Abstract
Fluorescent spatial sequencing brings next-generation sequencing into a new realm capable of identifying nucleic acids in the cell’s natural environment. For the first time, scientists are able to multiplex the assignment of specific locations to hundreds of transcriptional targets and lay the foundation for understanding how genetic changes control the fate of each cell within the tissue microenvironment. In this perspective, we discuss the capabilities of fluorescent spatial sequencing in the context of other spatial imaging technologies and describe how these new technologies offer a data-rich, multiomic solution to many research applications. Fluorescent spatial sequencing has opened options for exploring many fundamental questions in biology, helping us gain a better understanding of cell and tissue development and disease progression.
Keywords: brain atlas, molecular images, FISSEQ
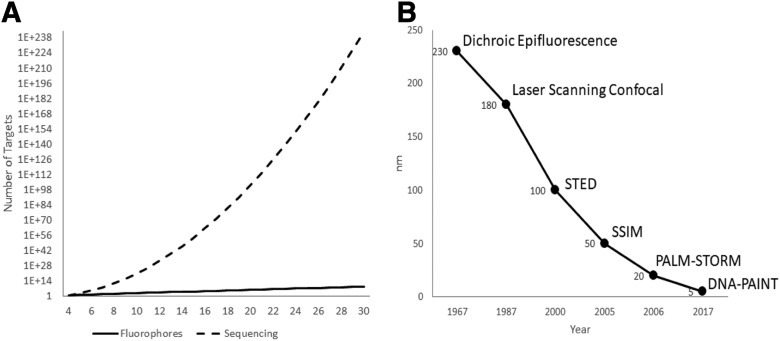
Biomolecular characterization at the cellular level provides critical information about cell structure and function. The spatial organization of these biomolecules at the tissue level determines cell development, maturation, and pathogenesis. Spatially profiling biomolecules at the cellular level, however, is a challenging problem. Prior to the development of spatial sequencing technologies, researchers were faced with nonquantitative and low-throughput approaches, such as immunohistochemistry and in situ hybridization. These methods are limited by a linear readout based on the number of fluorescent probes employed in each imaging cycle and the total number of imaging cycles performed in each experiment. In addition, they have finite multiplexing capabilities due to spectral overlap of available fluorophores. Fluorescent spatial sequencing, in contrast, benefits from an exponential increase in the number of targets per imaging cycle based on the sequence space interrogated, resulting in an increasingly multiplexed target library as technologies such as sequencing-by-synthesis, sequencing-by-ligation, and sequencing-by-hybridization continue to mature (Fig. 1A). This benefit is realized regardless of whether the targets are subjected to amplification or are sequenced through traditional de novo modalities or through approaches that employ targeted barcoding schemes. Bulk sequencing, by contrast, provides a population average and eliminates all spatial context to the acquired data. Even single-cell sequencing approaches suffer from low dynamic range, low multiplexing capabilities, and data integration challenges across genomic, transcriptomic, and proteomic data.1
FIGURE 1.
Flourescence spatial sequencing addresses near-infinite targets and molecular resolution. A) Increasing the number of fluorophores to address targets is vastly underpowered compared with sequencing. B) Light microscope has acheived finer resolutions over time, routinely beating IMS.
Optical imaging approaches to molecular detection face technical limits as well. Using imaging mass spectrometry (IMS), the highest spatial resolution routinely achievable with today’s off-the-shelf technology is about 10 microns, and custom research instrumentation is required if resolutions of 1 micron or less are required.2 At higher spatial resolutions, the sensitivity per pixel decreases as a result of the smaller area acquired at the expense of increased acquisition time and data file size.
