Abstract
Bacterial cell wall synthesis is an essential process in bacteria and one of the best targets for antibiotics. A critical step on this pathway is the export of the lipid-linked cell wall monomer, Lipid II, by its transporter MurJ. The mechanism by which MurJ mediates the transbilayer movement of Lipid II is not understood because intermediate states of this process have not been observed. Here we demonstrate a method to capture and detect interactions between MurJ and its substrate Lipid II by photocrosslinking and subsequent biotin-tagging. We show that this method can be used to covalently capture intermediate transport states of Lipid II on MurJ in living cells. Using this strategy we probed several lethal arginine mutants and found that they retain appreciable substrate-binding ability despite being defective in Lipid II transport. We propose that Lipid II binding to these residues during transport induces a conformational change in MurJ required to proceed through the Lipid II transport cycle. The methods described to detect intermediate transport states of MurJ will be useful for characterizing mechanisms of inhibitors.
GRAPHICAL ABSTRACT
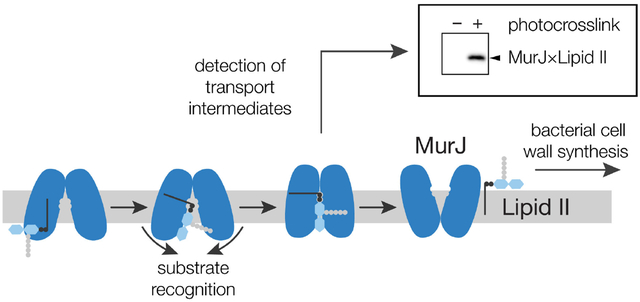
The bacterial cell wall is a crosslinked polymer assembled from a lipid-linked building block, Lipid II, which is made in the cytoplasm.1 Before it can be used to build the cell wall, Lipid II must first be flipped across the cytoplasmic membrane (Figure 1a). The protein responsible for flipping Lipid II is MurJ.2–3MurJ belongs to a large superfamily of transporters found in all domains of life, but their transport mechanism is not understood.4–5 The general model for how transporters work is by cycling between conformational states, which allows alternating access of the substrate to either side of the membrane.6 We have previously shown that MurJ requires the membrane potential to complete this conformational cycling.7–8 Binding of Lipid II to purified MurJ has been quantified,9 but no flipping assay for purified protein has been reported and no intermediate transport states of Lipid II bound to MurJ have been observed.10–12
Figure 1.
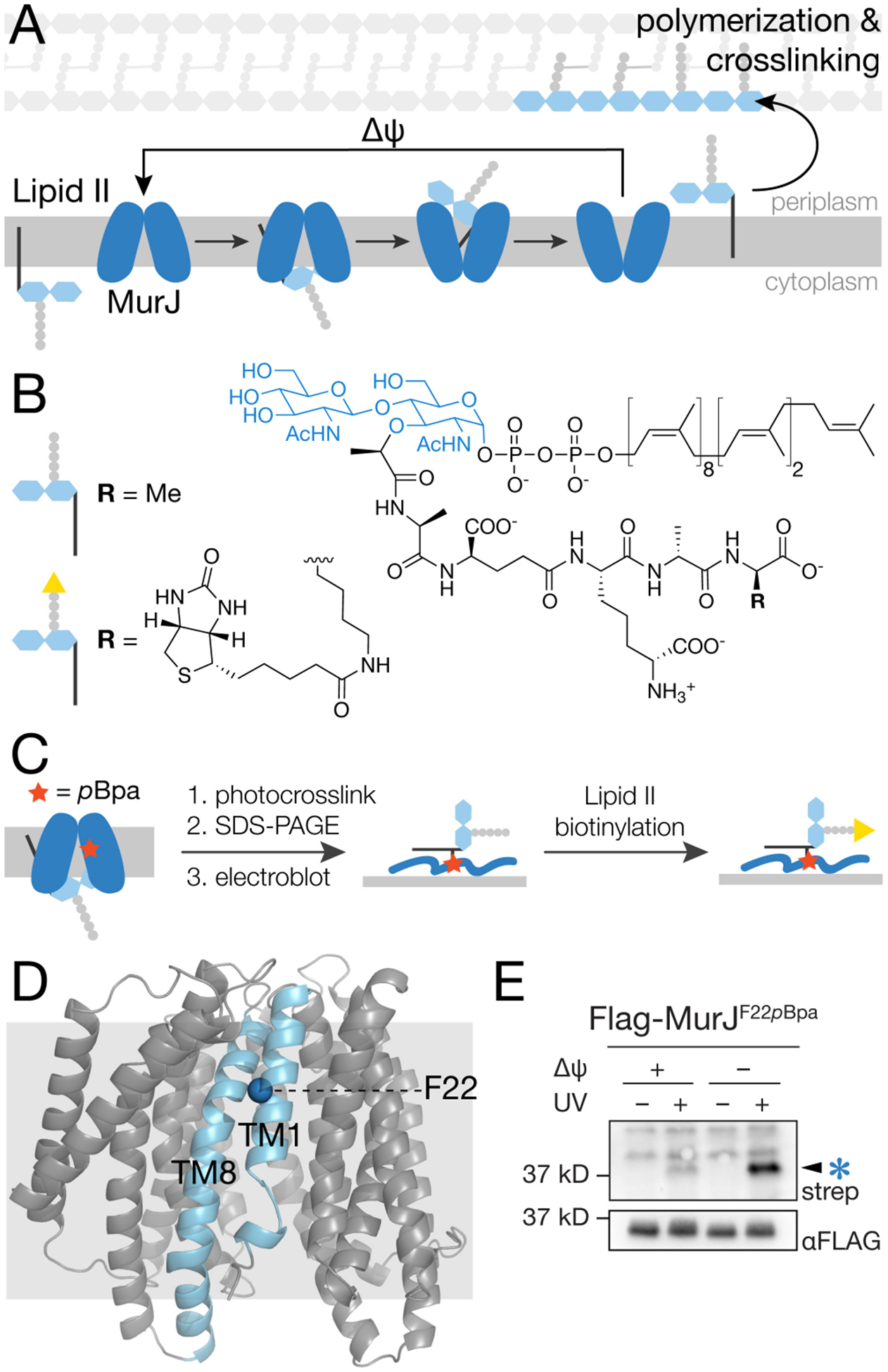
