Abstract
8-Oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) is the most common marker of oxidative stress and its accumulation within the genome has been associated with major human health issues such as cancer, aging, cardiovascular and neurodegenerative diseases. The characterization of the different genomic sites where 8-oxodG accumulates and the mechanisms underlying its formation are still poorly understood. Using OxiDIP-seq, we recently derived the genome-wide distribution of 8-oxodG in human non-tumorigenic epithelial breast cells (MCF10A). Here, we identify a subset of human promoters that accumulate 8-oxodG under steady-state condition. 8-oxodG nucleotides co-localize with double strand breaks (DSBs) at bidirectional and CG skewed promoters and their density correlate with RNA Polymerase II co-occupancy and transcription. Furthermore, by performing OxiDIP-seq in quiescent (G0) cells, we found a strong reduction of oxidatively-generated damage in the majority of 8-oxodG-positive promoters in the absence of DNA replication. Overall, our results suggest that the accumulation of 8-oxodG at gene promoters occurs through DNA replication-dependent or -independent mechanisms, with a possible contribution to the formation of cancer-associated translocation events.
INTRODUCTION
The 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), an oxidatively modified form of the 2′-deoxyguanosine, is the most common marker of oxidative stress (1–3). Accumulation of 8-oxodG correlates with major human health issues such as cancer, aging, cardiovascular and neurodegenerative diseases (1–3). The 8-oxodG is a premutagenic lesion which mispairs with 2′-deoxyadenosine (dA) (3,4). In particular, when the 8-oxodG-dA mismatch is left un-repaired, G:C to A:T transversion mutations may arise during DNA replication (3,4).
The 8-oxodG is preferentially repaired by the OGG1 glycosylase/AP (apurinic/apyrimidinic) lyase-initiated BER (Base Excision Repair) pathway (4,5). OGG1, specifically recognizes and removes the 8-oxodGs from the sugar backbone by cleavage of the N-glycosidic bond and creates an abasic site. This site is subsequently incised by either the OGG1 intrinsic AP lyase activity, that creates 3′ α, β unsaturated aldehyde and 5′-phosphate termini, or by the AP endonuclease 1, APE1, resulting in a single strand DNA (ssDNA) break. Then, the repair process carries on with two alternative ways: (i) the short patch BER with the insertion of a new base by DNA Polymerase beta; (ii) or the long patch BER, which creates a 5′ overhanging ‘flaps’ that is removed by FEN1 (flap structure-specific endonuclease 1). The repair process is finally completed with the ligation of the nicked strand by the DNA ligase I (short patch) or DNA ligase III (long patch) (4,5).
Recent studies demonstrate that the accumulation of 8-oxodG in the G-rich promoters of different genes can stimulate transcription via the BER pathway, inducing a transition in the DNA structure that in turn leads to a G-quadruplex conformation (6,7). Other reports show that the 2′-deoxyguanosine can be oxidized by the hydrogen peroxide produced by the Lysine demethylase 1, LSD1, activity and this in turn determines the recruitment of OGG1 (8–12). Therefore, the ability of 8-oxodG to activate transcription suggests that this oxidatively-modified base may function as a regulatory or epigenetic marker in gene expression processes (13–18).
However, there is a lack of information concerning how cells maintain their steady-state levels of oxidatively-damaged DNA. This knowledge is fundamental for understanding how cells respond to oxidative damage and repair their oxidatively-damaged DNA regions. Several strategies to map endogenous 8-oxodGs have been recently reported by different laboratories (19–24): (i) Burrow's laboratory developed OG-Seq, which uses chemistry to label 8-oxodG with biotin for affinity purification, to identify the 8-oxodG sites in the mouse genome (19); (ii) Wu et al. used Click-code-seq, which incorporates an alkylated nucleotide at the oxidation sites following excision of oxidative damage via click chemistry, to map 8-oxodG in the Saccharomyces cerevisiae genome (20); (iii) Poetsch et al. developed AP-seq, which uses an aldehyde reactive probe (ARP) to label apurinic-sites and 8-oxodG with biotin for affinity, for genome-wide mapping of apurinic sites and 8-oxodGs (21); (iv) Boldogh's laboratory performed chromatin immunoprecipitation-coupled sequencing (ChIP-seq) to analyze the stimulus-induced binding of OGG1, a dedicated reader of 8-oxodG within chromatinized DNA (22); (v) Fang and Zou described the enTRAP-seq protocol, that, by taking advantage of the DNA-trapping ability of a peculiar OGG1 mutant, is able to identify guanosine oxidation in mouse embryonic fibroblasts (23); vi) we recently developed OxiDIP-seq to isolate oxidized DNA fragments in mammalian cells using an 8-oxodG-selective antibody (24). All these strategies, along with their unique benefits and pitfalls, have been recently reviewed in (25).
Collectively, these studies demonstrate that 8-oxodG residues are not uniformly distributed across the genome, suggesting the existence of genomic regions where the formation of oxidatively-damaged DNA is more frequent and/or its repair is less efficient.
Here, we report the characterization of a subset of human promoters accumulating endogenous 8-oxodG in human epithelial cells. We found that oxidatively-damaged promoters show characteristic features of genomic instability (such as CG skewness, G4 structures, R-loops, bidirectional promoters) and that the accumulation of 8-oxodG correlates with RNAPII occupancy and nascent transcription. Oxidatively-damaged promoters tend to accumulate both single and double strand breaks (SSBs and DSBs), as revealed by the co-occurrence of PARP1 and γH2AX, respectively. Oxidized promoters are characterized by recruitment of XRCC4, a marker of the NHEJ pathway, and are associated with translocation breakpoints in breast cancer. Notably, the majority of oxidized promoters showed reduced levels of the oxidatively modified nucleobase upon growth arrest, suggesting the existence of both DNA replication-dependent and -independent events that are responsible for the 8-oxodG formation at gene promoters.
MATERIALS AND METHODS
Cell culture and treatments
MCF10A cells were cultured in 1:1 mixture DMEM-F12 supplemented with 5% horse serum, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 20 ng/ml epidermal growth factor, 100 ng/ml cholera enterotoxin and incubated at 37°C in a humidified atmosphere with 5% CO2 (33). MCF10A-AsiSI cells were arrested in G0 by growth for 2 days in minimal medium containing 1:1 mixture DMEM-F12 supplemented with 5% horse serum (26).
Flow cytometry analysis and 8-oxodG genomic quantification
DNA profiles were analyzed as follows: cells were fixed in 70% ethanol at −20°C, then stained in hypotonic solution of PBS, 50 μg/ml propidium iodide, 50 μg/ml and 0.00125% Nonident-P40 for 30 min at room temperature. For Ki67 quantification cells were, after fixing, permeabilized with 0.1% Triton X-100/PBS and blocked in 5% bovine serum albumin/PBS. Cells were incubated with the primary antibody anti-Ki67 and with the secondary antibody Alexa647 donkey anti-goat (Invitrogen) before propidium iodide staining. For 8-oxodG quantification, cells were fixed and permeabilized as indicated for Ki67, treated with 50 μg/ml RNAse incubated with anti-8-oxodG (Millipore, 1:200 diluted 5% bovine serum albumin/PBS) and the secondary antibodies Alexa488 anti-goat before propidium iodide staining. Cytofluorimetric acquisition and analysis were performed on a Becton Dickinson FACScalibur flow cytometer using FACSDiva and Cyflogic for analysis.
OxiDIP assays in G0 MCF10A cells
The 8-oxodG-enriched genomic fragments from G0 arrested MCF10A cells were obtained as previously described (24). Briefly, genomic DNA was extracted by using Dneasy Blood&Tissue kit (Cat. no. 69504, Qiagen). 10 μg of genomic DNA per immuno-precipitation were sonicated in 100 μl TE buffer (100 mM Tris–HCl pH 8.0, 0.5 M EDTA pH 8.0) using Bioruptor. 4 μg of fragmented DNA were denatured and immuno-precipitated overnight at 4°C with 4 μl of polyclonal antibodies against 8-oxodG (AB5830 Millipore) in a final volume of 500 μl IP buffer (110 mM NaH2PO4, 110 mM NaH2PO4 pH 7.4, 0.15 M NaCl, 0.05% Triton X-100, 100 mM Tris–HCl pH 8.0, 0.5 M EDTA pH 8.0). Then, 50 μl Dynabeads Protein G (Cat. No. 10003D, ThermoFisher Scientific, previously saturated with 0.5% bovine serum albumin diluted in PBS) was added for 3 h at 4°C, under constant rotation, and washed three times with 1 ml washing buffer (110 mM NaH2PO4, 110 mM Na2HPO4 pH 7.4, 0.15 M NaCl, 0.05% Triton X- 100). The immune-complexes were then disrupted by incubation in 200 μl lysis buffer (50 mM Tris–HCl pH 8, 10 mM EDTA pH 8, 1% SDS, 0.5 mg/ml Proteinase K) for 4 h at 37°C, and 1 h at 52°C following addition of 100 μl lysis buffer. MinElute PCR purification kit immuno-precipitated was used to purify the ssDNA (Cat. No. 28004, Qiagen) in a final volume of 72 μl EB buffer (provided in the kit). All the steps of OxiDIP-seq protocol, including the washes of the immunocomplexes, were carried out in low-light conditions, and 50 μM N- tert-butyl-α-phenylnitrone (stock solution: 28 mM in H2O; B7263, Sigma) was added to each Dneasy Blood&Tissue buffer, IP and washing buffers, to preserve the oxidized DNA. The recovered ssDNA was converted to dsDNA using Random Primers DNA Labeling System (Cat. No. 18187-013, ThermoFisher Scientific).
