Abstract
The Dictyostelium Intermediate Repeat Sequence 1 (DIRS-1) is the name-giving member of the DIRS order of tyrosine recombinase retrotransposons. In Dictyostelium discoideum, DIRS-1 is highly amplified and enriched in heterochromatic centromers of the D. discoideum genome. We show here that DIRS-1 it tightly controlled by the D. discoideum RNA interference machinery and is only mobilized in mutants lacking either the RNA dependent RNA polymerase RrpC or the Argonaute protein AgnA. DIRS retrotransposons contain an internal complementary region (ICR) that is thought to be required to reconstitute a full-length element from incomplete RNA transcripts. Using different versions of D. discoideum DIRS-1 equipped with retrotransposition marker genes, we show experimentally that the ICR is in fact essential to complete retrotransposition. We further show that DIRS-1 produces a mixture of single-stranded, mostly linear extrachromosomal cDNA intermediates. If this cDNA is isolated and transformed into D. discoideum cells, it can be used by DIRS-1 proteins to complete productive retrotransposition. This work provides the first experimental evidence to propose a general retrotransposition mechanism of the class of DIRS like tyrosine recombinase retrotransposons.
INTRODUCTION
Retrotransposons are mobile genetic elements that amplify in eukaryotic genomes by a copy-and-paste mechanism (1). Diverse transposable elements constitute about 10% of the 34 Mb genome of Dictyostelium discoideum (2,3). DIRS-1 represents the most abundant retrotransposon in the D. discoideum genome with 40 intact and 200–300 fragmented copies covering ∼3.3% of the genome (2). While the activity of mobile genetic elements is considered a major source of genetic variability (4), random transposition can negatively impact the fitness of host cells (5), and this appears particularly relevant in gene-dense genomes like that of D. discoideum (3). Two general strategies have emerged in different hosts against this threat: restriction of integration at ‘safe’ genomic sites, and restriction of mobility by means of regulation of retrotransposon gene expression (6). Both strategies have been implemented to regulate DIRS-1 retrotransposition in D. discoideum: First, the positions of DIRS-1 copies in the genome appear confined to centromeres (7), which can be considered safe genomic sites because of their heterochromatic status, as experimentally shown by the association of DIRS-1 sequences with the heterochromatin-associated histone modification H3K9me3 (7). Second, DIRS-1 is under post-transcriptional control by the endogenous RNA-dependent RNA polymerase (RdRP) RrpC (8) and the Argonaute protein AgnA (9), as part of the RNA-mediated gene silencing pathways of the amoeba.
In general, retrotransposons (retroelements) can be classified into five distinctive orders (10): the long terminal repeat (LTR) retroelements, the non-LTR retroelements with the orders long and short interspersed elements (LINE and SINE), the Penelope (PLE) retrotransposons and the DIRS elements (11), also called tyrosine recombinase (YR) retroelements (12). As this name suggests, the group is characterized by the presence of tyrosine recombinases instead of an integrase (INT) or endonuclease (13,14). Usually, these enzymes mediate site-specific genomic integration of retrotransposons (15). YR retroelements form four families (11,12,16) and all of them feature a similar composition (Figure 1A), however with differently arranged open reading frames (ORFs). These comprise, next to the name-giving YR, a putative GAG protein (GAG), a reverse transcriptase (RT) and an RNase H (RH). Distinct from classical LTR elements, DIRS-1 sequences are flanked by inverted terminal repeats and these LTRs are non-identical in sequence. However, their outer sequences are repeated (adjacent to each other) in the internal complementary region (ICR) of the element further described below. PAT-like elements and Ngaro elements are distinct from these as they are surrounded by a series of direct repeats of which three are repeated adjacent to each other. DIRS and PAT elements encode additionally a methylase with unknown function (11), and some Ngaro elements encode additionally a hydrolase (17). Finally, the recently described VIPER retroelements (16) also belong to the YR class, and here the coding region is bordered by LTR derived from short interspersed repetitive element (SIRE).
Figure 1.
The DIRS1 group elements and a proposed replication cycle of DIRS-1. (A) Schematic structure of the four clades of DIRS1 group retrotransposons encoding tyrosine recombinases, consisting of DIRS-1 PAT-like, Ngaro-like and VIPER. Terminal repeats are represented by boxed triangles, either as inverted left and right long terminal repeats (lLTR and rLTR, respectively) or as split direct repeats (A1 to B2). The internal complementary (ICR) is shown with inverted triangles. The arrows represent the encoded proteins or protein domains: group specific antigen (GAG), tyrosine recombinase (YR), reverse transcriptase (RT), RNase H (RH), methyltransferase (MT) and hydrolase (HYD). (B) Schematic representation of the mechanism, by which DIRS-1 retrotransposons restore their full sequence (4814 bp) form its sub-genomic transcript (ca. 4500 nt). The simplified scheme of DIRS-1 with the lLTR and rLTR, respectively and the ICR, according to Cappello et al. (18). The transcription start site (TS) is shown, and red dotted lines indicate the beginning and the end of the transcript. Open reading frames (A) are omitted here for clarity. The circular junction at the ICR is shown magnified to the right for the restored full-length single-stranded circular element. For details cf. main text.
DIRS-1 from D. discoideum is the first described YR retroelement (18), yet not much is known about its replication cycle. However, its characteristic sequence features and an early analysis of primary transcripts allowed Cappello, Cohen and Lodish to propose a theoretical model of its retrotransposition cycle (18), which is summarized next. The left LTR lies upstream of the putative transcription start site (TS, Figure 1B) and has been suggested to be the promoter for DIRS-1 transcription (19). The sense mRNA transcript derived from complete, 4814 bp long genomic DIRS-1 copies is a 4500 nt-long sequence that lacks most of the left LTR and almost the entire right LTR, as shown experimentally (19). In the first step of the hypothetical replication cycle, the mRNA is proposed to be reverse transcribed into single-stranded cDNA (Figure 1B), but it is not known which sequence may serve as the primer in this process. Starting from the hypothetical single-stranded cDNA, Cappello et al. suggested that the missing terminal sequences would be reconstituted in two steps. First, the majority of the left LTR sequence is restored by using residual sequences of the right LTR as template. Second, the missing parts of the full DIRS-1 sequence are restored by intramolecular annealing of the LTR fragments with the ICR, followed by DNA synthesis. (Figure 1B). That ICR sequence of 88 bp is exact complementary to the extremities of both, left and right LTRs (18,20). In detail, the 5′ end of left LTR (position L, Figure 1B) is complementary to the first 33 bases of the ICR (position L’, Figure 1B), and the 3′ end of right LTR (position R, Figure 1B) is complementary with the remaining 55 final bases of ICR (position R’, Figure 1B). In view of the universal 5′-3′ directionality of DNA polymerization, this final restoration of the complete DIRS-1 element requires further rearrangements. In the model, a ligation step is thought to result in a circle junction representing exactly the ICR with the fragments of the two inverted LTRs. The resulting single-stranded circular DNA would then be converted to a double-stranded circular DNA, which would be ready to be incorporated into the genome by means of the DIRS-1-encoded YR activity. The specificity of the YR has not been investigated yet experimentally, however, as pointed out by Cappello, Cohen and Lodish many of the DIRS-1 seems to be incorporated preferentially into pre-existing DIRS-1 sequences (18). So far, this coherent model (Figure 1B) was largely untested experimentally, although substantial bioinformatic analyses by Goodwin and Poulter are in general support of the model (11,17,20,21).
Recently, Nellen et al. described a novel DIRS-1-related extrachromosomal cDNA that accumulated exclusively in the cytoplasm of AgnA-deficient D. discoideum strains (9). Further characterization of this sub-genomic DNA revealed that it corresponded to an almost complete antisense strand of the retrotransposon. In this fragment, only the sequence of the left LTR was missing. Such a cDNA would indeed represent a central intermediate in the DIRS-1 retrotransposition cycle as proposed by Cappello et al. (Figure 1B), whose model predicts that the missing sequence of DIRS-1 is restored by self-complementarity to the ICR. This was thought to lead to a circular intermediate, from which a double stranded, retrotransposition competent element could be generated (18). Here we set out to experimentally investigate the DIRS-1 retrotransposition model proposed by Cappello et al. (18). Using genetically traceable versions of DIRS-1, we show that the retroelement undergoes a full retrotransposition cycle in rrpC and agnA gene deletion strains of D. discoideum, for which the ICR sequence of DIRS-1 is in fact indispensable. Further, we show that the extrachromosomal cDNA is a single-stranded, mainly linear molecule, which serves as a true intermediate of the DIRS-1 replication cycle.
