Highlights
-
•
We developed a diagnostic to detect all CoVs from the four main genera.
-
•
This assay can detect and identify all previously recognized CoVs and any future related CoVs that may emerge.
-
•
The assay was highly specific and sensitive in detecting CoVs, and performed well on different sample types.
Keywords: Coronavirus, Emerging, Infectious diseases, SARS-CoV-2, COVID-19
Abstract
Background
During the past two decades, three novel coronaviruses (CoVs) have emerged to cause international human epidemics with severe morbidity. CoVs have also emerged to cause severe epidemics in animals. A better understanding of the natural hosts and genetic diversity of CoVs are needed to help mitigate these threats.
Objective
To design and evaluate a molecular diagnostic tool for detection and identification of all currently recognized and potentially future emergent CoVs from the Orthocoronavirinae subfamily.
Study design and Results
We designed a semi-nested, reverse transcription RT-PCR assay based upon 38 published genome sequences of human and animal CoVs. We evaluated this assay with 14 human and animal CoVs and 11 other non-CoV respiratory viruses. Through sequencing the assay's target amplicon, the assay correctly identified each of the CoVs; no cross-reactivity with 11 common respiratory viruses was observed. The limits of detection ranged from 4 to 4 × 102 copies/reaction, depending on the CoV species tested. To assess the assay's clinical performance, we tested a large panel of previously studied specimens: 192 human respiratory specimens from pneumonia patients, 5 clinical specimens from COVID-19 patients, 81 poultry oral secretion specimens, 109 pig slurry specimens, and 31 aerosol samples from a live bird market. The amplicons of all RT-PCR-positive samples were confirmed by Sanger sequencing. Our assay performed well with all tested specimens across all sample types.
Conclusions
This assay can be used for detection and identification of all previously recognized CoVs, including SARS-CoV-2, and potentially any emergent CoVs in the Orthocoronavirinae subfamily.
1. Background
The family Coronaviridae consists of the largest single-stranded RNA viruses that infect humans, other mammals, and birds [1,2]. The Coronaviridae is comprised of two subfamilies, Letovirinae, and Orthocoronavirinae. The Letovirinae subfamily includes only one previously recognized virus (partial genome) which was detected in an ornate chorus frog (Microhyla fissipes). The subfamily Orthocoronavirinae is comprised of many diverse viruses which are divided into four genera based on the criteria of the International Committee on Taxonomy of Viruses [3] including Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV).
This century has recorded an increase in emerging zoonotic CoVs. Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, swine acute diarrhea syndrome coronavirus (SADS-CoV) in 2016, and most recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019 [4]. Notably, SARS-CoV, MERS-CoV and SARS-CoV-2 infect humans, while SADS-CoV causes disease only in pigs [5,6].
While the other zoonotic CoV outbreaks have been largely controlled, attempts at containing SARS-CoV-2 have not been successful. Most experts would agree that this has been due, in part, to the sparse availability of diagnostic assays. Hence, currently, there is a massive, worldwide effort to provide diagnostic tests for SARS-CoV-2 infections. According to the Foundation for Innovative New Diagnostics website [7], as of April 9, 2020 there were more than 470 molecular and immunological assays in the development pipeline. However, few diagnostics are available or being developed to detect other animal CoVs that may jump species to man. Hence, in this study we sought to design and evaluate develop a new molecular diagnostic tool to detect most CoVs, including those that have been previously identified and those that may be discovered in the future.
2. Objectives
In this work we sought to develop and test a semi-nested, RT-PCR assay that targets the conserved RNA-dependent RNA polymerase (RdRp) genome region common to all members of the Orthocoronavirinae, including SARS-CoV-2.
3. Study design
In designing the assay, we selected genome sequences of 38 representative viruses from α-, β-, γ-, and δ- CoV genera that were available in the National Center for Biotechnology Information (NCBI) GenBank database.
