SUMMARY
Many chaperones promote nascent polypeptide folding followed by substrate release through ATP-dependent conformational changes. Here we show cryoEM structures of Gα subunit folding intermediates in complex with full-length Ric-8A, a unique chaperone-client system in which substrate release is facilitated by guanine nucleotide binding to the client G protein. The structures of Ric-8A-Gαi and Ric-8A-Gαq complexes reveal that the chaperone employs its extended C-terminal region to cradle the Ras-like domain of Gα, positioning the Ras core in contact with the Ric-8A core while engaging its switch2 nucleotide binding region. The C-terminal α5 helix of Gα is held away from the Ras-like domain through Ric-8A core domain interactions, which critically depend on recognition of the Gα C terminus by the chaperone. The structures, complemented with biochemical and cellular chaperoning data, support a folding quality control mechanism that ensures proper formation of the C-terminal α5 helix before allowing GTP-gated release of Gα from Ric-8A.
Graphical Abstract
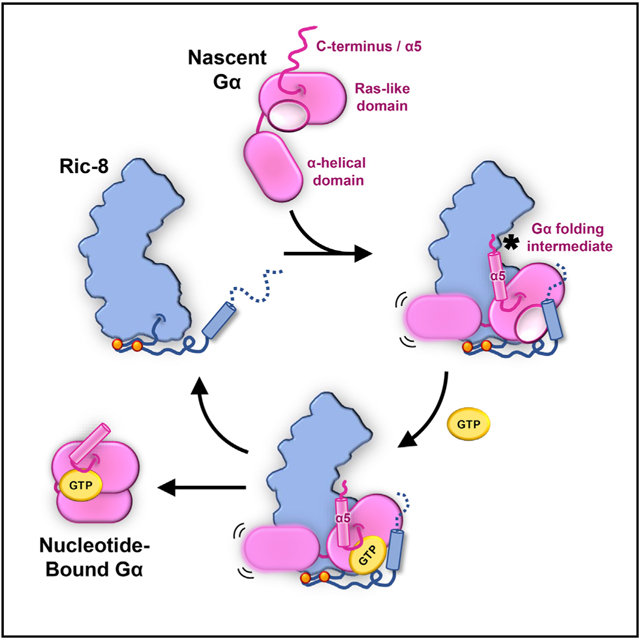
In Brief
Seven et al. present cryoEM structures of Gα subunit folding intermediates in complex with their universal chaperone, Ric-8. Ric-8 forms key interactions with the Ras domain to prepare GTP-gated release of Gα subunits from their chaperone. The structures, complemented by biochemical and cellular experiments, reveal a folding quality control mechanism.
INTRODUCTION
Heterotrimeric G proteins, composed of Gα, Gβ, and Gγ subunits, relay the vast majority of intracellular signaling mediated by G protein-coupled receptors (GPCRs), the largest and most diverse class of membrane proteins in eukaryotes. GPCRs respond to a remarkable array of extracellular stimulants, including light, ions, hormones, odorants, neurotransmitters, and natural chemicals, and in turn couple primarily to G proteins to facilitate the exchange of GDP for GTP on the Gα subunit (Hilger et al., 2018). Nucleotide exchange promotes the functional dissociation of Gα-GTP from the Gβγ obligate dimer, leading to downstream signaling through binding to effectors such as adenylyl cyclase, phospholipase C, and ion channels. G proteins are classified based on sequence homology of the Gα subunit into four main sub-classes: Gαi/o, Gαq/11, Gαs/olf, and Gα12/13, each comprised of multiple isoforms (Wilkie et al., 1992). Accordingly, GPCRs display distinct selectivity profiles for heterotrimeric G proteins, thereby orchestrating precise cellular pathways and signaling outcomes.
The central signaling role of heterotrimeric G proteins, along with their ability to undergo extensive conformational changes during highly tuned receptor coupling and dissociation, has co-evolved with quality control mechanisms for their structural and functional integrity. Recent work has shown that multiple chaperones are necessary for proper folding, assembly, and localization of heterotrimeric G proteins. Chaperonin-containing tailless complex polypeptide-1 (CCT) (Lukov et al., 2005, 2006) and dopamine receptor-interacting protein 78 (DRiP78) (Dupré et al., 2007) were proposed to facilitate the folding of Gβ and Gγ subunits, respectively, while the chaperone phosducin-like protein-1 (PhLP-1) is subsequently involved in the formation of the Gβγ obligate heterodimer (Lukov et al., 2005, 2006). Resisance to inhibitors of cholinesterase-8 (Ric-8) was initially discovered as a gene that positively influenced G protein signaling pathways in a Caenorhabditis elegans mutagenesis screen (Miller et al., 1996; Nguyen et al., 1995). Ric-8 proteins are now known to fold nascent Gα subunits prior to G protein heterotrimer formation. The initial in vitro work identified Ric-8 as a non-receptor guanine exchange factor (GEF) for Gα proteins (Chan et al., 2011b; Tall et al., 2003), but more recent studies revealed that Ric-8 proteins are molecular chaperones for nascent Gα subunits (Chan et al., 2013; Gabay et al., 2011), thereby explaining the positive influence of their activities on G protein signaling (Papasergi et al., 2015). Besides facilitating Gα folding, the observed Ric-8 GEF activity for Gα subunits (Chan et al., 2011a; Tall et al., 2003; Van Eps et al., 2015) has raised the possibility that these chaperones may also be involved in alternative modes of Gα subunit activation or reamplification of GPCR signaling.
Ric-8 proteins are evolutionarily conserved from fungi to humans (Li et al., 2010; Miller et al., 2000; Papasergi et al., 2015; Wright et al., 2011), although no homologs have been reported in plants and baker’s yeast. However, Arr4/Get3 proteins in yeast, structurally unrelated to Ric-8, are shown to be important for the biogenesis and signaling of Gα subunits and act as non-receptor GEFs (Lee and Dohlman, 2008). There are two isoforms of Ric-8 identified in vertebrates: Ric-8A, which acts within the biosynthetic pathway of the Gαi/o, Gαq/11, and Ga12/13 sub-classes, and Ric-8B, which primarily acts on the Gαs/olf subfamily (Chan et al., 2013; Gabay et al., 2011; Nagai et al., 2010). The isotypes Gαq/11, Ga12/13, and Gαolf appear to have the most stringent requirements for Ric-8 in order to be correctly folded and trafficked to the plasma membrane (Chan et al., 2011a; Gabay et al., 2011). Recent crystal structures of a truncated form of Ric-8A alone or in complex with a peptidomimetic of the C-terminal α5 helix of the G protein transducin (Gt) showed that the chaperone is composed of a combination of armadillo (ARM) and Huntington, Elongation Factor 3, PR65/A, TOR1 (HEAT) repeats. The ARM/HEAT repeats form a crescent-shaped core domain, with its concave surface engaging the α5 helix peptidomimetic (Srivastava et al., 2019; Zeng et al., 2019). However, the lack of structural information on full-length Ric-8-Ga complexes has hindered our understanding of the chaperone activity and the mechanism of Gα release from Ric-8, which presumably takes place upon successful Gα folding. Part of the challenge in attaining high-resolution information on folding intermediates is the intrinsic difficulties in obtaining stable complexes that are suitable for structural studies. In general, reported crystal structures of chaperone-client complexes lack high-resolution features of the client protein, thereby limiting the characterization of chaperoning mechanisms at large.
To delineate the critical structural components of the chaperoning and GEF activity of Ric-8 toward Gα protein subunits, we sought to visualize these complexes by cryoEM. To this end, we purified or reconstituted native folding intermediates of two Gα subunits, Gαq and Gαi1, in complex with Ric-8A in the absence of guanine nucleotides. CryoEM maps of Ric-8A-Gαq and Ric-8A-Gαi1 at near-atomic resolution provide views of a unique chaperone-client complex and illustrate a striking chaperone mechanism that is employed by Ric-8A to stabilize critical elements for guanine nucleotide coordination by Ga. Mutagenesis coupled to both nucleotide exchange assays and a cellular chaperoning readout indicates that the recognition of the most C-terminal residue of Gα by Ric-8 provides a critical checkpoint for GTP binding to the G protein subunit and its subsequent release from the chaperone. Collectively, these results suggest a quality control mechanism underlying the chaperone and GEF activity of Ric-8 on Gα subunits.
RESULTS
CryoEM Structures of Ric-8A in Complex with Gαi1 and Gαq
For our studies, we employed wild-type full-length Gαq or Gαi1 proteins and full-length Ric-8A, including its potentially unstructured regions, such as the Ric-8A C-terminal tail, which is required for chaperone activity (Oner et al., 2013; Papasergi-Scott et al., 2018; Thomas et al., 2011). We chose to study these complexes without any stabilizing agent such as antibody fragments or crosslinkers, aiming to preserve both the native structure as well as the conformational flexibility between the chaperone and the client Gα subunit. It has been previously shown that site-specific phosphorylation of Ric-8A is critical for both its chaperone and GEF activity (Papasergi-Scott et al., 2018). To achieve these post-translational modifications, we utilized a baculovirus system to express Ric-8A in insect cells, a strategy that has been shown to produce largely phosphorylated chaperone at positions S435, T440, S522, S523, and S527 (Papasergi-Scott et al., 2018). In the case of Ric-8A-Gαi1, a complex stable enough for cryoEM imaging was generated by in vitro reconstitution from purified Ric-8A and myristoylated Gαi1 proteins, recombinantly expressed in insect cells and E. coli, respectively (Figure S1). Apyrase was added to hydrolyze the GDP released from Gαi1 upon forming a complex with Ric-8A in order to promote formation of a stable nucleotide-free complex. Due to the lower expression level and protein quality of Gαq compared to Gαi1 in the absence of chaperone and Gβγ, we isolated the Ric-8A-Gαq complex after co-expression of Ric-8A and Gα protein in insect cells. The complex was purified by utilizing an N-terminal GST tag on Ric-8A that was subsequently cleaved by TEV protease treatment. Prior to GST protein purification, apyrase was added to the lysed insect cell suspension for maximal stabilization of the Ric-8A-Gαq complex by hydrolyzing free guanine nucleotides. After GST purification and protease cleavage, the Ric-8A-Gαq complex was further purified by size-exclusion chromatography in order to separate it from free Ric-8A and GST protein. Both purification strategies resulted in stable complexes that were monodisperse as assessed by size-exclusion chromatography and negative stain EM visualization (Figure S1). From these samples, we obtained cryoEM maps of Ric-8A-Gαq and Ric-8A-Gαi1 with a global indicated resolution of 3.5Å and 4.1Å, respectively (Figure 1; Figures S1–S3, S4A, S4B, and Table S1). The structured portion of these assemblies is only ~65 kDa, reinforcing the feasibility of conventional cryoEM for challenging proteins or complexes in the range of 50–100 kDa. These cryoEM maps enabled the modeling of Ric-8A between residues 1–482 in the Ric-8A-Gαi1 complex and residues 1–451 in the Ric-8A-Gαq complex, while the backbone of the C-terminal region (458–484) was traced with poly-alanine due to the poorer quality of the Ric-8A-Gαq map in the region 451–458. Nevertheless, both models included a relatively long stretch of the C-terminal region of Ric-8A (up to residue 482 or 484), which was not observed previously. The majority of the Ras-like domain residues of both Gαi1 (residues 32–54 and 193–354) and Gαq (residues 217–359) were also unambiguously modeled in these maps. Although the nucleotide binding regions of Gα are mostly ordered, we do not observe any density for a guanine nucleotide, consistent with the purification of both complexes in the presence of the nucleotide-hydrolyzing enzyme Apyrase and the known stability of Ric-8-Gα nucleotide-free complex. The absence of nucleotide destabilizes the closed conformation of Gα in which the α-helical domain is packed against the Ras-like domain. Thus, in both Ric-8A-Gαi1 and Ric-8A-Gαq complexes, the α-helical domain opens up and becomes flexible in relation to the rest of the complex, akin to its behavior in nucleotide-free G proteins in complex with a GPCR (Rasmussen et al., 2011; Van Eps et al., 2015; Westfield et al., 2011). Accordingly, the general region occupied by the α-helical domain was masked out in our high-resolution map refinements for Ric-8A-Gαi1 and Ric-8A-Gαq complexes. However, in lower resolution unmasked reconstructions during initial steps of image processing, we observed partial density for the α-helical domain, jutted away from the nucleotide binding site in an average position that is rotated ~90° compared to the nucleotide-bound structures of Gαi1 or Gαq (Figures 1E and 1F). Furthermore, the N-terminal helix of both G proteins is not resolved in the Ric-8A complexes, in agreement with its dynamic and unfolded state in the absence of Gβγ, as shown in previous crystal structures and by solution NMR studies of Gαi1 (Goricanec et al., 2016; Maly and Crowhurst, 2012; Medkova et al., 2002).
Figure 1. CryoEM Structures of Ric-8A-Ga Complexes.
