Abstract
Black soldier fly (BSF; Hermetia illucens L.) larvae can convert fresh pig manure into protein and fat-rich biomass, which can then be used as aquafeed for select species. Currently, BSF is the only approved insect for such purposes in Canada, USA, and the European Union. Pig manure could serve as a feed substrate for BSF; however, it is contaminated with zoonotic pathogens (e.g., Staphylococcus aureus and Salmonella spp.). Fortunately, BSF larvae inhibit many of these zoonotic pathogens; however, the mechanisms employed are unclear. We employed RNAi, qRT-PCR, and Illumina MiSeq 16S rDNA high-throughput sequencing to examine the interaction between two immune genes (Duox in Duox-reactive oxygen species [ROS] immune system and TLR3 in the Toll signaling pathway) and select pathogens common in pig manure to decipher the mechanisms resulting in pathogen suppression. Results indicate Bsf Duox-TLR3 RNAi increased bacterial load but decreased relative abundance of Providencia and Dysgonomonas, which are thought to be commensals in the BSF larval gut. Bsf Duox-TLR3 RNAi also inactivated the NF-κB signaling pathway, downregulated the expression of antimicrobial peptides, and diminished inhibitory effects on zoonotic pathogen. The resulting dysbiosis stimulated an immune response by activating BsfDuox and promoting ROS, which regulated the composition and structure of the gut bacterial community. Thus, BsfDuox and BsfTLR3 are important factors in regulating these key gut microbes, while inhibiting target zoonotic pathogens.
Introduction
Hermetia illucens L. (Diptera: Stratiomyidae) is a saprophytic insect whose larvae (BSFL) consume a wide range of organic wastes and convert them into biomass [1]. BSFL consuming livestock waste, such as pig manure, inhibit many associated zoonotic pathogen loads. For example, Liu et al. (2008) [2] determined BSFL can reduce Escherichia coli in dairy manure. Furthermore, Lalander et al. (2015) [3] discovered that BSFL reduce Salmonella spp. as well as viruses in organic wastes.
The mechanisms allowing BSFL to inhibit these zoonotic pathogens have been investigated. Park et al. (2015) [4] characterized an H.illucens defensin-like peptide which has activity against Gram-positive bacteria. Elhag et al. (2017) [5] identified seven gene fragments responsible for the production of three types of antimicrobial peptides. And, Zdybicka-Barabas et al. (2017) [6] determined E. coli-challenged BSFL had increased phenoloxidase, lysozyme and anti-Gram-positive bacterium activity.
At a much broader scale, insects rely on innate defense reactions to inhibit pathogens by producing antimicrobial peptides, phenoloxidase and H2O2. In Drosophila (Diptera: Drosophilidae), the Toll signaling pathway is mainly induced by Gram-positive bacteria and fungi [7]. In the sea urchin, Strongylocentrotus intermedius, petidoglycan and PolyI: C, but not LPS or ZOA, increased the expression of SiTLR11, which activated antifungal responses [8]. And, with mosquitoes (Diptera: Culicidae), the intestinal microbiota inhibits the development of Plasmodium spp. and other human pathogens through activation of the insect’s basal immunity [9].
Toll-like receptors (TLRs) are proteins present in cellular membranes that are capable of recognizing invading foreign bodies (sentinel cells). They are a type I membrane receptor with an extracellular amino terminus and a conserved cytoplasmic region. TLRs recognize specific molecular structures associated with microbial pathogens, which serve to active innate and adaptive immune responses. With routine microbial burdens, such as those found in the absence of infection, the Toll pathway is at low activation levels. However, acute pathogenic bacterial infection transiently increases nuclear factor kappa B (NF-κB)-dependent innate immune signaling.
The insect gut immune system produces microbicidal ROS by dual oxidase (Duox) to restrict the proliferation of invading microorganisms. In addition, ROS is involved in regulating the healing process of intestinal trauma in insects and also functions as a signaling molecule to initiate other self-balancing signaling pathways [10]. The intestinal bacterial community also is associated with host immunity and bacteriostasis. The microbiota modulates anti-pathogen effects of some immune genes plausibly through activating basal immunity [9]. For example, in the oriental fruit fly, Bactrocera dorsalis, (Hendel) (Diptera: Tephritidae) ROS, which is induced by the BdDuox gene; a gene that plays a key role in intestinal bacterial community homeostasis [11].
ROS serves as an important immune mechanism for many insects against pathogenic microorganisms, such as bacteria, fungi, entomopathogenic viruses, and parasites [12]. For example, when mosquitoes are exposed to Enterobacter spp., known to naturally occur in mosquitoes, they are less likely to be infected by Plasmodium parasites. ROS activation is suspected to serve as a primary mechanism inhibiting development of the pathogen in situ [13].
The Duox regulatory pathway also contributes to maintaining gut–microbe homeostasis in insects [14]. Gut membrane-associated proteins, such as Mesh, regulate Duox expression through an arrestin-mediated MAPK/JNK/ERK phosphorylation cascade and play an important role in controlling the proliferation of gut bacteria. Expression of both Mesh and Duox is correlated with the gut bacterial microbiome, which, in mosquitoes, increases dramatically soon after acquisition of a blood meal [15].
Recent surveys of BSF gut microbiota revealed a diverse community dominated by Bacteroidetes and Proteobacteria [16,17]. The microbiota of the anterior midgut of the BSF contained the greatest microbial diversity, which gradually decreased distally; however, in contrast, bacterial load increased. The native gut microbiota (i.e., indigenous) of a number of insects has been determined to provide immunity against select pathogens [18,19,20]. For example, the microbial community of the red flour beetle Tribolium castaneum, Herbst, (Coleoptera:Tenebrionidae) offers protection against Bacillus thuringiensis bv tenebrionis [21]. Experiments on silkworm Bombyxmori, L., (Lepidoptera: Bombycidae) demonstrated that lactic acid bacteria in the gut enhance host resistance against Pseudomonas aeruginosa. Despite the involvement of native gut microbiota in combating infections, the manner by which the immune system of BSFL regulates gut microbiota homeostasis to suppress zoonotic pathogens remains unknown.
In this study, we examined the antimicrobial activity of immune genes dual oxidase (BsfDuox) and TLR 3 (BsfTLR3) in BSFL, which represent two classic immune pathways. We explored the, (1) reduction in pathogen loads in pig manure by BSFL, (2) expression profile of immune genes BsfDuox and BsfTLR3 in BSFL, (3) expression profile of immune genes after oral pathogenic bacterial challenge, (4) whether suppression of zoonotic pathogens by BSFL is reduced after BsfDuox-TLR3 RNA interference, and (5) the dynamic change in intestinal bacterial community after RNAi of Duox-TLR3 genes and their relationship with the suppression of zoonotic pathogens.
Material and methods
Rearing and dissection of H. illucens L.
The colony of BSF (Wuhan strain) used in this study was located at the State Key Laboratory of Agricultural Microbiology of HZAU. BSFL were reared for 10 days at 27°C and 70%-80% relative humidity on an artificially sterilized feed (75 g of wheat bran, 75 g of corn flour, and 350 g of water mixed and then autoclaved at 121°C for 15 min). Third instars were surface-sterilized prior to use. After a 70% ethanol wash for one minute, the BSFL is washed three times in sterile water for one minute. Approximately 60 larvae from each experiment were dissected in sterile distilled water with a sterilized tweezers under a stereo microscope [22]. The dissected guts were transferred to a 2 ml sterile grind tube containing PBS(0.01 M, 1ml) and ground guts into homogenate. The homogenate was used for the different experiments.
Zoonotic pathogenic bacteria assay in pig manure conversion
One hundred of surface-sterilized BSFL at 8–10 days old were placed into 100 g of fresh pig manure collected from a facility located near the university. In addition, the control group consisted of 100g pig manure. After interfering with the genes of Duox and TLR3, the transformation experiments were also carried out. 105 CFU/g Salmonella spp. and 106 CFU/g S. aureus, as well as 100 Duox-TLR3 RNAi-injected larvae, were added to the 100 g sterilized feed. All treatments were replicated three times. The method for detecting targeted zoonotic pathogens was based on the National Standards of China GB4789.10–2010, in which Staphylococcus aureus was detected in 8 days [23]. The methods are described as follows. The selective medium (purchased from Qingdao Hope Bio-technology Co., Ltd) for S. aureus was prepared by placing 6.3 g of Baird-Parker agar in 95 ml of distilled water, heated to a boil until completely dissolved, autoclaved at 121°C for 15 min, and agitated well after sterilization, thereby preventing agar from depositing at the bottom and solidifying. The medium was then cooled to 50°C, 5 ml of potassium citrate-potassium yolk enrichment solution added, gently agitated, and poured into a plastic plate(diameter:90 mm). To count S. aureus colonies, we first collected manure samples (5 g) at different times after larval introduction (0, 2, 4, 6, and 8 days). Samples were mixed with 45 ml of sterile physiological saline in a 100 ml sterile glass conical flask and agitated for 16 min at 180 rpm. The sample was stored at room temperature (RT) for 5 min prior to the upper layer be sampled. Each sample was diluted using 0.9% saline solution. For each dilution, 0.1 ml was mixed with the appropriate selective medium and plated. Pathogen counts were determined using selective plates, following incubation of plates at 37°C for 48 h. S. aureus colonies exhibited a diameter of 2–3 mm, gray to black appearance, light-colored edge, turbid zone, and transparent ring on the outer layer. The mean S. aureus count of three plates was expressed as logCFU/g.
