Abstract
Background
There is a well-documented lack of rapid, low-cost tuberculosis (TB) drug resistance diagnostics in low-income settings across the globe. It is these areas that are plagued with a disproportionately high disease burden and in greatest need of these diagnostics.
Methods
In this study, we compared the performance of Light Forge, a microfluidic high-resolution melting analysis (HRMA) prototype for rapid low-cost detection of TB drug resistance with a commercial HRMA device, a predictive “nearest-neighbor” thermodynamic model, DNA sequencing, and phenotypic drug susceptibility testing (DST). The initial development and assessment of the Light Forge assay was performed with 7 phenotypically drug resistant strains of Mycobacterium tuberculosis (M.tb) that had their rpoB gene subsequently sequenced to confirm resistance to Rifampin. These isolates of M.tb were then compared against a drug-susceptible standard, H37Rv. Seven strains of M.tb were isolated from clinical specimens and individually analyzed to characterize the unique melting profile of each strain.
Results
Light Forge was able to detect drug-resistance linked mutations with 100% concordance to the sequencing, phenotypic DST and the “nearest neighbor” thermodynamic model. Researchers were then blinded to the resistance profile of the seven M.tb strains. In this experiment, Light Forge correctly classified 7 out of 9 strains as either drug resistant or drug susceptible.
Conclusions
Light Forge represents a promising prototype for a fast, low-cost diagnostic alternative for detection of drug resistant strains of TB in resource constrained settings.
Keywords: High-resolution melting analysis, Drug resistance, Mycobacterium tuberculosis, Real-time PCR, Molecular diagnostics
Impact Statement
The information presented in this study is primarily positioned to benefit TB-infected individuals from resource limited regions such as Sub-Saharan Africa and South East Asia where affordable and accessible diagnostics are required. The evidence presented in this manuscript illustrates that combining DNA-melting analysis with microfluidics can be a foundational formulation for a diagnostic that will be cheap, parallel, and accessible in various clinical settings.
Introduction
Tuberculosis (TB) is a deadly infectious disease with 1.6 million deaths reported in 2017 (1). The highest disease burden is seen in Africa and South East Asia, low income regions, often with poor healthcare delivery (2). The inability to control infectious diseases adequately in these areas is rooted in poor diagnosis and treatment. In TB, this leads to increased infectivity, transmission, morbidity, and mortality. Further compounding this problem is the significant rise in the number of drug-resistant strains of Mycobacterium tuberculosis (M.tb), which are associated with higher morbidity and mortality rates (3). It would be possible to provide more efficient healthcare delivery by increasing access to more economical diagnostic devices. If M.tb infection and drug resistance profiles can be detected at a significantly lower cost to public health systems, treatment would be initiated earlier, with a substantial attainable decrease in disease incidence (4). Unfortunately, despite increased academic and commercial interest in point-of-care diagnostics, few commercially available devices have managed to effectively deliver to this underserved demographic.
Creating diagnostics within this context requires an adept appreciation of the unique challenges and limitations in the development, production, and marketing of a diagnostic test for the developing world (5). Notably, the most important considerations are device affordability and turnaround time (6). Microfluidic technology can reduce the cost of diagnosis by precisely manipulating minute fluid volumes in parallel, thereby reducing the overall consumption of reagents whilst increasing diagnostic throughput. Engineering and refining these fine networks of micro-plumbing allows integration of many functional components onto a single device (7). A sample can be partitioned into several independent fluid circuits in an efficient manner, allowing multiple assays to be carried out in parallel with minimal end-user intervention. Another cost saving strategy for TB diagnostic devices is the utilization of high-resolution melting analysis (HRMA), a post polymerase chain reaction (PCR) method used to detect single nucleotide polymorphisms (SNPs) (8). It has the advantage of being a single step as well as a closed tube assay that allows for a rapid and reliable examination of PCR product. This method has been used to detect drug-resistant M.tb, with performance metrics comparable to the “gold standard” M.tb phenotypic drug susceptibility testing (9, 10). HRMA has been reported to cost USD $0.30 per reaction, but performing it at volumes consistent with microfluidics will reduce reagent costs by approximately 1,000 (11). We leveraged the efficiency and simplicity afforded by a microfluidics platform with the simple, linear workflow of HRMA to create Light Forge, a functional and foundational blueprint for a low-cost TB diagnostic for resource-limited settings.
In this study, we address the following research questions: (1) Are the performance characteristics of Light Forge comparable to a commercial HRMA device, phenotypic drug susceptibility testing, a predictive nearest-neighbor thermodynamic model, and Sanger sequencing? (2) Can Light Forge provide similar specificity to traditional Sanger sequencing when blinded samples are analyzed? To answer these questions, we obtained seven rifampicin-resistant strains with known mutations in the rifampicin (RIF) resistance determining region (RRDR) rpoB. Custom primers were designed and the amplicons were melted to quantify the melting temperature compared to H37Rv. RIF resistance was selected as an appropriate target as sequence aberrations in and around the 81 bp fragment of the RRDR accounting for 95% of TB drug resistant cases (12, 13). A similar approach was adopted for the subsequent blinded phase of the study.