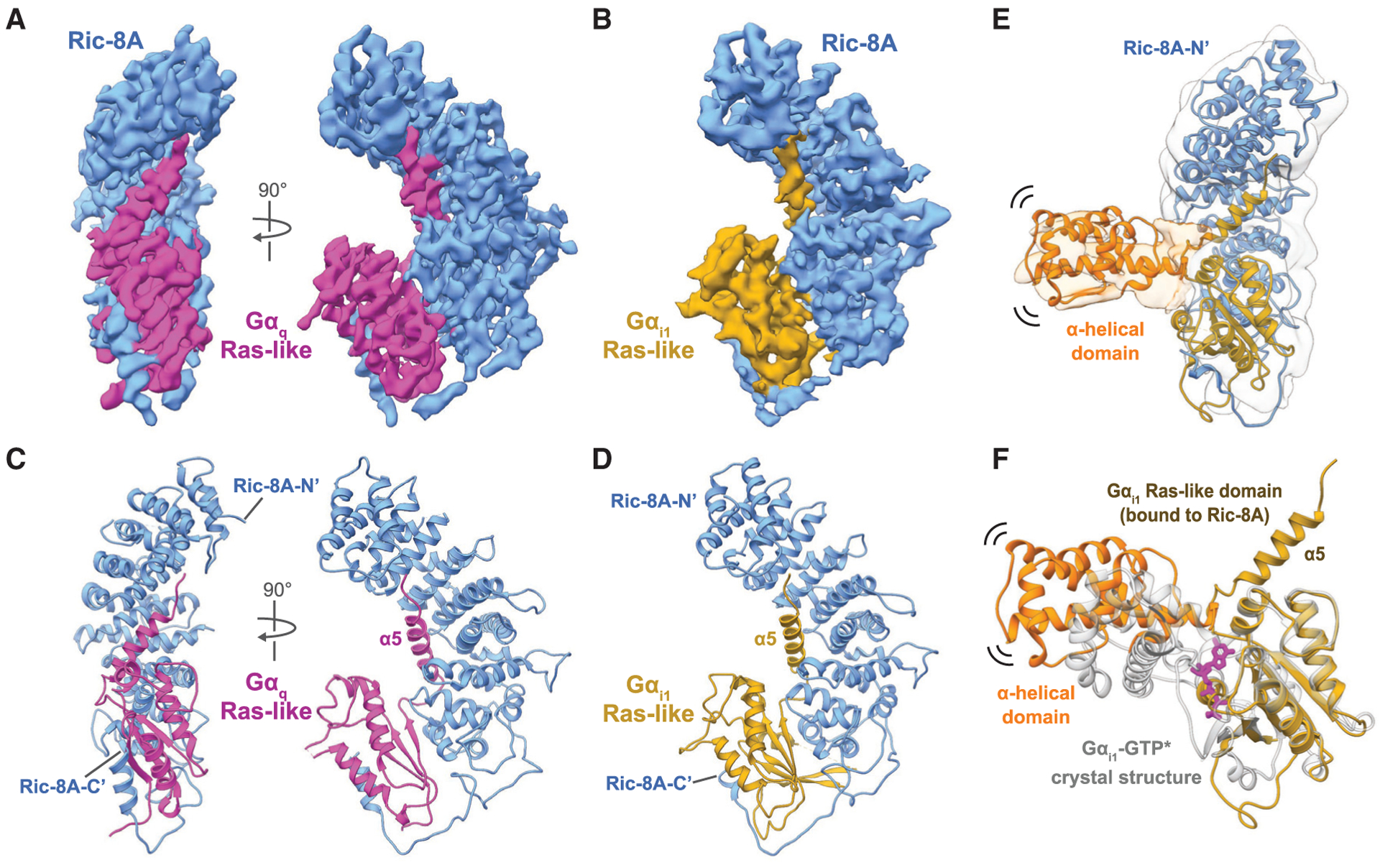
(A and B) Orthogonal views of the cryoEM maps of full-length Ric-8A (blue) bound to (A) full-length Gaq (magenta) at a global resolution of 3.5Å, or (B) Gαi1 (gold) at a global resolution of 4.1Å
(C and D) Ribbon diagrams of (C) Ric-8A-Gaq and (D) Ric-8A-Gαi1 models.
(E) A low-resolution map of the Ric-8A-Gαi1 complex including density for the Gαi1 α-helical domain that is positioned distal to the Ras-like domain.
(F) Comparison of the Gαi1-GTPγS crystal structure (gray; PDB: 1GIA) with the Ric-8A-Gαi1 complex (Ras-like domain: gold, α-helical domain: orange). Gαi1 adopts a distinct conformation from the canonical GTP-bound state.
The core domain of Ric-8A (residues 1–421), comprised of a combination of nine ARM and HEAT repeats (R1–R9), adopts a crescent-shaped conformation that is similar to the one observed in recent crystal structures of truncated Ric-8A alone (Srivastava et al., 2019; Zeng et al., 2019) (Figure S4C) or in complex with the Gαt α5 helix peptidomimetic (Srivastava et al., 2019) (Figure S4D), with an RMSD of 0.9–1Å. This observation suggests that the Ric-8A core domain maintains an overall stable conformation in the presence and absence of Gα. Interestingly, however, the core domain of Ric-8A adopts a slightly more compact configuration in its complex with Gαq compared to Gαi1 (Figure S4B). Furthermore, the bulk of the observable Ric-8A C terminus (residues 421–482) becomes ordered in the presence of Gαi1 or Gαq. This region largely maintains a coil structure, which encapsulates the Ras-like domain of Gαq or Gαi1, before forming an α-helix most likely comprised of residues 474–484 for Gαq and 474–481 for Gαi1 (Figure 2). The last C-terminal 40 residues of Ric-8A (490–530), predicted to include a coil/β-strand and an a-helix/coil structure (Figure S4E), have been shown to be critical for the chaperone function of Ric-8A in vivo, but are not required for in vitro GEF activity (Oner et al., 2013; Papasergi-Scott et al., 2018; Thomas et al., 2011). In our maps, this region appears disordered yet lies within the vicinity of the α-helical domain of Gα, raising the possibility that the 40 C-terminal residues may engage the flexible α-helical domain.
Figure 2. Interfaces in the Ric-8A-Gαi1 Complex.
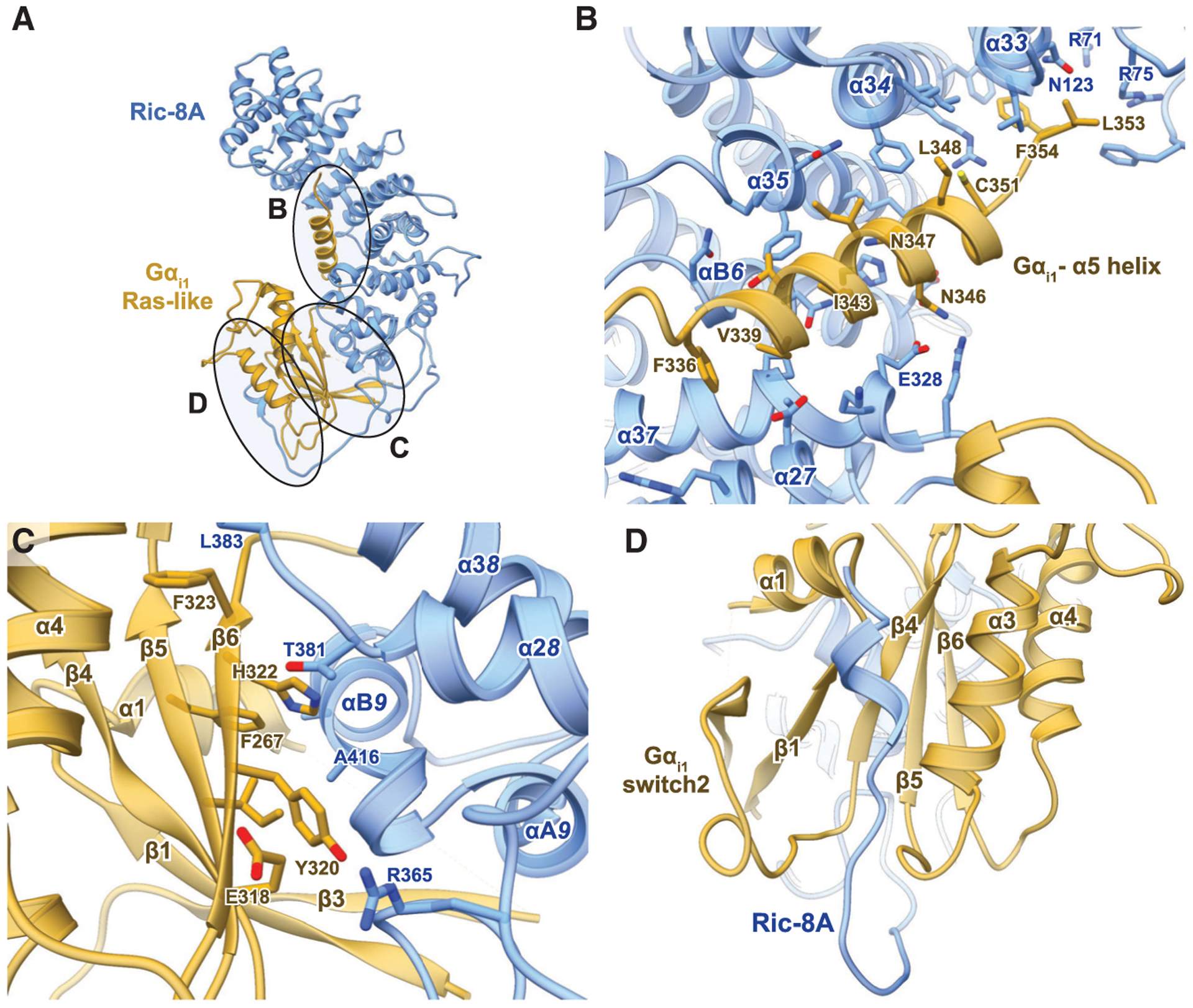
(A) Ric-8A binds to Gα through three interfaces.
(B) The α5 helix of Gαi1 binds to the concave surface of the Ric-8A N-terminal domain. Within the Ric-8A structure, “a” denotes the α-helical secondary structure followed by the repeat identifier of an individual ARM (1,2, or 3) or HEAT (A or B) helix and ending with the global ARM/HEAT repeat number from the N to C terminus of Ric-8A.
(C) The b5 and b6 strands of Gαi1 bind against the last two ARM/HEAT repeats of Ric-8A.
(D) A helical element comprising residues 472–480 of Ric-8A inserts between the switch2 motif and a3 helix of Gαi1.
Chaperone-Client Interactions in Ric-8A-Gα Structures
The cryoEM structures of Ric-8A-Gαi1 and Ric-8A-Gαq reveal three primary interaction surfaces between the chaperone and Gα Ras-like domain (Figure 2A). One interface involves an extensive interaction of the C-terminal α5-helix of Gα with the concave surface of the ARM/HEAT repeats (Figure 2B). A second interface is formed by the β4,βb5, and β6 strands of Gα with the R8-R9 ARM/HEAT repeats and a loop formed by residues 453–459 of the extended C terminus of Ric-8A (Figure 2C). The third interface is formed by the C-terminal helix of Ric-8A, which is in contact with switch2, the P loop, and the α3 helix of the Gα Ras-like domain (Figure 2D).
A comparison of the Gα Ras-like domain conformation in the Ric-8A-Gα maps and nucleotide-bound Gα crystal structures reveals several notable differences. The most distinct difference is the position of the α5 helix and the very C-terminal residues of Gαi1 and Gαq, which interact with the inner concave surface of the ARM/HEAT core domain of Ric-8A. In both complexes, the α5 helix is completely detached from the Ras-like domain and rotated by more than 90° away from its position in the Gα nucleotide-bound conformation (Figures 1E and 1F). The α5 helix is connected to the α6 strand by a loop containing the conserved TCAT motif, a critical element with residues that coordinate the guanine base of GDP or GTP bound to the Ras-like domain. Because of the displacement of the α5 helix (Figure 3A), the TCAT is positioned 3–5Å away from its nucleotide coordination site (Figure 3B). The positioning of the TCAT motif of Gαi1 and Gαq in complex with Ric-8A is similar to its configuration in nucleotide-free heterotrimeric Gαi1-Gbg and Gα11-Gβγ (Gα11 is a close homolog of Gαq) proteins in complex with GPCRs (Figures S5A and S5B). Of note, the displacement of the α5 helix from the Ras-like domain core may be promoted by the second helix of the final HEAT repeat (αB9) of Ric-8. Structural alignment of the Ras-like domains shows that the αB9 helix in the Ric-8A-bound Gαi1 structure occupies the position of the α5 helix in the GTPγS-bound Gαi1 structure (Figures S5C and S5D).
Figure 3. Guanine Nucleotide-free Gα Stabilized by Ric-8A Adopts an Open Conformation.