Sample processing to perform the counts was performed in the same way for Salmonella spp. as for S. aureus. Briefly, triplicate plating of the previous samples described was used following GB 4789.4–2016 [24] (Standards Press of China, 2010). The medium used for isolating Salmonella spp. growth(Qingdao Hope Bio-technology Co.,Ltd) was prepared by adding 5 g of bismuth sulfite agar to 100 ml of distilled water that was then stirred and boiled for 1 min, cooled to 50°C-55°C, plated, and used the next day. The mean Salmonella spp. (CFU/g) of triplicate plates was determined as logCFU/g(Standards Press of China, 2016).
Total RNA isolation and cDNA synthesis
Total RNA was isolated from black soldier fly at different developmental stages, including eggs, first-fourth instars, adults after mating (13–15 days after eclosion) using RNA extract reagent kit (Invitrogen, Thermofisher, USA).Taking 100 steriled (90% alcohol wash for 1 minute and distilled water wash for 1 minute) eggs in the egg-laying carton of the adult BSF and grind them in trizol extraction kit to extract RNA(Invitrogen, Thermofisher, USA) according to the manufacturer instructions. In addition, total RNA was isolated from different organs and tissues such as gut and the whole body. In addition, total RNA was isolated from RNAi-Duox-TLR3 and RNAi-egfp groups. The experiments were performed three times. RNA quality was analyzed by NanoDrop 2000 spectrophotometry (ThermoFisher Scientific Inc., Waltham, MA, USA) at 260 nm. 1 μg of total RNA was reversely transcribed into first-strand cDNA by using Hiscipt 1st stand cDNA synthesis kit (Nanjing Vazyme Biotechnology Co., Ltd, Nanjing, China).
Sequence analysis of full-length BsfDuox and BsfTLR3
The genome of BSF was sequenced and assembled. The genome size of BSF was 1.1 Gb, which is relatively large among the Diptera that has been sequenced [25]. The BsfDuox and BsfTLR3 genes were searched in the BSF genome database by using the Duox protein sequence and TLR3 protein sequence of the fruit fly. The highest similarity sequence of protein was the protein sequence of Duox and TLR3 of BSF. Specific primers for BsfDuox and BsfTLR3 were designed on the basis of the fragment sequence searched from BSF genome database using Premier5.0 software. The transmembrane domains in BsfDuox and BsfTLR3 were identified using TMHMM online software (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and the structural domains of BsfDuox and BsfTLR3 were predicted using the simple modular architectural research tool (SMART, version 7.0, http://smart.embl-heidelberg.de/).
Microbial oral infection
Third-instar fed an artificial diet supplement with 5% sucrose solution containing concentrated microbe solution (1 × 108 colony-forming units (CFUs) per ml).The microorganisms used in this study were zoonotic pathogens, S. aureus and Salmonella spp. from the State Key Laboratory of Agricultural Microbiology of HZAU. Bacteria used for oral infection were grown in lysogeny broth medium at 28°C and 180 rpm. Exponential microbial culture (OD600 = 1.0) was used for all the experiments as previously described [26] and centrifuged for 15 s (8,000 g), then adjusted to the final concentration with aseptic distilled water. The resulting bacterial counts in each sample were adjusted to approximately 1×108 CFU/ml by aseptic distilled water. We did a preliminary experiment initially to establish appropriate methods. The sample of different dilutions were applied on LB plates after overnight culture at 37°C, the number of bacteria on the plate was counted to calculate the number of acquired bacteria by per larvae, then measure the turbidimetric OD value of the sample by spectrophotometer, it will take several days. For formal experiment, the bacterial turbidimetric OD value of all the sample was measured by spectrophotometer refer to the turbidimetric OD value of the preliminary experiments results.
Third-instars (8 days old) were fed a 1 ml solution containing concentrated microbes(approximately 1×108 CFU/ml). The larvae fed with a 5% sucrose diet only served as the control. For the analyses of BsfDuox and BsfTLR3 gene expression and ROS level changes after oral infection, the gut samples of different treatments were collected at different times post-oral infection(POI).
Real-time quantitative PCR (qRT-PCR) analysis
In all cases of gene expression analysis, three independent cohorts of third-instar BSFL were collected for RNA extraction and cDNA synthesis. In the present work, genes, such as Duox, TLR3, egfp, drosal, cecropin, ubiquitin, dif and stomoxyn were investigated for their expression level at the third-instar stage. The primers for these gene was shown in S1 Table. qRT-PCR was performed using a Bio-Rad CFX system (Bio-Rad, Hercules, CA, USA) with a 384-well plate. Each PCR mixture consisted of 7.8 μl of SYBR Green Mix (Hiscipt 1st stand cDNA synthesis kit, Nanjing Vazyme Biotechnology Co., Ltd, Nanjing, China), 10 nM of each primer, and 2 μl of cDNA (diluted 1:10). The amplification program consisted of pre-incubation at 95°C for 3 min, followed by 39 cycles of denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 72°C for 20 s. After the fluorescence quantitative PCR was over, the dissolution curve was analyzed to ensure specific amplification followed by 40 cycles started at 65°C for 10 s with a 0.5°C increase for 5 s each cycle until 95°C. Real-time fluorescence quantitative PCR results were measured by 2-ΔΔCt method as described previously [27]. All the samples were analyzed in triplicate, and the levels of the detected mRNA determined by cycling threshold analysis were normalized using β-actin as the control, The target gene expression is presented as the relative expression levels after normalization. The loads of total bacteria were quantified by qRT-PCR using 16S rRNA gene-specific primers (F 5’-ACTCCTACGGGAGGCAGCAG and R 5’-ATTACCGCGGCTGCTGG) [28] and normalized by using β-actin as the control via a previously described method [29]. PCR mixture consisted of 7.8 μl of SYBR Green Mix (Hiscipt 1st stand cDNA synthesis kit, Nanjing Vazyme Biotechnology Co., Ltd, Nanjing, China), 10 nM of each primer, and 2 μl of cDNA (diluted 1:10). The amplification program was the same as described above. All the samples were analyzed using ANOVA method.
Measurement of intestinal ROS
The intestine of individual third-instar larvae was hand-dissected in PBS(0.01 M, 1ml). Three independent cohorts of three intestines were used for each measurement. The dissected intestines were ground with PBS solution(0.01 M, 1ml) in the grinder. The quantification of ROS was completed according to the corresponding kit instructions provided by the Institute of Nanjing Jiancheng Bioengineering. In brief, dissected intestines were weighed using a weighing meter, then was added nine times volume of 0.9% saline, centrifuged the sample at 5,000 g for 5 min, the supernatant was further used for the colorimetric quantitative determination of diffused ROS. The absorbance values of each tube were measured under 405 nm by using UV756 (756 spectrophotometer, Shanghai Opal Instrument Co., Ltd.). The spectrophotometer was preheat for 30 min or more and adjust the wavelength to 405 nm, using distilled water adjust to zero. We detected ROS after a 30-min incubation of the diluted supernatant with the working solution I, II, III, IV(Volume ratio:10:0:1:2:10). Sample was added working solutionI, II, III and centrifugated (4000 g, 25°C, 10 min) then discarded the supernatant. Adding the working solution IV to solve the precipitation and take 200 μl to the colorimetric dish to determine the absorbance value .ΔOD = determined OD-referenced OD. The protein concentration of each tube was determined by NanoDrop2000 (ThermoFisher Scientific Inc., Waltham, MA, USA). The ROS value of each tube was calculated using the following formula: ROS concentration = (measured OD value−blank OD value) × 163 / (Standard OD value−blank OD value) × sample protein concentration.
Double-stranded RNA (dsRNA) synthesis and delivery by injection
The BsfDuox and BsfTLR3 sequence fragments were amplified with PCR with premier 5.0 to design of specific primers conjugated with the T7 RNA polymerase promoter. The primer pairs used in dsRNA synthesis are shown in S1 Table.1 μg PCR product was used as the template for dsRNA synthesis utilizing the T7 Ribomax Express RNAi System (Promega, Madison, WI, USA). The dsRNA was isopropanol-precipitated overnight, resuspended in RNase-free H2O, and quantified at 260 nm by using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific Inc., Waltham, MA, USA) before microinjection. The quality and integrity of dsRNA were determined by agarose gel electrophoresis. The injection condition was set to Pi of 300 hpa and Ti of 0.3 s using an Eppendorf micromanipulation system (Microinjector for cell biology, FemtoJet 5247, Hamburg, Germany). Gene silencing experiments of Duox-TLR3 RNAi and egfp RNAi larvae were performed by injecting 1 μl of 2 μg/μl dsDuox-RNA and dsTLR3-RNA solution as well as dsegfp-RNA into the abdomen of each larva.
Isolation of bacterial DNA from the gut and high-throughput sequencing
Total bacterial genomic DNA samples from the intestines of 21 individuals were extracted using Fast DNASPIN extraction kits (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions and stored at −20°C prior to further analysis. The quantity and quality of extracted DNA were measured using a NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. PCR amplification of the bacterial 16S rDNA genes to assess the microbial diversity of the BSFL at the V3–V4 regions was performed using 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). These primers were designed to contain a 7 nt barcode sequence for multiple samples. PCR was performed in a total reaction volume of 25 μl. The PCR conditions were as follows: initial denaturation at 95°C for 5 min; 30 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 2.5 min. PCR amplicons were purified with Agencourt AMPure beads (Beckman Coulter, Indianapolis, IN, USA) and quantified using the PicoGreen dsDNA assay kit (Invitrogen, Carlsbad, CA, USA). Purified PCR product libraries were quantified by Qubit dsDNA HS Assay Kit, amplicons were pooled in equal amounts, and pair-end 2 × 300 bp sequencing was performed using the Illumina Miseq PE2500 platform with MiSeq Reagent Kit version 3 (Shanghai Personal Biotechnology Co., Ltd., Shanghai, China). A total of 21 gut samples were subjected to high-throughput sequencing, including three gut samples of 8- to 10-day-old larvae with no treatment of RNAi and 18 gut samples of the egfp RNAi- and Duox-TLR3 RNAi-treated larvae from 4, 8, and 12 days post-RNAi (DPR).