3.1. Phylogenetic analysis of CoVs
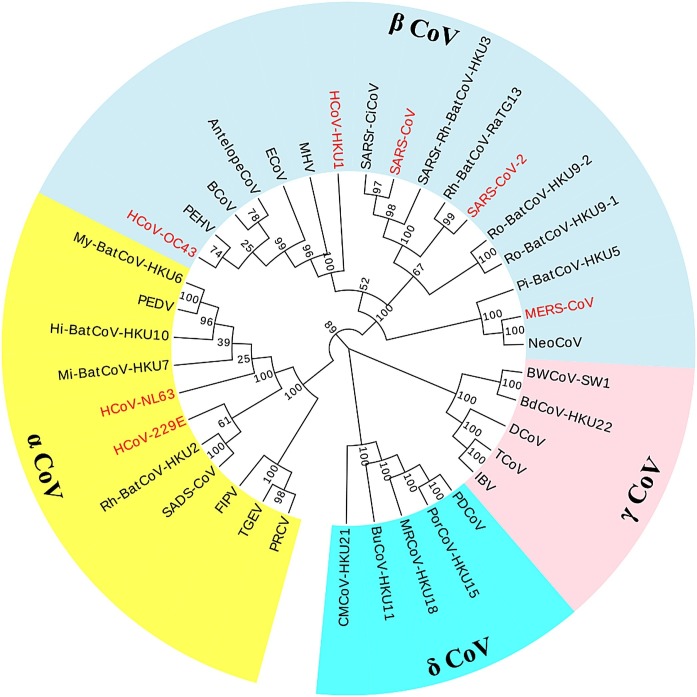
Since the RdRP is the most conserved region among the Orthocoronavirinae, this region was chosen for designing RT-PCR primers to all members of this subfamily. As a first step, RdRp sequences were extracted from NCBI and multiple alignments were prepared using the Clustal W program with default parameters implemented in MEGA 7 (http://www.megasoftware.net). Phylogenetic trees of the RdRp genes were then constructed using the neighbor-joining method with 1000 bootstrap replicates also using MEGA 7 (Fig. 1 ). The GenBank accession numbers of these viruses are recorded in Table S1.
Fig. 1.
The phylogenetic tree of 38 coronaviruses with nucleotide sequences of RNA-dependent RNA polymerase, constructed by the neighbor-joining method using MEGA 7. The four coronavirus genera are grouped in shades of yellow (Alphacoronavirus), lightblue (Betacoronavirus), pink (Gammacoronavirus), and cyan (Deltacoronavirus). Seven human coronaviruses are marked in red.
3.2. Degenerate primers design
The 38 CoV RdRp gene sequences from GenBank were next aligned using Geneious software (Biomatters Ltd, Auckland, New Zealand). Highly conserved regions were visually identified and three degenerate primers were then designed using Geneious software to be broadly reactive across all Orthocoronavirinae. All primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). For first round RT-PCR, the external primers (pan-CoV_outF and pan-CoV_R) were used to amplify a 670–673 bp product; for the second round semi-nested PCR, an internal primer (pan-CoV_inF) was used with pan-CoV_R for amplification of an internal 599–602 bp product. The primer sequences used for each amplification are shown in Table S2.
3.3. Reference viral strains and clinical specimens used in evaluating the assay
A panel of 25 viral isolates and positive clinical specimens harboring a total of 23 unique respiratory viruses were selected (Table 1 ). The 25 samples were acquired from the American Type Culture Collection (ATCC) and from previously studied clinical or field collection specimens at our One Health Laboratories at Duke University (Durham, NC, USA) and Duke-NUS Medical School (Singapore). Fourteen human and animal CoV reference strains or molecularly-positive samples that were archived at Duke University, served as positive controls.
Table 1.