Materials and Methods
Mycobacteria tuberculosis Isolates
Eight DNA samples were used to develop the assay for subsequent experiments. These were obtained from 7 rifampicin-resistant laboratory strains (Kzn605, R35, R271, Tkk-01-0039, Tkk-01-0043, Tkk-01-0050, Tkk-01-0062), and H37Rv, used as a drug-susceptible standard throughout the study. The strains were sourced from Medical Microbiology, School of Laboratory Medicine and Medical Sciences at the Nelson R. Mandela School of Medicine (Durban, South Africa) and Dr. Alex Pym’s laboratory at the African Health Research Institute (AHRI) (Durban, South Africa). The phenotypic drug-susceptibility profile of all the test strains confirmed rifampicin resistance.
To further validate the performance of the assay, genomic DNA from 9 clinical M. tuberculosis isolates (Tkk-01-0011, Tkk-01-0030, Tkk-01-0032, Tkk-01-0061, Tkk-01-0078, Tkk-03-0082, Tkk-04-0006, Tkk-04-0030, Tkk-04-0048) were also sourced from Dr. Alex Pym’s laboratory at AHRI. The drug susceptibility profile of each of these strains was revealed after the Light Forge blinded study was concluded.
DNA Extraction
DNA was extracted from heat-killed cultures using the hexadecyltrimethylammonium bromide (CTAB) method (14).
Sanger Sequencing of the rpoB Gene Region
PCR amplicons of the rpoB gene for each strain were sequenced by Inqaba Biotech Industries, using primer sets identical to the primers in the high-resolution melting (HRMA) assay (Table 1).
Table 1.
Primer sequences used for PCR amplification, Sanger sequencing and subsequent HRMA of the rpoB gene’s region known to determine rifampicin resistance (15).
| Primer | Sequence | Annealing temperature(°C) | Product size(bp) | Nucleotide position | Accession number |
|---|---|---|---|---|---|
| rpoB-F | CGCGATCAAGGAGTTCTTC | 65 | 118 | 2339 to 2357 | L27989.1 |
| rpob-R | TGACAGACCGCCGGGCCC | 2456 to 2439 |
Real-Time PCR and High-Resolution Melting Analysis (HRMA) with Light Cycler®96
Polymerase chain reaction (PCR) was performed by first preparing a master mix through adding 25 µL of 2X Xtreme™ Buffer (Novagen), 10 µL of dNTPs (Novagen, ), 5 µl of LC Green (Idaho Technology Inc.), 0.3 µM of each Forward and Reverse primer, 1 µL of KOD Xtreme™ Hot Start DNA polymerase (Novagen, Toyobo) and 5 µL of template DNA containing 200 ng of the DNA in a final volume of 50 µL. PCR was performed using the Light Cycler®96 (Roche Diagnostics, Switzerland). The initial denaturation temperature was 95 °C for 300 s, followed by 35 cycles of 95 °C for 10 s, 60 °C for 10 s, 72 °C for 10 s (fluorescence readout step). A nontemplate control (NTC) was included in all experiments, in which PCR Grade water (Life Technologies) was substituted for the DNA template.
For the HRMA, the following profile was used: 95 °C for 60 s, 40 °C for 60 s, 65 °C for 1 s then at a 0.07 °C/s ramp rate, acquiring 15 readings every degree until 97 °C. Difference plots were generated using H37Rv as the baseline signal. The test readings from the samples were then normalized to this standard. The total reaction time was 70 min.
Light Forge Microfluidic Chip Design and Fabrication
The Light Forge PCR microfluidic chip was fabricated out of the silicone elastomer polydimethylsiloxane (PDMS) (General Electric RTV 615) using multi-layer soft lithography, as described previously (16). Up to 20 independent PCR reactions can run in parallel on each chip (Fig. S1).
Real-Time PCR and High-Resolution Melting with Light Forge
The real-time PCR and HRMA steps on the chip were captured using the Light Forge software, which was developed in house by Dr. Frederick Balagaddé. This software uses a feedback system that allows acquisition of fluorescence signals at easily programmable temperatures, whilst displaying the reaction progress.
The PCR master mix was prepared using 30 µL of 2X Xtreme™ Buffer (Novagen), 15 µL of dNTPs (Novagen), 9 µL of LC Green (Idaho Technology Inc.), 0.46 µM of each primer (Life Technologies), 3 µL of KOD Xtreme™ Hot Start DNA polymerase (Novagen), 0.08%(v/v) 1% Tween 20, and 3 µL of template DNA(∼6ng) to make a total volume of 75 µL. For the 20-reactor chip, 8 reactors were used for H37Rv, 8 for the test strains (R35, Kzn 605). The final 4 reactors contained the nontemplate controls.
The thermal cycling was performed using a G-STORM GS1 (Somerten) thermocycler modified to house the microfluidic device. The fluorescence signal was acquired in real-time using an Olympus MVX10 (New York) Macro zoom microscope (Fig. S2). The thermal profile was 99 °C for 8 minutes, followed by 35 cycles of 99 °C for 65 s, 60 °C for 115 s, 74.5 °C for 130 s. The HRMA was performed by increasing the temperature from 75 °C to 94 °C at a ramp rate of 0.5 °C/s with 0.25 °C increments for each step. This resulted in a total reaction time of 180 min (Fig. S3).
Nearest-Neighbor (NN) Thermodynamic Model
The data from our experiments were compared to the theoretical relative melting temperature differences predicted by the mathematical nearest-neighbor (NN) thermodynamic model (17). This model predicts the melting temperature () of a DNA strand based on the cumulative standard enthalpies () and standard entropies () of neighboring duplex base pairs as well as the total oligonucleotide strand concentration () as shown by Equation 1 below.
| (Eq. 1) |
Using the sequencing information from Sanger sequencing, the () between the reference strain (H37Rv) and the test strains used for the initial evaluation were computed. This model was also used to compute the expected () of the blinded samples. The computed prediction error of the NN model was ±1.2 0C (18). The SNPs observed in resistant strains of M.tb result in temperature deviations ranging from (<0.1 to 1.4 0C) (19). Thus, the model cannot be used for predicting exact differences but remains useful in predicting whether the SNPs lead to a positive or negative .