The raw data have been upload to NCBI under BioprojectID PRJNA600829 and SRA under SRP247530. Data analysis was conducted by the bioinformatics software called Quantitative Insights into Microbial Ecology (QIIME, v. 1.8.0) [30]. We get clean sequence reads by removing of the primer sequence, truncation of sequence reads less than an average quality of 20 over a 30 bp sliding window based on the Phred algorithm, and removal of sequences that had a length of <150 bp, as well as sequences that contained mononucleotide repeats of >8 bp [31]. These strict criteria resulted in nearly 94% of the reads being retained.
FLASH (Fast Length Adjustment of Short reads)was used to extend the length of short reads by overlapping paired-end reads for genome assemblies [32]. First, we sorted exactly the same sequence of clean reads according to their abundance and filtered out the singletons and used Usearch tool to cluster under 0.97 similarity. After chimera detection the remaining high-quality sequences were clustered into operational taxonomic units(OTUs) by UCLUST [33]. Selecting the highest abundance sequence from each OTU Library as the representative sequence by using default parameters. OTU taxonomic classification was conducted by BLAST search of the representative sequences set against the 16S database of known species (RDP, http://rdp.cme.msu.edu) [34] using the best hit [35]. Reads that did not match a reference sequence at 97% identity were discarded. Bioinformatics and sequence data analyses were mainly performed using QIIME and R packages (version 3.2.0). Using QIIME software calculates the alpha diversity index of a sample including richness and diversity indices (observed species [Sobs], Chao, abundance-based coverage estimator [ACE], and Shannon) and dissimilarity matrices (Bray–Curtis and weighted UniFrac) [36,37].
Statistical analysis
Results are shown as the average ± SEM of three independent biological samples. Each experiment was performed three times. Comparison between the two independent samples were performed with student's t-test. Multiple comparisons were conducted by one-way ANOVA (GraphPad Software, La Jolla, CA, USA). Significant level was set at p < 0.05. The graphs were also made using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
Results
BSFL significantly inhibited pathogenic bacteria in the conversion of pig manure
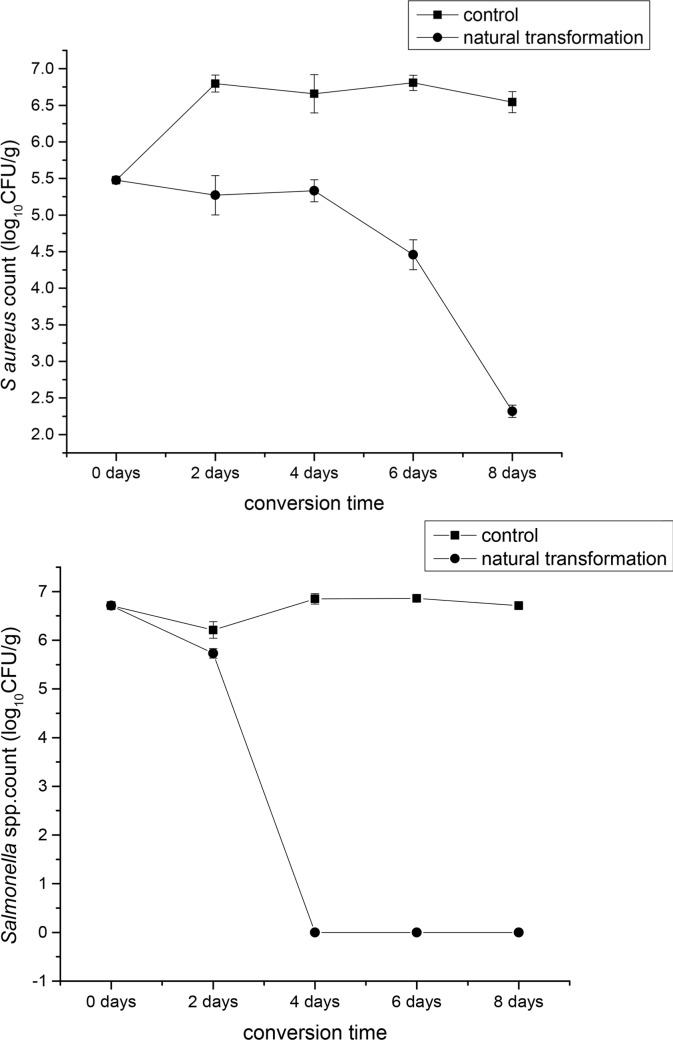
We investigated the inhibitory action of BSFL in the natural conversion of pig manure. BSFL significantly reduced S. aureus and Salmonella spp. counts in pig manure from day 2 through day 8. The number of S. aureus and Salmonella spp. was 5.47 log CFU/g and 6.71 log CFU/g on day zero, respectively. On the eighth day after conversion, the number of S. aureus and Salmonella spp. reduced to 2.31 log CFU/g and 1 log CFU/g, respectively. BSFL exhibited the greatest success in reducing S. aureus from days 2 until days 8. The inhibition rate increased significantly from the fourth to the sixth day, and reached the peak on the eighth day while the number of S. aureus in pig manure decreased to the lowest. The inhibition rate of the BSFL to Salmonella spp. reached the highest from the second day to the fourth day, after which the inhibition rate dropped to 0 until the eighth day (Fig 1).
Fig 1.
Mean Staphylococcus aureus (A) and Salmonella spp. (B) log CFU/g (mean ± SEM) during 8 days conversion in pig manure with 8-d-old black soldier fly larvae (control) and stored at 27°C, 60%-70%RH, and a photoperiod of 16:8 (L:D) h in a growth chamber.
Attenuation of bacteriostasis after Duox-TLR3 RNAi
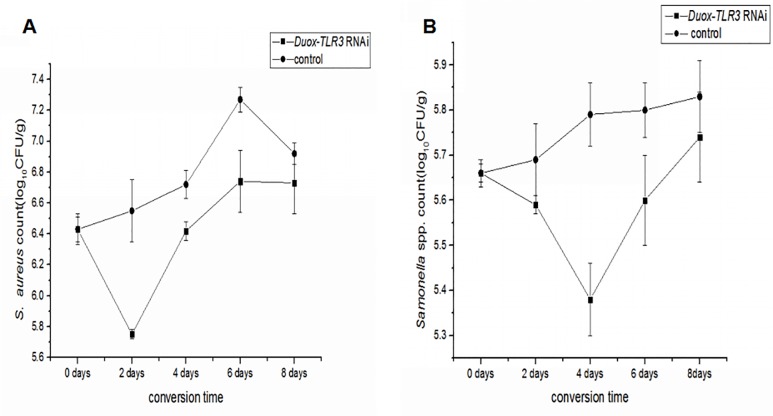
After RNAi of Duox-TLR3 in BSFL, we simulated the experiment of pig manure conversion by BSFL using the sterilized feed. By compared with pig manure, the use of sterilized feed(add only S.aureus and Salmonella spp.) can reduce the interference of other pathogenic bacteria(see materials and methods). On day 0 and 8th of conversion, the S. aureus count was 6.43 log CFU/g and 6.73 log CFU/g, respectively, and the Salmonella spp. count was 5.66 log CFU/g and 5.74 log CFU/g, respectively. In the Duox-TLR3 RNAi group, the BSFL were also able to inhibit S.aureus on day 0 to day 2 of conversion. The inhibitory effect on S. aureus was reduced from 2th day to 8th day. Meanwhile, the BSFL were also able to inhibit Salmonella spp from day 0 to day 4 of conversion. The inhibitory effect on Salmonella spp. was reduced from the 4th to 8th day. Thus, the BSFL induced a diminished effect on pathogen inhibition in Duox-TLR3 RNAi-injected larvae compared with the natural pig manure transformation conditions (Fig 2).
Fig 2.
Mean Staphylococcus aureus (A) and Salmonella spp. (B). log CFU/g± SD during 8 days conversion in Duox-TLR3 RNAi larvae group with (control) 8-d-old black soldier fly larvae and stored at 27°C, 60–70%RH, and a photoperiod of 16:8 (L:D) h in a growth chamber.
Sequence analysis and expression profiles of BsfDuox and BsfTLR3
We searched the BsfDuox and BsfTLR3 cDNA from the genome database of the BSF. BsfDuox was 4.2 kb and encoded 1540 amino acids (S1 Fig), whereas BsfTLR3 was 1.134 kb and encoded 378 amino acids (S2 Fig). Structural analyses demonstrated that BsfDuox consists of FAD-binding and NAD-binding domains and ferric-reduct domain typical of all members of electronic delivery system responsible for H2O2 generation, as well as extracellular peroxidase homology domain (PHD). The PHD shows myeloperoxidase activity, thereby enabling the conversion of H2O2 to HOCl in the presence of chloride ion. PHD of Duox is vital for the host immune defense system (S3 Fig). BSFL have one TLR3 gene, which primes the immune response. Structural analyses demonstrated that TLR3 had three Leucine-rich repeat domains and was responsible for the protein binding and cell signaling functions (S4 Fig). The structure of BsfDuox and BsfTLR3 was similar with the previously reported Duox and TLR3 of B. dorsalis, suggesting that they may have the same function.
The BsfDuox gene was highly expressed in the egg, first, second, and fourth instar as well as adult stages, but it was weakly expressed in the third-instar larval stage (S5A Fig). The BsfTLR3 gene was highly expressed in the egg, first, second, third, and fourth instar stages, but it was weakly expressed in the adult stage (S5B Fig). We speculated that the two genes have different expression level to meet their different functions at different stages of BSF development.
