Abstract
Linker histones H1 are principal chromatin components, whose contribution to the epigenetic regulation of chromatin structure and function is not fully understood. In metazoa, specific linker histones are expressed in the germline, with female-specific H1s being normally retained in the early-embryo. Embryonic H1s are present while the zygotic genome is transcriptionally silent and they are replaced by somatic variants upon activation, suggesting a contribution to transcriptional silencing. Here we directly address this question by ectopically expressing dBigH1 in Drosophila S2 cells, which lack dBigH1. We show that dBigH1 binds across chromatin, replaces somatic dH1 and reduces nucleosome repeat length (NRL). Concomitantly, dBigH1 expression down-regulates gene expression by impairing RNApol II binding and histone acetylation. These effects depend on the acidic N-terminal ED-domain of dBigH1 since a truncated form lacking this domain binds across chromatin and replaces dH1 like full-length dBigH1, but it does not affect NRL either transcription. In vitro reconstitution experiments using Drosophila preblastodermic embryo extracts corroborate these results. Altogether these results suggest that the negatively charged N-terminal tail of dBigH1 alters the functional state of active chromatin compromising transcription.
INTRODUCTION
Linker histones H1 constitute an evolutionarily conserved family of chromosomal proteins that play an important structural role in regulating chromatin compaction and higher order chromatin organization (see (1) for a recent review). In metazoan species, histones H1 usually exist as multiple variants, some of which are specifically expressed in the germline (reviewed in (2)). For instance, four of the eleven mice/human H1 isoforms are germline specific, of which three are expressed in males (H1T, HILS1 and H1T2) and one in females (H1oo). Female-specific variants usually accumulate in the oocyte and are retained during early embryogenesis (2). In comparison to most metazoa, H1 complexity in Drosophila is much reduced since it contains a single somatic dH1 variant (3–5), which is ubiquitously expressed throughout development, and a single germline specific variant dBigH1, which is expressed in both the female and male germlines, and it is retained in the early embryo (6,7). Embryonic H1s persist as long as the zygotic genome remains transcriptionally silent, being replaced by somatic variants when transcription begins during zygotic genome activation (ZGA) (reviewed in (2)). In Drosophila, dBigH1 is present during early embryogenesis until ZGA onset at cellularization (6). At this stage, dBigH1 is replaced by somatic dH1 in somatic cells, whereas it is retained in the primordial germ cells (PGC) (6), which remain transcriptionally silent.
These observations suggest that dBigH1, and embryonic H1s in general, are general transcriptional regulators that contribute to silencing. Linker histones H1 have been usually associated with transcription repression, but somatic H1s are readily detected across expressed genes (8–14). In this regard, it has been reported that somatic H1s can even enhance the synergism between transcription factors (15). In contrast, in the presence of dBigH1, chromatin appears to be transcriptionally silent, suggesting that dBigH1 enhances transcriptional silencing. Here, we analyze the mechanisms of action of dBigH1. For this purpose, we have performed ectopic expression experiments in Drosophila S2 cells, which lack dBigH1. These experiments confirm the contribution of dBigH1 to transcriptional silencing, identifying the acidic N-terminal ED-domain as responsible for its negative effect on transcription.
MATERIALS AND METHODS
Antibodies and recombinant proteins
αdBigH1 antibodies are described in (6). αdH1 antibodies were a gift from Dr J. Kadonaga and are described in (16). αRpb3 antibodies were a gift from Dr J. Zeitlinger and are described in (17). The rest of antibodies used in these experiments were commercially available: αPol IIoser5 (Abcam, ab5131), αH3Kac (Millipore 06-599), αH3K36me3 (Cell Signaling, 9715), αH3 (Abcam, ab9050), αH4 (Abcam, ab10158) and αFLAG (Sigma F3165).
Recombinant His-tagged dBigH1 constructs were expressed in Escherichia coli BL21-LysS cells using pET30b(+) expression vectors (Novagen), and purified using Ni-NTA columns (BioRad) by conventional methods.
Stable S2 cell lines and transgenic flies
Stable S2 cell line expressing dBigH1::FLAG under the control of a Cu2+-inducible promoter is described in (7). Stable lines expressing dBigH1ΔED::FLAG and dBigH1ΔNTD::FLAG were generated identically as the dBigH1::FLAG-expressing line.
w1118 and daughterless(da)-GAL4 flies were obtained from the Bloomington Stock Center. Transgenic flies carrying UAS-dBigH1 construct are described in (6). Transgenic flies carrying UAS-dBigH1ΔCTD, UAS-dBigH1ΔED and UAS-dBigH1ΔNTD were obtained by specific site-directed integration of the appropriate constructs into ZH-51D att line. All Drosophila stocks were maintained at 25°C on standard media.
Expression of dBigH1 constructs in S2 cells and salivary glands
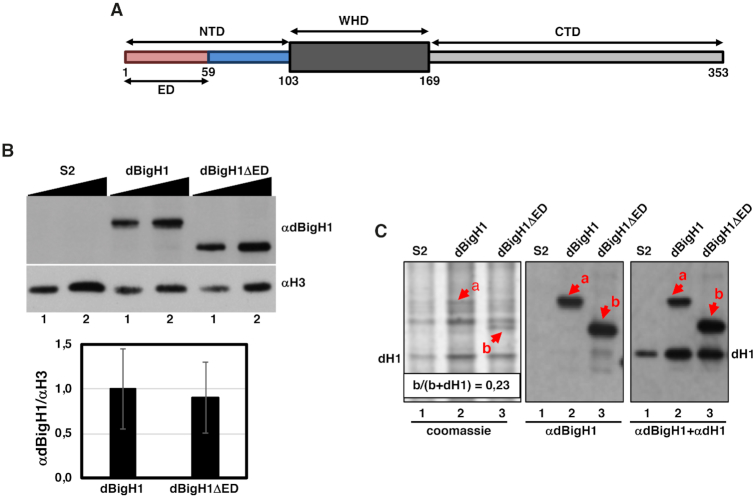
In S2 cells, expression of the dBigH1::FLAG, dBigH1ΔED::FLAG, dBigH1ΔNTD::FLAG constructs was induced with 1mM CuSO4 for 24 h. To determine the extent of binding to chromatin of the different constructs, crosslinked chromatin was prepared as for ChIP (see below), diluted in PLB and analyzed by WB using αdBigH1 (1:10,000), αFLAG (1:2500), αH3 (1:2500) and αH4 (1:5000) antibodies. For quantitative analysis, WBs were digitalized using a GS-800 Calibrated laser densitometer (BioRad) and analyzed using Fiji software (18). From these analyses we estimated that chromatin abundance of the full-length dBigH1::FLAG, and the truncated dBigH1ΔED::FLAG and dBigH1ΔNTD::FLAG forms was similar (Figure 1B and Supplementary Figure S7A). To determine the actual abundance of the various dBigH1 constructs, total histones were prepared by acid extraction of purified nuclei in 0.2N HCl overnight in a rotating wheel at 4°C. Acid extracts were analyzed by Coomassie staining and WB using αdBigH1 (1:10,000) and αdH1 (1:20,000) antibodies (Figure 1C). Quantitative analysis of Coomassie stained gels using Fiji software, showed that dBigH1ΔED::FLAG accounted for ∼23% of total linker histones. The levels of full-length dBigH1::FLAG could not be directly determined since it overlapped with an unrelated species present in control non-induced cells. Similarly, dBigH1ΔNTD::FLAG expression levels could neither be determined directly since it overlapped with dH1 (Supplementary Figure S7B). However, considering that chromatin abundance of the full-length dBigH1::FLAG and the truncated dBigH1ΔNTD::FLAG forms was similar to that of dBigH1ΔED::FLAG (Figure 1B and Supplementary Figure S7A), we estimated that all three forms were expressed to a similar extent, accounting for ∼20–25% of total linker histones.
Figure 1.