MATERIALS AND METHODS
Strains
The Dictyostelium strains analyzed in this study comprise the previously published Ax2 wildtype (22), drnB– (23), rrp– (24) and Argonaute mutants (9).
Oligonucleotides
DNA oligonucleotides (Sigma-Aldrich) used in this study are listed in Supplementary Table S1.
Design of DIRS-1bsr
The mbsrI gene was initially designed to follow retrotransposition of the TRE5-A retrotransposon in D. discoideum cells (25). The mbsrI sequence was used to design a traceable DIRS-1 sequence referred to as DIRS-1bsr. A virtual sequence covering a full-length, presumably retrotransposition-competent DIRS-1 (Supplementary Figure S1A) was assembled from GenBank entry M11339. The mbsrI retrotransposition marker was inserted at position 4464 of the DIRS-1 sequence, between the ICR and the right LTR, and surrounded by suitable restrictions sites. Further, to facilitate sequence modifications, two restriction sites (AgeI and PstI, separated by 5 bp spacer) were introduced between left LTR and ORF1 (Supplementary Figure S1B). The DIRS-1bsr sequence was chemically synthesized and cloned into the pGH vector (Celtek Genes; Celtec Bioscience, LLC).
Generation of traceable DIRS-1 constructs
The cloning of modified versions of DIRS-1bsr is summarized in Supplementary Figure S1. The retrotransposition marker mhygI was designed based on the sequence of the hygromycin resistance gene (hyg) present in D. discoideum expression vector pDM358 (26). The hyg sequence was codon-optimized for translation in D. discoideum and fused with 302 bp of upstream sequence derived from the actin15 promoter found in pDM358. The 74 bp intron derived from the S17 gene (25) was inserted into the hyg gene after codon 9 in the reverse orientation, yielding hygI. To generate DIRS-1hyg, the mbsrI gene was removed from the DIRS-1bsr element by digestion with SacII and re-ligation of the plasmid (Supplementary Figure S1C). The resulting construct ‘DIRS-1 without resistance cassette’ served as cloning intermediate for the generation of further genetically traceable DIRS-1 constructs, as detailed in Supplementary Figure S1C–H.
Dictyostelium cell culture
All strains were grown in HL5 medium (Formedium) supplemented with glucose at constant temperature of 22°C and continuous light. Usually the medium was supplemented with prokaryote-specific antibiotics (50 μg/l ampicillin, 0.25 μg/l amphotericin B, 10 ml penicillin–streptomycin, 10 000 U/ml of penicillin and 10 g/ml of streptomycin). The media used for cultivation of transformed cells with respective plasmids were additionally supplemented with G418 (10 μg/ml), blasticidin (10 μg/ml) or hygromycin B (30 μg/ml).
Isolation of DIRS-1 extrachromosomal cDNA
A fully confluent Petri dish (ca. 2 × 107Dictyostelium cells) was washed with Sørensen phosphate buffer (2 mM Na2HPO4, 15 mM KH2PO4, pH 6.0, adjusted with H3PO4). Next, cells were harvested and processed using a plasmid minipreparation kit suitable for plasmid extraction from Escherichia coli (GeneElute™ HP Plasmid Miniprep Kit, Sigma-Aldrich). The kit was used according to the manufacturer's description and the final elution was performed using 50 μL water.
Detection of DIRS-1 extrachromosomal cDNA (native and semi-denaturing conditions) by Southern blotting
Samples (10–15 ul) eluted from the miniprep column were mixed with 6× Loading Dye (Thermo Scientific) and loaded on a 1.4% agarose gel containing 1x MOPS (20 mM MOPS, 5 mM NaAc, 1 mM EDTA, pH 7.0). For semi-denaturing conditions, 10 μl of DNA samples were mixed with 2 μl of 10× MOPS, 2 μl of 37% formaldehyde, 10 μl of formamide and denatured at 70°C for 5 min. After cooling down, samples were loaded on 1.2% agarose gels containing 1× MOPS. The samples were run together with the GeneRuler™ 1 kb DNA ladder (Thermo Scientific), which served in this experiment as reference, rather than a precise measure of molecular weight. As a consequence of the DNA isolation protocol, no loading control can be shown and the normalization relies on using the same amount of starting material. After capillary transfer to a nylon membrane (Amersham Biosciences HybondTM-NX), the DNA was cross-linked by UV irradiation (0.5 J/cm2). Prehybridization and hybridization were carried out in Church buffer (27). The radioactive probe was generated using random primed labeling of DNA, using a DIRS-1 specific PCR product as template (28). The blot was washed for 1 h in Wash I solution (2× SSC, 0.1% w/v SDS) and for 1 h in Wash IV (0.1× SSC, 0.1% w/v SDS) at 60°C. Hybridization signals were detected after exposure to an image plate and read-out by phosphorimaging (FLA-3000, Fujifilm).
Nuclease treatment and detection of DIRS-1bsr extrachromosomal cDNA
The extracted extrachromosomal cDNA sample (plasmid miniprep kit, as described before) was subjected to digestion with RNase A (Sigma-Aldrich), DNase I, exonuclease I, exonuclease III or S1 nuclease (Thermo Scientific). For each set-up, 5 μl of the extracted DNA were digested in a final volume of 10 μl according to the manufacturers' instructions. To avoid the formation of any cDNA secondary structures, samples were denatured for 5 min at 95°C and snap cooled on ice, before adding the respective enzyme. The digestion reaction was performed for at least 1 h and followed by heat inactivation of the respective enzymes. Of each digest, 5 μl were used in PCR reactions using cassette specific primer pairs (Supplementary Table S1).
Retotransposition assay
The retrotransposition assay in D. discoideum cells using the mbsrI selection marker has been described previously. To detect retrotransposition of DIRS-1bsr and DIRS-1hyg elements, D. discoideum strains were co-transformed using electroporation (29) with 10 μl of transformation mix that contained 10 μg of plasmid carrying the genetically tagged version of DIRS-1 and 10 μg of the pISAR plasmid (30). After selection with G418 (10 μg/ml), stable transformants were mixed and pools of cells were cultured until they reached 80–90% confluence. Half of the cells were used for isolation of genomic DNA (gDNA) (31) and generation of spores (32). The second half was cultured in medium supplemented with the antibiotics allowing to select for retrotransposition events, namely blasticidin (10 μg/ml) or hygromycin B (30 μg/ml). After selection, resistant clones from one Petri dish were split into two aliquots. The first served as a material for gDNA isolation, whereas the second was used for the generation of spores. All experiments were repeated at least twice. The retrotransposition events were monitored with pairs of mbsrI or mhygI exon-specific primers (Supplementary Table S1).
Crystal violet staining
Cells plated on a Petri dish were washed with 20 ml Sørensen phosphate buffer. After draining of the liquid, cells were covered with 5 ml of ready-to-use Crystal Violet solution (Pro-Lab Diagnostics) and incubated at RT. After 20 min, the dye was removed and remaining liquid was washed away by immersing the Petri dish 3× in 1 l of tap water. The stained cells were documented using an office scanner.
Quantitative PCR (qPCR) analysis of extrachromosomal cDNA
For quantitative analysis, qPCR was performed on an Mx3000P system (Agilent) using the PowerUp™ SYBR™ Green Master Mix kit (Applied Biosystems) and following the manufacturer's instructions. Primer sequences are listed in Supplementary Table S1. All measurements were carried out in triplicate. Relative quantification of either exonuclease I, or S1 nuclease digested molecules was determined by the ΔCT method (33) using non-digested samples as reference. The paired t-test was performed using GraphPad Prism version 7.0a for Mac (GraphPad Software, La Jolla, CA, USA).
