Abstract
STUDY QUESTION
Can an artificial intelligence (AI)-based model predict human embryo viability using images captured by optical light microscopy?
SUMMARY ANSWER
We have combined computer vision image processing methods and deep learning techniques to create the non-invasive Life Whisperer AI model for robust prediction of embryo viability, as measured by clinical pregnancy outcome, using single static images of Day 5 blastocysts obtained from standard optical light microscope systems.
WHAT IS KNOWN ALREADY
Embryo selection following IVF is a critical factor in determining the success of ensuing pregnancy. Traditional morphokinetic grading by trained embryologists can be subjective and variable, and other complementary techniques, such as time-lapse imaging, require costly equipment and have not reliably demonstrated predictive ability for the endpoint of clinical pregnancy. AI methods are being investigated as a promising means for improving embryo selection and predicting implantation and pregnancy outcomes.
STUDY DESIGN, SIZE, DURATION
These studies involved analysis of retrospectively collected data including standard optical light microscope images and clinical outcomes of 8886 embryos from 11 different IVF clinics, across three different countries, between 2011 and 2018.
PARTICIPANTS/MATERIALS, SETTING, METHODS
The AI-based model was trained using static two-dimensional optical light microscope images with known clinical pregnancy outcome as measured by fetal heartbeat to provide a confidence score for prediction of pregnancy. Predictive accuracy was determined by evaluating sensitivity, specificity and overall weighted accuracy, and was visualized using histograms of the distributions of predictions. Comparison to embryologists’ predictive accuracy was performed using a binary classification approach and a 5-band ranking comparison.
MAIN RESULTS AND THE ROLE OF CHANCE
The Life Whisperer AI model showed a sensitivity of 70.1% for viable embryos while maintaining a specificity of 60.5% for non-viable embryos across three independent blind test sets from different clinics. The weighted overall accuracy in each blind test set was >63%, with a combined accuracy of 64.3% across both viable and non-viable embryos, demonstrating model robustness and generalizability beyond the result expected from chance. Distributions of predictions showed clear separation of correctly and incorrectly classified embryos. Binary comparison of viable/non-viable embryo classification demonstrated an improvement of 24.7% over embryologists’ accuracy (P = 0.047, n = 2, Student’s t test), and 5-band ranking comparison demonstrated an improvement of 42.0% over embryologists (P = 0.028, n = 2, Student’s t test).
LIMITATIONS, REASONS FOR CAUTION
The AI model developed here is limited to analysis of Day 5 embryos; therefore, further evaluation or modification of the model is needed to incorporate information from different time points. The endpoint described is clinical pregnancy as measured by fetal heartbeat, and this does not indicate the probability of live birth. The current investigation was performed with retrospectively collected data, and hence it will be of importance to collect data prospectively to assess real-world use of the AI model.
WIDER IMPLICATIONS OF THE FINDINGS
These studies demonstrated an improved predictive ability for evaluation of embryo viability when compared with embryologists’ traditional morphokinetic grading methods. The superior accuracy of the Life Whisperer AI model could lead to improved pregnancy success rates in IVF when used in a clinical setting. It could also potentially assist in standardization of embryo selection methods across multiple clinical environments, while eliminating the need for complex time-lapse imaging equipment. Finally, the cloud-based software application used to apply the Life Whisperer AI model in clinical practice makes it broadly applicable and globally scalable to IVF clinics worldwide.
STUDY FUNDING/COMPETING INTEREST(S)
Life Whisperer Diagnostics, Pty Ltd is a wholly owned subsidiary of the parent company, Presagen Pty Ltd. Funding for the study was provided by Presagen with grant funding received from the South Australian Government: Research, Commercialisation and Startup Fund (RCSF). ‘In kind’ support and embryology expertise to guide algorithm development were provided by Ovation Fertility. J.M.M.H., D.P. and M.P. are co-owners of Life Whisperer and Presagen. Presagen has filed a provisional patent for the technology described in this manuscript (52985P pending). A.P.M. owns stock in Life Whisperer, and S.M.D., A.J., T.N. and A.P.M. are employees of Life Whisperer.
Keywords: assisted reproduction, embryo quality, IVF/ICSI outcome, artificial intelligence, machine learning
Introduction
With global fertility generally declining (GBD, 2018), many couples and individuals are turning to assisted reproduction procedures for help with conception. Unfortunately, success rates for IVF are quite low at ~20–30% (Wang and Sauer, 2006), placing significant emotional and financial strain on those seeking to achieve a pregnancy. During the IVF process, one of the critical determinants of a successful pregnancy is embryo quality, and the embryo selection process is essential for ensuring the shortest time to pregnancy for the patient. There is a pressing motivation to improve the way in which embryos are selected for transfer into the uterus during the IVF process.
Currently, embryo selection is a manual process involving assessment of embryos by trained clinical embryologists, through visual inspection of morphological features using an optical light microscope. The most common scoring system used by embryologists is the Gardner Scale (Gardner and Sakkas, 2003), in which morphological features such as inner cell mass (ICM) quality, trophectoderm quality and embryo developmental advancement are evaluated and graded according to an alphanumeric scale. One of the key challenges in embryo grading is the high level of subjectivity and intra- and inter-operator variability that exists between embryologists of different skill levels (Storr et al., 2017). This means that standardization is difficult even within a single laboratory, and impossible across the industry as a whole. Other complementary techniques are available for assisting with embryo selection, such as time-lapse imaging, which continuously monitors the growth of embryos in culture with simple algorithms that assess critical growth milestones. Although this approach is useful in determining whether an embryo at an early stage will develop through to a mature blastocyst, it has not been demonstrated to reliably predict pregnancy outcomes, and therefore is limited in its utility for embryo selection at the Day 5 time point (Chen et al., 2017). Additionally, the requirement for specialized time-lapse imaging hardware makes this approach cost prohibitive for many laboratories and clinics, and limits widespread use of the technique.
The objective of the current clinical investigation was to develop and test a non-invasive artificial intelligence (AI)-based assessment approach to aid in embryo selection during IVF, using single static two-dimensional images captured by optical light microscopy methods. The aim was to combine computer vision image processing methods and deep learning to create a robust model for analysis of Day 5 embryos (blastocysts) for prediction of clinical pregnancy outcomes. This is the first report of an AI-based embryo selection method that can be used for analysis of images taken using standard optical light microscope systems, without requiring time-lapse imaging equipment for operation, and which is predictive of pregnancy outcome. Using an AI screening method to improve selection of embryos prior to transfer has the potential to improve IVF success rates in a clinical setting.
A common challenge in evaluating AI and machine learning methods in the medical industry is that each clinical domain is unique, and requires a specialized approach to address the issue at hand. There is a tendency for industry to compare the accuracy of AI in one clinical domain to another, or to compare the accuracy of different AI approaches within a domain that assess different endpoints. These are not valid comparisons as they do not consider the clinical context, nor the relevance of the ground-truth endpoint used for the assessment of the AI. Caution needs to be taken to understand the context in which the AI is operating and the benefit it provides in complement with current clinical processes. One example presented by Sahlsten et al. (2019) described an AI model that detected fundus for diabetic retinopathy assessment with an accuracy of over 90%. In this domain, the clinician baseline accuracy is ~90%, and therefore an AI accuracy of >90% is reasonable and necessary to justify clinical relevance. Similarly, in the field of embryology, AI models developed by Khosravi et al. (2019) and Kragh et al. (2019) showed high levels of accuracy in classification of blastocyst images according to the well-established Gardner scale. This approach is expected to yield a high accuracy, as the model simply mimics the Gardner scale to predict a known outcome. While this method may be useful for standardization of embryo classification according to the Gardner scale, it is not in fact predictive of pregnancy success as it is based on a different endpoint. Of relevance to the current study, an AI model developed by Tran et al. (2019) was in fact intended to classify embryo quality based on clinical pregnancy outcome. This study did not report percentage accuracy of prediction, but instead reported a high level of accuracy for their model IVY using a receiver operating characteristic (ROC) curve. The AUC for IVY was 0.93 for true positive rate versus false positive rate; negative predictions were not evaluated. However, the datasets used for training and evaluation of this model were only partly based on actual ground-truth clinical pregnancy outcome—a large proportion of predicted non-viable embryos were never actually transferred, and were only assumed to lead to an unsuccessful pregnancy outcome. Thus, the reported performance is not entirely relevant in the context of clinical applicability, as the actual predictive power for the presence of a fetal heartbeat has not been evaluated to date.
