Absract
Since SARS-CoV-2 spreads rapidly around the world, data have been needed on the natural fluctuation of viral load and clinical indicators associated with it. We measured and compared viral loads of SARS-CoV-2 from pharyngeal swab, IgM anti-SARS-CoV-2, CRP and SAA from serum of 114 COVID-19 patients on admission. Positive rates of IgM anti-SARS-CoV-2, CRP and SAA were 80.7%, 36% and 75.4% respectively. Among IgM-positive patients, viral loads showed different trends among cases with different severity, While viral loads of IgM-negative patients tended to increase along with the time after onset. As the worsening of severity, the positive rates of CRP and SAA also showed trends of increase. Different CRP/SAA type showed associations with viral loads in patients in different severity and different time after onset. Combination of the IgM and CRP/SAA with time after onset and severity may give suggestions on the viral load and condition judgment of COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, IgM, C-reactive protein, Serum amyloid a, Viral load
Dear Editor
I'm sending a original article entitled “Association of Viral Load With Serum Biomakers Among COVID-19 Cases”, which we would like to submit for publication in VIROLOGY. No part of this article has been published or submitted elsewhere, and no conflict of interest exists in the submission of this manuscript.
In this article, we investigated the natural fluctuation of viral load among COVID-19 cases before antiviral therapy and the serum biomarkers associated with it. We found the viral loads had different trends and peaked at different time among COVID-19 patients. Combining the detection of IgM anti-SARS-CoV-2, C-reactive protein and Serum amyloid A may give suggestions on the viral load and condition judgment of COVID-19 patients at different stages of illness. The changes of viral load among patients of different clinical severity and different time after onset also can be tracked and used to inform public health policies to prevent the spread of SARS-CoV-2.
The findings may facilitate the prevention and control of COVID-19.
1. Introduction
SARS-CoV-2 has been the focus of worldwide attention. In February, the World Health Organization has declared it a public health emergency of international concern. Within three months, as of March 31st, SARS-CoV-2 has spreaded around the world with 750890 confirmed cases and 36405 deaths (O. novel-coronavirus-20, 2019). The number is still increasing.
Since the discovery of SARS-CoV-2, scientists and doctors from various countries have done a lot of work in the past three months. The genome variations (Zhu et al., 2019; Phan, 2020), epidemiological (Chen et al., 2020) and clinical characteristics (Wang et al., 2020) and other investigative findings (Ahmed et al., 2020) have been previously described, but the natural fluctuation of viral loads among different COVID-19 cases before antiviral therapy were still not well presented (Kam et al., 2020; Kim et al., 2020; Zou et al., 2020; Pan et al., 2020; To et al., 2020). The clinical indicators associated with the load changes at different stages of the illness are also largely unknown.
Here, we explored the association of viral load with IgM anti-SARS-CoV-2 (IgM), C-reactive protein (CRP) and Serum amyloid A (SAA) among COVID-19 patients on admission, to find the clues that may facilitate the prevention and treatment of this infectious disease.
2. Methods
2.1. Samples and data collection
As of February 4th, samples collected on admission from Jiangsu Province of China were sent to our laboratory to confirm the infection of SARS-CoV-2. Real-time reverse-transcriptase polymerase-chain-reaction (RT-PCR) assay (BioGerm, China) was used to detect SARS-CoV-2 nucleic acids from pharyngeal swab samples. Only laboratory confirmed cases were included in this study. Clinical data were obtained from patients’ medical records. According to the severity of pneumonia based on radiologic assessments (NHC China), these patients were classified into non-pneumonia group, pneumonia group and severe pneumonia group in this study. The days after onset was the interval between the date of sampling when on admission and the date of onset. All data were cross-checked.
2.2. Serum biomakers detection
IgM Anti-SARS-CoV-2, CRP and SAA were measured with dry fluorescence immunoassay (Lansionbio, China) from the serum of the COVID-19 cases. According to the manufacturer's instruction, IgM concentrations ≥0.04 AU/mL, CRP concentrations >10 μg/mL, and SAA concentrations ≥10 μg/mL were judged as positive and marked as IgM(+), CRP(+) and SAA(+) respectively. Each sample was confirmed twice.