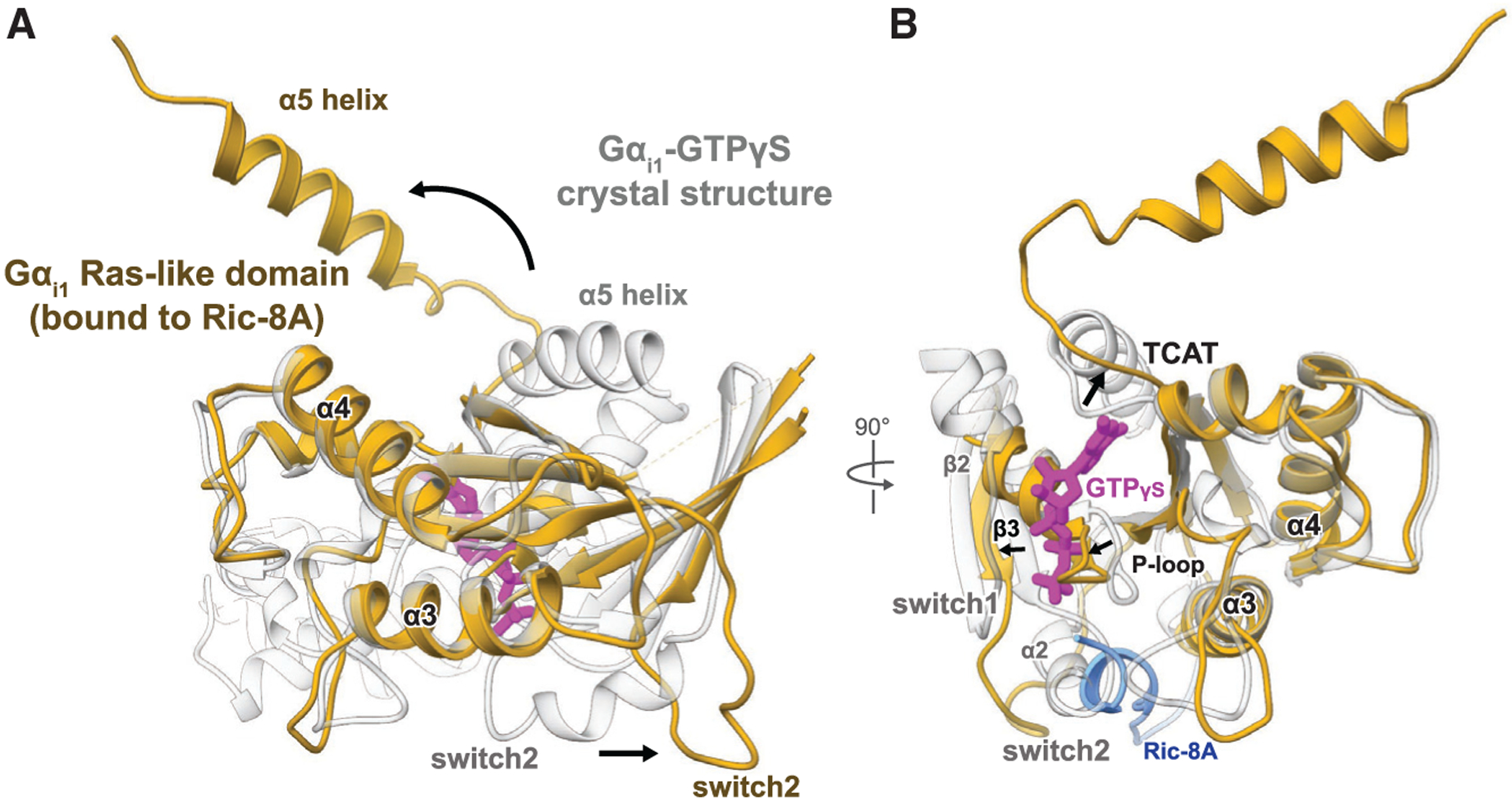
Comparison of the Gαi1 structures bound to the GTP analog 5′-guanosine-diphosphate-monothiophosphate (GTPgS) (PDB: 1GIA) and to Ric-8A. Gαi1 adopts a distinct conformation when bound to Ric-8A (gold) from that of the canonical GTP bound state (white).
(A) The α5 helix in the Ric-8A-Gαi1 complex is rotated more than 90 away from its position in the Gαi1-GTPγS crystal structure. The Gα switch2 motif is displaced outward compared to its conformation in the Ga-GTPγS structure. The helical segment of Ric-8A interacting with switch2 and the a3 helix of Gα is colored blue, and the rest of Ric-8A was omitted for clarity.
(B) The β6-a5 loop containing the TCAT motif is displaced 3–5Å outward from its position for guanine nucleotide coordination (thick arrow), and the interface between the α-helical and Ras-like domain is disordered together with the a2 helix and b2 strand.
Another distinct difference in the Ras-like domain involves the conformation of the switch2 motif and α2 helix that are also important for Gα nucleotide binding. The switch2/α2 elements are mostly disordered in the Ric-8A-Gαi1 structure, while in the map of the Ric-8A-Gαi1 complex, we can trace their density away from their corresponding conformations in the Gα GTP-bound state (Figures 3A and 3B). This positioning is induced by a C-terminal helix of Ric-8A (474–484), which is sandwiched between the switch2 motif and α3 helix, potentially forming interactions with the P loop and α2 and α3 helices.
The switch1 motif of Gα appears disordered in both of our Ric-8A complex structures, potentially due to the conformational flexibility of the α-helical domain. Additionally, the β2 and β3 strands together with α1 helix of the Ras-like domain are partially ordered in the Ric-8A-Gαq structure and modestly more ordered in the Ric-8A-Gαi1 structure. Notably, the map densities corresponding to these secondary structure elements in both the Gαi1 and Gαq complexes consistently displayed variability across several 3D classes. Although this region is too limited in size to be effectively resolved through particle classification, this observation suggests that the underlying elements are flexible when Gα is engaged with Ric-8A (Figure S3C).
Role of Phosphorylation of Ric-8A-Gα Complexes
The extended C-terminal coil region of Ric-8A includes two patches of phosphorylated residues, one closer to the core ARM/HEAT domain (S435 and T440) and another at the C terminus (S522, S523, and S527) (Figure S4E). Phosphorylation of S435 and T440 are critical for Ric-8A function and highly conserved in both mammalian Ric-8 isoforms (Ric-8A and Ric-8B) and across species (Figure 4D; Figure S6A) (Papasergi-Scott et al., 2018). However, these phosphorylated residues were not represented in the recent crystal structures of Ric-8A (Srivastava et al., 2019; Zeng et al., 2019). In our cryoEM maps, phosphorylated S435 and T440 were both resolved. These critical phosphorylated residues interact with a highly positively charged patch within Ric-8A formed by R345, R348, K349, K352, and R405 (Figure 4). Notably, the phosphate group of S435 in our structures occupies the same position as a reported sulfate ion, which is chemically similar to phosphate, in the crystal structure of truncated Ric-8A alone (Figure S4C) (Zeng et al., 2019). Neither S435 nor T440 residues are directly involved in the interaction between Ric-8A and Gαi1/Gαq. However, the interaction of the phosphates with the positive patch of Ric-8A anchors an extended coil of Ric-8A closer to the core ARM/HEAT domain. In this conformation, a loop formed by Ric-8A residues 451–461 packs against the Ric-8A core domain itself, which interacts with the Gα Ras-like domain to stabilize the interaction between the Gα β4, β5, and β6 strands with the R8-R9 ARM/HEAT repeats. The effect of T440 phosphorylation was shown to be much more pronounced than S435, presumably because it further stabilizes the coil region in closer proximity to the core, enhancing both the GEF and chaperone activity of Ric-8A (Papasergi-Scott et al., 2018). The interaction of the phosphorylated residues with the positively charged patch of Ric-8A also increases its overall thermal stability in the absence of Gα (Figure S6B), suggesting that it promotes a more compact and stable protein conformation.
Figure 4. Ric-8A Phosphorylated Residues S435 and T440 Stabilize the R8-R9 ARM/HEAT Interaction with the Ras-like Domain Core.
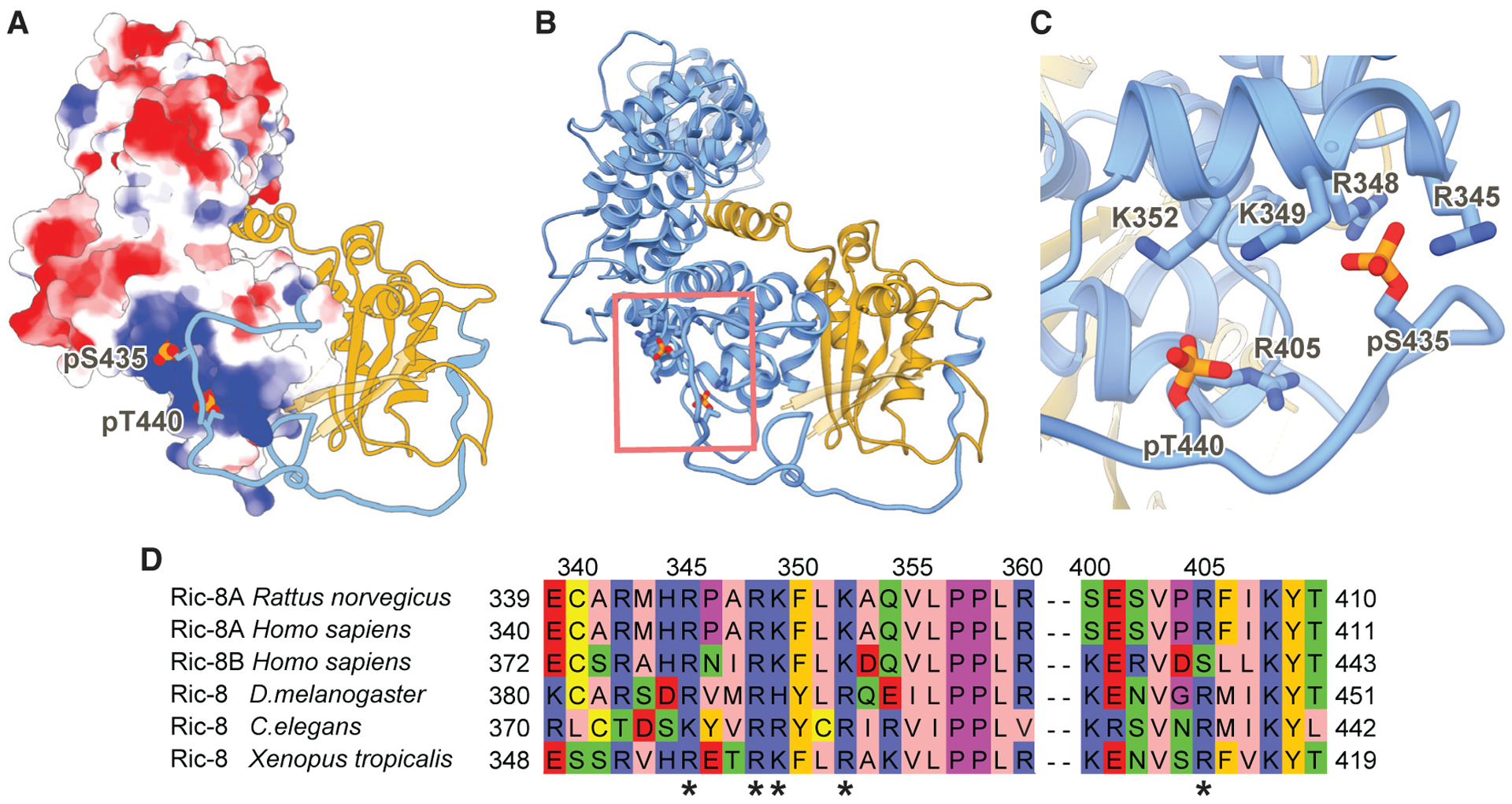
(A) Electrostatic surface representation of Ric-8A 1–421 (8 to +8 kT/e electrostatic potential is colored from red to blue respectively). Ric-8A residues 422–482 and Gαi1 are shown as ribbon models in blue and gold, respectively. Phosphorylated residues S435 and T440 are shown as spheres.
(B) Ribbon diagram of Ric-8A 1–482 (blue) and Gαi1 (gold) of the phosphorylated, full-length Ric-8A-Gαi1 complex. Phosphorylated residues S435 and T440 are shown as sticks.
(C) Phosphorylated residues S435 and T440 bind to conserved positively charged residues of Ric-8A.
(D) Sequence alignment of Ric-8 residues interacting with the phosphorylated S435 and T440 sites. The positively charged interacting residues (*) are highly conserved in both mammalian Ric-8 isoforms and across species.
Gα Subtype Selectivity by Ric-8A and Ric-8B
To construct a framework for understanding the molecular underpinnings of the G protein isotype specificity of Ric-8A, we analyzed the intermolecular contacts of the Ric-8A-Gαi1 and the Ric-8A-Gαq structures. Our analysis reveals that the majority of contact residues at the primary interfaces between Ric-8A and the two G protein isotypes are well conserved. This includes the interface between the C-terminal α5 helices of Gαi1 and Gαq and the concave surface of Ric-8A. In this region, a number of highly conserved hydrophobic residues (L353, L348, I343, V339, and F336; residue numbering based on Gαi1) mediate conserved hydrophobic and side-chain packing interactions with Ric-8A (Figure 5; Figure S7). The coupling of Ric-8A to Gαi1 and Gαq is further promoted by key polar interactions that include an H-bond between N347 in Gαi1 (N352 in Gαq) and H273 in Ric-8A and contacts between the last C-terminal carboxyl group of the G protein (F354 and V359 in Gαi1 and Gαq, respectively) and Ric-8A residues R71, R75, and N123 (Figure 5; Figure S7). Even though the last C-terminal residue is not conserved between the two G protein isotypes, the common apolar and polar contacts, described above, similarly position the α5 helices within the concave surface of Ric-8A.
