Abstract
Microbial biosynthetic gene clusters are a valuable source of bioactive molecules. However, because they typically represent a small fraction of genomic material in most metagenomic samples, it remains challenging to deeply sequence them. We present an approach to isolate and sequence gene clusters in metagenomic samples using microfluidic automated plasmid library enrichment. Our approach provides deep coverage of the target gene cluster, facilitating reassembly. We demonstrate the approach by isolating and sequencing type I polyketide synthase gene clusters from an Antarctic soil metagenome. Our method promotes the discovery of functional-related genes and biosynthetic pathways.
INTRODUCTION
Microbes represent a rich source of bioactive molecules with valuable properties, many of which are yet to be discovered (1). The biosynthesis of a given molecule is accomplished using specific enzymes, the genes for which are often grouped as a biosynthetic gene cluster in the host genome (2). By exploiting this genomic architecture, shotgun sequencing of metagenomic samples has discovered a variety of novel gene clusters and their associated molecules (3). However, since most metagenomes contain diverse mixtures of DNA from many organisms, the gene clusters most likely to be sequenced are those that are most abundant, and often least interesting (4). While deeper sequencing can increase coverage of rare gene clusters, the process remains inefficient, expensive, and computationally challenging (4).
Nucleic acid enrichment allows target DNA molecules to be isolated from a sample, focusing the sequencing on them, and thereby reducing costs and improving data quality. While effective in many scenarios, current PCR and hybridization approaches commonly recover 5–10 kb target molecules per probe, and often require thousands of probes to recover a full region of interest; in addition to being laborious and expensive, this approach can fail when dealing with repetitive or homologous sequences (5–8). Recently, CRISPR–Cas9 methods have enabled long-region target enrichment. However, these methods require multiple carefully chosen restriction enzymes to prevent cutting of the target regions, and sufficient sequence knowledge to design guide RNAs, limiting their value for gene clusters spanning tens of kilobases of unknown sequences (9–12). Alternatively, bacterial artificial chromosome (BAC) or fosmid screening can return full-length, high-quality gene clusters (13). These circular plasmids can carry hundred-kilobase length fragments of DNA from a metagenome as unique inserts stored in individual bacteria (14–16). To isolate the target plasmid, the library of billions of cells is split into hundreds of aliquots, and each is tested by PCR for the insert. If an aliquot contains the insert, it is selected for further dilution and subculture, and the process is repeated until a pure colony with the target is obtained (13,17). Although this approach yields pure target DNA yielding high quality sequence data, it requires multiple rounds and hundreds of PCR assays, and thus is laborious and expensive. To enhance our ability to sequence novel gene clusters in metagenomic samples, a new method that simplifies, speeds, and lowers the cost of plasmid library screening is needed.
In this paper, we present Microfluidic Automated Plasmid Library Enrichment (MAPLE), an approach that automates, accelerates, and miniaturizes plasmid library screening. MAPLE performs the typical steps of plasmid library screening, but on single bacterial colonies in reactors a millionth the normal volume, and at screening throughputs of thousands per second. MAPLE screens each colony for a target insert and returns its DNA, which is then sequenced. As with conventional plasmid screening, MAPLE can isolate any insert of interest with appropriate design of PCR primers, thereby enhancing targeted metagenomic sequencing and isolation of rare targets.
MATERIALS AND METHODS
Metagenomic library and cell culture
Two metagenomic libraries are used in this work. The first one is obtained from Dr. Wenjun Zhang's lab. Streptomyces sparsogenes (ATCC25498) genome (NZ_MAXF00000000.1) (18) fragments are inserted into the pCC2FOS™ vector (Epicentre) and transformed into Escherichia coli EPI300™ cells. A total of 1.5 × 104 clones with ∼40 kb inserts have been generated. Library cells are propagated in 2xYT media (Research Products International) supplemented with 25 μg/ml chloramphenicol at 37°C overnight, and then 1 ml of the culture further supplemented with 1× Fosmid Autoinduction Solution (Epicentre) is used for cell encapsulation in droplets. The second library is obtained from the Canadian MetaMicroBiome Library (19). The Arctic Tundra 2 (2ATN) metagenomic DNA sample is comprised of ∼6 × 104 unique clones, with each ∼31 kb random insert carried by the pJC8 within E. coli HB101 cells. Library cells are propagated in 2xYT media supplemented with 15 μg/ml Tetracycline at 37°C overnight, and 1 ml of the culture is used for cell encapsulation in droplets.
Fabrication of microfluidic devices
The microfluidic devices are fabricated from photoresist masters. The masters are made by spinning the SU-8 photoresist (Microchem) onto a 3-inch silicon wafer (University Wafer). The choice of SU-8 and spin speed is determined by the channel height of the microfluidic device. In this work, we spin SU-8 3025 at 2500 rpm on a SCS G3P-8 Spin Coater (Specialty Coating Systems) for the drop maker device; SU-8 3025 at 2500 rpm for the first layer of the merger device and SU-8 3010 at 1000 rpm for the second layer; SU-8 3025 at 2000 rpm for the sorter device. After baking at 95°C for 45 min, the photoresist on the wafer is exposed to ultraviolet light for 1 min over photolithography masks (CAD/Art Services) with patterns of microfluidic channels. The wafer is then baked at 95°C for 5 min, cooled to room temperature and then developed for 15 min in propylene glycol monomethyl ether acetate (PGMEA, Sigma Aldrich). When fabricating the two-layer merger device, before the development, we expose the wafer to ultraviolet light again for 1 min over the second photolithography mask and then bake it at 95°C for 5 min. After development, the wafer is then rinsed with fresh PGMEA and isopropanol, before being dried by blowing with air. After baking at 65°C for 1 h to remove solvent, the wafer with channel patterns is stored as the photoresist master.