2.3. Viral loads detection
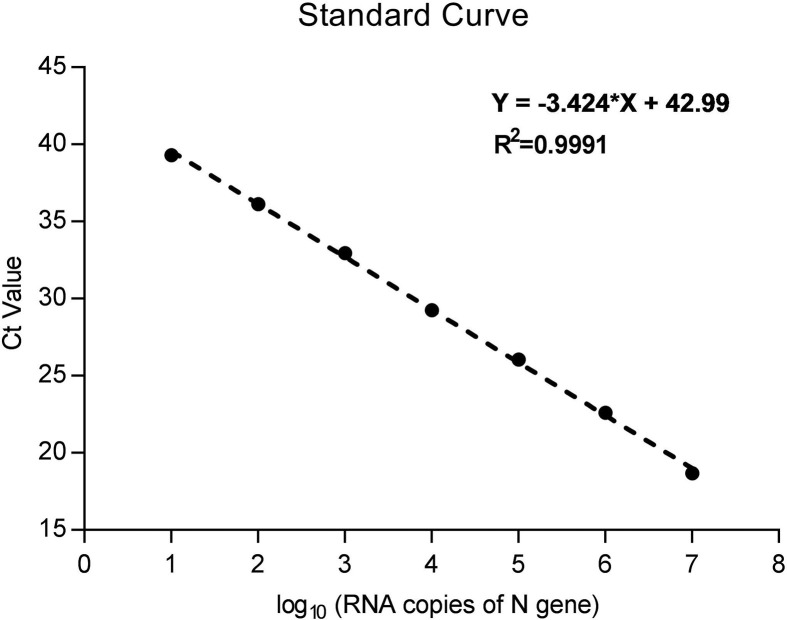
The viral loads of SARS-CoV-2 were measured as the copy number of the N gene from pharyngeal swabs of the COVID-19 cases. Briefly, based on the SARS-CoV-2 genome sequence in Genbank (Accession Number: MN908947), SARS-CoV-2 N gene was amplified with primers containing T7 promoter sequence: 5′- ACTCGTTAATACGACTCACTATAGGGAAAGGCCAACAACAACAAGG-3’ (Forward) and 5′- AGTCTGCGGTAAGGCTTGAGTT-3’ (Reverse), and in vitro transcribed with T7 RNA polymerase (TaKaRa, China), then purified and quantified, and finally ten-fold diluted ranging from 107 to101 RNA copies/μL to make RNA standards (Ge et al., 2013). The N gene from pharyngeal swabs of the COVID-19 cases was amplified (Chu et al., 2020) along with the RNA standards (TaKaRa, China). The viral loads were calculated from mean Ct values of three repeats from each patient and the standard curve generated from the RNA standards (Supplementary Figure S1).
2.4. Statistical analysis
Continuous variables with normal distribution were analyzed with one way ANOVA or student t-test. Other continuous variables were analyzed with Kruskal-Wallis test. Categorical variables were analyzed using Chi-square test, and Fisher exact test when the data were limited. SPSS 19.0 software and GraphPad 7.0 were used for statistical analysis.
3. Results
3.1. Demographic and laboratory findings
Of all 125 cases with both serum and pharyngeal swab collected on admission in this study, 11(8.8%) with hemolytic serum were excluded. In the final 114 patients, 32 cases were categorized into non-pneumonia group, 74 into pneumonia group and 8 into severe pneumonia group on admission (Table 1 ). The median age was 43.5 years, and 48.3% were females, with a median time after onset of 4 days. Age and days after onset differed significantly among the three groups (both P < 0.05). The positive rates of IgM anti-SARS-CoV-2, CRP and SAA were 80.7%, 36% and 75.4% respectively. Along with the worsening of severity, the positive rates of CRP and SAA tended to increase with significant differences in CRP (P < 0.01).
Table 1.
