liu, Bittner and Wang 2015). Many of these additional (p)ppGpp targets share a common theme of being GTPases or GTP-binding proteins with binding pockets for other guanine nucleotides that likely provide a natural binding site for (p)ppGpp. In addition to the GTPases or GTP-binding proteins, (p)ppGpp has been shown to target E. coli lipid and amino acid metabolism enzymes (Merlie and Pizer 1973; Polakis, Guchhait and Lane 1973; Stein and Bloch 1976; Heath, Jackowski and Rock 1994; Kanjee et al.2011; Kanjee, Ogata and Houry 2012). Whether (p)ppGpp binds and modulates similar enzymes in Mtb remains an open question for further study, but it is possible that (p)ppGpp effects on Mtb physiology are much more complex than our current understanding. In particular, our current understanding of how the Mtb stringent response promotes pathogenesis is based on transcriptional responses that are lost in the Δrelmtb mutant. Importantly, there exists a clear precedent for (p)ppGpp interacting with and affecting the activity of proteins post-translationally, so a purely transcriptome-level approach may miss key mechanistic details of the stringent response. Nonetheless, it is clear that by some mechanism (p)ppGpp metabolism is important for survival during stress, antibiotic treatment and virulence in animal models, highlighting the stringent response as a key mediator of Mtb pathogenesis.
Figure 4.
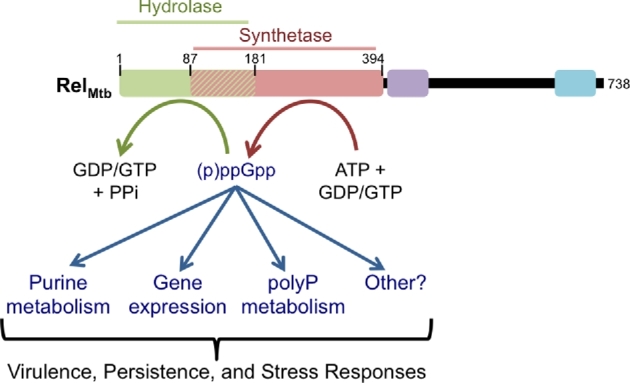
Summary of the effects of (p)ppGpp in Mtb. The RelMtb synthetase domain transfers the 5΄-β,γ-pyrophosphate from ATP to the 3΄-OH of GDP or GTP to synthesize (p)ppGpp, which can then be hydrolyzed by the RelMtb hydrolase domain. (p)ppGpp accumulates in response to a number of stresses, including starvation and oxidative stress, leading to alterations in purine metabolism, gene expression and polyP metabolism. It is also possible that there are other unidentified targets of (p)ppGpp in Mtb. These effects of (p)ppGpp accumulation contribute to enhanced virulence, persistence and stress responses.
Table 2.
Molecular targets of (p)ppGpp that have been investigated in Mycobacterium tuberculosis (Mtb).
| Molecular targets | Cellular process | Effects in Mtb | References |
|---|---|---|---|
| RNAP | Transcription | Inhibited at high [(p)ppGpp] in vitro | China et al. (2012), Tare, China and Nagaraja (2012) and Tare, Mallick and Nagaraja (2013) |
| GMK, Guanlyate Kinase | Purine metabolism | Insensitive to (p)ppGpp | Liu et al. (2015) |
| PPX1 and PPX2, Polyphosphatases | Polyphosphate metabolism | Inhibited by (p)ppGpp in vitro | Choi et al. (2012) and Chuang et al. (2015) |
Table 3.