BsfDuox and BsfTLR3 genes were induced upon infection
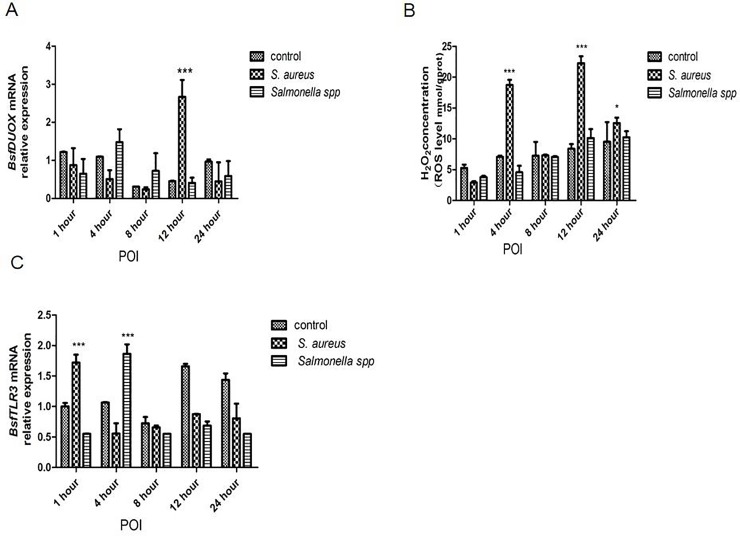
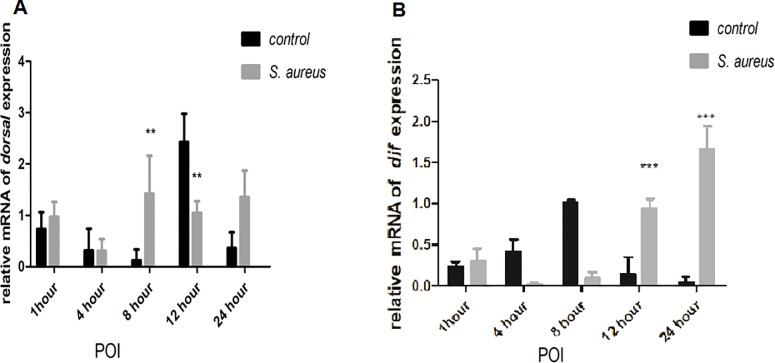
To investigate the role of BsfDuox and BsfTLR3 in the immune system response, we detected BsfDuox and BsfTLR3 gene expression in the gut upon oral infection with S. aureus and Salmonella spp. More specifically, S. aureus induced a 4.05-times in BsfDuox gene expression with a peak at 12 h POI (Fig 3A). Consistent with this, ROS level was increased at 12 h (Fig 3B). Salmonella spp. did not activate the Duox gene at any point significantly. Salmonella spp. and S. aureus differently induce the expression of Duox gene. Thus, BsfDuox gene expression may be regulated by factors secreted by S. aureus. To date, uracil secreted by bacteria is the only bacterial ligand [38] that can activate Duox activity. Our results showed that S. aureus induced a 1.76-fold increase in BsfTLR3 gene expression at 1 h POI, whereas Salmonella spp. induced a 1.75-fold increase in BsfTLR3 gene expression at 4 h POI. Thus, gram-positive and gram-negative bacteria could induce TLR3 gene expression (Fig 3C). This study also showed that oral S. aureus infection to BSFL could induce the expression of the toll-like receptor pathway nucleic acid transcription factors Dif and Dorsal to mediate antibacterial activity (Fig 4).
Fig 3. The response of the BsfDuox gene and BsfTLR3 gene in the gut during oral infection.
(A) Expression levels of BsfDuox at different time points in whole guts (without Malpighian tubules) after oral infection with S. aureus and Salmonella spp. (B) Expression levels of BsfTLR3 at different time points in whole guts (without Malpighian tubules) after oral infection with S. aureus and Salmonella spp. (C) The total intestinal ROS levels were quantified with flies at different time points after oral infection. Data are representative of three independent experiments (mean ± SEM). Statistical comparison was based on Student’s t–test (*p< 0.05,**p< 0.01, ***p< 0.001). Different letters indicate a significant difference in BsfDuox expression and BsfTLR3 expression and ROS levels among the oral infection with Salmonella spp. or S. aureus.
Fig 4.
(A) The expression level of Bsf dorsalgene at different time points after oral infection of S. aureus (B) The expression level of Bsf dif gene at different time points after oral infection of S. aureus.Values are the mean ± SEM of three independent experiments. Statistical comparison was based on Student’s t–test(*p< 0.05,**p< 0.01, ***p< 0.001 with Student’S t–test).
Sequential difference between the intestinal Duox-ROS/Toll signaling pathway after zoonotic pathogen challenge
We studied the temporal differentiation of the immune response of Duox-ROS immunity and Toll signaling pathway by feeding the zoonotic pathogen S. aureus to larvae. Infected by zoonotic pathogens could induce the immune response of the Duox-ROS system and Toll signaling pathway in a short time.
S. aureus induced a 2.65-fold increase in ROS in the host intestine after 4 h (Fig 3B) but a two-fold increase in Bsfdorsal gene in the Toll signaling pathways after 8 h (Fig 4A). The effector gene Dif increased by six-fold after 12 h POI compared with the control (Fig 4B). Thus, the immune response of the Duox-ROS system to zoonotic pathogen challenge was earlier than that of the Toll pathway.
BsfDuox-TLR3 regulated gut bacterial density
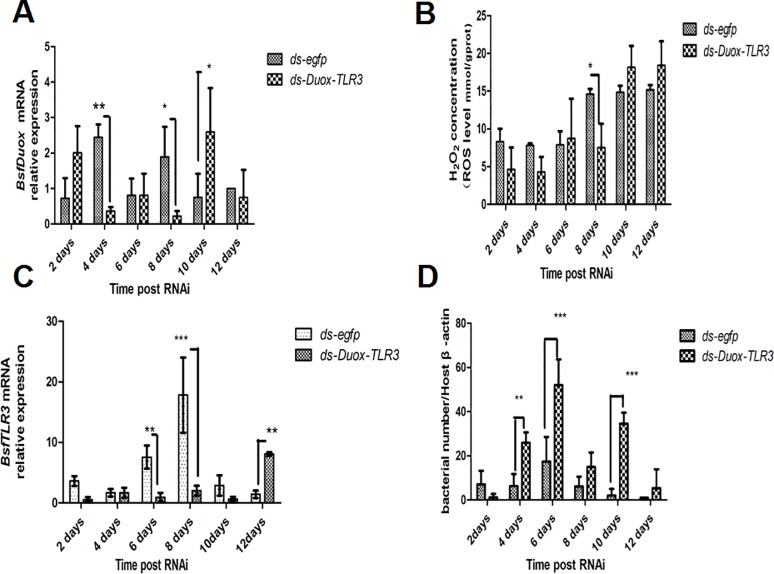
On the basis of the above results (Fig 3), we concluded that the BsfDuox and BsfTLR3 genes were activated differentially with zoonotic pathogens. Immune genes Duox and TLR3 are expressed to control gut microbiota. It was not H2O2 the only measurable bioactive compound controlling the microbial load. Subsequently, we tested the change in composition of different intestinal symbionts that reacted to the silencing of the Duox-TLR3 gene by injecting larvae with ds-Duox-TLR3. The level of BsfDuox gene transcript was inhibited at 4–8 days, BsfDuox gene expression varied from 85% to 88% compared with the egfp RNAi control (Fig 5A). However, BsfDuox expression increased in Duox-TLR3 RNAi-treated larvae at 10 days and then returned to the basal expression level at 12 days. The level of ROS followed the same pattern with a decrease of 48%-57% as compared with the egfp RNAi control at 4–8 days and with an increase of 21% at 10–12 days (Fig 5B). The level of BsfTLR3 expression was inhibited at 6–10 days, BsfTLR3 gene expression varied from 77% to 88% compared with the egfp RNAi control. BsfTLR3 gene expression was 4.48 times higher in Duox-TLR3 RNAi-treated larvae at 12 days than in the egfp RNAi control (Fig 5C). Interestingly, the analysis of the overall bacterial density by qRT-PCR showed that Duox-TLR3 RNAi larvae had more bacteria at 4–10 days compared with the control. The bacteria load returned to the wild-type level at 12 days (Fig 5D).
Fig 5. Interference effects of RNAi of the BsfDuox-TLR3 gene.
(A) The expression level of the BsfDuox gene at different time points after injecting ds-Duox-TLR3 and ds-egfp. (B) The expression level of the BsfTLR3 gene at different time points after injecting ds-Duox-TLR3 and ds-egfp. (C) The total intestinal ROS levels at different time points. (D) The total gut bacterial load at different time points. Data are representative of three independent experiments (mean ± SEM). Statistical comparison was based on Student’s t–test (*p<0.05, **p<0.01,***p< 0.001).
