Abstract

Collagen is the most abundant protein in humans and the major component of human skin. Collagen mimetic peptides (CMPs) can anneal to damaged collagen in vitro and in vivo. A duplex of CMPs was envisioned as a macromolecular mimic for damaged collagen. The duplex was synthesized on a solid support from the amino groups of a lysine residue and by using olefin metathesis to link the N termini. The resulting cyclic peptide, which is a monomer in solution, binds to CMPs to form a triple helix. Among these, CMPs that are engineered to avoid the formation of homotrimers but preorganized to adopt the conformation of a collagen strand exhibit enhanced association. Thus, this cyclic peptide enables the assessment of CMPs for utility in annealing to damaged collagen. Such CMPs have potential use in the diagnosis and treatment of fibrotic diseases and wounds.
Introduction
The collagen triple helix is the most abundant structure adopted by a biopolymer within the human body. There, collagen comprises one-third of the total protein, accounts for three-fourths of the dry weight of skin, and is the most prevalent component of the extracellular matrix.1
Collagen is damaged in fibrotic and other diseases and in wounds. Collagen mimetic peptides (CMPs) can anneal to damaged collagen.2 Such annealing could allow for the delivery of diagnostic or therapeutic agents that are conjugated to the CMP. Indeed, we have used such CMP conjugates to anneal fluorescent dyes,3 a growth factor,4 and even a sunscreen5 to natural collagen. Damaged collagen in a human body is, however, a complex target that confounds physicochemical analyses, complicating the assessment of therapeutic potential for new CMP designs.
We sought to develop a molecular mimic of damaged collagen. We envisioned doing so with a collagen “duplex”, that is, two cross-linked CMPs. Several double-stranded duplexes have been synthesized and used to form collagen triple helices.6 In most of these precedents, one or both ends of the peptides are free, allowing for the assembly of many possible complexes. To make a duplex more amenable to rigorous analyses and more biomimetic, we sought to tether two parallel strands at both termini (Figure 1). This design not only minimizes the conformational entropy of the duplex, but also mimics natural collagen fibers that have a disrupted triple helix, as might be found in damaged collagen. Here, we describe the creation of a “host” and report on its interaction with “guest” strands, that is, CMPs.
Figure 1.

Scheme showing the molecular mimicry of damaged collagen by a cyclic “host” of two parallel collagen strands. A collagen mimetic peptide with a conjugated pendant “X” (purple) is shown annealing to the damaged collagen.
Experimental Methods
General
Boc-flp–OH and Fmoc-flp–OH were from OmegaChem (Lévis, Québec). (Here, “flp” refers to (2S,4S)-4-fluoroproline.) Boc-Flp–OH, Fmoc-Hyp–OH, and other amino acid derivatives, resins, Fmoc-OSu, and HOBt were from Chem-Impex International (Wood Dale, IL). (Here, “Flp” refers to (2S,4R)-4-fluoroproline and “Hyp” refers to (2S,4R)-4-hydroxyproline.) DIC and 4-methylpiperidine were from Oakwood Chemical (Estill, SC). Streptavidin-coated fluorescent blue particles were product SVFP-106805 from Spherotech (Lake Forest, IL). 6-Aminohexanoic acid and all other reagents were from Sigma–Aldrich (St. Louis, MO) and were used without further purification.
DMF was dried with a Glass Contour system from Pure Process Technology (Nashua, NH). In addition, DMF was passed through an associated isocyanate “scrubbing” column to remove any amines. Water was purified with an Arium Pro system from Sartorius (Göttingen, Germany).
The phrase “concentrated under reduced pressure” refers to the removal of solvents and other volatile materials with a rotary evaporator at water-aspirator pressure (<20 Torr) while maintaining a water bath below 40 °C. Residual solvent was removed from samples with a high vacuum (<0.1 Torr).
All procedures were performed in air at ambient temperature (∼22 °C) and pressure (1.0 atm) unless indicated otherwise.
Instrumentation
Solid-phase peptide synthesis was performed with a Liberty Blue Peptide Synthesizer from CEM (Matthews, NC). Synthetic peptides were purified by HPLC with a Prominence instrument from Shimadzu (Kyoto, Japan) equipped with a VarioPrep 250/21 C18 column from Macherey–Nagel (Düren, Germany). Molecular mass was determined by matrix-assisted laser desorption/ionization–time-of-flight (MALDI–TOF) mass spectrometry on an α-cyano-4-hydroxycinnamic acid or sinapic acid matrix with a microflex LRF instrument from Bruker (Billerica, MA). Purity analyses were performed with an Acquity UPLC H-Class system from Waters that was equipped with an Acquity photodiode array detector, Acquity quaternary solvent manager, Acquity sample manager with a flow-through needle, Acquity UPLC BEH C18 column (2.1 × 50 mm, 1.7 μm particle size), and Empower 3 software. 1H and 13C NMR spectra were acquired with an Avance III 400 spectrometer from Bruker. Sedimentation equilibrium experiments were performed with an XL-A analytical ultracentrifuge and An-60 Ti rotor from Beckman Coulter (Brea, CA) at the Biophysics Instrumentation Facility of the University of Wisconsin–Madison (UW BIF). Beads were imaged using a Eclipse Ti inverted confocal microscope from Nikon (Melville, NY) at the Biochemistry Optical Core of the University of Wisconsin–Madison. Flow cytometry was performed with an Accuri Flow Cytometer with C-Sampler from BD (San Jose, CA) at the UW BIF. CD data were acquired with a Model 420 CD spectrophotometer from Aviv Biomedical (Lakewood, NJ) at the UW BIF.
Small-Molecule Synthesis
Fmoc-6-aminohexanoic Acid
6-Aminohexanoic acid (1.00 g, 7.62 mmol) was dissolved in a saturated aqueous solution of NaHCO3 (50 mL). In a separate flask, Fmoc-OSu (2.82 g, 8.38 mmol) was dissolved in dioxane (50 mL). The two solutions were combined, and the reaction mixture became cloudy and was stirred for 16 h. The mixture was then concentrated under reduced pressure. The residue was dissolved in EtOAc and washed with aqueous 1.0 M HCl and brine. The organic layer was dried over Na2SO4(s), decanted, and concentrated under reduced pressure. Crude product was purified by chromatography on silica-gel, eluting with EtOAc (40% v/v) and acetic acid (1% v/v) in hexanes to yield Fmoc-6-aminohexanoic acid (2.56 g, 95%) as a white solid. HRMS–ESI (m/z): [M + H]+ calcd, 354.17; found, 354.17. 1H NMR (400 MHz, MeOD, δ): 7.78 (d, J = 7.5 Hz, 2H), 7.63 (d, J = 7.5 Hz, 2H), 7.37 (t, J = 7.4 Hz, 2H), 7.33–7.26 (m, 2H), 4.33 (d, J = 6.8 Hz, 2H), 4.18 (t, J = 6.9 Hz, 1H), 3.08 (t, J = 7.0 Hz, 2H), 2.27 (t, J = 7.4 Hz, 2H), 1.60 (p, J = 7.5 Hz, 2H), 1.49 (p, J = 7.1 Hz, 2H), 1.33 (p, J = 10.1, 6.0 Hz, 2H). 13C NMR (101 MHz, MeOD, δ): 157.49, 143.95, 141.20, 127.34, 126.71, 124.75, 119.50, 66.11, 47.13, 40.17, 33.71, 29.18, 25.97, 24.45.