Expression of dBigH1 constructs used in these experiments. (A) Schematic representation of the domain organization of dBigH1. NTD, N-terminal domain; WHD, winged helix domain; CTD, C-terminal domain; ED, acidic domain. Numbers indicate aa position in the primary sequence. (B) The abundance of the indicated dBigH1 constructs in crosslinked chromatin prepared from control S2 cells and cells expressing full-length dBigH1::FLAG and the truncated dBigH1ΔED::FLAG form was determined by WB with αdBigH1 antibodies. Increasing amounts of chromatin are analyzed (lanes 1 and 2). αH3 antibodies were used for normalization. Quantitative analysis of the results is shown in the bottom (N = 3; error bars are SD; two tailed t-test, P-value = 0.247). (C) Total acid extracts prepared from control S2 cells (lane 1) and cells expressing full-length dBigH1::FLAG (lane 2) and the truncated dBigH1ΔED::FLAG (lane 3) forms were analyzed in Coomassie-blue stained gels (left) and by WB with αdBigH1 (center) and αdBigH1+αdH1 (right) of the gel in the left. Bands corresponding to dBigH1::FLAG (a) and the truncated dBigH1ΔED::FLAG (b) forms are indicated. The band corresponding to dH1 is also indicated. Note that dBigH1::FLAG overlaps with an unrelated species present in control S2 cells. The proportion of dBigH1ΔED::FLAG (band b) respect to total linker histones (b+dH1) is indicated.
For expression in salivary glands homozygous flies carrying the various dBigH1 constructs described above were crossed to homozygous da-GAL4 flies at 25°C.
ChIP experiments
For ChIP experiments, 108 cells were collected and fixed with 1.8% of formaldehyde for 10 min at room temperature by gently mixing. Cross-linking was stopped adding glycine to a final concentration of 125mM. After 5 min, cells were spun down for 2 min at 1500g and washed with 5 ml of PBS. Then, cells were resuspended in 10 ml of 10 mM HEPES pH 7.9, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100 and incubated for 10 min in a wheel at 4°C. Cells were spun down again and resuspended in 10 ml of 10 mM HEPES pH 7.9, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.01% Triton X-100, incubated for 10 min in a rotating wheel at 4°C and spun again for 2 min at 1500g. Later, cells were lysed in 5 ml of TE (10 mM Tris–HCl pH 8, 1 mM EDTA) with 1% SDS. Chromatin was washed three times with 5 ml of TE, resuspended in TE, 1 mM PMSF, 0.1% SDS and sonicated in a Bioruptor Pico sonifier (Diagenode) to obtain fragments of 200–500 bp. After sonication, lysates were adjusted to 1% Triton X-100, 0.1% sodium deoxycholate (DOC), 140 mM NaCl, incubated for 10 min in a rotating wheel at 4°C and chromatin was recovered by centrifugation. For each experiment, 400 μl of chromatin was used for the immunoprecipitation (IP) and 40 μl for the input sample. IPs were carried out in RIPA buffer (140 mM NaCl, 10 mM Tris–HCl pH 8, 1 mM EDTA, 1% Triton X-100, 0.01% SDS, 0.1% DOC). A pre-clearing step was carried out in a rotating wheel for 1 h at 4°C with 30 μl of 50% (v/v) protein A-Sepharose CL4B beads (GE Healthcare, 17-0780-01) previously blocked with RIPA–1%BSA. Then, the corresponding antibodies were added and incubated overnight at 4°C in a rotating wheel. IPs were performed by adding 40 μl of 50% (v/v) protein A-Sepharose CL4B beads previously blocked with RIPA–1%BSA and incubating in a rotating wheel for 3 h at 4°C. Beads were washed five times for 5 min in 1 ml of RIPA buffer, once for 5 min in 250 mM LiCl, 10 mM Tris–HCl pH 8, 1 mM EDTA, 0.5% NP-40, 0.5% DOC and twice for 5 min in TE. Beads were then resuspended in 40 μl of TE, DNase-free RNaseA was added at 0.25 μg/ml to the IPs and input samples, and incubated for 30 min at 37°C. To purify the DNA, samples were adjusted to 1% SDS, 0.1 M NaHCO3, 0.2 mg/ml Proteinase K and incubated overnight at 65°C. DNA was purified by phenol-chloroform extraction. Antibodies used were αdBigH1 (1 μl), αH1 (1 μl), αRpb3 (3.5 μg), αPol IIoser5 (2 μg), αH3K36me3 (2 μg) and αH3Kac (2 μg).
For ChIP-seq experiments, all experiments were done in duplicate. Libraries were prepared from 3 to 10 ng of DNA using the NEBNext Ultra II DNA Library Prep kit for Illumina (NEB, E7645S) according to manufacturer's instructions. Libraries were then subjected to PCR amplification (10 cycles) and cluster generation for subsequent sequencing. For ChIP-qPCR experiments, IPs were analyzed according to the ΔΔCt method. Primers used in these experiments are described in Supplementary Table S1.
Expression profiling and RT-qPCR analysis
For expression analysis, total RNA was isolated from 107 cells using the RNeasy Mini Kit (Qiagen) and cDNAs were generated from 25 ng of total RNA and subjected to PCR amplification (17 cycles) using the TransPlex® Complete Whole Transcriptome Amplification Kit (Sigma, WTA2) according to manufacturer's instructions. 8 μg of amplified cDNAs were fragmented and labeled using the GeneChip Human Mapping 250K Nsp Assay kit (Affymetrix, 900766) according to manufacturer's instructions. Labeled cDNAs were then hybridized in GeneChip Drosophila Genome arrays 2.0 from Affymetrix (Thermo Fisher, 900531) for 16 h at 45°C. After incubation, the arrays were processed in the GeneChip Fluidics Station 450 from Affymetrix and scanned in GSC3000 System from Affymetrix.
For RT-qPCR analysis, total RNA was isolated from 107 cells using the RNeasy Mini Kit (Qiagen). For cDNA synthesis, 1 μg of total RNA was used and qPCRs were run in triplicate in at least 4 independent experiments. Expression data were normalized to Rpl32 and analyzed using ΔΔCt method. Primers used in these experiments are described in Supplementary Table S1.
Bioinformatics and biostatistics analyses
Quality control of raw ChIP-Seq data was assessed with the FastQC tool version 0.11 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Then, FastQ files were aligned against the dm3 genome using Bowtie 0.12.5 (19), allowing for two mismatches (-n 2) and reporting only unique hit alignments (-m 1). Afterwards, potential PCR over-amplification artifacts were assessed and removed using sambamba 0.5.1 (20). Number of uniquely aligned reads were checked to be within the ENCODE recommended guidelines (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3431496/). A summary of these ChIP-seq quality control data is presented in Supplementary QC Data. Binary tracks for samples in TDF format for visual assessment of sample signal were generated with IGVTools 2 (21). In order to determine whole genome distribution, peak calling for IP vs their respective input sample was performed with MACS 1.4.2 (22), setting the option –g dm to account for Drosophila melanogaster genome size, and leaving the rest of options as default. Unless otherwise specified, all downstream analyses were performed using R3.4.4 and Bioconductor (23). Additional assessment of immunoprecipitation, group separation and biological replicate correlation was performed via generation of Gini/SSD coverage inequality indexes, PCA-like MDS plots and coverage correlation heatmaps using htSeqTools 1.26.0 (24). Lorenz curves for assessing coverage distribution in the analyzed samples were generated with the ineq package version 0.2–13 using genome bins of 1000bp. Per-sample gene level quantification in log2RPKM and log2RPKM(IP/Input) ratios were generated with the countOverlaps function from the GenomicRanges package version 1.30.3, and using the UCSC Drosophila melanogaster dm3 annotation. Raw metagene profiles were generated from normalized coverage ratio values with the regionsCoverage function from the htSeqTools package. Whole-genome epigenetic profile Multidimensional Scaling maps were generated with the chroGPS package version 2.0 (25,26). The pausing index (PI) of total RNApol II (Rpb3) was calculated as described elsewhere (27,28). In brief, Rpb3 occupancy (average log2RPKM(IP/input) ratio between replicates) was computed at the TSS-region (defined as TSS ± 250 bp) and the CDS (defined as TSS + 250 bp to TTS – 250 bp) for each gene longer than 1 kb and, then, the PI was determined as the TSS/CDS ratio of Rpb3 occupancy.