RESULTS
Extrachromosomal DIRS-1 cDNA accumulates in rrpC deletion strains
In a previous study, the accumulation of the extrachromosomal DIRS-1 cDNA in the agnA– strain had been correlated with a down-regulation of DIRS-1-related siRNAs and an up-regulation of DIRS-1 sense transcripts (9). These phenotypes had already been observed in strains missing the rrpC gene (8). We therefore tested whether the extrachromosomal cDNA can also be detected in rrpC kock-out strains, or in other available RdRP knock-out mutants (24). In addition, the deletion strain of the drnB gene, which encodes a dicer-like nuclease, was analyzed. DrnB has been previously reported to play a role in miRNA maturation and the transitivity of RNA silencing signals (23,34–37). As controls, the previously studied agnA–, agnB– and agnA–/agnB– deletion strains were included in our analysis (9). DNA was isolated analogous to the study in which the extrachromosomal DIRS-1 cDNA had been discovered (9), and DNA samples were subjected to Southern blotting after semi-denaturing gel electrophoresis. All rrpC deletion strains displayed a signal similar to that previously observed in the agnA– strains (Figure 2B), indicating the presence of the extrachromosomal DIRS-1 cDNA. The intensities of the signals in the rrpC deletion strains resembled that detected in the double mutant agnA–/agnB–, which had been previously shown to be significantly reduced compared to agnA–, while no signal was not detectable in the agnB– strain (9). Like in the Ax2 wildtype strain, our analysis did not reveal the presence of the extrachromosomal DIRS-1 cDNA in any of the gene deletion strains of RdRPs RrpA and RrpB alone or in combination. Likewise, no extrachromosomal DIRS-1 cDNA was observed in the drnB mutant (Figure 2B). When DNA samples were analyzed by Southern blotting after native gel electrophoresis, the signal strengths in the respective deletion strains were similar to those observed in the first experiments. However, an additional band with reduced electrophoretic mobility was detected (Supplementary Figure S2), suggesting alternative conformations of the extrachromosomal DIRS-1 cDNA. When the samples were chemically and thermally denatured prior to gel loading (Figure 2B), these alternative conformations were not discernable, suggesting that they are readily interconvertible.
Figure 2.

Extrachromosomal DIRS-1 DNA accumulates in rrpC– strains. (A) Schematic representation of the DIRS-1 retrotransposon (4814 bp). For details cf. main text and Figure 1A. Underneath, OLB marks the position of a body-labelled PCR product used for Southern blotting. Positions qP1 and qP2 indicate the amplicons used for qPCR analysis. (B) Southern Blot analysis with DNA samples from the indicated Dictyostelium strains separated under semi-denaturing conditions during gel electrophoresis, followed by transfer to a nitrocellulose membrane and hybridization with the OLB probe. Marker is the GeneRuler™ 1 kb DNA ladder, which served in this experiment as an estimate, rather than a precise measure of molecular weight.
Genetically tagged DIRS-1 is mobilized in all rrpC deletion strains
The presence of the extrachromosomal cDNA of DIRS-1 in the rrpC– and agnA– strains strongly suggested that the retrotransposon is mobilized in these particular strains. Indeed, Southern blotting of genomic DNA prepared after long-term culture of the rrpC mutant cells suggested an increased genomic copy number of DIRS-1 with time (8). However, the high copy number of complete and fragmented DIRS-1 sequences in the Dictyostelium genome made a detailed analysis of DIRS-1 mobilization by Southern blotting challenging. Therefore we adopted a retrotransposition assay established in mammals and yeast (38,39), in which we applied a previously established retrotransposition marker gene (25) to study DIRS-1 retrotransposition in D. discoideum.
The DIRS-1 retrotransposition assay was performed by co-transforming Dictyostelium strains with two plasmids (Figure 3): The first plasmid, pISAR, conferred resistance to G418 (G418R) and allowed for the selection of transformed cells. This second plasmid carried a genetically traceable version of DIRS-1, which allowed us to enrich cultures for cells with new retrotransposition events via blasticidin selection. The retrotransposition marker consists of a blasticidin resistance gene (bsr) that is disrupted by a reverse intron (= bsrI). In this first version of traceable DIRS-1 retrotransposons (DIRS-1bsr), the bsrI gene is inserted in the reverse orientation relative to the transcription direction of the DIRS-1 element (= mbsrI) between the ICR region and the right LTR of DIRS-1 (Figure 3A). As such, DIRS-1bsr transformed into the Dictyostelium genome (referred to as ‘master elements’) cannot confer resistance to blasticidin due to the presence of the intron that disrupts the bsr reading frame. After transcription of DIRS-1bsr the intron of the mbsrI marker is removed from the primary transcript of the retrotransposon by splicing. If a full retrotransposition cycle is completed, the newly integrated element (referred as a ‘copy element’) will express a functional mbsr gene that confers resistance to blasticidin. Retrotransposition can be analyzed in blasticidin-resistant clones by PCR on genomic DNA. The corresponding PCR products of master and copy elements (510 and 436 bp, respectively) differ by the size of the intron (74 bp) (Figure 3A, B).
Figure 3.
Retrotransposition of DIRS-1bsr in D. discoideum. (A) Schematic presentation of the DIRS-1bsr retrotransposition assay. The mbsrI selection marker was positioned between the ICR and the rLTR of DIRS-1 at position 4465. The mbsrI gene is under the control of an actin15 promoter (A15P) and an actin8 terminator (A8T). The DIRS-1bsr bearing plasmid and a G418 resistant plasmid (pISAR) were co-transformed into D. discoideum strains. Uptake of plasmids was monitored by selection with G418. Upon integration of the plasmid, transformed cells harbour a DIRS-1bsr sequence, referred to as ‘master element’. This does not confer blasticidin resistance to the transformed cells, because the bsr gene is inactivated by a reverse intron. After a complete retrotransposition cycle the DIRS-1bsr is integrated into the genome (referred as ‘copy element’) and allows for the expression of a functional bsr mRNA, thus adding resistance to blasticidin to the cells. Position qBS indicates the amplicon used for qPCR analysis. (B) Experimental procedure for detection of DIRS-1 retrotransposition. The Dictyostelium strain under investigation is co-transformed with the pISAR plasmid and the plasmid containing the genetically traceable DIRS-1bsr. The neomycin phosphotransferase encoded on pISAR confers resistance to G418 (G418R). G418-resistant cells are analyzed by PCR to detect transformed cells harbouring the master element. Next, correctly transformed cells are subjected to selection with blasticidin (BS). Resistance to BS indicates successful retrotransposition in respective strains, and the presence of the copy element is assessed by subsequent PCR. (C) Representative example of BSR colony formation upon the retrotransposition assay using the DIRS-1bsr construct in Ax2 WT and rrpC– strains. Cells were stained with crystal violet eight days after starting blasticidin selection (D) PCR verification of the retrotransposition assay with DIRS-1bsr. PCR Analysis of genomic DNA to determine the structure of the mbsrI gene after the selection with G418 (upper panel) and subsequently with BS (lower panel). Clones marked as ‘+’ survived the indicated antibiotic, and clones marked with ‘–’ died. PCR was performed using specific primers #2212 and #2213. The upper band (510 bp) was derived from stably integrated master elements, and the lower band (436 bp) was derived from newly retrotransposed copies (copy element).
The first set of strains investigated with this DIRS-1 retrotransposition assay included various RdRP mutants and the drnB– strain. Ax2 wildtype cells were used as negative control. For all strains, G418R transformants were obtained, indicating successful transformation of cells with pISAR. Because no selection marker was present on the plasmid carrying the DIRS-1bsr, we confirmed transformation of master elements by PCR (Figure 3D, upper panel). Sequencing of the PCR product confirmed the identity of the 510 bp part of the mbsrI sequence (Supplementary Figure S3), indicating successful transformation also of the second plasmid. Next, we determined whether DIRS-1bsr was mobilized in the co-transformed strains by subjecting cells to blasticidin selection. Blasticidin-resistant clones were only recovered from strains in which the rrpC gene had been deleted (Figure 3C). Interestingly, also the drnB mutant did not survive the blasticidin treatment, suggesting that DrnB is not involved in suppressing DIRS-1 retrotransposition. Cells surviving blasticidin selection were pooled and genomic DNA was isolated and subjected to PCR analysis using mbsr-specific primers. This revealed the presence of two bands (Figure 3D, lower panel), suggesting that the surviving cells not only contained the master element (510 bp), but also the spliced, intron-free version of the mbsrI cassette, which is indicative of productive DIRS-1 retrotransposition. The 436 bp band was isolated and sequenced, which confirmed removal of the intron (Supplementary Figure S4). In summary, the data indicated that the mbsrI-tagged DIRS-1 construct was suitable to trace retrotransposition events in Dictyostelium, and that amongst the tested strains only those lacking RrpC allowed for the completion of a full retrotransposition cycle.
