Abstract
Biological and chemical DNA fragmentation generates DNA molecules with a variety of termini, including blunt ends and single-stranded overhangs. We have developed a Next Generation Sequencing (NGS) assay, XACTLY, to interrogate the termini of fragmented DNA, information traditionally lost in standard NGS library preparation methods. Here we describe the XACTLY method, showcase its sensitivity and specificity, and demonstrate its utility in in vitro experiments. The XACTLY assay is able to report relative abundances of all lengths and types (5′ and 3′) of single-stranded overhangs, if present, on each DNA fragment with an overall accuracy between 80–90%. In addition, XACTLY retains the sequence of each native DNA molecule after fragmentation and can capture the genomic landscape of cleavage events at single nucleotide resolution. The XACTLY assay can be applied as a novel research and discovery tool for fragmentation analyses and in cell-free DNA.
INTRODUCTION
The termini of fragmented DNA contain clues that can reflect their molecular history and the fragmentation processes that created them (1–5). To characterize DNA termini via high-throughput sequencing, DNA molecules must first be converted into sequencing libraries, which requires ligation of DNA adapters (6). To prepare the template for ligation, most library preparation protocols end-repair the termini of each double-stranded DNA (dsDNA) molecule, commonly using T4 DNA polymerase to fill in 5′ overhangs and digest 3′ overhangs (7). This step renders all input DNA molecules uniformly ready for T/A or blunt-end adapter ligation while erasing the original state of DNA termini. A variation of this procedure uses Tn5 transposase to cleave the DNA template and deliver adapters (8), again erasing the native state of the termini of the input DNA molecules.
For next-generation sequencing (NGS) applications like genome assembly, the loss or alteration of DNA termini is inconsequential. There are cases, however, where fragmentation profiles observed in vitro at DNA termini can reveal the precise effects of DNA digestion or cleavage. For example, blunt ends or overhanging termini can result from breakage following deliberate mechanical or enzymatic manipulation (e.g. restriction digest, sonication, acoustic shearing). In vivo, DNA breaks are formed during both normal and pathological physiological processes, such as from damaging ionizing radiation or, more commonly, through the activation of nucleases during cell-death. Upon cell-death, genomic DNA is degraded and released where it can later be recovered from biofluids as short, cell-free DNA fragments. In particular, the apoptotic endonuclease DFF40/CAD has been shown to cleave dsDNA leaving a mixture of blunt ends and one base pair 5′ overhangs (3). Early necrotic cells, on the other hand, reportedly leave DNA with only 5′ overhangs (2).
Traditional approaches to detect fragmentation and the resulting DNA termini that follow cell death or other nuclease activity rely on standard molecular biology methods (1,2,9–11), e.g. gel electrophoresis/comet assays (10,12,13), in situ ligation and PCR (1), and DNA labeling such as TUNEL (14). These methods are widely available and can, in a few cases, provide nucleotide resolution at termini, but are generally low-throughput and require targeting predefined loci or specific overhang types. Furthermore they often lack sequence information and genome context for the breakpoints.
While NGS methods like BLESS (15), BLISS (15), and EndSeq (16,17) are high-throughput and can be performed without a priori knowledge of cleavage sites, they primarily focus on identifying where double-stranded DNA breaks (DSBs) occur in situ in the genomes of living or cultured cells. These are important questions for investigating how cells repair DNA damage, how the accumulation of DSBs contributes to genomic instability, the effects of programmable nucleases on DNA duplexes (18), or in the case of gene-editing tools, off-target binding sites (19). However, these NGS methods integrate DNA end-repair steps into their library preparation workflows prior to sequencing adapter ligation. The end-repair step may biochemically erase the precise nucleotide position of the break and more importantly never indicates the type of DNA termini produced by the cleavage mechanism(s), 3′ or 5′ overhangs, for example. Furthermore, these methods are not intended for naked DNA substrates that have undergone cleavage, for example, in cell-free or in vitro environments.