The pan-CoV assay was evaluated against a panel of known coronaviruses and other unrelated human respiratory virus pathogens.
| Virus Name | CoV genus | Type | Source |
|---|---|---|---|
| Human coronavirus 229E | α CoV | Lab strain | NR-470; ATCC |
| Human coronavirus NL63 | α CoV | Lab strain | NR470; BEI Resources |
| Human coronavirus OC43 | β CoV | Lab strain | Betacoronavirus 1; ATCC |
| Human coronavirus HKU | β CoV | Fragment | HKU1_PC; IDT |
| SARS-CoV-2 | β CoV | RNA | USA-WA1 isolate; BEI |
| MERS-CoV | β CoV | Plasmid | pUCIDT-MERS-CoV; IDT |
| SARS-CoV | β CoV | Plasmid | pUCIDT-SARS-CoV; IDT |
| SARS-CoV-2 | β CoV | Fragment | SARS-CoV-2_PC; IDT |
| Canine coronavirus | α CoV | Lab strain | UCD1; NR-868; BEI Resources |
| Porcine respiratory coronavirus | α CoV | Lab strain | NR-43,286; BEI Resources |
| Transmissible gastroenteritis virus | α CoV | Lab strain | TGEV; BEI Resources |
| Bovine coronavirus | β CoV | Lab strain | 020-BDV; NVSL |
| Infectious bronchitis virus | γ CoV | Lab strain | Massachusetts; BEI Resources |
| Porcine deltacoronavirus | δ CoV | Lab strain | 026-PDV; NVSL |
| Human adenovirus | NA | Isolate | Clinical sample derived |
| Human adenovirus | NA | Isolate | Clinical sample derived |
| Human enterovirus 71 | NA | Lab strain | BrCr; ATCC |
| Human parainfluenza virus | NA | Lab strain | HPIV1/FRA/29,221,106/2009; BEI Resources |
| Human rhinovirus | NA | Lab strain | 1059; ATCC |
| Influenza virus A H1N1 | NA | Lab strain | A/California/07/2009 (H1N1) pdm09; IRR |
| Influenza virus A H3N2 | NA | Lab strain | A/Switzerland/9,715,293/2013 (H3N2); IRR |
| Influenza virus B | NA | Lab strain | B/Phuket/3073/2013 (yamagata lineage); IRR |
| Influenza virus C | NA | Lab strain | C/Taylor/1233/1947; BEI Resources |
| Respiratory syncytial virus A | NA | Lab strain | A 1997/12−35; BEI Resources |
| Respiratory syncytial virus B | NA | Lab strain | B1 BPR-348−00; BEI Resources |
NA = not associated with CoV; CDC = Centers for Disease Control and Prevention; IDT = Integrated DNA Technologies, Inc.; NVSL = National Veterinary Services Laboratories; BEI = Biodefense and Emerging Infections Research Resources Repository; ATCC = American Type Culture Collection; IRR = Influenza Reagents Resource.
Additionally, a total of 382 human clinical or animal field specimens were screened with the pan-CoV assay, including 192 human swabs (collected from hospitalized pneumonia patients at Sibu and Kapit Hospitals of Sarawak, Malaysia) [8], 81 poultry swabs (collected from a live poultry market in Hanoi, Vietnam) [9], and 109 pig slurry samples (collected from pig farms in North Carolina, USA) [10]. These samples were supplemented with bioaerosol samples (n = 31) collected using the National Institute for Occupational Safety and Health’s (NIOSH, Morgantown, West Virginia, USA) model BC 251 two-stage bioaerosol sampler from a large poultry market in Vietnam [9]. Finally, we compared the pan-CoV assay’s performance to the US Center for Disease Control and Prevention’s 2019-nCoV RT-PCR assay [11] against a set of clinical specimens from COVID-19 patients.