Preparation of OxiDIP sequencing libraries
Library preparation was performed as described in (24) using 2 ng of DIP or Input DNA. Prior to sequencing, libraries were quantified using Qubit dsDNA HS Assay Kit (Invitrogen) and quality-controlled using Agilent Bioanalyzer. 50 bp single-end sequencing was performed using Illumina HiSeq 2000 platform according to standard operating procedures.
Read processing and identification of 8-oxodG-enriched regions in G0 MCF10A cells
Reads were quality checked and filtered with NGS-QC Toolkit (27). Alignments were performed with BWA (28) to hg18 reference genome using default parameters. SAMtools (29) and Bedtools (30) were used for filtering and format conversion steps. The identification of peaks from uniquely mapped reads, after removal of PCR duplicates, was performed using MACS (31) (P < 1e–5 and fold enrichment > 7) and DNA Input was used as control. UCSC genome browser was employed for data visualization. Uniquely mapped reads of the 8-oxodG signal were normalized over genomic input (log2 8-oxodG/Input ratio) using the bamCompare tool from Deeptools suite (32), with SES method (33) as scaling factor. This should account for GC content sequencing bias, which would affect the pull-down samples and the inputs alike, as well as for the bias linked to different amounts of DNA. Metagene analysis and heatmaps were generated using the computeMatrix and plotHeatmap tools from Deeptools suite with default parameters. Signal profile plots were derived using R starting from the matrices generated by the computeMatrix tool.
ChIP-seq, ATAC-seq and DRIP-seq analysis
OGG1 ChIP-seq data was downloaded from GEO (GSE89017); ChIP-seq of PARP1, Pol II-S5P, Pol II-S2P, TOPIIB, γH2AX, NBS1, XRCC4 and RAD51 were downloaded from GEO (GSE93040); ChIP-seq of H3K4me3, H3K4me1, H3K9ac, H3K27ac were from GEO (GSE85158). ChIP-seq reads were quality checked and filtered with NGS-QC-Toolkit. Alignments to hg18 reference genome were performed with BWA using default parameters. SAMtools and Bedtools were used to perform filtering steps and format conversions. Uniquely mapped reads of each signal were normalized over genomic input (log2 ChIP/Input ratio) using the bamCompare tool from Deeptools suite (32), using SES method (33) as scaling factor as described above.
Metagene analysis and heatmaps were generated using computeMatrix and plotHeatmap tools from Deeptools suite with default parameters, signal profile plots were derived using custom script in R starting from the matrices generated by the computeMatrix tool.
Preprocessed and normalized data for ATAC-seq and DRIP-seq were publicly available from GEO (GSE89013 and GSE45530, respectively; Supplementary Table S5). Genomic coordinates were converted to hg18 reference coordinates using UCSC liftover tool. Metagene analysis was performed using computeMatrix tool from the Deeptools suite with default parameters, while plots were generated using custom script in R starting from the data matrix produced by computeMatrix tool.
GRO-seq analysis
GRO-seq data was obtained from Array-Express (E-MTAB-742). FASTQ files were aligned to hg18 reference genome using Bowtie (34) with default parameters and allowing a maximum of two mismatches for the identification of uniquely mapping regions. GRO-seq read quantifications were performed using HTSeq (35) and transcription levels of the human genes were converted into transcripts per million (TPM) of mapped reads. Bidirectional transcription at TSSs was determined analyzing the GRO-seq signal with the computeMatrix tool from the Deeptools suite with default parameters and R was used for plots generation.
RNA-seq analysis
Publicly available RNA-seq data for three quiescent (G0) MCF10A cells (SRR2061414, SRR2061415, SRR2061416) and for two growing MCF10A cells (SRR5136806, SRR5136807) were downloaded from (NCBI-SRA) using SRA toolkit (36) and aligned against hg19 genome reference using STAR aligner (37) with default parameters. For each aligned profile, read counting was performed by mapping aligned reads over hg19 exons and summarizing counts at the gene level using the SummarizeOverlaps function from GenomicAlignments R package. Genes having <1 mapped read (2767) among all samples were discarded from successive analyses. Differential expression analysis between quiescent and growing cells was performed with DESeq function from DESeq2 R package (38), followed by FDR correction for multiple testing. We considered a gene as significantly differentially expressed (DE gene) between G0 and growing cells if the corrected P-value was <0.05 and the absolute log fold change was >1.
BLISS analysis
Data for BLISS (Breaks Labeling In Situ and Sequencing) analyses was obtained from (39) with SRA codes SRA136789 and SRA136790. Illumina single end fastq files were downloaded using SRA toolkit. The variable part of the Unique Molecular Identifiers (UMIs) sequences, constituted by the first eight bases of each sequenced read, was extracted from the reads and annotated on the fastq headers using Umi_tools (40) extract command from the UMI-tools suite. Reads were then quality trimmed using Trimmomatic (41) and the fixed part of the UMI identifier (CATCACGC) was removed using Cutadapt with a 20% error tolerance. Reads were then aligned to hg18 reference genome using BWA-aln with default options. Finally, Umi_tools dedup command was used to remove true PCR duplicates from aligned files. The BLISS signal was then summarized using 100 bp bins for each promoter and the promoter-level signal was defined as the maximum BLISS value computed overall the bins of each promoter by using the computeMatrix tool from Deeptools suite and custom R scripts. Since the BLISS signal is particularly skewed toward extremely high values in sub-centromeric regions, compared to its genomic distribution, we removed sub-centromeric promoters in successive analyses of this particular signal.
Reference, damaged and control gene/promoter datasets
The 21074 human genes used in this study were obtained from the hg18 Refseq genes catalogue removing all of the transcripts having the same genomic coordinates (‘chromosome’, ‘start’ and ‘end’) and keeping those showing alternative TSSs. Genes <2.5 kb in length were also removed. The promoter regions were then defined as the 1 kb regions flanking the TSSs of the above selected (21 074) human genes. Bedtools suite was used to intersect the identified 8-oxodG high-confidence peaks (24) with the above described promoters. The genes containing at least one 8-oxodG high-confidence peak in their promoter region were defined as 8-oxodG-positive (oxidized) promoters in growing cells. Control genes were defined as genes with negligible levels of 8-oxodG at promoter regions in growing cells. In particular, the 8-oxodG signal was binned over the considered human promoters using a 100 bp bin size. Then, the promoter level of 8-oxodG signal was defined as the maximum 8-oxodG value computed overall the bins of each promoter by using the computeMatrix tool from Deeptools suite with default parameters and custom R scripts. Finally, the subset of control genes was defined as the genes where the value of 8-oxodG promoter signal over the Input DNA was lower than 1-fold, corresponding to the bottom 15% of the overall 8-oxodG promoter signal distribution.
Comparison of oxidized promoters identified in G0 and growing cells
Starting from the set of peaks identified from cells in the G0 condition we defined a set of (n = 811) G0 promoters by following the same procedure as for the growing cells (see previous paragraph).
We defined the class of growing-specific promoters as the promoters marked as oxidized in growing cell condition only and the set of G0-specific promoters as the promoters marked as oxidized in G0 condition only. Common oxidized promoters were defined as the intersection between G0 oxidized promoters and growing oxidized promoters. We analyzed the presence of DE genes between growing and G0 condition in these two classes by counting how many genes from the class of common oxidized promoters and the class of growing-specific promoters were also marked as differentially expressed between growing and G0 cells (∼11% and ∼9% respectively).
GC content, CG skew and G4 analysis/enrichment at the promoter regions
For the quantification of GC content, the hg18.gc5Base track was retrieved from UCSC and the average GC content was assessed at oxidized and control promoters using the computeMatrix tool from the Deeptools suite with default parameters.
To investigate the relationship between GC content and 8-oxodG levels, oxidized promoters were analyzed at two resolution levels by first dividing the corresponding regions in bins of 50 bp (Figure 2C) or 10 bp (Figure 2D) and then calculating the average GC content per bin using the computeMatrix tool from the Deeptools suite with default parameters.