Statistical Analysis
A two-way t-test was used to evaluate the significance of the differences, where a P value of 0.05 or less was considered statistically significant (GraphPad Prism 7).
Results
rpoB Gene Mutations in Clinical M.tb Strains
Seven strains with rifampicin resistance and a reference H37Rv wild-type strain (rifampicin sensitive) were selected for the initial evaluation of the Light Forge. The 118 bp rpoB gene target of each of the isolates was Sanger-sequenced prior to HRMA. All the 7 sequence variants demonstrated SNPs associated with resistance to rifampicin. A total of 5 sequence SNPs present in 7 strains were identified, including Kzn 605 (533 T → C and 516 A → G), R35 (533 T → C), R271 (531 C → T), Tkk-01-0062 (516 A → T), Tkk-01-0050 (526 C → T), Tkk-01-0043 (531 C → T), and Tkk-01-0039 (526 C → T).
Real-Time PCR and HRMA for Drug Susceptibility Testing on Test Strains
The rpoB PCRs for 7 clinical strains were run on the Light Cycler®96 and were compared to the Light Forge system (using the microfluidic chip shown in Fig. 1). Fig. 2 illustrates the fluorescence imaging of the Light Forge reactors at the 1st (A) and 25th (B) thermal cycles during real-time PCR. The A panel in Fig. 2 is a fluorescent image at 60 °C at the first PCR cycle, while the B panel is the fluorescent image at 60 °C after 25 PCR cycles. Each color-coded chamber contains ∼1.5 nL volume of PCR master mix. This is a representative layout using a clinical mutant isolate (Tkk-01-0050) and the wild type reference control H37Rv used in the subsequent experiments. The test strain was amplified in 8 out of 20 reactors (red border), the reference wild type strain (H37Rv) was amplified in 8 out of 20 reactors (blue border), while the nontemplate controls were amplified in 4 of 20 reactors (yellow border). At the end of the amplification, the Light Forge analysis was immediately initiated to melt the amplicons and detect mutations. Mutations were identified as melting temperature () deviations relative to H37Rv. Each individual line indicates a melt-curve profile for an individual reactor.
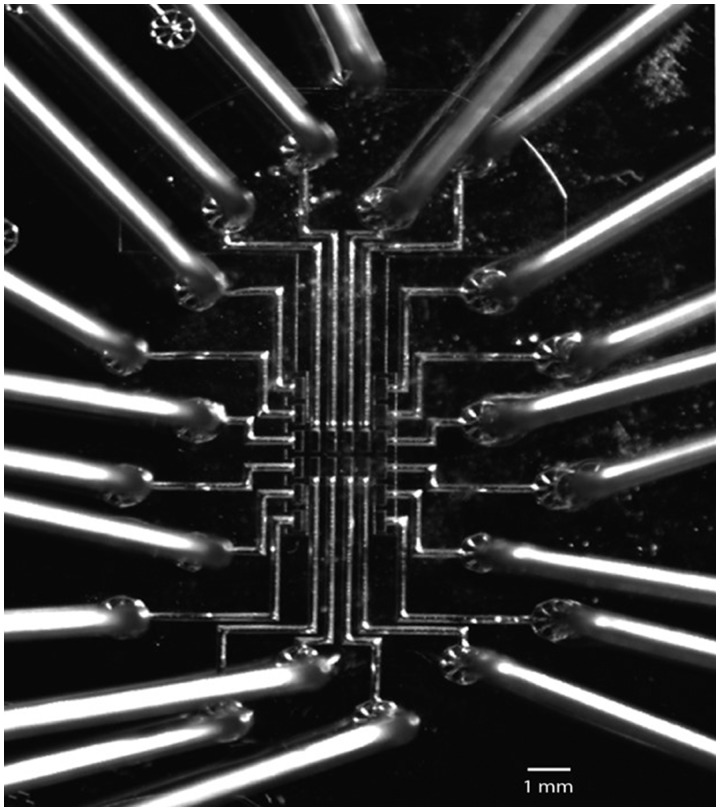
Fig. 1.
The Light Forge microfluidic chip. Optical micrograph of the Light Forge microfluidic chip/cartridge for real-time PCR and high-resolution melting analysis of amplified samples. Scale bar, 1 mm. The chip/cartridge is made up of an elastomeric transparent polymer called polydimethylsiloxane and has 20 reactors, each linked to an independent input port. These reactors are 500 µm long, 200 µm wide, and 150 µm high, translating into a volume of 1.5 nL. The PCR master mix was loaded at 5 psi until the PCR chambers were full.