Demographic and laboratory Characteristics among different severity cases.
| Characteristics | All cases | Non-pneumonia | Pneumonia | Severe Pneumonia | P value |
| (n = 114) | (n = 32) | (n = 74) | (n = 8) | ||
| Age, -yrs | |||||
| Median (range) | 43.5 (6–79) | 43.5 (6–75) | 41.5 (7–79) | 59 (34–75) | 0.044 |
| Days after onset, -days | |||||
| Median (range) | 4 (0–12) | 3 (0–8) | 4 (0–12) | 5 (2–12) | 0.034 |
| Female sex | |||||
| No. (%) | 55/114 (48.3) | 15/32 (46.9) | 36/74 (48.6) | 4/8 (50) | 0.981 |
| IgM anti-SARS-CoV-2 | |||||
| Positive – No. (%) | 92/114 (80.7) | 27/32 (84.4) | 58/74 (78.4) | 7/8 (87.5) | 0.68 |
| C-reactive protein | |||||
| Positive – No. (%) | 41/114 (36.0) | 5/32 (15.6) | 31/74 (41.9) | 5/8 (62.5) | 0.009 |
| Serum amyloid A | |||||
| Positive – No. (%) | 86/114 (75.4) | 20/32 (62.5) | 58/74 (78.8) | 8/8 (100%) | 0.054 |
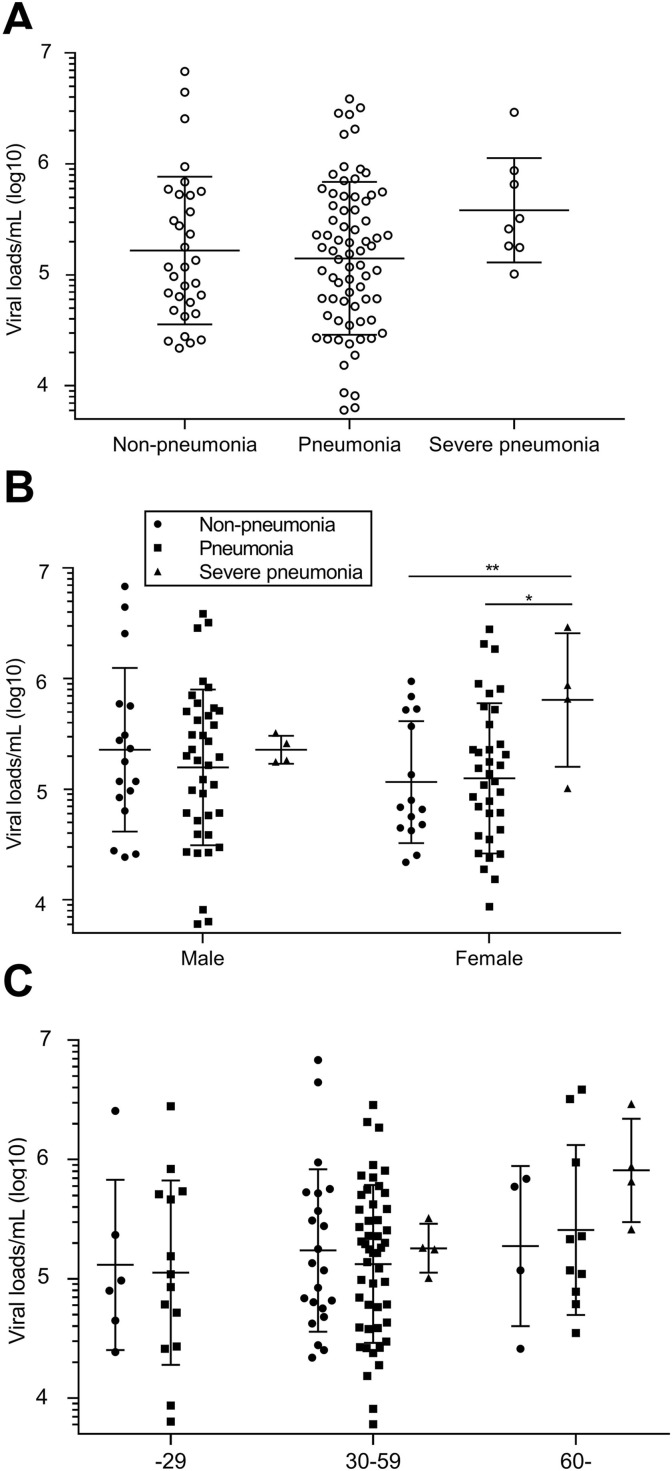
As shown in Fig. 1 A, the mean viral load/mL (log10) was lower in pneumonia cases (5.15), followed by non-pneumonia cases (5.22), and highest in severe pneumonia cases (5.58), but no significant differences were found. Also, no statistical significance was found between male and female cases with the same severity (mean, 5.36, 5.20 and 5.36 for male cases; 5.06, 5.10 and 5.81 for female case, respectively, in Fig. 1B). But within the female cases, the mean viral load in severe pneumonia patients was higher and significantly differed from non-pneumonia patients (P < 0.001) and pneumonia patients (P < 0.05). Mean viral loads tended to increase along with the age of patients, and no severe pneumonia patient was found younger than 29 years old in this study, but all the differences were not significant (Fig. 1C).