The microfluidic devices are made by pouring a polydimethylsiloxane solution (PDMS, Dow Corning, Sylgard 184) with an 11:1 polymer-to-crosslinker ratio over the master and then cured at 65°C overnight. The devices are extracted with a metal scalpel, and punched with a 0.75 mm biopsy punch (World Precision Instruments, catalog no. 504529) to create holes at each fluid inlet or outlet. Devices are bonded to a glass slide after plasma treatment and the channels are made hydrophobic by treatment with Aquapel (PPG Industries) (20).
Cell encapsulation and culture in droplets
Escherichia coli cells are suspended in media and cell concentration is calculated by manual cell counting under a microscope to determine cell number per droplet. The cell solution is diluted as needed to ensure the appropriate Poisson loading before being transferred to a 1 ml syringe. Another 3 ml syringe is loaded with HFE 7500 fluorinated oil (3 M) with 2% (w/w) PEG-PFPE amphiphilic block copolymer surfactant (Ran Biotechnologies). Both syringes are placed on syringe pumps (New Era) and connected via PTFE microtubing (Fisher Scientific) to the microfluidic flow-focusing (21) drop maker device with nozzle dimensions of 30 μm. The pumps are controlled by a custom Python script (available at GitHub: https://github.com/AbateLab/Pump-Control-Program) to pump HFE 7500 at 800 μl/h and cell solution at 400 μl/h. 30 μm droplets are generated and collected in a 1 ml syringe and incubated at 37°C overnight to allow cells to grow into colonies in droplets.
Droplet merging and colony PCR in droplets
TaqMan primers for the S. Sparsogenes library are as follows: Primer1: 5′-CGA GGT CCT TCT CGT TCA C-3′, Primer2: 5′-ATC GAC AAG TAC CGC ATC AC-3′, Probe: 5′-6-FAM/AGC AGC AGC/ZEN/ATG TCC TCC CA/IABkFQ/-3′. Primers for the Antarctic soil library are as follows (22): Primer1: 5′-GGR TCN CCI ARY TGI GTI CCI GTI CCR TGI GC-3′, Primer2: 5′-MGI GAR GCI YTI CAR ATG GAY CCI CAR CAR MG-3′. All primers and probes are purchased from Integrated DNA Technologies (IDT). PCR reagents contain 1× Platinum™ Multiplex PCR Master Mix (Thermo Fisher), 1 μM of each primer, 0.25 μM of TaqMan probe (if used), 2.5% PEG6K, and 1% Tween 20. 500 μL of PCR reagents are loaded into a 1 mL syringe and then connected to the droplet merger device via PTFE microtubing. HFE 7500 with 2% surfactant is used as spacer oil and droplet generation oil. The electrode and moat channels are filled with 2 M NaCl solution. The moat channel prevents stray fields from causing unintended droplet coalescence at other locations on the device (23). The droplets containing cell colonies are reinjected into the droplet merger device. The flow rates are as follows: colony droplets 50 μl/h, spacer oil 400 μl/h, PCR reagents 300 μL/h, droplet generation oil 600 μl/h. The dimensions and flow rates of this device are configured to produce 45 μm PCR droplets. To merge with the colony droplet, the droplet pairs flow into the merging zone and electrode is charged with an alternating current (AC) voltage (3 V, 58 kHz). After collecting the merged droplets in PCR tubes, the oil underneath the emulsion is replaced with FC-40 fluorinated oil (Sigma Aldrich) containing 5% surfactant to enhance droplet stability during thermocycling. The emulsion is transferred to a thermocycler (Bio-Rad) for droplet PCR with the following program: 92°C for 3 min, followed by 40 cycles of 92°C for 30 s, 55°C for 30 s, 72°C for 30 s. 1× SYBR GREEN I in HFE 7500 oil is used to stain positive droplets when there is no TaqMan probe in the PCR reagents.
Droplet sorting and DNA recovery
After thermocycling, the oil underneath the emulsion is replaced with HFE 7500 fluorinated oil with 2% surfactant, transferred to a 1 ml syringe and reinjected into the microfluidic droplet sorter at a flow rate of 50 μl/h. The flow rates of spacer and bias oil are both 1000 μl/h. All the oil used is HFE 7500 with 0.1% surfactant and loaded in a 10 ml syringe. A 10 mL syringe connected to the outlet of waste channel is constantly withdrawing with a flow rate of –1000 μl/h. The electrode and moat channels are filled with 2 M NaCl solution. The droplet fluorescence is excited with a 473 nm laser (CNI lasers) and a custom LabVIEW code (available at GitHub: https://github.com/AbateLab/sorter-code) detects the fluorescence signal in real time. The fluorescence signal of positive droplets falls in the user-defined range which triggers the dielectrophoretic sorting pulse output with a 40 kHz, 1 kV signal to the electrode and pulls the droplet to the sorted channel (23,24). 200–500 droplets are collected for the downstream next-generation sequencing (NGS). For the non-sorted control sample in Figure 2, all droplets are gated in the LabVIEW code and thereafter sorted.1H,1H,2H,2H-Perfluoro-1-octanol (final concentration 20% (v/v), Sigma Aldrich) and 10 μl water is added to the droplets and then mixed to break emulsion (20). The tube is centrifuged briefly and the aqueous droplet floating on top is transferred to a new clean tube.