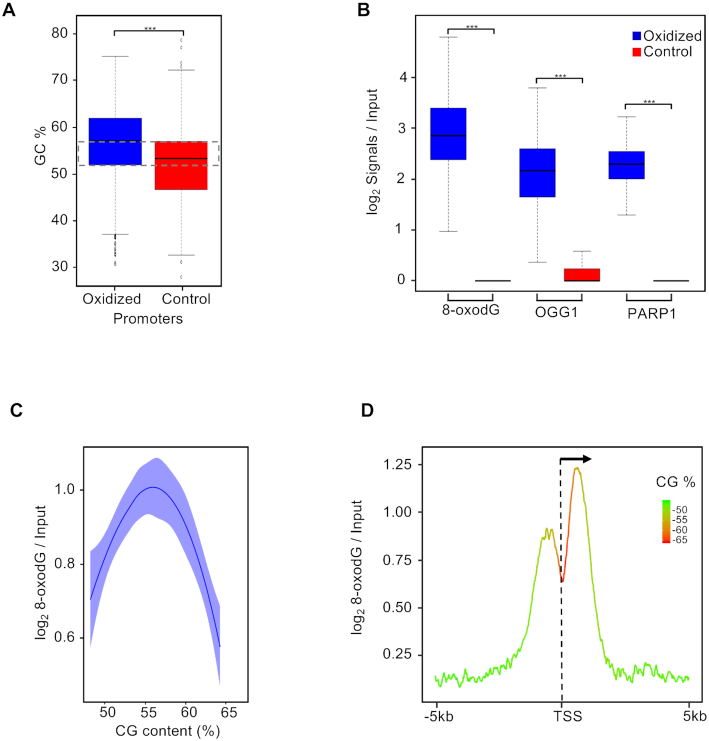
Figure 2.
(A) Box plot showing the GC content (%) distribution measured at oxidized (blue) and control (red) promoters (Bonferroni adjusted pairwise t-test; ***P = 2.2e−16). The dashed rectangle indicates the selected region of the box plots where the oxidized and control promoters have comparable GC content (52–57%); (B) Box plot showing the distribution of the normalized signal of 8-oxodG, OGG1 and PARP1 measured at oxidized (blue) and control (red) promoters with comparable GC content (52–57%) as indicated (Bonferroni adjusted pairwise t-test; ***P < 2.2e–16). (C) Plot showing dependencies between 8-oxodG and GC content at oxidized promoters with a 50 bp resolution; (D) mean-density profile of the normalized 8-oxodG signal and average GC content (color code, as indicated) ±5 kb from TSS of genes with oxidized promoters.
Bedtools was used to determine the overlap between the data sets of i) previously identified human positive CG skew promoters (42), ii) G4-containing regions (43) and iii) oxidized or control promoters. Statistical enrichment was determined by using Fisher's exact test.
Fusion breakpoints association analysis
We referred to 2822 fusion transcripts identified in patients with breast cancer containing 5147 unique fusion breakpoints (44). Translocation breakpoints have been previously reported (39) to prevalently map within introns, hence we tested the association between the oxidized promoter (±2.5 kb from the TSS) introns and the introns-containing breakpoints. The genomic position of hg18 introns was downloaded from the UCSC Genome Browser database. Bedtools suite was used to measure the overlap between datasets and Fisher's exact test was used to test the enrichment of fusion breakpoints in oxidized and control promoters. We considered the set PI of (n = 35 202) unambiguous (5′-3′ splicing site, SS) UCSC hg18 promoter introns. We then defined the set of breakpoints plus (PI B+) introns as those introns from PI containing at least one fusion breakpoint in the 3bp region flanking their 3′ or 5′ splicing site and the set of breakpoints minus (PI B−) introns as their complement to PI. To test the enrichment for breakpoints events among the introns localized in oxidized promoters, we defined the oxidation plus (PI O+) introns as the subset of introns from PI containing at least one 8-oxodG peak and oxidation minus introns (PI O−) as their complement to PI. The enrichment of PI B+ introns for PI O+ was tested using Fisher's exact test.
To test the relative enrichment for breakpoints events between introns localized in oxidized promoter and introns localized in control promoters, we defined the OxiProm introns as the subset of introns from PI located in an oxidized promoter and ContrProm introns as the subset of introns from PI located in at least one control promoter. We then counted the number of introns marked as PI B+ (PI B−) in the OxiProm and ContrProm set respectively and tested their relative enrichment using Fisher's Exact test.
Promoter classification based on transcription direction
Promoters were classified in Convergent and Divergent promoters as follows.
Let Pa and Pb be the genomic coordinates of a pair of TSSs, str(Pa) and str(Pb) respectively be the strand (plus or minus) were Pa and Pb lay, then the couple of promoters (Pa, Pb) was defined as:
Convergent promoters, if str(Pa) = minus and str(Pb) = plus and Pa − Pb > 0 or if str(Pa) = plus and str(Pb) = minus and Pa − Pb < 0;
Divergent promoters, if str(Pa) = minus and str(Pb) = plus and Pa − Pb < 0 or if str(Pa) = plus and str(Pb) = minus and Pa − Pb > 0.
Bedtools was used to determine the overlap between each promoter class and statistical enrichment was determined with Fisher's exact test.
Statistical analysis
Linear correlations between Pol II-S5P/Pol II-S2P/OGG1 signal, transcription levels, and 8-oxodG signals, as well as between biological replicates of Input and OxiDIP-seq experiments, were tested by means of Pearson's correlation test on the gene loci of the oxidized promoters using multiBamSummary and plotCorrelation tools from the Deeptools suite with default parameters. Heatmaps were generated with R. Statistical significance of the observed differences in transcription levels and gene lengths between the gene clusters were evaluated by means of two tailed t-test with heteroskedasticity assumption. Fisher's exact test was used to test the statistical significance of the distribution of each promoter class among the identified gene clusters. Mean value and standard deviation of each genomic signal at the promoter/gene loci was calculated with computeMatrix tool from Deeptools suite, with default parameters, while the standard error was calculated in R. This study was conducted using 0.05 as the significance threshold; all statistical analyses were performed with R version 3.5.
RESULTS
8-oxodG, OGG1 and PARP1 co-localize at the promoter regions of human genes
We recently reported the genome-wide distribution of 8-oxodG in human non-tumorigenic epithelial breast cells (MFC10A) using OxiDIP-seq, a highly sensitive methodology that combines immunoprecipitation, by efficient anti–8-oxodG antibodies, with high-throughput sequencing and found 8-oxodG enrichment within RNAPII promoter regions (24).
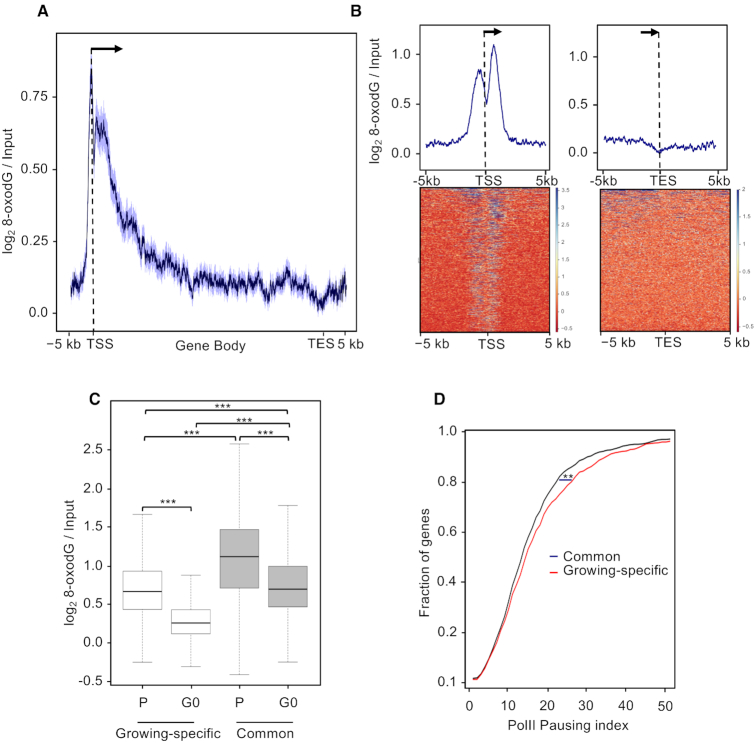
In order to identify 8-oxodG-positive promoters, we intersected the dataset of 8-oxodG high-confidence peaks (52 298) with promoter regions of RefSeq human genes (n = 21 074) and identified 1456 promoters (hereafter called ‘oxidized promoters’; Supplementary Table S1) containing a total of 1459 8-oxodG peaks. Analysis of both raw and normalized 8-oxodG signals over the corresponding gene loci (n = 1456), namely from 5 kb upstream of the Transcription Start Site (TSS) to 5 kb downstream of the transcription end site (TES), confirmed the enrichment of 8-oxodG within the promoter region (Supplementary Figure S1A, S1B and Figure 1A, respectively) and showed 8-oxodG enrichment also within the gene body. This finding is consistent with our previous observations reporting one-third of oxidized promoters (n = 449/1456) to be associated with oxidatively-damaged gene bodies (24). Similar analyses performed in the ±5 kb region flanking the TSS and the ±5 kb region flanking the TES of the same genes (n = 1456) showed a promoter-specific bimodal distribution of 8-oxodG peaking at about +600 and −600 bp from the TSS (Figure 1B).