Boc-Flp-OBn
Boc-Flp–OH (2.5 g, 10.7 mmol) was dissolved in DMF. Solid Cs2CO3 (1.5 g, 10.7 mmol) was added, and the reaction mixture was stirred for 10 min. Benzyl bromide (1.27 mL, 10.7 mmol) was added dropwise, and the mixture was stirred for 16 h. The mixture was then concentrated under reduced pressure. Crude product was purified by chromatography on silica gel, eluting with EtOAc (10% v/v) in hexanes to yield product (2.26 g, 65%). HRMS–ESI (m/z): [M + H]+ calcd, 324.15; found, 324.16. 1H NMR (400 MHz, CDCl3, δ): 7.35 (d, J = 4.5 Hz, 5H), 5.32–5.19 (m, 1H), 5.19–5.14 (m, 1H), 5.14–5.04 (m, 1H), 4.58–4.38 (m, 1H), 3.99–3.75 (m, 1H), 3.61 (ddt, J = 36.1, 13.0, 3.9 Hz, 1H), 2.70–2.47 (m, 1H), 2.20–1.97 (m, 1H), 1.53–1.31 (m, 9H), which are consistent with literature values for this known compound.7
Boc-flp-Flp-OBn
Boc-Flp-OBn (2.26 g, 6.99 mmol) was dissolved in 4 N HCl (8.0 mL), and the reaction mixture was stirred for 30 min. The reaction mixture was then concentrated under reduced pressure. The residue was dissolved in DMF. DIEA (4.87 mL, 27.96 mmol) was added dropwise. Solid PyBrOP (3.91 g, 8.39 mmol) and Boc-flp–OH (1.79 g, 7.69 mmol) were added, and the reaction mixture was stirred for 16 h. The residue was concentrated under reduced pressure, taken up in EtOAc, and washed successively with 1.0 M HCl (2×), saturated aqueous NaHCO3 (2×), and brine (2×). The organic layer was dried over Na2SO4(s), filtered, and concentrated under reduced pressure to yield crude product (3.96 g), which was carried forward without further purification. HRMS–ESI (m/z): [M + H]+ calcd, 439.20; found, 439.20. 1H NMR (400 MHz, CDCl3, δ): 7.35 (d, J = 4.0 Hz, 6H), 5.32–5.10 (m, 4H), 4.55 (dt, J = 20.6, 8.4 Hz, 1H), 4.06–3.77 (m, 4H), 3.66 (ddt, J = 36.6, 13.0, 3.5 Hz, 1H), 2.62 (dddt, J = 28.3, 16.6, 8.0, 1.8 Hz, 1H), 2.22–1.99 (m, 2H), 1.85 (ddt, J = 71.3, 13.3, 6.7 Hz, 1H), 0.94 (d, J = 6.7 Hz, 3H), 0.85 (dd, J = 6.7, 2.3 Hz, 4H). 13C NMR (101 MHz, CDCl3, δ): 172.16, 172.04, 155.00, 154.56, 135.47, 135.28, 128.65, 128.58, 128.52, 128.42, 128.33, 128.16, 92.65, 91.86, 90.86, 90.08, 77.25, 71.88, 71.83, 67.09, 67.02, 60.40, 57.72, 57.48, 53.58, 53.35, 53.15, 52.93, 37.77, 37.54, 36.71, 36.48, 28.36, 28.17, 27.98, 27.86, 19.03, 18.96, 18.92, 14.22.
Fmoc-Gly-flp-Flp-OBn
Crude Boc-flp-Flp-OBn (3.96 g) was dissolved in 4 N HCl (8.0 mL), and the reaction mixture was stirred for 30 min. The mixture was then concentrated under reduced pressure. The residue was dissolved in DCM. DIEA (3.65 mL, 20.97 mmol) was added dropwise to the resulting solution. Solid Fmoc-Gly-OPfp (3.24 g, 6.99 mmol) was added, and the reaction mixture was stirred for 16 h. The residue was concentrated under reduced pressure, taken up in EtOAc, and washed successively with 1.0 M HCl (2×), saturated aqueous NaHCO3 (2×), and brine (2×). The organic layer was dried over Na2SO4(s), filtered, and concentrated under reduced pressure. Crude product was purified by chromatography on silica gel, eluting with dichloromethane, followed by a methanol flush to yield product (3.70 g, 83%). HRMS–ESI (m/z): [M + NH4]+ calcd, 635.23; found, 635.27. 1H NMR (400 MHz, CDCl3, δ): 7.75 (d, J = 7.5 Hz, 2H), 7.59 (dd, J = 7.5, 3.2 Hz, 2H), 7.43–7.26 (m, 9H), 5.86–5.48 (m, 1H), 5.44–5.02 (m, 4H), 4.85–4.67 (m, 2H), 4.45–4.02 (m, 4H), 4.03–3.60 (m, 3H), 2.62 (ddd, J = 20.3, 14.1, 8.1 Hz, 1H), 2.56–2.22 (m, 2H), 2.08 (dddd, J = 40.9, 18.3, 9.1, 4.8 Hz, 1H), 1.82 (s, 2H). 13C NMR (101 MHz, CDCl3, δ): 171.14, 168.75, 167.23, 156.27, 143.88, 141.26, 135.40, 128.58, 128.39, 128.29, 127.69, 127.10, 125.21, 119.95, 91.95, 77.25, 67.21, 57.96, 57.13, 47.08, 43.41, 35.31, 34.48, 34.26.
Fmoc-Gly-flp-Flp–OH
Fmoc-Gly-flp-Flp–OH (3.70 g, 5.98 mmol) was dissolved in methanol (25 mL). The head space was purged with N2(g). Pd/C (10% w/w, 0.64 g) was added, and the flask was capped with a septum. H2(g) was added via a balloon. The reaction was monitored by thin-layer chromatography and observed to be complete at 6 h. The reaction mixture was filtered through diatomaceous earth and concentrated under reduced pressure. Crude product was purified by chromatography on silica gel, eluting with 1% v/v acetic acid and 20% v/v methanol in EtOAc to yield product (2.95 g, 93%). HRMS–ESI (m/z): [M – H]− calcd, 526.52; found, 526.18. 1H NMR (400 MHz, MeOD, δ): 7.81 (d, J = 7.6 Hz, 2H), 7.74–7.64 (m, 2H), 7.48–7.28 (m, 4H), 5.49–5.03 (m, 3H), 4.72–4.20 (m, 4H), 4.20–3.43 (m, 5H), 2.85–2.27 (m, 3H), 2.26–2.06 (m, 1H). 13C NMR (101 MHz, CDCl3, δ): 177.16, 173.49, 172.00, 160.94, 147.71, 145.13, 132.21, 131.59, 131.14, 130.97, 130.78, 128.99, 123.79, 96.63, 96.29, 94.84, 94.48, 71.07, 68.30, 61.94, 61.35, 50.96, 46.99, 39.48, 39.26, 38.24, 38.03.