Figure 5. Comparison of the Interactions between Ric-8 and the Gα α5 Helices.
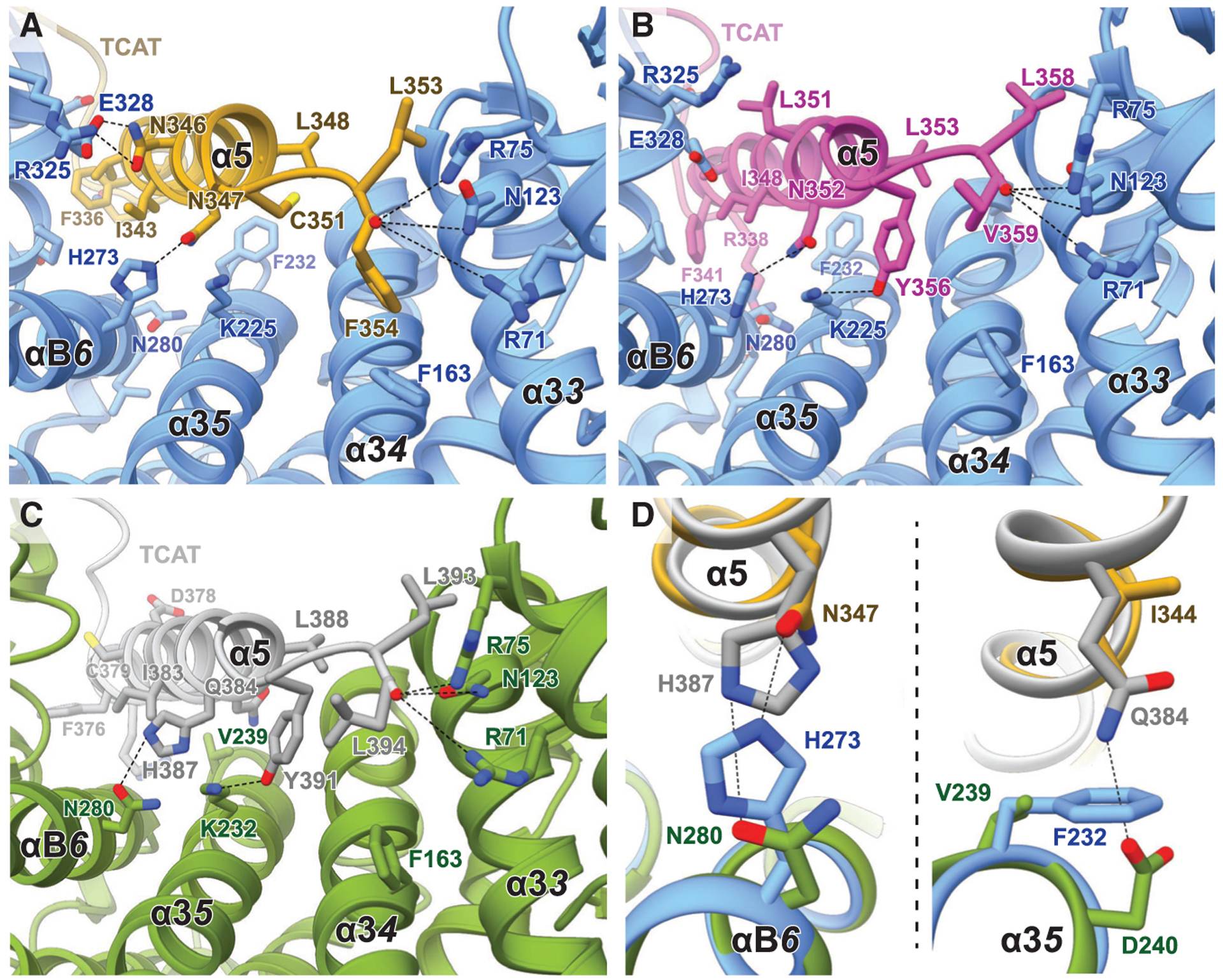
(A–C) CryoEM structures of (A) Ric-8A-Gαi1 (blue and yellow), (B) Ric-8A-Gαq (blue and magenta), and (C) the computational model of Ric-8B-Gαi1 (green and gray).
(D) Overlay of the side chains from the models Ric-8A-Gαi1 and Ric-8B-Gαs models that are potentially important for Ric-8 specificity.
Despite the overall similarity of the interfaces between Gαi1 or Gαq and Ric-8A, there are a few different contact residues in the α5 helix of the two G protein isotypes. Notably, these variations are well accommodated by Ric-8A through reorientation of side chains, the formation of alternative backbone or side-chain interactions, and conformational changes to adjust the size of the binding grove within the concave surface of the protein. Collectively, these variations result in a larger contact surface between the last 23 C-terminal residues of Gαq and Ric-8A (2,780Å2) compared to the Ric-8A-Gαi1 structure (2,201Å2). In contrast to Gαq, Gαi1 can be folded efficiently when expressed in bacteria that do not have a Ric-8 homolog (Linder et al., 1990). Thus, the tighter interaction between Ric-8A and Gαq, in comparison to Gαi1, might provide an explanation for the strict dependency of Ric-8A for Gαq folding and the lesser dependence of Gαi1. Furthermore, the larger contact surface may be a reason for the slightly more compact conformation of the chaperone in the Ric-8A-Gαq complex than in the Ric-8A-Gαi1 structure (Figure S4B). This suggests that Ric-8A is able to adjust the size of its G protein binding groove to accommodate the α5 helices of different G protein isotypes of the Gi/o, Gq/11, and G12/13 subfamilies, providing an explanation for the observed range of dependencies of various Gα subtypes for Ric-8 in cells.
In addition, we generated a homology model of the Ric-8B-Gαs complex and compared it with the Ric-8A complex structures to better understand G protein selectivity between the two Ric-8 subtypes. This comparison shows that important interactions are shared between the G protein complexes formed by the two Ric-8 subtypes. We observe significant similarities in side-chain interactions within the Ric-8A-Gαq and Ric-8B-Gαs complexes, which might explain the previously observed modest binding of Gαq to Ric-8B (Chan et al., 2011a). However, Ric-8A shows no GEF or chaperone activity for Gαs, suggesting that the sequence variations in the α5 helices of Gαi1, Gαq, and Gαs must contribute to the G protein selectivity of the two Ric-8 subtypes. Indeed, superposition of the Ric-8B-Gαs model and Ric-8A-Gα complex structures shows that differences in the amino acid composition of the α5 helix of Gαs are not compatible with the formation of key interactions with Ric-8A (Figure 5; Figure S7). Furthermore, side chain variations in Gαs presumably create some clashes and repulsions that might prevent binding to Ric-8A. Compared to the Ric-8A-Gα structures, the most pronounced difference in the predicted Ric-8B-Gαs complex is a swap of the side chains of N347 (Gαi1) and H273 (Ric-8A). In Gαs, N347, which is highly conserved in G proteins that bind to Ric-8A, is replaced by a histidine (H387). This alteration in Gαs would prevent the formation of the critical polar contact with H273 in Ric-8A and would create a clash between the two side chains (Figure 5D). In contrast, Ric-8B contains an asparagine (N280) at the corresponding position to H273 in Ric-8A that may enable a polar interaction with H387 of Gαs. An additional steric clash between Ric-8A and the α5 helix of Gαs might be caused between the glutamine residue Q384 of Gαs and F232 of Ric-8A. Ric-8B, on the other hand, possesses a smaller side chain (V239) at the homologous position, which is more compatible with Gαs binding (Figures 5C and 5D). Thus, subtype specificity of Ric-8 for G protein isotypes is not only encoded by the sequences of the α5 helices of the G proteins, as previously proposed (Srivastava et al., 2019), but is also mediated by differences in the G protein binding residues of Ric-8 subtypes that are crucial for the formation of specific contacts with different G protein isotypes (Figure 5; Figure S7).
Ric-8A Maintains Gαi1 and Gαq in a Quasi-folded State Primed for GTP Binding
The cryoEM structures of Ric-8A-Gαi1 and Ric-8A-Gαq reveal that the Gα subunit is partially folded, with key elements required for guanine nucleotide binding positioned with subtle differences in comparison to guanine nucleotide-bound conformations. The ARM/HEAT repeats and the C-terminal helix of Ric-8A interact with the α5 helix and the switch2 motif of Gα, respectively, to position the TCAT motif, the P loop, and switch2 in relatively close proximity to their guanine nucleotide-coordinated conformation, while the extended loop region of Ric-8A wraps around and stabilizes the Ras-like domain of Gα against the chaperone’s core. These structural features suggest that the observed Ric-8-bound Gα conformations are primed to engage GTP, which would lead to its release from the chaperone, as it has been shown biochemically (Chan et al., 2011a; Johnston et al., 2008; Tall et al., 2003). By chaperoning critical regions for nucleotide binding, such as the switch2 motif and the α-helix/TCAT-motif module of Gα, Ric-8 presumably ensures proper formation and positioning of the nucleotide binding elements before GTP binding and dissociation of the G protein subunit Gα from Ric-8. The initial guanine nucleotide binding may be facilitated through P loop interactions with the nucleotide phosphates; the three phosphates of GTP can form stronger interactions than the two phosphates of GDP, which could also explain the GTP preference for Gα release. Of note, GTP has a concentration that is 10-fold higher than GDP inside the cell (Traut, 1994), thereby potentially driving the chaperone activity of Ric-8 to evolve as a GTP-gated folding machine. Importantly, displacement of Gα switch2 also protects Gα from premature engagement of the Gβγ heterodimer. Alignment of the Ras-like domain in the structures of the Ric-8A-Gαi1 complex and the Gi1 protein heterotrimer reveals that switch2 is not adopting its partially α-helical conformation in the region that interacts with the Gβ subunit (Figure S5E). Thus, the placement of the Ric-8A C-terminal region prepares Ras-like domain elements for GTP binding while prohibiting premature Gβγ engagement (Tall et al., 2003).
Considering these observations, one emerging question is what the trigger is for GTP-gated Gα release from Ric-8A. To further probe this question and delineate the mechanism of chaperone and GEF activity of Ric-8A, we tested the role of key Ric-8 regions observed in our structures for effect on Gαi1 stimulated GDP release Gαi1 (Figure 6A). In our assays, intrinsic Gαi1 GDP release was enhanced by phosphorylated full-length Ric-8A. A Ric-8A truncation that includes the Ric-8A C-terminal region but lacks the entire core domain (MBP-Ric-8A 423–530) stabilized the GDP-bound state of Gαi1, suggesting that the C-terminal extended loop of Ric-8A can interact with Gα switch2, and potentially the alpha helical domain, without requiring an interaction between its ARM/HEAT repeats and the α5 helix of the Gα subunit. Assuming that the α5 helix maintains the same position as observed in the Gαi1-GDP crystal structure (Coleman and Sprang, 1998), this observation suggests that proper TCAT motif coordination with nucleotide stabilizes bound GDP even without proper switch2 motif positioning. In support of this interpretation, the switch2 region of Ras-like domain is not ordered in GDP-bound Gαi1 crystal structures (Raw et al., 1997; Rensland et al., 1995; Wittinghofer and Vetter, 2011). Strikingly, we observe that phosphorylated full-length Ric-8A promotes a comparable stabilization of the GDP bound state of Gαi1 that lacks only the C-terminal residue F354 (Gαi-DF354), even though this mutant G protein possesses a similar intrinsic GDP release rate as wild-type Gαi1. The importance of the C-terminal residue of Gαi1 (F354) is indicated from our structures and the previously reported structure of Ric-8A with the C-terminal Gα transducin α5 helix peptidomimetic (Srivastava et al., 2019). Ric-8A makes several interactions with the C-terminal residues of Gαi1 and Gαq including the coordination of the C-terminal carboxyl group by Ric-8A R71, R75, and N123 and an additional π-π interaction with F163, as described above.
Figure 6. The Chaperone and GEF Activity of Ric-8A Requires Recognition of the Gα C Terminus.