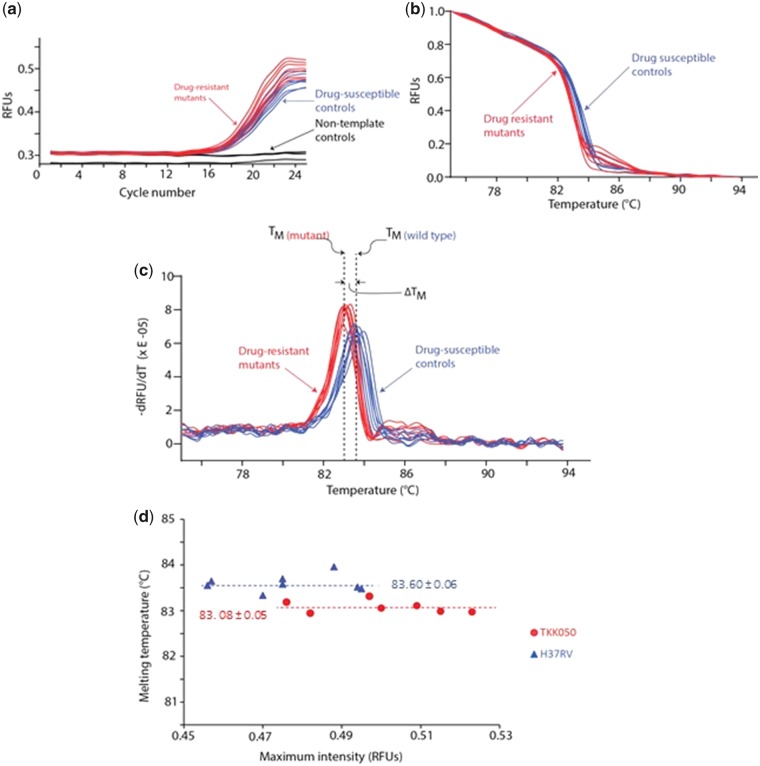
Fig. 2.
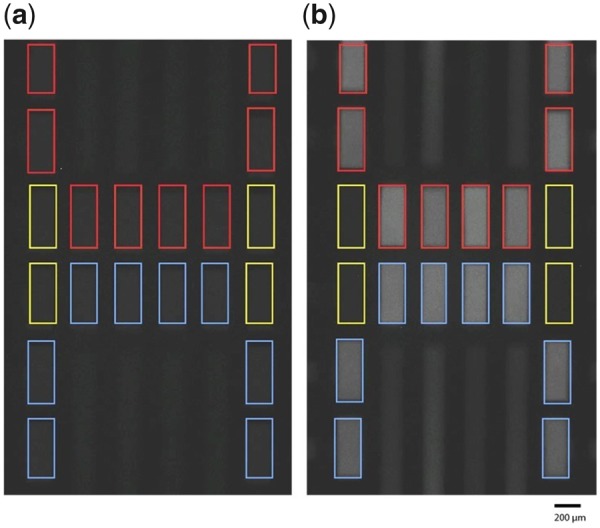
DNA quantification and high-resolution melt analysis in Light Forge PCR reactors. The Light Forge chip/cartridge imaging area is shown in greater detail, demonstrating the visual accumulation of fluorescence signal (within each of the 20 reactors, linked to independent input ports) as the Light Forge system performs real-time PCR and High-Resolution Melting Analysis. An increased fluorescence signal intensity is demonstrated with an increase in PCR cycle number. Fluorescence images of Light Forge PCR reactors of the 1st (a) and 25th(b) thermal cycles during real-time PCR. The reactors in the image have been demarcated with color-coded boundaries depending on the genomic DNA amplified: drug-resistant mutants [Tkk-01-0050 (526 C→T), red]; drug-susceptible strains (H37Rv, blue); and nontemplate controls (yellow). The amount of double-stranded DNA in each reactor corresponds to the level of fluorescence of LC Green—a reporter double-stranded DNA intercalating dye in the master mix. LC Green binds specifically to double-stranded DNA and fluoresces brightly when it binds. Unless specified otherwise, all images were taken during the 60 °C step of each thermal cycle. We were detecting fluorescence—in real-time via a custom program that runs on a temperature feedback loop—at 470-520 nm, the emission wavelength range of the reporter double stranded DNA saturating dye LC Green.
The real-time PCR and HRMA are graphically illustrated in Fig. 3 a and b respectively. The HRMA profiles of H37Rv were used as the standard to which the 7 clinical strains were then compared. Mutant melting curves could be distinguished from the wild type melting curve in the normalized graphs (Fig. 3b) but were best differentiated in the negative 1st derivative plot (-dRFU/dT) shown in Fig. 3c. The average melting temperature was computed for both the test strain and H37Rv as shown in Fig. 3d. There was 100% concordance of the results from Light Cycler®96 and Light Forge.
Fig. 3.
The real-time PCR and HRMA data from the Light Forge system testing the rpoB gene of the [Tkk-01-0050 (526 C→T)] clinical strain. (a) Real-time PCR amplification curves for the rpoB region. These data were all acquired simultaneously using the Light Forge microfluidic chip. For each reactor, a single data point was acquired during the 60 °C step. The relative fluorescence units (RFUs) corresponding to the concentration of double-stranded DNA was plotted against the PCR cycle number. Eight real-time amplification curves for drug-resistant [Tkk-01-0050 (526 C→T)] mutant strain are shown in red, 8 curves for the drug-susceptible controls strains (H37Rv) in blue and four curves of the no template control reactors in black. Both drug-susceptible control and drug-resistant mutant strain reactors increased by more than 0.15 RFUs to over 0.45 RFUs, whereas the amplification signal for the nontemplate controls remained relatively suppressed. (b) Normalized high-resolution melt (HRM) curves for the rpoB PCR amplicons of the drug-resistant mutant strain [Tkk-01-0050 (526C→T), red] and the drug-susceptible control strain (H37Rv, blue). (c) Negative first derivatives plots revealing the melt peaks. The melting temperature (TM) of each strain was determined by taking the mean of the melt peak of every replicate for each individual strain. The average melting temperature for the drug-susceptible strain was 83.60 ± 0.06 °C and that of the drug-resistant mutant was 83.08 ± 0.05 °C. The relative difference of the melt peaks of these strains () provides important clues to the nature of the mutation in the drug-resistant genome. (d) Peak intensity as a function of melting temperature. The average melting temperatures for the drug-resistant mutant (Tkk-01-0050, red circles) and drug-susceptible control (H37Rv, blue triangles) were 83.60 ± 0.06 °C and 83.08 ± 0.05 °C respectively. The melting temperatures were independent of the maximum fluorescence intensity (double-stranded DNA concentration) and thus independent of the amplification efficiency in each reactor.