Molecular targets of (p)ppGpp that have been identified in other bacteria but not yet explored in mycobacteria.
| Cellular process | Molecular targets | References |
|---|---|---|
| DNA replication | DnaG | Wang, Sanders and Grossman (2007) and Maciag et al. (2010) |
| Translation | IF2, EF-G, EF-Tu, EF-Ts, Obg, RsgA | Legault, Jeantet and Gros (1972), Rojas et al. (1984) and Persky et al. (2009) |
| Nucleotide metabolism | AprT, HprT, GprT, Upp, GuaB, PurA, GdpP | Gallant, Irr and Cashel (1971), Hochstadt-Ozer and Cashel (1972), Fast and Sköld (1977), Pao and Dyess (1981) and Rao et al. (2010) |
| Amino acid metabolism | HisG, LdcI, LdcC, SpeC | Kanjee et al. (2011) |
| Lipid metabolism | PlsB, PgsA, AccA, AccD, FabA, FabZ, | Merlie and Pizer (1973), Polakis, Guchhait and Lane (1973) and Stein and Bloch (1976) |
| Central metabolism | GdhA, Pcc | Pao and Dyess (1981) and Maurizi and Rasulova (2002) |
| Glycogen biosynthesis | GlgC | Dietzler and Leckie (1977) |
FUNDING
CLS is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award and National Institutes of Health (NIH) grants GM107544 and AI111696. JP is supported by the National Institute of General Medicine of the National Institutes of Health (NIGMS) Cell and Molecular Biology Training Grant GM007067 and the Stephen I. Morse Graduate Fellowship.
Conflict of interest. None declared.
REFERENCES
- Abranches J, Martinez AR, Kajfasz JK et al. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol 2009;191:2248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 1998;23:469–72. [DOI] [PubMed] [Google Scholar]
- Arenz S, Abdelshahid M, Sohmen D et al. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res 2016;44:6471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 2011;6:e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarbock A, Avarbock D, Teh J-S et al. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 2005;44:9913–23. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Avarbock A, Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry 2000;39:11640–8. [DOI] [PubMed] [Google Scholar]
- Avarbock D, Salem J, Li LS et al. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 1999;233:261–9. [DOI] [PubMed] [Google Scholar]
- Bag S, Das B, Dasgupta S et al. Mutational analysis of the (p)ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. Arch Microbiol 2014;196:575–88. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R et al. Bacterial persistence as a phenotypic switch. Science 2004;305:1622–5. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol 2001;305:689–702. [DOI] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J Bacteriol 2008;190:1224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts JC, Lukey PT, Robb LC et al. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 2002;43:717–31. [DOI] [PubMed] [Google Scholar]
- Bhatt K, Gurcha SS, Bhatt A et al. Two polyketide-synthase-associated acyltransferases are required for sulfolipid biosynthesis in Mycobacterium tuberculosis. Microbiology 2007;153:513–20. [DOI] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 2013;21:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Fernández IS, Gordiyenko Y et al. Ribosome-dependent activation of stringent control. Nature 2016;534:277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier CJ, Takaki KK, Larson RP et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 2014;505:218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 1969;244:3133–41. [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 1969;221:838–41. [DOI] [PubMed] [Google Scholar]
- China A, Mishra S, Tare P et al. Inhibition of Mycobacterium tuberculosis RNA polymerase by binding of a Gre factor homolog to the secondary channel. J Bacteriol 2012;194:1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol 2001;11:694–700. [DOI] [PubMed] [Google Scholar]
- Choi MY, Wang Y, Wong LL et al. The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLoS One 2012;7:e42561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholo MC, Mothiba MT, Fourie B et al. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother 2017;72:338–53. [DOI] [PubMed] [Google Scholar]
- Chuang Y-M, Bandyopadhyay N, Rifat D et al. Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. mBio 2015;6:e02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y-M, Belchis DA, Karakousis PC. The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. mBio 2013;4:e00039–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y-M, Dutta NK, Hung C-F et al. Stringent response factors PPX1 and PPK2 play an important role in Mycobacterium tuberculosis metabolism, biofilm formation, and sensitivity to isoniazid in vivo. Antimicrob Agents Chemother 2016;60:6460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JL, Kraus CN, Boshoff HIM et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA 2003;100:10026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzler DN, Leckie MP. Regulation of ADP-glucose synthetase, the rate-limiting enzyme of bacterial glycogen synthesis, by the pleiotropic nucleotides ppGpp and pppGpp. Biochem Biophys Res Commun 1977;77:1459–67. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC et al. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 2010;74:171–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Pal RR, Bag S et al. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene. Mol Microbiol 2009;72:380–98. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol 2006;61:564–72. [DOI] [PubMed] [Google Scholar]
- Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 2013;110:6554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymann C, Homuth G, Scharf C et al. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol 2002;184:2500–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast R, Sköld O. Mechanism of Uracil Uptake Regulation in Escherichia coli B. Biol Chem 1977;252:76–204. [PubMed] [Google Scholar]
- Gallant J, Irr J, Cashel M. The Mechanism of Amino Acid Control of Guanylate and Adenylate Biosynthesis. J Biol Chem 1971;246:5812–16. [PubMed] [Google Scholar]
- Ganguly N, Giang PH, Basu SK et al. Mycobacterium tuberculosis 6-kDa early secreted antigenic target (ESAT-6) protein downregulates lipopolysaccharide induced c-myc expression by modulating the extracellular signal regulated kinases 1/2. BMC Immunol 2007;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sureka K, Ghosh B et al. Phenotypic heterogeneity in mycobacterial stringent response. BMC Syst Biol 2011;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale RS, Saxena P, Chopra T et al. Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat Prod Rep 2007;24:267–77. [DOI] [PubMed] [Google Scholar]
- Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis 2004;84:29–44. [DOI] [PubMed] [Google Scholar]
- Gropp M, Strausz Y, Gross M et al. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol 2001;183:570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke LD, Shivers RP, Sonenshein AL. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol 2008;190:798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci 1973;70:1564–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KS et al. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Micro 2015;13:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Hovey R, Kane J et al. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J Bacteriol 2006;188:2134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zahrt TC. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J Bacteriol 2005;187:202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB). J Biol Chem 1994;269:26584–90. [PubMed] [Google Scholar]
- Hochstadt-Ozer J, Cashel M. The Regulation of Purine Utiliztion in Bacteria. J Biol Chem 1972;247:7067–72. [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke H et al. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 2004;117:57–68. [DOI] [PubMed] [Google Scholar]
- Jain V, Saleem-Batcha R, Chatterji D. Synthesis and hydrolysis of pppGpp in mycobacteria: a ligand mediated conformational switch in Rel. Biophys Chem 2007;127:41–50. [DOI] [PubMed] [Google Scholar]
- Jain V, Saleem-Batcha R, China A et al. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci 2006;15:1449–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Sujatha S, Ojha AK et al. Identification and characterization of rel promoter element of Mycobacterium tuberculosis. Gene 2005;351:149–57. [DOI] [PubMed] [Google Scholar]
- Kanjee U, Gutsche I, Alexopoulos E et al. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J 2011;30:931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol 2012;85:1029–43. [DOI] [PubMed] [Google Scholar]
- Karakousis PC, Yoshimatsu T, Lamichhane G et al. Dormancy phenotype displayed by extracellular mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 2004;200:647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Minami S, Rubin E et al. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2011;2:e00100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg LG, Lee J-H, Bishai WR et al. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 2010;202:1397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krásný L, Tiserová H, Jonák J et al. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol 2008;69:42–54. [DOI] [PubMed] [Google Scholar]
- Kriel A, Bittner AN, Kim SH et al. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 2012;48:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Petchiappan A, Singh A et al. R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol Microbiol 2016;102:168–82. [DOI] [PubMed] [Google Scholar]
- Kuzmine I, Gottlieb PA, Martin CT. Binding of the priming nucleotide in the initiation of transcription by T7 RNA polymerase. J Biol Chem 2003;278:2819–23. [DOI] [PubMed] [Google Scholar]
- Legault L, Jeantet C, Gros F. Inhibition of in vitro protein synthesis by ppGpp. FEBS Lett 1972;27:71–5. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Lin VK, Nascimento MM et al. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol 2007;65:1568–81. [DOI] [PubMed] [Google Scholar]