DNA sequencing
For shotgun sequencing, 1 ng of metagenomic library plasmid DNA extracted using FosmidMAX™ DNA Purification Kit (Lucigen) is used for library preparation with the Nextera XT library kit. For amplicon sequencing, 5 ng of metagenomic library plasmid DNA is used as template following the 16S Metagenomic Sequencing Library Preparation protocol (Illumina) using primers for the keto-synthase gene as follows: Primer1: 5′-GTC TCG TGG GCT CGG GGR TCN CCI ARY TGI GTI CCI GTI CCR TGI GC-3′, Primer2: 5′-TCG TCG GCA GCG TCM GIG ARG CIY TIC ARA TGG AYC CIC ARC ARM G-3′. For DNA recovered from sorted droplets, the NGS library is made using half the amount of reagents described in the Nextera XT library kit (Illumina). The quality of sequencing libraries is assayed by Bioanalyzer (Agilent) using a High Sensitivity DNA Assay. Sequencing is performed with a MiSeq sequencer (Illumina). A 150-cycle MiSeq Reagent Kit v3 is used for the S. sparsogenes sample yielding 48 375 540 total sequencing reads. A 600-cycle kit is used for the Antarctic soil sample yielding 34 496 030 total sequencing reads.
Data analysis
Reads are aligned to the S. sparsogenes reference genome using Bowtie2 (25). Genome coverage at each position is calculated using SAMtools (26). Reads are de novo assembled using SPAdes (27) with default parameters. Reference DNA and protein sequences corresponding to the key modules in a type I polyketide synthase (PKS) gene cluster—including 1000 ketosynthase (KS), 372 acyl carrier protein (ACP), 143 acyltransferase (AT), 198 enoyl reductase (ER), 140 ketoreductase (KR), 1001 dehydratase (DH), and 113 thioesterase (TE) sequences—are downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov). DNA sequences are used for reads alignment and protein sequences are used for building phylogenetic tree using MUSCLE v3.8.31 (28) and iTOL (29). Reference DNA sequences of known and complete type I PKS gene clusters are downloaded from two databases: 600 sequences are from The Minimum Information about a Biosynthetic Gene cluster (MIBiG) (2) and 1831 from NCBI. Chemical structures are generated by PubChem Sketcher v2.4 (https://pubchem.ncbi.nlm.nih.gov/edit2/index.html). Contigs are annotated by the RAST app in Kbase (30). Gene pathways are analyzed by KEGG database (31).
RESULTS
The workflow of MAPLE
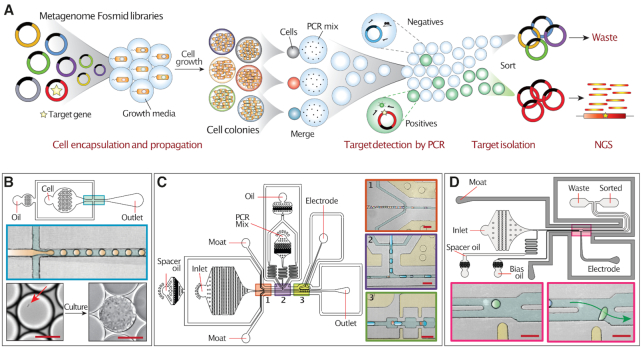
The MAPLE workflow comprises four steps, single cell colony formation, target detection by PCR, plasmid isolation by sorting, and sequencing of isolated plasmids (Figure 1A). Like conventional plasmid library screening, MAPLE exploits the storage of the metagenome as high-molecular weight plasmids (BACs or fosmids) in a suspension of living bacteria. Each bacterium contains a plasmid carrying a unique insert from the metagenome that can be tens of kilobases long; many plasmids are large enough to contain complete gene clusters. Moreover, individual cells harboring a BAC or fosmid can be cultured to provide ample DNA for sequencing.
Figure 1.
MAPLE workflow and associated microfluidic devices. (A) Overview of MAPLE workflow. (B) The droplet maker device used for encapsulating single cells. Top, device schematic; middle, enlarged view of the cross junction where droplets are generated; bottom, image of a single bacterium (red arrow) in the droplet before culture (left) and resulting colony after incubation (right). Scale bar = 20 μm. (C) Droplet merger device. Left, device schematic. Right, inserts showing magnified views of the three numbered regions. Insert 1, reinjection of close-packed droplets containing cell colonies spaced out by oil flow. Insert 2, pairing of colony droplets (orange) with PCR reagent droplets (blue) at a ∼1:1 ratio. Insert 3, entrance of droplet pairs into merging zone for electro-coalescence. Scale bar = 100 μm. (D) Droplet sorter device used for sorting fluorescently positive droplets. Top, device schematic. Bottom, inserts showing the junction where droplets are sorted. If a droplet passing the laser (light spot) has a fluorescence signal exceeding the threshold, the electrode (yellow bar) activates, applying a dielectrophoretic force to pull it into the ‘sorted’ channel. Scale bar = 100 μm.
The first step in MAPLE is to encapsulate and culture individual cells from the library in picoliter droplets, generating millions of genetically distinct pico-colonies. This is accomplished using a cross-junction droplet generator (Figure 1B) that encapsulates single cells in droplets at kilohertz rates through a plug-squeeze mechanism (32) following a Poissonian process (33). The generated emulsion is incubated so that isolated single cells can expand into pico-colonies within each picoliter droplets (34) (Figure 1B, lower). The colonies provide ample DNA for sequencing, while minimizing biases common with in vitro amplification methods, such as Multiple Displacement Amplification (MDA) (35).