3.4. Amplification conditions for pan-CoV assay
The protocol consisted of a semi-nested RT-PCR strategy with two amplification steps. A first RT-PCR amplification round was conducted using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). Each 25μL first step reaction mixture contained 4μL RNA samples, 12.5μL 2X Reaction Mix (containing 0.4 mM of each dNTP, 3.2 mM MgSO4), 0.4μM of each primer (pan-CoV_outF and pan-CoV_R), a 0.4μL volume SuperScript™ III RT/Platinum™ Taq Mix, and nuclease-free water to obtain a final volume of 25μL. The thermal profile included incubation at 55 °C for 30 min for reserve transcription, followed by incubation at 94 °C for 2 min to inactivate the reverse transcriptase enzyme. Following the reverse transcription, amplification was performed in the CFX PCR System (Bio-Rad, USA) according to the following program: 30 cycles of 94 °C for 15 s, 53.4 °C for 30 s and 68 °C for 1 min; a final extension at 68 °C for 5 min; and holding at 4 °C. The conventional PCR for second step was carried out in 25μL volume containing 1μL of first RT-PCR product, 2.5μL 10X PCR Buffer (no magnesium chloride), 0.75μL 50 mM magnesium chloride, 0.5μL dNTP mix (dATP, dCTP, dGTP and dTTP), 1μL of each primer (pan-CoV_inF and pan-CoV_R), 0.25μL Platinum™ Taq DNA Polymerase, and enough nuclease-free water for a final volume of 25μL for each reaction. Finally, 599–602 bp RdRp amplicons were obtained by thermocycling using the following parameters: 94 °C for 5 min; 35 cycles of 94 °C for 15 s, 54.5 °C for 30 s and 72 °C for 1 min; and a final extension of 72 °C for 10 min. To avoid possible contamination, positive and negative controls were contained in each run of the assay. The PCR products were kept at 4 °C until analysis by gel electrophoresis. Amplification products were electrophoresed and visualized on a 2 % agarose gel together with a 100 bp DNA ladder at 120 V for 30 min. Sanger sequencing for positive samples was performed by Eton Bioscience Inc (Eton Bioscience, Raleigh, NC, USA).
3.5. Additional study methods
Additional study methods such as RNA extraction methods, and construction of the recombinant plasmids used in testing are described in the supplemental materials.
4. Results
4.1. The pan-CoV assay
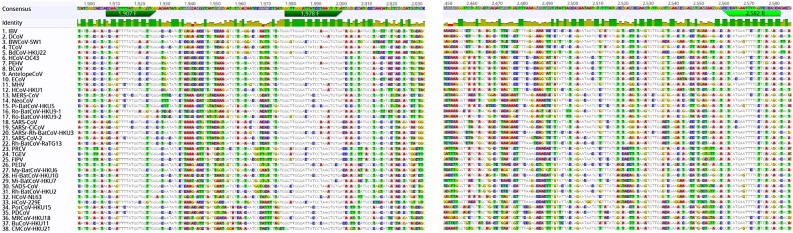
Through alignment of 38 RdRp gene sequences from human and animal CoVs, the region containing the most conserved sequences was identified (Fig. 2 ). Two degenerate primer pairs were constructed (Table S2) with binding sites within this region and a semi-nested PCR protocol was developed. Amplicons obtained by the RT-PCR assay were checked by agarose gel electrophoresis. No primer dimers were detected and no false positive results were observed in the CoV negative control samples. Amplicons of expected size were obtained by the optimized semi-nested RT-PCR protocol with all CoV-positive samples tested.
Fig. 2.
Multiple alignment of the of 38 available RdRp gene segments of coronaviruses, with nucleotide sequences and positions for the primers of the pan-CoV assay.