Photocrosslinking of MurJF22pBpa in cells produces an adduct that is detected using a method that specifically labels Lipid II (a) Lipid II is transported from the cytoplasm to the periplasm by MurJ via conformational changes driven by membrane potential (Δψ). (b) Structure of E. coli Lipid II and its biotinylated derivative. (c) Schematic showing the method to detect MurJ-Lipid II adducts. MurJ containing the unnatural amino acid pBpa (red star) is UV-irradiated to photocrosslink bound Lipid II. After SDS-PAGE and electroblotting, the Lipid II crosslinked to MurJ is biotinylated by D-amino acid exchange using PBP4, enabling detection with streptavidin-HRP.17 (d) Crystal structure of E. coli MurJ in the inward-facing conformation (PDB 6CC4)10 showing location of F22 in transmembrane helix 1 (TM1). (e) A UV-dependent band (arrow) was detected in a streptavidin-HRP blot after irradiating cells expressing MurJF22pBpa and labeling to detect Lipid II as in (c). Intensity of this band increased substantially when Lipid II was pre-accumulated on the inner leaflet of the cytoplasmic membrane (compare lanes 2 and 4). To accomplish this, cells were treated with 100 μM CCCP for 5 min and then diluted 100-fold to restore membrane potential just prior to irradiation. See also Figures S1–S3.
Transporters are challenging to study because they do not alter the chemical structure of their cargo; starting material and product are differentiated only by their topology with respect to the membrane. Lipid flippases are particularly challenging because the initial and final states are tethered to the same membrane and are not easily separable. We have previously shown that in vivo photocrosslinking can be used to monitor transport of lipopolysaccharide (LPS) as it moves through its transport machine.13 The transport machine was engineered to contain unnatural photocrosslinkable amino acids that crosslinked to lipopolysaccharide during transport.14 The crosslinks to lipopolysaccharide were detected using anti-LPS antibodies. We wanted to use a similar photocrosslinking strategy to monitor intermediate states of MurJ; however, no antibodies to Lipid II exist, so detecting crosslinked adducts has not been possible.15–16 Here we developed a method to detect Lipid II covalently bound to MurJ. Using this method, we can observe intermediate transport states of Lipid II on MurJ in living cells, which establishes that MurJ is directly responsible for transport. Finally, using these tools we have shown that Lipid II still binds to MurJ lacking three amino acids that are critical for function.