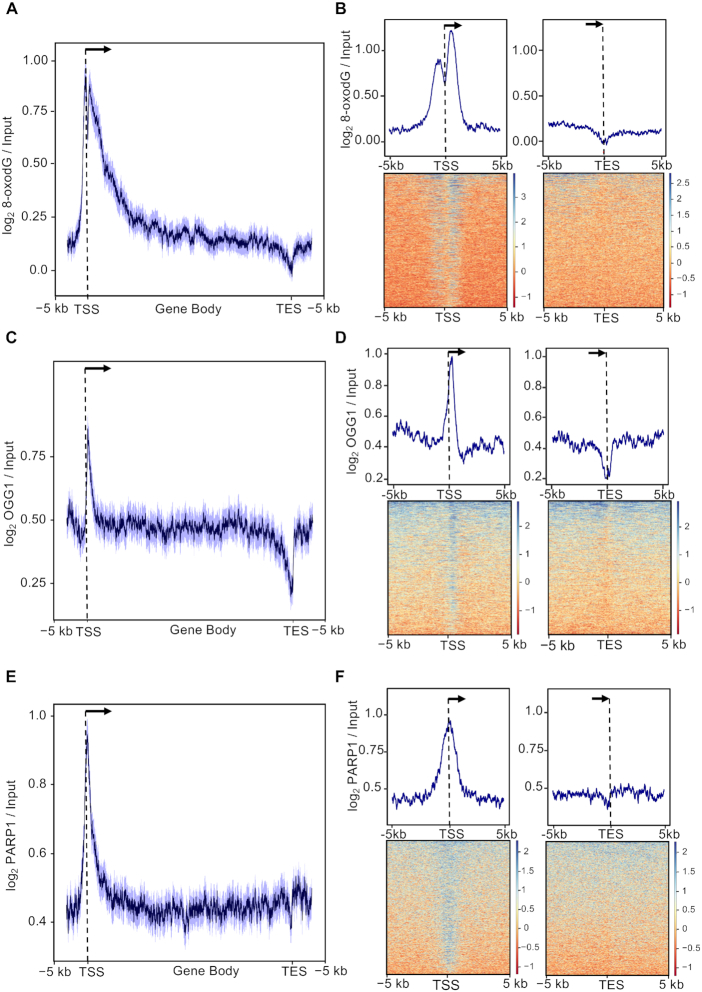
Figure 1.
(A) Distribution of mean 8-oxodG signal profile normalized over the input DNA at gene loci (−5 kb from the transcription start site, TSS, to +5 kb from transcription end site, TES) within oxidized promoters identified in MCF10A cells. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue shaded area. The arrow indicates the direction of transcription. (B) Mean-density profile (top) and heatmap (bottom) of the normalized 8-oxodG signal ±5 kb from TSS (left) or ±5 kb from TES (right) of genes with oxidized promoters. Transcribed region and direction of transcription (arrow), as indicated. (C, D and E, F) Same as in (A) for OGG1 (C, D) and PARP1 (E,F) signal.
We then investigated the presence of OGG1 at oxidized promoters, using a publicly available ChIP-seq dataset (22). First, we measured the OGG1 signal at the MCF10A 8-oxodG peaks (n = 52 298) and their respective flanking regions, and found: (i) highly correlating OGG1 and 8-oxodG levels (Pearson correlation test, r = 0.80, P = 2.2e−16, Supplementary Figure S1C) and (ii) OGG1 enrichment at 8-oxodG peaks (Supplementary Figure S1C). Consistently, analysis of OGG1 signal over the genes marked by oxidized promoters (n = 1456) showed a promoter-specific OGG1 peak that is slightly shifted downstream of the TSS (Figure 1C, D and Supplementary Figure S1D).
Since previous studies showed that PARP1 protein is: (i) a negative modulator of BER pathway when inhibited (45); (ii) a molecular nick-sensor in the repair induced by oxidatively-generated DNA damage (46,47) and (iii) a general DSB sensor (48), we analyzed the distribution of PARP1 signal (39) over the genes with oxidized promoters (n = 1456) and found a clear peak at their TSS (Figure 1E, F). In addition, OGG1 and PARP1 signals showed high correlation (Pearson correlation test, r = 0.85, Supplementary Figure S1E). Finally, we compared oxidized promoters with those showing negligible 8-oxodG levels (Controls; n = 410; Supplementary Figure S2A and B) and found co-presence of OGG1 and PARP1 only in the oxidized set.
All together, these data show that 8-oxodG accumulation at gene promoters is associated with OGG1 and PARP1 recruitment.
Relationship between GC content and 8-oxodG deposition at promoter regions
In order to investigate the role of GC content on 8-oxodG accumulation at gene promoters, we first measured the GC composition of both oxidized and control promoters (Figure 2A) and observed a marginal reduction of GC content in control promoters when compared with oxidized ones (44% versus 48%, median value). Then, we selected two subsets of oxidized and control promoters showing the same GC content (52–57%, indicated by dashed rectangle in Figure 2A) and analyzed the 8-oxodG, OGG1 and PARP1 signals. As shown in Figure 2B, a specific enrichment of all the analyzed signals was found only for oxidized promoters.
Concomitant analysis of GC content and 8-oxodG signal at the oxidized promoter regions (divided in 50 bp window size, Figure 2C) showed that 8-oxodG levels increase as the GC content arises from 50 to 56% and then sharply drop down when GC content arises to 65%. Furthermore, a higher resolution (10 bp windows size) analysis of GC content and 8-oxodG signal at the oxidized promoters (Figure 2D and Supplementary Figure S3) revealed very different profiles at the TSS: while the GC content reached the highest levels (65%) in the promoter region, the 8-oxodG enrichment dropped down, thus generating two peaks, with the peak downstream of the TSS showing the strongest signal.
Thus, it appears that GC content is not the sole determinant of 8-oxodG distribution at gene promoters, suggesting the existence of epigenetic mechanisms involved in the 8-oxodG accumulation in the TSS region.
8-oxodG-positive promoters show genetic features typically associated with genome instability
To investigate on the genetic features associated with oxidized promoters, we analyzed the occurrence of G quadruplexes (G4) and GC skew in oxidized and control promoters.
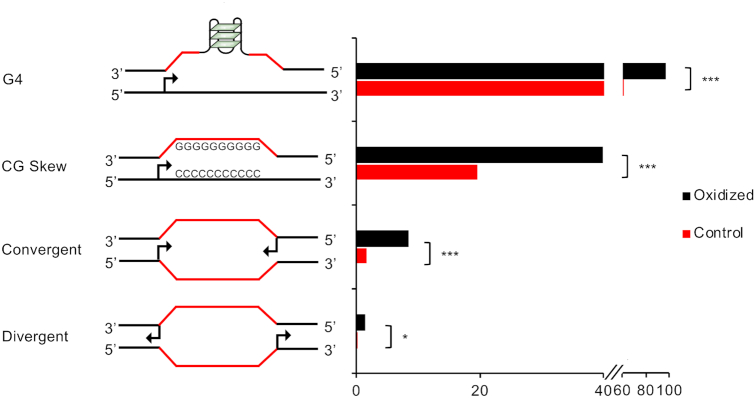
We found that, when compared to controls, oxidized promoters were enriched both in G4 structures (76% versus 96%; P < 2.2e−16) and in skewed CpG island (CGI) promoters (18% versus 40%; P < 2.2e−16) (Figure 3).
Figure 3.
Bar plot reporting the percentage of oxidized and control promoters containing G4 structures, GC skew, convergent or divergent closely spaced (<2.5 kb) TSSs, as indicated (*** P < 2.2e−07, * P = 0.04, Fisher's exact test).
Additionally, oxidized promoters were enriched in bidirectional promoters, either convergently or divergently oriented (P = 2.2e−7 and P = 4.0e−2, respectively; Figure 3), that were previously associated with transcription-related frailty (39,49–54).
In conclusion, oxidized promoters show genetic features predisposing to genomic instability (39,42,43,47,48,55–58).