Peptide Synthesis
Peptides were prepared by automated solid-phase peptide synthesis. Fmoc-deprotection was achieved by treatment with 4-methylpiperidine (20% v/v) in DMF. Tripeptides, amino acids, and small-molecule carboxylic acids (5 equiv) were activated by using DIC and HOBt. Peptides were cleaved from the resin with 96.5:2.5:1.0 TFA/H2O/TIPSH (5 mL), precipitated from diethyl ether at −80 °C, and isolated by centrifugation. Peptides were purified by preparative HPLC using a gradient of 10–50% B (single strands) or 40–65% (hosts) over 50 min (A: H2O containing 0.1% v/v TFA; B: acetonitrile containing 0.1% v/v TFA). The purity of each peptide was assessed to be >95% by UPLC.
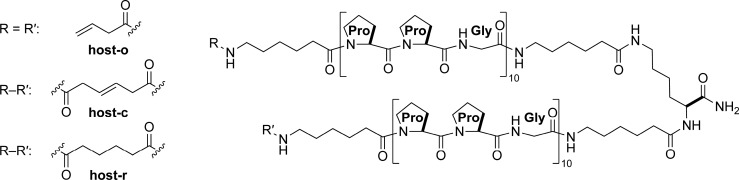
Host-o
The open host was synthesized by first doing a 0.05 mmol coupling of Fmoc-Lys(Fmoc)–OH to TGT S RAM resin (0.22 mmol/g). Next, Fmoc-6-aminohexanoic acid was single-coupled at a 0.10 mmol scale. Additional amino acid and small-molecule carboxylic acid additions were done at a 0.10 mmol scale and double-coupled. MALDI (m/z): [M + H]+ calcd, 5757.05; found, 5757.07. A 0.05 mmol scale synthesis afforded 7.6 mg (2.7%) of host-o after purification.
Host-c
The closed host was synthesized by olefin metathesis on the N-terminal 4-butenoic acids of the open next on resin, following a procedure similar to that for the stapling of peptide side chains.8 Peptide bound to resin was added to a Schlenk flask. The resin was then dried for at least 3 h on a high-vacuum manifold. Then, the flask was purged rigorously with N2(g). While under N2(g), the resin was preswelled in 2.5 mL of dry CH2Cl2 for at least 15 min prior to the addition 0.5 mL of a 2.5 mM solution of the Grubbs G2 catalyst (C46H65Cl2N2PRu)9 in dry CH2Cl2 using standard Schlenk techniques. The reaction flask was equipped quickly with an oven-dried reflux condenser, purged with N2(g), and heated in an oil bath at 40 °C for 36 h under N2(g). After 36 h, another 0.5 mL aliquot of the Grubbs G2 catalyst solution was added, and the flask was heated at 40 °C for another 36 h under N2(g). Dry solvent was added over the course of the reaction to maintain at least 3 mL of CH2Cl2. The reaction mixture was then allowed to cool to room temperature and filtered. The resin was washed with DCM to remove any remaining catalyst. MALDI (m/z): [M + H]+ calcd, 5729.02; found, 5728.55. A 0.05 mmol scale synthesis afforded 7.0 mg (2.4%) of host-c after purification.
Host-r
Following olefin metathesis, the resin was filtered to remove catalyst and a small sample was taken to confirm the generation of host-c by MALDI mass spectrometry. The resin was returned to a Schlenk flask and suspended in DCE. Again, following a literature precedent,10 Umicore M2 (46.5 mg, 0.05 mmol) and Et3SiH (0.80 mL, 9.4 mmol) were added to the flask. The vessel was capped with a septum, and the reaction mixture was heated to 60 °C for 72 h. The resin was washed with DCM to remove any remaining catalyst. MALDI (m/z): [M + H]+ calcd, 5731.03; found, 5731.67. A 0.05 mmol scale synthesis afforded 9.8 mg (3.4%) of host-r after purification.
Host-o–Biotin and Host-r–Biotin
Biotin was conjugated to the Nε-amino group of a lysine residue installed near the C-terminus of host-o and host-r. Specifically, Fmoc-Lys(Boc)–OH was coupled to TGT S RAM resin (0.22 mmol/g) followed by a (Gly-Ser)3 sequence synthesized by the addition of Fmoc-protected amino acids. The remainder of the host-o and host-r syntheses followed as described above.
After cleavage from the resin, crude host-o (5.3 mg, 0.84 μmol) was dissolved in 500 μL of DMSO. A solution of biotin–NHS ester (4.0 mg, 11.7 μmol) and DIEA (0.1 mL, 0.57 mmol) in 500 μL of DMSO was added, and the mixture was allowed to react for 12 h. The host-o–biotin conjugate was purified by HPLC. MALDI (m/z): [M + H]+ calcd, 6543.38; found, 6541.63. A 0.05 mmol scale synthesis afforded 0.4 mg (0.52%) of host-o–biotin after purification.
After cleavage from the resin, crude host-r (4.3 mg, 0.68 mmol) was dissolved in 500 μL of DMSO. A solution of biotin–NHS ester (6.7 g, 19.6 μmol) and DIEA (0.1 mL, 0.57 mmol) in 500 μL of DMSO was added, and the mixture was allowed to react for 12 h. The host-r–biotin conjugate was purified by HPLC. MALDI (m/z): [M + H]+ calcd, 6517.37; found, 6519.48. A 0.05 mmol scale synthesis afforded 0.2 mg (0.14%) of host-r–biotin after purification.
(flp-Hyp-Gly)7
(flp-Hyp-Gly)7 was synthesized by the addition (double-coupling) of Fmoc-protected amino acids to preloaded Fmoc-Gly-Wang resin (0.65 mmol/g). MALDI (m/z): [M + H]+ calcd, 2015.80; found, 2015.10. A 0.05 mmol scale synthesis afforded 6.4 mg (6.4%) of (flp-Hyp-Gly)7 after purification.
(flp-Flp-Gly)7
(flp-Flp-Gly)7 was synthesized by using Fmoc-Gly-flp-Flp–OH tripeptide and Fmoc-flp–OH and Fmoc-Flp–OH monomers. Six segment condensations of tripeptide were followed by the addition of each monomer, with each addition being double-coupled on preloaded Fmoc-Gly-Wang resin (0.65 mmol/g). The peptide was then cleaved from the resin and purified by HPLC. MALDI (m/z): [M + H]+ calcd, 2028.77; found, 2028.75. A 0.05 mmol scale synthesis afforded 8.3 mg (8.1%) of (flp-Flp-Gly)7 after purification.