We previously developed methods to isolate Lipid II from bacteria and label it with biotin by exchanging the terminal DAla for biotin-D-Lys (Figure 1b, S1–S2), allowing its detection by streptavidin immunoblot.17–19 We wondered if this method could be used to detect adducts between Lipid II and MurJ (Figure 1c). When we irradiated cells expressing functional MurJ containing p-benzoyl-L-phenylalanine (pBpa) in place of F22, which is located in a region of MurJ proposed to allow access of Lipid II into the cavity (Figure 1d)11, we were unable to detect any signal. Speculating that Lipid II was inaccessible when crosslinked to folded MurJ, we instead performed the biotinylation reaction after SDS-PAGE separation of MurJ and transfer of the denatured protein to a PVDF western blot membrane (Figure 1c). We were only able to observe a weak UV-dependent signal at ~37 kD, the expected molecular weight of MurJ (Figure 1e, lane 2), suggesting that normally the residence time of Lipid II in an intermediate transport state on MurJ is too short to efficiently crosslink. To increase the amount of Lipid II capture, we sought methods to accumulate Lipid II at the transporter. To do this, we first treated cells with the protonophore CCCP (carbonyl cyanide m-chlorophenyl hydrazone) to dissipate the membrane potential, which stops the transport cycle, leading to Lipid II accumulation on the inner leaflet of the cytoplasmic membrane.7 After diluting CCCP to restore membrane potential and MurJ function, we UV-irradiated cells. Under this condition, we observed a substantial increase in the UV-dependent biotinylated species (Figure 1e, lane 4).
We established that the biotinylated species was the crosslinked adduct MurJ×Lipid II using several different experiments. First, when we treated cells with fosfomycin (fos), which inhibits formation of Lipid II (Figure S4), we found that the UV-dependent signal at 37 kD did not appear (Figure 2a). This result showed that the biotinylated species was dependent on Lipid II biosynthesis. Second, we performed a pulldown against Flag-MurJ after crosslinking treatment and subjected the purified protein to the biotinylation procedure. We observed a single band at the same molecular weight as seen for the crude lysates (Figure 2b), showing that the UV-dependent biotinylated species is a MurJ adduct. Third, we found that the UV-dependent band did not appear when PBP4, the transpeptidase used to catalyze exchange of the terminal D-Ala for biotin-D-Lys in Lipid II on the western blot membrane, was omitted from the reaction (Figure 2c).17 Finally, if after transfer the PVDF membrane was pretreated with DacA, a carboxypeptidase that removes the terminal DAla from the Lipid II stem peptide,20 the band did not appear in the presence of PBP4 (Figure 2d). These latter experiments show that the crosslinked species contains Lipid II. Taken together, these experiments establish that the detected species is biotin-labeled Lipid II crosslinked to MurJ.
Figure 2.
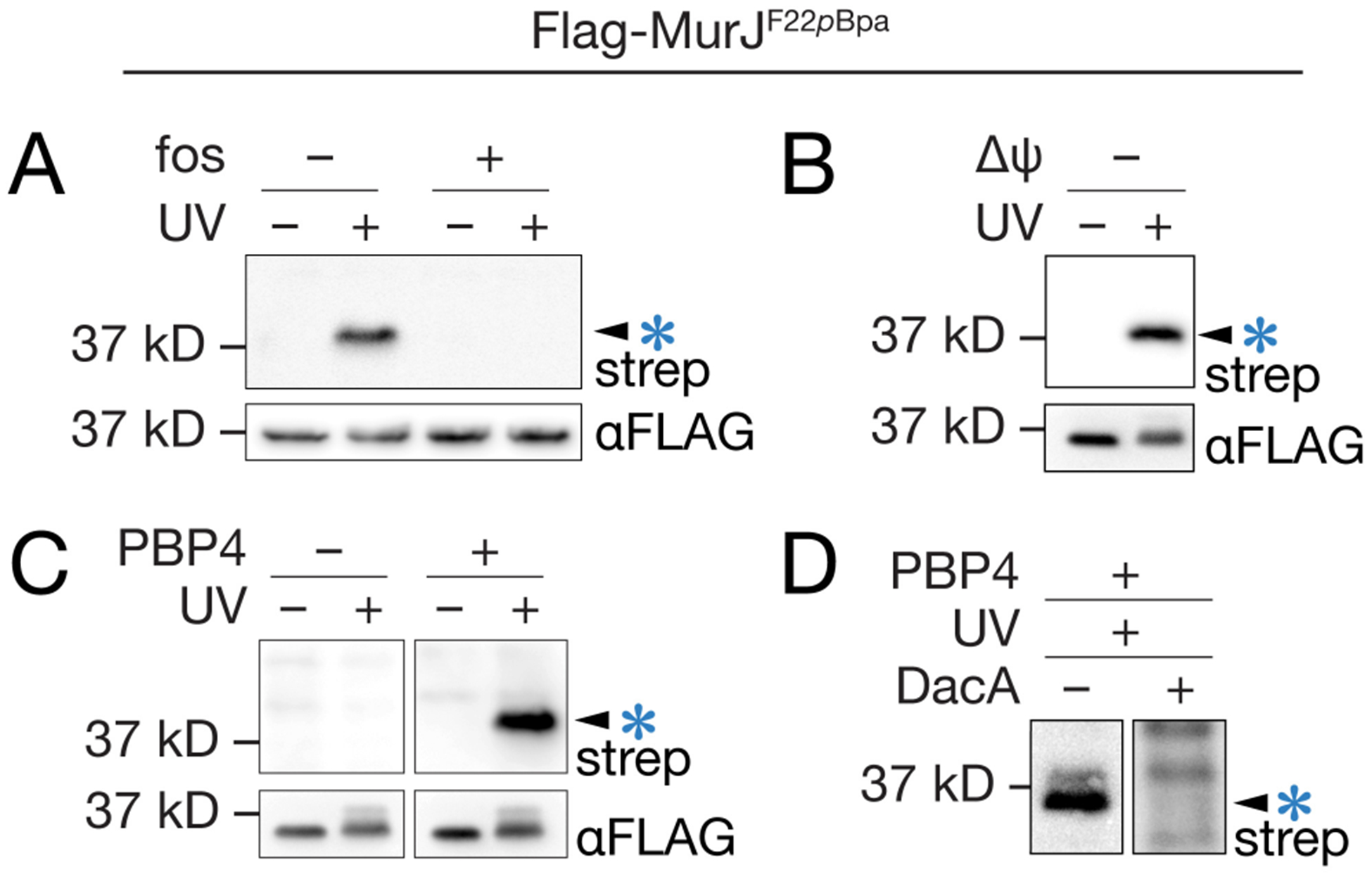