8-oxodG-positive promoters are associated with R-loops and bidirectional transcription
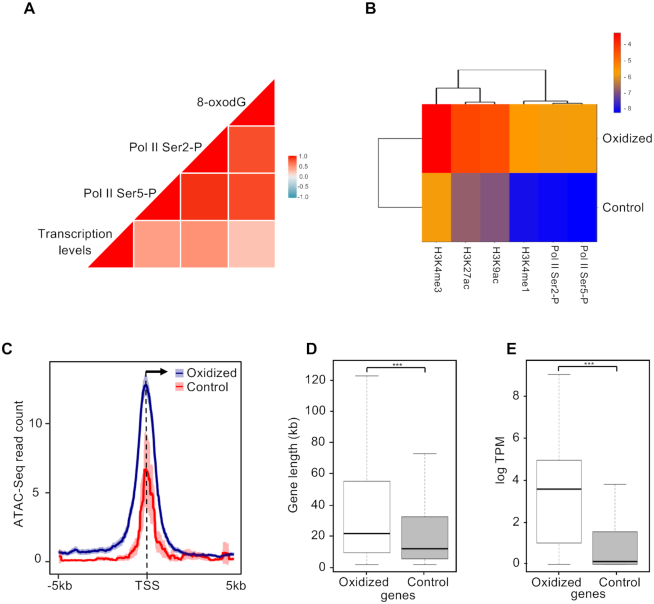
We next asked whether 8-oxodG accumulation is associated with transcriptional activity. In particular, we compared the 8-oxodG signals to either the ChIP-seq signals of Ser5- and Ser2-phosphorylated isoforms of RNA Polymerase II (Pol II-S5P and Pol II-S2P), or gene transcription measured by global run-on sequencing GRO-seq. 8-oxodG levels showed high-to-moderate correlation with Pol II-S5P and S2P (r = 0.9) and transcription levels (r = 0.3) (Figure 4A). Moreover, oxidized promoters were found to be associated with open chromatin markers (H3K4me3, H3K27ac and H3K4ac; Figure 4B), as previously described in (21). Consistently, oxidized promoters showed higher chromatin accessibility, as determined by the analysis of ATAC-seq data (59) (Figure 4C), and higher levels of Pol II-S5P and S2P (Figure 4B and Supplementary Figure S4) than the control promoters. Moreover, genes associated with oxidized promoters, which are longer than control genes (average length of ∼54.2 kb and ∼27.8 kb, respectively; P = 1.0e−09; Figure 4D), showed higher transcription levels than control ones (median transcription levels of 3.6 and 0.1 logTPM, respectively, P < 2.2e−16; Figure 4E).
Figure 4.
(A) Correlation plot reporting Pearson correlation coefficients between transcription (GRO-seq data), 8-oxodG (Oxi-DIP-seq signal), Pol II-S5P and Pol II-S2P (ChIP-seq signals) at genes with oxidized promoters. (B) Heatmap reporting levels of H3K4me3, H3K27ac, H3K9ac, H3K4me1, Pol II-S5P and Pol II-S2P (log2 signal/Input DNA) at the oxidized and control promoters. (C) Mean-density profile of normalized ATAC-seq signal ±5 kb from the TSS of oxidized (blue) and control (red) promoters. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue and light red shaded areas. The arrow indicates the direction of transcription. (D) Box plot showing the distribution of the lengths (bp) of genes with oxidized and control promoters respectively (Bonferroni adjusted pairwise t-test; ***P = 1.0e−9). (E) Box plot showing the transcription levels (GRO-seq; logTPM) distribution of genes with oxidized and control promoters (Bonferroni adjusted pairwise t-test; ***P < 2.2e–16).
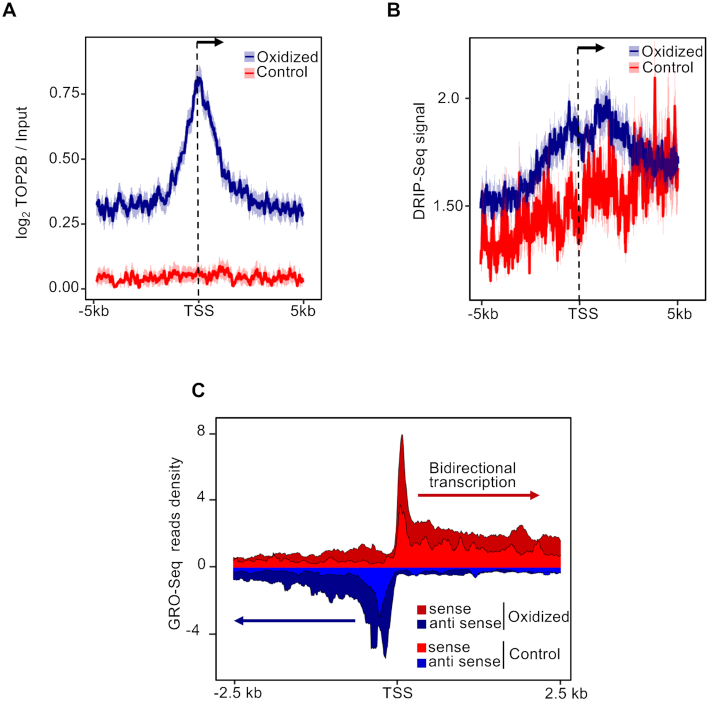
Since transient nicks generated by 8-oxodG removal have been proposed to serve as entry points for Topoisomerase IIB (TOPIIB), that in turn favors the accommodation of the transcription initiation complex (8), we measured TOPIIB levels at oxidized and control promoters, and found that TOPIIB accumulates specifically at the TSS of the formers (Figure 5A and Supplementary Figure S4).
Figure 5.
(A) Mean-density profile of normalized TOPIIB signals ±5 kb from the TSS of oxidized (blue) and control (red) promoters. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue and light red shaded areas. The arrow indicates the direction of transcription. (B) Mean-density profile of DRIP-seq signal ±5 kb from the TSS of oxidized (blue) and control (red) promoters. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue and light red shaded areas. The arrow indicates the direction of transcription. (C) GRO-seq read density ±2.5 kb from the TSS of both sense (red) and antisense (blue) transcripts, within oxidized (dark red and dark blue) and control (light red and light blue) promoters.
Transcription from skewed CGI promoters leads to the formation of stable RNA:DNA hybrid structures typically found at R-loops (60). To investigate the occurrence of RNA:DNA hybrids at oxidized promoters, we analyzed publicly available DRIP-seq signal (42). Oxidized promoters showed higher frequency of endogenous RNA:DNA hybrids than controls (Figure 5B). Notably, peaks of RNA:DNA hybrids, co-localizing with the 8-oxodG peaks at these promoters (compare Figures 1 and 4B), were visible both upstream and downstream of the TSS, thus suggesting the presence of bidirectional transcription, which was confirmed by analysis of GRO-seq data (Figure 5C).
In conclusion, both genetic and epigenetic data show that oxidized promoters are characterized by features typically associated with genomic instability.
DSB formation and DDR activation co-occur at 8-oxodG-enriched promoters
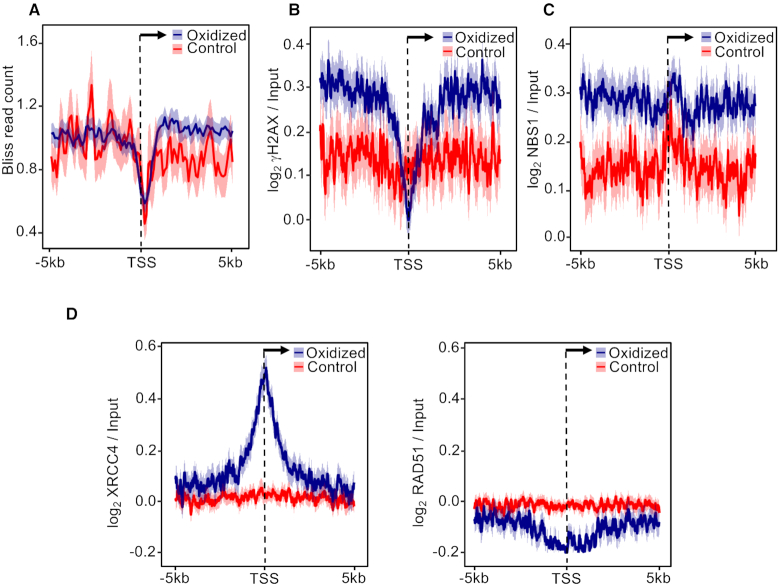
We previously demonstrated that oxidation of guanines in MCF10A cells strongly correlates with H2AX phosphorylation, a marker of DNA Damage Response (DDR) activation, within the gene body of long transcribed genes (24). Since closely opposed oxidative lesions produce clustered SSBs in both DNA strands, thus leading to DSB formation (55,61–63), we asked if endogenous DSBs and markers of activated DDR accumulate at oxidized promoters. In particular, we measured the levels of γH2AX and NBS1 (as markers of DDR activation) and of BLISS signal (as a marker of DSB occurrence) (39). Strikingly, oxidized promoters showed higher BLISS signal than the controls specifically in the region downstream of the TSS (Figure 6A). Consistently, oxidized promoters showed higher γH2AX (Figure 6B and Supplementary Figure S4) and NBS1 (Figure 6C) signals than controls, thus suggesting that accumulation of 8-oxodG at these promoters is associated with DSB formation and DDR activation.