Here, we describe a simple ligation-based NGS library approach, XACTLY, that provides comprehensive information about the native-state of the termini of extracted fragmented DNA. By omitting the standard DNA end-repair step, XACTLY libraries encode the type of break at each molecule terminus using custom sequencing adapters. The end result of XACTLY is a high-throughput NGS assay that provides nucleotide resolution of DNA fragmentation across the genome, requiring no knowledge of expected cleavage patterns. Our method for generating Illumina-compatible dsDNA sequencing libraries introduces into the sequencing adapter a unique identifier that encodes the overhang type (3′, 5′ or blunt), length, and sequence at the DNA termini. Here, we first demonstrate the accuracy of XACTLY using (i) a population of control oligos with known single-stranded overhangs and (ii) DNA digestion products from specific restriction enzymes. We then describe the distribution of native termini of dsDNA fragments produced by common methods of enzymatic and mechanical shearing using the Diagenode Bioruptor, DNaseI, and Micrococcal nuclease. Finally, using XACTLY, we show that common procedures for collecting human blood vary in their ability to protect circulating cell-free DNA fragments from degradation by nucleases present in the blood.
MATERIALS AND METHODS
DNA templates for XACTLY experiments
Synthetic control oligos
Synthetic control oligos (Supplementary Table S1) were designed using a random sequence generator at 50% GC content. Sequences matching any known organism in public databases were removed. Each control molecule (n = 12) is a unique 50 bp sequence of double-stranded DNA with one blunt-end, and one 3′ or 5′ single-stranded overhang of random sequence, 1–6 nucleotides in length. Because each control is a unique sequence, it serves as its own barcode indicating the structure of the oligo. Oligos were synthesized using standard desalting purification and duplexed by Integrated DNA Technologies (IDT); all random nucleotides were ‘hand-mixed’ to reduce synthesis bias. Control oligos were pooled together in an equimolar ratio.
NA12878 genomic DNA (gDNA)
NA12878 gDNA was purchased from the Coriell Institute for Medical Research, was prepared for XACTLY ligation in several ways. Mechanical shearing: NA12878 was sheared to an average length of 350 bp using a Bioruptor Pico (Diagenode) and manufacturer's instructions. Sheared DNA was then size selected from 200 to 600 bp using a Pippen Prep dye free 2% gel (Sage Sciences) following manufacturer's instructions. Restriction enzyme digest: 1 μg of NA12878 was digested in a 50 μl reaction using 10 units of MluCI (New England Biolabs) at 37°C for 1 h. Digested DNA was purified using 2× AMPure beads (Beckman Coulter) following manufacturer's instructions. After purification DNA was size-selected from 200 to 600 bp using a Pippen Prep dye free 2% gel (Sage Sciences) and manufacturer's instructions. Enzymatic shearing: DNase I: 1 μg of NA12878 was digested in a 50 μl reaction using 0.01 units of DNase I (New England Biolabs) at 37°C for 10 min and stopped with 0.1 mM EDTA; DNA was purified as above. Micrococcal nuclease: 1 μg of NA12878 was digested in a 50 μl reaction using 2 units of Micrococcal nuclease (New England Biolabs) at 37°C for 5 min and stopped with 0.1 mM EDTA; DNA was purified as above.
Human plasma extraction and cell-free DNA preparation
Whole blood from deidentified donors was obtained for in vitro investigational use from the Stanford Blood Center in Palo Alto, CA. Blood was drawn into one of several tube types (Supplementary Table S2). Blood plasma was extracted from whole blood by spinning the blood collection tubes at 1800 g for 10 min at 4°C. Without disturbing the cell layer, the supernatant was transferred to microcentrifuge tubes under sterile conditions in 2 ml aliquots and spun again at 16 000 g for 10 min at 4°C to remove cell debris and stored at −80°C as 1 ml aliquots. cfDNA was extracted from 1 ml plasma using the QiaAmp ccfDNA kit (Qiagen) following manufacturer's protocol. Purified cfDNA was measured for double-stranded DNA (dsDNA) concentration using the Quant-iT high sensitivity dsDNA Assay Kit and a Qubit Fluorometer (ThermoFisher). Purified cfDNA was analyzed for size distribution using the Agilent TapeStation 4200 and associated D1000 and D5000 high sensitivity products. Cell-free DNA was prepared for XACTLY ligation by dephosphorylation followed by 5′ phosphorylation using the protocol detailed below (Preparing DNA termini for adapter ligation).