Affymetrix data from Drosophila2 arrays for dBigH1-expressing and control mock-induced cells were normalized with R and Bioconductor using RMA background correction, quantile normalization and RMA summarization to obtain probeset expression estimates (29). This method of data normalization is based on the assumption that there is not a general change in gene expression between the analyzed experimental conditions, with most genes suffering no big expression changes. In our case, raw intensity values before RMA did not present a global shift in expression in a consistent manner, except for a weak global decrease detected in one of the replicates (Supplementary QC Data). Next, we used limma (29) to determine differentially expressed probesets in dBigH1-expressing vs control cells, using a Benjamini-Hochberg adjusted P < 0.1 and a |FC| > 1.5. The GSEA pre-ranked algorithm (30) was used to identify significantly enriched and depleted KEGG pathways as provided by the org.Dm.eg.db package (November 2014), using all probesets in the Drosophila genome ranked by mean log2FC, summarized at gene level using the annotation from the drosophila2.db package version 2.8.1.
ChIP-seq and arrays data are deposited at NCBI GEO (GSE127227 and GSE103292).
NRL determination
For NRL determination, 2 × 107 cells of each condition were grown and treated with 1 mM CuSO4 for 24 h. The cells were collected washed twice with PBS and fixed with 1.1% of formaldehyde for 10 min at room temperature by gently mixing. Cross-linking was stopped adding glycine to a final concentration of 125 mM. After 5 min, cells were spun down for 8 min at 1000g at 4°C and washed twice with 10 ml of cold PBS before pelleting and flash freezing in liquid nitrogen. For MNase digestion, the pellet was resuspended in PBS, 0.1% Triton X-100 (PBS-TX). The digestion was performed at 37°C in a volume of 400 μl PBS–TX with 4 × 106 cells per digestion time. CaCl2 was adjusted to 1 mM, cells were pre-warmed and, after adding 0.8 U of MNase (Sigma-Aldrich), incubated for increasing time from 30 s to 5 min. Digestion was stopped in ice by adding EDTA and EGTA to a final concentration of 20 mM each. To purify the DNA, samples were adjusted to 10 mM Tris HCl pH 8, 0.4% SDS, 0.4 mg/ml Proteinase K and incubated overnight at 65°C. DNA was extracted by phenol-chloroform and precipitated with ethanol. Later the samples were treated with RNase A for 30 min at 37°C, ran in 2% agarose gels at 90 V for 5 h and stained with ethidium bromide. Images were analyzed using Fiji software and the size of each fragment was determined from the maximum of the corresponding size distribution using MW markers. To determine the apparent NRL, the size of fragments containing increasing number of nucleosomes, from mono- to hexanucleosomes, was plotted against the number of nucleosomes and the apparent NRL calculated from the slope of the corresponding regression curve. To compare NRLs of the different conditions, samples showing similar extent of digestion were analyzed.
In vitro chromatin reconstitution and transcription experiments
Drosophila embryo extract DREX was prepared in Exb50 buffer (10 mM HEPES pH 7.6, 50 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA pH 8, 10 mM β-glicerophosphate, 10% glycerol) from preblastodermic embryos, as described in (31). To prepare dBigH1-depleted DREX, αBigH1 antibodies were coupled to protein A magnetic beads and incubated with DREX for five rounds of 45 min each at 4°C. The extent of depletion was determined by WB (Supplementary Figure S4B). Chromatin reconstitution experiments were performed as described in (31), using 600 ng of a pAc5.1-V5-His plasmid (Invitrogen) carrying an EGFP reporter gene under the control of the Drosophila actin5C promoter. After reconstitution, chromatin was precipitated with 15 mM of MgCl2 and resuspended in Exb50. The extent of chromatin assembly was determined by MNase digestion as described in (31). For in vitro transcription assays, 12 μl of the reconstituted chromatin were incubated for 60 min at 30°C with 10 μl Buffer-C90 (20 mM Tris–HCl pH 7.8, 1 mM EDTA, 1 mM DTT, 10% glycerol, 90 mM NaCl), 20 μl Rxn mix (20 mM Tris pH 8.3, 5 mM MgCl2, 3 mM DTT, 25 mM rNTP mix, 6.25% PEG 8000) and 14 μl of HeLa nuclear extract (CILBiotech) as described in (15). Transcription was stopped by incubation at 39°C for 15 min in stop-mix (200 mM NaCl, 20 mM EDTA, 1% SDS, 12 μl glycogen, 0.12 μg/μl Proteinase K). Then, RNA was purified using the RNeasy Mini Kit (Qiagen) and cDNA was prepared using the Transcriptor First Strand cDNA Synthesis Kit (Roche) with oligo (dT)18 primers. The amount of GFP mRNA was quantified by RT-qPCR using primers against GFP (Supplementary Table S1) and normalized with respect to the total amount of chromatin template used in the assay determined by qPCR with the same primers before transcription. For each experimental condition, three independent biological replicates assembled in the same dBigH1-depleted DREX (Supplementary Figure S4B) were analyzed.
Immunostaining experiments in salivary glands
For immunostaining, salivary glands from L3 larvae were dissected in Cohen solution (10 mM MgCl2, 25 mM glycerol-3-phosphate, 3 mM CaCl2, 10 mM KH2PO4, 0.5% NP-40, 30 mM KCl, 160 mM sucrose) and fixed in 0.74% formaldehyde in PBS for 2 min. Glands were then incubated for 3 min and squashed in 45% acetic acid, 0.62% formaldehyde. Preparations were washed for 5 min in PBS-T (PBS, 0.05% Tween 20) and incubated with αdBigH1 (1:400) antibodies in PBS-T, 1%BSA overnight at 4°C. Preparations were washed three times for 5 min in PBS-T and incubated with the secondary antibody for 2 h at room temperature. Slides were mounted in Mowiol (Calbiochem-Novabiochem) containing 0.2 ng/μl DAPI (Sigma) and visualized on a Nikon Eclipse E1000 microscope.
RESULTS
Ectopically expressed dBigH1 binds across chromatin and down-regulates gene expression
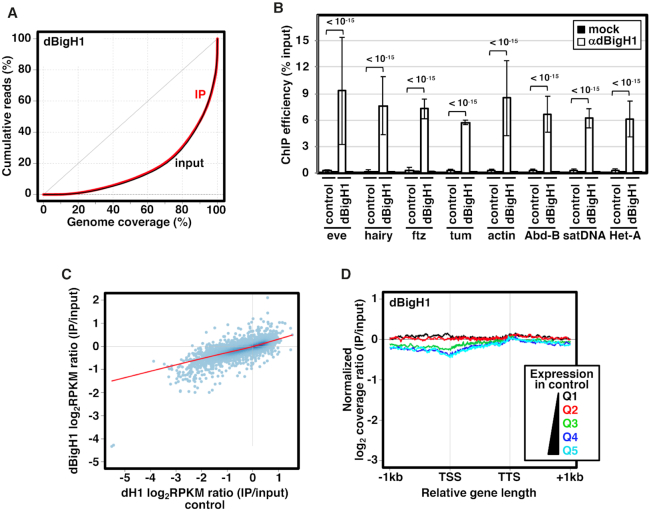
To ectopically express dBigH1 in Drosophila S2 cells we used a stable cell line expressing a dBigH1::FLAG construct under the control of a Cu2+-inducible promoter (Figure 1) (7). Upon induction, dBigH1 accounted for ∼23% of total linker histones (Figure 1B and C) (see Materials and Methods). ChIP-seq analyses using αdBigH1 antibodies showed that ectopically expressed dBigH1 bound broadly across chromatin since similar genomic inequality Lorenz’ curves were observed in the immunoprecipitated (IP) and input samples (Figure 2A). ChIP-qPCR experiments confirmed these results since dBigH1 was detected at multiple randomly selected genomic regions, including repetitive DNA elements (Figure 2B). We observed that, though moderately, the genomic distribution of ectopically expressed dBigH1 in S2 cells positively correlated with those observed in embryos (6) (Pearson's correlation coefficient, r = 0.239) and testes (7) (Pearson's correlation coefficient, r = 0.331) (Supplementary Figure S1). Interestingly, in S2 cells, deposition of ectopically expressed dBigH1 strongly correlated with dH1 content in control non-induced cells since genes containing high dBigH1 levels in induced cells corresponded to genes with high dH1 content in non-induced cells (Pearson's correlation coefficient, r = 0.796) (Figure 2C). Along genes, linker histones H1 are usually depleted in the region around the transcription start-site (TSS) of highly expressed genes and their occupancy increases progressively along the transcription unit (CDS) (8–14) (see also Supplementary Figure S9B). On the other hand, no such depletion is detected in low expressed genes that, overall, have higher H1 content than highly expressed genes (8–14) (see also Supplementary Figure S9B). The dBigH1 coverage profile showed similar features (Figure 2D), though, in comparison to dH1 (Supplementary Figure S9B), the depletion at TSS in highly expressed genes, as well as the differences in dBigH1 content between high and low expressed genes, were less pronounced. We also performed mock ChIP-seq experiments with αdBigH1 antibodies in control non-induced cells to assess the background level of non-specificity. Although, like in induced cells, genomic inequality of the mock IP samples was similar to the inputs (Supplementary Figure S2A), the background αdBigH1 coverage profile along genes was markedly different (Supplementary Figure S2B). ChIP-qPCR experiments confirmed these results since, in comparison to the signal observed in induced cells, background αdBigH1 signal in control non-induced cells was equally negligible at both TSS and CDS of several genes (Supplementary Figure S3A). ChIP-qPCR experiments also showed that, at TSS of highly expressed genes, the occupancy of dBigH1 was higher in comparison to dH1 and similar to the occupancy at CDS (Supplementary Figure S3B, left). Instead, in low expressed genes, dH1 and dBigH1 showed similar high relative occupancy at TSS (Supplementary Figure S3B, right). ChIP-qPCR experiments also confirmed that dBigH1 occupancy was similar in high and low expressed genes (Supplementary Figure S3C, center), while dH1 was generally more abundant in low expressed genes (Supplementary Figure S3C, left).