The photocrosslinked adduct is MurJ×Lipid II. (a) Cells treated with the Lipid II biosynthesis inhibitor fosfomycin (fos) did not produce a crosslinked adduct. (b) Immunopurification of Flag-MurJF22pBpa with α-Flag resin prior to SDS-PAGE, biotinylation and immunoblotting confirmed that the 37 kD band contains MurJ. (c) When the Lipid II labeling enzyme PBP4 was omitted from the procedure shown in Figure 1c, the MurJ adduct was not detected. (d) When electroblots were pretreated with the D,D-carboxypeptidase DacA, which cleaves the terminal D-Ala residue from Lipid II, the MurJ adduct was not detected. See also Figures S1, S4–S9.
We asked whether the species detected after accumulating Lipid II by blocking MurJ represented a physiologically-relevant transport state. If so, we reasoned that both Lipid II abundance and abundance of the crosslinked adduct should return to baseline cellular levels in a time-dependent fashion after MurJ was re-activated. We examined two reversible conditions to block MurJ activity and monitored both Lipid II levels7 and MurJ×Lipid II crosslinking. In one condition, we used CCCP to stall MurJ in an outward-open conformation7–8. We then monitored Lipid II crosslinking to MurJF22pBpa as a function of time after diluting cells to a sub-inhibitory CCCP concentration to allow the membrane potential to be re-established and MurJ conformational cycling to resume (Figure 3a,b, Figure S11). Lipid II levels remained elevated for five minutes after washout (Figure 3b, top), but returned to baseline by ten minutes. This time-dependent change in cellular Lipid II levels was mirrored in MurJ×Lipid II crosslink intensities (Figure 3b, bottom; note that crosslinking is performed for 5 min after the indicated washout times). Importantly, the increase of adduct formation in these conditions is specific to MurJ, as no crosslinking of Lipid II to the unrelated inner membrane protein GlpT could be detected (Figure S13)21. In the other condition, we used a thiol-labeling strategy to inactivate MurJ engineered to contain a Cys residue on the periplasmic apex of the cavity (MurJA29C).3, 7 We treated cells containing MurJF22pBpa/A29C with MTSES (2-sulfonatoethyl methanethiosulfonate) to sterically block conformational cycling3, 11 and then reversed the block by treatment with the reducing agent dithiothreitol (DTT) (Figure 3c,d). Similarly to the first condition, Lipid II levels were initially elevated, but returned to baseline within a few minutes of DTT treatment (Figure 3d, top). As above, the MurJ×Lipid II crosslink intensity mirrored the rise and fall of cellular Lipid II levels (Figure 3d, bottom). We observed an increase in MurJ×Lipid II adduct formation under conditions of MTSES treatment without added DTT (Figure 3d, bottom, lanes 1 & 2), but the adduct increased further when DTT was present during the 5 minute photocrosslinking before returning to baseline (Figure 3d, bottom, lanes 2 and 3). The crosslinked adducts in the inactivated MurJ (Figure 3d, bottom, lane 2) presumably result from the large accumulation of substrate at the stalled transporter (Figure 3d, top, lane 2). The simplest interpretation of these observations is that the increased crosslinking in lane 3 compared to lane 2 (Figure 3d, bottom) reflects reactivation of MurJ transport activity. Because Lipid II levels and abundance of MurJ×Lipid II adducts return to baseline in the same time frame after the MurJ block is lifted, it follows that the captured MurJ×Lipid II species represents an on-pathway intermediate in Lipid II transport.