Control oligo-blood spike experiments
Approximately 40 ml of whole blood was collected in four blood collection tubes (10 ml each) from a single donor (Supplementary Table S2). Blood from each collection tube was divided into three equal aliquots. To evaluate the effect of blood nucleases on DNA termini, a pool of our control oligos (1 pmol total per ml of whole blood) was added to aliquoted blood collection tubes under sterile conditions. In the case of serum tubes, because coagulation initiates from the time of blood draw, the clot was separated at the start of the experiment and the control oligo pool was added to 1 ml of the supernatant prior to serum preparation. Water and 1X PBS pH7.4 were used as negative controls, substituting for whole blood. The blood product-oligo mixtures (and negative controls) were incubated for 0, 4 or 24 h. Immediately following each time point, blood plasma extraction was performed as above. cfDNA extractions were performed from each spiked plasma sample using the Qiagen QiaAmp ccfDNA kit. The bead binding buffer, proteinase K and magnetic bead volumes were scaled according to the input plasma volume. DNA termini preparation of control-spiked cfDNA was performed as described below, followed by XACTLY adapter ligation, nick repair and amplification.
Ethics statement
This work is not considered human subjects research under the HHS human subjects regulations (45 CFR Part 46).
XACTLY library preparation
Preparing XACTLY sequencing adapters
Each XACTLY adapter contains Illumina sequencer-specific priming sites and a Unique-End-Identifier (UEI)—a barcode sequence that indicates the length and identity (5′ or 3′) of the overhang, if any, present in the original molecule (Supplementary Table S3). The XACTLY adapters were synthesized using standard desalting purification and duplexed by Integrated DNA Technologies (IDT). For purposes of this study the 13 XACTLY adapters include six with 3′ overhangs (1–6 nt in length), six with 5′ overhangs (1–6 nt in length), and a single blunt adapter (i.e. no overhang). XACTLY adapters were not phosphorylated and thus are discouraged from forming dimers. All 13 duplexed XACTLY adapters were pooled in equimolar ratio and prepared for ligation by terminal dephosphorylation using the following 20 μl reaction: 1pmol of pooled XACTLY adapters, 10 units of rapid Shrimp Alkaline Phosphatase (New England Biolabs), 1× CutSmart Buffer, incubated at 37°C for 30 min followed by a 10-min heat inactivation at 65°C. Multiple dephosphorylation reactions were combined over a single QIAquick Nucleotide Removal column (Qiagen) and purified according to manufacturer's instructions. XACTLY adapter molarity was calculated using DNA concentration (Qubit Fluorometric Quantitation) and double-stranded base pair length. Purified XACTLY adapters could be used directly and/or stored at −20°C.
Preparing template DNA termini for adapter ligation
The termini of template DNA molecules, including control oligos, were prepared for adapter ligation. Up to 1 pmol, DNA ends were dephosphorylated in a 20 μl reaction using rapid Shrimp Alkaline Phosphatase (rSAP) (New England Biolabs) and 1× CutSmart buffer incubated at 37°C for 30 min followed by a 10-min heat inactivation at 65°C. DNA was then 5′ phosphorylated by bringing the heat-inactivated 20 μl rSAP reaction up to 40 μl using 20 units of T4 Polynucleotide Kinase (New England Biolabs), and a 5% final concentration of PEG 8000. The phosphorylation reaction was carried out at 37°C for 30 min followed by a 30-min heat inactivation step at 65°C.