Fig. 1.
Viral loads among patients with different disease severity, sex and age. A, Viral loads among patients with different disease severity; Viral loads among patients with different sex; Viral loads among patients with different age. Error bars mean SD. Statistical significances are marked as: *, P < 0.05; **,P < 0.01. Viral loads among IgM anti-SARS-Cov-2 positive and negative Patients.
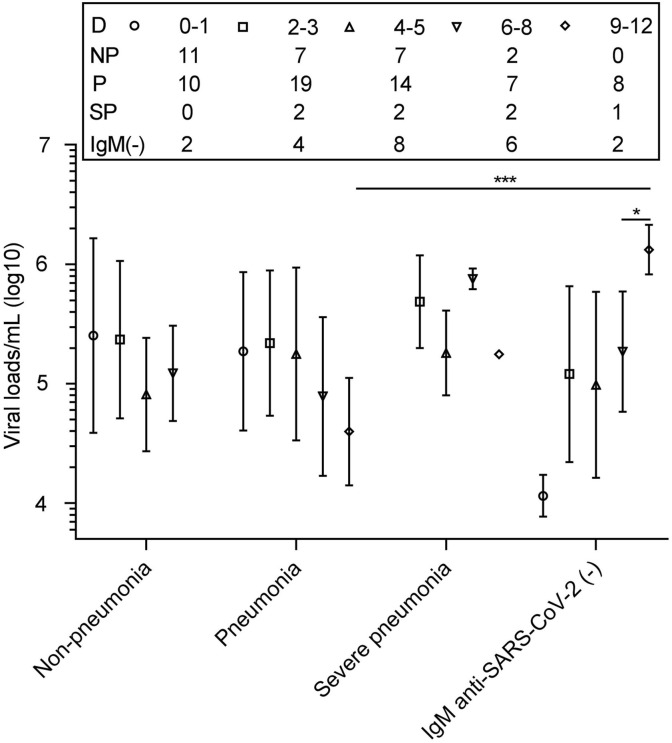
According to the time after onset, we divided the 114 patients into five groups (day 0–1, day 2–3, day 4–5, day 6–8 and day 9–12). For the IgM(+) patients, the mean viral load/mL (log10) in non-pneumonia cases was 5.27 in day 2–3 group, dropped from 5.40 in day 0–1 group. The value decreased to 4.91 in day 4–5, and increased to 5.09 in day 6–8 (Fig. 2 ). The value in pneumonia cases slightly increased to 5.34 in day 2–3 group from 5.27 in day 0–1. It was maintained at 5.25 in day 4–5, decreased to 4.89 in day 6–8 group, and showed a lower level in day 9–12 group (4.60). The value in severe cases maintained high among the groups (all>5.25). For the IgM(−) patients, it tended to increase along with the increasing days. Significance was found when compared it in day 9–12 with in day 6–9 (P < 0.05). The two IgM(−) patients day 9–12 were both with pneumonia, and their mean viral load/mL (log10) also differed significantly from IgM(+) pneumonia patients in day 9–12 (P < 0.001).
Fig. 2.
Viral loads among IgM anti-SARS-Cov-2 positive and negative Patients. The number of patients tested on each day is shown above the plot. Datapoints indicate mean. Error bars mean SD. Statistical significances are marked as: *, P < 0.05; ***,P < 0.001. Association of CRP/SAA with SARS-Cov-2 Viral loads.