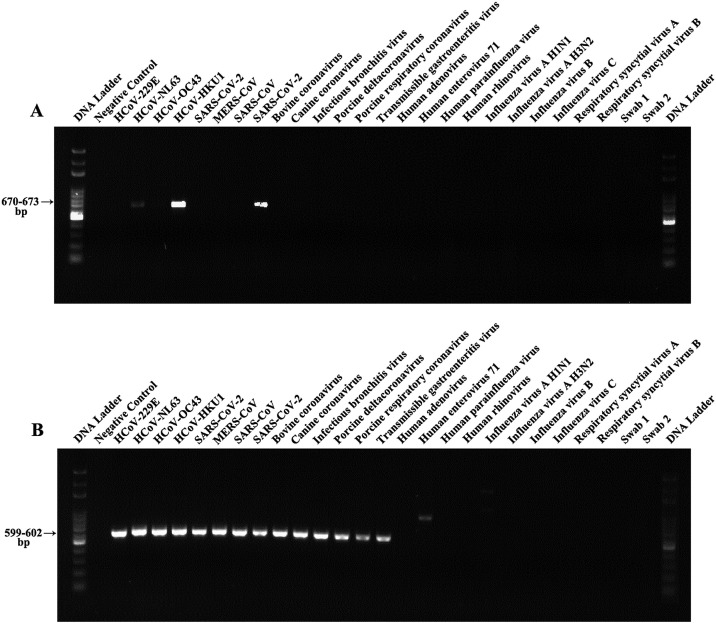
4.2. Validation of assay specificity, sensitivity and reproducibility
To validate the specificity of the pan-CoV assay, a wide range of different CoVs were prepared for templates. The assay specificity was further determined by attempting to amplify nucleic acid from non-specific pathogens like common respiratory viruses. Our assay yielded an amplicon of the expected size (599–602 bp) from the well-characterized control and reference CoV strains (Fig. 3 ). The results showed that the pan-CoV assay could accurately identify different CoVs without cross-reactivity with other non-target pathogens, demonstrating a high specificity for the primers used in this study (Fig. 3). Furthermore, Sanger sequencing confirmed the amplification of expected sequences from the tested samples (Table S3). Some other CoVs (except for the tested CoVs) were not available for testing, but in silico alignment of the targeted region in the RdRp gene of these species demonstrated a sufficient primer match to predict successful amplification.
Fig. 3.
Specificity analysis of pan-CoV assay using a panel of respiratory viral species. (A) Results of first amplification round (reverse transcription PCR). (B) Results of the conventional PCR for second step. Swabs 1 and 2 were collected from healthy people.
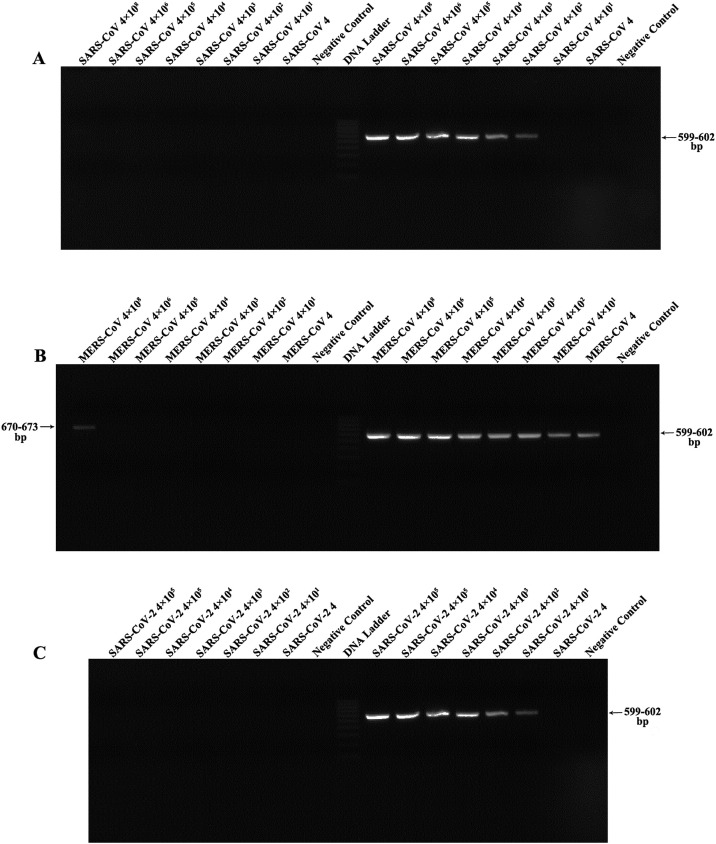
To determine the limits of detection (LOD) of this assay, 10-fold dilution series of templates (plasmids and RNA) were tested, each performed in triplicate. The LOD was defined as the lowest concentration that was detected in all triplicate reactions. Under general semi-nested RT-PCR conditions, our results revealed that the assay was able to detect bands in gels at dilutions ranging from 4 to 4 × 102 copies/reaction for corresponding plasmids (SARS-CoV with 4 and MERS-CoV with 4 × 102 copies/reaction). In addition, RNA extracted from SARS-CoV-2 (USA-WA1 isolate) infected cells with known concentration (with a pre-determined titer of 4.8 × 107 genome copies/mL, data not shown) was used to determine the LOD of this assay. These data revealed that the pan-CoV assay could detect SARS-CoV-2 RNA at 4 × 101 copies per reaction (Fig. 4 ).
Fig. 4.
Sensitivity tests of pan-CoV assay using serial dilutions of RNA and plasmid containing RdRp sequences of coronaviruses. (A) Sensitivity of pan-CoV assay using SARS-CoV plasmid. (B) Sensitivity of pan-CoV assay using mers-CoV plasmid. (C) Sensitivity of pan-CoV assay using SARS-CoV-2 RNA. The left and right sides of the DNA ladder are the results of first and second step of pan-CoV assay, respectively.