- Lenaerts AJ, Hoff D, Aly S et al. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother 2007;51:3338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Tumasz S et al. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 2012;26:2634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Bittner AN, Wang JD. Diversity in (p)ppGpp metabolism and effectors. Curr Opin Microbiol 2015;24:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Myers AR, Pisithkul T et al. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell 2015;57:735–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Bah E, Madireddy R et al. Ribosome•RelA structures reveal the mechanism of stringent response activation. Elife 2016;5:17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag M, Kochanowska M, Łyzeñ R et al. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 2010;63:61–7. [DOI] [PubMed] [Google Scholar]
- Manganelli R, Voskuil MI, Schoolnik GK et al. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 2001;41:423–37. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Rasulova F. Degradation of L-glutamate dehydrogenase from Escherichia coli: Allosteric regulation of enzyme stability. Arch Biochem Biophys 2002;397:206–16. [DOI] [PubMed] [Google Scholar]
- Merlie JP, Pizer LI. Regulation of phospholipid synthesis in Escherichia coli by guanosine tetraphosphate. J Bacteriol 1973;116:355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov V, Sineva E, Zhang L et al. Allosteric effector ppGpp potentiates the inhibition of transcript initiation by DksA. Mol Cell 2018;69:828–839e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdeshwar MS, Chatterji D. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol 2012;194:4003–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa A et al. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol 2008;67:291–304. [DOI] [PubMed] [Google Scholar]
- Natori Y, Tagami K, Murakami K et al. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J Bacteriol 2009;191:4555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha AK, Baughn AD, Sambandan D et al. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 2008;69:164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha AK, Mukherjee TK, Chatterji D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun 2000;68:4084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst MJ, Gross JM, Brozna JP et al. Inhibition of macrophage priming by sulfatide from Mycobacterium tuberculosis. J Immunol 1988;140:634–40. [PubMed] [Google Scholar]
- Pang JM, Layre E, Sweet L et al. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J Bacteriol 2012;194:715–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao CC, Dyess BT. Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim Biophys Acta 1981;677:358–62. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 2004;118:311–22. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA 2005;102:7823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky NS, Ferullo DJ, Cooper DL et al. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 2009;73:253–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis SE, Guchhait RB, Lane MD. Stringent control of fatty acid synthesis in Escherichia coli. Possible regulation of acetyl coenzyme A carboxylase by ppGpp. J Biol Chem 1973;248:7957–66. [PubMed] [Google Scholar]
- Primm TP, Andersen SJ, Mizrahi V et al. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 2000;182:4889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NN, Roberts MF, Torriani A. Amount and chain length of polyphosphates in Escherichia coli depend on cell growth conditions. J Bacteriol 1985;162:242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, See RY, Zhang D et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 2010;285:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M, Serror P, Wong KW et al. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 2001;15:1093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JP, Ojha AK. Mycobacterial biofilms. Microbiol Spectrum 2014;2:MGM2-0004-2013. [DOI] [PubMed] [Google Scholar]
- Rojas AM, Ehrenberg M, Andersson SGE et al. ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. MGG Mol Gen Genet 1984;197:36–45. [DOI] [PubMed] [Google Scholar]
- Ross W, Sanchez-Vazquez P, Chen AY et al. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell 2016;62:811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P et al. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 2013;50:420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee J-H et al. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 2009;23:236–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajish M, Kalayil S, Verma SK et al. The significance of E X DD and R X KD motif conservation in Rel proteins. J Biol Chem 2009;284:9115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajish M, Tiwari D, Rananaware D et al. A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins. J Biol Chem 2007;282:34977–83. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Banerjee SK, Banerjee R et al. Polyphosphate kinase 1, a central node in the stress response network of Mycobacterium tuberculosis, connects the two-component systems MprAB and SenX3-RegX3 and the extracytoplasmic function sigma factor, sigma E. Microbiology 2013;159:2074–86. [DOI] [PubMed] [Google Scholar]
- Shum KT, Lui ELH, Wong SCK et al. Aptamer-mediated inhibition of Mycobacterium tuberculosis polyphosphate kinase 2. Biochemistry 2011;50:3261–71. [DOI] [PubMed] [Google Scholar]