(Pro-Pro-Gly)7
(Pro-Pro-Gly)7 was synthesized by the addition of Fmoc-protected amino acids to preloaded Fmoc-Gly-Wang resin (0.65 mmol/g). MALDI (m/z): [M + H]+ calcd, 1777.02; found, 1776.87. A 0.05 mmol scale synthesis afforded 18.0 mg (20.2%) of (Pro-Pro-Gly)7 after purification.
(Pro-Ile-Gly)7
(Pro-Ile-Gly)7 was synthesized by the addition of Fmoc-protected amino acids to preloaded Fmoc-Gly-Wang resin (0.65 mmol/g). MALDI (m/z): [M + H]+ calcd, 1890.31; found, 1890.30. A 0.05 mmol scale synthesis afforded 9.6 mg (10.2%) of (Pro-Ile-Gly)7 after purification.
Fluorescein–CMP
Ac-Lys-(Ser-Gly)3-(Pro-Pro-Gly)7 was synthesized by the addition of Fmoc-protected amino acids to preloaded Fmoc-Gly-Wang resin (0.65 mmol/g) and then cleaved from the resin. 5(6)-Carboxyfluorescein (112.4 mg, 0.30 mmol), HATU (104.8 mg, 0.28 mmol), and DIEA (100 μL, 0.57 mmol) were incubated for 15 min in 500 μL of DMSO. This solution was added to a solution of crude Ac-Lys-(Ser-Gly)3-(Pro-Pro-Gly)7 (74.2 mg) in 500 μL of DMSO, and the mixture was allowed to react for 12 h. MALDI (m/z): [M + Na]+ calcd, 2759.20; found, 2760.47. A 0.05 mmol scale synthesis afforded 3.5 mg (2.5%) of Ac-Lys(fluorescein)-(Ser-Gly)3-(Pro-Pro-Gly)7 after purification.
Fluorescein–d-CMP
Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 was synthesized by amino acid addition on preloaded Fmoc-Gly-Wang resin (0.65 mmol/g). Fmoc-deprotection was achieved by treatment with 4-methylpiperidine (20% v/v) in DMF. The amino acid monomer (5 equiv) was converted to an active ester by using DIC and HOBt. The peptide was then cleaved from the resin. A solution of 5(6)-carboxyfluorescein (9.7 mg, 25.7 μmol), HATU (9.1 mg, 23.9 μmol), and DIEA (100 μL, 0.57 mmol) in 500 μL of DMSO was allowed to react for 15 min. A solution of crude Ac-Lys-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 (36.0 mg) dissolved in 500 μL of DMSO was added to the solution and allowed to react for 12 h. The peptide was then purified by HPLC. MALDI (m/z): [M + Na]+ calcd, 2759.20; found, 2760.47. A 0.05 mmol synthesis afforded 2.5 mg (1.8%) of Ac-Lys(fluorescein)-(Ser-Gly)3-(d-Pro-d-Pro-Gly)7 after purification.
Biotin–Fluorescein
Fmoc-Lys(Boc)–OH was coupled first to TGT S RAM resin (0.22 mmol/g) followed by a (Gly-Ser)3 sequence that was synthesized by the addition of Fmoc-protected amino acids. The resulting decapeptide was treated on-resin with biotin–NHS ester (5 equiv). The peptide was then cleaved from the resin and deprotected. A solution of 5(6)-carboxyfluorescein (9.5 mg, 25.2 μmol), HATU (8.7 mg, 22.8 μmol), and DIEA (100 μL, 0.57 mmol) in 500 μL DMSO were incubated for 15 min. This solution was added to a solution of crude biotin-(Gly-Ser)3-Lys-NH2 (12.3 mg) in 500 μL of DMSO, and the mixture was allowed to react for 12 h. The biotin–fluorescein conjugate was then purified by HPLC. MALDI (m/z): [M + H]+ calcd, 1162.41; found, 1162.52. A 0.05 mmol synthesis afforded 1.5 mg (2.5%) of biotin-(Gly-Ser)3-Lys(fluorescein)-NH2 after purification.
Analytical Ultracentrifugation
A host (100 μL of a 45 μM solution) and matching buffer (110 μL) were placed in a cell with an Epon 12 mm double-sector charcoal-filled centerpiece from Beckman Coulter. Experiments were run at 4 °C for more than 7 days at speeds of 20000, 26000, 34000, and 42000 rpm, and gradients recorded at 235 nm were monitored until superimposable 4 h apart. Equilibrium gradients at 4 °C were modeled as single and multiple noninteracting species through nonlinear least-squares fits to the gradient data, using a buffer density of 1.00037 g/mL and partial specific volumes of 0.7471 and 0.7460 mL/g calculated based on amino acid and functional group content for host-o and host-r, respectively.11 Nonsedimenting baseline-attenuance was applied during data analysis, which was performed with programs written by D. R. McCaslin (UW BIF) for IGOR PRO software from WaveMetrics (Lake Oswego, OR).
Confocal Microscopy and Flow Cytometry
A 200 μL suspension of streptavidin-coated fluorescent blue particles was added to 200 μL of a 58 μM solution of biotinylated host-o or host-r in H2O, and the mixture was agitated for 9 h. Beads were pelleted by centrifugation at 12000 rpm on a benchtop centrifuge for 2 min. The supernatant was removed, and the beads were resuspended in 200 μL of H2O and stored at 4 °C.
A 10 μL aliquot of suspended beads was treated with Ac-Lys(fluorescein)-(Ser-Gly)3-(Pro-Pro-Gly)7 such that the final concentration of added peptide was 60 μM. Mixtures were allowed to anneal by heating the samples to 65 °C and cooling slowly to 4 °C at a rate of −12 °C per hour. At 4 °C, beads were pelleted, the supernatant was removed, and the beads were resuspended in water. This process was repeated three times to wash the beads. Finally, beads were resuspended in 300 μL of water. For confocal images, beads (3 μL) were spotted on a microscope slide and allowed to dry. For flow cytometry, resuspended beads were imaged with 488 and 640 nm lasers by using 530/30 and 675/25 nm filters, respectively.
Circular Dichroism Spectroscopy
Solutions of peptide were prepared in 50 mM HOAc. Many experiments in this study involve mixtures of hosts and guest peptides, and preparation of equimolar mixtures of the components is of vital importance. To ensure equimolar mixtures, relative concentrations were determined by integrating the absorbance of peptides at 218 nm during UPLC, and concentrations of each peptide were calculated based on the absorbance of 180 μM (Pro-Pro-Gly)10 and the assumption that the extinction coefficient of all Xaa-Yaa-Gly repeats were identical at 218 nm. Analyte solutions were prepared such that the concentration of individual collagen strands was 180 μM. For example, solutions contained 60 μM of a double-stranded host and 60 μM of a (Xaa-Yaa-Gly)7 peptide. To facilitate formation of the most stable complex, analyte solutions were heated to 65 °C, then cooled to 4 °C at a rate of −12 °C per hour. Samples were left at 4 °C for at least 48 h before data acquisition.
