Abstract
Background: Severe acute respiratory syndrome (SARS) is a newly discovered disease caused by a novel coronavirus. The present study studied the longitudinal profile of antibodies against SARS‐coronavirus (SARS‐CoV) in SARS patients and evaluated the clinical significance of these antibodies.
Methods: Two methods, ELISA and indirect immunofluorescent assay, were used for the detection of the anti‐SARS‐CoV IgG and IgM in 335 serial sera from 98 SARS patients. In 18 patients, serum antibody profiles were investigated and antibody neutralization tests were performed from 7 to 720 days after the onset of symptoms.
Results: The ratios of positive IgG/IgM by ELISA were 0/0, 45.4/39.4, 88.6/71.4, 96/88, 100/48.6, 100/30.9, 100/17.1, 100/0 per cent, respectively, on 1–7, 8–14, 15–21, 22–28, 29–60, 61–90, 91–180 and 181–720 days after the onset of symptoms. Antibodies were not detected within the first 7 days of illness, but IgG titre increased dramatically on day 15, reaching a peak on day 60, and remained high until day 180 from when it declined gradually until day 720. IgM was detected on day 15 and rapidly reached a peak, then declined gradually until it was undetectable on day 180. Neutralizing viral antibodies were demonstrated in the convalescence sera from SARS patients.
Conclusion: The persistence of detectable IgG antibodies and neutralizing viral antibodies for up to 720 days suggest that SARS patients may be protected from recurrent SARS‐CoV infection for up to 2 years.
Keywords: antibody, antibody neutralization test, severe acute respiratory syndrome
INTRODUCTION
Severe acute respiratory syndrome (SARS) was first reported in Guangdong Province of the People's Republic of China in November, 2002 and it swept through more than 30 countries and regions during the first half of 2003. It was a life‐threatening pandemic, affecting more than 8000 people worldwide with a case‐fatality rate as high as 9.6%. 1 , 2 , 3 Studies showed that SARS was a rapidly progressive, atypical pneumonia caused by a novel coronavirus. 4 , 5 , 6 , 7 Since the discovery of SARS‐coronavirus (SARS‐CoV), laboratory diagnosis of the infection has become an important part of patient management, contact tracing and epidemiological studies. Recent studies have demonstrated that >95% of patients with SARS mount an antibody response during convalescence. 8 , 9 , 10 , 11 The most widely used current methods for the detection of antibodies against SARS‐CoV in acute and convalescent phase sera are ELISA and indirect immunofluorescence assay (IFA). 4 , 12 The aim of this study was to investigate the longitudinal profile of antibodies against SARS‐CoV in SARS patients during the course of illness using ELISA and IFA and to assess the effect of convalescent serum from SARS patients on SARS‐CoV using the virus neutralization test. The intention was to gain a greater understanding of the immune response in humans following SARS infection.
METHODS
Subjects
Between December 2002 and June 2003, 98 patients with SARS, comprising 43 men and 55 women, aged 20–75 years (mean 37.8 ± 12.2 years), were enrolled in the Guangzhou Institute of Respiratory Diseases in Guangzhou, China. The average duration of hospitalization was 23.1 ± 12.3 days. All clinical specimens collected were studied retrospectively. Blood samples were taken from patients in the hospital and only 18 SARS patients completed follow up. Serial blood samples were taken on days 7, 15, 30, 60, 90, 180, 270, 360, 450, 540 and 720 from the onset of illness. This study had the approval of the ethics committee of the Guangzhou Institute of Respiratory Diseases. All SARS patients in this study met the clinical criteria for diagnosis as recommended by World Health Organization. 12
Clinical treatment
As soon as a diagnosis of SARS was established, all patients were treated with a combination of antibiotics (cephalosporin and erythromycin) and antiviral agents (ribavirin or traditional Chinese medicine). Subsequently, patients whose fever persisted for more than 3 days or who showed a progressive deterioration in their CXR (79.6%), received methylprednisonlone (2–4 mg/kg/day, 14–28 days (17 ± 7 days)). Patients who then showed no clinical improvement or SaO2 <93% after high‐flow oxygen (5 L/min) therapy (38.8%), were subject to CPAP or bi‐level positive airway pressure (BiPAP) via a nasal mask to improve oxygen supply and prevent alveolar collapse. Patients (14.3%) who failed to respond to this intervention were intubated for intermittent positive pressure ventilation (IPPV) (pressure‐control ventilation (PCV)/pressure‐regulated volume‐control (PRVC) + PEEP), with sedation during ventilation.