Fluorescence microscopy routinely improves on the image resolution achieved using optical methods, but it is not without its own constraints. Fluorescence imaging is bound by the diffraction limit, proportional to the wavelengths used for imaging and inversely proportional to the numerical aperture of the magnifying objective. Several far-field techniques have been developed, such as Photo-Activated Localization Microscopy (PALM), Stochastic Optical Reconstruction Microscopy (STORM), Stimulated Emission Depletion (STED), Saturated Structured Illumination Microscopy (SSIM), and DNA-Points Accumulation for Imaging in Nanoscale Topography (DNA-PAINT), that temporally control the selective emission of multiple fluorophores to reconstruct an image of greater resolution than the diffusion limit alone can support.3 Expansion microscopy focuses on imaging isotropically expanded specimens in a controlled gel matrix followed by reconstruction of the original sample dimensions.4 These techniques have led the industry in finer resolutions capable of capturing tissue-scale images, with DNA-PAINT achieving sub–5-nanometer resolution (Fig. 1B). However, these superresolution approaches require costly equipment upgrades and/or significantly increased acquisition times to capture macro-level structures. With targeted probes, the sensitivity is single molecule and is unaffected by pixel size, in contrast to IMS, in which more abundant molecular species can interfere with less common species.
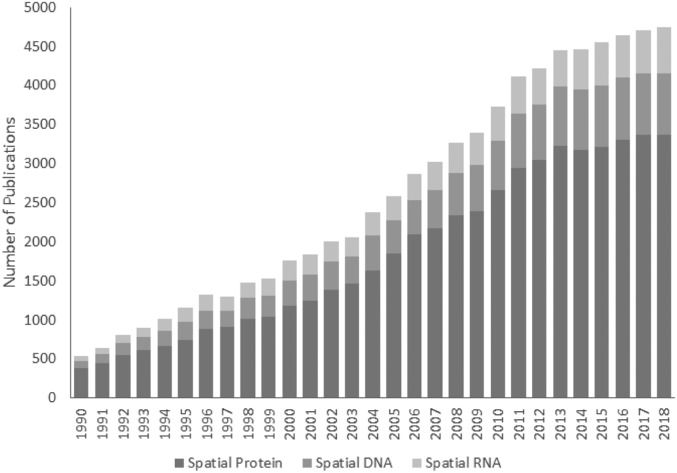
With the initial development of next-generation sequencing on microscopy devices, it was a natural transition to combine fluorescence imaging and sequencing into 1 platform. Indeed, the field has witnessed an explosion in interest as evidenced by the number of spatial-omics publications over the past 3 decades (Fig. 2). This synergy is best exemplified by fluorescent in situ sequencing (FISSEQ).5,6 FISSEQ provides a powerful, multiomic tool capable of combining in situ detection of RNA, DNA, proteins, and small molecules in the intact architecture of a single biologic specimen. Next-generation sequencing libraries are generated in the native tissue, immobilizing all nucleic acids for interrogation by de novo or targeted chemistries. Proteins and small molecules can be detected by oligo-conjugated scaffolds, opening the door to engineered natural molecules (e.g., antibodies, single-chain variable Fragment (scFv), adnectins, fynomers) and antibody mimetics (e.g., affibodies, affimers, alphabodies, nanobodies) to elicit highly specific recognition of target antigens.7 The nucleic acid conjugates are similarly spatially preserved, allowing for a simultaneous, universal sequencing readout for all biomolecules. The data produced by FISSEQ complement existing histology workflows and provide unparalleled insights into spatial genetic processes underlying disease biology, therapeutic delivery, and mechanisms of action (Fig. 3).
FIGURE 2.
The number of publications on the topic of spatial-omics obtained from MEDLINE trend using key words “spatial” with either “protein,” “DNA,” or “RNA.” 11
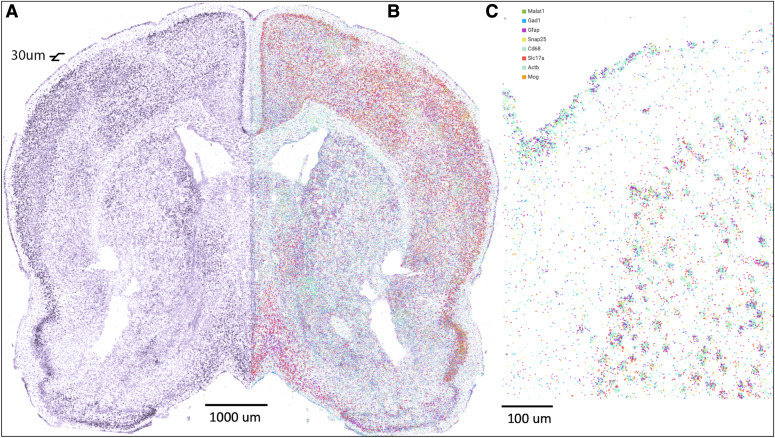
FIGURE 3.