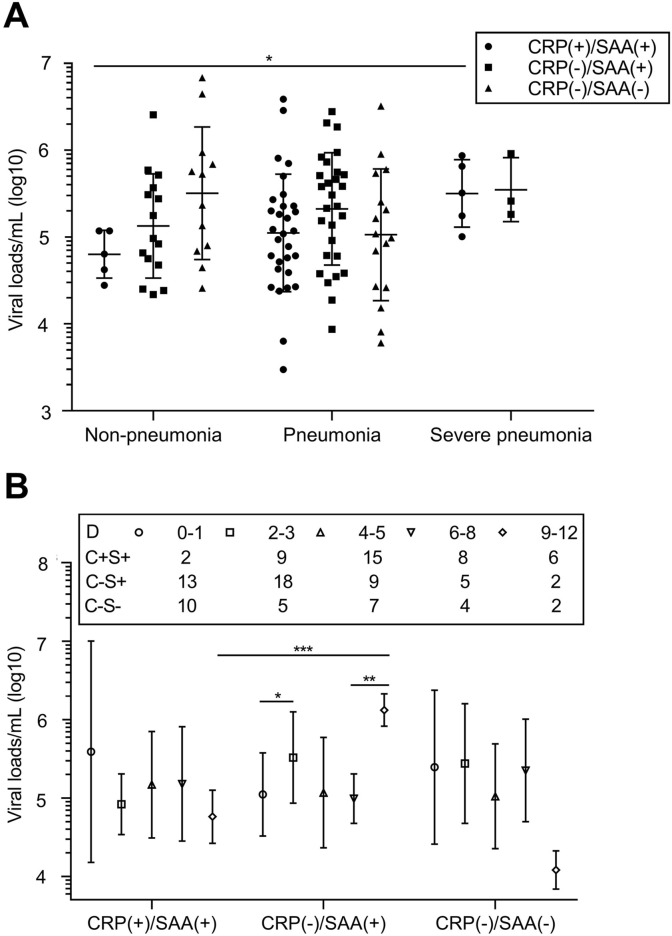
Besides 1 case of CRP(+)/SAA(−), the other 113 cases were divided into three groups, CRP(+)/SAA(+) (n = 41), CRP(−)/SAA(+) (n = 44) and CRP(−)/SAA(−) (n = 28).
As shown in Fig. 3 A, The value of CRP(+)/SAA(+) non-pneumonia patients (4.80) was also lower than that of other groups, but no significantly different were found. Within CRP(+)/SAA(+) group, mean viral load/mL (log10) tended to increase along with the worsening of severity (mean, 4.80, 5.05 and 5.50 respectively; 4.80 vs 5.50, P < 0.05). No statistical differences were found between CRP(+)/SAA(+) group and CRP(−)/SAA(+) group of severe pneumonia patients (5.50 vs 5.55, P > 0.05). The value in pneumonia patients did not change much among the groups. Interestingly, no severe pneumonia case showed CRP(−)/SAA(−).
Fig. 3.
Association of CRP/SAA with SARS-Cov-2 Viral loads. A, Association of CRP/SAA type with Viral load among patients of different severity; B, Association of CRP/SAA type with Viral load among patients of different onset time, and the number of patients tested on each day is shown above the plot. Error bars mean SD. Statistical significances are marked as: *, P < 0.05; **, P < 0.01; ***,P < 0.001.
As shown in Fig. 3B, at the early stage after onset (0–1 days), mean viral loads/mL (log10) were higher in CRP(+)/SAA(+) and CRP (−)/SAA(−) groups (5.59 and 5.40 respectively). The trend of viral load in the CRP(−)/SAA(+) group was different from others. The value in CRP(−)/SAA(+) group of day 0–1 was 5.05, and rose to 5.52 on day 2–3 (P < 0.05). The value was dropped to 5.07 on day 4–5, and maintained at 4.99 on day 6–8. Then it increased about 10 fold in patients of day 9–12 from cases in day 6–8 (6.12 vs 5.00, P < 0.05), and about 100 fold more than that of patients 9–12 days after onset in CRP(−)/SAA(−) group (6.12 vs 4.08). The two CRP(−)/SAA(+) patients were both pneumonia cases with IgM (−), while the two CRP(−)/SAA(−) patients were both pneumonia cases with IgM (+).