CXR scores
The severity of CXR abnormalities were scored according to the extent of lesions in six zones on each chest film (upper, middle and lower zones on both lungs). They were evaluated and scored (mild = 1, moderate = 2, severe = 3), generating six subscores that were summed up into a composite score for each CXR. The score of a normal CXR was 0 and the maximum score was 18. Each CXR was independently scored by two individuals, a radiologist and a respiratory physician. Differences in scoring were resolved by consensus.
ELISA
ELISA was used to detect specific serum antibodies (IgG and IgM) against SARS virus in all patients as well as serial samples from 18 patients and was performed as described previously (SARS ELISA kits were donated by GBI Biotech, Beijing China). 13 Briefly, 96‐well polystyrene microtitre plates were coated with 10 µg/mL pure corona virus antigen (SARS‐CoV BJ01 strain, accession no. AY278488 in GenBank). Serum samples at 1:10 dilutions were added and incubated at 37°C for 30 min. Each well was aspirated and washed with rinsing buffer five times. A total of 100 µL antihuman IgG and IgM conjugate was added, incubated at 37°C for 20 min, and washed with a rinsing buffer five times. Substrate solution was added for colour development. The optical density at 450 nm (A450) of each well was measured within 30 min, using a microplate reader. The cut‐off limit was 0.13 + Anegative control, according to the manufacturer's instruction. A value higher than the cut‐off limit was considered positive.
Indirect immunofluorescent assay
Indirect immunofluorescence assay was performed as described previously (IFA kit were donated by Beijing Institute of Microbiology and Epidemiology and approved by State Drugs Administration of China). 14 Briefly, 20 µL diluted sera (1:20 or greater) were added onto slides containing monolayers of SARS‐CoV‐infected Vero cells (SARS‐CoV BJ01 strain) and non‐infected Vero cells, as a negative control. The slides were incubated at 37°C for 30 min, washed three times with phosphate buffered saline, and combined with FITC‐labelled anti‐human IgG or FITC‐labelled anti‐human IgM for 30 min. The slides were washed as before, and fluorescence was detected with a microscope. The data were analysed by SPSS 10.0 software (SPSS, Chicago, IL, USA) and the geometric means of sero‐positive samples were employed in all statistics.
Virus neutralization test
Neutralization tests were performed in a Biosafety level 3 laboratory. Virus stocks were produced by infecting Vero‐E6 cells with the SARS‐CoV (strain HZS2‐D, accession no. AY395004 in GenBank; the nucleotide sequences have been identified 15 ). Briefly, Vero‐E6 cells were cultured in an humidified atmosphere with 5% CO2 at 37°C until they formed a confluent monolayer. The cells were then inoculated with SARS‐CoV in MEM medium with 1% FCS (Sigma, Poole, UK) and incubated at 37°C for 48–72 h until a 75% cytopathic effect (CPE) was observed. The culture was frozen at −20°C and the cells were lysed by freeze/thawing three times. The supernatants containing the SARS‐CoV were collected and aliquoted for later use.
Viral titres were determined by the KARBER dilution assay. Vero‐E6 cells were infected with serial dilutions of the viral supernatants (1–10−8), in 2 × 8 wells. The titre of the virus stock used was 104.58 TCID50/25 µL.
The neutralization test was performed according to the following protocol: Sera were heat‐inactivated at 56°C for 30 min and diluted 1:10, 1:20, 1:40, 1:80, 1:160, 1:320, 1:640, 1:1280, 1:2560 and 1:5120. In total, 25 µL diluted serum was mixed with 25 µL 100TCID50/25 µL SARS‐CoV in quadruplicate in 96‐well microtitre plates, and incubated at 37°C for 2 h. In total, 100 µL (1 × 105 cells/mL) Vero‐E6 cells were then added and the mixture further incubated at 37°C, 5% CO2 for 7 days. CPEs were detected using an inverted microscope and the highest dilution of the serum at which 50% of the wells were protected from viral CPE was considered to be the neutralizing titre. Quality control was set up in parallel with the neutralization test. Blood samples from 10 healthy volunteers, four men and six women aged 17–58 years (mean 35.6 ± 12.2 year), served as normal controls. Thus, the assay consisted of (i) cell control (Vero‐E6 cells medium alone); (ii) virus control (Vero‐E6 cells mixed with virus medium); (iii) negative serum control (Vero‐E6 cells mixed with virus and normal serum); and (iv) positive serum control (Vero‐E6 cells mixed with virus and acute and convalescent infected serum). In each assay, a back‐titration of the test dilution of virus was also set up in quadruplicate.