CD spectra of peptides at 180 μM strand concentration were recorded at 4 °C with a 1 nm band-pass filter and an averaging time of 3 s in a 0.1 cm path-length quartz cuvette. For thermal denaturation experiments, the CD signal was monitored at 226 nm as the sample was heated at a rate of 12 °C/h in 3 °C steps. The value of Tm, which is the temperature at the midpoint of the thermal transition, was calculated by fitting the data to a Boltzmann sigmoidal curve with the program Prism from GraphPad (La Jolla, CA).
CD spectra of equimolar noninteracting host and guest mixtures, [θ]host+guest, were calculated based on the spectra of individual components by using the following equation:
| 1 |
where [θ]host and [θ]guest are the respective mean residue ellipticities for a host with nres,host residues and a guest with nres,guest residues.
Results
Design and Synthesis of a Host
We sought to design a host in which two parallel CMPs are tethered together at both ends. Because lysine has two amino groups that could serve to initiate synthesis, we began with lysine immobilized via its carboxyl group. The collagen triple helix forms with a one-residue stagger between its strands, which the distance between the α and ε amino groups of lysine helps to recreate. We reasoned that the linkers on both ends must be flexible to allow for proper folding. Accordingly, we condensed Fmoc-6-aminohexanoic acid spacer to both of the amino groups of lysine. Then, we used standard solid-phase peptide synthesis methods to add a (Pro-Pro-Gly)10 sequence to each amino group.
We considered flexibility as well in the design of the N-terminal linker, which would close the macrocycle. 6-Aminohexanoic acid spacers were attached at the N-termini of the two (Pro-Pro-Gly)10 peptides while still on the solid support. Initial attempts to close the macrocycle enlisted small molecules. Glutaric acid resulted in a mixture of products containing one or two amides but no detectable macrocycle. Similarly, adding a cysteine to each end and treating with dibromobimane12 provided a mixture of products containing one or two thioethers but no macrocycle. A cyclic duplex that has been optimized to anneal to endogenous single strands of collagen has been closed by forming a disulfide bond,13 but that methodology was unsuccessful in our hands. Finally, we sought to deploy peptide “stapling”8,14 by condensing 3-butenoic acid to the N termini (generating the open host) and using olefin metathesis on-resin with Grubbs G2 catalyst to form a macrocycle (the closed host). Mass spectrometry revealed a decrease of 28 amu, consistent with the loss of ethylene and successful tethering of the two ends (Figure S1). We reduced the nascent alkene to endow greater flexibility (the reduced host). The open, closed, and reduced hosts (host-o, host-c, and host-r, respectively) are depicted in Figure 2.
Figure 2.
Chemical structures of the open host (host-o), closed host (host-c), and closed, reduced host (host-r). The macrocyclic rings of host-c and host-r contain 221 atoms.
Analysis of Host Self-Association with Ultracentrifugation
To assess the aggregation state of host-o and host-r in solution, we subjected their solutions to analytical ultracentrifugation (AUC). Although sedimentation-equilibrium data fitted to a single-species model presents host-o as a dimer and host-r as a monomer in solution, introduction of a multimer in the models improves fits significantly by better representing the contributions from high-molecular weight (MW) species. Monomer–pentamer and monomer–trimer models best explain the high-MW species observed in host-o and host-r gradients, respectively (Figure S2). Monomer–multimer models for host-o and host-r data collected at 34,000 rpm exemplifies the abundance of each species across the gradients. The presence of high-MW species in the host-o sample results in an appreciable deviation from linearity (Figure 3A), while the host-r gradient remains linear with only minor deviation from sedimentation expected of a monomer (Figure 3B). Thus, AUC indicates multimerization of host-o, whereas host-r remains essentially in a monomeric state.
Figure 3.
Graphs of analytical ultracentrifugation data obtained at 34000 rpm for host-o and host-r. (A) Data for host-o fitted to a monomer + pentamer mode. (B) Data for host-r fitted to a monomer + trimer mode.
Analysis of Host·CMP Complex Formation with Fluorescence Spectroscopy
To reveal whether a triple helix forms between the hosts and a CMP, we employed fluorescence spectroscopy. Host-o and host-r were conjugated to biotin and then complexed to fluorescent beads that were coated with streptavidin. A fluorescein probe was tethered to a CMP, (Pro-Pro-Gly)7. Upon mixing, coinciding fluorescence would indicate association and, presumably, triple-helix formation (Figure 4A). Application of the positive control biotin–fluorescein, (Ser-Gly)3 conjugated to both biotin and fluorescein, reveals a green halo upon the red fluorescence of beads under confocal microscopy where the focal plane bisects the bead (Figure 4B). No fluorescent quenching was observed upon bead-labeling. The same pattern was apparent for host-o and host-r coated beads when mixed with the fluorescein–CMP conjugate. In contrast, treatment with fluorescein–CMP alone did not lead to green fluorescence (data not shown). Host-coated beads mixed with fluorescein–d-CMP, the enantiomeric peptide incapable of forming a triple helix with either host, showed a substantial reduction in bead-labeling (Figure S3). This reduction is indicative of the specific binding of fluorescein–CMP to hosts on the bead surface.
Figure 4.

Binding of immobilized host-o and host-r to a fluorescent CMP. (A) Scheme showing the experimental design. (B) Representative confocal microscopy images. Streptavidin-coated fluorescent beads (red) were treated with biotin-conjugated host-o or host-r and then incubated with fluorescein–CMP (green), which is Ac-Lys(fluorescein)-(Ser-Gly)3-(Pro-Pro-Gly)7. Also shown are images from a negative control of untreated beads and a positive control of beads treated with a biotin–fluorescein conjugate (green), which is biotin-(Gly-Ser)3-Lys(fluorescein)-NH2. Scale bar: 10 μm.
The qualitative results obtained with microscopy were quantified by examining beads with flow cytometry, where 10000 events were evaluated for each sample in a single run. Again, beads coated with both host-o and host-r showed substantial labeling upon incubation with the fluorescein–CMP conjugate (Figure 5). The fluorescence with host-r was greater than that with host-o, which is not closed on its N terminus. Moreover, the triple helix formed by host-r with a CMP has greater kinetic stability than does that with host-o, where agitation for 9 h diminished fluorescence.
Figure 5.
Quantification of binding of immobilized host-o and host-r to a CMP. Streptavidin-coated fluorescent beads were treated with host-o–biotin or host-r–biotin, and then with fluorescein–CMP, which is Ac-Lys(fluorescein)-(Ser-Gly)3-(Pro-Pro-Gly)7. Beads were also treated with biotin–fluorescein conjugate (which is biotin-(Gly-Ser)3-Lys(fluorescein)-NH2), host-o alone, or host-r alone. Values represent the percent of the sample population with fluorescein-labeling relative to that from treatment with biotin–fluorescein.
Analysis of Host·CMP Complex Formation with Circular Dichroism Spectroscopy
The collagen triple helix generates a distinct circular dichroism (CD) spectrum with maximum ellipticity near 226 nm.15 In our host·guest system, however, that diagnostic method is complicated by two factors. First, the hosts themselves have CD spectra like that of a triple helix (Figure 6A). Second, guest strands alone can form a homotrimeric helix as well as a host·guest complex, and those two triple helices are likely to have indistinguishable CD spectra. Nonetheless, interactions with a monomeric host (e.g., host-r) could reveal trends, especially if analyses are restricted to strands that do not form homotrimers. Accordingly, changes in CD signal upon mixing such strands with a host could be attributed to the formation of a host·guest triple helix.