Figure 6.
Mean-density profile of (A) BLISS, (B) γH2AX, (C) NBS1, (D) XRCC4 (left) and RAD51 (right) signals ±5 kb from the TSS of oxidized (blue) and control (red) promoters. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue and light red shaded areas. The arrow indicates the direction of transcription.
DDR activation allows the recruitment of DSB-repair proteins such as RAD51 (involved in homologous recombination) or XRCC4 (non-homologous end-joining, NHEJ) (64,65). Thus, we analyzed MCF10A RAD51 and XRCC4 ChIP-seq signals (39) at the oxidized and control promoters. Only XRCC4 was found specifically at the oxidized promoters (Figure 6D and Supplementary Figure S4), thus suggesting that endogenous DSBs associated with 8-oxodG are processed by the NHEJ pathway in these regions. Since NHEJ is considered an error-prone repair pathway (66), and MCF10A promoter introns (i.e. introns with their 5′ splice site, or SS, mapping <2.5 kb from the TSS) containing endogenous DSBs were recently shown to be enriched in translocation breakpoints identified in breast cancer patients (39), we asked whether oxidized promoters were also associated with these translocation events. For this purpose, we used a dataset containing 2822 fusion transcripts from breast cancer patients (44) and found a total 1654 breakpoint-positive promoter introns, with 549 containing at least one 8-oxodG peak (P < 2.2e−16, Supplementary Table S2; Fisher's exact test). Consistently, oxidized promoter introns were enriched in breakpoint-positive introns compared to control promoter introns (P = 4.7e−10; Fisher's exact test).
Collectively, these data indicate that a fraction of endogenous DSBs associated with oxidized promoters might derive from clustered SSBs formed during 8-oxodG processing and be repaired by NHEJ, thus contributing to the translocation events observed in breast cancers.
Spontaneous accumulation of 8-oxodG at promoters is associated with DNA replication and/or transcription
It has been shown that the binding of Origin Recognition Complex (ORC1), as well as the firing of DNA replication origins (ORIs), mainly occurs at the TSS of transcribed genes (67,68). Thus, we asked whether DNA replication, alone or in combination with transcription, contributes to the accumulation of DNA oxidation at the oxidized promoter regions. To test this hypothesis, we arrested the MCF10A cells in G1 phase (G0) by growth factors withdrawal, as confirmed by FACS analyses of both cell cycle and Ki67 proliferation markers (Supplementary Figures S5A and B). We then measured the global levels of 8-oxodG in cycling and quiescent (G0) MCF10A cells and found that the latter were characterized by a 2-fold reduction of 8-oxodG levels compared to growing cells (Supplementary Figure S5C).
As a next step, we performed OxiDIP-seq in quiescent (G0) MCF10A, using two biological replicates (Pearson correlation test, r = 0.98; Supplementary Figure S6A), and identified 23,641 8-oxodG high-confidence peaks (Supplementary Table S3).
To identify oxidized promoters in quiescent (G0) cells, we intersected the dataset of G0 8-oxodG high-confidence peaks (n = 23,641) with the promoter regions of human RefSeq genes and obtained 811 oxidized promoters (Supplementary Table S4) containing 832 high-confidence 8-oxodG peaks. Analysis of both raw and normalized 8-oxodG signals over gene loci confirmed the presence of DNA oxidation specifically within the promoter regions (Supplementary Figure S6B and Figure 7A, respectively). Furthermore, 8-oxodG signal at promoters showed the same TSS-centered bimodal distribution observed in cycling MCF10A cells (Figure 7B), with only a slight reduction of 8-oxodG signal. Interestingly, while 676 promoters showed persistent 8-oxodG signal upon growth factors withdrawal (i.e. oxidized promoters ‘common’ to growing and quiescent cells), the majority of the oxidized promoters identified at steady-state in the growing cells were lost in the G0 cells (54%; n = 780/1456; ‘growing-specific’ oxidized promoters), as confirmed by the strong 8-oxodG signal drop specifically observed at these sites upon growth arrest (Figure 7C).
Figure 7.
(A) Distribution of mean 8-oxodG signal profile normalized to the input DNA at gene loci (−5 kb from the transcription start site, TSS, to +5 kb from transcription end site, TES) with oxidized promoters identified in quiescent (G0) MCF10A cells. The 95% confidence interval (2 standard error) of the mean is indicated by the light blue shaded area. The arrow indicates the direction of transcription. (B) Mean-density profile (top) and heatmap (bottom) of the normalized 8-oxodG signal ±5 kb from TSS (left) or ±5 kb from TES (right) of genes with oxidized promoters. Transcribed region and direction of transcription (arrow), as indicated. (C) Box plot showing the normalized 8-oxodG signal measured at promoter regions of the growing-specific (white) and persistently (grey) oxidized promoters in proliferating (P) and quiescent (G0) MCF10A cells (Bonferroni adjusted pairwise t-test; ***P < 2.2e−16) (D) Fraction of genes with growing-specific (red) or persistently (black) oxidized promoters showing the indicated Pol II pausing index (calculated using the Pol II-S2P ChIP-seq data) in the proliferating MCF10A cells (Kruskal−Wallis test; **P = 2.5e−2).
We then asked whether genes associated with the oxidized promoters show changes in expression levels upon growth arrest. For this purpose, we measured expression levels in G0 and growing MCF10A cells and found that ∼90% of genes associated with either common or growing-specific oxidized promoters, did not show statistically significant changes in expression levels (see Methods). Interestingly, steady-state 8-oxodG levels at the common promoters were higher than those observed at the growing-specific ones (Figure 7C; compare P of common versus P of growing-specific) and showed smaller 8-oxodG signal drop upon growth arrest (Figure 7C; compare P versus G0 of common with P versus G0 of growing-specific). Altogether, these data suggest that 8-oxodG accumulation at promoters showing persistent 8-oxodG signal in G0 cells is mainly associated with replication-independent transcription-associated events.
RNA:DNA hybrids can form, and/or be stabilized, when the transcriptional elongation complexes are blocked, as it occurs at physiological Pol II pause sites at promoter regions of human genes (69), or following collisions with the slowed replication machinery (70). Since RNA:DNA hybrids are enriched at oxidized promoters, we first calculated the Pol II pausing index of the genes associated with oxidized promoters at the steady state, by computing the ratio of promoter to gene body signals of Pol II-S2P. Interestingly, the Pol II pausing index measured at the genes associated with the growing-specific oxidized promoters was higher than the genes with persistent oxidatively-damaged promoters (Figure 7D; P = 2.5e−2), suggesting a contribution of paused Pol II to the 8-oxodG levels observed in this loci. We then measured the occurrence of DNA replication origins, as revealed by ORC1 binding, at the growing-specific oxidized promoters, and found that they were enriched compared to common promoters (186 versus 130, P = 3.5e−2). Thus, frequent transcription-replication clashes might contribute to the oxidation levels observed at the growing-specific oxidized promoters, possibly as a consequence of the increased sensitivity to DNA oxidation of the persistent ssDNA formed at these sites.
Collectively, we identified two promoter classes which accumulate 8-oxodG through either DNA replication-dependent or replication-independent transcription-associated events.
DISCUSSION
In this work, using the MCF10A diploid human epithelial cells, we identified human promoters containing 8-oxodG peaks, as revealed by OxiDIP-seq, in unperturbed growth conditions. Promoter 8-oxodG enrichment showed a peculiar bimodal distribution, with two sharp peaks immediately upstream and downstream of the TSS, which correlated with RNAPII occupancy and nascent transcription.
In a previous study, Ding et al. (19) reported increased oxidative damage at promoter regions. However, Poetsch et al. (21) by re-analyzing the raw data found no evidence for such an increase. Therefore, we performed a similar re-analysis and, in accordance with Poetsch et al. (21), we found that the OG-seq dataset does not support the hypothesis of increased oxidative DNA damage in promoters (Supplementary Figure S7). These contradicting results may be explained by the different experimental systems and methodologies as recently discussed in (25).