Comparison of Light Forge HRMA with Sanger Sequencing, Roche Light Cycler®96 and the Nearest-Neighbor (NN) Model
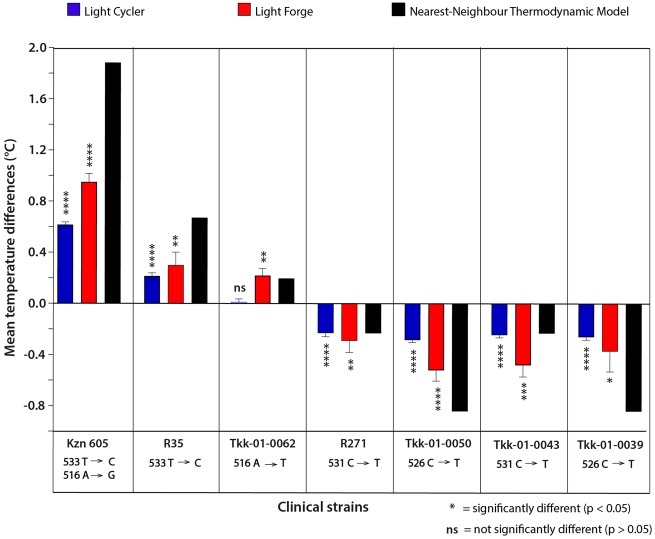
This section refers to Fig. 4. The sequencing data for Kzn 605 identified 2 positive class two mutations 533 T → C and 516 A → G within the rpoB region. The Light Forge system identified these Kzn 605 SNPs as having a melting temperature difference () of 0.95 ± 0.06 °C. The Light Cycler®96 detected a () of (0.61 ± 0.02 °C). The NN model predicted a positive () of +1.88 °C.
Fig. 4.
Comparison of melting temperature differences from Light Forge and Roche Light Cycler®96 HRMA systems alongside the NN thermodynamic model predictions. Relative melting temperature differences of the rpoB amplicon among 7 RIF-resistant strains and the reference RIF-susceptible strain H37Rv were plotted using data generated from Light Forge (red bars) and Light Cycler®96 (blue bars). The black bars represent the theoretical relative melting temperature differences predicted by the nearest neighbor (NN) thermodynamic model. The Light Forge data is consistent with Light Cycler®96 data as well as the theoretical model predictions. However, Light Forge successfully detected a low-energy class 4 mutation that eluded detection by Light Cycler®96 despite the nearest neighbor (NN) thermodynamic model having predicted that the mutation is detectable.
Sequencing the R35 isolate revealed the positive class one mutation 533 T → C within the rpoB region. Consistently, Light Forge detected a () of (0.30 ± 0.11 °C). For the same strain, Light Cycler®96 detected a () of (0.22 ± 0.02 °C). The NN model predicted a melting temperature of + 0.67 °C.
Sequencing the R271 isolate revealed the presence of a negative class one SNP 531 C → T within the rpoB region. Light Forge detected a () of −0.29 ± 0.09 °C. A () of −0.23 ± 0.03 °C was detected for the same strain using the Light Cycler®96. Consistent with the two systems, the NN model detected a () −0.23 °C.
The Light Forge system detected a () of (0.22 ± 0.05 °C) when the Tkk-01-0062 isolate was compared to the reference. Sequencing highlighted the presence of (1 positive) or (a positive) class four SNP 516 A → T. The Light Cycler®96 could not detect this SNP, shown by a () of 0.013 ± 0.02 °C. A () of + 0.20 °C was predicted by the NN model.
When the Tkk-01-0050 isolate was run in the Light Cycler®96 system, a () of −0.28 ± 0.02 °C was detected. This observation was consistent with sequencing findings, which denoted the presence of 1 negative energy class one SNP 526 C → T. Light Forge detected a () of −0.52 ± 0.08 °C. The NN model predicted a () of −0.84 °C, comparable with both the Light Cycler®96 and the Light Forge systems.
The sequencing data for the Tkk-01-0043 isolate identified one negative energy class one SNP 531 C → T. Light Forge system detected a () of −0.48 ± 0.09 °C, whereas the Light Cycler®96 system detected −0.24 ± 0.02 °C as the (). The NN model predicted a melting temperature difference of −0.23 °C.
Sanger sequencing the Tkk-01-0039 isolate identified 1 negative energy class one SNP 526 C → T. Consistently, the Light Forge system detected a () of −0.37 ± 0.16 °C when this isolate was compared to the reference, whereas the Light Cycler®96 system detected a () of −0.26 ± 0.02 °C. The NN model predicted a melting temperature difference of −0.84 °C, which agreed with both the Light Cycler®96 and the Light Forge systems.