The AI approach presented here is the first study of its kind to evaluate the true ability of AI for predicting pregnancy outcome, by exclusively using ground-truth pregnancy outcome data for AI development and testing. It is important to note that while a pregnancy outcome endpoint is more clinically relevant and informative; it is inherently more complex in nature due to patient and laboratory variability that impact pregnancy success rates beyond the quality of the embryo itself. The theoretical maximum accuracy for prediction of this endpoint based on evaluation of embryo quality is estimated to be ~80%, with the remaining 20% affected by patient-related clinical factors, such as endometriosis, or laboratory process errors in embryo handling, etc., that could lead to a negative pregnancy outcome despite a morphologically favorable embryo appearance (Annan et al., 2013). Given the influence of confounding variables, and the low average accuracy presently demonstrated by embryologists using traditional morphological grading methods (~50% in the current study, Tables I and II), an AI model with an improvement of even 10–20% over that of embryologists would be considered highly relevant in this clinical domain. In the current study, we aimed to develop an AI model that demonstrated superiority to embryologists’ predictive power for embryo viability, as determined by ground-truth clinical pregnancy outcome.
Table I.
Results of pilot study demonstrate feasibility of creating an artificial intelligence-based image analysis model for prediction of human embryo viability.
| Validation dataset | Blind Test Set 1 | Blind Test Set 2 | Total blind test dataset | |
|---|---|---|---|---|
| Composition of dataset | ||||
| Total image no. | 390 | 368 | 632 | 1000 |
| No. of positive clinical pregnancies | 70 | 76 | 194 | 270 |
| No. of negative clinical pregnancies | 320 | 292 | 438 | 730 |
| AI model accuracy | ||||
| Accuracy viable embryos (sensitivity) | 74.3% | 63.2% | 78.4% | 74.1% |
| Accuracy non-viable embryos (specificity) | 74.4% | 77.1% | 57.5% | 65.3% |
| Overall accuracy | 74.4% | 74.2% | 63.9% | 67.7% |
| Comparison to embryologist grading – viable versus non-viable embryos | ||||
| No. of images with embryologist grade | ND | 121 | 477 | 598 |
| AI model accuracy | ND | 71.9% | 65.4% | 66.7% |
| Embryologist accuracy | ND | 47.1% | 52.0% | 51.0% |
| AI model improvement | ND | 52.7% | 25.8% | 30.8% |
| No. times AI model correcta | ND | 42 | 106 | 148 |
| No. times embryologist correcta | ND | 12 | 42 | 54 |
| AI model fold improvement | ND | 3.5 x | 2.5 x | 2.7 x |
aImages where the AI model was correct and the embryologist was incorrect, and vice versa.
AI = artificial intelligence; ND = not done; No. = number.
Table II.
Results of the pivotal study demonstrate generalizability of the Life Whisperer AI model for prediction of human embryo viability across multiple clinical environments.
| Validation dataset | Blind Test Set 1 | Blind Test Set 2 | Blind Test Set 3 | Combined blind sets | |
|---|---|---|---|---|---|
| Composition of dataset | |||||
| No. of images | 193 | 280 | 286 | 1101 | 1667 |
| No. of positive clinical pregnancies | 97 | 141 | 180 | 334 | 655 |
| No. of negative clinical pregnancies | 96 | 139 | 106 | 767 | 1012 |
| AI model accuracy | |||||
| Accuracy viable embryos (sensitivity) | 76.3% | 72.3% | 73.9% | 67.1% | 70.1% |
| Accuracy non-viable embryos (specificity) | 53.1% | 54.7% | 54.7% | 62.3% | 60.5% |
| Overall accuracy | 64.8% | 63.6% | 66.8% | 63.8% | 64.3% |
| Comparison to embryologist grading – viable versus non-viable embryos | |||||
| No. of images with embryologist grade | ND | 262 | 0a | 539 | 801 |
| AI model accuracy | ND | 63.7% | ND | 57.0% | 59.2% |
| Embryologist accuracy | ND | 50.4% | ND | 46.0% | 47.4% |
| AI model improvement | ND | 26.4% | ND | 23.8% | 24.7% |
| No. times AI model correctb | ND | 71 | ND | 101 | 172 |
| No. times embryologist correctb | ND | 36 | ND | 42 | 78 |
| AI model fold improvement | ND | 2.0 x | ND | 2.4 x | 2.2 x |
| Comparison to embryologist grading – embryo ranking | |||||
| AI model ranking correctb | ND | 44.3% | ND | 38.8% | 40.6% |
| Embryologist ranking correctb | ND | 30.5% | ND | 27.6% | 28.6% |
| AI model improvement | ND | 45.2% | ND | 40.6% | 42.0% |
aEmbryologist scores were not available. bImages where the AI model was correct and the embryologist was incorrect, and vice versa.
ND = not done; No. = number.
Materials and Methods
Experimental design
These studies were designed to analyze retrospectively collected data for development and testing of the AI-based model in prediction of embryo viability. Data were collected for female subjects who had undergone oocyte retrieval, IVF and embryo transfer. The investigation was non-interventional, and results were not used to influence treatment decisions in any way. Data were obtained for consecutive patients who had undergone IVF at 11 independent clinics in three countries (the USA, Australia and New Zealand) from 2011 to 2018. Data were limited to patients who received a single embryo transfer with a Day 5 embryo, and where the endpoint was clinical pregnancy outcome as measured by fetal heartbeat at first scan. The clinical pregnancy endpoint was deemed to be the most appropriate measure of embryo viability as this limited any confounding patient-related factors post-implantation. Criteria for inclusion/exclusion were established prospectively, and images not matching the criteria were excluded from analysis.
For inclusion in the study, images were required to be of embryos on Day 5 of culture taken using a standard optical light microscope mounted camera. Images were only accepted if they were taken prior to PGS biopsy or freezing. All images were required to have a minimum resolution of 512 x 512 pixels with the complete embryo in the field of view. Additionally, all images were required to have matched clinical pregnancy outcome available (as detected by the presence of a fetal heartbeat on first ultrasound scan). For a subset of patients, the embryologist’s morphokinetic grade was available, and was used to compare the accuracy of the AI with the standard visual grading method for those patients.
Ethics and compliance
All patient data used in these studies were retrospective and provided in a de-identified format. In the USA, the studies described were deemed exempt from Institutional Review Board (IRB) review pursuant to the terms of the United States Department of Health and Human Service’s Policy for Protection of Human Research Subjects at 45 C.F.R. § 46.101(b) (IRB ID #6467, Sterling IRB). In Australia, the studies described were deemed exempt from Human Research Ethics Committee review pursuant to Section 5.1.2.2 of the National Statement on Ethical Conduct in Human Research 2007 (updated 2018), in accordance with the National Health and Medical Research Council Act 1992 (Monash IVF). In New Zealand, the studies described were deemed exempt from Health and Disability Ethics Committee review pursuant to Section 3 of the Standard Operating Procedures for Health and Disability Ethics Committees, Version 2.0 (August 2014).