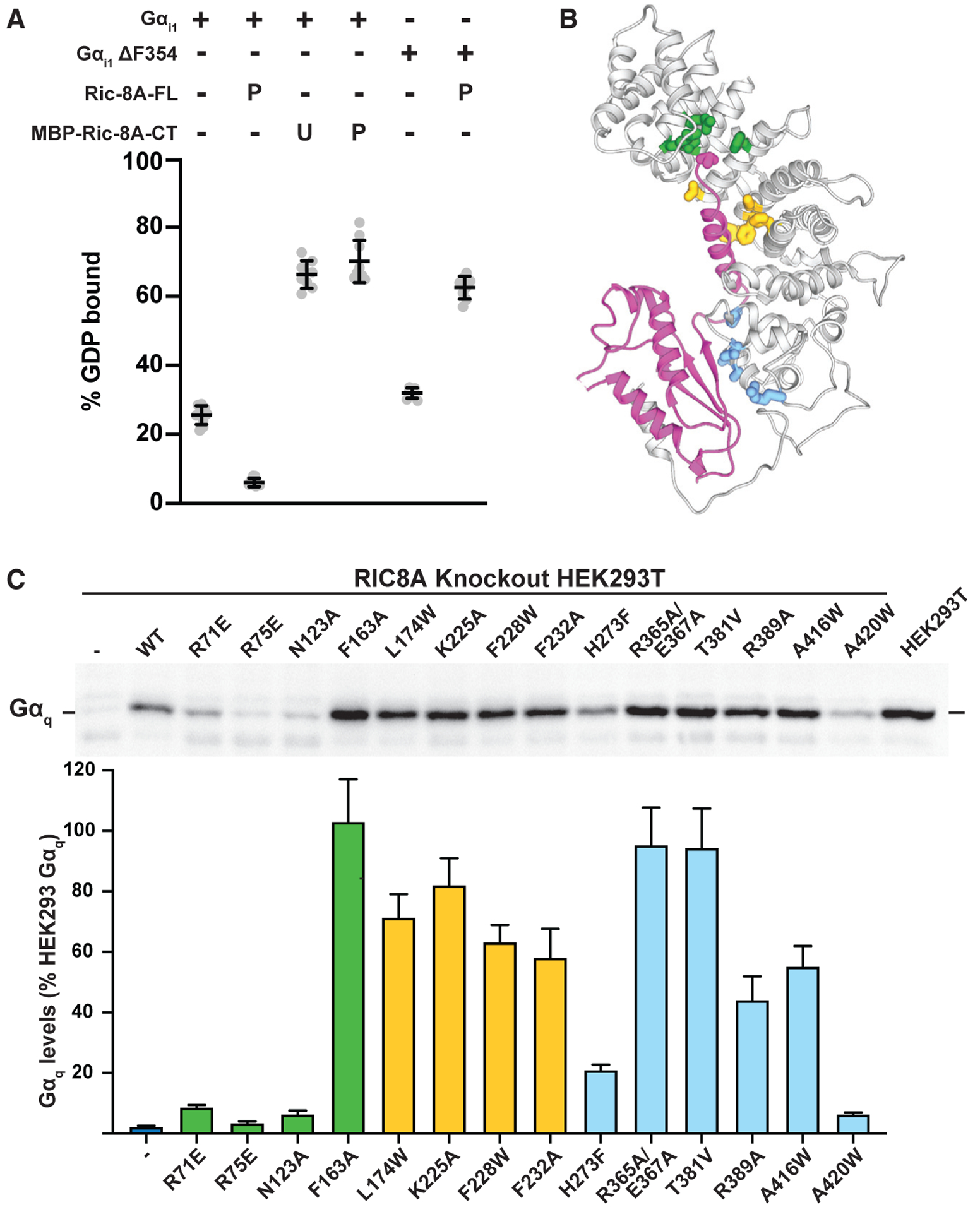
(A) Stabilization of the nucleotide-bound state of Gai1 by the Ric-8A C terminus and the importance of the α5 helix-Ric-8A interaction for the GEF activity of Ric-8A. The effect of the MBP-fused unphosphorylated “U” or phosphorylated “P” C terminus of Ric-8A (423–530) on Gα GDP release was tested with wild-type Gαi1 or Gαi1 D354 mutant proteins that were preloaded with 3H-GDP. The amount of bound 3H-GDP after 20 min incubation with or without Ric-8A was determined using a filter binding method and scintillation counting. Error bars are the mean ±SD, n R 3.
(B) Ribbon diagram of the Ric-8A-Gαq complex highlighting putative sites of interaction with the Gα C terminus (green), α5 helix (yellow), or Ras-like domain (blue).
(C) Knockout of RIC-8A in HEK293T cells results in a Gαq abundance defect due to Gα misfolding (Papasergi-Scott et al., 2018). The rescue of Gαq abundance upon stable expression of Ric-8A mutants was analyzed by quantitative immunoblotting of cell membrane samples. Representative western blot of membrane-bound Gαq is shown with quantification of Gaq levels as percentage of HEK293T Gαq abundance. Error bars are the mean ± SEM, n = 3.
To test the importance of individual residues of Ric-8A that interact with Gα, we utilized an established Ric-8A complementation assay in HEK293T cells in which RIC-8A was deleted using CRISPR/Cas9 technology (Papasergi-Scott et al., 2018). Devoid of Ric-8A, the cells have greatly diminished Gαq, Gα13, and Gαi protein abundance due to misfolding, which can be rescued to native levels by stable expression of functional Ric-8A (Figures 6A and 6B). Using this system, we created stable cell lines expressing Ric-8A mutated at predicted interaction sites identified from our structures. In cells expressing Ric-8A R71E, R75E, or N123A mutants, we observed a phenotype similar to RIC-8A knockout cells whereby the Gαq abundance defect was not rescued (Figures 6B and 6C; Figure S6C). These results indicate that the polar interactions of Ric-8A with the carboxy-terminus of the C-terminal Gα residue is a critical structural element for Ric-8 chaperone and GEF activities. Similarly, mutation of Ric-8A H273, which participates in hydrogen bonding interactions with the α5 helix and is predicted to be critical for Gα subtype selectivity (see above), significantly abrogates the Gαq abundance rescue. Single Ric-8A point mutations at other positions showed modest effects, with the exception of A420, which resides at the interface of Ric-8A with the β4-β5-β6 interface of the Gα Ras-like domain. Mutating A420 to a bulky tryptophan brings it into a direct clash with Gα F267, explaining its deleterious effect on chaperoning activity.
Our assays reveal that the residues of the Ric-8 chaperone that engage the C-terminal amino acid of the Gα Ras-like domain dramatically affect nucleotide-dependent release of Gα from Ric-8A. This indicates that binding of the C-terminal residue by the chaperone may be prerequisite for proper Gα folding. Moreover, binding of the α5 helix in the conformation observed in our structures positions the TCAT motif within the β6-α5 loop in close proximity to the nucleotide binding site. Of note, a synthesized 20-mer peptide of the α5 helix of Gαs did not adopt an α-helical structure in solution but a random coil conformation that was not able to bind to the β2-adrenergic receptor (Boyhus et al., 2018). This suggests that the α5 helix of G proteins might require the hydrophobic core interactions within the Ras-like domain or Ric-8 to form a helical structure. We therefore predict that binding of the nascent α5 helix region and C-terminal residue of Gα to the ARM/HEAT groove of Ric-8A might shield these hydrophobic residues and provide the environment required for folding of this critical region. Furthermore, the chaperoning of the α5 helix away from the Ras core structure would allow the Ras-like domain to fold before it could properly protect the α5 helix. Thus, we propose that engagement of the Gα C terminus by the chaperone serves as both a folding tool for critical regions across the protein and as a quality control check point to release properly folded Gα in response to high-affinity GTP binding.
DISCUSSION
The cryoEM structures of Ric-8A-Gαi1 and Ric-8A-Gαq complexes along with our in vitro and in vivo biochemical assays have led us to propose a model for the chaperoning and GEF activity of Ric-8 for Gα subunits (Figure 7). In this model, the C-terminal extended region of Ric-8 may engage the Ras-like domain of a nascent Gα polypeptide, possibly co-translationally, while also protecting it from premature Gβγ heterodimer engagement (Figure S5E). Phosphorylation of the Ric-8 C-terminal region creates interactions that serve to stabilize Gα against the ARM/HEAT repeats, creating a compact chaperoning cradle. In this configuration, the α-helical and Ras-like domain of Gα are kept at a distance to allow their independent folding. The Ras-like domain positioning also allows binding of its C terminus to a distinct position within the Ric-8 core, thereby protecting the amphipathic α5 helix and promoting its folding away from the Ras core domain. The recognition of the C-terminal Gα residue appears to provide a critical proof-reading mechanism that promotes the binding and folding of the α5 helix of Gα within the Ric-8 core. The α5 helix is a key element that undergoes conformational changes to engage a GPCR and to respond to the presence of GTP within the Ras-like domain. Thus, the Ric-8-Gα chaperone-client system has evolved to monitor the integrity of full-length Gα subunit production, with an emphasis on protecting a crucial Gα component. Upon Ric-8-assisted folding, the α5 helix can position the β6-α5 loop containing the TCAT motif in close proximity to engage GTP. Hence, when all nucleotide binding elements are properly folded and positioned, the coordinated engagement of GTP will pull the α5 helix away from the Ric-8 core and instead place it in its properly folded position where it is protected against the Ras-like domain. This compact, canonical configuration of the α5/Ras-like domain in the GTP-bound state likely favors Gα dissociation from Ric-8. GPCR-triggered guanine nucleotide exchange of Gα subunits within heterotrimeric G proteins is mainly carried out by perturbation of the β1-P loop region, a conformational change of the α5 helix and the associated displacement of the TCAT motif of Gα. Strikingly, Ric-8A uses a very similar strategy to stabilize the nucleotide-free form of Gα, where Ric-8A interacts with the β1 strand and the α5 helix of Gα, which orients the TCAT motif slightly outward from its nucleotide-bound conformation. Furthermore, in the Ric-8A-Gα complex, the α-helical domain is completely detached from the Ras-like domain, similarly to the Gα conformations observed in GPCR-G protein complexes. On the other hand, these domains are closed together in both the GDP- and GTP-bound conformations with the guanine nucleotide packed in between. Unlike GPCRs, Ric-8A additionally interacts with Gα switch2, which is critical for Gβγ binding to Gα. Considering these tailored interactions, together with the fact that, to date, Gα subunits are the only known folding clients for Ric-8, it emerges that Ric-8 is a uniquely evolved chaperone system for the biogenesis of Gα subunits.
Figure 7. Proposed Mechanism for Ric-8A Chaperoning of Gα.
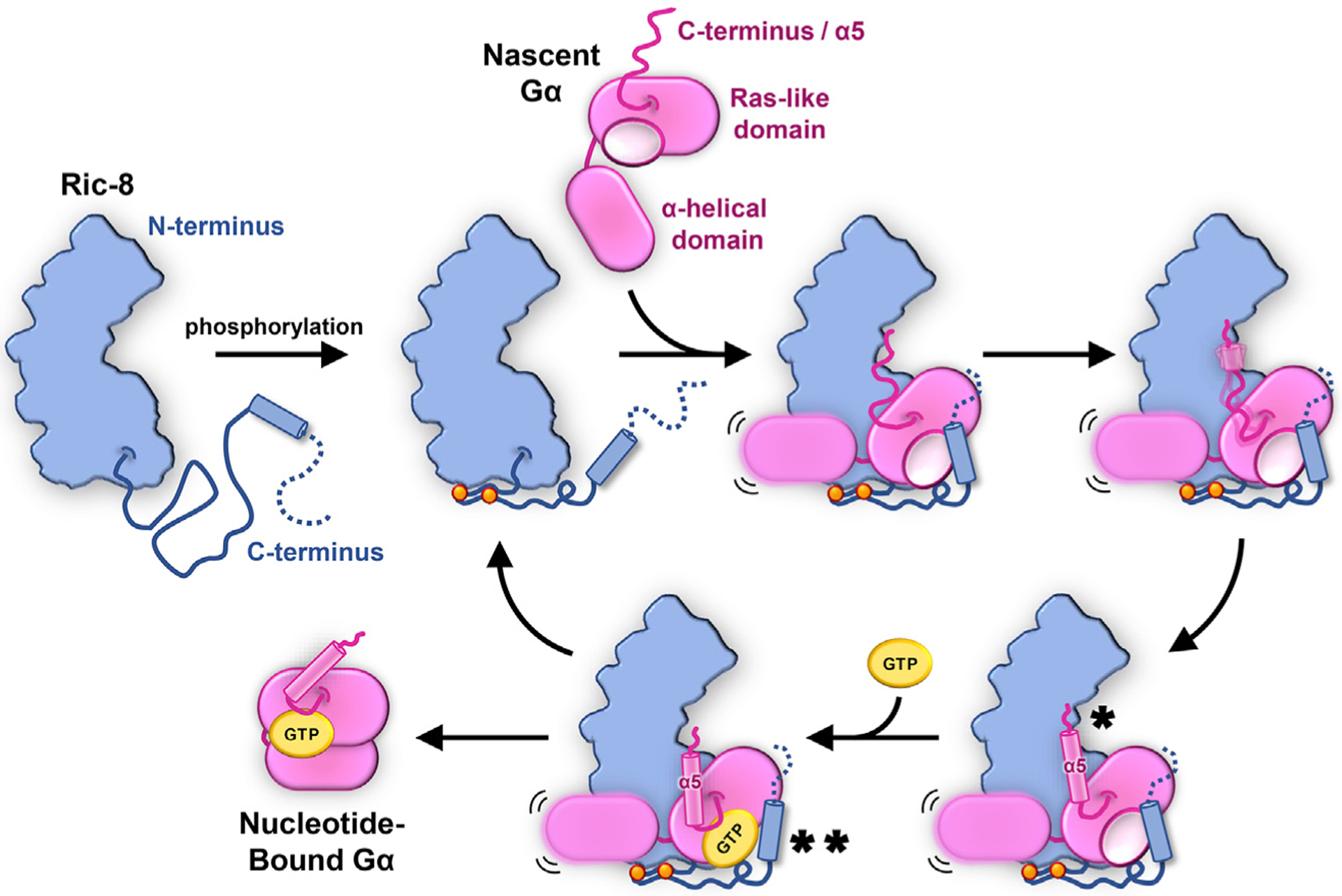
Dual phosphorylation of Ric-8 within a conserved acidic region (orange spheres) facilitates an intramolecular interaction that results in optimal positioning for Gα Ras-like domain engagement by Ric-8. Nascent, nucleotide-free Gα binds Ric-8 either co-translationally or shortly following release from the ribosome. Key interactions occur between the concave surface of the Ric-8 core and the a5-helix of Gα (*) and between a C-terminal helix of Ric-8 with Gα switch2 (**). The recognition of the final C-terminal residue of Gα by Ric-8 serves as a prerequisite for a5-helix folding (*) and positioning of the b6-a5 loop (TCAT motif) to engage GTP (**), thereby releasing properly folded Gα and freeing Ric-8 for subsequent rounds of chaperoning.