Figure 2.
Ectopic dBigH1 expression in S2 cells results in its binding across chromatin. (A) Lorenz curves showing coverage inequality for IP (red) and input samples (black) of dBigH1 ChIP-seq analyses in induced cells. (B) ChIP-qPCR analyses with αdBigH1 antibodies and preimmune serum (mock) at the indicated genomic regions in control and dBigH1-expressing cells (N = 2; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values are indicated). (C) Correlation of gene level dBigH1 content (log2RPKM IP/Input) in dBigH1-expressing cells and dH1 content (log2RPKM IP/Input) in control cells. (D) The normalized log2 coverage ratio IP/input of dBigH1 in dBigH1-expressing cells is presented along an idealized gene-length ± 1 kb for genes longer than 1 kb categorized according to their expression quantile in control cells (Q1-lowest to Q5-highest). TSS, transcription start site; TTS, transcription termination site.
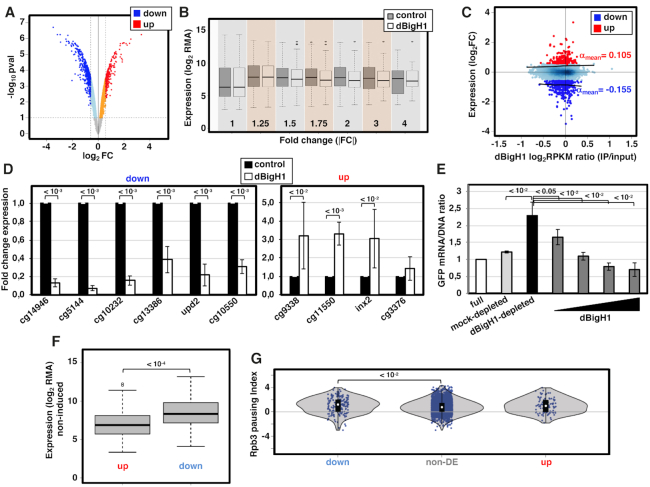
Next, we performed expression-profiling analyses to determine the effects of dBigH1 on gene expression. Upon dBigH1 expression, we detected 630 differentially expressed (DE) genes at fold-change |FC|>1.5 (Supplementary Table S2), accounting for ∼5% of the genes present in the array. Considering all genes, the proportion of up-regulated and down-regulated genes was roughly the same (9669 versus 9283 probesets, respectively) (Figure 3A). However, within the subset of DE genes, ∼68% were down-regulated (Figure 3A) and, overall, a global down-regulation was observed at |FC|>1.5 (Figure 3B). RT-qPCR experiments confirmed down-regulation of six randomly selected DE genes showing FC < –1.5 (Figure 3D, left) and, though weakly, the extent of down-regulation of the differentially down-regulated genes significantly correlated with their dBigH1 content (Pearson's correlation coefficient, r = 0.11; P-value = 0.0227) (Figure 3C). Regarding differentially up-regulated genes, RT-qPCR experiments confirmed up-regulation of three out of four genes tested (Figure 3D, right). However, up-regulation did not significantly correlate with dBigH1 content (Pearson's correlation coefficient, r = 0.09; P-value = 0.1737) (Figure 3C). Furthermore, permutation test analysis showed that the extent of up-regulation was significantly lower than the extent of down-regulation (permutation test; P-value = 0.007, B = 5000).
Figure 3.
dBigH1 down-regulates gene expression. (A) Volcano plot showing the change in expression of each individual gene in cells expressing dBigH1 in comparison to control mock-induced cells. Differentially down-regulated and up-regulated genes (|FC| > 1.5; Benjamini-Hochberg adjusted P-value < 0.1) are indicated in blue and red, respectively. (B) The normalized gene expression (log2RMA) in dBigH1-expressing cells and control mock-induced cells is presented for all genes classified according to their absolute fold-change (|FC|) expression upon dBigH1 expression. (C) Correlation of gene level dBigH1 content (log2RPKM IP/input) and expression FC upon dBigH1 expression (log2FC). Genes differentially up-regulated (red) and down-regulated (blue) are indicated. Non-DE genes are also indicated (light blue). Black lines are lowess regression fits for up-regulated and down-regulated genes; the average slopes (αmean) are indicated. (D) The fold change expression with respect to control mock-induced cells of six randomly selected down-regulated genes (left) and four randomly selected up-regulated genes (right) was determined by RT-qPCR in dBigH1-expressing cells (white) (N = 7; error bars are SD; two-tailed t-test, P-values are indicated). (E) The relative GFP mRNA/DNA ratios of a chromatin template carrying a GFP-reporter gene assembled in vitro in full (white), mock-depleted (light grey) and dBigH1-depleted DREX in the absence (black) and presence of increasing amounts of bacterially-expressed dBigH1 added during assembly (dark grey) (N = 3; error bars are SD; two-tailed t-test, P-values are indicated). (F) The expression (log2RMA) in control cells is presented for up-regulated and down-regulated genes (Wilcoxon rank sum test, P-value is indicated). (G) The pausing index of total RNApol II (Rpb3) in control cells is presented for down-regulated, non-DE and up-regulated genes (Wilcoxon rank sum test, P-values are indicated).
In vitro chromatin reconstitution experiments using Drosophila preblastodermic extracts (DREX) confirmed down-regulation induced by dBigH1. DREX has been extensively used in chromatin reconstitution experiments (32,33). DREX is enriched in dBigH1 (34) and chromatin reconstituted in DREX contains dBigH1 (Supplementary Figure S4A). To assess the effect of dBigH1 on transcription, a plasmid construct carrying a GFP-reporter was subjected to chromatin reconstitution in full DREX or after dBigH1 depletion using αdBigH1 antibodies (Supplementary Figure S4B), and the resulting reconstituted minichromosomes were used as template in in vitro transcription assays using HeLa extracts. MNase digestion showed that dBigH1-depleted DREX was as competent as full DREX for chromatin reconstitution (Supplementary Figure S4C). However, the GFP-reporter was transcribed to a significantly higher extent when the chromatin template was reconstituted in dBigH1-depleted DREX than in full or control mock-depleted DREX (Figure 3E). Furthermore, the addition of bacterially expressed recombinant dBigH1 during reconstitution with dBigH1-depleted DREX strongly reduced GFP transcription (Figure 3E).