We found a multi-layered relationship between 8-oxodG accumulation and GC content at gene promoters. The comparison analysis of the GC content between oxidized and control promoter showed very little differences, suggesting that the GC content could have only a marginal role in the different accumulation of 8-oxodG between these two promoter classes. However, a higher resolution analysis showed a rather complex association. Indeed, the 8-oxodG levels were strongly decreased within promoter regions with high GC content while promoter regions with similar GC content showed different 8-oxodG levels, thus suggesting a significant contribution of epigenetic mechanisms in the accumulation of oxidatively-modified nucleobases. Indeed, oxidatively-damaged promoters show an excess of G residues downstream of the TSS, on the non-template transcription strand (42,57,58). Such regions, when transcribed, are able to form peculiar secondary structures such as RNA:DNA hybrids as a consequence of the stable pairing between the G-rich nascent RNA and the C-rich template behind the progressing RNA Polymerase (71,72) and/or the paused RNAPII (57). The presence of stable RNA:DNA hybrids at oxidized promoters might lead to prolonged exposure of the G-rich unpaired strand to endogenous reactive oxygen species. In addition, the unpaired DNA strand can form G-quadruplexes (73,74), as shown by enrichment in G4 structures at oxidized promoters. Notably, CG skew, G4 and RNA:DNA hybrids have been shown to promote DNA damage and translocations as well as to hamper the transcription of specific genes (72,75).
Oxidatively-damaged promoters, which show bidirectional transcription (with the closest TSS within 2.5 kb, either upstream or downstream) and are associated with long genes, recruit TOPIIB. This is consistent with the requirement of Topoisomerases for resolution of the topological tension conferred by long transcripts or two Pol II complexes at closely spaced promoters (53,76,77). All these features are known to favor head-on RNAPII collision (51,78) or RNAPII pausing (50) that, due to the increased sensitivity of persistent ssDNA to the accumulation of nucleobase oxidation and/or other kinds of lesion (79), may contribute to DSBs formation and consequently to chromosomal translocations (49,80). Indeed, DSB formation and local genome instability at oxidized promoters may derive from the enzymatic activities of the base-excision repair (BER) pathway, as previously shown in human cells (61,62) and in Escherichia coli, where clusters of DNA damage (characterized by closely opposed base damages that are at least three nucleotides apart and the presence 8-oxodG) inhibit the BER process (81,82). Consistently, we observed recruitment of OGG1, one of the major DNA glycosylases/AP lyases involved in the BER excision activity, to the oxidatively-damaged promoters, suggesting that the cells are able to sense and repair these 8-oxodGs. Accumulation of unresolved SSB intermediates during the processing of 8-oxodGs (55,61–63) is also consistent with the observed recruitment of the nick sensor PARP1 at oxidized promoters. Consequently, processing of clusters of 8-oxodGs on both strands by BER enzymes may convert single strand lesions into DSBs, as shown by H2AX phosphorylation observed at oxidatively-damaged promoters. Together, our data suggest that the repair, perhaps initiated and not completed, of high local levels of endogenous 8-oxodG might contribute to the formation of DSBs via accumulation of unresolved SSBs intermediates and DDR activation.
Notably, the finding of XRCC4 recruitment at the 8-oxodG-positive promoters, in the absence of detectable enrichment of RAD51, strongly argues in favor of a preferential repair of DSBs occurring at these promoters via the NHEJ pathway, consistent with the observed association between 8-oxodG-positive introns and translocation breakpoints. However, since RNAPII is known to form a multiprotein complex with NHEJ proteins (83), and co-occurrence of paused RNAPII-S5P/TOPIIB/XRCC4 is a prerequisite for the generation of chromosomal translocations, we cannot determine the relative contribution of repair intermediates in the processing of clustered 8-oxodGs to the generation of chromosome translocations observed in human cells.
Overall, our data suggest that the repair of clustered 8-oxodG lesions could be a risky approach for the cell to take, at least at specific promoter regions where multiple repair pathways (e.g. BER and NHEJ) are likely to be simultaneously operating.
Finally, mapping of 8-oxodG-enriched regions in G0-arrested cells allowed the identification of two classes of oxidatively-damaged promoters. The majority of those observed at the steady state disappeared, or showed strong 8-oxodG signal reduction, upon growth arrest (growing-specific oxidized promoters), while the remaining, which showed the highest 8-oxodG signals at the steady state, underwent milder signal reduction (persistently oxidized promoters). Interestingly, however, the expression levels of the vast majority of genes associated with both promoter classes did not show significant changes upon growth arrest and this further support an epigenetic role of the 8-oxodGs in gene expression processes (13–18). Thus, while transcription seems to be the main contributing factor to the 8-oxodG accumulation observed at the persistently oxidized promoters, DNA replication-associated events (alone, or in combination with transcription) are responsible for the 8-oxodG accumulation at the growing-specific oxidized promoters. Indeed, all the genetic and epigenetic features of oxidized promoters favor blockage of transcriptional elongation and/or replication complexes, which in turn lead to the formation of persistent ssDNA, which is more sensitive to oxidation than dsDNA (79,84,85). Consistently, we found high levels of physiological Pol II pausing and ORC1 binding in a significant fraction of the growing-specific oxidized promoters, thus suggesting increased probability of DNA replication-transcription clashes.
In conclusion, our findings provide preliminary mechanistic insight into how oxidatively-generated DNA damage at gene promoters may contribute to genome instability in proliferating and postmitotic cells.
DATA AVAILABILITY
Sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE137929.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr E.V. Avvedimento and Dr M. Gottesman for helpful discussions and critical review of the manuscript.
Author contributions: F.G. and G.D.P. performed experiments. G.S., S.A. and S.C. carried out bioinformatics and statistical analyses. L.L. and P.G.P. supervised the study. S.A., B.M., G.I.D., L.L. and G.S. interpreted data and wrote the manuscript. S.A. and B.M. conceived and coordinated the work. All authors read and commented on the manuscript.
Notes
Present address: Giacomo Di Palo, Centre for Molecular Oncology, Barts Cancer Institute, Queen Mary University of London, John Vane Science Centre, London, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
POR Campania FESR 2014–2020 ‘SATIN’ grant (to S.A.); AIRC [IG13173 to B.M.]; EC-FP7-ERC-InMec: 341131 and AIRC IG 17: IG-2017-20162 (to P.G.P.); Computer resource for this study was supported to SA from ELIXIR ITA [ELIX prj10]. Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J.. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003; 17:1195–1214. [DOI] [PubMed] [Google Scholar]
- 2. Cooke M.S., Evans M.D.. 8-Oxo-deoxyguanosine: Reduce, reuse, recycle. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13535–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Valavanidis A., Vlachogianni T., Fiotakis C.. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Heal. Part C. 2009; 27:120–139. [DOI] [PubMed] [Google Scholar]
- 4. Bruner S.D., Norman D.P., Verdine G.L.. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000; 403:859–866. [DOI] [PubMed] [Google Scholar]
- 5. Robertson A.B., Klungland A., Rognes T., Leiros I.. Base excision repair: the long and short of it. Cell. Mol. Life Sci. 2009; 66:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fleming A.M., Zhu J., Ding Y., Burrows C.J.. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-Quadruplex is an on-off switch for transcription. ACS Chem. Biol. 2017; 12:2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fleming A.M., Zhu J., Ding Y., Burrows C.J.. Location dependence of the transcriptional response of a potential G-quadruplex in gene promoters under oxidative stress. Nucleic Acids Res. 2019; 47:5049–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E. V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008; 319:202–206. [DOI] [PubMed] [Google Scholar]
- 9. Amente S., Bertoni A., Morano A., Lania L., Avvedimento E.V., Majello B.. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010; 29:3691–3702. [DOI] [PubMed] [Google Scholar]
- 10. Amente S., Lania L., Avvedimento E.V., Majello B.. DNA oxidation drives Myc mediated transcription. Cell Cycle. 2010; 9:3002–3004. [DOI] [PubMed] [Google Scholar]
- 11. Zuchegna C., Aceto F., Bertoni A., Romano A., Perillo B., Laccetti P., Gottesman M.E., Avvedimento E. V., Porcellini A.. Mechanism of retinoic acid-induced transcription: Histone code, DNA oxidation and formation of chromatin loops. Nucleic Acids Res. 2014; 42:11040–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang S., Zhang J., Zhang Y., Wan X., Zhang C., Huang X., Huang W., Pu H., Pei C., Wu H. et al.. KDM1A triggers androgen-induced miRNA transcription via H3K4me2 demethylation and DNA oxidation. Prostate. 2015; 75:936–946. [DOI] [PubMed] [Google Scholar]
- 13. Li J., Braganza A., Sobol R.W.. Base excision repair facilitates a functional relationship between guanine oxidation and histone demethylation. Antioxid. Redox. Signal. 2013; 18:2429–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ba X., Bacsi A., Luo J., Aguilera-Aguirre L., Zeng X., Radak Z., Brasier A.R., Boldogh I.. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 2014; 192:2384–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zarakowska E., Gackowski D., Foksinski M., Olinski R.. Are 8-oxoguanine (8-oxoGua) and 5-hydroxymethyluracil (5-hmUra) oxidatively damaged DNA bases or transcription (epigenetic) marks. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2014; 764–765:58–63. [DOI] [PubMed] [Google Scholar]