Intra- and inter-assay variability were determined at high, medium and low copy number (106, 104 and 102) using the plasmids prepared above, which were serially diluted 10-fold to a final concentration between 1 and 108 copies/μL. For intra-assay variability, all samples were run in triplicate. For inter-assay variability, the assays were repeated three times individually on different days. The plasmids, with different dilutions, were successfully detected under different conditions and showed similar results among the assays. It is worth mentioning that this assay was also successfully implemented in the hands of a research team in Singapore (Fig. S1 in the supplemental materials).
4.3. Evaluation of RT-PCR assay performance with clinical samples
To assess the performance of our semi-nested RT-PCR assay with human clinical specimens, a total of 192 swab samples from patients with pneumonia were tested. Of these, nine (4.7 %) tested positive with the expected amplicon size (Table S4). Upon sequencing, four specimens had evidence of HCoV−OC43, four had evidence of canine-CoV, and one (1) had evidence of HCoV-NL63.
The assay was then evaluated with 190 animal samples. Among 81 poultry swab samples, we identified 25 (30.9 %) positive for CoV. All 81 poultry swab samples had previously tested positive for influenza A by real-time RT-PCR [9]. Sequencing of the CoV positive samples identified one avian CoV, two duck CoVs and 22 infectious bronchitis viruses (IBVs) (Table S4). Among 109 tested pig samples, one (0.92 %) tested positive for CoV. This sample was confirmed as positive for transmissible gastroenteritis virus (TGEV) by Sanger sequencing.
To determine whether the assay could be used to identify naturally occurring CoVs from the environment, we blindly screened 31 bioaerosol samples collected from a poultry market in Hanoi, Vietnam. Among the 31 bioaerosol samples tested, three (9.7 %) were positive for duck CoV, two (6.5 %) were positive for infectious bronchitis virus and one (3.2 %) was positive for avian CoV (Table S4). Negative control samples were negative in all tests, ruling out amplicon contamination as a source of the positive results.
Finally, the pan-CoV assay had 100 % agreement with the US CDC’s rRT-PCR assay for SARS-CoV-2 on two nasopharyngeal swabs, two saliva specimens, and one rectal swab from COVID-19 patients (Table S5).