DIRS-1 is mobilized in the agnA– strain, but not in the double mutant agnA–/agnB–
Given the similar phenotypes of rrpC– and agnA– strains with respect to reduced production of DIRS-1 siRNAs and increased full length transcripts and cDNAs of DIRS-1 sequences (8,9), we next set out to investigate the retrotransposition activity of DIRS-1 in the existing Argonaute gene deletion strains. Some of the available mutants (agnA– and agnA–/agnB–) were already resistant to blasticidin due to the homologous recombination knock-out procedure (9). Therefore, an alternative retrotransposition marker gene useful for DIRS-1 retrotransposition assays had to be established. To this end, we generated the DIRS-1hyg element, in which an intron-disrupted hygromycin resistance cassette (mhygI) was inserted between the left LTR and ORF1 of DIRS-1 (Figure 4A). Otherwise the principle of the assay was the same as described for the DIRS-1bsr element (Figure 3). In experiments to test the retrotransposition competence of the DIRS-1hyg element, the rrpC– strain served as a positive control and the Ax2 wildtype as negative control. As in the previous experiment, G418-resistant cell lines were established, and the presence of the DIRS-1hyg master element was confirmed by PCR on genomic DNA of G418-resistant transformants (Figure 4B) and sequencing of that product (Supplementary Figure S5). The transformants were subsequently cultivated in medium supplemented with hygromycin B. As expected, the Ax2 cells did not survive hygromycin selection, confirming that wildtype cells do not support DIRS-1hyg amplification. In contrast, hygromycin-resistant clones were readily obtained from rrpC– transformants, suggesting that DIRS-1hyg actively retrotransposed in these cells. Out of the three available Argonaute mutants, the agnA– strain supported DIRS-1hyg retrotransposition, as judged by the appearance of hygromycin-resistant clones and a corresponding PCR signal from these cells (Figure 4 and sequencing data in Supplementary Figure S6). In contrast, DIRS-1hyg retrotransposition was not observed in the agnB– mutant. DIRS-1hyg was unable to retrotranspose in the agnA–/agnB– double mutant, which was surprising because it would suggest that AgnB may be involved in the completion of DIRS-1 retrotransposition, at least in the background of de-repressed posttranscriptional silencing due to the lack of AgnA expression. Taken together, the data indicated that also the DIRS-1hyg construct was suitable to monitor the retrotransposition cycle of DIRS-1, and that both the rrpC– and agnA– strains support DIRS-1 retrotransposition, whereas amplification of the element is completely silenced in wildtype cells.
Figure 4.

Retrotransposition of DIRS-1hyg. (A) Schematic representation of the DIRS-1hyg construct design with an intron-disrupted hygromycin resistance cassette inserted between left LTR and ORF1 (GAG) of DIRS-1 at position 321. The workflow of the experiment proceeds analogous to that shown for DIRS-1bsr (for details see Figure 3). Position qHYG indicates the amplicon used for qPCR analysis. (B) PCR verification retrotransposition of DIRS-1hyg in the indicated strains. Analysis of genomic DNA to determine the structure of the mhygI gene after the selection with G418 and subsequently with hygromycin. PCR was performed using specific primers #2313 and #2314. The upper band (532 bp) was derived from master elements, and the lower band (458 bp) was derived from newly retrotransposed copies (copy element).
DIRS-1 requires its ICR for retrotransposition
The full-length transcript of DIRS-1 (4500 nucleotides) was shown experimentally (19) to lack part of the full-length genomically encoded canonical sequence (4814 bp). The extrachromosomal cDNA molecule was also shown to be incomplete, because it lacked 300 nucleotides corresponding to the left LTR (9). In theory, the full genomic sequence of DIRS-1 can be restored from the sub-genomic sequence, utilizing the features of the ICR region (18); see also the retrotransposition model in Figure 1B. To test this hypothesis experimentally, we generated a DIRS-1bsr derivative with the ICR deleted (for a detailed description of the cloning procedure, see Supplementary Figure S1). The DIRS-1bsr*[-ICR] and the control DIRS-1bsr* (Figure 5A) contained the mbsrI gene positioned between the left LTR and ORF1, analogous to the position of the cassette in the DIRS-1hyg construct (Figure 4). To discriminate it from the previously used DIRS-1bsr plasmid (Figure 3), we named the resulting construct DIRS-1bsr*.
Figure 5.
DIRS-1bsr variants reveal features necessary for retrotransposition and trans-action of DIRS-1 encoded proteins. (A) Overview of the DIRS-1bsr variants used in this study, with the parental DIRS-1bsr (Figure 2A), DIRS-1bsr*, DIRS-1bsr*[-ICR] lacking the ICR and the truncated trDIRS-1bsr* element encoding fragments of ORF2 and ORF3 only. (B) PCR analysis of genomic DNA from retrotransposition assay with two variants of the retroelement, DIRS-1bsr* and DIRS-1bsr*[-ICR]. (C) PCR analysis of genomic DNA from retrotransposition assays with two variants of the retroelement, DIRS-1bsr* and trDIRS-1bsr*. For further details regarding the assay, please refer to Figure 3. (D) PCR analysis of extrachromosomal cDNA extracted from the indicated strains during the retrotransposition assay and in different selection media, using the specific primer pair #2212 and #2213 (see Figure 3).
To test the retrotransposition competence of DIRS-1bsr*[-ICR] and DIRS-1bsr* elements, we co-transformed them with pISAR into wildtype and rrpC– cells. Retrotransposition assays were performed as described in Figure 3B. Only rrpC– cells transformed with the DIRS-1mbsrI* construct survived the BS treatment, whereas no blasticidin-resistant clones could be obtained with the DIRS-1mbsrI*[-ICR] element. PCR confirmed the presence of the spliced version of the mbsrI gene indicative of retrotransposition of the DIRS-1bsr* element, whereas the DIRS-1bsr*[-ICR] element was not active (Figure 5B).
DIRS-1 reverse transcriptase acts in trans
In LTR-retrotransposons, the primary transcript has a dual function. Firstly, it serves as mRNA for translation of the proteins required for retrotransposition. Secondly, the RNA is the template for reverse transcription and the generation of cDNA that is integrated into the genome (40–42). In this process, the retrotransposon proteins can act in two different modes. When acting in cis, the proteins interact with the RNA template that encodes them, either during or immediately after translation, thereby often promoting the formation of virus-like particles (VLPs) (43). In turn, proteins acting in trans do not discriminate between the RNA sequences from which they were transcribed and other (retrotransposon) RNAs. For example, trans-action of proteins encoded by the Ty1 retroelement has been demonstrated by Curcio and Garfinkel (44). Later, trans-action of proteins was also demonstrated by Xu and Boeke in their helper-donor assay (45). The authors co-transformed yeast cells with the helper Ty1 element, that is incapable to transpose but encodes functional proteins, and various truncated versions of Ty1 termed mini-Ty1. Using this set-up, the minimal sequence of a transposition-competent mini-Ty1 was determined to consist of 380 nucleotides of the 5′ terminus and 357 nucleotides of the 3′ terminus.
To test whether DIRS-1-encoded proteins can act in trans to mobilize DIRS-1 transcripts, we generated a truncated version of DIRS-1 (trDIRS-1bsr*), in which the region encoding ORF1 and about half of ORF2 and ORF3 were deleted (Figure 5A). As a modification of the set-up used by Xu and Boeke (45), we relied in our experiments on the activity of endogenous proteins derived from genomic DIRS-1 copies in the rrpC– strain, which we infer from the presence of endogenous extrachromosomal cDNA (Figure 2B). Even though this strain should possess the proteins required for retrotransposition, the trDIRS-1bsr* element did not confer blasticidin resistance in the rrpC– strain, unlike the parental DIRS-1mbsrI (Figure 5C).