We observed that, in comparison to non-DE or up-regulated genes down-regulated genes exhibited higher expression (Figure 3F) and stronger RNApol II pausing in control cells (Figure 3G). Gene ontology analyses showed that down-regulated genes mainly associated with KEGG pathways related to metabolic processes and housekeeping functions, while up-regulated ones correlate with more diverse functional pathways (Supplementary Figure S5A). We also generated whole-genome gene level epigenetic profile maps using Multidimensional Scaling with the chroGPS package (25,26). In these maps, all genes in the genome are represented in a two-dimensional space according to similarity between their epigenetic state and three main domains can be identified corresponding to active, Polycomb (PC) and HP1a chromatin (25). Down-regulated genes scattered throughout the map (Supplementary Figure S5B). Instead, we observed a lower proportion of up-regulated genes in the PC-chromatin domain (Supplementary Figure S5B).
The acidic ED-domain of dBigH1 is required for down-regulation
dBigH1 has the characteristic tripartite domain organization of linker histones, in which a central winged-helix domain (WHD) is flanked by unstructured N-terminal (NTD) and C-terminal (CTD) domains (35–37) (Figure 1A). The WHD and CTD of dBigH1 are relatively well-conserved with respect to somatic dH1 (6). In contrast, the NTD of dBigH1 contains an extra domain enriched in acidic E and D residues (6) (Figure 1A). As generally observed in linker histones (38,39), the positively charged CTD is required for dBigH1 binding to chromatin since, opposite to full length dBigH1, a truncated form missing the CTD did not bind to polytene chromosomes when ectopically expressed in salivary glands (Supplementary Figures S6A–C). On the other hand, truncated forms missing the acidic ED-domain or the entire NTD bound polytene chromosomes (Supplementary Figures S6D and E).
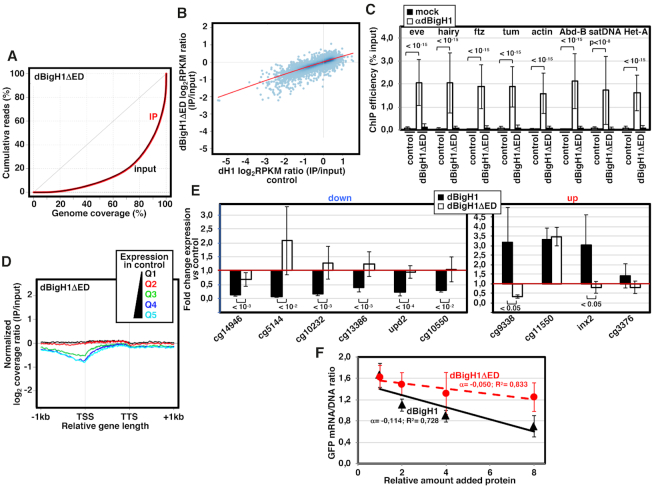
Next, we addressed the contribution of the extra ED-domain of dBigH1 to gene expression. For this purpose, we used a stable S2 cell line expressing a truncated dBigH1ΔED form, which was expressed to a similar level than full-length dBigH1 (Figure 1B). ChIP-seq analyses showed that, similar to full-length dBigH1, dBigH1ΔED was binding across chromatin (Figure 4A) and its deposition correlated with dH1 content in control non-induced cells (Pearson's correlation coefficient, r = 0.863) (Figure 4B). ChIP-qPCR experiments confirmed binding of dBigH1ΔED across chromatin (Figure 4C). However, at the gene level, the coverage profile of dBigH1ΔED showed a deeper depletion at TSS of highly expressed genes in comparison to full-length dBigH1 (Figure 4D). Regarding the effects on gene expression, we observed that deletion of the ED-domain in dBigH1ΔED abolished the down-regulation observed with full-length dBigH1 (Figure 4E, left). Furthermore, addition of dBigH1ΔED during in vitro reconstitution experiments using dBigH1-depleted DREX inhibited GFP expression to a weaker extent than addition of full-length dBigH1, as judged from the slopes of the inhibition curves obtained upon the addition of doubling amounts of the corresponding proteins (Figure 4F). Altogether these results suggest that the acidic ED-domain is required for down-regulation induced by dBigH1. The effects on up-regulated genes were less consistent (Figure 4E, right). Deletion of the entire NTD caused similar effects (Supplementary Figure S7).
Figure 4.
The acidic N-terminal ED-domain of dBigH1 is required for down-regulation. (A) Lorenz curves showing coverage inequality for IP (red) and input samples (black) of dBigH1ΔED ChIP-seq analyses in induced cells. (B) Correlation of gene level dBigH1ΔED content (log2RPKM IP/Input) in dBigH1ΔED-expressing cells and dH1 content (log2RPKM IP/Input) in control cells. (C) ChIP-qPCR analyses with αdBigH1 antibodies and preimmune serum (mock) at the indicated genomic regions in control and dBigH1ΔED-expressing cells (N = 2; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values are indicated). (D) The normalized log2 coverage ratio IP/input of dBigH1ΔED in dBigH1ΔED-expressing cells is presented along an idealized gene-length±1kb for genes longer than 1kb categorized according to their expression quantile in control cells (Q1-lowest to Q5-highest). TSS, transcription start site; TTS, transcription termination site. (E) The fold change expression with respect to control mock-induced cells of six randomly selected down-regulated genes (left) and four randomly selected up-regulated genes (right) was determined by RT-qPCR in dBigH1ΔED-expressing cells (white) and compared to dBigH1-expressing cells (black) (N≥4; error bars are SD; two-tailed t-test, P-values are indicated). (F) Inhibition curves showing the relative GFP mRNA/DNA ratio of a chromatin template carrying a GFP-reporter assembled in vitro in dBigH1-depleted DREX upon the addition of increasing amounts of bacterially-expressed dBigH1 (black) and dBigH1ΔED (red). The correlation coefficients (R2) and slopes (α) are indicated (error bars are SD; N = 3).
dBigH1 interferes with RNApol II binding
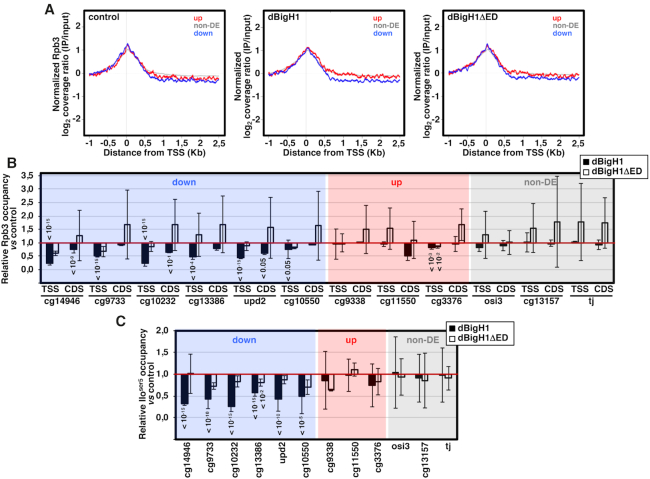
Next, we performed ChIP-seq experiments with αRpb3 antibodies to determine the effects of dBigH1 binding on the genomic distribution of RNApol II. We observed that, in comparison to non-DE and up-regulated genes, RNApol II coverage along down-regulated genes was reduced upon dBigH1 expression (Figure 5A). Actually, in comparison to control mock-induced cells, total RNApol II content of down-regulated genes tended to decrease upon dBigH1 expression (Supplementary Figure S8A). These defects depended on the acidic ED-domain since they were not observed when the truncated dBigH1ΔED form was expressed (Figure 5A and Supplementary Figure S8A). ChIP-qPCR experiments confirmed these results since, upon dBigH1 expression, several randomly selected down-regulated genes showed reduced Rpb3 occupancy at TSS and/or CDS, whereas expression of dBigH1ΔED showed no such general reduction (Figure 5B). Similar results were obtained when occupancy at TSS of the promoter-proximal active RNApol IIoser5 form was analyzed (Figure 5C). In the down-regulated genes, RNApol II occupancy was generally more reduced at TSS than at CDS (Figure 5B), suggesting an effect on RNApol II pausing. In this regard, ChIP-seq analysis showed that the pausing index (PI) of down-regulated genes tended to decrease upon dBigH1 expression (Supplementary Figure S8B). On the other hand, dBigH1 expression did not affect RNApol II occupancy in non-DE and up-regulated genes (Figure 5B and C, and Supplementary Figure S8A).