Colonies carrying sequences of interest are identified using digital droplet PCR. This is accomplished using another microfluidic device to fuse each colony-containing droplet with a second droplet carrying primers and PCR reagents (Figure 1C). The monodispersed colony droplets are flowed into the device as a close-pack and evenly spaced with oil (Figure 1C1). On another part of the device, PCR droplets are generated by a cross-junction (Figure 1C2). The outlets of the colony spacer and PCR droplet generator merge the different droplets into an interdigitated stream (Figure 1C2). Because smaller droplets flow faster in microchannels, the colony droplets catch up to the larger PCR droplets, forming pairs that flow into the toothed merging region (Figure 1C3) (36,37). The surrounding salt-water electrode channels (38) (Figure 1C3, upper yellow channel) are charged with an AC voltage that triggers droplet merging (36) and the resultant fused-droplets are collected into a PCR tube for thermocycling.
During PCR, thermolysis of the bacteria releases the plasmids into solution, where they can be amplified if a colony contains the target insert. PCR-positive drops become fluorescent either by TaqMan assay or SYBR Green staining (39); droplets lacking the target, by contrast, remain dim (40). At the conclusion of thermocycling, the fluorescence of a given droplet thus relates to whether it is positive for the target insert. To complete the screen, we sort out the positive droplets, which is achieved using a droplet sorter (Figure 1D) (20,23). The sorter functions by accepting and spacing a close-packed emulsion and using electrodes to deflect droplets between two outlet channels. To determine if a droplet is positive, its fluorescence intensity is measured upstream of the sorter. Droplets with a fluorescence falling within user-defined gates (Supplementary Figure S1) trigger the sorting electrode, inducing a dielectrophoretic force (36) that pulls them into the lower ‘collection’ channel (Figure 1D, lower right). If the droplet is non-fluorescent, the electrode remains inactive, and the droplet continues its default path into the ‘waste’ channel (41). Droplet sorting is possible at rates comparable to flow cytometry (kilohertz) (23), allowing millions of colonies to be screened per hour, compared to just a few hundred screened per day in well plates (42).
The final step in MAPLE is to recover the DNA from the sorted droplets for sequencing. This is accomplished by chemically rupturing the emulsion and extracting and sequencing the released DNA (20). The obtained data is processed using a custom bioinformatics pipeline (Figure 2A), which removes reads corresponding to the host genome (E. coli) and plasmid backbone before de novo assembly to generate contigs. Because MAPLE enriches the target gene and its surrounding genomic context, the contigs with high coverage are likely physically linked to the target gene.
Figure 2.
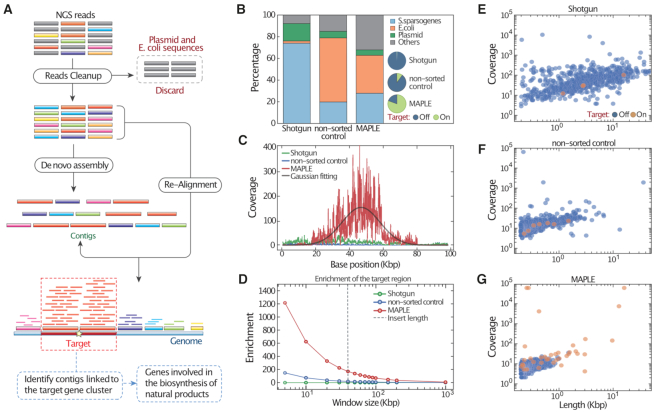
MAPLE allows enrichment of genomic regions associated with a target gene from a model fosmid library. (A) Illustration of bioinformatic analysis pipeline for processing MAPLE sequencing data. Raw reads aligning to the fosmid backbone and E. coli genome are removed prior to de novo assembly, and those remaining are aligned to contigs. Contigs with the highest depth are likely associated with the target gene cluster and thus selected for annotation. Genes identified from the selected contigs are potentially involved in the target biosynthesis pathway of natural products. (B) Stacked bar plot showing the proportion of raw reads mapping to the reference genome, for three aliquots of the sample processed differently: conventional metagenomics (Shotgun), unenriched droplet colonies (non-sorted control), and enriched colonies (MAPLE). The location of reads mapping to the S. sparsogenes genome are characterized in the pie chart inserts. ‘On-target’ denotes reads within an 80 kb window centered on the target gene, while ‘off-target’ refers to the rest of the genome. (C) Read coverage per million reads plotted over the target region. (D) Enrichment ratio as a function of the size of the quantitation window. The enrichment follows an approximately exponential decay, deflecting downward at the average insert size of the fosmid library, as indicated by the dashed grey line. (E–G) Scatter plots of distribution of de novo assembled contigs versus length and coverage. Contigs falling within or outside of the target region are differentiated by color.
Enriching the target genomic regions by MAPLE using a model library
To evaluate the effectiveness of MAPLE, we apply it to a model fosmid library comprising inserts from the S. sparsogenes genome (Figure 2) (18). For a target sequence, we choose the GlpA gene locus, generating appropriate primers and a TaqMan probe. As controls, we perform NGS directly on the plasmid library (‘Shotgun’) and on DNA processed through a control workflow where all droplets are collected regardless of the fluorescence signals (‘non-sorted control’). We compare the proportion of NGS reads from each sample aligning to the S. sparsogenes genome, E. coli genome, and plasmid backbone (Figure 2B). As expected, direct shotgun sequencing of purified plasmids yields reads mapping primarily to S. sparsogenes (74%) and plasmid (17%). The non-sorted control reads map primarily to E. coli (59%), the remaining to S. sparsogenes (20%) and plasmid (6%). The MAPLE sample recovers mostly S. sparsogenes (28%) and E. coli (35%) with a small fraction (5%) mapping to the plasmid. All reads failed to map to the listed references are assigned to the ‘other’ category, including sequences likely belong to the gap regions of the S. sparsogenes reference genome, extrachromosomal genetic elements of E. coli, and other sequences likely generated by errors from PCR, NGS and bioinformatic algorithms. E. coli DNA reads are expected in MAPLE data because, in addition to the target plasmids, the recovered droplets also contain the genome of host E. coli cells.