To investigate potential causes for the retrotransposition incompetence of the DIRS-1mbsrI*[-ICR] and the trDIRS-1mbsrI* constructs, we transformed them in the retrotransposition permissive background of the rrpC– strain, which we kept under G418 selection. Separately, we used the retrotransposition competent plasmids DIRS-1mbsrI and DIRS-1mbsrI* under G418 selection in the Ax2 wildtype cells as negative controls. The latter constructs were also transformed as positive controls in the rrpC– strain, which was initially kept in medium supplemented with G418 and later with G418 + blasticidin to detect newly transposed DIRS-1bsr elements. From these transformants, extrachromosomal cDNA was isolated as described (9) and analyzed by PCR instead of the earlier used Southern blotting approach (Figure 2C), because the use of the primer pair addressing the sequence surrounding the mbsrI cassette (Figure 3A) allowed to discriminate between the spliced and non-spiced form of the resistance cassette and thus directly indicate retrotransposition of DIRS-1bsr. As expected, no extrachromosomal DIRS-1bsr cDNA was observed in the wildtype strain (Figure 5D). In the rrpC– strain, we detected the spliced form of the mbsrI cassette of both the DIRS-1bsr and DIRS-1bsr* already after selection in G418 medium (e.g., prior to blasticidin-selection to enrich for retrotransposition events). Under these conditions, the DIRS-1bsr* element also generated an incompletely spliced version of the extrachromosomal cDNA. However, both strains contained after selection with G418 + blasticidin mainly the spliced form of the resistance cassette. PCR products were also obtained from the extrachromosomal cDNAs of the retrotransposition incompetent constructs DIRS-1bsr*[-ICR] and trDIRS-1bsr* in the rrpC– strain. The PCR products from the former were similar to the bands detected in the strain transformed with DIRS-1mbsrI* during the same antibiotic selection (G418). The results obtained with the DIRS-1mbsrI*[-ICR] construct (Figure 5A, B, D) would thus indicate that it cannot complete the full retrotransposition cycle, despite the fact that at least some molecules were spliced and reverse transcribed. This was distinct from the cDNA obtained from the trDIRS-1bsr* construct, where the PCR signal corresponded to only the non-spliced form (Figure 5D). This observation may explain why this construct did not generate blasticidin resistance in the retrotransposition assay (Figure 5C). Furthermore, this result suggested that the DIRS-1 encoded reverse transcriptase (Figure 2A), which is missing in trDIRS-1bsr*, can act in trans on the mRNA derived from that construct, resulting in the observed cDNA. Whether other DIRS-1-encoded proteins also act in trans cannot be answered with this assay because the non-spliced and thus dysfunctional blasticidin resistance cassette from this construct counteracts the observation of a full retrotransposition cycle.
Extrachromosomal cDNA of DIRS-1bsr is single-stranded and exists as predominantly linear molecules
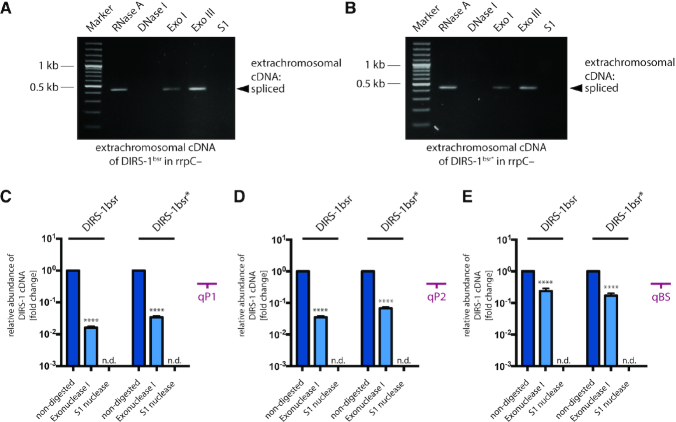
In a previous report the extrachromosomal DIRS-1 cDNA accumulating in the agnA– strain was shown to be single-stranded (9). However, the isolated cDNA was not affected by incubation with exonuclease I (Exo I), which should degrade ssDNA in the 3′→5′ direction. While this observation may in fact argue for a circular molecule, the authors suggested that modifications or secondary structures could be the reason for the insensitivity to Exo I (9). Thus, the question whether the cDNA is a circular or a linear molecule could not be answered unambiguously in that study. To investigate this further, we isolated extrachromosomal cDNAs of the DIRS-1bsr and DIRS-1bsr* elements and digested them with various nucleases, similar to the earlier experiments performed by Nellen and co-workers (9). As readout, we preferred to use PCR amplification to detect cDNA integrity rather than the less sensitive Southern blotting (Figure 5D). In line with earlier results (9), S1 nuclease, which targets single-stranded DNA (ssDNA), degraded the extrachromosomal cDNA (Figure 6A, B). Also, DNase I did, but RNase A did not degrade the DIRS-1 cDNA. Likewise, treatment with exonuclease III, which is specific for dsDNA, did not degrade the DIRS-1 cDNA. Interestingly, reduced amounts of PCR products of the extrachromosomal DIRS-1bsr cDNA were detectable after treatment with Exo I (Figure 6A, B). In order to quantify this reduction, we performed quantitative PCR (qPCR) analyses of Exo I digested samples extracted from the rrpC– strain transformed with DIRS-1mbsrI or DIRS-1mbsrI* and selected with G10/B10 medium. As control we used non-digested and S1 nuclease treated samples. Analyses using primers sets targeting the positions qP1 and qP2 (Figure 2A) revealed a 50- and 20-fold reduction of molecules compared with the non-digested samples (Figure 6C and D respectively). In the samples treated with S1 nuclease, DNA was fully degraded. To complete this analysis, we also monitored specifically the molecules derived from the traceable version of the retroelement using primers targeting position qBS in the blasticidin cassette (Figure 3A). Here, the amount of DIRS-1bsr and DIRS-1bsr* cDNA decreased ∼4-fold upon Exo I digestion, compared to the non-digested samples (Figure 6E). These results indicate that the extrachromosomal cDNA is predominantly linear. The smaller fraction that survived the Exo I treatment may represent circular molecules. This can inferred from the experimental procedures used for cDNA isolation, where proteins that might have protected cDNA ends against Exo I digestion were removed and the cDNA was denatured before nuclease treatment to prevent the formation of protective secondary structures at the cDNA ends. It is also remarkable that nuclease treatment of extrachromosomal DIRS-1bsr cDNAs extracted from the rrpC– strain transformed with DIRS-1mbsrI*[-ICR] or trDIRS-1mbsrI* revealed the same behavior as seen for DIRS-1bsr and DIRS-1bsr* (Supplementary Figure S7). In an attempt to consolidate this with the reported insensitivity of the extrachromosomal cDNA to Exo I in the agnA– strain (9), we expanded the experiments to DIRS-1hyg isolated from that strain. Also this showed similar sensitivity to Exo I (Supplementary Figure S8), and thus identical behavior in both strains, rrpC– and agnA–. In summary, these data indicate that the extrachromosomal cDNA derived from all genetically traceable versions of DIRS-1 exists as a mixture of single-stranded molecules, of which the majority is linear and only a minor fraction may be circular.
Figure 6.
Features of extrachromosomal DIRS-1bsr cDNA. Extrachromosomal cDNA samples were extracted from rrpC– strains transformed with DIRS-1bsr (A) or DIRS-1bsr* (B), and were cultivated in G418/BS10 medium that selects for strains with mobilized retrotransposon. The samples were treated separately with RNase A, DNase I, Exonuclease I (Exo I), Exonuclease III (Exo III) and S1 nuclease (S1). After digestion and heat inactivation of enzymes, the samples were analyzed by PCR with the primers #2212 and #2213 binding within the region of mbsrI cassette (binding position in Figure 2A). Quantification of endogenous (C and D) and mbsrI-tagged versions (E) of DIRS-1 extrachromosomal cDNA after treatment with Exonuclease I and S1 nuclease, relative to the non-digested sample. Samples were extracted from rrpC– strains transformed with DIRS-1bsr and DIRS-1bsr* upon cultivation in G418/BS10 medium that selects for strains with mobilized retrotransposons. The cDNA abundance was monitored by qPCR using primers targeting positions qP1, qP2 and qBS (Figures 2A and 3A) and is shown in logarithmic scale relative to the non-digested sample (set to 1). Each qPCR reaction was run in triplicate. Error bars: mean with S.D. Statistics: paired t test: P < 0.0001 (****); n.d. = not detected.