Notably, the chaperoning activity of Ric-8 has parallels to other well-described chaperone systems, including the ubiquitous HSP70 foldase. In both cases, upon successful folding of client protein, a nucleotide switch is employed to release the folded client from the chaperone (Figure 7) (Bukau and Horwich, 1998; Mayer and Bukau, 2005). In the case of the HSP70 system, HSP40 nucleotide exchange factors facilitate exchange of ATP for ADP on HSP70 in order to alter the conformation of HSP70 to favor client protein dissociation (Bukau and Horwich, 1998; Mayer and Bukau, 2005). By contrast, Ric-8 proteins are not ATPases, are not known to directly consume cellular energy during Gα subunit folding, and do not use an ADP-ATP switch to release folded Gα. Rather, the nucleotide switch feature is provided by the Gα client protein itself, which likely serves as the final checkpoint for the production of properly folded G protein. As a molecular chaperone, Ric-8A cradles the majority of the Gα Ras-like domain to facilitate its proper folding and development of the Gα guanine nucleotide binding pocket (Figures 1 and 7). When Gα is capable to competently bind GTP (i.e., is folded) it switches to its active conformation that has low affinity for Ric-8 and the complex dissociates (Figure 7). An appealing future direction of our work is to examine the prospect that Ric-8 may serve as a co-translational Gα subunit chaperone. The finding that the extreme C-terminal residue of Gα is required for folded Gα release from Ric-8 provides a timing or checkpoint mechanism during translation to ensure that the Ras-like domain core is properly folded prior to productive GTP binding and release. A lesser known fact of various isotypes of Gα subunits purified from bacteria or without the presence of Ric-8 is that a substantial portion of the protein preparations exist in a soluble, monomeric form that is inactive as far as the ability to bind guanine nucleotide, e.g., Gαq. This inactive, soluble form of Gα has been proposed to be a nucleotide-free, alternatively folded bottleneck conformation. The function of Ric-8 may therefore be to disfavor such inactive conformational bottlenecks and provide the chaperone activity to favor folding of active Gα.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Georgios Skiniotis (yiorgo@stanford.edu).
All the reagents generated in this study will be made available on request but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rat GST-TEV-Ric-8A was expressed in Tni (Trichoplusia ni) cells infected with recombinant baculovirus (pFastBac, Invitrogen). Tni cells were obtained from Expression Systems, LLC. Rat GST-TEV-Ric-8A together with human Gαq were expressed in Tni (Trichuplusia ni) cells infected with two recombinant baculoviruses (pFastBac, Invitrogen). Human 6His-3C-Gαi1 WT or 6His-3C-Gαi1 ΔF354 were expressed in Rosetta 2(DE3) cells (Merck Millipore). Human Gαi1 (6His between M119 and T120) was expressed in JM109 (DE3) cells (Promega). MBP-Ric-8A (423–530) was expressed in Rosetta 2 (DE3) cells. The HEK293T cell line (CRL-11268) was obtained from the American Type Culture Collection (ATCC). Crispr/Cas9 procedures were used previously to delete RIC-8A in the HEK293T cells (Papasergi-Scott et al., 2018). 3X-FLAG-Ric-8A point mutant plasmids were transfected and selected for stable expression with Hygromycin B.
METHOD DETAILS
Expression and purification of myristoylated Gαi1 for Ric-8A complex formation
Myristoylated Gαi1 was expressed and purified as described previously (Mumby and Linder, 1994) with some modifications. In brief, a hexa-histidine tag was inserted between amino acid residues M119 and T120 of the wild-type human Gαi1. The modified GNAI1 gene was cloned into a pETDuet-1 vector (Sigma Aldrich) together with the NMT1 gene that encodes for the N-myristoyl transferase from Saccharomyces cerevisiae. Both proteins were coexpressed in JM109 (DE3) cells (Promega) grown in Terrific Broth medium. After the cells reached an OD600 of 0.6, protein expression was induced by addition of 0.5 mM IPTG together with 30 mM of myristic acid. The cells were harvested after 15 hours induction at room temperature and resuspended in lysis buffer (50 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 50 μM GDP, 5 mM beta-mercaptoethanol, 5 mM imidazole, lysozyme, and protease inhibitors). Cells were disrupted by sonification using a 50% duty cycle, 70% power for four times 45 s. Intact cells and cell debris were subsequently removed by centrifugation and the supernatant was incubated with Ni-chelated Sepharose (GE healthcare) resin for 1.5 h at 4°C. The Ni-chelated Sepharose resin was washed multiple times in batch with lysis buffer supplemented with 20 mM imidazole and then loaded into a wide-bore glass column. The protein was eluted with lysis buffer containing 200 mM imidazole and dialyzed overnight in dialysis buffer (50 mM Tris pH 7.5, 100 mM sodium chloride, 1 mM EDTA, 2 mM dithiothreitol (DTT), 10 μM GDP). To isolate myristoylated Gαi1, the dialyzed protein was further purified by hydrophobic interaction chromatography using a HiPrep 16/10 Phenyl HP column (GE Healthcare). For this purpose, 3.6 M ammonium sulfate ((NH4)2SO4) was added dropwise to the protein to adjust the concentration to 1.2 M. Precipitated protein was removed by centrifugation and filtration and the protein was loaded onto a HiPrep 16/10 Phenyl HP column equilibrated with buffer A (50 mM Tris pH 7.5, 100 mM sodium chloride, 1 mM EDTA, 2 mM DTT, 10 mM GDP, 1.2 M (NH4)2SO4. The column was washed with 5 column volumes of buffer A before the protein was eluted with a linear gradient 0%–100% of buffer B (50 mM Tris pH 7.5, 100 mM sodium chloride, 1 mM EDTA, 2 mM DTT, 10 μM GDP) over 45 min. The myristoylated protein was concentrated and dialyzed overnight in buffer containing 20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 100 mM TCEP, and 10 mM GDP. After concentrating the protein, it was flash frozen in liquid nitrogen and stored at ~80°C until further use.
Formation and purification of the Ric-8A-Gαi1 complex
Phosphorylated Ric-8A protein purified from insect cells was mixed with myristoylated Gαi1 at a stoichiometric ratio of 1:1.5 and incubated for 90 min at room temperature. To stabilize the complex, 2.5 U of Apyrase was added to enzymatically digest GDP. After additional incubation for 90 min at room temperature, the complex was concentrated and further purified by size exclusion chromatography to remove monomeric Gαi1 and Ric-8A. For this purpose, the protein was loaded on a Superdex 200 Increase 10/300 GL column equilibrated with SEC buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 100 μM TsCEP). The complex peak was collected and concentrated to 15 mg/mL for cryoEM imaging.
Expression and purification of the Ric-8A-Gαq complex
The glutathione S-transferase (GST) tag, followed by a tobacco etch virus (TEV) protease site, was added to the N terminus of unmodified rat Ric-8A as described (Chan et al., 2011a). This construct as well as untagged, wild-type human Gαq were cloned into pFastBac1 expression vectors and recombinant baculoviruses were prepared using the Bac-to-Bac baculovirus expression system (Thermo Fisher). Trichoplusia ni cells (Expression Systems) were co-infected with both viruses at a ratio of 1:5 (GST-TEV-Ric-8A:Gαq) at a cell density of 3.0 × 106 cells per mL. Cultures were harvested 48 hours post infection and incubation at 27 C with gentle rotation. Cells were lysed in CHAPS buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM DTT, 11 mM CHAPS, protease inhibitors, 1 mM magnesium chloride, and 5 U Apyrase) by Dounce homogenization followed by sonification using a 50% duty cycle, 70% power for four times 45 s. The soluble fraction was incubated with Glutathione resin (Genscript) for 1 hour at 4 C. The Glutathione resin was washed several times in batch with CHAPS buffer and then loaded into a wide-bore glass column. The complex was eluted with CHAPS buffer supplemented with 10 mM reduced glutathione and dialyzed overnight at 4 C in dialysis buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM DTT, and 11 mM CHAPS). During dialysis, TEV protease (1:5 w/w) was added to the dialysis cassette to cleave off the amino-terminal GST-tag from Ric-8A. The cleaved GST tag and uncleaved fractions were removed by loading the protein onto Glutathione resin at 4 C. The flow through containing Ric-8A-Gαq complex, was concentrated and further purified by size-exclusion chromatography using a Superdex 200 Increase 10/300 column equilibrated with SEC buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 100 uM TCEP). The monomeric complex peak was collected and re-run on a Superdex 200 Increase 10/300 GL column to remove oligomers and finally concentrated to approximately 15 mg/mL for cryoEM imaging.
Expression and purification of non-myristoylated Gαi1 protein and the Gαi1-ΔF354 mutant
Human Gαi1 subunit and Gαi1-ΔF354 with an amino-terminal 6x histidine tag followed by a rhinovirus 3C protease side were expressed in Rosetta 2 (DE3) cells (Merck) using pET21a. Cells were grown in Terrific Broth to OD600 of 0.6, and protein expression was induced by addition of 0.5 mM IPTG. After 15 h of incubation at room temperature, cells were harvested and resuspended in lysis buffer (50 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 50 mM GDP, 5 mM beta-mercaptoethanol, 5 mM imidazole, lysozyme, and protease inhibitors). Cells were disrupted by sonification using a 50% duty cycle, 70% power for four times 45 s. Intact cells and cell debris were subsequently removed by centrifugation and the supernatant was incubated with Ni-chelated Sepharose (GE healthcare) resin for 1.5 h at 4 C. The Ni-chelated Sepharose resin was washed multiple times with lysis buffer in batch and then loaded into a wide-bore glass column, and protein was eluted with lysis buffer containing 200 mM imidazole. The eluted protein was dialyzed overnight in dialysis buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 20 μM GDP, 5 mM beta-mercaptoethanol, and 5 mM imidazole). The amino terminal histidine tag was cleaved by adding 1:1000 w/w 3C protease into the dialysis bag. Uncleaved protein, cleaved histidine tag, and 3C protease were subsequently removed by incubation with Ni-NTA resin for 45 min at 4 C. The resin was loaded into a wide-bore glass column and the flow-through containing the Gα subunit was collected. The protein was concentrated and run on a Superdex 200 10/300 GL column in SEC buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 20 μM GDP, and 100 uM TCEP).
Purification of His8-MBP-Ric-8A(423–530) for GDP release assay
The DNA encoding residues 423 to 530 of the C terminus of Ric-8A was cloned into a pMal vector to generate a construct containing a N-terminal octahistidine tag followed by the maltose binding protein and Ric-8A(423–530) (His8-MBP-Ric-8A(423–530)). Rosetta 2 (DE3) cells (Merck) transformed with pMal-His8-MBP-Ric-8A(423–530) were grown in Terrific Broth medium at 37 C to an OD600 of 1. For protein expression, the cells were induced with 0.5 mM IPTG for 15 hours at room temperature. After harvesting the cultures, cells were disrupted by sonification using a 50% duty cycle, 70% power for four times 45 s in buffer containing 50 mM HEPES pH 7.5, 100 mM sodium chloride, 5 mM beta-mercaptoethanol, 5 mM imidazole, protease inhibitors, and lysozyme. Intact cells and cell debris were subsequently removed by centrifugation and the supernatant was incubated with Ni-chelated Sepharose (GE healthcare) resin for 1.5 h at 4 C. The Ni-chelated Sepharose resin was washed 3 times in batch using wash buffer (50 mM HEPES pH 7.5, 100 mM sodium chloride, 5 mM beta-mercaptoethanol, protease inhibitors, and 20 mM imidazole). After loading the resin into a wide-bore glass column, it was washed with 10 column volumes of wash buffer before the protein was eluted in wash buffer supplemented with 200 mM imidazole. The eluted protein was concentrated and further purified by size-exclusion chromatography using a Superdex 200 Increase 10/300 GL column and SEC buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, and 100 uM TCEP) to remove oligomers and free MBP. The monomeric protein was collected, concentrated, and flash frozen in liquid nitrogen for further usage.
In vitro phosphorylation of MBP-Ric-8A (423–530)
Purified His8-MBP-Ric-8A(423–530) (4 mg) was phosphorylated with 60 μg of Casein Kinase II (CKII) (New England Biolabs Inc.) for 2 hours at 30 C in 4 mL of phosphorylation buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 5 mM EDTA, 100 uM TCEP, 10 mM magnesium chloride, and 250 μM ATP. The phosphorylated protein was further purified by size-exclusion chromatography and phosphorylation was analyzed with Pro-Q Diamond Phosphoprotein Gel Stain (Thermo Fisher).