These studies were not registered as a clinical trial as they did not meet the definition of an applicable clinical trial as defined by the ICMJE, that is, ‘a clinical trial is any research project that prospectively assigns people or a group of people to an intervention, with or without concurrent comparison or control groups, to study the relationship between a health-related intervention and a health outcome’.
Viability scoring methods
For the AI model, an embryo viability score of 50% and above was considered viable, and below 50% non-viable. Embryologist’s scores were provided for Day 5 blastocysts at the time when the image was taken. These scores were placed into scoring bands, which were roughly divided into ‘likely viable’ and ‘likely non-viable’ groups. This generalization allowed comparison of binary predictions from the embryologists with predictions from the AI model (viable/non-viable). The scoring system used by embryologists was based on the Gardner scale of morphokinetic grading (Gardner and Sakkas, 2003) for the quality of the ICM and the trophectoderm of the embryo, indicated by a single letter (A–E). Also included was either a numerical score or a description of the embryo’s stage of development toward hatching. Numbers were assigned in ascending order of embryo development as follows: 1 = start of cavitation, 2 = early blastocyst, 3 = full blastocyst, 4 = expanded blastocyst and 5 = hatching blastocyst. If no developmental stage was given, it was assumed that the embryo was at least an early blastocyst (>2). The conversion table for all clinics providing embryologists scores is provided in Supplementary Table SI. For embryologist’s scores, embryos of 3BB or higher grading were considered viable, and below 3BB considered non-viable.
Comparisons of embryo viability ranking were made by equating the embryologist’s assessment with a numerical score from 1 to 5 and, similarly, dividing the AI model inferences into five equal bands labeled 1 to 5 (from the minimum inference to the maximum inference). If a given embryo image was given the same rank by the AI model and the embryologist, this was noted as a ‘concordance’. If, however, the AI model provided a higher rank than the embryologist and the ground-truth outcome was recorded as viable, or the AI model provided a lower rank than the embryologist and the ground-truth outcome was recorded as non-viable, then this outcome was noted as ‘model correct’. Similarly, if the AI model provided a lower rank than the embryologist and the ground-truth outcome was recorded as viable, or the AI model provided a higher rank and the outcome was recorded as non-viable, this outcome was noted as ‘embryologist correct’.
Computer vision image processing methods
All image data underwent a pre-processing stage, as outlined below. These computer vision image processing methods were used in model development, and incorporated into the final AI model.
-- Each image was stripped of its alpha channel to ensure that it was encoded in a 3-channel format (e.g. RGB). This step removed additional information from the image relating to transparency maps, while incurring no visual change to the image. These portions of the image were not used.
Each image was padded to square dimensions, with each side equal to the longest side of the original image. This process ensured that image dimensions were consistent, comparable and compatible for deep learning methods, which explicitly require square dimension images as input, while also ensuring that no key components of the image were cropped.
Each image was RGB color normalized, by taking the mean of each RGB channel, and dividing each channel by its mean value. Each channel was then multiplied by a fixed value of 100/255, in order to ensure the mean value of each image in RGB space was (100, 100, 100). This step ensured that color biases among the images were suppressed, and that the brightness of each image was normalized.
Each image was then cropped so that the center of the embryo was in the center of the image. This was carried out by extracting the best ellipse fit from an elliptical Hough transform, calculated on the binary threshold map of the image. This method acts by selecting the hard boundary of the embryo in the image, and by cropping the square boundary of the new image so that the longest radius of the new ellipse is encompassed by the new image width and height, and so that the center of the ellipse is the center of the new image.
Each image was then scaled to a smaller resolution prior to training.
For training of selected models, images underwent an additional pre-processing step called boundary-based segmentation. This process acts by separating the region of interest (i.e. the embryo) from the image background, and allows masking in order to concentrate the model on classifying the gross morphological shape of the embryo.
Finally, each image was transformed to a tensor rather than a visually displayable image, as this is the required data format for deep learning models. Tensor normalization was obtained from standard pre-trained ImageNet values, mean (0.485, 0.456, 0.406) and standard deviation (0.299, 0.224, 0.225). Figure 1 shows an example embryo image carried through the first six pre-processing steps described above.
Figure 1.
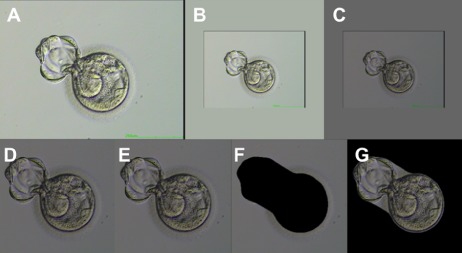
Sample image of human embryo with pre-processing steps applied in order. The six main pre-processing steps, prior to transforming the image into a tensor format, are illustrated. (A) The input image is stripped of the alpha channel. (B) The image is padded to square dimensions. (C) The color balance and brightness levels are normalized. (D) The image is cropped to remove excess background space such that the embryo is centered. (E) The image is scaled in resolution for the appropriate neural network. (F–G) Segmentation is applied to the image as a pre-processing step for portion of the neural networks. An image with the inner cell mass (ICM) and intra-zona cavity (IC) masked is shown in (F) and an image with the ICM/IC exposed is shown in (G). Images were taken at 200x magnification.
Embryo images obtained for this study were divided into training, validation and blind dataset categories by randomizing available data with the constraint that each dataset was to have an even distribution of examples across each of the classifications (i.e. the same ratio of viable to non-viable embryos). For model training, the images in the training dataset were additionally manipulated using a set of augmentations. Augmentations are required for training in order to anticipate changes to lighting conditions, rotation of the embryo and focal length so that the final model is robust to these conditions from new unseen datasets. The augmentations used in model training are as follows:
Rotations: Images were rotated a number of ways, including 90 degree rotations, and also other non-90 degree rotations where the diagonal whitespace in the square image due to these rotations was filled in with background color using the OpenCV (version 3.2.0; Willow Garage, Itseez, Inc. and Intel Corporation; 2200 Mission College Blvd. Santa Clara, CA 95052, USA) ‘Border_Replicate’ method, which uses the pixel values near the image border to fill in whitespace after rotation.
Reflections: Horizontal or vertical reflections of the image were also included as training augmentations.
Gaussian blur: Gaussian blurring was applied to some images using a fixed kernel size (with a default value of 15).
Contrast variation: Contrast variation was introduced to images by modifying the standard deviation of the pixel variation of the image from the mean, away from its default value.
Random horizontal and vertical translations (jitter): Randomly applied small horizontal and vertical translations (such that the blastocyst did not deviate outside the field of view) were used to assist the model in training invariance to translation or position in the image.
Random compression or jpeg noise: While uncompressed file formats are preferred for analysis (e.g. ‘png’ format), many embryo images are provided in the common compressed ‘jpeg’ format. To control for compression artifacts from images of jpeg format, jpeg compression noise was randomly applied to some images for training.
Model architectures considered
A range of deep learning and computer vision/machine learning methods were evaluated in training the AI model as follows. The most effect deep learning architectures for classifying embryo viability were found to be residual networks, such as ResNet-18, ResNet-50 and ResNet-101 (He et al., 2016), and densely connected networks, such as DenseNet-121 and DenseNet-161 (Huang et al., 2017). These architectures were more robust than other types of models when assessed individually. Other deep learning architectures including InceptionV4 and Inception-ResNetV2 (Szegedy et al., 2016) were also tested but excluded from the final AI model due to poorer individual performance. Computer vision/machine learning models including support vector machines (Hearst, 1998) and random forest (Breiman, 2001) with computer vision feature computation and extraction were also evaluated. However, these methods yielded limited translatability and poorer accuracy compared with deep learning methods when evaluated individually, and were therefore excluded from the final AI model ensemble. For more information see the Model selection process section.
Loss functions considered
The following quantities were evaluated to select the best model types and architectures:
-- Model stabilization: How stable the accuracy value was on the validation set over the training process.