RESULTS
IgG and IgM antibodies against SARS‐CoV
Anti‐SARS‐CoV IgG and IgM were studied in 335 serial sera from 98 clinically diagnosed SARS patients. The results in Table 1 show that IgG and IgM could not be detected within the first 7 days. IgG antibody was first detected by ELISA on the 10th day after onset of the disease. Both IgG and IgM antibodies were present in nearly half of the patients in the second week after the onset of illness. The positive titre for IgM kept increasing up to 1 month and then declined gradually until it was undetectable at >180 days. IgG titre reached its maximum after a month and remained higher than IgM. Detection by ELISA showed that the IgG/IgM antibody titres for samples collected during days 1–7, 8–14, 15–21, 22–28, 29–60, 61–90, 91–180 and 181–720 after the onset of the disease were 0/0, 45.4/39.4, 88.6/71.4, 96/88, 100/48.6, 100/30.9, 100/17.1, 100/0 per cent, respectively. Similar results were obtained using the IFA.
Table 1.
Detection of IgG and IgM positive rate by ELISA and indirect immunofluorescent assay (IFA) relative to days from the onset of symptoms
| Days from onset of symptoms | No. of samples | IgG (n (%)) | IgM (n (%)) | ||
|---|---|---|---|---|---|
| ELISA | IFA | ELISA | IFA | ||
| 1–7 | 23 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 8–14 | 33 | 15 (45.4) | 15 (45.4) | 13 (39.4) | 14 (42.4) |
| 15–21 | 35 | 31 (88.6) | 30 (85.7) | 25 (71.4) | 26 (74.3) |
| 22–28 | 50 | 48 (96.0) | 49 (98.0) | 44 (88.0) | 40 (80.0) |
| 29–60 | 37 | 37 (100) | 37 (100) | 18 (48.6) | 18 (48.6) |
| 61–90 | 42 | 42 (100) | 42 (100) | 13 (30.9) | 14 (33.3) |
| 91–180 | 41 | 41 (100) | 41 (100) | 7 (17.1) | 7 (17.1) |
| 181–720 | 74 | 74 (100) | 74 (100) | 0 (0.0) | 0 (0.0) |
| Total | 335 | 288 | 288 | 120 | 119 |
Because the SARS‐CoV antibody serological tests were not available at the beginning of SARS outbreak, only 23 and 33 blood samples on day 1–7 and day 8–14, respectively, which were already collected and stored during hospitalization, were measured. Eight patients died between 7 and 14 days because of adult respiratory distress syndrome complicated with multiple organ failure. Another 15 patients were not contactable following discharge from the hospital and not all patients returned for blood samples to be taken at specified time points. There were only 18 patients who completed sampling required for the study.
Profile of CXR in SARS patients
The daily temperature and CXR scores of the 18 patients with complete blood samples (described above) are shown in Figure 1. All patients presented with fever and temperatures reached a peak on day 2 then declined and returned to normal by day 10 in most cases. CXR abnormalities were localized or patchy and/or spotty pulmonary infiltrates on one or both sides during the first week, maximal on day 10–12, and almost completely resolved by day 21. By day 15, when serum IgG and IgM became strongly positive, most patients’ CXR abnormalities had improved and almost resolved.
Figure 1.
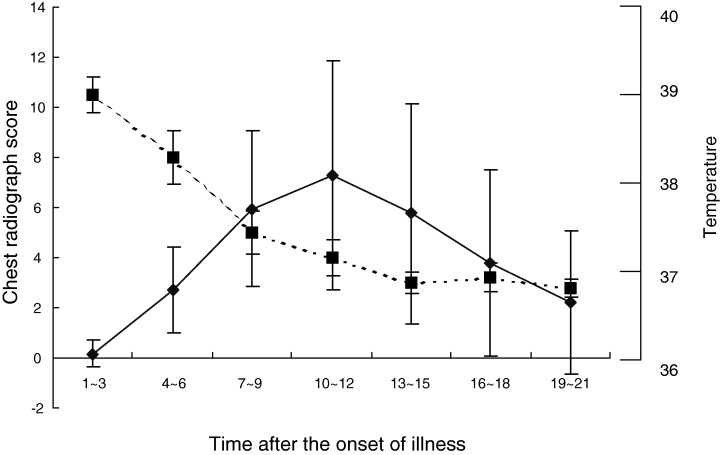
Temperature and CXR severity scores in 18 SARS patients. (—◆—) Chest radiograph score; (‐ ‐ ‐▪‐ ‐ ‐ ) temperature (average over 3 days).