If MAPLE performs as expected, the obtained reads should map to S. sparsogenes and the target locus. To confirm this, we inspect the sequence results for loci surrounding the target. Because the average fosmid insert is ∼40 kb, enriched reads should fall within an ∼80 Kbp window centered on the target. For reads mapping to the S. sparsogenes genome, >80% from MAPLE fall within this window, compared to 0.7% for Shotgun and 10.2% for non-sorted samples (Figure 2B, pie charts). The MAPLE coverage map has a Gaussian-like distribution centered at the target gene locus in the window (Figure 2C). By contrast, the shotgun and non-sorted samples have uniform coverage with a very low sequencing depth. Integrating the distributions and taking the ratio for target to off-target, we estimate fold-enrichment within a given window size. The enrichment by MAPLE peaks at >1200× within 5 kb window that includes genomic context close to the target gene and drops with the window increasingly covering genomic context far away from the target gene until reaches background (∼1×) by 1 Mb (Figure 2D); the Shotgun sample remains ∼1× regardless of window size, indicating uniform coverage over the genome, as expected for unbiased sequencing. The non-sorted control exhibits slight enrichment due to PCR amplicons from a small portion of target plasmids (Figure 2D).
An essential step in many metagenomic studies is contiguous sequence (contig) assembly from raw read data; this, ultimately, provides information about the relationships between genes and their roles in gene clusters (43). MAPLE should improve assembly quality of the target region because it allows long target fragments from a metagenome to be enriched and sequenced deeply. To investigate this, we process the S. sparsogenes data through available assemblers (27). After De novo assembly, the Shotgun sample yields contigs with a wide range of lengths, from 200 bp to >10 kb, with only 4 of 693 contigs mapping to the 80 Kbp region (Figure 2E). MAPLE yields a higher proportion (37 of 684) of target region contigs, indicating improved assembly of the target gene's genomic context (Figure 2F and G). Importantly, read-mapping (Figure 2A) indicates that MAPLE’s longest and most deeply covered contigs relate to the target region; this is not the case for the two controls (Figure 2E–G). We have also performed MAPLE on another fosmid library constructed with a different Streptomyces genome, and observed a Gaussian-like coverage map over a 70 kb target region centered at the target gene. After assembly, contigs with the highest coverage map to the target region with 99% alignment, illustrating the consistency of MAPLE (Supplementary Figure S2),
Enriching the target PKS gene clusters from an Antarctic soil metagenomic library by MAPLE
Our experiments with the S. sparsogenes genome library demonstrate that MAPLE allows enrichment of long DNA fragments containing a target sequence, and their high-coverage sequencing. To illustrate this, we use MAPLE to characterize microbial polyketide synthesis in an Antarctic soil metagenome. Polyketides are an important and diverse class of natural products with pharmaceutical value as antibiotics and chemotherapeutics (44). A signature enzyme within polyketide encoding biosynthetic gene clusters is the type I polyketide synthase (PKS). All type I PKS biosynthetic gene clusters consist of a repeating structure of modular ketosynthase (KS), acyltransferase (AT), acyl carrier protein (ACP), dehydratase (DH), methyltransferase (MT), ketoreductase (KR), enoyl reductase (ER) and thioesterase (TE) domains. (Figure 3A).
Figure 3.
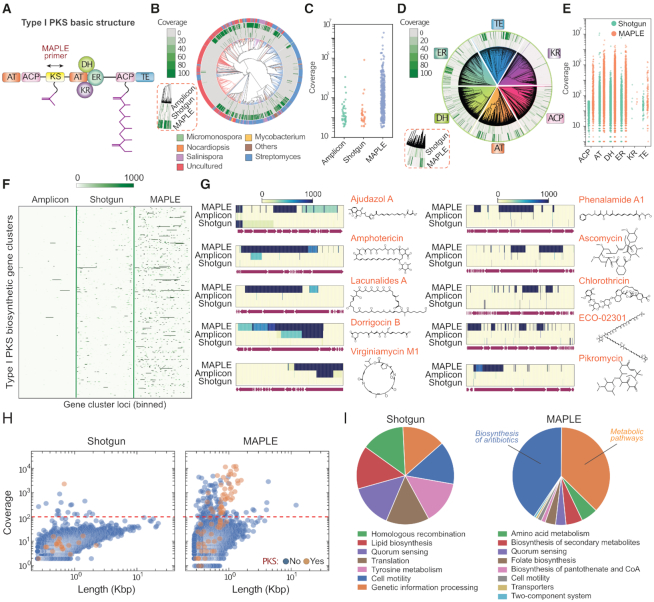
MAPLE enriches PKS gene clusters and associated pathways from an Antarctic soil metagenomic library. (A) Basic modular structure of type I PKS gene cluster. The arrow indicates MAPLE primers targeting the KS gene. AT: acyltransferase; ACP: acyl carrier protein; KS: keto-synthase; DH: dehydratase; ER: enoyl-reductase; KR: keto-reductase; TE: thioesterase. (B) Reads mapping to the phylogenetic tree of KS genes from the NCBI database. Methods are shown in the red dashed box. Branch colors and outermost ring indicate genus from which a KS domain's gene cluster originates, with labels at the bottom. (C) Identification and coverage of KS genes. Each dot represents an identified KS gene. (D) Reads mapping to phylogenetic trees of the six other genes in PKS gene clusters. Methods are shown in the red dashed box. (E) Identification and coverage of six other genes in PKS gene clusters. Each dot represents an identified gene. (F) Heatmap illustrating coverage of selected PKS gene clusters. The x-axis is normalized positions of every gene cluster, with intensity indicating sequencing depth at that position. (G) Heatmaps for ten representative gene clusters for the synthesis of antibiotics with corresponding chemical structures on the right. (H) Scatter plots illustrating the distribution of de novo assembled contigs versus length and reads coverage. Contigs with and without PKS annotations are differentiated by color. The red dashed line indicates the threshold to separate on- and off-target contigs in MAPLE. (I) Pie charts comparing the distribution of the pathways identified by the two methods.