Adapter ligation and nick repair
XACTLY ligation consisted of an initial ligation step and a subsequent nick repair ligation step prior to standard NGS library amplification and indexing. First, 0.05 pmol of substrate DNA (control/NA12878/cfDNA) was combined with 1 pmol of XACTLY adapters in a 60 μl ligation reaction with 800 units of T4 DNA ligase (New England Biolabs) and 1× T4 DNA Ligase Buffer, and incubated at 20°C for 1 h, followed by either a 2× AMPure clean for control oligos, or a 1.2× AMPure clean for NA12878 or cfDNA. After DNA purification, DNA was again phosphorylated with 20 units of T4 Polynucleotide Kinase (New England Biolabs) and 1× T4 DNA ligase buffer in a 48.8 μl reaction and incubated at 37°C. After 30 min, 480 units of T4 DNA ligase was added to the reaction and the temperature reduced to 20°C for 15 min. Nick repair was followed by a 2× AMPure bead clean and elution in 20 μl of low TE (10 mM Tris pH 8, 0.1 mM EDTA).
Library amplification and indexing for Illumina sequencing
For Index PCR, 10 μl of purified XACTLY ligated DNA was combined with 1× Kapa HiFi HotStart ReadyMix (Kapa Biosystems) and 0.4 mM final concentrations of the Illumina-compatible IS4 primer and a single indexing primer, as described in (7), in a 50 μl reaction and amplified using the following thermal cycling conditions: 3 min at 98°C for initial denaturation followed by 15 cycles for control/NA12878 or 18 cycles for cfDNA at 98°C for 20 s, 68°C for 30 s, 72°C for 30 s, and finally an elongation step of 1 min at 72°C. After index PCR, DNA was purified with either a 1.5× AMPure clean for control oligos, or a 1.2× AMPure clean for NA12878/cfDNA. For each sequencing DNA library, final molarity estimates were calculated using fragment length distribution and dsDNA concentration (Agilent Tapestation 4200 and Qubit Fluorometric Quantitation unit). Samples were then pooled and run 2 × 150 bp cycles on an Illumina MiSeq benchtop sequencer (following manufacturer's instructions) to a depth of ∼100 000 read-pairs per sample. Step-by-step instructions for XACTLY library preparation method are detailed in the Supplemental Protocol.
Informatic analysis
Read processing
Mapping UEI-barcoded read pairs poses a bioinformatic challenge when template molecules, plus the 7-nt UEIs, are shorter than the sum of the lengths of the forward and reverse reads. The challenge of mapping short fragments exists because each read can extend through its mate's UEI sequence and possibly beyond into the Illumina adapter sequence. Standard practice in studies where short template molecules are expected, such as in the field of ancient DNA, is simultaneously to remove adapter sequences and merge reads (20). Specifically, the process collapses forward and reverse reads into single sequences based on sequence similarity and a minimum amount of overlap while trimming ends of reads that match known Illumina adapter sequences (see SeqPrep https://github.com/jstjohn/SeqPrep). When UEIs are present, however, a merged read that is shorter than or equal to the read length will have a 7-nt UEI on both ends, one of which will be reverse-complemented. The reverse-complemented UEI from R2 has the potential to interfere with read mapping. For this reason, we truncated each forward and reverse read wherever its mate's reverse-complemented UEI sequence was found.
For each read, we first checked for the presence of a known UEI at the start of the forward and reverse read in each pair. UEIs were allowed to contain up to one ‘N’ base, but no other base mismatches were allowed. If both reads had a known UEI sequence, we then checked whether reads merged by searching each sequence for the reverse complement of its mate's UEI. If neither read met this criterion, both reads were output unchanged, since a read can only include adapter sequence if it extends through its mate's UEI sequence. If both reads contained their mate's reverse-complemented UEI sequence, and the positions at which the mates’ UEIs were encountered matched, then both reads were truncated at that position. If the positions did not match, indicating an artifact such as a chimera, both reads were discarded. Across all control oligo experiments, an average of 3.3% of reads per library were discarded this way, compared to 4.3% discarded for lacking a known UEI sequence.