Figure 6.
Circular dichroism spectra of hosts (A), guest CMPs (B), and host·CMP complexes (C). Calculated spectra for noninteracting mixtures of host-r and CMPs are shown (dashed gray lines) together with acquired spectra for host-r·CMP complexes (red lines). Spectra were obtained in 50 mM HOAc at 4 °C.
The CMPs (flp-Hyp-Gly)7,3b,16 (flp-Flp-Gly)7,17 (Pro-Pro-Gly)7,17 and (Pro-Ile-Gly)718 cannot form stable homotrimeric triple helices, exhibit low CD signal, and are ideal for testing the utility of host-r as a mimic for damaged collagen (Figure 6B). In contrast to their spectra alone, a collagen signature is apparent in their mixtures with host-r (Figure 6C). To determine the extent of interaction between host-r and each guest, we used data on individual species to calculate the CD spectra where the components of the mixture do not interact. The calculated spectra explain the CD signal observed for mixtures of host-r with (Pro-Pro-Gly)7 and (Pro-Ile-Gly)7 (Figure 6C). In contrast, (flp-Hyp-Gly)7 and (flp-Flp-Gly)7 interact cooperatively with host-r, producing significantly higher triple-helix signal than each species can independently contribute. In comparison, host-o·guest complexes do not yield similar levels of cooperativity (Figure S4).
The results from temperature-denaturation experiments are consistent with the spectral analysis above, and also highlight the impact of macrocycle formation on host structure. Although similar folded states can be imagined for all hosts, macrocycle formation and flexibility limit their conformations upon denaturation. This is clearly reflected in their denaturation profiles (Figure S5A). Whereas a distinct transition is apparent for host-o (Tm = 45.6 °C), an extremely shallow transition is apparent for the highly constrained host-c and increases after reduction to host-r. This trend is consistent with that observed in CD spectra (Figure 6A). Furthermore, the thermal transitions for host-r·guest complexes become increasingly recognizable when a more cooperative guest is selected (Figure S5B), consistent with our analysis of host·guest cooperativity (Figure 6C).
The peptides (flp-Hyp-Gly)7, (flp-Flp-Gly)7, and (Pro-Pro-Gly)7 were used previously as invasive strands to deliver cargo to damaged collagen.3−5 (Pro-Hyp-Gly)7 has also been employed for this purpose.19 Unlike the other peptides, however, (Pro-Hyp-Gly)7 readily forms homotrimers at room temperature, complicating its therapeutic use. As anticipated, application of (Pro-Hyp-Gly)7 to host-r does not enhance triple-helical content (Figure S6).
Discussion
A collagen duplex could be an effective mimic of damaged collagen (Figure 1). To optimize a duplex for this purpose, we synthesized two parallel strands of (Pro-Pro-Gly)10 from the amino groups of an immobilized lysine residue. The N-terminal closing of this acyclic duplex, host-o, was achieved by olefin metathesis using the Grubbs G2 catalyst. Reduction of the ensuing alkene afforded a cyclic duplex, host-r (Figure 2). This simple modification appears to be critical, as it results in a dramatic increase in CD signal (Figure 6A).
Analytical ultracentrifugation revealed that host-r exists primarily as a monomer in solution (Figure 3B). Yet, it exhibits triple-helical character by CD spectroscopy, despite its topological inability to form a triple helix. This finding is consistent with our recent discovery that two singly cross-linked CMPs adopt a collagen-like structure even in the absence of a third strand.20
Although host-r exhibits a lower triple-helix signal than does host-o (Figure 6A), the benefits of macrocycle formation outweigh any accompanying structural penalty. Closing the macrocycle reduces multimerization (Figure 3), improves the cooperativity of host·guest association (cf.: Figures 6C and S4), and enables better retention of a CMP on a host-coated bead surface (Figure 5). The kinetically stable interaction of such beads with (Pro-Pro-Gly)7 is especially interesting, as this CMP displays little cooperativity with host-r. CD spectra calculated for noninteracting species are likely overestimations due to higher concentrations used to obtain data for individual species. Perhaps more importantly, the bead-retention experiment is unique in being a measurement done on a solid support, which can enhance ligand association as compared to solution-state measurements.21 Interestingly, (Pro-Pro-Gly)7 has been used as an invasive strand to deliver cargo to wound beds, which similarly display damaged collagen on a 3-D surface.3a,4,5
Different CMPs were examined as “guests”–third strands. Of those, (flp-Hyp-Gly)7 and (flp-Flp-Gly)7 are superior at promoting the formation of a triple helix with host-r (Figure 6C). Their ability to form a stable triple helix can be attributed to the preorganization endowed by their flp residues (which adopt a requisite Cγ-endo ring pucker and ϕ ∼ −75° dihedral angle) and Hyp and Flp residues (which adopt a requisite Cγ-exo ring pucker and smaller ϕ ∼ −60° dihedral angle).1 Interestingly, the interaction of host-r with (flp-Hyp-Gly)7 produces a greater rise in CD signal than that with (flp-Flp-Gly)7, even though Flp-to-Hyp substitutions at the Yaa position of a (Pro-Yaa-Gly)7 peptide lowers thermostability.22 This finding is consistent with our recent discovery that (flp-Hyp-Gly)7 is an optimal strand for annealing to damaged collagen in vitro and ex vivo.3b Hence, we conclude that host-r provides a reliable mimic of damaged collagen.
Although all CMPs tested are deficient in homotrimer formation, they are so due to different factors. The peptides, (flp-Hyp-Gly)7 and (flp-Flp-Gly)7, feature residues with high triple-helical propensity, but cannot form triple helices due to severe steric hindrance between Xaa = flp and Yaa = Flp or Hyp residues on neighboring strands.3b,7 In contrast, (Pro-Pro-Gly)7 and (Pro-Ile-Gly)7 lack the preorganization of their counterparts, and their triple helices are not stable above 4 °C despite being unhindered by sterics. Thus, (Pro-Pro-Gly)7 and (Pro-Ile-Gly)7 are not well-positioned for cooperative interactions with host-r, which is corroborated by our findings (Figure 6C). Interestingly, we also observe similar uncooperative behavior in mixtures of host-r and (Pro-Hyp-Gly)7, a peptide that forms stable homotrimers up to 36 °C (Figure S5).23 Together, our results point to engineered guest peptides, and especially to (flp-Hyp-Gly)7, as the optimal CMPs for targeting collagen damage.
Conclusions
Host-r is a macrocycle that contains two collagen-mimetic peptides and forms a collagen triple helix with a third collagen-mimetic peptide. Its development provides opportunities for new types of analyses. Historically, the rigorous analysis of triple-helix formation has been complicated by the process being termolecular. For example, hysteresis often confounds analyses of the unfolding–refolding transition with three strands.24 As the basis of a bimolecular rather than a termolecular system, host-r could provide clarity as well as new insight.