Profile of IgG and IgM against SARS‐CoV in SARS patients over a 2‐year period
Geometric mean values of IgG and IgM antibody titres (determined by IFA test) for the available serum specimens from 18 SARS patients, at various intervals over a 2‐year period, were calculated and plotted against the number of days from the onset time of symptoms (Fig. 2). Both IgG and IgM were not detectable on day 7. On day 15, IgG titres increased and peaked on day 60, with an average of 1:670, then plateaued up to day 180, and declined gradually. At day 720, all 18 SARS patients, from whom serum samples were available, were still positive for IgG. However, a few patients’ IgG antibodies titres declined dramatically on day 540 and 720 and the average titre was close to the cut‐off value for positivity (1:10). In contrast, a significantly lower titre of IgM was found with a peak of 1:61.2 on day 30, which then declined and became undetectable after day 180 and 270. Similar results were obtained with the ELISA.
Figure 2.
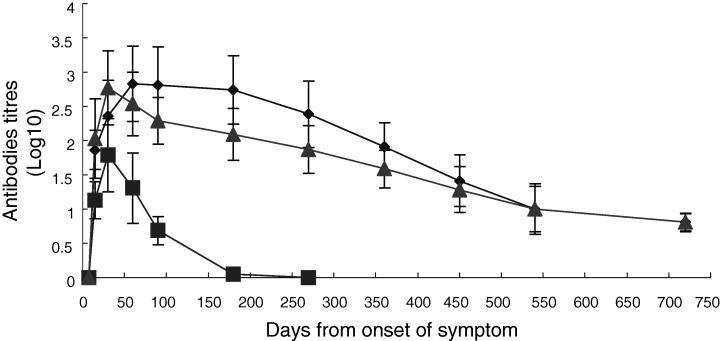
Geometric means of immunofluorescent antibodies titres for IgG (◆) and IgM (▪) and neutralizing viral antibodies titres (▴) in 18 SARS patients. IgG and IgM are detected by indirect immunofluorescence assay and total Ig was detected by the antibody neutralization test.
Neutralizing capacity of convalescent sera
To assess viral neutralization potential against subsequent infection in convalescent patients, neutralizing antibodies were assayed in 18 SARS patients’ sera collected 7, 15, 30, 60, 90, 180, 270, 360, 450, 540 and 720 days after the onset of symptoms. It was found that, in all 18 patients, neutralizing antibodies were not detectable on day 7. On day 15, neutralizing antibodies rose and then peaked on day 30 with an average of 1:590. After this initial surge, neutralizing antibodies dropped gradually. At days 540 and 720, one of the 18 SARS patients had no detectable neutralizing antibodies, and the remaining patients had a low titre, with an average of 1:10 (Fig. 2). Furthermore, it was found that neutralizing antibodies were not detectable in normal sera from 10 healthy persons, indicating that the virus cannot be inactivated by normal serum components.
DISCUSSION
In total, 335 clinical blood specimens from 98 SARS patients were analysed and 75 patients who were in the convalescent phase following successful treatment of the disease were followed up. All patients in the study demonstrated seroconversion to SARS‐CoV, thereby confirming the diagnosis. IgG and IgM could not be detected within the first 7 days of illness. The earliest seroconversion occurred on day 10 after the onset of the disease. IgM antibodies were detected later than IgG antibodies, which may be because of the earlier development of IgG antibodies or the differential sensitivities of the class‐specific ELISAs. IgG antibodies were significantly elevated on day 15, peaked on day 60, and remained high beyond day 180. IgM antibodies were detected and peaked on day 15 and disappeared by day 90.
Almost all patients with serological evidence of SARS coronavirus infection developed pneumonic change on CXR. Most patients improved clinically and CXRs had resolved to a large extent by day 15 when serum antibodies began to rise or peaked, suggesting that these antibodies may be important in eliminating or neutralizing the CPEs of the SARS‐CoV.