Validation of Light Forge with Blinded Samples
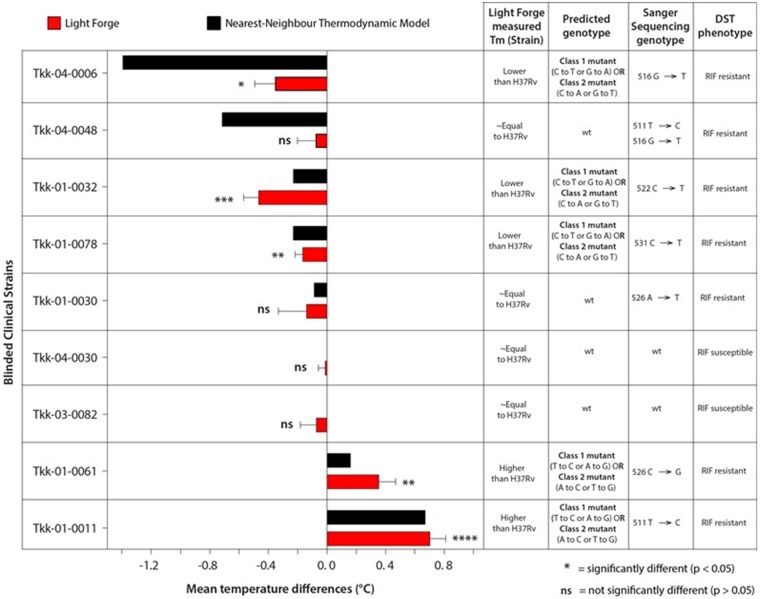
For further validation of the Light Forge assay, 9 blinded samples were analyzed. The () melt temperature of the blinded samples was compared to H37Rv. Four (44.4%) of the 9 blinded isolates showed melting curve profiles that were synonymous to that of H37Rv (Fig. 5). However, 5 (55.5%) of the 9 blinded isolates were distinguishable from the H37Rv melting curve profile. In comparison to the culture-based RIF-susceptibility test (DST), Light Forge correctly identified 7 out of 9 of the isolates with 71.43% sensitivity and 100% specificity. The discrepancy was noted for Tkk-04-0048, which had a drug-susceptible profile on the Light Forge system but showed resistance in culture, confirmed with sequencing by the presence of a double mutation in the drug-resistance determining region of the rpoB. The isolate Tkk-01-0030 also showed discrepancy with a drug-susceptible profile on the Light Forge but resistance in culture, confirmed with sequencing by the presence of an (A to T) change.
Fig. 5.
Precision and reproducibility comparisons of Light Forge phenotypic resistance prediction on blinded TB clinical isolates using HRMA. These results were reproducible by two independent Light Forge experiment repeats with very small standard deviations.
Discussion
Detecting drug-resistant strains of TB remains vital for the timely clinical management of the disease, with potential to significantly reduce transmission. This proof of concept (POC) study details the design, development and preliminary performance evaluation of Light Forge, a microfluidic device to detect RIF resistance linked mutations in M.tb.
In the initial phase of the study, 7 RIF-resistant strains of M.tb were used to bench mark the performance of Light Forge to the Roche Light Cycler®96. As shown in Fig. 4, Light Forge showed 100% concordance with Sanger sequencing compared to the 86% demonstrated by the Light Cycler®96. The same trend was observed when Light Forge was compared to the nearest neighbor (NN) thermodynamic model. An interesting observation from the panel was that the commercial device did not detect the presence of the mutation in the Tkk-01-0062 strain. This strain harbored a class 4 transversion (516 A → T), which is difficult to detect as it results in a melting temperature difference of less than 0.4 °C (15). This was consistent with a study that showed that HRMA had diminished ability to detect trans versions when the reaction volume increased 5-fold (20). The higher sensitivity is achieved through rapid heat transfer in microfluidic-based PCR due to the small reaction mass and the higher surface to volume ratio of the small reactor, leading to a more uniform temperature distribution (21).
To circumvent subjectivity of the study (22), the second phase of the experiments was performed with the scientists blinded to the resistance profile for all nine clinical isolates. Light Forge detected the presence of mutations in 7/9 isolates as shown in Fig. 5 (5 RIF-R and 3 RIF-S), when compared to sequencing and drug susceptibility profiles. Two RIF-resistant strains, Tkk-04-0048 and Tkk-01-0030, were erroneously detected as wild type. Tkk-04-0048 contained double mutations at positions (511 T → C and 516 G → T). These mutations were undetectable as the increase in melting temperature due to the mutation at position 511 was offset by a decrease in melting temperature by the mutation at position 516. This observation is not unique to this study, as it has been reported that co-occurrence of class 3 and 4 mutations within the same amplicon leads to minimal melting temperature deviation (23). Tkk-01-0030 contained a single mutation (526 A → T). This specific mutation was located between two cytosine (C) nucleotides as shown in supplemental Fig. S4. The mutation appears to lead to minimal deviation in the melting temperature in comparison to H37Rv, consistent with the observations that (A/T) transversion have miniscule impact on the melting profile of the amplicons (24–26). As stated previously, we speculate that reducing the volume of the microfluidic reactors by between 5- to 20-fold could potentially enhance the resolution power of the HRMA assay.