Rather than storing all merged read pairs as collapsed sequences, we kept them as truncated read pairs, so that UEI sequences of mates would not interfere with mapping to reference genomes. For the sake of our control oligo experiments, in which relatively short known sequences were expected, we also stored collapsed sequences for read pairs that merged using our criteria. For such sequences, we allowed the bases within the merged region to contain at most one mismatch (the chosen base at mismatching positions was the base with the higher quality, or a random base in the case of a tie).
To reduce the risk of contamination of our sequencing data by the Illumina sequencing control DNA—phiX—due to index misassignment, we first aligned all of our raw data to the phiX genome using bwa mem (21) with default parameters. Across all experiments, reads aligning to the phiX genome comprised on average 0.28% of the data. We extracted reads that did not map to phiX (samtools) (22) and used these for downstream analyses.
Limiting to reverse reads
Because we found that overhanging adapters were less reliable when encountered on forward (P5) rather than reverse (P7) reads (see Results section on Accuracy), our analyses ignored forward reads that began with an overhanging adapter. Blunt adapters were allowed on both the forward and reverse reads. In all cases, this filtering step was applied only when computing the results of experiments, that is, all reads were included when processing, merging, and aligning, but overhanging adapters on forward reads were not allowed to affect results.
Accuracy, precision and recall measurements in control oligo experiments
When processing control oligos we expected all properly formed sequences to merge using our criteria (see Read processing above), except in cases where control oligos chained together. We defined three ways of assessing control oligo experiments, accuracy, precision and recall.
To measure accuracy, we evaluated how reliably each UEI ligates to its correct target. We generated 17 replicate libraries using an equimolar pool of control oligos containing overhangs corresponding to the overhangs types and lengths available in the UEI adapter pool. Per library accuracy is measured as the proportion of correct ligation events within that library considering only UEIs from reverse reads. The overall accuracy is averaged over 17 libraries.
To measure precision, we computed the proportion of UEI sequences that were ligated to the correct end of the control oligo with the matching overhang. In this case, we did not exclude the ends of control oligos that formed chains, thereby assessing all DNA end available for UEI adapter ligation. For every paired-end read (truncated as described in Read Processing above, but not merged), we aligned the sequence following the UEI to a reference sequence containing all control oligo sequences, separated by runs of ‘N’ bases equal to the length of the longest overhang. The best alignment, allowing up to one mismatch and with ‘N’ matching any base, was used to determine the correct control oligo sequence. Only non-chimeric alignments, i.e. within the coordinates of a single control oligo sequence were considered. Precision was then defined as the proportion of reads for which the UEI at the beginning of the read was followed by the correct type of control oligo end, in the correct orientation.
To measure recall, we computed the proportion of control oligo ends that were correctly identified using our adapters. First, all reads that merged using our criteria (see Read Processing above) were considered. Next, we constructed a reference sequence consisting of all control oligo sequences and their reverse complements, separated by runs of ‘N’ bases equal in length to the longest control oligo overhang. To determine the control oligo type of each merged read, we aligned merged reads to this reference sequence using the Edlib C++ sequence alignment library (23), allowing gaps at the beginning and end of the read in the alignment and allowing up to one base mismatch, letting ‘N’ match any base with no penalty. If the best alignment fell within the coordinates of a single control oligo sequence (a non-chimeric alignment), that control oligo was chosen as the correct sequence. A control oligo was considered correct if the barcode for the correct overhang was ligated to the expected overhang end of the oligo and the barcode for blunt adapters was ligated to the opposite end.
Nucleotide composition of overhang sequence
When assessing the base composition of overhang sequences, we required that all adapters be ligated to the correct type of control oligo. We considered all bases between the end of a UEI sequence and the beginning of a control oligo sequence to be the true sequence of the overhang.
Human DNA data processing
Paired-end reads that remain after filtering were truncated if necessary (see Read Processing above) and aligned to the hg19 human reference genome downloaded from the UCSC genome browser (24). We used bwa aln and bwa sampe (25) with default parameters for alignment, skipping the UEI sequences at the beginning of the reads (-B parameter). Duplicate reads were then removed using samtools rmdup. We counted as mapped only reads that were in proper pairs with a minimum map quality of 20 (samtools view –c –f66 –q20), except in the case of the restriction enzyme experiments, in which we removed the requirement for proper pairing (samtools view –c –f64 –q20) due to the possibility of chaining fragments causing chimeric alignments.