Figure 5.
The acidic N-terminal ED-domain of dBigH1 perturbs RNApol II binding. (A) The normalized log2 coverage ratio IP/input of Rpb3 in control mock-induced cells (left), dBigH1-expressing cells (center) and dBigH1ΔED-expressing cells (right) are presented as a function of the distance to TSS for up-regulated genes (red), down-regulated genes (blue) and non-DE genes (gray). (B) The relative Rpb3 occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at the indicated genomic regions in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 2; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (C) As in B but for the promoter-proximal RNApol IIoser5 form at TSS of the indicated genes (N = 3 (dBigH1), 2 (dBigH1ΔED); error bars are SD; Mixed linear model Benjamini–Hochberg adjusted P-values with respect to mock-induced cells are indicated).
dBigH1 replaces somatic dH1 and perturbs the pattern of histone modifications
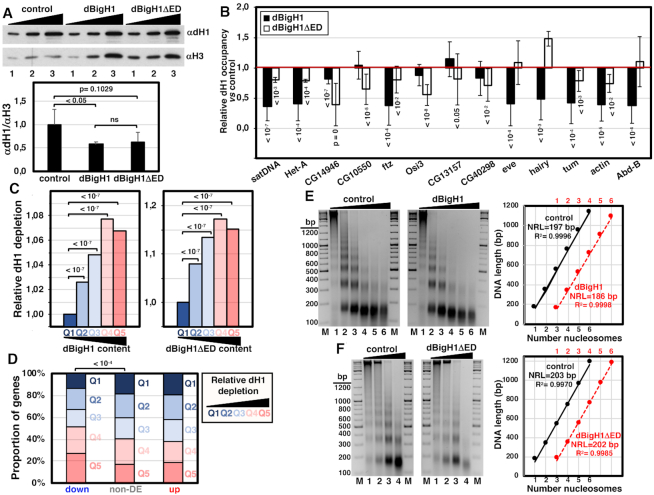
WB analysis showed that the levels of chromatin-bound dH1 decreased upon expression of full-length dBigH1 or the truncated dBigH1ΔED form (Figure 6A). This reduction occurred broadly across chromatin since ChIP-seq analyses performed with αdH1 showed similar Lorenz’ inequality curves for the IP samples obtained from control cells and cells expressing dBigH1 or dBigH1ΔED (Supplementary Figure S9A). Consistently, ChIP-qPCR experiments showed that dH1 occupancy tended to decrease at multiple genomic locations, including repetitive DNA elements (Figure 6B). Analysis of ChIP-seq data showed that the extent of dH1 depletion increased as a function of increasing dBigH1/dBigH1ΔED content. For these analyses, we grouped genes into five quantiles according to their dBigH1/dBigH1ΔED content and determined for each quantile the average dH1 content fold-change upon dBigH1/dBigH1ΔED expression. We observed that genes having high dBigH1/dBigH1ΔED content showed higher dH1 depletion relative to genes with low dBigH1/dBigH1ΔED content (Figure 6C). Furthermore, we observed that, in comparison to non-DE and up-regulated genes, down-regulated ones were enriched in genes showing high dH1 depletion upon dBigH1 expression (Figure 6D). Altogether these results suggest that both dBigH1 and dBigH1ΔED replace somatic dH1 to a similar extent. In this regard, in comparison to control mock-induced cells, we observed that dBigH1 expression reduced the overall nucleosome repeat length (NRL) by ∼10 bp (Figure 6E; see also Supplementary Figure S10A), while it was not altered upon dBigH1ΔED expression (Figure 6F; see also Supplementary Figure S10B). In addition, at the gene level, the relative dH1 depletion at TSS of highly expressed genes was reduced in dBigH1-expressing cells, in comparison to control and dBigH1ΔED-expressing cells (Supplementary Figure S9B).
Figure 6.
dBigH1 replaces dH1. (A) WB analysis with αdH1 antibodies of crosslinked chromatin prepared from control mock-induced cells and cells expressing full-length dBigH1 and the truncated dBigH1ΔED forms. Increasing amounts of chromatin are analyzed (lanes 1–3). αH3 antibodies were used for normalization. Quantitative analysis of the results is shown in the bottom (N = 3; errors bars are SD; two tailed t-test, P-values are indicated). (B) The relative dH1 occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at the indicated genomic regions in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 3; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (C) The average extent of dH1 depletion is presented for all genes categorized according to their dBigH1 (left) and dBigH1ΔED (right) content quantile in dBigH1-expressing and dBigH1ΔED-expressing cells (Q1-lowest to Q5-highest), normalized respect to Q1 (Wilcoxon rank-sum test, Benjamini–Hochberg adjusted P-values are indicated). (D) The proportion of genes categorized according to their relative dH1 depletion (Q1-lowest to Q5-highest) is presented for down-regulated, up-regulated and non-DE genes (Chi-square test, P-values are indicated). (E) MNase digestion for increasing time (lanes 1–6) of nuclei obtained from control mock-induced cells (left) and dBigH1-expressing cells (right). Lanes M correspond to MW markers (the sizes in bp are indicated). Quantitative analysis of the results is shown in the right where the size in bp of fragments containing increasing number of nucleosomes, from mono- to hexanucleosomes, are plotted against the number of nucleosomes for samples showing equivalent extent of digestion (lanes 2 in the left). The correlation coefficients (R2) and slopes, which correspond to the apparent NRL, are indicated. (F) As in E but for control mock-induced cells (left) and dBigH1ΔED-expressing cells (right).
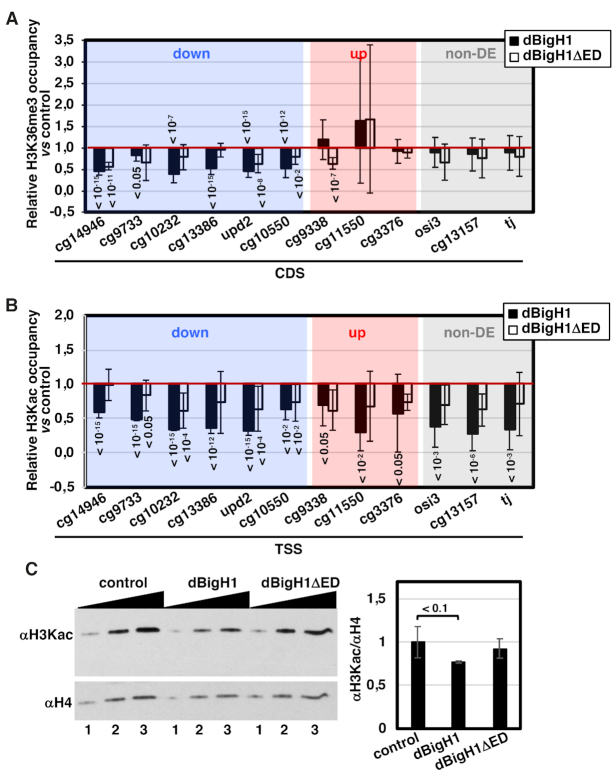
Transcriptional activity correlates with specific patterns of histone modifications (reviewed in (40)). In particular, transcriptionally active genes are enriched in H3K36me3 along the CDS, a modification associated with transcription elongation. In this regard, we observed that dBigH1 expression reduced H3K36me3 levels at CDS of down-regulated genes (Figure 7A). This effect correlated well with the effect on transcription since it was not observed in non-DE and up-regulated genes, and it was much weaker when dBigH1ΔED was expressed (Figure 7A). Histone K-acetylation is also a hallmark of active genes that generally occurs at promoters and correlates with transcription initiation. Similar to the effect on H3K36me3, we observed that expression of dBigH1 reduced H3 K-acetylation (H3Kac) levels at promoters of down-regulated genes (Figure 7B). However, in contrast to the effect on H3K36me3, this effect was also observed at promoters of non-DE and up-regulated genes (Figure 7B). Furthermore, WB analyses showed that total H3Kac levels decreased upon dBigH1 expression (Figure 7C). Altogether these results suggest that dBigH1 expression impaired H3Kac globally across chromatin. This effect mostly depended on the acidic ED-domain since dBigH1ΔED expression did not significantly decrease total H3Kac levels (Figure 7C) and, in comparison to full-length dBigH1, it had a weaker effect on H3Kac levels at promoters (Figure 7B).