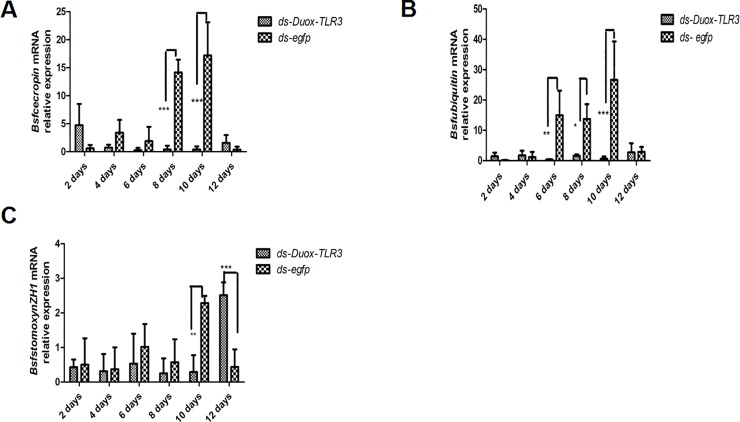
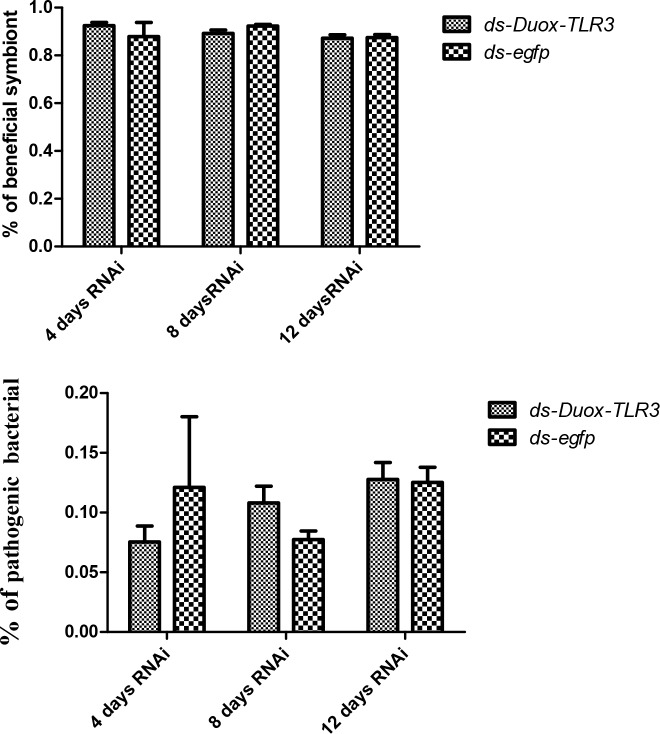
BSFL feeding on a diet containing high bacterial loads could induce the production of an expanded spectrum of antimicrobial peptides (AMPs) [39]. However, the expression of AMP was also down-regulated after interfering with two immune genes. The AMP level of cecropin expression in Duox-TLR3 RNAi larvae decreased by 26.14–36 times compared with that in the control at 8 and 10 days (Fig 6A). The AMP level of ubiquitin expression in Duox-TLR3 RNAi larvae decreased by 8.71–41.55 times compared with that in the control at 6, 8, and 10 days (Fig 6B). The AMP level of stomoxyn ZH1 expression in Duox-TLR3 RNAi larvae decreased by 7.93 times compared with that in the control at 10 days (Fig 6C). The results of 16s DNA sequencing of the intestinal tract of BSF showed that the beneficial symbiont bacteria in Duox-TLR3 RNAi-injected larvae were 3.4% lower than that in egfp RNAi-injected larvae [40] (Fig 7A), whereas the pathogenic bacteria in Duox-TLR3 RNAi larvae was less than the control group at 4 days while increased by 44.8% compared with that in egfp RNAi larvae at 8 days post-RNAi (DPR; Fig 7B). However, these differences were not statistically significant. The depletion of these two immune genes in BSFL reduced AMP and ROS production, leading to the decrease in symbiont bacteria and increase in pathogenic bacteria.
Fig 6.
The expression level of antimicrobial peptide genes (A) Bsf cecropin, (B) Bsf ubiquitin, (C) Bsf stomoxynZH1, in black soldier fly larvae following the Duox and TLR3 RNA interference. Data are representative of three independent experiments (mean ± SEM). Statistical comparison was based on Student’s t–test (*p<0.05, **p<0.01, ***p< 0.001).
Fig 7.
(A) The beneficial symbiont in Duox-TLR3 RNAi larvae and egfp RNAi larvae at 4,8,12 days post dsRNA interfence (B) The pathogenic bacteria in Duox-TLR3 RNAi and egfp RNAi larvae at 4, 8, 12 days post dsRNA interfence. Values are the mean ± SEM of three independent experiments.
The interference of BsfDuox-TLR3 gene reduced the ROS and AMP expression levels, resulting in an increase in the number of intestinal bacteria. We attributed the low BsfDuox and BsfTLR3 gene expression levels and ROS levels observed at 4–8 days to the increase in bacteria at a time when RNAi was effective. Low levels of gene expression after interference led to an increase in the total number of bacteria, as well as an increase in the number of pathogenic bacteria, which can stimulate gene expression to return to normal after interference that is the activities that take the observed number of days to recover normal activity. Conversely, high levels of BsfDuox gene expression and ROS at 10 days resulted from the decrease in bacteria at a time when RNAi was ineffective. Thus, the larval innate immune genes BsfDuox and BsfTLR3 were in regulating the homeostasis of the gut bacterial and in controlling the bacterial load in the midgut, and exposure to increased pathogens resulted in the increased production of some of these anti-pathogenic factors.
BsfDuox-TLR3 regulated the intestinal bacterial community homeostasis
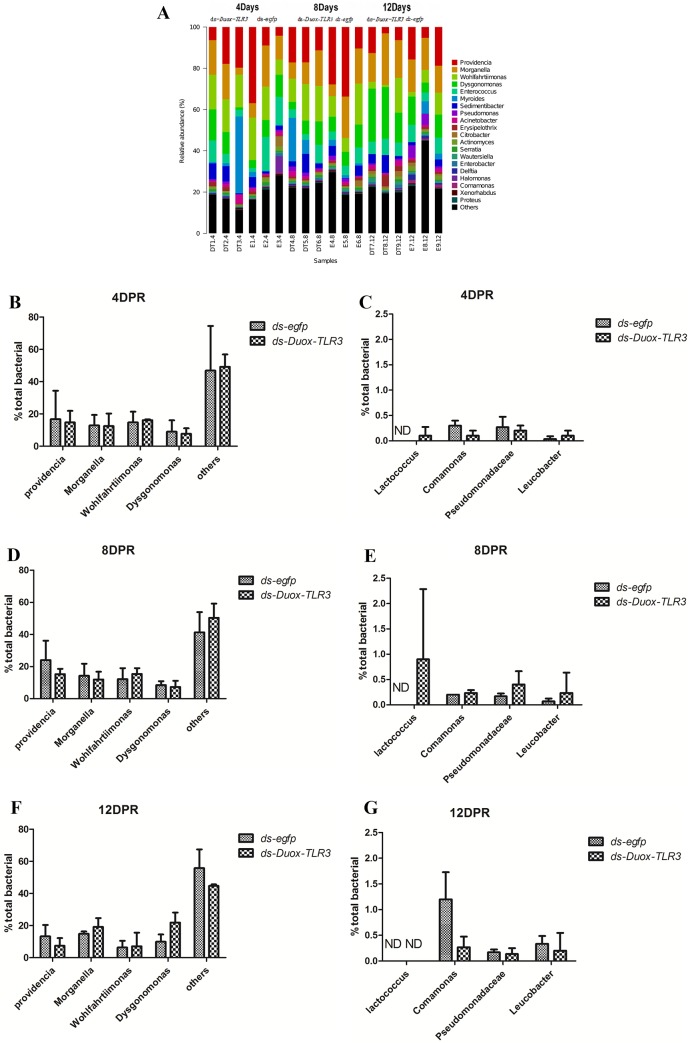
BsfDuox and BsfTLR3 regulated bacterial community density but whether BsfDuox and BsfTLR3 affect the gut bacterial community composition remains unknown. The bacterial composition in egfp RNAi-treated and Duox-TLR3 RNAi-treated larvae was investigated by MiSeq Illumina high-throughput sequencing. The rarefaction curves moved toward saturation representing the bacterial community well (S6A Fig). Rank abundance curve showed a rich species composition (S6B Fig). Overall, five bacterial phyla were detected in BSF samples, namely, Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and TM7, which composed 63.2%, 18.7%, 16.4%, 1.29%, and 0.41% of the bacterial communities in the gut, respectively. Differences between the bacterial community of egfp RNAi and Duox-TLR3 RNAi samples were calculated using the UniFrac metrics, which measures phylogenetic dissimilarities between microbial communities [41]. Genus- and species-level profiling histograms and principal coordinate analyses based on weighted UniFrac demonstrated great variation in the composition of the gut microbial community upon Duox-TLR3 RNAi and egfp RNAi treatment, especially on days 4 and 8 post-dsRNA injection (Fig 8A). Principal coordinate analysis showed separation of Duox-TLR3 RNAi- and egfp RNAi-treated samples along the major component 1(pc1) and major component 2 (pc2) axes, which explained 44.01% and 21.08% of data variation (S7 Fig), respectively. The analysis of the control egfp RNAi samples further confirmed that the intestinal bacterial composition was rich in diversity, and the major genus members of the gut community of BSF were Providencia, Morganella, Wohlfahrtiimonas, and Dysgonomonas, whereas the minor members were Lactococcus, Comamonas, Pseudomonadaceae, and Leucobacter(Fig 8A). At 4th day, a decrease of 12% and 14.8% abundance of Providencia and Dysgonomonas in Duox-TLR3 RNAi larvae compared with that of control, respectively. However, the relative abundance of others indigenous bacteria increased by 1.17 times in the Duox-TLR3 RNAi larvae than in the control larvae (Fig 8B). Bacterium Lactococcus with low relative abundance increased to detectable levels in the Duox-TLR3 RNAi larvae compared with that in the control larvae, which in egfp group could not reach the minimum level for detection because of their relatively low abundance.The minor group bacterium Leucobacter also has certain increase in Duox-TLR3 RNAi group.(Fig 8C). On the 8th day after dsRNA injection, the relative percentage of Leucobacter bacteria in the Duox-TLR3 RNAi was still significantly higher than that of the control group, The relative proportion of Lactococcus bacteria was similar to that of 4th day after dsRNA injection. However, on the 12th day after dsRNA injection, the relative proportion of bacteria in the Duox-TLR3 RNAi group recovered to the same level as that in the egfp control group except to the Comamonas bacteria in the egfp RNAi group higher than those in the Duox-TLR3 RNAi group. Thus, the larvae with reduced BsfDuox and BsfTLR3 gene expression displayed a significant difference in bacterial community composition, possibly because of variations in the resistance of bacteria to ROS killing activity. At 8th day, the abundance of Pseudomonadaceae, a family of bacteria with low relative abundance in Duox-TLR3 RNAi larvae increased significantly by two fold compared with that in the controls. The relative abundance of Comamonas at 8th day was similar with that at 4th day in Duox-TLR3 RNAi larvae and was higher than that of egfp RNAi group(Fig 8E), which may be due to the reduced ROS and AMP production levels. After ds-Duox-TLR3 injection at 12th day, disordered intestinal bacterial communities stimulated the expression of the BsfDuox gene and the production of ROS to suppress non-symbionts. Finally, bacteria taxa composition in Duox-TLR3 RNAi-treated larvae at 12th day return to wild-type, excluding Dysgonomonas and Morganella, which remained high in Duox-TLR3 RNAi-treated larvae. Therefore, the Duox-TLR3 gene had a pivotal role in regulating the structure of the bacterial community in BSFL (Fig 8F). In the present study, the gene interference of Duox-TLR3 resulted in a decrease in the number of beneficial symbiotic bacteria Providencia and Dysgonomonas and an increase in the number of conditional pathogenic bacteria Pseudomonadaceae, leading to reduced resistance to pathogenic bacteria in the environment.