-- Model transferability: How well the accuracy on the training data correlated with the accuracy on the validation set.
-- Prediction accuracy: Which models provided the best validation accuracy, for both viable and non-viable embryos, the total combined accuracy and the balanced accuracy, defined as the weighted average accuracy across both class types of embryos.
In all cases, use of ImageNet pretrained weights demonstrated improved performance of these quantities.
Loss functions that were evaluated as options for the model’s hyper-parameters included cross entropy (CE), weighted CE and residual CE loss function. The accuracy on the validation set was used as the selection criterion to determine a loss function. Following evaluation, only weighted CE and residual CE loss functions were chosen for use in the final model, as these demonstrated improved performance. For more information see the Model selection process section.
Deep learning optimization specifications
Multiple models with a wide range of parameter and hyper-parameter settings were trained and evaluated. Optimization protocols that were tested to specify how the value of the learning rate should be used during training included stochastic gradient descent (SGD) with momentum (and/or Nesterov accelerated gradients), adaptive gradient with delta (Adadelta), adaptive moment estimation (Adam), root-mean-square propagation (RMSProp) and limited-memory Broyden–Fletcher–Goldfarb–Shanno (L-MBFGS). Of these, two well-known training optimization strategies (optimizers) were selected for use in the final model; these were SGD (Rumelhart et al., 1986) and Adam (Kingma and Ba, 2014). Optimizers were selected for their ability to drive the update mechanism for the network’s weight parameters to minimize the objective/loss function.
Learning rates were evaluated within the range of 1e-5 to 1e-1. Testing of learning rates was conducted with the use of step scheduler, which reduces the learning rate during the training progress. Learning rates were selected based on their ability to stably converge the model toward a minimum loss function. The dropout rate, an important technique for preventing over-training for deep learning models, was tested within the range of 0.1 to 0.4. This involved probabilistically dropping out nodes in the network with a large number of weight parameters to prevent over-fitting while training. For more information see the Model selection process section.
Each deep neural network used weight parameters obtained from pre-training on ImageNet, with the final classifier layer replaced with a binary classifier corresponding to non-viable and viable classification. Training of AI models was conducting using PyTorch library (version 0.4.0; Adam Paszke, Sam Gross, Soumith Chintala and Gregory Chanan; 1601 Willow Rd, Menlo Park, CA 94025, USA), with CUDA support (version 9; Nvidia Corporation; 2788 San Tomas Expy, Santa Clara, CA 95051, USA), and OpenCV (version 3.2.0; Willow Garage, Itseez, Inc. and Intel Corporation; 2200 Mission College Blvd. Santa Clara, CA 95052, USA).
Individual models were trained and evaluated separately using a train-validate cycle process as follows:
-- Batches of images were randomly sampled from the training dataset and a viability outcome predicted for each embryo image.
-- Results for each image were compared to known outcomes to compute the difference between the prediction and the actual outcome (loss).
-- The loss value was then used to adjust the model’s weights to improve its prediction (backpropagation), and the running total accuracy was assessed.
-- This process was repeated thousands of times until the loss was reduced as much as possible and the value plateaued.
-- When all batches in the training dataset had been assessed (i.e. 1 epoch), so that the entire training set had been covered, the training set was re-randomized, and training was repeated.
-- After each epoch, the model was run on a fixed subset of images reserved for informing the training process to prevent over-training (the validation set).
-- The train-validate cycle was carried out for 2–100 epochs until a sufficiently stable model was developed with low loss function. At the conclusion of the series of train-validate cycles, the highest performing models were combined into a final ensemble model as described below.
Model selection process
Evaluation of individual model performance was accomplished using a model architecture selection process. Only the training and validation sets were used for evaluation. Each type of prediction model was trained with various settings of model parameters and hyper-parameters, including input image resolution, choice of optimizer, learning rate value and scheduling, momentum value, dropout and initialization of the weights (pre-training).
After shortlisting model types and loss functions using the criteria established in the preceding sections, models were separated into two groups: first, those that included additional image segmentation, and second those that required the entire unsegmented image. Models that were trained on images that masked the ICM, exposing the zona region, were denoted as zona models. Models that were trained on images that masked the zona (denoted ICM models), and models that were trained on full-embryo images, were also considered in training. A group of models encompassing contrasting architectures and pre-processing methods was selected in order to maximize performance on the validation set. Individual model selection relied on two criteria, namely diversity and contrasting criteria, for the following reasons:
-- The diversity criterion drives model selection to include different model’s hyper-parameters and configurations. The reason is that, in practice, similar model settings result in similar prediction outcomes and hence may not be useful for the final ensemble model.
-- The contrasting criterion drives model selection with diverse prediction outcome distributions, due to different input images or segmentation. This approach was supported by evaluating performance accuracies across individual clinics. This method ensured translatability by avoiding selection of models that performed well only on specific clinic datasets, thus preventing over-fitting.
The final prediction model was an ensemble of the highest performing individual models (Rokach, 2010). Well-performing individual models that exhibited different methodologies, or extracted different biases from the features obtained through machine learning, were combined using a range of voting strategies based on the confidence of each model. Voting strategies evaluated included mean, median, max and majority mean voting. It was found that the majority mean voting strategy outperformed other voting strategies for this particular ensemble model. This voting strategy gave the most stable model across all datasets and was therefore chosen as the preferred model.
The final ensemble model includes eight deep learning models of which four are zona models and four are full-embryo models. The final model configuration used in this study is as follows:
-- One full-embryo ResNet-152 model, trained using SGD with momentum = 0.9, CE loss, learning rate 5.0e-5, step-wise scheduler halving the learning rate every 3 epochs, batch size of 32, input resolution of 224 x 224 and a dropout value of 0.1.
-- One zona model ResNet-152 model, trained using SGD with momentum = 0.99, CE loss, learning rate 1.0e-5, step-wise scheduler dividing the learning rate by 10 every 3 epochs, batch size of 8, input resolution of 299 x 299 and a dropout value of 0.1.
-- Three zona ResNet-152 models, trained using SGD with momentum = 0.99, CE loss, learning rate 1.0e-5, step-wise scheduler dividing the learning rate by 10 every 6 epochs, batch size of 8, input resolution of 299 x 299, and a dropout value of 0.1, one trained with random rotation of any angle.
-- One full-embryo DenseNet-161 model, trained using SGD with momentum = 0.9, CE loss, learning rate 1.0e-4, step-wise scheduler halving the learning rate every 5 epochs, batch size of 32, input resolution of 224 x 224, a dropout value of 0 and trained with random rotation of any angle.
-- One full-embryo DenseNet-161 model, trained using SGD with momentum = 0.9, CE loss, learning rate 1.0e-4, step-wise scheduler halving the learning rate every 5 epochs, batch size of 32, input resolution of 299 x 299, a dropout value of 0.
-- One full-embryo DenseNet-161 model, trained using SGD with momentum = 0.9, Residual CE loss, learning rate 1.0e-4, step-wise scheduler halving the learning rate every 5 epochs, batch size of 32, input resolution of 299 x 299, a dropout value of 0 and trained with random rotation of any angle.
The architecture diagram corresponding to ResNet-152, which features heavily in the final model configuration, is shown in Figure 2. A flow chart describing the entire model creation and selection methodology is shown in Figure 3. The final ensemble model was subsequently validated and tested on blind test datasets as described in the results section.
Figure 2.