In addition, we note that human type-I collagen, which is the most abundant protein in the extracellular matrix and connective tissue,1 is composed of two α1[I] strands and one α2[I] strand. In collagen fibrils, a Gly-Phe-Hyp-Gly-Glu-Arg sequence clusters integrins on the surface of endothelial cells and promotes wound healing.25 Disruption of the triple helix in this region could be especially deleterious to wound repair. Host-r variants composed of two copies of the GAOGPSGARGERGFOGERGVQGPOGPAGPR sequence of human α1[I] strands (where “O” refers to Hyp) could lead to the discovery of CMPs that enable new therapeutic interventions.13b
Acknowledgments
We are grateful to Dr. Darrell R. McCaslin and Dr. Martha M. Vestling for facility support and contributive discussions and to Prof. Clark R. Landis for use of his glovebox. A.J.E. was supported by a Chemistry–Biology Interface (CBI) Training Grant T32 GM008505 (NIH). This work was supported by Grant R56 AR044276 (NIH).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.0c00103.
Figures S1–S6, NMR spectra, and UPLC traces (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Shoulders M. D.; Raines R. T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Siebler C.; Erdmann R. S.; Wennemers H. From azidoproline to functionalizable collagen. Chimia 2013, 67, 891–895. 10.2533/chimia.2013.891. [DOI] [PubMed] [Google Scholar]; b Chattopadhyay S.; Raines R. T. Collagen-based biomaterials for wound healing. Biopolymers 2014, 101, 821–833. 10.1002/bip.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wahyudi H.; Reynolds A. A.; Li Y.; Owen S. C.; Yu S. M. Targeting collagen for diagnostic imaging and therapeutic delivery. J. Controlled Release 2016, 240, 323–331. 10.1016/j.jconrel.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Shekhter A. B.; Fayzullin A. L.; Vukolova M. N.; Rudenko T. G.; Osipycheva V. D.; Litvitsky P. F. Medical applications of collagen and collagen-based materials. Curr. Med. Chem. 2019, 26, 506–516. 10.2174/0929867325666171205170339. [DOI] [PubMed] [Google Scholar]
- a Chattopadhyay S.; Murphy C. J.; McAnulty J. F.; Raines R. T. Peptides that anneal to natural collagen in vitro and ex vivo. Org. Biomol. Chem. 2012, 10, 5892–5897. 10.1039/c2ob25190f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Dones J. M.; Tanrikulu I. C.; Chacko J. V.; Schroeder A. B.; Hoang T. T.; Gibson A. L. F.; Eliceiri K. W.; Raines R. T. Optimization of interstrand interactions enables burn detection with a collagen-mimetic peptide. Org. Biomol. Chem. 2019, 17, 9906–9912. 10.1039/C9OB01839E. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Song J. Y.; Pineault K. M.; Dones J. M.; Raines R. T.; Wellik D. M.. Hox genes maintain critical roles in the adult skeleton. Proc. Natl. Acad. Sci. U. S. A., 10.1073/pnas.1920860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S.; Guthrie K. M.; Teixeira L.; Murphy C. J.; Dubielzig R. R.; McAnulty J. F.; Raines R. T. Anchoring a cytoactive factor in a wound bed promotes healing. J. Tissue Eng. Regener. Med. 2016, 10, 1012–1020. 10.1002/term.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison A. J.; Raines R. T. A pendant peptide endows a sunscreen with water-resistance. Org. Biomol. Chem. 2018, 16, 7139–7142. 10.1039/C8OB01773E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Koide T.; Homma D. L.; Asada S.; Kitagawa K. Self-complementary peptides for the formation of collagen-like triple helical supramolecules. Bioorg. Med. Chem. Lett. 2005, 15, 5230–5233. 10.1016/j.bmcl.2005.08.041. [DOI] [PubMed] [Google Scholar]; b Kotch F. W.; Raines R. T. Self-assembly of synthetic collagen triple helices. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 3028–3033. 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yamazaki C. M.; Asada S.; Kitagawa K.; Koide T. Artificial collagen gels via self-assembly of de novo designed peptides. Biopolymers 2008, 90, 816–823. 10.1002/bip.21100. [DOI] [PubMed] [Google Scholar]; d Yamazaki C. M.; Kadoya Y.; Hozumi K.; Okano-Kosugi H.; Asada S.; Kitagawa K.; Nomizu M.; Koide T. A collagen-mimetic triple helical supramolecule that evokes integrin-dependent cell responses. Biomaterials 2010, 31, 1925–1934. 10.1016/j.biomaterials.2009.10.014. [DOI] [PubMed] [Google Scholar]; e Tanrikulu I. C.; Raines R. T. Optimal interstrand bridges for collagen-like biomaterials. J. Am. Chem. Soc. 2014, 136, 13490–13493. 10.1021/ja505426g. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Delsuc N.; Uchinomiya S.; Ojida A.; Hamachi I. A host-guest system based on collagen-like triple-helix hybridization. Chem. Commun. 2017, 53, 6856–6859. 10.1039/C7CC03055J. [DOI] [PubMed] [Google Scholar]
- Hodges J. A.; Raines R. T. Stereoelectronic and steric effects in the collagen triple helix: Toward a code for strand association. J. Am. Chem. Soc. 2005, 127, 15923–15932. 10.1021/ja054674r. [DOI] [PubMed] [Google Scholar]
- Blackwell H. E.; Clemons P. A.; Schreiber S. L. Exploiting site-site interactions on solid support to generate dimeric molecules. Org. Lett. 2001, 3, 1185–1188. 10.1021/ol015647i. [DOI] [PubMed] [Google Scholar]
- Scholl M.; Ding S.; Lee C. W.; Grubbs R. H. Synthesis and activity of a new generation of ruthenium-based olefin metathesis catalysts coordinated with 1,3-dimesityl-4,5-dihydroimidazol-2-ylidene ligands. Org. Lett. 1999, 1, 953–956. 10.1021/ol990909q. [DOI] [PubMed] [Google Scholar]
- Jida M.; Betti C.; Schiller P. W.; Tourwe D.; Ballet S. One-pot isomerization-cross metathesis-reduction (ICMR) synthesis of lipophilic tetrapeptides. ACS Comb. Sci. 2014, 16, 342–351. 10.1021/co500020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchschlag H.; Zipper P. Calculation of the partial volume of organic compounds and polymers. Ultracentrifucation 1994, 94, 20–39. 10.1007/BFb0115599. [DOI] [Google Scholar]