Exogenously applied extrachromosomal cDNA of DIRS-1 is able to transpose in the rrpC– strain
In all experiments, the accumulation of the extrachromosomal DIRS-1bsr cDNA in certain RNAi knock-out strains was symptomatic for cells in which DIRS-1 was mobilized (with the notable exception of the agnA–/agnB– double knock out). However, this may not necessarily mean that this molecule is a true intermediate of DIRS-1 retrotransposition (Figure 1B). Alternatively, it may well represent a non-functional dead-end by-product of DIRS-1 amplification. To determine whether the extrachromosomal DIRS-1bsr cDNA is a functional retrotransposition intermediate, we extracted extrachromosomal cDNA of DIRS-1bsr and DIRS-1bsr* from rrpC– transformants after selection for retrotransposition events in blasticidin-containing medium. The isolated material was used to transform rrpC– and wildtype Ax2 cells by electroporation. Transformed cells were allowed to recover for ca. 6 h before genomic DNA was extracted from one half the cells for PCR analysis, while the other half of cells was further cultivated in medium supplemented with blasticidin. PCR analysis of genomic DNA after transformation (Figure 7A) showed that the extrachromosomal cDNA of DIRS-1bsr and DIRS-1bsr* was present in the transformed rrpC– cells. This was not the case in the Ax2 wildtype strain, and this transformant did not survive the subsequent blasticidin treatment. Most likely, the absence of endogenous DIRS-1 proteins in the Ax2 wildtype did not allow for propagation of the elements. Instead, blasticidin-resistant clones were obtained from rrpC– transformants, and the presence of the spliced form of mbsrI was confirmed by PCR (Figure 7B). As the transformed single-stranded cDNA is unlikely to serve as template for transcription, the blasticidin resistance of these rrpC– transformants would indicate that the cDNA entered a new transposition cycle. This is likely due to the endogenous DIRS-1 proteins that are available in the rrpC– strain, but not in the Ax2 wildtype. The inability of the transformed cDNA to confer blasticidin resistance to wildtype cells also indicated that the cDNA was not incorporated into the genome by alternative, retrotransposition-independent mechanisms, such as homologous recombination. Rather, the obtained results suggest that the extrachromosomal DIRS-1 element transformed in the rrpC– strain was recognized by endogenous DIRS-1 proteins. It therefore does not represent a dead-end by-product, but an integral part of the DIRS-1 retrotransposition cycle (Figure 1B).
Figure 7.

Exogenously applied DIRS-1 extrachromosomal cDNA confers blasticidin resistance. (A) PCR analysis of genomic DNA from strains 6 h after transformation with extrachromosomal cDNA derived from two variants of retroelement, DIRS-1bsr and DIRS-1bsr*. (B) PCR analysis of genomic DNA extracted from the same strains having undergone the retrotransposition assay with two variants of the retroelement, DIRS-1bsr and DIRS-1bsr*. PCR analyses were carried out using the specific pair of primers #2212 and #2213.
DISCUSSION
DIRS-1bsr is mobilized in distinct RNAi-deficient strains
In this study, a well-established retrotransposition assay (25) was adapted to investigate the retrotransposition activity of DIRS-1 in D. discoideum. This allowed for the first time to monitor the mobility of the class of DIRS-1-like elements that encode a tyrosine recombinase instead of a classical integrase. Our results show that DIRS-1 retrotransposition is facilitated only in RrpC-deficient strains and in an agnA mutant. RrpC and AgnA were previously described as proteins that play an important role in the generation of secondary siRNAs of DIRS-1 during post-transcriptional gene silencing (PTGS) (8,9). The single deletions of the RdRPs RrpA and RrpB genes have no obvious effect on DIRS-1 siRNA and mRNA level (8). This is consistent with the observation that DIRS-1 is not actively transposing in these strains (Figure 2). Both proteins, RrpA and RrpB share 94% protein sequence identity, and might have a similar, yet unknown function, which would be distinct from that of RrpC—at least concerning DIRS-1 amplification. For example, they may be involved in transcriptional gene silencing, as exemplified by the sole RdRP Rdp1 of S. pombe. This enzyme is the main component of the RNA directed RNA polymerase complex (RDRC) that directly interacts with the RNA-induced transcriptional silencing (RITS) complex (46). Both complexes are localized in the nucleus and coordinate RNAi-mediated heterochromatin assembly within the region of centromeric repeats and the mating type locus (47,48). Also the six Arabidopsis RdRP homologues, RDR1–6, exhibit a broad range of functions, including RNA-directed DNA methylation (RdDM) and antiviral response (summarized in (49)).
DIRS-1 was not mobilized in mutants of the Argonaute protein AgnB and the Dicer-like protein DrnB. As reported previously, DrnB is involved in miRNA maturation (23,34–37) and does not participate in DIRS-1 regulation (8). Surprisingly, DIRS-1 was also not actively transposing in the agnA–/agnB– double mutant. While the lack of the AgnA protein activity clearly leads to DIRS-1 proliferation ((9) and Figure 4), the role of AgnB in DIRS-1 regulation remains unclear. The agnA–/agnB– double mutant lacks DIRS-1 related siRNAs similar to agnA– cells and is characterized by moderate accumulation of DIRS-1 mRNA (9). Moreover, the amount of the extrachromosomal cDNA detected in the agnA–/agnB– mutant corresponds to that found in the rrpC mutant (Figure 2). Thus, it would be expected that DIRS-1 is mobilized in the double mutant, but this is not the case (Figure 4). This suggests that AgnB plays a role in the completion of the retrotransposition cycle of DIRS-1 and not in its posttranscriptional silencing. As such, AgnB might act in processing bodies (P-bodies), non-membrane-bound cytoplasmatic foci (50), which contain proteins implied in RNA decay (51), and which are also sites of retrovirus and retroelement replication. In humans, for example, P-bodies containing the Argonaute Ago2 and the helicase DDX6 facilitate capsid assembly of the retrovirus HIV-1 (52,53). Components of P-bodies found in yeast are essential during the assembly of Ty1 virus-like particles (54,55). Moreover, the Dhh1 and Xrn1 proteins, other components of P-bodies, are necessary for yeast Ty3 retrotransposition (56). Although P-bodies have not yet been defined experimentally in Dictyostelium, a recent report suggested that GFP-tagged and overexpressed AgnB protein localizes in the cytoplasm and in the nucleus (9). Thus, further studies will be required to analyze whether that localization of AgnB is functionally related to its presumed role in the completion of the DIRS-1 retrotransposition cycle.
The replication cycle of DIRS-1 requires the ICR
The retrotransposition assay using a full-length, genetically tagged DIRS-1bsr faithfully mimics the retrotransposition of the natural DIRS-1 from D. discoideum. This assumption is supported by the long experimental history of using retrotransposition marker genes in yeast and mammals (38,39) and also by studies of the non-LTR retrotransposon TRE5-A using the mbsrI gene in D. discoideum (25). One of the unique structural features of DIRS-1-like elements that discriminates them from other retrotransposons is the presence of the ICR near the right LTR (18) (Figure 1B). In the retrotransposition model put forward by Cappello et al., this sequence is the template to restore the complete sequence of the retroelement from its shorter subgenomic transcript, which results eventually in a circular DNA double-strand (18). A comparison of DIRS elements in different species has resulted in the intriguing observation that species-specific trinucleotide sequences, present at the circular junction of the element's termini, might be used to define integration sites (11). The site-specific action of the DIRS YR would result in new integrations, in which the element is flanked by the respective trinucleotide sequence at either end, one resulting from the element and one from the target site. As such, circular and double-stranded DIRS elements would indeed represent a necessary replication intermediate for de novo integration. This, in turn necessitates correctly restored ends of the elements at the ICR. Here, we have shown that the ICR sequence is indeed essential for retrotransposition. The DIRS-1bsr*[-ICR] element exhibited during selection with G418-containing medium a pattern of single-stranded extrachromosomal cDNAs (Supplementary Figure S7) similar to that of the parental construct DIRS-1mbsrI* (Figure 6) with spliced and non-spliced forms of the resistance cassette (Figure 5D). This indicates that an mRNA was successfully expressed from the DIRS-1bsr*[-ICR] element and converted to single-stranded cDNA, which implies no role of the ICR in the initiation of the reverse transcription. So far, is not known how the process of copying the mRNA into cDNA is started and no primer related to this has been determined. However, Poulter and Goodwin noted an internal hairpin at the 3′ end of the mRNA (positions 4792–4805) that could in fact serve to prime the reverse transcription (20). Either way, without the ICR, the DIRS-1 element was not able to complete the retrotransposition cycle and to generate blasticidin-resistant clones. This indicates that the ICR sequence is indeed essential for DIRS-1 retrotransposition, but in a process downstream of first-strand reverse transcription. As the ICR is conserved among the DIRS-1 like elements (57), this notion likely holds true for the entire class of retrotransposons.