GDP release assay
The GDP release assay was performed with purified full-length Ric-8A or MBP-Ric-8A(423–530) protein in the unphosphorylated or phosphorylated state, respectively, and Gαi1. For this purpose, Gαi1 in 20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 100 μM TCEP, and 5 μM GDP was diluted to 0.4 μM in GDP-loading buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 1 mM EDTA pH 8.0, 100 μM TCEP) and 3H-GDP (39.8 Ci/mmol, Perkin Elmer) was added to a final concentration of 2.5 μM. After incubation for 60 min at room temperature, the GDP release was initiated upon the addition of Ric-8A proteins (0 or 1 mM) in reaction buffer (20 mM HEPES pH 7.5, 100 mM sodium chloride, 1 mM magnesium chloride, 100 mM TCEP, and 100 μM GTPγS). At 0 and 20 min, 500 μL of ice-cold wash buffer (20 mM HEPES pH 7.5, 150 mM sodium chloride, 20 mM magnesium chloride, 30 uM aluminum sulfate, 5 mM sodium fluoride) was added to 20 uL of the reaction and the mixture was immediately filtered using a microanalysis filter holder (EMD Milipore) and pre-wet mixed cellulose filters (25 mm, 0.22 μm). The filter was washed three times with 500 μL ice-cold wash buffer and dried for at least 1 h at room temperature. The amount of radioactivity that remained bound to the filters was determined by liquid scintillation spectrometry. GDP release data were analyzed using GraphPad Prism.
Fluorescence polarization spectroscopy
FITC-labeled Ric-8A (504–530) peptide with phosphoserines at positions 522, 523, and 527 was obtained by custom peptide synthesis from Tufts University Core Facility and dissolved to an approximate concentration of approximately 1mM in 100 mM HEPES pH 7.4. The exact concentration was then determined by diluting the stock appropriately to be in a linear absorbance range and by measuring the FITC absorbance at 495 nm. Using the chosen dilution factor and the calculated extinction coefficient at pH 7.5 of 84000 cm−1 M−1, the exact stock concentrations was calculated. Fluorescence polarization spectroscopy measurements were performed and analyzed based on published guidelines (Moerke, 2009; Rossi and Taylor, 2011). To perform saturation binding experiments, two-fold dilution series in 20 mM HEPES pH 7.4, 100 mM sodium chloride, 100 μM TCEP, 0.01% (w/v) DDM and 20 μM GDP of unlabeled Gαi1, Gαs, and α-helical domain of Gαi1 (Gαi1-AHD) were made, yielding twelve samples with final concentrations ranging from 592 μM to 0.07 μM, 550 μM to 0.07 μM, and 1008 μM to 0.1 μM, respectively. Each of these samples was incubated with FITC-labeled peptide for 1 hour at room temperature prior to measurements, and for each dataset, a control sample containing no protein was included to measure the free anisotropy of FITC-labeled peptide. The FITC-labeled Ric-8A (504–530) was used at a final concentration of 4 nM in all assays. Measurements were performed in quintuplicate in a 384-well plate on a Tecan Infinite M1000 (Tecan Life Sciences), using an excitation wavelength of 470 nm, an emission wavelength of 525 nm and excitation, and emission bandwidths of 5 nm. The obtained data was fit using “One Site – Total” nonlinear regression in GraphPad Prism.
CryoEM data acquisition and processing
3.5 μl of Ric-8A-Gαi or Ric-8A-Gαq (5mg/mL) supplemented with 0.05% (w/v) β-octyl glucoside detergent was applied to freshly glow-discharged gold holey carbon grids (Quantifoil, Au-R1.2/1.3) under 100% humidity. Excess sample was blotted away for 1.5 s at 20 C, and the grids were subsequently plunged-frozen using a Vitrobot Mark IV (Thermo Fisher Scientific).
Ric-8A-Gα protein complexes were imaged at cryogenic temperatures with a Titan Krios G2 (ThermoFisher Scientific) transmission electron microscope with a post column energy filter (20mV slit width) operated at 300 kV on a Gatan K2 Summit direct electron camera at a magnification of × 130,000 (1.06Å/pixel) for Ric-8A-Gαi1 and × 165,000 (0.82Å/pixel) for Ric-8A-Gαq. 3095 micrographs, dose fractioned over 50 frames, for Ric-8A-Gαi1, and 7560 micrographs, dose fractioned over 48 frames, for Ric-8A-Gαi1 complexes, were recorded with a dose rate of 1.3 e/Å2 (7.3 e/pixel/second) using counted mode with a defocus range of 1.0–2.0 μm using SerialEM (Mastronarde, 2005). Beam induced anisotropic image motion on the micrographs was corrected using MotionCor2 (versions; 1.2.1 and 1.2.6) (Zheng et al., 2017) with dose weighting. Contrast transfer function values of each micrograph was initially determined using both CtfFind4 (Rohou and Grigorieff, 2015) and Gctf 1.06 (Zhang, 2016) and further refined using Relion3 (Zivanov et al., 2018). Auto-picked particle projections, 4,723,551 for Ric-8A-Gαq and 4,960,082 for Ric-8A-Gαi1, were subjected to several rounds of two-dimensional reference-free alignment to exclude damaged particles and contaminations using Relion3 (Zivanov et al., 2018). The initial models for both maps were obtained using Relion3 stochastic gradient descent algorithm implemented from cryoSPARC (Punjani et al., 2017). The particles in the well-defined 2D classes were further processed with several rounds of 3D classification to remove particles populating poorly defined classes. Initial 3D classifications yielded 424,000 high quality particle projections for Ric-8A-Gαq and 727,000 for Ric-8A-Gαi1. The core part of both Ric-8A-Gα complexes were stable across several 3D classifications, in contrast to the C-terminal extended region of Ric-8A that displayed variability. The most complete maps were obtained by selection of particle sets belonging to classes with clear density around the C-terminal extended loop of Ric-8A. Around 70,000 and 153,000 particles for Ric-8A-Gαq and Ric-8A-Gαi1, respectively, were refined independently to obtain maps with global indicated resolution of 3.5 and 4.1Å, respectively, using the FSC 0.143 criterion. The masks used for final refinements were generated with Relion3 and excluded the α-helical domain. Local resolution maps were generated with Relion3.1. Validation of cryoEM maps and models was performed with PHENIX (Liebschner et al., 2019). Three-dimensional orientation distribution is assessed by 3D-FSC curves generated using the 3DFSC tool (Tan et al., 2017). UCSF Chimera was used for map/model visualizations and figure preparation (Pettersen et al., 2004).
Model building and refinement
Initial models of Ric-8A-Gαi1 and Ric-8A-Gαq were built by rigid-body fitting of the Ric-8A (residues 1–423) (PDB: 6NMG) (Zeng et al., 2019), as well as the Ras-like domain of Gαi1 (PDB: 1GP2) (Wall et al., 1995) and Gαq (PDB: 3AH8) (Nishimura et al., 2010) into the cryoEM maps. Ric-8A residues 424–482 of the Ric-8A-Gαi1 and 424–484 of the Ric-8A-Gαq complex, as well as the b6-a5 loop and α5 helix of Gαi1 and Gαq, were built by de novo modeling. For most of Ric-8A, the map quality was sufficient for building and assign side chains, except for the C-terminal residues 452–484 in the Ric-8A-Gαq complex that did not allow unambiguous assignment of the registration of amino acid residues. As a result, residues 452–457 were not included in the final model and residues 458–484 were built as a poly alanine model. The map quality in this region was better in the Ric-8A-Gαi1 structure and allowed building of the Ric-8A model up to residue 482 including side chains. The Ras-like domain of Gαi1 was modeled between residues 32–54 and 193–354 due to the poor density of the alpha helical domain (residues 63–176) and adjacent loops that was only visible in low-pass filtered maps. For Gαq, we were only able to build a model for residues 217–359 due to the weaker cryoEM map density in the N-terminal region compared to Gαi1. Side chains and loop residues without sufficient EM density were stubbed or completely deleted.
The starting models were then subjected to iterative rounds of manual refinement in Coot (Emsley et al., 2010) and real space refinement in Phenix (Adams et al., 2011). The final models were visually inspected for general fit to the maps and the geometry was analyzed using MolProbity (Williams et al., 2018) as part of the Phenix software suite. The final refinement statistics for both models are summarized in supplemental data (Table S1). All molecular graphics figures were prepared with UCSF Chimera (Pettersen et al., 2004). Residue numbering for the interaction profiles between Ric-8A and Gα subunits (Figure S7) are based on Gαi1 and the “common Gα numbering (CGN) system in superscript” (Flock et al., 2015).
Generation of a homology model of Ric-8B-Gαs
A homology model of the Ric-8B-Gas complex was generated by using SWISS-MODEL (Waterhouse et al., 2018). For this purpose, we used the Ric-8A-Gαq structure as a template, including Ric-8A residues 1–451 and the poly-alanine model of the C-terminal tail (residues 458–484) as well as Gαq 217–359. For the target sequences we used the Uniprot entries P63092 (human Gαs) and Q9NVN3 (human Ric-8B).
Protein Thermal Stability Assays
Thermal stability of indicated Ric-8A proteins were analyzed by a SYPRO Orange fluorescent dye-based denaturation assay using a Bio-Rad C1000 – CFX96 Real-Time PCR detection system. SYPRO Orange dye was added at 1000x dilution in samples containing 0.5mg/mL Ric-8A 1–452 or full-length with or without phosphorylation (Buffer: 20 mM HEPES pH:7.4, 140 mM KCl, 5% glycerol and 1mM TCEP). Fluorescence of SYPRO orange is measured between 10 to 80 C (30 s per 1 C increments) using the SYBR channel on the CFX96 plate reader.
Cell culture and cell line generation
Wild-type and RIC-8A knockout HEK293T cell lines were cultured in DMEM with 10% (v/v) FBS. The RIC-8A knockout cell line was generated using CRISPR technology (Papasergi-Scott et al., 2018) RIC-8A knockout cell lines with stably-expressed 3X FLAG tagged Ric-8A point mutants were generated by transfection of pcDNA3.1-Hygro-3XFlag-TEV-Ric-8A point mutant plasmids and selection with 300 μg/mL Hygromycin B.
Membrane preparation and Immunoblotting
Cells were washed and harvested in PBS containing protease inhibitor cocktail (23 μg/mL phenylmethylsulfonyl fluoride, 21 μg/mL Na-p-tosyl-L-lysine-chloromethyl ketone, 21 μg/mL L-1-p-tosylamino-2-phenylethyl-chloromethyl ketone, 3.3 mg/mL leupeptin, and 3.3 mg/mL lima bean trypsin inhibitor). Cells were suspended in 20 mM HEPES, pH 8.0, 150 mM NaCl, 2 mM MgCl2 and 1 mM EDTA, and protease inhibitor cocktail and lysed using a nitrogen cavitation device (Parr Industries). The lysates were centrifuged at 750 g and the supernatants were centrifuged at 100,000 g for 30 min. The pelleted membranes were suspended in 2X SDS sample buffer. The amido black protein assay was used to quantify total protein in all samples (Schaffner and Weissmann, 1973). The lysates were resolved by 12% SDS-PAGE, transferred to nitrocellulose and immunoblotted with a Gq/11α antibody (Millipore 06–709).
Multiple sequence alignment and secondary structure prediction
The multiple sequence alignment for Ric-8 was done using Clustal Omega program (Madeira et al., 2019) and displayed using Jalview software (Waterhouse et al., 2009). The following species and their corresponding reference sequences are used for the RIC-8 multiple sequence alignment; Ric-8A Rattus norvegicus (NP_001093990.1), Ric-8A Homo sapiens (NP_001273063.1), Ric-8B Homo sapiens (NP_001317074.1), Ric-8 Drosophila melanogaster (NP_001285048.1), Ric-8 Caenorhabditis elegans (NP_001023561.1) and Ric-8 Xenopus tropicalis (NP_989159.1).
The secondary prediction for Ric-8A was done using PSIPRED 4.0 (Jones, 1999), visualized by Jalview (Waterhouse et al., 2009) and the previously known phosphorylation sites are manually added to its corresponding figure.
QUANTIFICATION AND STATISTICAL ANALYSIS
GDP dissociation was quantified by liquid scintillation spectrometry using a scintillation counter (Beckman). The counts after 20 min incubation time were normalized to the counts at time point zero to calculate the percentage of bound GDP: (counts at t20): (counts at t0). Experiments were run at least in triplicate and error bars are the mean ± SD.