Figure 7.
dBigH1 perturbs the pattern of histone modifications. (A) The relative H3K36me3 occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at CDS of the indicated genes in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 3; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (B) The relative H3Kac occupancy with respect to control mock-induced cells was determined by ChIP-qPCR at TSS of the indicated genes in dBigH1-expressing cells (black) and dBigH1ΔED-expressing cells (white) (N = 2; error bars are SD; Mixed linear model Benjamini-Hochberg adjusted P-values with respect to mock-induced cells are indicated). (C) In the left, WB analysis with αH3Kac antibodies of crosslinked chromatin prepared from control mock-induced cells and cells expressing full-length dBigH1 and the truncated dBigH1ΔED forms. Increasing amounts of chromatin are analyzed (lanes 1–3). αH4 antibodies were used for normalization. Quantitative analysis of the results is shown in the right (N = 3; error bars are SD; two tailed t-test, P-values are indicated).
DISCUSSION
Here, we have addressed the mechanism of dBigH1 action in transcription regulation using ectopic dBigH1 expression experiments in S2 cells. Though weakly, the genomic distribution of ectopically expressed dBigH1 positively correlates with those observed in embryos and testes, where dBigH1 is naturally expressed, suggesting that the mechanisms governing dBigH1 deposition might be partially conserved in S2 cells. Our results suggest that binding of dBigH1 negatively affects transcription. Upon dBigH1 expression, more than two-thirds of the DE-genes were down-regulated. This effect was probably underestimated since, though only in one replicate, we observed a global decrease in gene expression that, considering the methodology used for normalization, could hamper identification of differentially down-regulated genes. This down-regulation occurred at the transcriptional level, as down-regulated genes showed reduced RNApol II content. Conversely, RNApol II content of up-regulated genes was not increased upon dBigH1 expression, suggesting that the observed up-regulation was not transcriptional. Moreover, in vitro experiments showed that dBigH1 inhibited transcription of a chromatin template. Consistent with the negative effect on transcription, dBigH1 expression specifically decreased H3K36me3 levels at CDS of down-regulated genes.
Our results show that dBigH1 replaces dH1. In our experiments, dBigH1 binding to chromatin was likely taking place in the absence of DNA replication, as dBigH1 induction was sustained for 24h and, during this time, cell density did not increase noticeably. Whether dBigH1 deposition involves active dH1 replacement remains to be determined. In vitro, incubation of purified nuclei with DREX results in binding of dBigH1 to chromatin without dH1 displacement (32), suggesting that the replacement observed in S2 cells responds to an active process. Along the same lines, we observed that dBigH1 was preferentially deposited at regions enriched in dH1. Replacement of somatic H1s by embryonic H1s has been reported in nuclear transfer experiments (41–45) and NAP-1 has been shown to be involved in both B4/H1M deposition and somatic H1s removal in Xenopus (46,47). Further work is required to determine the mechanisms regulating dBigH1 deposition.
The acidic ED-domain of dBigH1 is required to inhibit transcription since expression of the truncated dBigH1ΔED form, which also replaced somatic dH1, did not down-regulate gene expression either affected RNApol II loading or H3Kac levels. The presence of the negatively charged acidic ED-domain in dBigH1 is peculiar as histones are highly positively charged. It is possible that, due to the negative charge of the ED-domain, the structural organization of chromatin is compromised in the presence of dBigH1. Actually, the overall NRL changed upon dBigH1 expression, but not when dBigH1ΔED was expressed. Interestingly, although the ED-domain of dBigH1 is not conserved outside of the Drosophila genus (6), embryonic H1s are generally more acidic than somatic ones (reviewed in (2)). In this regard, it was shown that both the Xenopus B4/H1M and the mammalian H1oo embryonic linker histones alter chromatin organization and dynamics (41,44,48–53). An altered chromatin organization would perturb access to chromatin and/or functioning of chromatin remodelers/modifiers and transcription factors that, ultimately, would affect RNApol II loading and transcription. In fact, regardless of the actual transcriptional outcome, dBigH1 expression globally affected H3Kac. In contrast, we reported earlier that incubation of purified nuclei with DREX, which also results in dBigH1 binding, increased H3Kac levels (32). However, it is important to note that the increase in H3Kac levels observed in this case was independent of dBigH1 binding (32).
It might be argued that the down-regulation observed upon dBigH1 expression is a consequence of increased global linker histones content. However, similar or even higher levels of expression of the truncated dBigH1ΔED and dBigH1ΔNTD forms did not affect transcription. Moreover, binding of dBigH1 is compensated by removal of dH1, thus total linker histones content is not greatly increased.
dBigH1 binding affected expression of a relatively small subset of genes. This may reflect the fact that in our experimental setup, dBigH1 accounted for only 20–25% of total linker histones. Thus, from this point of view, affected genes appear to correspond to a subset of genes more sensitive to dBigH1 levels. In this regard, we observed that down-regulated genes had strong RNApol II pausing, which tended to decrease upon dBigH1 expression. In addition, though dBigH1 binding reduced H3Kac globally, only the down-regulated genes were transcriptionally affected. Interestingly, impairing RNApol II pausing generally down-regulates gene expression (27,54,55), while reduced H3Kac levels preferentially affects expression of highly paused genes (56). These observations suggest that the higher sensitivity to dBigH1 expression of down-regulated genes is likely due to the way their transcription is regulated.
In summary, we have presented here direct evidence supporting that the acidic N-terminal tail of the embryonic dBigH1 linker histone of Drosophila compromises transcription by altering the functional epigenetic state of active chromatin. Other embryonic H1s might share similar properties since they are generally more acidic than their somatic counterparts.