Splicing of the intron is influenced by the positioning of the resistance cassette
Both active versions of the tagged retroelement, DIRS-1bsr and DIRS-1bsr* were mobilized despite the different positions of the resistance cassette in the DIRS-1 element (Figures 3 and 4). Nevertheless, the extrachromosomal cDNAs of the tagged elements differed, because a population of reverse transcribed products with a non-spliced form of the mbsrI cassette was observed from the DIRS-1mbsrI* construct, but not from DIRS-1mbsrI (Figure 5D). In DIRS-1mbsrI*, the resistance cassette is embedded between the left LTR and ORF1 (Figure 5A). Transcripts of retroelements often bind proteins and other factors facilitating their replication cycle (58). As such, the presence of the non-spliced form might indicate a competition of proteins involved in different processes, namely splicing and retrotransposition. A presumed occupancy of factors involved in retrotransposition at the 5′ end of DIRS-1 transcripts might also restrict the access of the RNAi machinery. This would help to explain why this segment of DIRS-1 exclusively is void of siRNAs (8,9), which otherwise are found asymmetrically distributed over the rest of the retroelement.
We also observed the non-spliced form for the extrachromosomal cDNA generated from the trDIRS-1mbsrI* construct, that also contains the mbsrI cassette near the 5′ end of DIRS-1 (Figure 5). This observation can easily serve to explain why this strain does not generate blasticidin resistant clones, as the retained intron prevents the expression of the BS resistance. Thus, even in the case of successful DIRS-1 mobilization, which we cannot observe, the resulting strain would not survive the blasticidin treatment.
DIRS-1 proteins act in trans
Retrotransposon mRNAs have a dual function in serving as templates for protein synthesis and reverse transcription (59,60); reviewed in (61). In these processes, proteins can act in trans, as exemplified by the proteins transcribed from Ty1, which can encapsidate the RNA molecule derived from any Ty1 mRNA, regardless of whether they had served as template (44). In contrast, cis-acting proteins bind the same mRNA from which they originated, either during or immediately after translation, and finally leading to the mRNA’s encapsidation (62). The results from the modified ‘helper/donor’ assay (63), in which we used the trDIRS-1mbsrI* construct that lacks the coding capacity for the GAG, RT and RNase H proteins, indicated that the endogenous DIRS-1 reverse transcriptase can act in trans (Figure 5). Furthermore, the successful transformation of the rrpC– strain with the extrachromosomal cDNA of genetically traceable DIRS-1 (Figure 7) suggests that also the other endogenous proteins that mediate retrotransposition in that strain can act in trans. From the blasticidin sensitivity of the wildtype strain (Figure 7), we exclude the possibility that these DNA molecules have been incorporated into the genome of the rrpC– strain via other, non-retrotransposition related mechanisms, e.g. homologous recombination.
The extrachromosomal cDNA of DIRS-1
Treatment with different nucleases showed that the extrachromosomal cDNAs generated from endogenous and traceable versions of DIRS-1 exist as a mixture of single-stranded linear and circular molecules, not only in the rrpC– strain, but also in the agnA– strain (Figure 6, Supplementary Figures S7 and S8). This is consistent with the interpretation of experimental data by Nellen et al., who previously suggested that the extrachromosomal cDNA might be linear despite surviving the Exo I treatment (9). According to the same authors, this particle is a molecule of ∼4500 nt that almost corresponds in size to the original genomic copy (4814 bp). We attempted to amplify the junction of the circular molecules by inverted PCR, which, however, did not yield PCR products (data not shown). This might be explained by the complexity of the sample with linear and circular molecules that furthermore exhibit variable sizes and conformations (Figure 2 and Supplementary Figure S2). Additionally, the structure of the proposed circular junction (Figure 1A) might be complex due to its self-complementarity and thus not be susceptible to PCR amplification. A large fraction of DIRS-1 elements in the D. discoideum genome is fragmented (2), and they might give rise to the shorter extrachromosomal cDNAs we observe (Figure 2B). If these molecules were susceptible to intramolecular ligation, but not further processed into a double strand, they might accumulate, giving rise to the Exo I surviving molecules. It is tempting to speculate that the complete DIRS-1 extrachromosomal cDNAs, in contrast to the shorter ones, might be rapidly converted into double-stranded circular cDNA followed by immediate integration. This scenario would alternatively explain the lack of PCR products covering the circular junction: complete DIRS-1 circular cDNA molecules would be immediately integrated and not available for PCR amplification, and incomplete versions would lack the sequences required for PCR.
Functionally, our observation that isolated extrachromosomal DIRS-1 cDNA transformed into D. discoideum cells can be used as substrates for de novo integrations by endogenous DIRS-1-related proteins, strongly suggests that the cDNA is a true replication intermediate of the DIRS retrotransposition model (Figure 1B) originally proposed by Cappello et al. (18).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the most helpful comments on the manuscript by our colleagues Drs Monica Hagedorn, Fredrik Söderbom and Wolfgang Nellen. Furthermore, we thank the anonymous reviewers for their comments, which significantly improved the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [HA3459/7-1 to C.H., WI1142/14-1 to T.W.]; Tönjes-Vagt-Stiftung (TVS) [TVS-XXXV to C.H.]. Funding for open access charge: TVS and Jacobs University Bremen gGmbH funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Craig N.L., Craigie R., Gellert M., Lambowitz A.M.. Mobile DNA II. 2002; American Society of Microbiology. [Google Scholar]
- 2. Glöckner G., Szafranski K., Winckler T., Dingermann T., Quail M.A., Cox E., Eichinger L., Noegel A.A., Rosenthal A.. The complex repeats of Dictyostelium discoideum. Genome Res. 2001; 11:585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eichinger L., Pachebat J.A., Glöckner G., Rajandream M.A., Sucgang R., Berriman M., Song J., Olsen R., Szafranski K., Xu Q. et al.. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005; 435:43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodier J.L., Kazazian H.H. Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008; 135:23–35. [DOI] [PubMed] [Google Scholar]
- 5. Orgel L.E., Crick F.H.. Selfish DNA: the ultimate parasite. Nature. 1980; 284:604–607. [DOI] [PubMed] [Google Scholar]
- 6. Malicki M., Iliopoulou M., Hammann C.. Retrotransposon domestication and control in Dictyostelium discoideum. Front. Microbiol. 2017; 8:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubin M., Fuchs J., Graf R., Schubert I., Nellen W.. Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis. Nucleic Acids Res. 2010; 38:7526–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiegand S., Meier D., Seehafer C., Malicki M., Hofmann P., Schmith A., Winckler T., Földesi B., Boesler B., Nellen W. et al.. The Dictyostelium discoideum RNA-dependent RNA polymerase RrpC silences the centromeric retrotransposon DIRS-1 post-transcriptionally and is required for the spreading of RNA silencing signals. Nucleic Acids Res. 2014; 42:3330–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boesler B., Meier D., Förstner K.U., Friedrich M., Hammann C., Sharma C.M., Nellen W.. Argonaute proteins affect siRNA levels and accumulation of a novel extrachromosomal DNA from the Dictyostelium retrotransposon DIRS-1. J. Biol. Chem. 2014; 289:35124–35138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wicker T., Sabot F., Hua-Van A., Bennetzen J.L., Capy P., Chalhoub B., Flavell A., Leroy P., Morgante M., Panaud O. et al.. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007; 8:973–982. [DOI] [PubMed] [Google Scholar]
- 11. Goodwin T.J., Poulter R.T.. The DIRS1 group of retrotransposons. Mol. Biol. Evol. 2001; 18:2067–2082. [DOI] [PubMed] [Google Scholar]
- 12. Eickbush T.H., Jamburuthugoda V.K.. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008; 134:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malik H.S., Eickbush T.H.. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol. 1999; 73:5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finnegan D.J. Transposable elements: how non-LTR retrotransposons do it. Curr. Biol. 1997; 7:R245–248. [DOI] [PubMed] [Google Scholar]
- 15. Haren L., Ton-Hoang B., Chandler M.. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 1999; 53:245–281. [DOI] [PubMed] [Google Scholar]