Fig 8. BsfDuox-TLR3gene regulates the composition and structure of gut bacterial community.
(A) Taxonomic breakdown at the genus level grouped by ds-egfp and ds-Duox-TLR3 treatments. (B-G) Relative abundance of different bacterial taxa after injecting ds-Duox-TLR3and ds-egfp at 4, 8 and 12 Day. Data are representative of three independent experiments (mean+s.e.m.). Statistical comparison was based on Student’s t–test (*p<0.05). ND, not detected.
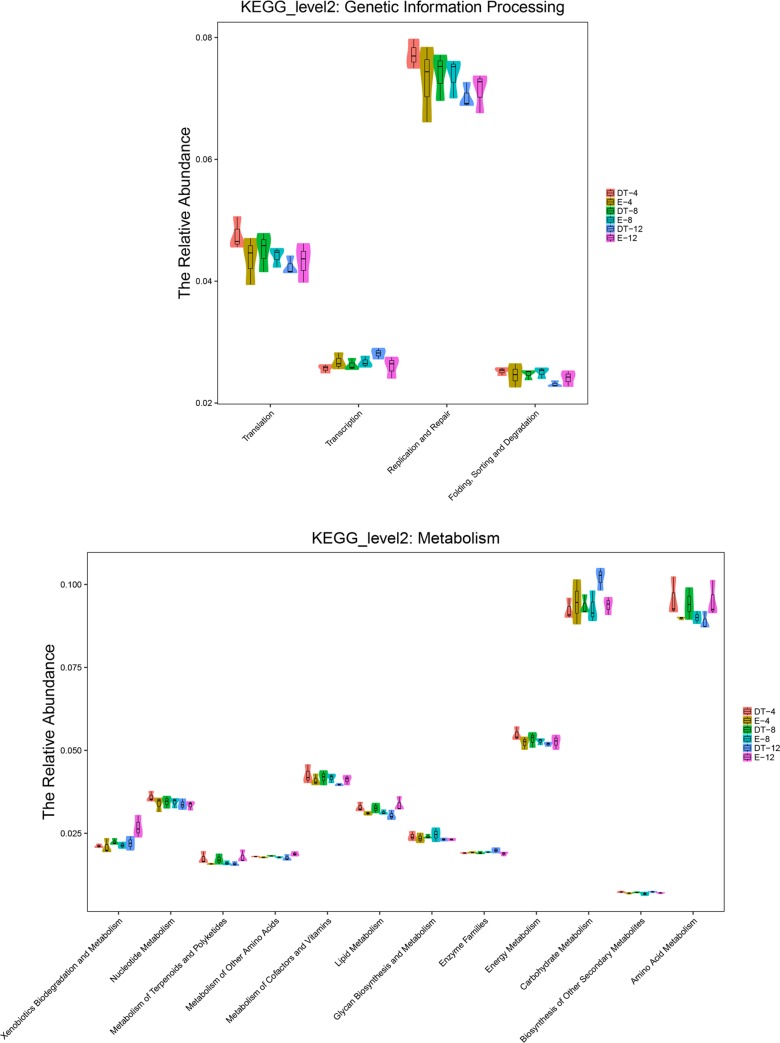
With the use of PICRUST, different RNAi treatments associated with functional potentials was predicted. The intestinal microbiota of Duox-TLR3 RNAi-treated larvae was more enriched with genes involved in replication, repair, infectious diseases, carbohydrate metabolism, and amino acid metabolism but less enriched with genes involved in the biosynthesis of other secondary metabolism and enzyme family compared with that of egfp RNAi-treated larvae (Fig 9A and 9B).
Fig 9. All of the predicted KEGG metabolic pathways are shown at the second hierarchical level and grouped by major functional categories.
(A) KEGG_level2: Genetic information processing. (B) KEGG_level2: Matabolism.
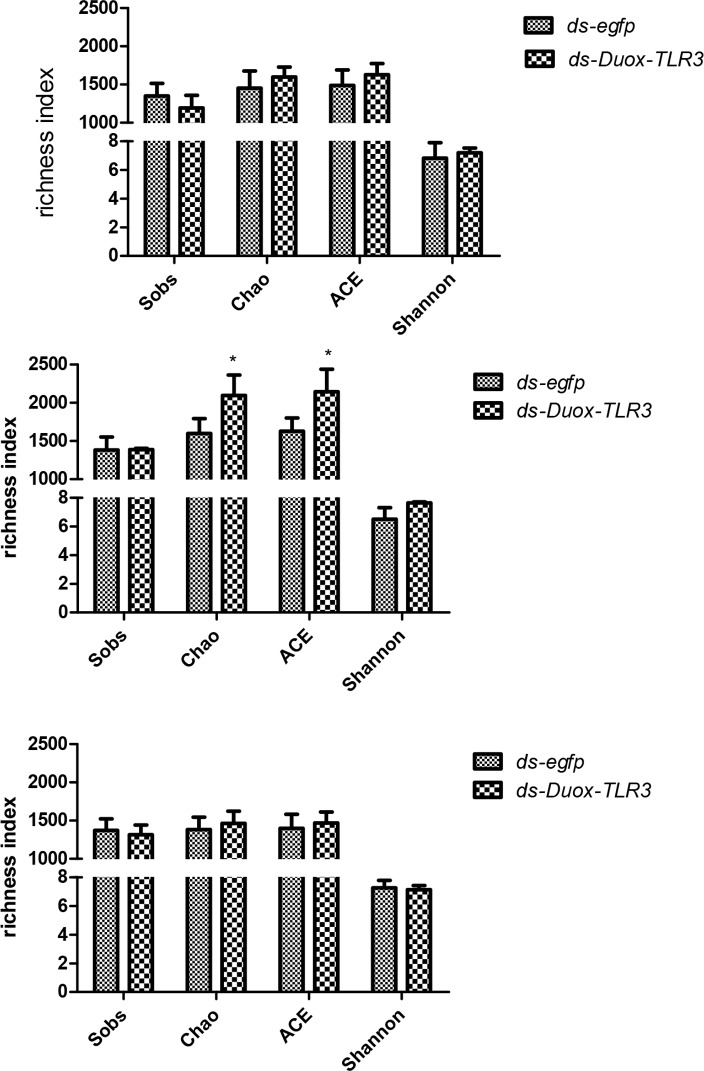
Microbial community richness was altered by BsfDuox-TLR3 RNAi
The effect of Duox-TLR3 RNAi on microbial diversity was investigated by a standardized approach that evaluated community richness. At 4th day. the Sobs (1192.3 ± 165.7), Chao(1598.9 ± 128.9), ACE(1628.1 ± 145.6), and Shannon(7.1 ± 0.3) indices of the intestinal microorganisms in Duox-TLR3 RNAi larvae were the same as the Sobs(1351.3 ± 164.7), Chao(1452.8 ± 223.5), ACE(1488.4 ± 202.1), and Shannon(6.8 ± 1.0) indices in the control larvae(Fig 10A). Meanwhile, the richness metrics of Chao and ACE in Duox-TLR3 RNAi larvae significantly increased at 8th day. In the control larvae, 1387 Sobs were identified, which was almost identical to those in Duox-TLR3 RNAi larvae. The Chao, ACE, and Shannon indices increased by 31.17%, 31.6%, and 17.4%, respectively, in the Duox-TLR3 RNAi larvae compared with those in the control larvae at 8th day (Fig 10B).
Fig 10. Effects of RNAi of the BsfDuox-TLR3gene on diversity metrics.
Richness measured as observed operational taxonomic units (OTUs; Sobs), Chao, ACE and Shannon indices of gut bacterial communities from different treatment at three timepoints. (A) 4 DPR. (B) 8 DPR. (C) 12 DPR. Data are representative of three independent experiments (mean ± SEM). Statistical comparison was based on Student’s t–test (p).
Finally, no significant difference was noted in the Sobs(1316.0 ± 125.9 vs. 1370.7 ± 151.3), Chao(1464.4 ± 158.8 vs. 1381.3 ± 163.4), ACE(1470.2 ± 142.9 vs. 1398.3 ± 185.1), and Shannon(7.1 ± 0.3 vs. 7.3 ± 0.5) indices between Duox-TLR3 RNAi and egfp RNAi larvae at 12th day(Fig 10C). Thus, BsfDuox-TLR3 gene silencing by RNAi led to increased bacterial diversity at 8th day compared with the control, possibly because of the low solubility of ROS and AMP.