Figure 3.
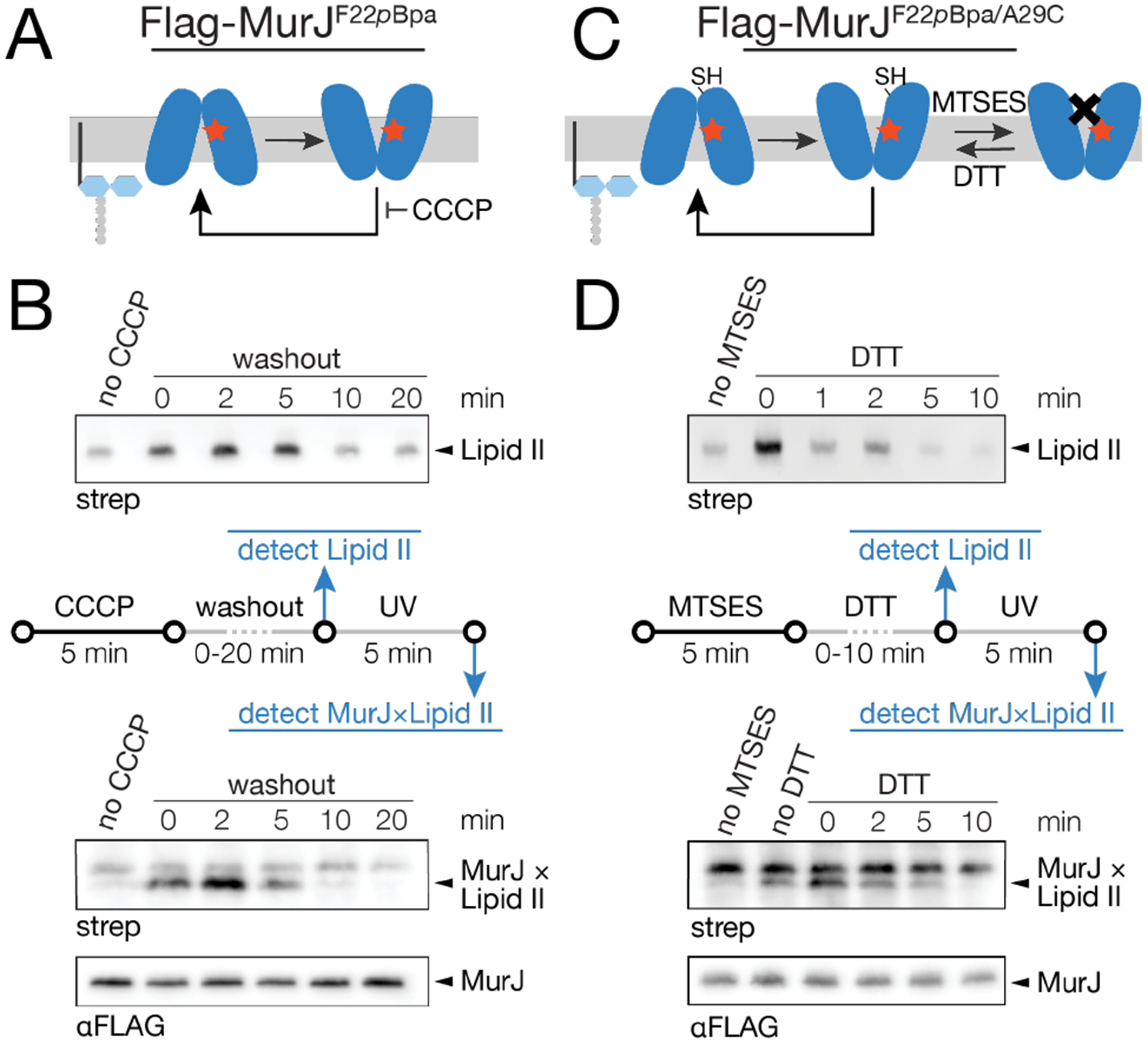
The captured MurJ×Lipid II species represents an on-pathway intermediate in Lipid II transport. (a) The protonophore CCCP inhibits MurJ cycling. (b) Cells were pre-treated with 100 μM CCCP for 5 min, then diluted 100-fold to wash out CCCP and restore MurJ function. After 0 – 20 min, Lipid II levels were assessed as previously described7. Cells were then UV-irradiated for 5 min and assessed for MurJ×Lipid II formation. (c) The MurJA29C variant is inactivated by the thiol-labeling probe MTSES. This inactivation is reversed by the reducing agent DTT. (d) Cells were pre-treated with 400 μM MTSES for 5 min then diluted 10-fold into media containing 10 mM DTT to restore MurJ function. After 0 – 10 min, Lipid II levels were assessed. Cells were then UV-irradiated for 5 min and assessed for MurJ×Lipid II content. See also Figures S1, S10–S13.
Having established that we can detect increased crosslinking to functional MurJ, we sought to investigate MurJ residues previously found to be critical for cell viability. Substrate recognition by MurJ is thought to be a key step in facilitating its transport cycle, and three arginine residues located midway through the central cavity of MurJ (R18, R24, R270) have been proposed to bind the pyrophosphate of the Lipid II headgroup (Figure 4a,b).10–12, 22–24 Because F22 is nestled between residues R18, R24, and R270, we reasoned that a cross-linker at this position would allow us to probe the contribution of this