5. Discussion
Detection of CoVs has most often been made by electron microscopy, cell culture, serological assays, or molecular methods. Microscopy requires expensive equipment and technical expertise. Although cell culture has been considered the gold standard for CoV detection with high specificity [12], its time requirements and low sensitivity limit its clinical application. Serologic assessments are fraught with low sensitivity and occasional cross-reactivity [13]. While fast and sensitive [14], molecular assays target previously recognized CoVs and may miss the detection of a novel CoV [15].
Our pan-CoV assay overcomes these problems. It employs a semi-nested RT-PCR approach using two pairs of degenerate primers that universally amplify CoVs within four genera. We recognize that semi-nested RT-PCR methods are more complex and offer additional risk of assay contamination. However, we argue that effective management of the laboratory environment can reduce such risk.
As the COVID-19 pandemic is currently surging, pathogen discovery of novel CoVs now seems even more important than before. All HCoVs come from two main genera (α-CoV and β-CoV) and are thought to have come from animals, specifically bats [[16], [17], [18], [19]]. It seems clear that large-scale surveillance is needed to fully understand the role played by γ-CoVs and δ-CoVs in the emergence of human CoVs. Hence, this assay might be especially useful in detecting CoVs that are potentially pathogenic to humans from all four genera. Interestingly, we found viruses that are genetically similar to canine-CoV in four human pneumonia samples. We are in the process of further investigating these findings with attempts at culture and next generation sequencing (NGS).
With the advent of NGS technology [20], numerous CoV strains of four genera have been identified via the increased surveillance of wild animal species. NGS has helped elucidate the evolutionary history of CoVs using a large amount of data generated by unbiased sequencing. However, some disadvantages limit the number of samples that can be sequenced. Although it becomes less expensive each consecutive year, the cost is still very high and the procedure includes time-consuming complicated library preparations and data processing. Hence, we posit that combined screening strategies are the most cost-effective way to identify new CoVs. We recommend employing novel coronavirus surveillance among people whose occupations or recreation cause them to have prolonged or frequent exposure to animals. We recommend screening them periodically, when healthy, for novel CoV nasal carriage and whenever they develop an acute respiratory illness. This surveillance might first include employing our pan-CoV assay and other pan-viral assays. When a novel virus is detected, the surveillance team might look for that same virus among the workers’ animals. Having confirmed a novel virus in both an animal worker and his or her animals, further investigations might be employed through cell culture, full genome sequencing, and if indicated, seroepidemiology of both the worker and the animals.
Novel CoVs also have caused tremendous morbidity among livestock. Currently, six CoVs have caused significant morbidity in pigs [5]. These include four α-CoVs [porcine epidemic diarrhea virus (PEDV), porcine respiratory coronavirus (PRCV), transmissible gastroenteritis virus (TGEV), and the recently emerged swine acute diarrhea syndrome coronavirus (SADS-CoV)], one β-CoV [porcine hemagglutinating encephalomyelitis virus (PHEV)], and a δ-CoV [porcine deltacoronavirus (PDCoV)]. Among them, SADS-CoV may have originated from bat CoVs and PDCoV from a sparrow CoV [5], indicating that pigs are the likely complex reservoir for containing these emerging different CoVs. Given its ability to detect CoVs from four CoV genera, our pan-CoV assay can be used to detect novel CoVs in animal herds.
It should be noted that δ-CoVs in particular have an affinity to jump species. A recent study demonstrated that poultry are susceptible to infection with PDCoV [21] and that PDCoV also infects human cell lines [22]. So, we should be particularly concerned when a novel CoV is detected at the human-animal interface.
6. Conclusions
We have designed and evaluated a pan-CoV assay that can detect all known human CoVs from four genera. This assay may be used as a diagnostic tool to identify all previously recognized human CoVs, including SARS-CoV-2 and related viruses, as well as any future emergent CoVs from the Orthocoronavirinae subfamily.