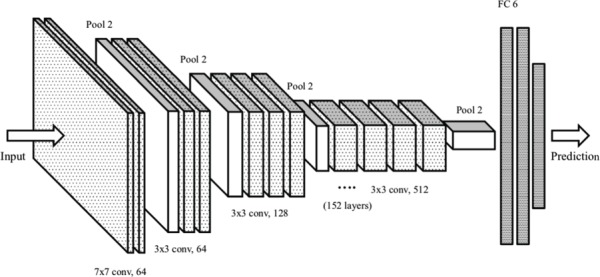
Example illustration of ResNet-152 neural network layers. The layer diagram from input to prediction for a neural network of type ResNet-152, which features prominently in the final Life Whisperer artificial intelligence (AI) model, is shown. For the 152 layers, the number of convolutional layers (‘conv’) are depicted, along with the filter size, which is the receptive region taken by each convolutional layer. Two-dimensional maxpooling layers (‘pool’) are also shown, with a final fully connected (FC) layer, which represents the classifier, with a binary output for prediction (non-viable and viable).
Figure 3.
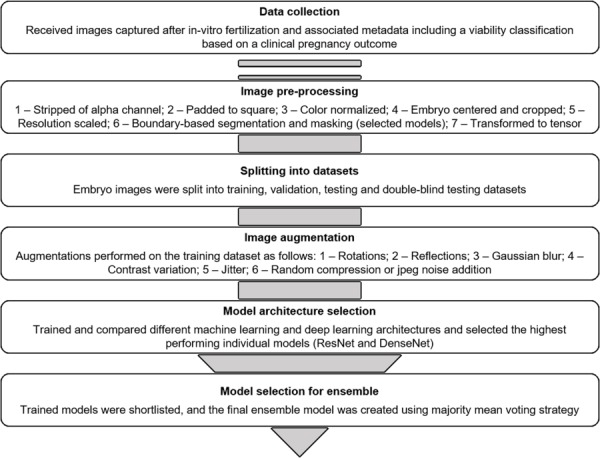
Flow chart for model creation and selection methodology. The model creation methodology is depicted beginning from data collection (top). Each step summarizes the component tasks that were used in the development of the final AI model. After image processing and segmentation, the images were split into datasets and the training dataset prepared by image augmentation. The highest performing individual models were considered candidates for inclusion in the final ensemble model, and the final ensemble model was selected based using majority mean voting strategy.
Statistical analysis
Measures of accuracy used in the assessment of model behavior on data included sensitivity, specificity, overall accuracy, distributions of predictions and comparison to embryologists’ scoring methods. For the AI model, an embryo viability score of 50% and above was considered viable, and below 50% non-viable. Accuracy in identification of viable embryos (sensitivity) was defined as the number of embryos that the AI model identified as viable divided by the total number of known viable embryos that resulted in a positive clinical pregnancy. Accuracy in identification of non-viable embryos (specificity) was defined as the number of embryos that the AI model identified as non-viable divided by the total number of known non-viable embryos that resulted in a negative clinical pregnancy outcome. Overall accuracy of the AI model was determined using a weighted average of sensitivity and specificity, and percentage improvement in accuracy of the AI model over the embryologist was defined as the difference in accuracy as a proportion of the original embryologist accuracy (i.e. (AI_accuracy—embryologist_accuracy)/embryologist_accuracy).
For these analyses, embryologist scores corresponding to blastocyst assessment at Day 5 were provided, that is, their assessment was provided at the same point in time as when the image was taken. This ensured that the time point for model assessment and the embryologist’s assessment were consistent. Note that the subset of data that includes corresponding embryologist scores was sourced from a range of clinics, and thus the measurement of the embryologist grading accuracy varied across each clinic, from 43.9% to 55.3%. This is due to the variation in embryologist skill, and statistical fluctuation of embryologist scoring methods across the dataset. In order to provide a comparison that ensured the most representative distribution of embryologist skill levels, all embryologist scores were considered across all clinics, and combined in an unweighted manner, instead of considering accuracies from individual clinics. This approach therefore captured the inherent diversity in embryologist scoring efficacy.
The distributions of prediction scores for both viable and non-viable embryo images were used to determine the ability of the AI model to separate true positives from false negatives, and true negatives from false positives. AI model predictions were normalized between 0 and 1, and interpreted as confidence scores. Distributions were presented as histograms based on the frequency of confidence scores. Bi-modal distributions of predictions indicated that true positives and false negatives, or true negatives and false positives, were separated with a degree of confidence, meaning that the predictive power of the model on a given dataset was less likely to have been obtained by chance. Alternatively, slight asymmetry in a unimodal Gaussian-like distribution falling on either side of a threshold indicated that the model was not easily able to separate distinct classes of embryo.
A binary confusion matrix containing class accuracy measures, i.e. sensitivity and specificity, was also used in model assessment. The confusion matrix evaluated model classification and misclassification based on true positives, false negatives, false positives and true negatives. These numbers were depicted visually using tables or ROC plots where applicable.
Final model accuracy was determined using results from blind datasets only, as these consisted of completely independent ‘unseen’ datasets that were not used in model training or validation. In general, the accuracy of any validation dataset will be higher than that of a blind dataset, as the validation dataset is used to guide training and selection of the AI model. For a true, unbiased measure of accuracy, only blind datasets were used. The number of replicates used for determination of accuracy was defined as the number of completely independent blind test sets comprising images that were not used in training the AI model. Double-blind test sets, consisting of images provided by clinics that did not provide any data for model training, were used to evaluate whether the model had been over-trained on data provided by the original clinics.
Results
Datasets used in model development
Model development was divided into two distinct studies. The first study consisted of a single-site pilot study to determine the feasibility of creating an AI model for prediction of embryo viability, and refine the training techniques and principles to be adopted for a second multi-site study. The first study, or pilot study, was performed using a total of 5282 images provided by a single clinic in Australia, with 3892 images used for the training process. The AI model techniques explored in the pilot study were then further developed in a second, pivotal study to determine generalizability to different clinical environments. The pivotal study used a total of 3604 images provided by 11 clinics from across the USA, Australia and New Zealand, with a total of 1744 images of Day 5 embryos used for training the Life Whisperer AI model presented in this article.
Images were split into defined datasets for model development in each study, which included a training dataset, a validation dataset and multiple blind test sets. Figure 4 depicts the number and origin of images that were used in each dataset in both the pilot and pivotal studies. A significant proportion of images in each study were used in model training, with a total of 3892 images used in the pilot study, and a further 1744 images used in the pivotal study. AI models were selected and validated using validation datasets, which contained 390 images in the pilot study and 193 in the pivotal study. Accuracy was determined using blind datasets only, comprising a total of 1000 images in the pilot study, and 1667 images in the pivotal study. Two independent blind test sets were evaluated in the pilot study; these were both provided by the same clinic that provided images for training. Three independent blind test sets were evaluated in the pivotal study. Blind Test Set 1 comprised images from the same clinics that provided images for training. Blind Test Sets 2 and 3 were, however, provided by completely independent clinics that did not provide any data for training. Thus, Blind Test Sets 2 and 3 represented double-blinded datasets as relates to AI computational methods.
Figure 4.
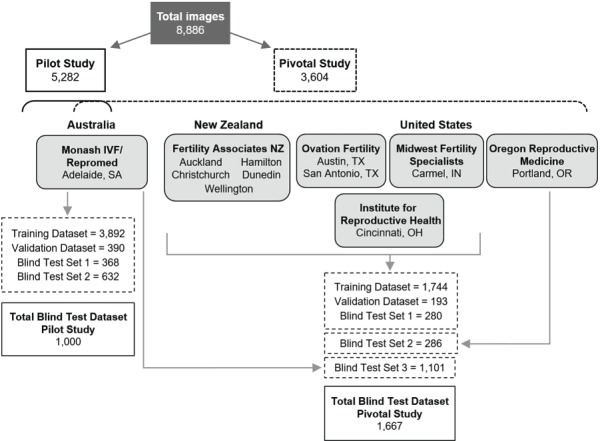
Image datasets used in AI model development and testing. A total of 8886 images of Day 5 embryos with matched clinical pregnancy outcome data were obtained from 11 independent IVF clinics across the USA, Australia and New Zealand. The pilot (feasibility) study to develop the initial AI model utilized 5282 images from a single clinic in Australia. This model was further developed in the pivotal study, which utilized an additional 3604 images from all 11 clinics. Blind test sets were used to determine AI model accuracy.