- Singal B, Balakrishna AM, Nartey W et al. Crystallographic and solution structure of the N-terminal domain of the Rel protein from Mycobacterium tuberculosis. FEBS Lett 2017;591:2323–37. [DOI] [PubMed] [Google Scholar]
- Singh P, Rao RN, Reddy JRC et al. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep 2016;6:21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Singh M, Arora G et al. Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J Bacteriol 2013;195:2839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Kuipers OP, Veening J-W. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Micro 2006;4:259–71. [DOI] [PubMed] [Google Scholar]
- Sreejit G, Ahmed A, Parveen N et al. The ESAT-6 protein of Mycobacterium tuberculosis interacts with beta-2-microglobulin (β2M) affecting antigen presentation function of macrophage. PLoS Pathog 2014;10:e1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings CL, Glickman MS. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect 2010;12:1091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings CL, Stephanou NC, Chu L et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 2009;138:146–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JP, Bloch KE. Inhibition of E. coli beta-hydroxydecanoyl thioester dehydrase by ppGpp. Biochem Biophys Res Commun 1976;73:881–4. [DOI] [PubMed] [Google Scholar]
- Sun J, Hesketh A, Bibb M. Functional analysis of relA and rshA, two relA/spoT homologues of Streptomyces coelicolor A3(2). J Bacteriol 2001;183:3488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Dey S, Datta P et al. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 2007;65:261–76. [DOI] [PubMed] [Google Scholar]
- Sureka K, Ghosh B, Dasgupta A et al. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One 2008;3:e1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Sanyal S, Basu J et al. Polyphosphate kinase 2: a modulator of nucleoside diphosphate kinase activity in mycobacteria. Mol Microbiol 2009;74:1187–97. [DOI] [PubMed] [Google Scholar]
- Syal K, Flentie K, Bhardwaj N et al. Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob Agents Chemother 2017;61:e00443–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K, Joshi H, Chatterji D et al. Novel pppGpp binding site at the C-terminal region of the Rel enzyme from Mycobacterium smegmatis. FEBS J 2015;282:3773–85. [DOI] [PubMed] [Google Scholar]
- Tare P, China A, Nagaraja V. Distinct and contrasting transcription initiation patterns at Mycobacterium tuberculosis promoters. PLoS One 2012;7:e43900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tare P, Mallick B, Nagaraja V. Co-evolution of specific amino acid in sigma 1.2 region and nucleotide base in the discriminator to act as sensors of small molecule effectors of transcription initiation in mycobacteria. Mol Microbiol 2013;90:569–83. [DOI] [PubMed] [Google Scholar]
- Thayil SM, Morrison N, Schechter N et al. The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLoS One 2011;6:e28076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo S, Kumamoto K, Hirooka K et al. Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis. J Bacteriol 2010;192:1573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen H-T et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 2008;68:1128–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional Control of Elongation of DNA Replication by (p)ppGpp. Cell 2007;128:865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Barnes PF, Dobos-Elder KM et al. ESAT-6 inhibits production of IFN-gamma by Mycobacterium tuberculosis-responsive human T cells. J Immunol 2009;182:3668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Zimmermann M, Goodwin MB et al. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog 2011;7:e1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 1996;64:2062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Stallings CL. Essential roles for mycobacterium tuberculosis rel beyond the production of (p)ppGpp. J Bacteriol 2013;195:5629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexselblatt E, Katzhendler J, Saleem-Batcha R et al. ppGpp analogues inhibit synthetase activity of Rel proteins from Gram-negative and Gram-positive bacteria. Bioorg Med Chem 2010;18:4485–97. [DOI] [PubMed] [Google Scholar]
- Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I et al. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog 2012;8:e1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Aravind L, Grishin NV et al. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res 1999;9:689–710. [PubMed] [Google Scholar]
- Wong K-W, Jacobs WR. Postprimary tuberculosis and macrophage necrosis: is there a big ConNECtion? mBio 2016;7:e01589–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xie J-P. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol Cell Biochem 2011;352:1–10. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yew WW, Barer MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother 2012;56:2223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Rubin EJ. Feast or famine: the host-pathogen battle over amino acids. Cell Microbiol 2013;15:1079–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 2013;50:430–6. [DOI] [PMC free article] [PubMed] [Google Scholar]