These findings suggest that the IgM antibodies to SARS‐CoV persist for a much shorter period of time than IgG, as would be expected. The prolonged production of IgG suggests that, not only does it play a principal role in the humoral immune response against acute SARS‐CoV infection, but also plays a crucial part in cleaning up residual virus during the recovery phase. Shi et al. have similarly shown that the activity of anti‐SARS‐CoV IgG remains active on day 210 after onset. 11 Recently, Woo et al. reported on the use of recombinant SARS‐CoV, nucleocapsid protein, ELISA‐based antibody tests for the serodiagnosis of SARS‐CoV pneumonia. 16 They showed that all convalescent patients (from whom serum samples were available) were still positive for anti‐nucleocapsid protein IgG antibody on day 240 following the onset of illness. This study is the first to describe the longitudinal profile of antibodies against SARS‐CoV in SARS patients up to 720 days after the onset of illness. The patients’ IgG titres still remained positive on day 720 following the onset of symptoms, although their average titre was close to the cut‐off value for positivity (1:10).
The virus neutralization test is a sensitive and specific assay applicable to the identification of virus‐specific antibody in animals and humans. The novel finding in this study is the demonstration of neutralizing antibodies in the convalescent sera of SARS patients by the inhibition of CPEs of SARS‐CoV on Vero‐E6 cells. All patients but one showed positive neutralization effects on day 540 and 720 after the onset of illness. This is an important observation as the presence of these antibodies may be necessary for the recovery of the patients as well as for the prevention of reinfection by SARS‐CoV. However, this protective effect may not be permanent and may decline or even disappear over a period of 2 years, suggesting an appropriate vaccine may be necessary at that time.
ACKNOWLEDGEMENTS
The authors are grateful to research funding from the National High Technology Research and Development Program of China (863 Program) 2003AA208107; the Municipal Scientific Committee of Guangzhou (grant No. 200364719).
REFERENCES
- 1. Tsang KW, Ho PL, Ooi GC et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003; 348: 1977–85. [DOI] [PubMed] [Google Scholar]
- 2. Booth CM, Matukas LM, Tomlinson GA et al. Clinical features and short‐term outcomes of 144 patients with SARS in the Greater Toronto area. JAMA 2003; 289: 2801–9. [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization. Summary of SARS Cases by Country from 1 November 2002 to 7 August 2003. World Health Organization, http://who.int/csr/sars/country/2003_08_15/en/. 11 November 2003. [Google Scholar]
- 4. Drosten C, Gunther S, Preiser W et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003; 384: 1967–76. [DOI] [PubMed] [Google Scholar]
- 5. Ksiazek TG, Erdman D, Goldsmith CS et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003; 348: 1953–66. [DOI] [PubMed] [Google Scholar]
- 6. Marra MA, Jones SJ, Astell CR et al. The genome sequence of the SARS‐associate coronavirus. Science 2003; 300: 1399–404. [DOI] [PubMed] [Google Scholar]
- 7. Rota PA, Oberste MS, Monroe SS et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003; 300: 1394–9. [DOI] [PubMed] [Google Scholar]
- 8. Peiris JSM, Chu CM, Cheng VCC et al. Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet 2003; 361: 1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li G, Chen X, Xu A. Profile of specific antibodies to the SARS‐associated coronavirus. N. Engl. J. Med. 2003; 349: 508–9. [DOI] [PubMed] [Google Scholar]
- 10. Rainer TH, Cameron PA, DeVilliers S et al. Evaluation of WHO criteria for identifying patients with severe acute respiratory syndrome out of hospital: prospective observational study. BMJ 2003; 326: 1354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi YL, Wan ZY, Li LH et al. Antibody responses against SARS‐coronavirus and its nucleocaspid in SARS patients. J. Clin. Virol. 2004; 31: 66–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peiris JSM, Lai ST, Poon LLM et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen WJ, Xu ZY, Mu JS et al. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)‐associated coronavirus infection. J. Med. Microbiol. 2004; 53: 435–8. [DOI] [PubMed] [Google Scholar]
- 14. Si BY, Yan B, Yu M. Development of IFA method for detecting antibodies of SARS coronavirus. Med. J. Chin. People's Liberat. Army 2003; 28: 699–700. [Google Scholar]
- 15. The Chinese SARS Molecular Epidemiology Consortium. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004; 303: 1666–9. [DOI] [PubMed] [Google Scholar]
- 16. Woo PC, Lau SK, Wong BH et al. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus. Clin. Diagn. Lab. Immunol. 2004; 11: 665–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