arginine motif to MurJ mechanism. We individually mutated these residues to alanine in a MurJF22pBpa background and assessed crosslinking to MurJ in the presence of a wild-type, non-crosslinking murJ allele. These single arginine mutants showed similar levels of crosslinked adducts as wild-type MurJ; even the triple mutant showed appreciable crosslinking (Figure 4c). Notably, mutants at these positions retain the ability to localize to sites of newly-synthesized Lipid II,21 consistent with our observation that they are competent for substrate binding. We further assessed Lipid II crosslinking in these mutants upon reactivation of MurJ. In contrast to wild-type MurJ, the arginine mutants retain bound Lipid II after restoration of membrane potential (Figure 4d, compare with Figure 3b; Figure S15). Because these mutants still bind Lipid II but are dramatically impaired in flipping, we conclude that these essential, conserved arginine residues are required for controlling movement of Lipid II through MurJ. The most likely way in which they might do this would be by facilitating conformational changes in the helices in response to substrate recognition in the cavity.12, 22–23 Destabilizing the inward-open state upon substrate binding would promote cycling between inward- and outward-open states (Figure 4e). Probing MurJ-Lipid II interactions beyond F22, however, will be needed to test the proposed model against alternative ones, such as impaired substrate release or transporter cycling.
Figure 4.