- 16. Lorenzi H.A., Robledo G., Levin M.J.. The VIPER elements of trypanosomes constitute a novel group of tyrosine recombinase-enconding retrotransposons. Mol. Biochem. Parasitol. 2006; 145:184–194. [DOI] [PubMed] [Google Scholar]
- 17. Goodwin T.J., Poulter R.T.. A new group of tyrosine recombinase-encoding retrotransposons. Mol. Biol. Evol. 2004; 21:746–759. [DOI] [PubMed] [Google Scholar]
- 18. Cappello J., Handelsman K., Lodish H.F.. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985; 43:105–115. [DOI] [PubMed] [Google Scholar]
- 19. Cohen S.M., Cappello J., Lodish H.F.. Transcription of Dictyostelium discoideum transposable element DIRS-1. Mol. Cell Biol. 1984; 4:2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poulter R.T., Goodwin T.J.. DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet. Genome Res. 2005; 110:575–588. [DOI] [PubMed] [Google Scholar]
- 21. Poulter R.T.M., Butler M.I.. Tyrosine Recombinase Retrotransposons and Transposons. Microbiol Spectr. 2015; 3:doi:10.1128/microbiolspec.MDNA3-0036-2014. [DOI] [PubMed] [Google Scholar]
- 22. Watts D.J., Ashworth J.M.. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 1970; 119:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Avesson L., Reimegård J., Wagner E.G.H., Söderbom F.. MicroRNAs in Amoebozoa: deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012; 18:1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiegand S., Kruse J., Gronemann S., Hammann C.. Efficient generation of gene knockout plasmids for Dictyostelium discoideum using one-step cloning. Genomics. 2011; 97:321–325. [DOI] [PubMed] [Google Scholar]
- 25. Siol O., Spaller T., Schiefner J., Winckler T.. Genetically tagged TRE5-A retrotransposons reveal high amplification rates and authentic target site preference in the Dictyostelium discoideum genome. Nucleic Acids Res. 2011; 39:6608–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Veltman D.M., Akar G., Bosgraaf L., Van Haastert P.J.. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009; 61:110–118. [DOI] [PubMed] [Google Scholar]
- 27. Church G.M., Gilbert W.. Genomic sequencing. Proc. Natl Acad. Sci. U.S.A. 1984; 81:1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith D.R. Random primed labeling of DNA. Methods Mol. Biol. 1993; 18:445–447. [DOI] [PubMed] [Google Scholar]
- 29. Gaudet P., Pilcher K.E., Fey P., Chisholm R.L.. Transformation of Dictyostelium discoideum with plasmid DNA. Nat. Protoc. 2007; 2:1317–1324. [DOI] [PubMed] [Google Scholar]
- 30. Maniak M., Nellen W.. pISAR, a tool for cloning genomic sequences adjacent to the site of vector integration. Nucleic Acids Res. 1989; 17:4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pilcher K.E., Gaudet P., Fey P., Kowal A.S., Chisholm R.L.. A general purpose method for extracting RNA from Dictyostelium cells. Nat. Protoc. 2007; 2:1329–1332. [DOI] [PubMed] [Google Scholar]
- 32. Fey P., Kowal A.S., Gaudet P., Pilcher K.E., Chisholm R.L.. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007; 2:1307–1316. [DOI] [PubMed] [Google Scholar]
- 33. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kruse J., Meier D., Zenk F., Rehders M., Nellen W., Hammann C.. The protein domains of the Dictyostelium microprocessor that are required for correct subcellular localization and for microRNA maturation. RNA Biol. 2016; 13:1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meier D., Kruse J., Buttlar J., Friedrich M., Zenk F., Boesler B., Förstner K.U., Hammann C., Nellen W.. Analysis of the microprocessor in Dictyostelium: The role of RbdB, a dsRNA binding protein. PLoS Genet. 2016; 12:e1006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiegand S., Hammann C.. The 5′ spreading of small RNAs in Dictyostelium discoideum depends on the RNA-dependent RNA polymerase RrpC and on the dicer-related nuclease DrnB. PLoS One. 2013; 8:e64804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao Z., Kjellin J., Hoeppner M.P., Grabherr M., Soderbom F.. Global characterization of the Dicer-like protein DrnB roles in miRNA biogenesis in the social amoeba Dictyostelium discoideum. RNA Biol. 2018; 15:937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dombroski B.A., Feng Q., Mathias S.L., Sassaman D.M., Scott A.F., Kazazian H.H. Jr, Boeke J.D.. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol. Cell Biol. 1994; 14:4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boeke J.D., Garfinkel D.J., Styles C.A., Fink G.R.. Ty elements transpose through an RNA intermediate. Cell. 1985; 40:491–500. [DOI] [PubMed] [Google Scholar]
- 40. Garfinkel D.J., Boeke J.D., Fink G.R.. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985; 42:507–517. [DOI] [PubMed] [Google Scholar]
- 41. Mellor J., Malim M.H., Gull K., Tuite M.F., McCready S., Dibbayawan T., Kingsman S.M., Kingsman A.J.. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985; 318:583–586. [DOI] [PubMed] [Google Scholar]
- 42. Eichinger D.J., Boeke J.D.. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988; 54:955–966. [DOI] [PubMed] [Google Scholar]
- 43. Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V.. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001; 21:1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Curcio M.J., Garfinkel D.J.. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics. 1994; 136:1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu H., Boeke J.D.. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol. Cell Biol. 1990; 10:2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D.. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004; 119:789–802. [DOI] [PubMed] [Google Scholar]
- 47. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D.. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004; 303:672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ekwall K. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell. 2004; 13:304–305. [DOI] [PubMed] [Google Scholar]
- 49. Willmann M.R., Endres M.W., Cook R.T., Gregory B.D.. The functions of RNA-Dependent RNA polymerases in Arabidopsis. Arabidopsis Book. 2011; 9:e0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sheth U., Parker R.. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003; 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parker R., Sheth U.. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007; 25:635–646. [DOI] [PubMed] [Google Scholar]
- 52. Reed J.C., Molter B., Geary C.D., McNevin J., McElrath J., Giri S., Klein K.C., Lingappa J.R.. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 2012; 198:439–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bouttier M., Saumet A., Peter M., Courgnaud V., Schmidt U., Cazevieille C., Bertrand E., Lecellier C.H.. Retroviral GAG proteins recruit AGO2 on viral RNAs without affecting RNA accumulation and translation. Nucleic Acids Res. 2012; 40:775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dutko J.A., Schafer A., Kenny A.E., Cullen B.R., Curcio M.J.. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 2005; 15:661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Havecker E.R., Gao X., Voytas D.F.. The diversity of LTR retrotransposons. Genome Biol. 2004; 5:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Irwin B., Aye M., Baldi P., Beliakova-Bethell N., Cheng H., Dou Y., Liou W., Sandmeyer S.. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 2005; 15:641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piednoel M., Goncalves I.R., Higuet D., Bonnivard E.. Eukaryote DIRS1-like retrotransposons: an overview. BMC Genomics. 2011; 12:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Checkley M.A., Mitchell J.A., Eizenstat L.D., Lockett S.J., Garfinkel D.J.. Ty1 gag enhances the stability and nuclear export of Ty1 mRNA. Traffic. 2013; 14:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wilhelm M., Heyman T., Boutabout M., Wilhelm F.X.. A sequence immediately upstream of the plus-strand primer is essential for plus-strand DNA synthesis of the Saccharomyces cerevisiae Ty1 retrotransposon. Nucleic Acids Res. 1999; 27:4547–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mellor J., Fulton A.M., Dobson M.J., Roberts N.A., Wilson W., Kingsman A.J., Kingsman S.M.. The Ty transposon of Saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985; 13:6249–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pachulska-Wieczorek K., Le Grice S.F., Purzycka K.J.. Determinants of genomic RNA Encapsidation in the Saccharomyces cerevisiae long terminal repeat retrotransposons Ty1 and Ty3. Viruses. 2016; 8:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Doucet A.J., Hulme A.E., Sahinovic E., Kulpa D.A., Moldovan J.B., Kopera H.C., Athanikar J.N., Hasnaoui M., Bucheton A., Moran J.V. et al.. Characterization of LINE-1 ribonucleoprotein particles. PLos Genet. 2010; 6:e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bolton E.C., Coombes C., Eby Y., Cardell M., Boeke J.D.. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA. 2005; 11:308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.