In total, 52.5% of all images in the blind datasets had embryologist grades available for comparison of outcome. Note that embryologist’s grades were not available for Blind Test Set 2 in the pivotal study; therefore, n = 2 for both studies in comparison of AI model accuracy to that of embryologists.
Pilot feasibility study
Table I shows a summary of results for the pilot study presented according to dataset (validation dataset, individual blind test sets and combined blind test dataset). In this study, negative pregnancies were found to outweigh positive pregnancies by approximately 3-fold. Sensitivity of the Life Whisperer AI model for viable embryos was 74.1%, and specificity for non-viable embryos was 65.3%. The greater sensitivity compared to specificity was to be expected, as it reflects the intended bias to grade embryos as viable that was introduced during model development. Overall accuracy for the AI model was 67.7%.
For the subset of images that had embryologist’s scores available, the AI model provided an average accuracy improvement of 30.8% over embryologist’s grading for viable/non-viable predictions (P = 0.021, n = 2, Student’s t test). The AI model correctly predicted viability over the embryologist 148 times, whereas the embryologist correctly predicted viability over the model 54 times, representing a 2.7-fold improvement for the AI model.
The AI model developed in this pilot study was used as a basis for further development in the pivotal study described below.
Model accuracy and generalizability
The results of the pivotal study are presented in Table II. In this study, the distribution of negative and positive pregnancies was more even than in the pilot study, with negative pregnancies occurring ~50% more often than positive pregnancies (1.5-fold increase compared to 3-fold increase in the pilot study).
After further development using data from a range of clinics, the Life Whisperer AI model showed a sensitivity of 70.1% for viable embryos, and a specificity of 60.5% for non-viable embryos. This was relatively similar to the initial accuracy values obtained in the pilot study, although values were marginally lower—this was not unexpected due to the introduction of inter-clinic variation into the AI development. Note that while the sensitivity in Blind Test Set 3 was ~5–7% lower than that of Blind Test Sets 1 and 2, the specificity was ~8% higher than in those datasets, making the overall accuracy comparable across all three blind test sets. The overall accuracy in each blind test set was >63%, with a combined overall accuracy of 64.3%.
Binary comparison of viable/non-viable embryo classification demonstrated that the AI model provided an average accuracy improvement of 24.7% over embryologist’s grading (P = 0.047, n = 2, Student’s t test). The AI model correctly predicted viability over the embryologist 172 times, whereas the embryologist correctly predicted viability over the model 78 times, representing a 2.2-fold improvement for the AI model. Comparison to embryologist’s scores using the 5-band ranking system approach showed that the AI model was correct over embryologists for 40.6% of images, and incorrect compared to embryologist’s scoring for 28.6% of images, representing an improvement of 42.0% (P = 0.028, n = 2, Student’s t test).
Confusion matrices showing the total number of true positives, false positives, false negatives and true negatives obtained from embryologist grading methods and the AI model are shown in Figure 5. By comparing the embryologist and AI model results, it is clear that the embryologist accuracy overall is significantly lower, even though the sensitivity is higher. This is most likely due to the fact that embryologist scores are typically high for the sub-class of embryos that have been implanted, and therefore there is a natural bias in the dataset toward embryos that have a high embryologist score. While alteration of the embryologist threshold score of ‘3BB’ above which embryos are considered ‘likely viable’ does not result in greater embryologist accuracy, a significant proportion of the embryos considered, (114 + 121)/(134 + 128) = 89.7%, were graded equal to or higher than 3BB. While the AI Model showed a reduction in true positives compared to the embryologist, there was a significant improvement in specificity. The AI model demonstrated an excess of 60% for both sensitivity and specificity, while still retaining a bias toward high sensitivity, in order to minimize the number of false negatives.
Figure 5.
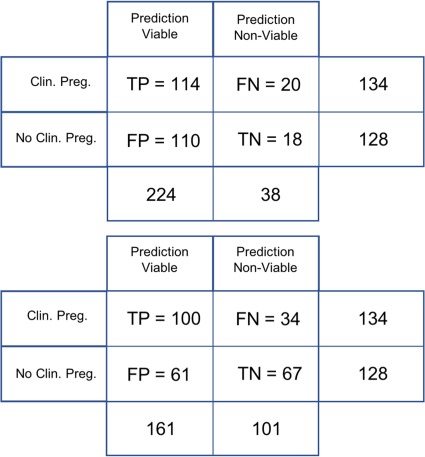
Confusion matrix of the pivotal study for embryologist and AI model grading. True positives (TP), false positives (FP), false negatives (FN) and true negatives (TN) are shown. The embryologists’ confusion matrix is depicted on the top panel, and the AI model’s confusion matrix is depicted on the bottom panel. The embryologists’ overall accuracy is significantly lower, despite a relatively higher sensitivity, due to the enhanced specificity of the AI model’s predictions. Clin. Preg. = clinical pregnancy; No Clin. Preg. = no clinical pregnancy.
A visual representation of the distribution of rankings from embryologists and from the AI model is shown in Figure 6. The histograms differ from each other in the shape of their distribution. There is a clear dominance in the embryologist’s scores around a rank value of 3, dropping off steeply for lower scores of 1 and 2, which reflects the tendency of embryologists to grade in the average to above average range. By comparison, the AI model demonstrated a smaller peak for rank values of 3, and larger peaks for rank values of 2 and 4. This reflects the AI model’s ability to distinctly separate predictions of viable and non-viable embryos, suggesting that the model provides a more granular scoring range across the different quality bands.
Figure 6.
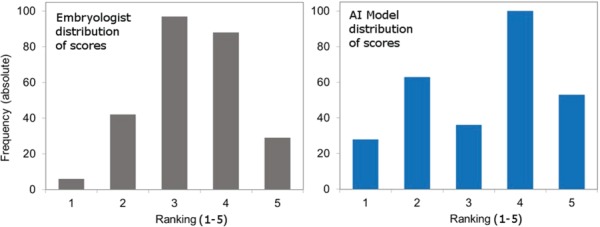
Distribution of viability rankings demonstrates the ability of the AI model to distinctly separate viable from non-viable human embryos. The left panel depicts the frequency of embryo viability rankings according to embryologist’s scores, and the right panel depicts the frequency of viability rankings according to AI model predictions. Results are shown for Blind Test Set 1. Y-axis = % of images in rank; x-axis = ranking band (1 = lowest predicted viability, 5 = highest predicted viability).
As a final measure of AI model performance, the distributions of prediction scores for viable and non-viable embryos were graphed to evaluate the ability of the model to separate correctly from incorrectly identified embryo images. The histograms for distributions of prediction scores are presented in Figure 7. The shapes of the histograms demonstrate clear separation between correctly and incorrectly identified viable or non-viable embryo images.
Figure 7.