Whilst Light Forge has the potential to contribute significantly to improving health care delivery systems, particularly those in low income settings, it has limitations arising from its design based on molecular testing of TB and its reliance on microfluidic technology. First, HRMA can detect melting temperature aberrations that can be misinterpreted as the presence of mutations, but which do not confer any phenotypic drug resistance (including silent mutations). Thus, the assay can erroneously predict that a strain with the mutation (516 G → C) is drug resistant, contrary to its drug-sensitive profile (27). A similar occurrence was reported when a silent mutation at codon 514 of the rpoB gene was misclassified as drug resistant using a commercial genotyping kit (28). This is the major reason why sequencing and DST remain more precise tools for asserting clinical resistance of strains (29). Nevertheless, with proper execution, Light Forge could be a useful tool for screening TB patients.
Light Forge was designed using principles adopted from microfluidic large-scale integration (MLSI), which allow for several hundred to thousands of reactors on a single device (30). Some limitations of this study are that few isolates were used, and that only DNA from pure clinical cultures but no primary specimens such as blood or sputum were used. In addition, the assay would have benefited from a comparison with the well-established GeneXpert MTB/RIF Assay. However, these limitations do not detract from the value and significance of the POC Light Forge findings. To our knowledge, the Light Forge chip represents one of the few iterations that were performed at nanoliter scale. There are several modifications that can be incorporated into Light Forge to address possible errors that could emanate from heterogeneity of M.tb populations, which may be the case if primary specimens are used. We, therefore, propose a future Light Forge platform that interrogates a single copy of DNA in each reactor by combining limiting dilution and MLSI to acquire melting point data for each single genome amplicon. Further advancements are required for Light Forge to transition from proof-of-concept to a commercially viable product. The run time of 180 min requires reduction to make it feasible in the clinical setting, an improvement which will be easily implemented with more robust hardware. It is encouraging that several devices have completed the transition from the diagnostic development pipeline to commercial point-of-care microfluidic devices for tasks such as blood analysis and nucleic acid quantification, as well as identification of pathogens such as M.tb (Cepheid’s GeneXpert System) (31).
Light Forge successfully detected 14 out of 16 samples based on their drug-resistance profiles. Whilst this is a positive step towards creating an accurate low-cost test for TB drug resistance, the assay needs to account for difficult-to-detect mutations such as double mutations and silent mutations, as well as work toward the final development of a version ready for clinical testing. Possible improvements include use of a 21 megapixel camera, a fluorescent lamp, a thermal block, and a simple computer interface to create a cost-sensitive device similar to that developed by Hatch and colleagues (32). Creating a device that is easy for an end user to operate will allow rapid integration within affected countries. A simplistic design will also reduce the cost of purchase and maintenance of the device. Prior to implementation, Light Forge should be validated using a larger number of isolates as well as biological specimens from patients with and without HIV from different geographical regions (33).
Supplementary Material
Nonstandard Abbreviations
TB, tuberculosis; HRMA, high-resolution melting analysis; DST, drug susceptibility testing; M.tb, Mycobacterium tuberculosis; SNP, single nucleotide polymorphism; PCR, polymerase chain reaction; RIF, rifampicin; RRDR, rifampicin resistance determining region; AHRI, Africa Health Research Institute; NTC, non-template control; PDMS, polydimethylsiloxane; NN, nearest neighbor thermodynamic model; RFU, relative fluorescence units; TM, melting temperature; ΔTM, melting temperature change; ΔHo, standard enthalpy change; ΔSo, standard entropy change; Ct, total oligonucleotide strand concentration; RIF-R, rifampicin resistant; RIF-S, rifampicin susceptible; A, adenine; T, tyrosine; G, guanine; C, cytosine; POC, proof of concept.
Disclaimer: The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
I.M. Mbano, T. Mandizvo, J. Rogich, and F.K. Balagaddé developed the concepts and designed the study. JR was involved in the development and implementation of the technology. I.M. Mbano and T. Mandizvo performed the experiments. All the authors were involved in the analysis and interpretation of the results. I.M. Mbano, T. Mandizvo wrote the manuscript with feedback from all the authors.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Authors’ disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared. Consultant or Advisory Role: None declared. Stock Ownership: None declared.
Honoraria: None declared. Research Funding: The Howard Hughes Medical Institute (http://www.hhmi.org/), The National Research Foundation (https://www.nrf.ac.za/) NFS13091238686, The Cannon Collins Educational and Legal Assistance Trust (https://www.canoncollins.org.uk/ and The University of KwaZulu-Natal College of Health Sciences. The Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant # DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant # 107752/Z/15/Z] and the UK government. Expert Testimony: I.M. Mbano, Biophysical Society; T. Mandizvo, Biophysics in the Understanding, Diagnosis, and Treatment of Infectious Diseases, International Workshop on Microsystems Technologies for African Health. Patents: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments: We thank Dr. Alex S. Pym for M.tb clinical strains as well as helpful discussions. Further, we acknowledge the mechanical and computational assistance received from Engineer, Yashveer Ramlakhan.
Glossary
List of abbreviations
- TB
tuberculosis
- HRMA
high-resolution melting analysis
- DST
drug susceptibility testing
- M.tb,
Mycobacterium tuberculosis
- SNP
single nucleotide polymorphism
- PCR
polymerase chain reaction
- RIF
rifampicin
- RRDR
rifampicin resistance determining region
- AHRI
Africa Health Research Institute
- NTC
non-template control
- PDMS
polydimethylsiloxane
- NN
nearest neighbor thermodynamic model
- RFU
relative fluorescence units
- TM
melting temperature
melting temperature change
standard enthalpy change
standard entropy change
total oligonucleotide strand concentration
- RIF-R
rifampicin resistant
- RIF-S
rifampicin susceptible
- A
adenine
- T
tyrosine
- G
guanine
- C
cytosine
Contributor Information
Ian M Mbano, Email: ian.mbano@ahri.org.