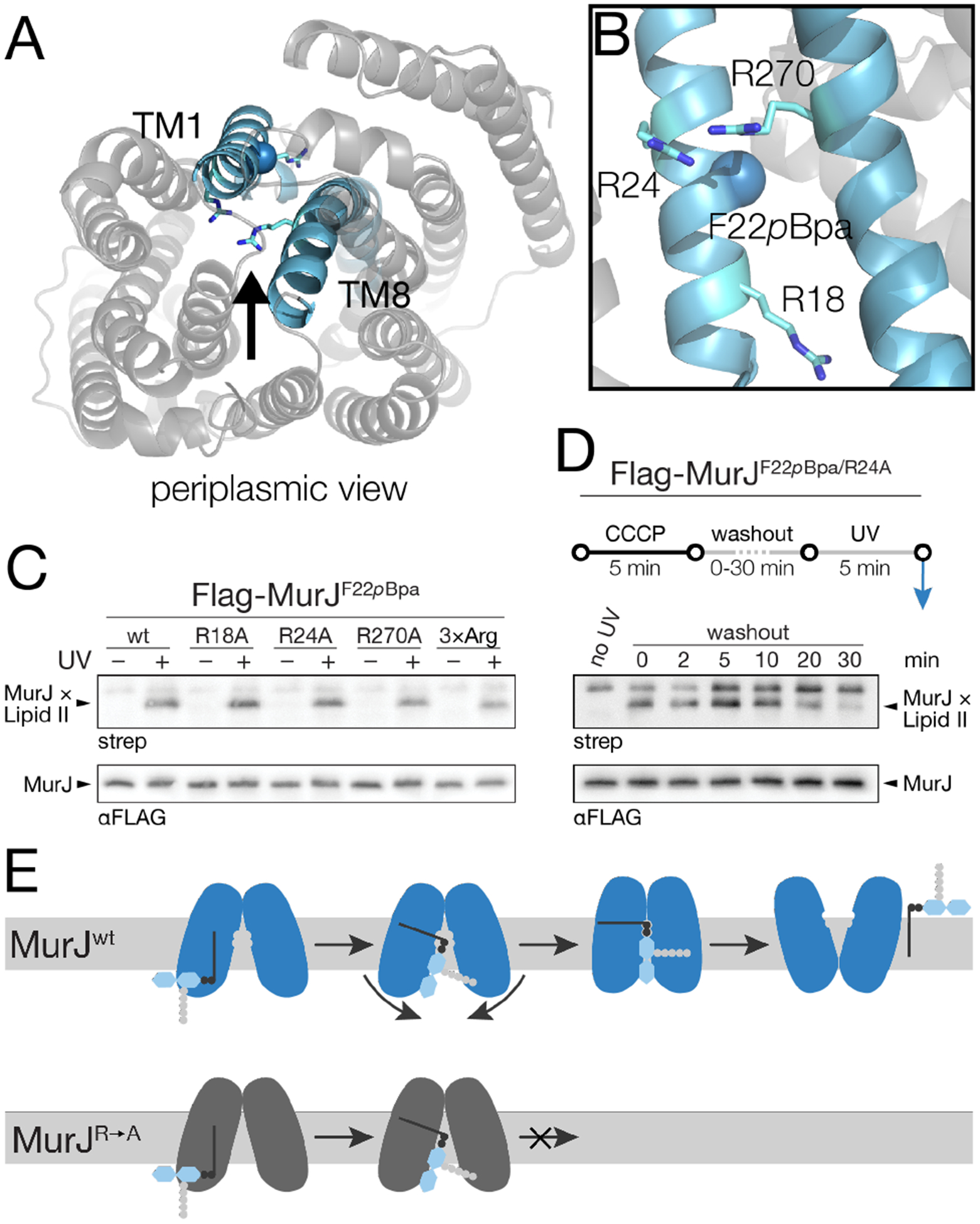
Lipid II recognition by three essential arginine residues triggers a conformational change in MurJ required for transport. (a) Crystal structure of E. coli MurJ in an inward-facing conformation (PDB ID code 6CC4), viewed from the periplasm. (b) TM1 and TM8 of MurJ as viewed from the central cavity (a, arrow). Essential residues R18, R24 and R270 surround photocrosslinking site F22pBpa. (c) Cells harboring both a wild-type murJ allele and Flag-MurJF22pBpa containing one or three R-to-A mutations were assessed for MurJ×Lipid II formation. (d) Cells in c were assessed for MurJ×Lipid II formation upon restoration of membrane potential as described in Figure 3b. (e) Proposed model depicting the role of Lipid II headgroup recognition by MurJ. The lipid tail of Lipid II first associates with MurJ near F22. Subsequent head-group recognition by R18, R24, and R270 mediates conformational rearrangement into an outward-facing state from which the substrate can be released. See also Figure S14–S15.
In this paper we have demonstrated a method to capture and detect transport intermediates of Lipid II on MurJ, the peptidoglycan flippase. We first accumulate Lipid II in the cytoplasm by reversibly blocking its transport and then probe its interaction with MurJ using photocrosslinking after relieving the block. In lieu of antibodies, which do not exist for Lipid II, we developed a D-amino acid exchange method to install a biotin in the crosslinked adduct. We have shown that we can detect on-pathway intermediates of MurJ and have used this capability to show that substituting essential residues in the cavity does not eliminate binding of Lipid II to MurJ at the entry gate. We anticipate that the ability to visualize Lipid II transport intermediates will not only enable dissection of the MurJ transport mechanism, but will allow characterization of MurJ inhibitors.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Institutes of Health (R01 GM100951 to N.R.; R01 GM076710 and R01 AI148752 to D.K. and S.W.).