DATA AVAILABILITY
GEO IDs: GSE127227 and GSE10329 In house scripts are provided upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Drs J. Kadonaga and J. Zeitlinger for antibodies. P.C.-C. and P.B. acknowledge receipt of a ‘Severo Ochoa’ FPI fellowship from MINECO, and MT of a ‘La Caixa’ fellowship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BFU2015-65082-P, PGC2018-094538-B-100]; Generalitat de Catalunya [SGR2014-204, SGR2017-475]; European Community FEDER program (to F.A.); MEC ‘Centro de Excelencia Severo Ochoa 2013–2017’ [SEV-2012-0208]; MINECO [SAF2016-75006-P to M.B.]; ‘Centre de Referència en Biotecnologia’ of the Generalitat de Catalunya. Funding for open access charge: MINECO.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fyodorov D.V., Zhou B.R., Skoultchi A.I., Bai Y.. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018; 19:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pérez-Montero S., Carbonell A., Azorín F.. Germline-specific H1 variants: the “sexy” linker histones. Chromosoma. 2016; 125:1–13. [DOI] [PubMed] [Google Scholar]
- 3. Mariño-Ramírez L., Hsu B., Baxenavis A.D., Landsman D.. The histone database: a comprehensive resource for histones and histone fold-containing proteins. Proteins. 2006; 62:838–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagel S., Grossbach U.. Histone H1 genes and histone clusters in the genus Drosophila. J. Mol. Evol. 2000; 51:286–298. [DOI] [PubMed] [Google Scholar]
- 5. Bayona-Feliu A., Casas-Lamesa A., Carbonell A., Climent-Cantó P., Tatarski M., Pérez-Montero S., Azorín F., Bernués J.. Histone H1: lessons from Drosophila. Biochim. Biophys. Acta. 2016; 1859:526–532. [DOI] [PubMed] [Google Scholar]
- 6. Pérez-Montero S., Carbonell A., Morán T., Vaquero A., Azorín F.. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell. 2013; 26:578–590. [DOI] [PubMed] [Google Scholar]
- 7. Carbonell A., Pérez-Montero S., Climent-Cantó P., Reina O., Azorín F.. The germline linker histone dBigH1 and the translational regulator Bam form a repressor loop essential for male germ stem cell differentiation. Cell Rep. 2017; 21:3178–3189. [DOI] [PubMed] [Google Scholar]
- 8. Braunschweig U., Hogan G.J., Pagie L., van Steensel B.. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009; 28:3635–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Izzo A., Kamieniarz-Gdula K., Ramírez F., Noureen N., Kind J., Manke T., van Steensel B., Schneider R.. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 2013; 3:2142–2154. [DOI] [PubMed] [Google Scholar]
- 10. Li J.Y., Patterson M., Mikkola H.K., Lowry W.E., Kurdistani S.K.. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012; 8:e1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao K., Lailler N., Zhang Y., Kumar A., Uppal K., Liu Z., Lee E.K., Wu H., Medrzycki M., Pan C. et al.. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet. 2013; 9:e1003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Millán-Ariño L., Islam A.B., Izquierdo-Bouldstridge A., Mayor R., Terme J.M., Luque N., Sancho M., López-Bigas N., Jordan A.. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014; 42:4474–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nalabothula N., McVicker G., Maiorano J., Martin R., Pritchard J.K., Fondufe-Mittendorf Y.N.. The chromatin architectural proteins HMGD1 and H1 bind reciprocally and have opposite effects on chromatin structure and gene regulation. BMC Genomics. 2014; 15:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayor R., Izquierdo-Bouldstridge A., Millán-Ariño L., Bustillos A., Sampaio C., Luque N., Jordan A.. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J. Biol. Chem. 2015; 290:7474–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koop R., Croce Di, Beato M.. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 2003; 22:588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayona-Feliu A., Casas-Lamesa A., Reina O., Bernués J., Azorín F.. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 2017; 8:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shao W., Zeitlinger J.. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017; 49:1045–1051. [DOI] [PubMed] [Google Scholar]
- 18. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P.. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015; 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. et al.. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004; 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Planet E., Attolini C.S.-O., Reina O., Flores O., Rossell D.. htSeqTools: high-throughput sequencing quality control, processing and visualization in R. Bioinformatics. 2012; 28:589–590. [DOI] [PubMed] [Google Scholar]
- 25. Font-Burgada J., Reina O., Rossell D., Azorín F.. chroGPS, a global chromatin positioning system for the functional analysis and visualization of the epigenome. Nucleic Acids Res. 2014; 42:2126–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reina O., Azorin F., Stephan-Otto Attolini C.. chroGPS2: differential analysis of epigenome maps in R. 2019; bioRxiv doi:14 May 2019, preprint: not peer reviewed 10.1101/425892. [DOI]
- 27. Kessler R., Tisserand J., Font-Burgada J., Reina O., Coch L., Attolini C.S., Garcia-Bassets I., Azorín F.. dDsk2 regulates H2Bub1 and RNA polymerase II pausing at dHP1c complex target genes. Nat. Commun. 2015; 6:7049. [DOI] [PubMed] [Google Scholar]
- 28. Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K.. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007; 39:1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carvalho B.S., Irizarry R.A.. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonte E., Becker P.B.. Preparation of chromatin assembly extracts from preblastoderm Drosophila embryos. Methods Mol. Biol. 1999; 119:187–194. [DOI] [PubMed] [Google Scholar]
- 32. Becker P.B., Tsukiyama T., Wu C.. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 1994; 44:207–223. [DOI] [PubMed] [Google Scholar]
- 33. Becker P.B., Wu C.. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell Biol. 1992; 12:2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Šatović E., Font-Mateu J., Carbonell A., Beato M., Azorín F.. Chromatin remodeling in Drosophila preblastodermic embryo extract. Sci. Rep. 2018; 8:10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chapman G.E., Hartman P.G., Bradbury E.M.. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The isolation of the globular and non-globular regions of the histone H1 molecule. Eur. J. Biochem. 1976; 61:69–75. [DOI] [PubMed] [Google Scholar]
- 36. Allan J., Hartman P.G., Crane-Robinson C., Aviles F.X.. The structure of histone H1 and its location in chromatin. Nature. 1980; 288:675–679. [DOI] [PubMed] [Google Scholar]
- 37. Ramakrishnan V., Finch J.T., Graziano V., Lee P.L., Sweet R.M.. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993; 362:219–223. [DOI] [PubMed] [Google Scholar]
- 38. Th’ng J.P., Sung R., Ye M., Hendzel M.J.. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 2005; 280:27809–27814. [DOI] [PubMed] [Google Scholar]
- 39. Hendzel M.J., Lever M.A., Crawford E., Th’ng J.P.. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004; 279:20028–20034. [DOI] [PubMed] [Google Scholar]
- 40. Schones D.E., Zhao K.. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008; 9:179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jullien J., Astrand C., Halley-Stott R.P., Garrett N., Gurdon J.B.. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl Acad. Sci. U.S.A. 2010; 107:5483–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao S., Chung Y.G., Parseghian M.H., King G.J., Adashi E.Y., Latham K.E.. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 2004; 266:62–75. [DOI] [PubMed] [Google Scholar]
- 43. Jullien J., Miyamoto K., Pasque V., Allen G.E., Bradshaw C.R., Garrett N.J., Halley-Stott R.P., Kimura H., Ohsumi K., Gurdon J.B.. Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Mol. Cell. 2014; 55:524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teranishi T., Tanaka M., Kimoto S., Ono Y., Miyakoshi K., Kono T., Yoshimura Y.. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev. Biol. 2004; 266:76–86. [DOI] [PubMed] [Google Scholar]
- 45. Yun Y., Zhao G.M., Wu S.J., Li W., Lei A.M.. Replacement of H1 linker histone during bovine somatic cell nuclear transfer. Theriogenology. 2012; 78:1371–1380. [DOI] [PubMed] [Google Scholar]
- 46. Kepert J.F., Mazurkiewicz J., Heuvelman G.L., Tóth K.F., Rippe K.. NAP1 modulates binding of linker histone H1 to chromatin and induces an extended chromatin fiber conformation. J. Biol. Chem. 2005; 280:34063–34072. [DOI] [PubMed] [Google Scholar]
- 47. Shintomi K., Iwabuchi M., Saeki H., Ura K., Kishimoto T., Ohsumi K.. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:8210–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nightingale K., Dimitrov S., Reeves R., Wolffe A.P.. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996; 15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 49. Saeki H., Ohsumi K., Aihara H., Ito T., Hirose S., Ura K., Kaneda Y.. Linker histone variants control chromatin dynamics during early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:5697–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ura K., Nightingale K., Wolffe A.P.. Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J. 1996; 15:4959–4969. [PMC free article] [PubMed] [Google Scholar]
- 51. Becker M., Becker A., Miyara F., Han Z., Kihara M., Brown D.T., Hager G.L., Latham K., Adashi E.Y., Misteli T.. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol. Biol. Cell. 2005; 16:3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Godde J.S., Ura K.. Dynamic alterations of linker histone variants during development. Int. J. Dev. Biol. 2009; 53:215–224. [DOI] [PubMed] [Google Scholar]
- 53. Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006; 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gilchrist D.A., Nechaev S., Lee C., Ghosh S.K., Collins J.B., Li L., Gilmour D.S., Adelman K.. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008; 22:1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang X., Hang S., Prazak L., Gergen J.P.. NELF potentiates gene transcription in the Drosophila embryo. PLoS One. 2010; 5:e11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Boija A., Mahat D.B., Zare A., Holmqvist P.H., Philip P., Meyers D.J., Cole P.A., Lis J.T., Stenberg P., Mannervik M.. CBP regulates recruitment and release of promoter-proximal RNA Polymerase II. Mol. Cell. 2017; 68:491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEO IDs: GSE127227 and GSE10329 In house scripts are provided upon request.