Author contributions
LX designed and tested the pan-CoV assay at Duke University, and drafted the manuscript. GCG conceived the study, guided the work, and helped draft the manuscript. DDE offered expert advice in laboratory experiments and edited the manuscript. RAB and KK and helped draft the manuscript. NAA and RAB assisted LX with laboratory assessments. KKC and STT evaluated the assay at Duke-NUS Medical School. ESB, VNB, STT conducted the field studies that provided specimens for the pan-CoV assay’s assessment. All authors reviewed and approved of the manuscript.
CRediT authorship contribution statement
Leshan Xiu: Methodology, Validation, Investigation, Writing - original draft. Raquel A. Binder: Writing - original draft, Validation, Writing - review & editing. Natalie A. Alarja: Validation, Resources. Kara Kochek: Writing - original draft. Kristen K. Coleman: Validation, Writing - review & editing. Son T. Than: Validation. Emily S. Bailey: Resources, Writing - review & editing. Vuong N. Bui: Resources. Teck-Hock Toh: Resources, Writing - review & editing. Dean D. Erdman: Writing - review & editing. Gregory C. Gray: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors have declared that no competing financial interests exist.
Acknowledgements
We give special thanks to Dr. Gregory Sempowski, Director of the Duke Global Health Research Building, a.k.a. Regional Biocontainment Laboratory (RBL), division of the Duke Human Vaccine Institute for providing the SARS-CoV-2 RNA. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: Genomic RNA from SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52285.
Acknowledgments
Funding
This study was supported by Professor Gray’s discretionary funding at Duke University and a grant from Duke University’s Clinical Translational Science Institute.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104391.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ICTV: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/222/coronaviridae (Accessed on April 9, 2020).
- 4.Guarner J. Three emerging coronaviruses in two decades. Am. J. Clin. Pathol. 2020;153:420–421. doi: 10.1093/ajcp/aqaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.FindDx: https://www.finddx.org/covid-19/pipeline/?section=immunoassays#diag_tab (Accessed on April 9, 2020).
- 8.Toh T.H., Hii K.C., Fieldhouse J.K., Ting J., Berita A., Nguyen T.T. High prevalence of viral infections among hospitalized pneumonia patients in equatorial Sarawak. Malaysia. Open Forum Infect Dis. 2019;6 doi: 10.1093/ofid/ofz074. ofz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui V.N., Nguyen T.T., Nguyen-Viet H., Bui A.N., McCallion K.A., Lee H.S. Bioaerosol sampling to detect avian influenza virus in Hanoi’s largest live poultry market. Clin. Infect. Dis. 2019;68:972–975. doi: 10.1093/cid/ciy583. [DOI] [PubMed] [Google Scholar]
- 10.Bailey ES, Borkenhagen LK, Choi JY, Greer AE, Gray GC. A Feasibility Study of Conducting Surveillance for Swine Pathogens in Swine Slurry in North Carolina Swine Farms. Under journal review. [DOI] [PMC free article] [PubMed]
- 11.CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel: https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2 (Accessed on April 18, 2020).
- 12.Hudu S.A., Alshrari A.S., Syahida A., Sekawi Z. Cell culture, technology: enhancing the culture of diagnosing human diseases. J. Clin. Diagn. Res. 2016;10:DE01–5. doi: 10.7860/JCDR/2016/15837.7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan T., Pinsky B.A. Current and future molecular diagnostics for ocular infectious diseases. Curr. Opin. Ophthalmol. 2016;27:561–567. doi: 10.1097/ICU.0000000000000311. [DOI] [PubMed] [Google Scholar]
- 15.Xiu L., Zhang C., Wu Z., Peng J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front. Microbiol. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91 doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11 doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boley P.A., Alhamo M.A., Lossie G., Yadav K.K., Vasquez-Lee M., Saif L.J. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis. 2020;26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A. 2018;115:E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.