4. Discussion
IgM is a specific indicator produced early of infectious diseases, and can be used for early diagnosis (Zhang et al., 2020a) and has some protective effects. CRP is a non-specific indicator for the early stage of infection, mainly but not limited to bacterial diseases (Yao et al., 2019). Current research has found that this indicator is related to COVID-19 severe cases (Guan et al., 2020). SAA is another non-specific indicator for the early stage of infection, mainly but not limited to viral diseases (Zhang et al., 2019). The association between this indicator and the COVID-19 has not been reported (Wang et al., 2020; Zhang et al., 2020b). In this study, both CRP and SAA showed trends of increase along with the worsing of severity. Although statistical significances were only found in CRP, our results suggest both CRP and SAA have some indication of the severity of the disease.
Previous studies have shown that these indicators have directive effects on the infection and condition judgment of other viral diseases (Vollmer et al., 2016; Moutachakkir et al., 2017; Piedra et al., 2017; Yuan et al., 2015). This study also reveals the relationship between these indicators and viral load among COVID-19 patients and provides clues for the prevention and control of the disease.
A higher viral load of SARS-CoV-2 in upper respiratory epithelial cells indicates a higher risk to transmit this virus. In this study, we found the natural fluctuation of SARS-CoV-2 viral load had different trends among COVID-19 patients before antiviral treatments in the early stage of illness. Pneumonia cases maintained high viral loads on days 0–5 after onset. However, IgM(+) non-pneumonia patients had higher viral loads within 0–3 days after onset. More attention showed be payed to such patients because their symptoms are not obvious. Patients with CRP(+)/SAA(+) and CRP(−)/SAA(−) also presented higher viral load within 0–1 days after onset. It is suggested that using these indicators may help us to predict the transmitting risk of the patients.
The viral load of IgM(−) patients did not decrease with the increase of onset time, instead, it showed an upward trend. This may reflect the antiviral effect of the IgM antibodies. However, the viral load of IgM(+) patients in severe cases remained high 12 days after onset, which might indicate that the antiviral effect of IgM antibodies is weak, or there are other factors involved in the early resistance to the virus. A high viral load at this time also indicated the patient's condition might be not in a good direction, and an effective antiviral therapy was needed.
CRP(+) might have different suggestive effects on the condition of different types of patients. The viral load of CRP(+)/SAA(+) non-pneumonia patients was significantly lower than that of CRP(+)/SAA(+) severe pneumonia patients, and it was also lower than that of other types of non-pneumonia patients. For severe pneumonia patients, the viral load of CRP(+)/SAA(+) cases was about the same as that of CRP(−)/SAA(+) patients. The relationship should be studied in patients after admission, which gave clues that the elevation of CRP in severe pneumonia patient might indicate the viral load of the patient is still at a high level, and the treatment plan may need to be changed to control the viral load.
Compared with 62.5% of severe pneumonia patients had CRP(+)/SAA(+), we did not find any severe cases in CRP(−)/SAA(−) group. We also found that in CRP(−)/SAA(−) group, the two patients of 9–12 days after onset had a relative viral load about 10 fold lower than that of patients of 6–8 days. Considering that the two patients was both IgM(+) and the antiviral effect of IgM, CRP(−)/SAA(−) with IgM(+) might indicate that the patients' condition is relatively optimistic. Interestingly, in CRP(−)/SAA(+) group, another two patients with an onset of 9–12 days had a relative viral load about 10 fold higher than that of patients with onset of 6–8 days. Since the two patients were both negative for IgM antibodies, CRP(−)/SAA(+) with IgM(−) might indicate that the patients' condition were not in good directions. Due to our sample size is not big enough, the viral load of CRP(−)/SAA(−) with IgM(−) and CRP(−)/SAA (+) with IgM(+) patients was not observed at 9–12 days. However, these clues still allow us to see the possibility of judging the condition of COVID-19 patients by combining IgM and CRP/SAA. If these hypotheses are tested with a larger population on and after admission, combining these three indicators will facilitate the judgment and treatment of COVID-19 patients.