To capture the largest number of KS variants and their associated gene clusters, we use a previously reported degenerate primer set targeting the KS gene (22). We compare sequencing results from conventional approaches (amplicon and direct shotgun sequencing) and MAPLE. KS sequences exhibit high diversity across the tree of life, having a rich and complex lineage, as illustrated centrally in Figure 3B. Direct shotgun sequencing of the bulk metagenome detects the fewest KS genes (32 out of 1000) with relatively low coverage (only 3 hits with a coverage over 1000-fold). Amplicon sequencing, in theory, should best resolve all KS domains by virtue of focusing all the sequencing power on a small region (6). Notably, however, MAPLE outperforms amplicon sequencing (43 hits, only 3 with a coverage over 1000-fold), recovering the highest number of unique KS genes (366 out of 1000) and yielding the deepest coverage (170 hits with a coverage over 1000-fold). (Figure 3B, C and Supplementary Figure S3A, S3B). For the genes detected by both MAPLE and shotgun sequencing, 96% from MAPLE have deeper coverage than shotgun sequencing, and 100% in MAPLE have deeper coverage than amplicon sequencing (Supplementary Figure S3B). This is likely due to MAPLE’s use of compartmentalized droplet amplification that, unlike bulk PCR, is insensitive to sequence and concentration dependent amplification biases that can result in variant drop out (45,46). We also find that most identified PKS genes in the arctic soil library are from either Streptomyces or currently uncultured microbes (outer-most ring, Figure 3B).
When applied to a diverse library containing inserts from many species, MAPLE affords the ability to query for a ‘keyword’ sequence to recover all physically connected sequences. This can be used to investigate how a specific gene or gene class is used across a metagenome in a variety of gene clusters. To illustrate this, we plot the distribution of recovered genes known to be associated with the PKS pathway (Figure 3A), finding that these genes are highly enriched with MAPLE compared to shotgun sequencing of unenriched DNA (Figure 3D and E). For the genes detected by both shotgun sequencing and MAPLE, most in MAPLE have higher coverage, with many obtaining >100x deeper coverage (Supplementary Figure S3C). We also map our data against previously identified PKS gene clusters (Figure 3F). Reads from amplicon sequencing map to the fewest (63 out of 441) and smallest portion of each gene cluster, since this method only sequences the region between the primers (6). Shotgun sequencing obtains reads from more gene clusters (318 out of 441), but most are poorly covered. By contrast, MAPLE shows the best coverage for the largest number of gene clusters (362 out of 441), with many reads covering long contigs (Supplementary Figure S4A). For the gene clusters detected by both MAPLE and shotgun sequencing, ∼70% have deeper coverage using MAPLE. This increases to 100% when comparing MAPLE to amplicon sequencing (Supplementary Figure S4B). Together, these results show that MAPLE outperforms these conventional methods. Analysis of ten representative gene clusters with known natural products provides a closer look at how well gene clusters are covered by MAPLE compared to the other methods (Figure 3G). For example, shotgun sequencing only detects a small part of the Ajudazol gene cluster, and almost none of the others. Amplicon sequencing only achieves good coverage in the amplicon region for Dorrigocin and Virginiamycin.
A principal advantage of MAPLE over other methods is its ability to associate a target gene with other genes in the same cluster, even if they are not in the same contig. This is valuable because such associative neighborhood information may be used to infer the function of a gene cluster, even if a contig spanning all portions of a gene cluster is unavailable (47). To illustrate this, we perform de novo assembly for both the shotgun and MAPLE sequenced data. While shotgun sequencing tends to yield longer contigs, MAPLE obtains more contigs mapping to PKS gene clusters (Figure 3H), indicating MAPLE’s ability to facilitate de novo assembly of novel variants. We map reads to the contigs and extract all having a sequencing depth of >100× within the shotgun and MAPLE sequenced samples (Figure 3H, red dotted line); this corresponds to 18 contigs for the shotgun data and 136 contigs for MAPLE. We aggregate all identified genes for selected contigs, annotate them from the database based on homologous genes, assign to them metabolism pathways (31), and plot the results as a pie chart (Figure 3I). For the shotgun data, we observe no clear pattern, with identification of seven genes for seven random cellular pathways. By contrast, when using MAPLE, a pattern of biosynthetic and metabolic pathways emerges, with identification of >60 genes for biosynthesis of antibiotics and secondary metabolites, including members from the type I PKS gene cluster ketosynthase (KS), acyltransferase (AT), dehydratase (DH), and acyl carrier protein (ACP) (44). We also discover many genes that physically link and therefore may be functionally related to PKS gene clusters, for example: genes homologous to amino acid and lipid metabolism; transporter genes that may act as multidrug efflux pumps for the antibiotic synthesized by neighboring PKS genes; genes for quorum sensing; transposases for gene cluster horizontal transfer. Many of these genetic elements have been previously reported to correlate with PKS biosynthetic gene clusters (48–51). This demonstrates that MAPLE allows a metagenome to be queried for a gene sequence to recover all information physically associated with it, analogous to extracting all sentences containing a keyword from a book. The resultant ‘targeted metagenome’ provides deep and comprehensive information about the gene, the contexts in which it is used, and other genes it is associated with. This should aid in characterizing a gene's use across a metagenome. Moreover, for genes of unknown function, the rich information on other genes it is associated with, some of which may have known functions, may provide guiding information to infer a function for the target gene.