Spatial sequencing of a 30-μm section of adult mouse brain reveals both structure and expression. A) Pseudostructure stain using nucleus bound long-non coding RNA (lncRNA) Malat1target pulled from FISSEQed sample. B) FISSEQ of the same sample with all 8 genes represented. C) Detailed view of (B), showing clear demarcations of layer 1 and 2/3 and clusters of neuronal cell bodies.
FISSEQ facilitates many exciting future spatial applications. Targeted capture and sequencing of long, nonfragmented DNA permits phasing of long arrays of nearly identical sequences (i.e., centromeres, rRNA) supporting the completion of many complex genomes. Integration of various single-cell omic data at organ scale allows us to map the connectome in the complete brain, creating a comprehensive map of all neural connections and insight into the structural connectivity of the mammalian nervous system.8,9 Using engineered guide RNA with Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) CRISPR associated protein 9 (Cas9) to introduce trackable genetic changes, FISSEQ can be used to trace the lineage of a cell from early zygote formation through complete organogenesis, creating a temporal record of cell differentiation.10 The ability to apply several complementing technologies to these complex, large-scale research questions will accelerate our understanding of systems biology faster than any singular technology can on its own.
CONCLUSIONS
Spatial sequencing has realized significant gains in deciphering the complex environment of the cell. With the massive multiplexity of Next-Generation Sequencing (NGS) and high resolution of fluorescence microscopy, spatial sequencing technologies, such as FISSEQ, simultaneously detect thousands of RNA, DNA, proteins, and molecules in situ. RNA FISSEQ enables powerful whole-transcriptome sequencing or flexible targeted sequencing for measuring single-cell gene expression, splice variation, and expressed sequence variants. DNA FISSEQ detects single-cell chromosomal conformation and structural variation, such as copy-number variation (CNV), and epigenetic changes that are causative to late-stage disease onset, like Alzheimer’s disease. Protein FISSEQ provides rich morphologic context to RNA and DNA localizations and establishes a framework for whole-organ reconstruction. The ability to multiplex detection of molecular classes in single cells within the native tissue organization has tremendous power to advance life sciences research, drug development, and clinical diagnostics and is the next big leap in omic profiling.
ACKNOWLEDGMENTS
The authors would like to acknowledge the U.S. National Institutes of Health, National Institute of Neurological Disorders and Stroke (Grant 1R01NS112716-01A1 to R.E.K. and G.M.C.) for their support of their work.
REFERENCES
- 1.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358:64–69. [DOI] [PubMed] [Google Scholar]
- 2.Caprioli RM. Imaging mass spectrometry: a perspective. J Biomol Tech. 2019;30:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godin AG, Lounis B, Cognet L. Super-resolution microscopy approaches for live cell imaging. Biophys J. 2014;107:1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassie AT, Zhao Y, Boyden ES. Expansion microscopy: principles and uses in biological research. Nat Methods. 2019;16:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Daugharthy ER, Scheiman J, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Daugharthy ER, Scheiman J, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X, Yang YP, Dikici E, Deo SK, Daunert S. Beyond antibodies as binding partners: the role of antibody mimetics in bioanalysis. Annu Rev Anal Chem (Palo Alto Calif). 2017;10:293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Sun YC, Church GM, Lee JH, Zador AM. Efficient in situ barcode sequencing using padlock probe-based BaristaSeq. Nucleic Acids Res. 2018;46:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Woehrstein JB, Donoghue N, et al. Rapid sequential in situ multiplexing with DNA exchange imaging in neuronal cells and tissues. Nano Lett. 2017;17:6131–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalhor R, Kalhor K, Mejia L, et al. Developmental barcoding of whole mouse via homing CRISPR. Science. 2018;361:eaat9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corlan AD. Medline trend: automated yearly statistics of pubMed results for any query. Available at: http://dan.corlan.net/medline-trend.html. Accessed February 14, 2012.