- 16. Fleming A.M., Ding Y., Burrows C.J.. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleming A.M., Burrows C.J.. 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst). 2017; 56:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Olinski R., Gackowski D., Cooke M.S.. Endogenously generated DNA nucleobase modifications source, and significance as possible biomarkers of malignant transformation risk, and role in anticancer therapy. Biochim. Biophys. Acta - Rev. Cancer. 2018; 1869:29–41. [DOI] [PubMed] [Google Scholar]
- 19. Ding Y., Fleming A.M., Burrows C.J.. Sequencing the mouse genome for the oxidatively modified base 8-Oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017; 139:2569–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu J., McKeague M., Sturla S.J.. Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-Code-Seq. J. Am. Chem. Soc. 2018; 140:9783–9787. [DOI] [PubMed] [Google Scholar]
- 21. Poetsch A.R., Boulton S.J., Luscombe N.M.. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis 06 Biological Sciences 0604 Genetics. Genome Biol. 2018; 19:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hao W., Qi T., Pan L., Wang R., Zhu B., Aguilera-Aguirre L., Radak Z., Hazra T.K., Vlahopoulos S.A., Bacsi A. et al.. Effects of the stimuli-dependent enrichment of 8-oxoguanine DNA glycosylase1 on chromatinized DNA. Redox Biol. 2018; 18:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fang Y., Zou P.. Genome-wide mapping of oxidative DNA damage via engineering of 8-oxoguanine DNA glycosylase. Biochemistry. 2019; 59:85–89. [DOI] [PubMed] [Google Scholar]
- 24. Amente S., Di Palo G., Scala G., Castrignanò T., Gorini F., Cocozza S., Moresano A., Pucci P., Ma B., Stepanov I. et al.. Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res. 2019; 47:221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poetsch A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020; 18:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ambrosio S., Di Palo G., Napolitano G., Amente S., Dellino G.I., Faretta M., Pelicci P.G., Lania L., Majello B.. Cell cycle-dependent resolution of DNA double-strand breaks. Oncotarget. 2016; 7:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patel R.K., Jain M.. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012; 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T.. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic. Acids. Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diaz A., Park K., Lim D.A., Song J.S.. Normalization, bias correction, and peak calling for ChIP-seq. Stat. Appl. Genet. Mol. Biol. 2012; 11:doi:10.1515/1544-6115.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B., Trapnell C., Pop M., Salzberg S.. 2C- Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic. Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson ., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dellino G.I., Palluzzi F., Chiariello A.M., Piccioni R., Bianco S., Furia L., De Conti G., Bouwman B.A.M., Melloni G., Guido D. et al.. Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 2019; 51:1011–1023. [DOI] [PubMed] [Google Scholar]
- 40. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ginno P.A., Lim Y.W., Lott P.L., Korf I., Chédin F.. GC skew at the 59 and 39 ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013; 23:1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 44. Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G.W.. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015; 34:4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynolds P., Cooper S., Lomax M., O’Neill P.. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic. Acids. Res. 2015; 30:4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hegde M.L., Izumi T., Mitra S.. Oxidized base damage and single-strand break repair in mammalian genomes: Role of disordered regions and posttranslational modifications in early enzymes. Prog. Mol. Biol. Transl. Sci. 2012; 110:123–153.In. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morales J.C., Li L., Fattah F.J., Dong Y., Bey E.A., Patel M., Gao J., Boothman D.A.. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014; 24:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang G., Liu C., Chen S.-H., Kassab M.A., Hoff J.D., Walter N.G., Yu X.. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018; 46:3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D. et al.. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014; 159:1538–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mayer A., Di Iulio J., Maleri S., Eser U., Vierstra J., Reynolds A., Sandstrom R., Stamatoyannopoulos J.A., Churchman L.S.. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015; 161:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hobson D.J., Wei W., Steinmetz L.M., Svejstrup J.Q.. RNA polymerase II collision interrupts convergent transcription. Mol. Cell. 2012; 48:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pannunzio N.R., Lieber M.R.. Dissecting the roles of divergent and convergent transcription in chromosome instability. Cell Rep. 2016; 14:1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pannunzio N.R., Lieber M.R.. RNA polymerase collision versus DNA structural distortion: twists and turns can cause break failure. Mol. Cell. 2016; 62:327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Core L.J., Waterfall J.J., Lis J.T.. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008; 322:1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cannan W.J., Pederson D.S.. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell. Physiol. 2016; 231:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yan W.X., Mirzazadeh R., Garnerone S., Scott D., Schneider M.W., Kallas T., Custodio J., Wernersson E., Li Y., Gao L. et al.. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017; 8:15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kellner W.A., Bell J.S.K., Vertino P.M.. GC skew defines distinct RNA polymerase pause sites in CpG island promoters. Genome Res. 2015; 25:1600–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim N., Jinks-Robertson S.. Transcription as a source of genome instability. Nat. Rev. Genet. 2012; 13:204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y., Walavalkar N.M., Dozmorov M.G., Rich S.S., Civelek M., Guertin M.J.. Identification of breast cancer associated variants that modulate transcription factor binding. PLoS Genet. 2017; 13:e1006761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chédin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang N., Galick H., Wallace S.S.. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair (Amst.). 2004; 3:1323–1334. [DOI] [PubMed] [Google Scholar]
- 62. Yang N., Chaudhry M.A., Wallace S.S.. Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in γ-irradiated human cells. DNA Repair (Amst). 2006; 5:43–51. [DOI] [PubMed] [Google Scholar]
- 63. Blaisdell J.O., Wallace S.S.. Abortive base-excision repair of radiation-induced clustered DNA lesions in escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7426–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rogakou E.P., Boon C., Redon C., Bonner W.M.. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ghezraoui H., Piganeau M., Renouf B., Renaud J.B., Sallmyr A., Ruis B., Oh S., Tomkinson A.E., Hendrickson E.A., Giovannangeli C. et al.. Chromosomal translocations in human cells are generated by canonical nonhomologous End-Joining. Mol. Cell. 2014; 55:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G.. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013; 23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen Y.H., Keegan S., Kahli M., Tonzi P., Fenyö D., Huang T.T., Smith D.J.. Transcription shapes DNA replication initiation and termination in human cells. Nat. Struct. Mol. Biol. 2019; 26:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 71. Boque-Sastre R., Soler M., Oliveira-Mateos C., Portela A., Moutinho C., Sayols S., Villanueva A., Esteller M., Guil S.. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:201421197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aguilera A., García-Muse T.. Causes of genome instability. Annu. Rev. Genet. 2013; 47:1–32. [DOI] [PubMed] [Google Scholar]
- 73. Lam E.Y.N., Beraldi D., Tannahill D., Balasubramanian S.. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013; 4:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shrestha P., Xiao S., Dhakal S., Tan Z., Mao H.. Nascent RNA transcripts facilitate the formation of G-quadruplexes. Nucleic Acids Res. 2014; 42:7236–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Aguilera A., García-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 76. Joshi R.S., Piña B., Roca J.. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic. Acids. Res. 2012; 40:7907–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T. et al.. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013; 501:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saeki H., Svejstrup J.Q.. Stability, Flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell. 2009; 35:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Clark D.W., Phang T., Edwards M.G., Geraci M.W., Gillespie M.N.. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic. Biol. Med. 2012; 53:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pefanis E., Wang J., Rothschild G., Lim J., Chao J., Rabadan R., Economides A.N., Basu U.. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014; 514:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Demple B. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994; 63:915–948. [DOI] [PubMed] [Google Scholar]
- 82. Harrison L., Hatahet Z., Wallace S.S.. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999; 290:667–684. [DOI] [PubMed] [Google Scholar]
- 83. Chakraborty A., Tapryal N., Venkova T., Horikoshi N., Pandita R.K., Sarker A.H., Sarkar P.S., Pandita T.K., Hazra T.K.. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 2016; 7:13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sagelsdorff P., Lutz W.K.. Sensitivity of DNA and nucleotides to oxidation by permanganate and hydrogen peroxide. Arch. Toxicol. Suppl. 1987; 88:84–88. [DOI] [PubMed] [Google Scholar]
- 85. Cerutti P.A. Friedberg EC, Hanawalt PC. Measurement of Thymidine Damage Induced by Oxygen Radical Species. DNA repair. 1981; NY: 57–68.Decker. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE137929.