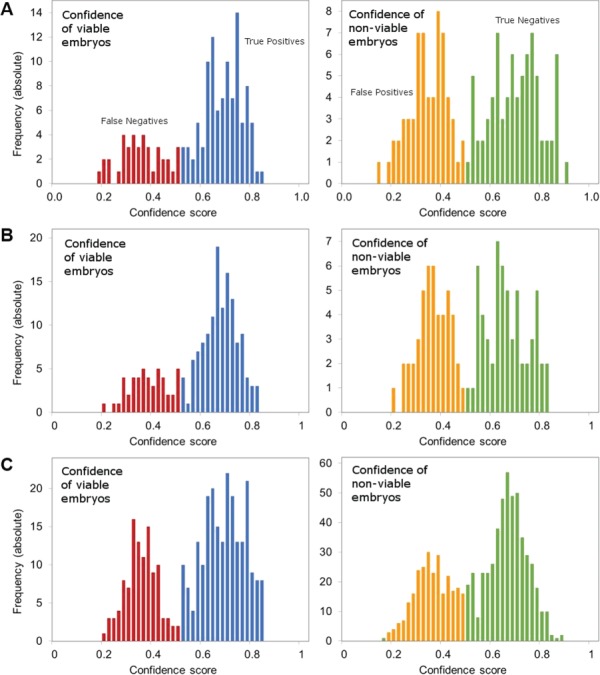
Distributions of prediction scores show the separation of correct from incorrect predictions by the AI model. Distributions of prediction scores are presented for Blind Test Set 1 (A), Blind Test Set 2 (B) and Blind Test Set 3 (C). The left panel in each set depicts the frequency of predictions presented as confidence intervals for viable embryos. True positives where the model was correct are marked in blue, and false negatives where the model was incorrect are marked in red. The right panel in each set depicts the frequency of predictions presented as confidence intervals for non-viable embryos. True negatives where the model was correct are marked in green, and false positives where the model was incorrect are marked in orange.
Discussion
In these studies, Life Whisperer used ensemble modeling to combine computer vision methods and deep learning neural network techniques to develop a robust image analysis model for the prediction of human embryo viability. In the initial pilot study, an AI-based model was created that not only matched the prediction accuracy of trained embryologists, but in fact surpassed the original objective by demonstrating an accuracy improvement of 30.8%. The AI-based model was further developed in a pivotal study to extend generalizability and transferability to multiple clinical environments in different geographical locations. Model accuracy was marginally lower on further development due to the introduction of inter-clinic variability, which may have affected efficacy due to varying patient demographics (age, health, ethnicity, etc.), and divergent standard operating procedures (equipment and methods used for embryo culture and image capture). Variation was also likely introduced due to embryologists being trained differently in embryo-scoring methods. However, the final AI model is both robust and accurate, demonstrating a significant improvement of 24.7% over the predictive accuracy of embryologists for binary viable/non-viable classification, despite variability of the clinical environments tested. The overall accuracy for prediction of embryo viability was 64.3%, which was considered relatively high given that research studies suggest a theoretical maximum accuracy of 80%, with ~20% of IVF cases thought to fail due to factors unrelated to embryo viability (e.g. operational errors, patient-related health factors, etc.).
Confusion matrices and comparison of the distribution of viability rankings highlighted the tendency for embryologists to classify embryos as viable, as it is generally considered preferable to allow a non-viable embryo to be transferred than to allow a viable embryo to be discarded. During development, the AI model was intentionally biased to similarly minimize the occurrence of false negatives; this was reflected in the slightly higher accuracy for viable embryos than non-viable embryos (70.1% and 60.5% for sensitivity and specificity, respectively). By examining the distribution of viability rankings for the AI model on the validation set, it was demonstrated that the model was able to distinctly separate predictions of viable and non-viable embryos on the blind test sets. Furthermore, graphical representation of the distribution of predictions for both viable and non-viable embryos demonstrated a clear separation of correct and incorrect predictions (for both viable and non-viable embryos, separately) by the AI model.
Machine learning methods have recently come into the spotlight for various medical imaging diagnostic applications. In particular, several groups have published research describing the use of either conventional machine learning or AI image analysis techniques to automate embryo classification. Two recent studies described conventional algorithms for prediction of blastocyst formation rather than clinical pregnancy. These studies achieved 76.4% (Segal et al., 2018) and >93% (Wong et al., 2010) accuracy, respectively, in their overall classification objectives, which included prediction of blastocyst formation based on Day 2 and/or Day 3 morphology and a number of other independent data points. However, it is important to note that blastocyst formation is not a reliable indicator of the probability of clinical pregnancy, and therefore the utility of this approach for prediction of pregnancy outcome is limited.
As discussed earlier, three recent studies described development of AI-based systems for classification of embryo quality (Khosravi et al., 2019; Kragh et al., 2019; Tran et al., 2019). All three studies utilized images taken by time-lapse imaging systems, which likely standardized the quality of images provided for analysis compared to those obtained by standard optical light microscopy. Khosravi et al. (2019) reported an accuracy of 97.5% for their model STORK in predicting embryo grade. Their model was not, however, developed to predict clinical pregnancy outcome. The high reported accuracy in this case may be attributed to the fact that the analysis was limited to classification of poor versus good quality embryos—fair quality embryos in between were excluded from analysis. Similarly, Kragh et al. (2019) reported accuracies of 71.9% and 76.4% for their model in grading embryonic ICM and trophectoderm, respectively, according to standard morphological grading methods. This was shown to be at least as good as the performance of trained embryologists. The authors also evaluated predictive accuracy for implantation, for which data were available for a small cohort of images. The AUC for prediction of implantation was not significantly different to that of embryologists (AUC of 0.66 and 0.64, respectively), and therefore, this model has limited ability for prediction of pregnancy outcome.
The approach taken by Tran et al. (2019) for development of their AI model IVY used deep learning to analyze time-lapse embryo images to predict pregnancy success rates. This study used 10 683 embryo images from 1648 individual patients throughout the course of the training and development of IVY, with 8836 embryos coded as positive or negative cases. Of note, although developed to predict pregnancy outcome, the IVY AI was trained on a heavily biased dataset of only 694 cases (8%) of positive pregnancy outcomes, with 8142 negative outcome cases (92%). Additionally, 87% (7063 cases) of the negative outcome cases were from embryos that were never transferred to a patient, discarded based on abnormal morphology considerations or aneuploidy, and therefore the ground-truth clinical pregnancy outcome cannot be known. The approach used to train the IVY AI only used ground-truth pregnancy outcome for a very small proportion of the algorithm training and thus has a heavy inherent bias toward the embryologist assessment for negative outcome cases. Although somewhat predictive of pregnancy outcome, the accuracy of the AI has not truly been measured on ground-truth outcomes of clinical pregnancy, and gives a false representation of the true predictive accuracy of the AI, which can only be truly assessed on an AI model that has been trained exclusively on known fetal heartbeat outcome data.
The works discussed above are not experimentally comparable with the current study, as they generally relate to different endpoints; for example, the prediction of blastocyst formation at Day 5 starting from an image at Day 2 or Day 3 post-IVF. While there is some benefit in these methods, they do not provide any power in predicting clinical pregnancy, in contrast to the present study evaluating the Life Whisperer model. Other studies have shown that a high level of accuracy can be achieved through the use of AI in replicating embryologist scoring methods (Khosravi et al., 2019; Kragh et al., 2019); however, the work presented here has shown that the accuracy of embryologist grading methods in predicting clinical pregnancy rates is in actuality fairly low. An AI model trained to replicate traditional grading methods to a high degree of accuracy may be useful for automation and standardization of grading, but it can, at best, only be as accurate as the grading method itself. In the current study, only ground-truth outcomes for fetal heartbeat at first scan were used in the training, validation and testing of the model. Given the nature of predicting implantation based on embryo morphology, which will necessarily be confounded by patient factors beyond the scope of morphological assessment, it would be expected that the overall accuracy of the Life Whisperer AI model would be lower than alternative endpoints but more clinically relevant. For the first time, this study presents a realistic measurement of AI accuracy for embryo assessment and a true representation of predictive ability for the pregnancy outcome endpoint. Given the relatively low accuracy for embryologists in predicting viability, as shown in this study (~50%), and a theoretical maximum accuracy of 80%, Life Whisperer’s AI model accuracy of ~65% represents a significant and clinically relevant improvement for predicting embryo viability in this domain.