Tawanda Mandizvo, Email: tawanda.mandizvo@ahri.org.
REFERENCES
- 1.World Health Organization. Global tuberculosis report 2018. Geneva: World Health Organization; 2018.
- 2.World Health Organization. World health statistics 2018: monitoring health for the SDGs, sustainable development goals. Geneva: World Health Organization; 2018.
- 3. Chung-Delgado K, Guillen-Bravo S, Revilla-Montag A, Bernabe-Ortiz A.. Mortality among MDR-TB cases: comparison with drug-susceptible tuberculosis and associated factors. PloS One 2015;10:e0119332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaramillo E. Guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: World Health Organization; 2008. [PubMed] [Google Scholar]
- 5. McNerney R. Diagnostics for developing countries. Diagnostics 2015;5:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peeling R, McNerney R. Increasing access to diagnostics through technology transfer and local production; 2011.
- 7. Mark D, Haeberle S, Roth G, Von Stetten F, Zengerle R.. Microfluidic lab-on-a-chip platforms: requirements, characteristics and applications. In: Microfluidics based microsystems. Berlin: Springer; 2010. p. 305–76. [DOI] [PubMed] [Google Scholar]
- 8. Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics; 2007. [DOI] [PubMed]
- 9. Lee AS, Ong DC, Wong JC, Siu GK, Yam W-C.. High-resolution melting analysis for the rapid detection of fluoroquinolone and streptomycin resistance in Mycobacterium tuberculosis. PLoS One 2012;7:e31934.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolff BJ, Thacker WL, Schwartz SB, Winchell JM.. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob Agents Chemother 2008;52:3542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ong DC, Yam W-C, Siu GK, Lee AS.. Rapid detection of rifampicin-and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J Clin Microbiol 2010;48:1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramaswamy S, Musser JM.. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tubercle Lung Dis 1998;79:3–29. [DOI] [PubMed] [Google Scholar]
- 13. Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T.. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother 1993;37:2054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR Jr. Genetic manipulation of Mycobacterium tuberculosis. Curr Protoc Microbiol 2007;6:10A.. [DOI] [PubMed] [Google Scholar]
- 15. Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL.. Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J Clin Microbiol 2011;49:3450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thorsen T, Maerkl SJ, Quake SR.. Microfluidic large-scale integration. Science 2002;298:580–4. [DOI] [PubMed] [Google Scholar]
- 17. SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci 1998;95:1460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watkins NE Jr, SantaLucia J Jr. Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res 2005;33:6258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 2004;50:1156–64. [DOI] [PubMed] [Google Scholar]
- 20. Pholwat S, Liu J, Stroup S, Gratz J, Banu S, Rahman SM, et al. Integrated microfluidic card with TaqMan probes and high-resolution melt analysis to detect tuberculosis drug resistance mutations across 10 genes. MBio 2015;6:e02273–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park S, Zhang Y, Lin S, Wang T-H, Yang S.. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv 2011;29:830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Day SJ, Altman DG.. Blinding in clinical trials and other studies. BMJ 2000;321:504.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramirez MV, Cowart KC, Campbell PJ, Morlock GP, Sikes D, Winchell JM, et al. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol 2010;48:4003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoek K, van Pittius NG, Moolman-Smook H, Carelse-Tofa K, Jordaan A, Van Der Spuy G, et al. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J Clin Microbiol 2008;46:1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pietzka AT, Indra A, Stöger A, Zeinzinger J, Konrad M, Hasenberger P, et al. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J Antimicrob Chemother 2009;63: 1121–7. [DOI] [PubMed] [Google Scholar]
- 26. Yin X, Zheng L, Liu Q, Lin L, Hu X, Hu Y, et al. High-resolution melting curve analysis for rapid detection of rifampin resistance in Mycobacterium tuberculosis: a meta-analysis. J Clin Microbiol 2013;51:3294–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakata N, Kai M, Makino M.. Mutation analysis of mycobacterial rpoB genes and rifampin resistance using recombinant Mycobacterium smegmatis. Antimicrob Agents Chemother 2012;56:2008–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alonso M, Palacios JJ, Herranz M, Penedo A, Menéndez Á, Bouza E, et al. Isolation of Mycobacterium tuberculosis strains with a silent mutation in rpoB leading to potential misassignment of resistance category. J Clin Microbiol 2011;49:2688–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moure R, Martín R, Alcaide F.. Silent mutation in rpoB detected from clinical samples with rifampin-susceptible Mycobacterium tuberculosis. J Clin Microbiol 2011;49:3722–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whitesides GM. The origins and the future of microfluidics. Nature 2006;442:368–373. [DOI] [PubMed] [Google Scholar]
- 31. Eicher D, Merten CA.. Microfluidic devices for diagnostic applications. Exp Rev Mol Diagn 2011;11:505–19. [DOI] [PubMed] [Google Scholar]
- 32. Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, et al. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 2011;11:3838–45. [DOI] [PubMed] [Google Scholar]
- 33. Mahilum-Tapay L, Laitila V, Wawrzyniak JJ, Lee HH, Alexander S, Ison C, et al. New point of care Chlamydia Rapid Test—bridging the gap between diagnosis and treatment: performance evaluation study. BMJ 2007;335:1190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.