Discussion
Intestinal microorganisms directly, or indirectly, affect the immunity of intestinal epithelium cells, and thus impacting the internal environment and development of the host. Microbial populations in the gut regulate host immunity by balancing between an efficient immune response to inhibit foreign pathogens from colonizing and proliferating [42]. Many immune genes of the host are involved in controlling gut microbes. By feeding BSFL zoonotic pathogenic bacteria it was found that the defense system of Duox-ROS plays an important role in the intestinal immune defense of BSFL. The silencing of the BSFL Duox gene resulted in a significant decrease in the level of ROS in the intestine, indicating that the ROS in the intestine of BSFL is mediated by Duox. The Toll signaling pathway plays an important role in the immune response of the BSFL system, zoonotic pathogenic bacteria can rapidly activate TLR3, Dif, and Drosal genes of toll signaling pathways. The zoonotic pathogens that activate Duox and TLR3 to produce ROS and AMPs that act to restrict the growth and proliferation of invading microorganisms [43, 44]. We determined toll pathway-mutant BSFL lacking AMP expression generally express reduced resistance and suffer lethal effects due to gut infection. By contrast, Duox-TLR3 inactivation leads to uncontrolled increase of bacteria in the gut as demonstrated in this study. That means the immune system relies mainly on two effector molecules, antimicrobial peptides and ROS, to inhibit the colonization of invasive microbes. With the advent of sequencing technology, many bacterial communities in the gut of various insects have been described. Indigenous bacterial community composition was of medium complexity in B. dorsalis, which is composed of Porphyromonadaceae, Streptococcaceae, and Sphingobacteriaceae and many unclasssifed bacteria. BdDuox silencing led to a decreased abundance of Enterobacteriaceae and Leuconostocaceae in the gut and overgrowth of minor pathobionts [11].
Compared with the intestinal bacteria community of B. dorsalis, that of the BSFL was relatively complex. The dominant symbiotic bacteria, Providencia, in the gut of BSF belonged to the family of Enterobacteriaceae. Enterobacteriaceae is a commonly found symbiotic taxon in insects, and specifically belonging to the γ-proteobacteria, a class that includes dominant symbiotic bacteria in many insect lineages [45]. Enterobacteriaceae, as dominant commensal bacteria in the intestinal tract of insects, may play an important role in improving host fitness by preventing the colonization of foreign pathogens [46] and contribute to nitrogen fixation [47]. Providencia spp. are gram-negative bacteria that belonged to the Enterobacteriaceae. Providencia spp. resist two kinds of antibiotics including colistin and tigecycline, making Providencia multidrug-resistant. Providencia spp. resistant to carbapenem antibiotics are increasingly reported. Meanwhile, Dysgonomonas, which belong to coccobacilli is a facultative anaerobic bacteria [48]. Dysgonomonas spp. is a major group represented in the gut microflora of BSFL, and Bactrocera tau. Dysgonomonas spp. isolated from human blood samples show antimicrobial susceptibility that directly inhibits competitors and potent pathogens from the same niche [49].
In this study, BSFL were determined to use the Duox-ROS immune system and Toll signaling pathway as a means of intestinal immune defense. Gene expression profiles of BsfDuox and BsfTLR3 at different development times indicated that they may have key functions in host development. The role in immune defense of Duox and TLR3 has been studied well, but their combined role in the regulation of intestinal microbial homeostasis and bacteriostasis is rarely reported. In this study, the silencing of the target gene BsfDuox-TLR3 was successfully achieved for 4 days. The expression level of the BsfDuox was downregulated at 4–8 days of treatment, whereas that of the BsfTLR3 was downregulated at 6–10 days of treatment. The maintenance of the RNAi effect in a short time is ideal for the study of changes in intestinal microflora homeostasis. Furthermore, silencing of the BsfDuox-TLR3 gene in larvae led to the increase in density and changes in composition and diversity of intestinal indigenous bacterial community. This finding was similar with the report on the inactivation of Duox and intestinal bacterial reproduction in B. dorsalis [11].
At the 10-12th day after dsRNA injection, the disordered intestinal bacterial community in turn stimulated BSFL Duox-TLR3 gene expression and ROS production to suppress the overgrowth of non-symbiotic bacteria. Finally, at the 12th day after dsRNA injection, BSFLDuox-TLR3 gene regulated the structure and composition of the host intestinal bacterial community to a normal level. The relative proportion of dominant intestinal commensal bacteria Dysgonomonas and Providencia decreased, whereas that of Lactococcus and Pseudomonadaceae increased in Duox-TLR3 RNAi-treated larvae at 4–8 days. Dysgonomonas and Providencia in the gut of BSF may enhance inhibition to Salmonella spp. and S. aureus, a significant decrease in Dysgonomonas and Providencia by Duox-TLR3 silencing led to an adverse effect on the host [50]. Meanwhile, Pseudomonadaceae is a minor component in the BSF gut and includes four pathogenic bacteria, namely, P. aeruginosa [51], Pseudomonas pertucinogena [52], Pseudomonas putida [53], and Pseudomonas fluorescens [54]. P. aeruginosa is a human pathogen associated with human skin, mucous membrane, intestinal tract, and upper respiratory tract. The increase in Pseudomonadaceae caused by silencing of the BsfDuox-TLR3 could cause toxicity to the host. Thus, silencing the BsfDuox-TLR3 gene in the intestines allowed the increase of harmful bacteria.
The result may suggest that the depletion of these two immune genes in BSFL resulted in a proliferation of the minor pathogenic bacteria, which may have proven the potential decrease in anti-environmental pathogenic bacteria immune responses.Therefore, BsfDuox and BsfTLR3 could regulate homeostasis by ensuring the stability of symbiotic bacteria and suppressing excessive growth of minor pathogen bacteria. Gut microbiome disorders are reminiscent of inflammatory bowel disease in humans, but intestinal symbiotes can also cause bowel disease under certain circumstance [55,56]. In humans, many intestinal mucosal inflammatory diseases arise from abnormal intestinal and microbial relationships [57]. Therefore, the study on the role of BSFL Duox-TLR3 gene in the homeostasis of intestinal bacterial community will provide reference for the further exploration of the maintenance mechanism of healthy intestinal microorganisms.
Overall, similar to humans, BSF larval intestine harbors a natural microbiota, participating in host metabolism and provide nutrients [58] as well as in the degradation of harmful substances [59] to protect the host from adverse factors such as natural enemies, parasites. In this study, a representative of the predominant gut immune gene Duox-TLR3 from BSF showed antimicrobial activity that directly prevented the emergence and overgrowth of minor pathobionts. The mode of protection against an encountered pathogen was possibly due to the persistent immune responses involving free radicals and antibacterial peptides. By interfering with the Duox-TLR3 gene, the intestinal bacterial richness and composition changed (P. putida, and P. fluorescens) to provoke chronic inflammation under dysbiosis conditions and weakened suppression on S. aureus and Salmonella spp.. Therefore, the natural bacterial flora is crucial to maintain the stable balance of intestinal microbial communities, resisting and eliminating foreign pathogenic bacteria for insect physiological ecology such as growth and development. Our results demonstrated that BsfDuox and BsfTLR3 could regulate the gut key bacteria Providencia and Dysgonomonas homeostasis to depress zoonotic pathogens.
Supporting information
(PDF)
(TIF)
(TIF)
(TIF)
(TIF)
(A) Rarefaction curve. (B) Rank-abundance curves.
(TIF)
The result showed a separation of ds-Duox-TLR3 and ds-egfp-treated samples along the first two axes, which explained 44.01% and 21.08% of the data variation, respectively. (A) 3Dscore plot of the ds-Duox-TLR3 RNAi and ds-egfp RNAi sample (B) 2D biplots on PC1–PC2 plane overlapping scores and loadings.
(TIF)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The raw data was upload to NCBI and SRA, under accession numbers PRJNA600829 and SRP247530.
Funding Statement
National Natural Science Foundation of China (31770136); National Key Technology R & D Program of China (2018YFD0500203).