The present study demonstrated that the Life Whisperer AI model provided suitably high sensitivity, specificity, and overall accuracy levels for prediction of embryo viability based directly on ground-truth clinical pregnancy outcome by indication of positive fetal cardiac activity on ultrasound. The model was able to predict embryo viability by analysis of images obtained using standard optical light microscope systems, which are utilized by the majority of IVF laboratories and clinics worldwide. AUC/ROC was not used as a primary methodology for evaluation of accuracy due to inherent limitations of the approach when applied to largely unbalanced datasets, such as those used in development of IVY (which used a dataset with a ~13:1 ratio of negative to positive clinical pregnancies) (Tran et al., 2019). Nevertheless, the ROC curve for the Life Whisperer AI model is presented for completeness in Supplementary Figure SI with results demonstrating an improved AUC for the AI model when compared to embryologist’s scores.
The unique power of the Life Whisperer AI model developed here lies in the use of ensemble modeling to combine computer vision image processing methods and multiple deep learning AI techniques to identify morphological features of viability that are not readily discernible to the human eye. The Life Whisperer AI model was trained on images of Day 5 blastocysts at all stages including early, expanded, hatching and hatched blastocysts, and as such it can be used to analyze all stages of blastocyst development. One potential limitation of the AI model as it currently stands is that it does not incorporate additional information from different days of embryo development. Emerging data using time-lapse imaging systems suggest that certain aspects of developmental kinetics in culture may correlate with embryo quality (Gardner et al., 2015). Therefore, it would be of interest to evaluate or modify the ability of the Life Whisperer AI model to extend to additional time points during embryo development. It would also be of interest to evaluate alternative pregnancy endpoints, such as live birth outcome, as fetal heartbeat is not an absolute indicator of live birth. However, it is important to note that the endpoint of live birth is additionally affected by patient-related confounding factors. The current investigation was performed with retrospectively collected data, and hence it will be of importance to collect data prospectively to assess real-world use of the AI model. Additional data collection and analysis is expected to further improve the accuracy of the AI.
The AI model developed here has been incorporated into a cloud-based software application that is globally accessible via the web. The Life Whisperer software application allows embryologists or similarly qualified personnel to upload images of embryos using any computer or mobile device, and the AI model will instantly return a viability confidence score. The benefits of this approach lie in its simplicity and ease of use; the Life Whisperer system will not require installation of complex or expensive equipment, and does not require any specific computational or analytical knowledge. Additionally, the use of this tool will not require any substantial change in standard operating procedures for IVF laboratories; embryo images are routinely taken as part of IVF laboratory standard procedures, and analysis can be performed at the time of image capture from within the laboratory to help decide which embryos to transfer, freeze or discard. The studies described herein support the use of the Life Whisperer AI model as a clinical decision support tool for prediction of embryo viability during IVF procedures.
Supplementary Material
Acknowledgements
The authors acknowledge the kind support of investigators and collaborating clinics for providing embryo images and associated data as follows: Hamish Hamilton and Michelle Lane, Monash IVF Group/Repromed (Adelaide SA, Australia); Matthew ‘Tex’ VerMilyea and Andrew Miller, Ovation Fertility (Austin TX and San Antonio TX, USA); Bradford Bopp, Midwest Fertility Specialists (Carmel IN, USA); Erica Behnke, Institute for Reproductive Health (Cincinnati OH, USA); Dean Morbeck, Fertility Associates (Auckland, Christchurch, Dunedin, Hamilton and Wellington, New Zealand); and Rebecca Matthews, Oregon Reproductive Medicine (Portland OR, USA).
Authors’ roles
M.V., J.M.M.H., D.P. and M.P. conceived the study and designed methodology. D.P. and M.P. were also responsible for project management and supervision of research activity. M.V., A.M. and A.P. provided significant resources. J.M.M.H., A.J., T.N. and A.P.M. were responsible for data curation, performing the research, formal analysis and software development. S.M.D. was involved in data visualization and presentation, and in writing the original manuscript draft. All the authors contributed to the review and editing of the final manuscript.
Funding
Life Whisperer Diagnostics, Pty Ltd is a wholly owned subsidiary of the parent company, Presagen Pty Ltd. Funding for the study was provided by Presagen with grant funding received from the South Australian Government: Research, Commercialisation and Startup Fund (RCSF). ‘In kind’ support and embryology expertise to guide algorithm development were provided by Ovation Fertility.
Conflict of interest
J.M.M.H., D.P. and M.P. are co-owners of Life Whisperer Diagnostics, Pty Ltd, and of the parent company Presagen, Pty Ltd. Presagen has filed a provisional patent for the technology described in this manuscript (52985P pending). A.P.M. owns stock in Life Whisperer, and S.M.D., A.J., T.N. and A.P.M. are employees of Life Whisperer.
References
- Annan JJ, Gudi A, Bhide P, Shah A, Homburg R. Biochemical pregnancy during assisted conception: a little bit pregnant. J Clin Med Res 2013;5:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine Learning 2001;45:5–32. [Google Scholar]
- Chen M, Wei S, Hu J, Yuan J, Liu F. Does time-lapse imaging have favorable results for embryo incubation and selection compared with conventional methods in clinical in vitro fertilization? A meta-analysis and systematic review of randomized controlled trials. PLoS One 2017;12: e0178720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Meseguer M, Rubio C, Treff NR. Diagnosis of human preimplantation embryo viability. Hum Reprod Update 2015;21:727–747. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Sakkas D. Assessment of embryo viability: the ability to select a single embryo for transfer—a review. Placenta 2003;24:S5–S12. [DOI] [PubMed] [Google Scholar]
- GBD Population and fertility by age and sex for 195 countries and territories, 1950–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018;392:1995–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 27–30 June, Piscataway NJ, US: Institute of Electrical and Electronics Engineers, 2016;770–778. [Google Scholar]
- Hearst MA. Support vector machines. IEEE Intell Syst 1998;13:18–28. [Google Scholar]
- Huang G, Liu Z, Maaten LVD, Weinberger KQ. Densely connected convolutional networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 21–26 July, Piscataway NJ, US: Institute of Electrical and Electronics Engineers, 2017;2261–2269. [Google Scholar]
- Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, Sigaras A, Lavery A, LAD C, Hickman C et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit Med 2019;2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D, Ba J. Adam: a method for stochastic optimization. Computing Research Repository (CoRR) 2014; abs/1412.6980. [Google Scholar]
- Kragh MF, Rimestad J, Berntsen J, Karstoft H. Automatic grading of human blastocysts from time-lapse imaging. Comput Biol Med 2019;115:103494. [DOI] [PubMed] [Google Scholar]
- Rokach L. Ensemble-based classifiers. Artif Intell Rev 2010;33:1–39. [Google Scholar]
- Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature 1986;323:533–536. [Google Scholar]
- Sahlsten J, Jaskari J, Kivinen J, Turunen L, Jaanio E, Hietala K, Kaski K. Deep learning fundus image analysis for diabetic retinopathy and macular edema grading. Sci Rep 2019;9:10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal TR, Epstein DC, Lam L, Liu J, Goldfarb JM, Weinerman R. Development of a decision tool to predict blastocyst formation. Fertil Steril 2018;109:e49–e50. [Google Scholar]
- Storr A, Venetis CA, Cooke S, Kilani S, Ledger W. Inter-observer and intra-observer agreement between embryologists during selection of a single Day 5 embryo for transfer: a multicenter study. Hum Reprod 2017;32:307–314. [DOI] [PubMed] [Google Scholar]
- Szegedy C, Ioffe S, Vanhoucke V. Inception-v4, inception-ResNet and the impact of residual connections on learning. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17), 4-9 February, Palo Alto CA, USA: AAAI Press, 2016;2017:4278–4284. [Google Scholar]
- Tran D, Cooke S, Illingworth PJ, Gardner DK. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod 2019;34:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sauer MV. In vitro fertilization (IVF): a review of 3 decades of clinical innovation and technological advancement. Ther Clin Risk Manag 2006;2:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, RA RP. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol 2010;28:1115–1121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