DISCUSSION
MAPLE combines living cells carrying long fragments of metagenomic DNA (fosmid/BAC libraries) with droplet microfluidic techniques. This allows accurate, unbiased amplification of the fragments by single-cell droplet culture, and efficient screening, recovery, and sequencing of plasmids. The result is superior sequencing data compared to conventional shotgun sequencing.
Because the MAPLE workflow's output is fosmids or BACs, the method is compatible with plasmid retransformation. This would extend the technique in two major ways. First, retransformation would allow MAPLE to stack over additional screening rounds, for which the enrichments will multiply, enabling the maximum enrichments to surpass 10 000-fold, which would facilitate the discovery and sequencing of very rare gene clusters that likely exist below current detection thresholds of metagenomic sequencing (52). Second, retransformation would also facilitate direct manipulation and integration of recovered gene clusters into production host cells, for example, to perform direct functional characterization (53).
While we have focused on PCR as the determining screening step in MAPLE, the droplet assay is generalizable to other assays, including chemical (54), enzymatic (55), and aptamer (56) assays. This should allow querying of the metagenome for a function of interest, rather than a sequence of interest (57). For example, MAPLE could be used to narrow a plasmid library down to all gene clusters associated with a specific drug resistance or the ability to catalyze a desired reaction, like the environmental degradation of waste plastics (58) or contaminants (59). MAPLE thus affords a significant improvement over conventional plasmid library screening that should impact our ability to query diverse mixtures of DNA to isolate and sequence those of most interest.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health (NIH) [1R01HG008978-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Harvey A.L., Edrada-Ebel R., Quinn R.J.. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015; 14:111–129. [DOI] [PubMed] [Google Scholar]
- 2. Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. et al.. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015; 11:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Charlop-Powers Z., Milshteyn A., Brady S.F.. Metagenomic small molecule discovery methods. Curr. Opin. Microbiol. 2014; 19:70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharon I., Banfield J.F.. Microbiology. Genomes from metagenomics. Science (New York, N.Y.). 2013; 342:1057–1058. [DOI] [PubMed] [Google Scholar]
- 5. Mertes F., Elsharawy A., Sauer S., van Helvoort J.M., van der Zaag P.J., Franke A., Nilsson M., Lehrach H., Brookes A.J.. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief Funct. Genomics. 2011; 10:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J.. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010; 7:111–118. [DOI] [PubMed] [Google Scholar]
- 7. Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., Lipkin W.I.. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio. 2015; 6:e01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gasc C., Peyret P.. Revealing large metagenomic regions through long DNA fragment hybridization capture. Microbiome. 2017; 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slesarev A., Viswanathan L., Tang Y., Borgschulte T., Achtien K., Razafsky D., Onions D., Chang A., Cote C.. CRISPR/Cas9 targeted CAPTURE of mammalian genomic regions for characterization by NGS. Sci. Rep. 2019; 9:3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hafford-Tear N.J., Tsai Y.-C., Sadan A.N., Sanchez-Pintado B., Zarouchlioti C., Maher G.J., Liskova P., Tuft S.J., Hardcastle A.J., Clark T.A. et al.. CRISPR/Cas9-targeted enrichment and long-read sequencing of the Fuchs endothelial corneal dystrophy–associated TCF4 triplet repeat. Genet. Med. 2019; 21:2092–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nachmanson D., Lian S., Schmidt E.K., Hipp M.J., Baker K.T., Zhang Y., Tretiakova M., Loubet-Senear K., Kohrn B.F., Salk J.J.. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS). Genome Res. 2018; 28:1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stevens R.C., Steele J.L., Glover W.R., Sanchez-Garcia J.F., Simpson S.D., O’Rourke D., Ramsdell J.S., MacManes M.D., Thomas W.K., Shuber A.P.. A novel CRISPR/Cas9 associated technology for sequence-specific nucleic acid enrichment. PLoS One. 2019; 14:e0215441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hover B.M., Kim S.-H., Katz M., Charlop-Powers Z., Owen J.G., Ternei M.A., Maniko J., Estrela A.B., Molina H., Park S. et al.. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018; 3:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim U.-J., Shizuya H., de Jong P.J., Birren B., Simon M.I.. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992; 20:1083–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hohn B., Collins J.. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980; 11:291–298. [DOI] [PubMed] [Google Scholar]
- 16. Shizuya H., Birren B., Kim U.-J., Mancino V., Slepak T., Tachiiri Y., Simon M.. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Owen J.G., Reddy B.V.B., Ternei M.A., Charlop-Powers Z., Calle P.Y., Kim J.H., Brady S.F.. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Zhou Q., Lou T., Wang S., Ruan H.. Draft genome sequence of broad-spectrum antibiotic sparsomycin-producing Streptomyces sparsogenes ATCC 25498 from the American Type Culture Collection. J. Glob. Antimicrob. Resist. 2017; 11:159–160. [DOI] [PubMed] [Google Scholar]
- 19. Neufeld J.D., Engel K., Cheng J., Moreno-Hagelsieb G., Rose D.R., Charles T.C.. Open resource metagenomics: a model for sharing metagenomic libraries. Stand. Genomic Sci. 2011; 5:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mazutis L., Gilbert J., Ung W.L., Weitz D.A., Griffiths A.D., Heyman J.A.. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2013; 8:870–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anna S.L., Bontoux N., Stone H.A.. Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett. 2003; 82:364–366. [Google Scholar]