- a Kim J.-S.; Raines R. T. Dibromobimane as a fluorescent crosslinking reagent. Anal. Biochem. 1995, 225, 174–176. 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]; b Sardi F.; Manta B.; Portillo-Ledesma S.; Knoops B.; Comini M. A.; Ferrer-Sueta G. Determination of acidity and nucleophilicity in thiols by reaction with monobromobimane and fluorescence detection. Anal. Biochem. 2013, 435, 74–82. 10.1016/j.ab.2012.12.017. [DOI] [PubMed] [Google Scholar]
- a Takita K. K.; Fujii K. K.; Kadonosono T.; Masuda R.; Koide T. Cyclic peptides for efficient detection of collagen. ChemBioChem 2018, 19, 1613–1617. 10.1002/cbic.201800166. [DOI] [PubMed] [Google Scholar]; b Takita K. K.; Fujii K. K.; Ishii K.; Koide T. Structural optimization of cyclic peptides that efficiently detect denatured collagen. Org. Biomol. Chem. 2019, 17, 7380–7387. 10.1039/C9OB01042D. [DOI] [PubMed] [Google Scholar]
- Blackwell H. E.; Sadowsky J. D.; Howard R. J.; Sampson J. N.; Chao J. A.; Steinmetz W. E.; O’Leary D. J.; Grubbs R. H. Ring-closing metathesis of olefinic peptides: Design, synthesis, and structural characterization of macrocyclic helical peptides. J. Org. Chem. 2001, 66, 5291–5302. 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- Fields G. B.; Prockop D. J. Perspective on the synthesis and application of triple-helical collagen-model peptides. Biopolymers 1996, 40, 345–357. . [DOI] [PubMed] [Google Scholar]
- Barth D.; Milbradt A. G.; Renner C.; Moroder L. A (4R)- or a (4S)-fluoroproline residue in position Xaa of the (Xaa-Yaa-Gly) collagen repeat severely affects triple-helix formation. ChemBioChem 2004, 5, 79–86. 10.1002/cbic.200300702. [DOI] [PubMed] [Google Scholar]
- Hodges J. A.; Raines R. T. Stereoelectronic effects on collagen stability: The dichotomy of 4-fluoroproline diastereomers. J. Am. Chem. Soc. 2003, 125, 9262–9263. 10.1021/ja035881z. [DOI] [PubMed] [Google Scholar]
- a Tamburro A. M.; Guantieri V.; Cabrol D.; Broch H.; Vasilescu D. Experimental and theoretical conformational studies on polypeptide models of collagen. Int. J. Pept. Protein Res. 1984, 24, 627–635. 10.1111/j.1399-3011.1984.tb03169.x. [DOI] [PubMed] [Google Scholar]; b Persikov A. V.; Ramshaw J. A. M.; Kirkpatrick A.; Brodsky B. Amino acid propensities for the collagen triple-helix. Biochemistry 2000, 39, 14960–14967. 10.1021/bi001560d. [DOI] [PubMed] [Google Scholar]
- a Wang A. Y.; Mo X.; Chen C. S.; Yu S. M. Facile modification of collagen directed by collagen mimetic peptides. J. Am. Chem. Soc. 2005, 127, 4130–4131. 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]; b Hwang J.; Huang Y.; Burwell T. J.; Peterson N. C.; Connor J.; Weiss S. J.; Yu S. M.; Li Y. In situ imaging of tissue remodeling with collagen hybridizing peptides. ACS Nano 2017, 11, 9825–9835. 10.1021/acsnano.7b03150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanrikulu I. C.; Westler W. M.; Ellison A. J.; Markley J. L.; Raines R. T. Templated collagen “double helices” maintain their structure. J. Am. Chem. Soc. 2020, 142, 1137–1141. 10.1021/jacs.9b07583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hintersteiner M.; Buehler C.; Auer M. On-bead screens sample narrower affinity ranges of protein-ligand interactions compared to equivalent solution assays. ChemPhysChem 2012, 13, 3472–3480. 10.1002/cphc.201200117. [DOI] [PubMed] [Google Scholar]; b Daniel C.; Roupioz Y.; Gasparutto D.; Livache T.; Buhot A. Solution-phase vs surface-phase aptamer-protein affinity from a label-free kinetic biosensor. PLoS One 2013, 8, e75419 10.1371/journal.pone.0075419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Holmgren S.; Taylor K.; Bretscher L.; Raines R. T. Code for collagen’s stability deciphered. Nature 1998, 392, 666–667. 10.1038/33573. [DOI] [PubMed] [Google Scholar]; b Holmgren S.; Bretscher L. E.; Taylor K. M.; Raines R. T. A hyperstable collagen mimic. Chem. Biol. 1999, 6, 63–70. 10.1016/S1074-5521(99)80003-9. [DOI] [PubMed] [Google Scholar]
- Bretscher L. E.; Jenkins C. L.; Taylor K. M.; DeRider M. L.; Raines R. T. Conformational stability of collagen relies on a stereoelectronic effect. J. Am. Chem. Soc. 2001, 123, 777–778. 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- a Davis J. M.; Bächinger H. P. Hysteresis in the triple helix-coil transition of type III collagen. J. Biol. Chem. 1993, 268, 25965–25972. [PubMed] [Google Scholar]; b Fischer G. Chemical aspects of peptide bond isomerisation. Chem. Soc. Rev. 2000, 29, 119–127. 10.1039/a803742f. [DOI] [Google Scholar]; c Mizuno K.; Boudko S. P.; Engel J.; Bächinger H. P. Kinetic hysteresis in collagen folding. Biophys. J. 2010, 98, 3004–3014. 10.1016/j.bpj.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Egli J.; Siebler C.; Köhler M.; Zenobi R.; Wennemers H. Hydrophobic moieties bestow fast-folding and hyperstability on collagen triple helices. J. Am. Chem. Soc. 2019, 141, 5607–5611. 10.1021/jacs.8b13871. [DOI] [PubMed] [Google Scholar]
- a Knight C. G.; Morton L. F.; Onley D. J.; Peachey A. R.; Messent A.; Smethurst P. A.; Tuckwell D. S.; Farndale R. W.; Barnes M. J. Identification in collagen type I of an integrin α2β1-binding site containing an essential GER sequence. J. Biol. Chem. 1998, 273, 33287–33294. 10.1074/jbc.273.50.33287. [DOI] [PubMed] [Google Scholar]; b Sweeney S. M.; DiLullo G.; Slater S. J.; Martinez J.; Iozzo R. V.; Lauer-Fields J. L.; Fields G. B.; Antonio J. D. S. Angiogenesis in collagen I requires α2β1 ligation of a GFP*GER sequence and possibly p38 MAPK activation and focal adhesion disassembly. J. Biol. Chem. 2003, 278, 30516–30524. 10.1074/jbc.M304237200. [DOI] [PubMed] [Google Scholar]; c Seo N.; Russell B. H.; Rivera J. J.; Liang X.; Xu X.; Afshar-Kharghan V.; Höök M. An engineered α1 integrin-binding collagenous sequence. J. Biol. Chem. 2010, 285, 31046–31054. 10.1074/jbc.M110.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Sipilä K. H.; Drushinin K.; Rappu P.; Jokinen J.; Salminen T. A.; Salo A. M.; Käpylä J.; Myllyharju J.; Heino J. Proline hydroxylation in collagen supports integrin binding by two distinct mechanisms. J. Biol. Chem. 2018, 293, 7645–7658. 10.1074/jbc.RA118.002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.