5. Conclusion
Combination of the IgM and CRP/SAA with some conventional clinical information, including time after onset and the type of severity, may give suggestions on the viral load and condition judgment of COVID-19 patients. More samples and researches are needed to confirm these clues. Taking into account these indicators had wide range of clinical applications, the confirmation of these clues will greatly help us to prevent and control the SARS-CoV-2 pandemic.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments:
We thank all our colleagues in Jiangsu Province for their supports on the data and sample collections and their dedication in the face of this outbreak. We greatly appreciate the kind assistance of Dr. Zhan Zhang and Dr. Yuancheng Li (Both from School of Public Health Nanjing Medical University) in statistical analysis.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.virol.2020.04.011.
Contributor Information
Baoli Zhu, Email: zhubl@jscdc.cn.
Lunbiao Cui, Email: lbcui@jscdc.cn.
Bin Wu, Email: jswb@jscdc.cn, wubinjscdc@163.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:figs1.
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa029. PMID: 32031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Wu B., Qi X., Zhao K., Guo X., Zhu Y. Rapid and sensitive detection of novel avian-origin influenza A (H7N9) virus by reverse transcription loop-mediated isothermal amplification combined with a lateral-flow device. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0069941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W-j, Ni Z-y, Hu Y., Liang W-h, Ou C-q, He J-x. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. PMID: 32109013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam K.Q., Yung C.F., Cui L., Lin Tzer Pin R., Mak T.M., Maiwald M. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America; 2020. A well infant with coronavirus disease 2019 (COVID-19) with high viral load. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S. Viral load kinetics of SARS-CoV-2 infection in first two patients in korea. J. Kor. Med. Sci. 2020;35(7):e86. doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutachakkir M., Lamrani Hanchi A., Baraou A., Boukhira A., Chellak S. Immunoanalytical characteristics of C-reactive protein and high sensitivity C-reactive protein. Ann. Biol. Clin. 2017;75(2):225–229. doi: 10.1684/abc.2017.1232. [DOI] [PubMed] [Google Scholar]
- NHC China. Revised Criteria for Diagnosis and Treatment of Novel Coronavirus Infection Pneumonia (Trial Version Fifth, Revised Version).
- https://www.who.int/emergencies/diseases/novel-coronavirus-2019 WHO. novel-coronavirus-2019. [cited; Available from:
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30113-4. PMID: 32105638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infection, genetics and evolution. journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra F.A., Mei M., Avadhanula V., Mehta R., Aideyan L., Garofalo R.P. The interdependencies of viral load, the innate immune response, and clinical outcome in children presenting to the emergency department with respiratory syncytial virus-associated bronchiolitis. PloS One. 2017;12(3) doi: 10.1371/journal.pone.0172953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. PMID: 32213337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer A.H., Gebre M.S., Barnard D.L. Serum amyloid A (SAA) is an early biomarker of influenza virus disease in BALB/c, C57BL/2, Swiss-Webster, and DBA.2 mice. Antivir. Res. 2016;133:196–207. doi: 10.1016/j.antiviral.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. Jama. 2020 doi: 10.1001/jama.2020.1585. PMID: 32031570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Zhang Y., Wu H. Regulation of C-reactive protein conformation in inflammation. Inflamm. Res. : official journal of the European Histamine Research Society [et al] 2019;68(10):815–823. doi: 10.1007/s00011-019-01269-1. [DOI] [PubMed] [Google Scholar]
- Yuan A., Li J., Liu P., Chen Z., Hou M., Wang J. Association of interleukin-6-572C/G gene polymorphism and serum or cerebrospinal fluid interleukin-6 level with enterovirus 71 encephalitis in Chinese Han patients with hand, foot, and mouth disease. Inflammation. 2015;38(2):728–735. doi: 10.1007/s10753-014-9983-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Sheng H., Li H., Wang R. Acute phase reactant serum amyloid A in inflammation and other diseases. Adv. Clin. Chem. 2019;90:25–80. doi: 10.1016/bs.acc.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q. Allergy; 2020. Clinical Characteristics of 140 Patients Infected by SARS-CoV-2 in Wuhan, China. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019 doi: 10.1056/NEJMoa2001017. 2020, PMID: 31978945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2001737. PMID: 32074444. [DOI] [PMC free article] [PubMed] [Google Scholar]