Western blot band intensities were quantified by pixel densitometry analysis using Adobe Photoshop. Images were inverted and band intensities were measured using the formula: (Mean pixel intensity * area) - (Mean pixel intensity blank * area). Western blots were run in triplicate and error bars are the mean ± SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FLAG M2 antibody | Millipore-Sigma | Cat# F3165; RRID: AB_259529 |
| Gαq/11 antibody | Millipore-Sigma | Cat# G4290; RRID: AB_259903 |
| Bacterial and Virus Strains | ||
| JM109(DE3) | Promega | Cat# P9801 |
| Rosetta 2(DE3) | Sigma | Cat# 71400 |
| DH5αT1R | Invitrogen | 12297016 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Benzamidine | Sigma | Cat# L2884 |
| Leupeptin | Sigma | Cat# L2884 |
| CHAPS | Anatrace | Cat# C316 |
| GDP | Sigma | Cat#7127 |
| ESF921 culture medium | Expression Systems | Cat# 96–001 |
| TCEP | Sigma | Cat# C4706 |
| [3H]-GDP | PerkinElmer | Cat# NET966250UC |
| IPTG | Goldbio | Cat# 12481 |
| Myristic acid | Sigma | Cat#M3128 |
| Lysozyme | Sigma | Cat# 62970 |
| Chelating Sepharose Fast Flow | GE Healthcare | Cat# 17057502 |
| DTT | Goldbio | Cat# DTT |
| Apyrase | NEB | Cat# M0398L |
| Glutathione Resin | Genscript | Cat# L00206 |
| Casein Kinase II (CKII) | NEB | Cat# P6010L |
| Guanosine 5’-O-(3-thiotriphosphate) (GTPyS) | Abeam | Cat#ab146662 |
| FITC-labeled Ric-8A (504–530) peptide | Analytical Core Facility, Tufts University | Custom Synthesis |
| Hygromycin B | Invitrogen | 10687010 |
| DMEM | Invitrogen | 11995065 |
| FBS, USDA | Invitrogen | 10437036 |
| Mixed cellulose membrane | EMD Millipore | Cat# GSWP02500 |
| HiPrep Phenyl HP 16/10 | GE Healthcare | Cat#29018184 |
| SYPRO Orange | Life Technologies Corporation | Cat# S6650 |
| Deposited Data | ||
| Ric-8A-Gαq coordinates | This paper | PDB: 6VU5 |
| Ric-SA-Gαi1 coordinates | This paper | PDB: 6VU7 |
| Ric-8A-Gαcq EM map | This paper | EMDB:EMD-21387 |
| Ric-SA-Gαn EM map | This paper | EMDB: EMD-21388 |
| Experimental Models: Cell Lines | ||
| Trichuplusia ni | Expression Systems | Cat# 94–002S |
| HEK293T | ATCC | CRL-11268 |
| HEK293T RIC-8A knockout | Papasergi-Scott et al. | N/A |
| Oligonucleotides | ||
| Refer to Table S2 | N/A | |
| Recombinant DNA | ||
| pFastbac-Gαq | This work | N/A |
| pFastbac-GST-TEV-Ric-8A | Chan etal., 2011a | N/A |
| DMal-His8-MBP-Ric-8A(423–530) | This work | N/A |
| pETDuet-1-Gαi1(6His between M119 and T120)/NMT1 | This work | N/A |
| pET21a-His6-3C-Gαi1 | This work | N/A |
| pET21a-His6–3C-Gαi1-ΔF354 | This work | N/A |
| 3X-FLAG-Ric-8A pcDNA3.1 Hygro | This work | N/A |
| Software and Algorithms | ||
| JCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| COOT | Emsley etal., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Mlolprobity | Williams etal., 2018 | http://molprobity.biochem.duke.edu/ |
| Phenix 1.17.1 | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| SWISS-MODEL | Waterhouse et al., 2018 | https://swissmodel.expasy.org/ |
| Adobe Photoshop 2020 | Adobe | https://www.adobe.com/ |
| Adobe Illustrator 2020 | Adobe | https://www.adobe.com/ |
| Relion3 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/ |
| CtfFind4 | Rohou and Grigorieff, 2015 | https://grigoriefflab.umassmed.edu/ctffind4 |
| Gctf | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/ |
| VlotionCor2 | Zheng etal., 2017 | https://emcore.ucsf.edu/ucsf-motioncor2 |
| 3DFSC | Liebschner et al., 2019 | https://3dfsc.salk.edu/ |
| Other | ||
| 3YPRO Orange | Life Technologies Corporation | Cat# S6650 |
Highlights.
CryoEM structures of Gαi1 and Gαq protein subunits in complex with chaperone Ric-8A
Ric-8 forms a cradle to accommodate the Ras-like domain of Gα
Gα C terminus binding to Ric-8 is a prerequisite for GTP-gated Gα release
Insights into G protein subtype binding specificity by Ric-8 isoforms
ACKNOWLEDGMENTS
We would like to thank Matthieu Masureel for fluorescent polarization assays, Rashed Chowdhury for initial data processing, Gozde Eskici and Michael J. Robertson for fruitful discussions, Elizabeth Ann Montabana and David A. Bushnell for assistance with cryoEM, and Wenxi Yu for technical support. Financial support was provided by the National Institutes of Health (Grants R01 NS092695 to G.S., R01NS028471 to B.K.K., T32-GM06841 and PhRMA Foundation predoctoral fellowship to M.M.P.-S., and GM088242 to G.G.T.)
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.086.
DECLARATION OF INTERESTS
B.K.K. is a co-founder of and consultant for ConfometRx, Inc.
DATA AND CODE AVAILABILITY
The atomic coordinates of the Ric-8A-Gαq and Ric-8A-Gαi1 have been deposited in the Protein Data Bank (PDB) under accession number PDB: 6VU5 and PDB: 6VU8, respectively. The cryoEM maps of Ric-8A-Gαq and Ric-8A-Gαi1 have been deposited in the Electron Microscopy Data Bank (EMDB) with the codes EMDB: EMD-21387 and EMDB: EMD-21388, respectively.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. (2011). The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyhus L-E, Danielsen M, Bengtson NS, Kunze MBA, Kubiak X, Sminia TJ, Løper JH, Tran PT, Lindorff-Larsen K, Rasmussen SGF, et al. (2018). Gs protein peptidomimetics as allosteric modulators of the beta(2)- adrenergic receptor. RSC Adv. 8, 2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, and Horwich AL (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Chan P, Gabay M, Wright FA, Kan W, Oner SS, Lanier SM, Smrcka AV, Blumer JB, and Tall GG (2011a). Purification of heterotrimeric G protein alpha subunits by GST-Ric-8 association: primary characterization of purified G alpha(olf). J. Biol. Chem 286, 2625–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Gabay M, Wright FA, and Tall GG (2011b). Ric-8B is a GTP-dependent G protein alphas guanine nucleotide exchange factor. J. Biol. Chem 286, 19932–19942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Thomas CJ, Sprang SR, and Tall GG (2013). Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein a subunits. Proc. Natl. Acad. Sci. USA 110, 3794–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DE, and Sprang SR (1998). Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment. Biochemistry 37, 14376–14385. [DOI] [PubMed] [Google Scholar]
- Dupré DJ, Robitaille M, Richer M, Ethier N, Mamarbachi AM, and Hébert TE (2007). Dopamine receptor-interacting protein 78 acts as a molecular chaperone for Ggamma subunits before assembly with Gbeta. J. Biol. Chem 282, 13703–13715. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock T, Ravarani CNJ, Sun D, Venkatakrishnan AJ, Kayikci M, Tate CG, Veprintsev DB, and Babu MM (2015). Universal allosteric mechanism for Gα activation by GPCRs. Nature 524, 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, and Tall GG (2011). Ric-8 proteins are molecular chaperones that direct nascent G protein a subunit membrane association. Sci. Signal 4, ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goricanec D, Stehle R, Egloff P, Grigoriu S, Plückthun A, Wagner G, and Hagn F (2016). Conformational dynamics of a G-protein a subunit is tightly regulated by nucleotide binding. Proc. Natl. Acad. Sci. USA 113, E3629–E3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger D, Masureel M, and Kobilka BK (2018). Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol 25, 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Afshar K, Snyder JT, Tall GG, Gönczy P, Siderovski DP, and Willard FS (2008). Structural determinants underlying the temperature-sensitive nature of a Galpha mutant in asymmetric cell division of Caenorhabditis elegans. J. Biol. Chem 283, 21550–21558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Lee MJ, and Dohlman HG (2008). Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr. Biol 18, 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yan X, Wang H, Liang S, Ma WB, Fang MY, Talbot NJ, and Wang ZY (2010). MoRic8 Is a novel component of G-protein signaling during plant infection by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact 23, 317–331. [DOI] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Ewald DA, Miller RJ, and Gilman AG (1990). Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli. J. Biol. Chem 265, 8243–8251. [PubMed] [Google Scholar]
- Lukov GL, Hu T, McLaughlin JN, Hamm HE, and Willardson BM (2005). Phosducin-like protein acts as a molecular chaperone for G protein betagamma dimer assembly. EMBO J. 24, 1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukov GL, Baker CM, Ludtke PJ, Hu T, Carter MD, Hackett RA, Thulin CD, and Willardson BM (2006). Mechanism of assembly of G protein betagamma subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J. Biol. Chem 281, 22261–22274. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, and Lopez R (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly J, and Crowhurst KA (2012). Expression, purification and preliminary NMR characterization of isotopically labeled wild-type human heterotrimeric G protein ai1. Protein Expr. Purif 84, 255–264. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mayer MP, and Bukau B (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova M, Preininger AM, Yu NJ, Hubbell WL, and Hamm HE (2002). Conformational changes in the amino-terminal helix of the G protein alpha(i1) following dissociation from Gbetagamma subunit and activation. Biochemistry 41, 9962–9972. [DOI] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, and Rand JB (1996). A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA 93, 12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, McManus JR, and Rand JB (2000). RIC-8 (Synembryn): a novel conserved protein that is required for G(q)alpha signaling in the C. elegans nervous system. Neuron 27, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ (2009). Fluorescence Polarization (FP) Assays for Monitoring Peptide-Protein or Nucleic Acid-Protein Binding. Curr. Protoc. Chem. Biol 1, 1–15. [DOI] [PubMed] [Google Scholar]
- Mumby SM, and Linder ME (1994). Myristoylation of G-protein alpha subunits. Methods Enzymol. 237, 254–268. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Nishimura A, Tago K, Mizuno N, and Itoh H (2010). Ric-8B stabilizes the alpha subunit of stimulatory G protein by inhibiting its ubiquitination. J. Biol. Chem 285, 11114–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, and Rand JB (1995). Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 140, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura A, Kitano K, Takasaki J, Taniguchi M, Mizuno N, Tago K, Hakoshima T, and Itoh H (2010). Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc. Natl. Acad. Sci. USA 107, 13666–13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oner SS, Maher EM, Gabay M, Tall GG, Blumer JB, and Lanier SM (2013). Regulation of the G-protein regulatory-Gai signaling complex by non-receptor guanine nucleotide exchange factors. J. Biol. Chem 288, 3003–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasergi MM, Patel BR, and Tall GG (2015). The G protein a chaperone Ric-8 as α potential therapeutic target. Mol. Pharmacol 87, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasergi-Scott MM, Stoveken HM, MacConnachie L, Chan PY, Gabay M, Wong D, Freeman RS, Beg AA, and Tall GG (2018). Dual phosphorylation of Ric-8A enhances its ability to mediate G protein α subunit folding and to stimulate guanine nucleotide exchange. Sci. Signal 11, eaap8113 10.1126/scisignal.aap8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). Cryo-SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011). Crystal structure of the b2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raw AS, Coleman DE, Gilman AG, and Sprang SR (1997). Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42–> Val mutant of Gialpha1. Biochemistry 36, 15660–15669. [DOI] [PubMed] [Google Scholar]
- Rensland H, John J, Linke R, Simon I, Schlichting I, Wittinghofer A, and Goody RS (1995). Substrate and product structural requirements for binding of nucleotides to H-ras p21: the mechanism of discrimination between guanosine and adenosine nucleotides. Biochemistry 34, 593–599. [DOI] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi AM, and Taylor CW (2011). Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc 6, 365–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W, and Weissmann C (1973). A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem 56 (2), 502–514. 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Gakhar L, and Artemyev NO (2019). Structural underpinnings of Ric8A function as a G-protein a-subunit chaperone and guanine-nucleotide exchange factor. Nat. Commun 10, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Krumins AM, and Gilman AG (2003). Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J. Biol. Chem 278, 8356–8362. [DOI] [PubMed] [Google Scholar]
- Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, and Lyumkis D (2017). Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CJ, Briknarová K, Hilmer JK, Movahed N, Bothner B, Sumida JP, Tall GG, and Sprang SR (2011). The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS ONE 6, e23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW (1994). Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem 140, 1–22. [DOI] [PubMed] [Google Scholar]
- Van Eps N, Thomas CJ, Hubbell WL, and Sprang SR (2015). The guanine nucleotide exchange factor Ric-8A induces domain separation and Ras domain plasticity in Gαi1. Proc. Natl. Acad. Sci. USA 112, 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, and Sprang SR (1995). The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, et al. (2011). Structural flexibility of the G alpha s alphα-helical domain in the beta2-adrenoceptor Gs complex. Proc. Natl. Acad. Sci. USA 108, 16086–16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie TM, Gilbert DJ, Olsen AS, Chen XN, Amatruda TT, Korenberg JR, Trask BJ, de Jong P, Reed RR, Simon MI, et al. (1992). Evolution of the mammalian G protein alpha subunit multigene family. Nat. Genet 1, 85–91. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittinghofer A, and Vetter IR (2011). Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem 80, 943–971. [DOI] [PubMed] [Google Scholar]
- Wright SJ, Inchausti R, Eaton CJ, Krystofova S, and Borkovich KA (2011). RIC8 is a guanine-nucleotide exchange factor for Galpha subunits that regulates growth and development in Neurospora crassa. Genetics 189, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B, Mou TC, Doukov TI, Steiner A, Yu W, Papasergi-Scott M, Tall GG, Hagn F, and Sprang SR (2019). Structure, Function, and Dynamics of the Galpha Binding Domain of Ric-8A. Structure 27, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