References
- 1.Rehman KU, Cai M, Xiao X, Zheng L, Wang H, Soomro AA, et al. Cellulose decomposition and larval biomass production from the co-digestion of dairy manure and chicken manure by mini-livestock (Hermetia illucens L.). J Environ Manage. 2017;196:458–65. Epub 2017/03/28. 10.1016/j.jenvman.2017.03.047 . [DOI] [PubMed] [Google Scholar]
- 2.Liu Q, Tomberlin JK, Brady JA, Sanford MR, Yu Z. Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ Entomol. 2008;37(6):1525–30. Epub 2009/01/24. 10.1603/0046-225x-37.6.1525 . [DOI] [PubMed] [Google Scholar]
- 3.Lalander CH, Fidjeland J, Diener S, Eriksson S, Vinnerås B. High waste-to-biomass conversion and efficient Salmonella spp. reduction using black soldier fly for waste recycling. Agro Sustain Dev. 2015;35(1):261–71. 10.1007/s13593-014-0235-4 [DOI] [Google Scholar]
- 4.Park SI, Kim JW, Yoe SM. Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae.Dev Comp Immunol. 2015;52(1):98–106. Epub 2015/05/10. 10.1016/j.dci.2015.04.018 . [DOI] [PubMed] [Google Scholar]
- 5.Elhag O, Zhou D, Song Q, Soomro AA, Cai M, Zheng L, et al. Screening, expression, purification and functional characterization of novel antimicrobial peptide genes from Hermetia illucens (L.). PLoS One. 2017;12(1):e0169582 Epub 2017/01/06. 10.1371/journal.pone.0169582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zdybicka-Barabas A, Bulak P, Polakowski C, Bieganowski A, Waśko A, Cytryńska M. Immune response in the larvae of the black soldier fly Hermetia illucens. Invert Surviv J. 2017;14:9–17. 10.1292/jvms.17-0236 . [DOI] [Google Scholar]
- 7.Peng J, Zipperlen P, Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Current Biology. 2005;15(18):1690–4. Epub 2005/09/20. 10.1016/j.cub.2005.08.048 . [DOI] [PubMed] [Google Scholar]
- 8.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–83. Epub 1996/09/20. 10.1016/s0092-8674(00)80172-5 . [DOI] [PubMed] [Google Scholar]
- 9.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5(5):e1000423 Epub 2009/05/09. 10.1371/journal.ppat.1000423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Gene Dev. 2009;23(19):2333–44. Epub 2009/10/03. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao Z, Wang A, Li Y, Cai Z, Lemaitre B, Zhang H. The dual oxidase gene BdDuox regulates the intestinal bacterial community homeostasis of Bactrocera dorsalis. ISME J. 2016;10(5):1037–50. 10.1038/ismej.2015.202 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science (New York, NY). 2010;327(5973):1644–8. Epub 2010/03/13. 10.1126/science.1184008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science (New York, NY). 2011;332(6031):855–8. Epub 2011/05/14. 10.1126/science.1201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira JH, Goncalves RL, Lara FA, Dias FA, Gandara AC, Menna-Barreto RF, et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog.2011;7(3):e1001320 Epub 2011/03/30. 10.1371/journal.ppat.1001320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao X, Yang L, Pang X, Zhang R, Zhu Y, Wang P, et al. A Mesh-Duox pathway regulates homeostasis in the insect gut. Nat Microbiol. 2017;2:17020 Epub 2017/03/02. 10.1038/nmicrobiol.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L, Crippen TL, Singh B, Tarone AM, Dowd S, Yu Z, et al. A survey of bacterial diversity from successive life stages of black soldier fly (Diptera: Stratiomyidae) by using 16S rDNA pyrosequencing. J Med Entomol. 2013;50(3):647–58. Epub 2013/06/28. 10.1603/me12199 . [DOI] [PubMed] [Google Scholar]
- 17.Bruno D,Bonelli M, De Filippis F,Di Lelio I, Tettamanti G, Casartelli M,et al. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions.Appl Environ Microb. 2019;85:e01864–18. 10.1128/AEM.01864-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wynants E, Frooninckx L, Crauwels S, Verreth C, De Smet J, Sandrock C, et al. Assessing the microbiota of black soldier fly larvae (Hermetia illucens) reared on organic waste streams on four different locations at laboratory and large scale. Micro Ecol. 2019; 77:913–930. 10.1007/s00248-018-1286-x [DOI] [PubMed] [Google Scholar]
- 19.Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog.2010;6(9):e1001097 Epub 2010/09/17. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14(1):82–91. Epub 2010/11/03. 10.1016/j.mib.2010.10.003 . [DOI] [PubMed] [Google Scholar]
- 21.Futo M, Armitage SA, Kurtz J. Microbiota Plays a Role in Oral Immune Priming in Tribolium castaneum. Front Microbiol. 2015;6:1383 Epub 2016/01/19. 10.3389/fmicb.2015.01383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Jin L, Zhang H. Comparison of the diversity of the bacterial communities in the intestinal tract of adult Bactrocera dorsalis from three different populations. J Appl Microbiol.2011;110(6):1390–401. Epub 2011/03/15. 10.1111/j.1365-2672.2011.05001.x . [DOI] [PubMed] [Google Scholar]
- 23.GB4789.10–2010. MoHPRC. Food microbiological examination: Staphylococcus aureus test. Peking: Standards Press of China.; 2010.
- 24.GB4789.4–2016. MoHPRC. Food microbiological examination:Salmonella test. Peking: Standards Press of China; 2016.
- 25.Zhan S, Fang G, Cai M, Kou Z, Xu J, CaoY, et al. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res.1–11. 10.1038/s41422-019-0252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, et al. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005;8(1):125–32. Epub 2004/12/29. 10.1016/j.devcel.2004.11.007 . [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 28.Guo X, Xia X, Tang R, Wang K. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe. 2008;14(4):224–8. Epub 2008/06/06. 10.1016/j.anaerobe.2008.04.001 . [DOI] [PubMed] [Google Scholar]
- 29.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. P Natl Acad Sci USA. 2009;106(37):15813–8. Epub 2009/10/07. 10.1073/pnas.0907722106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuleic Acids Res.1997;25(17):3389–402. Epub 1997/09/01. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. Epub 2010/04/13. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (New York, NY). 2006;312(5778):1355–9. Epub 2006/06/03. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics (Oxford, England). 2011;27(21):2957–63. Epub 2011/09/10. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England). 2010;26(19):2460–1. Epub 2010/08/17. 10.1093/bioinformatics/btq461 . [DOI] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb. 2007;73(16):5261–7. Epub 2007/06/26. 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res.1997;25(17):3389–402.Epub 1997/09/01. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemp PF, Aller JY. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. Fems Microbiol Ecol. 2004;47(2):161–77. Epub 2004/02/01. 10.1016/S0168-6496(03)00257-5 . [DOI] [PubMed] [Google Scholar]
- 38.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, et al. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell. 2013;153(4):797–811. Epub 2013/05/15. 10.1016/j.cell.2013.04.009 . [DOI] [PubMed] [Google Scholar]
- 39.Vogel H, Müller A, Heckel D G, Gutzeit H , Vilcinskas A. Nutritional immunology: diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev Comp Immunol.78, 141–148. 10.1016/j.dci.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 40.Jiang C L, Jin W Z, TaoX H, Zhang Q,Zhu J, Feng S Y et al. Black soldier fly larvae (Hermetia illucens) strengthen the metabolic function of food waste biodegradation by gut microbiome. Microb Biotechnol.12(3), 528–543. 10.1111/1751-7915.13393 PMCID:PMC6465238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371 Epub 2006/08/09. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–20. Epub 2008/05/13. 10.1038/nri2316 . [DOI] [PubMed] [Google Scholar]
- 43.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science (New York, NY). 2005;310(5749):847–50. Epub 2005/11/08. 10.1126/science.1117311 . [DOI] [PubMed] [Google Scholar]
- 44.Yi H-Y, Chowdhury M, Huang Y-D, Yu X-Q. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98(13):5807–22. 10.1007/s00253-014-5792-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paterson D. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Infect Control. 2006, 345: S20–S28. [DOI] [PubMed] [Google Scholar]
- 46.Behar A, Jurkevitch E, Yuval B. Bringing back the fruit into fruit fly-bacteria interactions. Mol Ecol. 2008;17(5):1375–86. Epub 2008/02/28. 10.1111/j.1365-294X.2008.03674.x . [DOI] [PubMed] [Google Scholar]
- 47.Behar A, Yuval B, Jurkevitch E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14(9):2637–43. Epub 2005/07/21. 10.1111/j.1365-294X.2005.02615.x . [DOI] [PubMed] [Google Scholar]
- 48.Wallace PL, Hollis DG, Weaver RE, Moss CW. Characterization of CDC group DF-3 by cellular fatty acid analysis. J Clin Microb. 1989;27(4):735–7. Epub 1989/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hironaga M, Yamane K, Inaba M, Haga Y, Arakawa Y. Characterization and antimicrobial susceptibility of Dysgonomonas capnocytophagoides isolated from human blood sample. Jpn J Infect Dis. 2008;61(3):212–3. Epub 2008/05/27. . [PubMed] [Google Scholar]
- 50.Jiang C L, Jin W Z, Tao X H, Zhang Q,Zhu J, Feng S Y et al. Black soldier fly larvae (Hermetia illucens) strengthen the metabolic function of food waste biodegradation by gut microbiome. Microb Biotechnol, 12(3), 528–543. 10.1111/1751-7915.13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors.EMBO J.2003;22(15):3803–15. Epub 2003/07/26. 10.1093/emboj/cdg366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai YF,Moribayashi A. Taxonomic implications of the lipid composition of Pseudomonas pertucinogena.1978Int J Syst Bacteriol.28(3):394–400 [Google Scholar]
- 53.Yamamoto S, Harayama S. PCR Amplification and Direct Sequencing of gyrB Genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microb. 1995;61(10):3768 Epub 1995/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duijff BJ, Gianinazzi-Pearson V, Lemanceau P. Involvement of the outer membrane lipopolysaccharides in the endophytic colonization of tomato roots by biocontrol Pseudomonas fluorescens strain WCS417r. New Phytol. 1997;135(2):325–34. Epub 02/01. 10.1046/j.1469-8137.1997.00646.x [DOI] [Google Scholar]
- 55.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8(3), 292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroen. 2010; 26(4), 327–331. 10.1097/MOG.0b013e328339536b [DOI] [PubMed] [Google Scholar]
- 57.Rokutan K, Kawahara T, Kuwano Y, Tominaga K, Nishida K, Teshima-Kondo S, et al. Nox enzymes and oxidative stress in the immunopathology of the gastrointestinal tract.Semin Immunopathol 2008. Vol. 30, No. 3, pp. 315–327. Springer-Verlag. 10.1007/s00281-008-0124-5 [DOI] [PubMed] [Google Scholar]
- 58.Thong-On A, Suzuki K, Noda S, Inoue J, Kajiwara S, Ohkuma M. Isolation and characterization of anaerobic bacteria for symbiotic recycling of uric acid nitrogen in the gut of various termites. Microbes Environ. 2012;27(2):186–92. Epub 2012/07/14. 10.1264/jsme2.ME11325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ceja-Navarro JA, Vega FE, Karaoz U, Hao Z, Jenkins S, Lim HC, et al. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 2015;6:7618 10.1038/ncomms8618 . [DOI] [PMC free article] [PubMed] [Google Scholar]