Footnotes
Supporting Information. Experimental procedures and strain information. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Lovering AL; Safadi SS; Strynadka NC Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem 2012, 81, 451–478. [DOI] [PubMed] [Google Scholar]
- (2).Ruiz N Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 15553–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Sham LT; Butler EK; Lebar MD; Kahne D; Bernhardt TG; Ruiz N Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014, 345, 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hvorup RN; Winnen B; Chang AB; Jiang Y; Zhou XF; Saier MH Jr. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem 2003, 270, 799–813. [DOI] [PubMed] [Google Scholar]
- (5).Ficici E; Zhou W; Castellano S; Faraldo-Gomez JD Broadly conserved Na(+)-binding site in the N-lobe of prokaryotic multidrug MATE transporters. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E6172–E6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Drew D; Boudker O Shared Molecular Mechanisms of Membrane Transporters. Annu. Rev. Biochem 2016, 85, 543–572. [DOI] [PubMed] [Google Scholar]
- (7).Rubino FA; Kumar S; Ruiz N; Walker S; Kahne DE Membrane Potential Is Required for MurJ Function. J. Am. Chem. Soc 2018, 140, 4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kumar S; Rubino FA; Mendoza AG; Ruiz N The bacterial lipid II flippase MurJ functions by an alternating-access mechanism. J. Biol. Chem 2019, 294, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bolla JR; Sauer JB; Wu D; Mehmood S; Allison TM; Robinson CV Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ. Nat. Chem 2018, 10, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zheng S; Sham LT; Rubino FA; Brock KP; Robins WP; Mekalanos JJ; Marks DS; Bernhardt TG; Kruse AC Structure and mutagenic analysis of the lipid II flippase MurJ from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 6709–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kuk AC; Mashalidis EH; Lee SY Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat. Struct. Mol. Biol 2017, 24, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kuk ACY; Hao A; Guan Z; Lee SY Visualizing conformation transitions of the Lipid II flippase MurJ. Nat Commun. 2019, 10, 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Okuda S; Freinkman E; Kahne D Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science. 2012, 338, 1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Chin JW; Martin AB; King DS; Wang L; Schultz PG Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sarkar S; Libby EA; Pidgeon SE; Dworkin J; Pires MM In Vivo Probe of Lipid II-Interacting Proteins. Angew. Chem. Int. Ed. Engl 2016, 55, 8401–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Beres SB Photoaffinity Labeling of Membrane Proteins Involved in the Synthesis of Peptidoglycan. Ph.D. Dissertation, Northwestern University, Evanston, Illinois, 1995. [Google Scholar]
- (17).Qiao Y; Lebar MD; Schirner K; Schaefer K; Tsukamoto H; Kahne D; Walker S Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc 2014, 136, 14678–14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Qiao Y; Srisuknimit V; Rubino F; Schaefer K; Ruiz N; Walker S; Kahne D Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat. Chem. Biol 2017, 13, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Welsh MA; Taguchi A; Schaefer K; Van Tyne D; Lebreton F; Gilmore MS; Kahne D; Walker S Identification of a Functionally Unique Family of Penicillin-Binding Proteins. J. Am. Chem. Soc 2017, 139, 17727–17730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nicholas RA; Strominger JL Relations between beta-lactamases and penicillin-binding proteins: beta-lactamase activity of penicillin-binding protein 5 from Escherichia coli. Rev. Infect. Dis 1988, 10, 733–738. [DOI] [PubMed] [Google Scholar]
- (21).Liu X; Meiresonne NY; Bouhss A; den Blaauwen T FtsW activity and lipid II synthesis are required for recruitment of MurJ to midcell during cell division in Escherichia coli. Mol. Microbiol 2018, 109, 855–884. [DOI] [PubMed] [Google Scholar]
- (22).Butler EK; Davis RM; Bari V; Nicholson PA; Ruiz N Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J. Bacteriol 2013, 195, 4639–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Butler EK; Tan WB; Joseph H; Ruiz N Charge requirements of lipid II flippase activity in Escherichia coli. J. Bacteriol 2014, 196, 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sham LT; Zheng S; Yakhnina AA; Kruse AC; Bernhardt TG Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP-family flippases. Mol. Microbiol 2018, 109, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