- 22. Parsley L.C., Linneman J., Goode A.M., Becklund K., George I., Goodman R.M., Lopanik N.B., Liles M.R.. Polyketide synthase pathways identified from a metagenomic library are derived from soilAcidobacteria. FEMS Microbiol. Ecol. 2011; 78:176–187. [DOI] [PubMed] [Google Scholar]
- 23. Sciambi A., Abate A.R.. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip. 2015; 15:47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang M., Bai Y., Sjostrom S.L., Hallström B.M., Liu Z., Petranovic D., Uhlén M., Joensson H.N., Andersson-Svahn H., Nielsen J.. Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. et al.. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Letunic I., Bork P.. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019; 47:W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S. et al.. KBase: The united states department of energy systems biology knowledgebase. Nat. Biotechnol. 2018; 36:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Romero P.A., Abate A.R.. Flow focusing geometry generates droplets through a plug and squeeze mechanism. Lab Chip. 2012; 12:5130–5132. [DOI] [PubMed] [Google Scholar]
- 33. Collins D.J., Neild A., deMello A., Liu A.Q., Ai Y.. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip. 2015; 15:3439–3459. [DOI] [PubMed] [Google Scholar]
- 34. Terekhov S.S., Smirnov I.V., Malakhova M.V., Samoilov A.E., Manolov A.I., Nazarov A.S., Danilov D.V., Dubiley S.A., Osterman I.A., Rubtsova M.P. et al.. Ultrahigh-throughput functional profiling of microbiota communities. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yilmaz S., Allgaier M., Hugenholtz P.. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods. 2010; 7:943–944. [DOI] [PubMed] [Google Scholar]
- 36. Ahn K., Kerbage C., Hunt T.P., Westervelt R.M., Link D.R., Weitz D.A.. Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl. Phys. Lett. 2006; 88:024104. [Google Scholar]
- 37. Lan F., Haliburton J.R., Yuan A., Abate A.R.. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat. Commun. 2016; 7:11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sciambi A., Abate A.R.. Generating electric fields in PDMS microfluidic devices with salt water electrodes. Lab Chip. 2014; 14:2605–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sukovich D.J., Lance S.T., Abate A.R.. Sequence specific sorting of DNA molecules with FACS using 3dPCR. Sci. Rep. 2017; 7:39385–39385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J., Bright I.J., Lucero M.Y., Hiddessen A.L., Legler T.C. et al.. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011; 83:8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agresti J.J., Antipov E., Abate A.R., Ahn K., Rowat A.C., Baret J.-C., Marquez M., Klibanov A.M., Griffiths A.D., Weitz D.A.. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farrar K., Donnison I.S.. Construction and screening of BAC libraries made from Brachypodium genomic DNA. Nat. Protoc. 2007; 2:1661–1674. [DOI] [PubMed] [Google Scholar]
- 43. Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N.. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017; 35:833–844. [DOI] [PubMed] [Google Scholar]
- 44. Staunton J., Weissman K.J.. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001; 18:380–416. [DOI] [PubMed] [Google Scholar]
- 45. Hori M., Fukano H., Suzuki Y.. Uniform amplification of multiple DNAs by emulsion PCR. Biochem. Biophys. Res. Commun. 2007; 352:323–328. [DOI] [PubMed] [Google Scholar]
- 46. Taly V., Kelly B.T., Griffiths A.D.. Droplets as microreactors for High-Throughput biology. ChemBioChem. 2007; 8:263–272. [DOI] [PubMed] [Google Scholar]
- 47. Overbeek R., Fonstein M., D’Souza M., Pusch G.D., Maltsev N.. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campelo A.B., Gil J.A.. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology. 2002; 148:51–59. [DOI] [PubMed] [Google Scholar]
- 49. Juhas M., van der Meer J.R., Gaillard M., Harding R.M., Hood D.W., Crook D.W.. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009; 33:376–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matilla M.A., Leeper F.J., Salmond G.P.. Biosynthesis of the antifungal haterumalide, oocydin A, in Serratia, and its regulation by quorum sensing, RpoS and Hfq. Environ. Microbiol. 2015; 17:2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alanjary M., Kronmiller B., Adamek M., Blin K., Weber T., Huson D., Philmus B., Ziemert N.. The antibiotic resistant target seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017; 45:W42–W48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lynch M.D., Neufeld J.D.. Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 2015; 13:217–229. [DOI] [PubMed] [Google Scholar]
- 53. Iqbal H.A., Low-Beinart L., Obiajulu J.U., Brady S.F.. Natural product discovery through improved functional metagenomics in streptomyces. J. Am. Chem. Soc. 2016; 138:9341–9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niu X., Gielen F., Edel J.B., deMello A.J.. A microdroplet dilutor for high-throughput screening. Nat. Chem. 2011; 3:437–442. [DOI] [PubMed] [Google Scholar]
- 55. Terekhov S.S., Smirnov I.V., Stepanova A.V., Bobik T.V., Mokrushina Y.A., Ponomarenko N.A., Belogurov A.A. Jr, Rubtsova M.P., Kartseva O.V., Gomzikova M.O. et al.. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2550–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abatemarco J., Sarhan M.F., Wagner J.M., Lin J.L., Liu L., Hassouneh W., Yuan S.F., Alper H.S., Abate A.R.. RNA-aptamers-in-droplets (RAPID) high-throughput screening for secretory phenotypes. Nat. Commun. 2017; 8:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Daniel R. The metagenomics of soil. Nat. Rev. Microbiol. 2005; 3:470–478. [DOI] [PubMed] [Google Scholar]
- 58. Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K.. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science (New York, N.Y.). 2016; 351:1196. [DOI] [PubMed] [Google Scholar]
- 59. Ghosal D., Ghosh S., Dutta T.K., Ahn Y.. Corrigendum: Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A Review. Front. Microbiol. 2016; 7:1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.