To count UEI types in mapped reads, we scanned through the BAM files using HTSLib's BAM parser (22) and obtained UEI sequences from the BC tag. Overhang sequences were obtained by taking a number of bases from the beginning of each read equal to the overhang length indicated by the UEI.
Downsampling data
To evaluate whether DNA termini profiles were affected by sequencing depth, we re-sequenced a library replicate of Bioruptor sonicated NA12878 gDNA (see DNA templates for XACTLY experiments above) on a Illumina NextSeq. From 6,757,000 read pairs, we down-sampled the raw sequencing data using seqtk sample to increasingly shallow sequencing depths: 5 million, 2 million, 1 million and 0.5 million and 0.1 million read pairs. We also included read data that were generated by sequencing the same library on an Illumina Miseq to a depth of ∼0.2 million reads. We then processed each of the down-sampled sequence files along with the original NextSeq and MiSeq files as described above in Human DNA data processing.
Control oligo spike-in experiments
Some sequencing libraries consisted of human DNA spiked with control oligos. To analyze these libraries, we first processed all sequencing reads as if the libraries contained only human DNA (see Human DNA data processing). Then, non-human sequences were extracted from the alignments to the human reference genome, by selecting unmapped reads and reads with map quality less than 10 (using a custom technique that can re-append barcodes to extracted read sequences). These reads, which consisted mostly of control oligos, were then processed the same way as other control oligo libraries.
RESULTS
Library construction
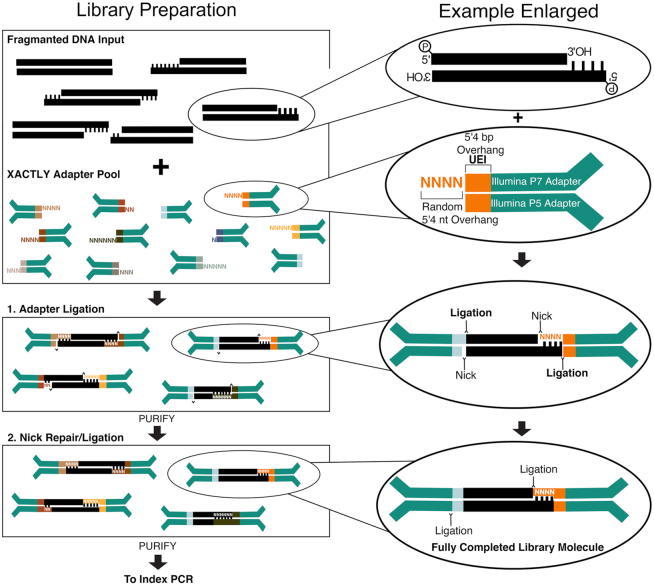
The XACTLY method assays fragmented and degraded dsDNA termini following a standard library preparation workflow, shown schematically in Figure 1. Each XACTLY adapter consists of three parts: the requisite P5/P7 Illumina-based sequencing and index priming sites, a 7 bp Unique End Identifier (UEI) – a barcode encoding the termini type, and a blunt end or a single-stranded overhang that hybridizes and ligates to the substrate's overhang, when present. The overhangs are synthesized with equal proportions of random sequence of length N, here up to six nucleotides (nt) long. XACTLY adapters are included in excess to ensure that every template dsDNA type has access to a compatible adapter. In this way, adapters are introduced in a competitive reaction that provides enough compatible sequences to hybridize with all possible sticky-ended template molecules. The overhangs of the XACTLY adapters create the potential for self-hybridization and ligation; consequently, the adapters are not phosphorylated to prevent adapter dimer formation.
Figure 1.
Schematic overview of the XACTLY method designed for use on Illumina sequencing platforms. Modified and dephosphorylated Illumina Y-adapters contain a 7nt-barcode – called a Unique End Identifier (short multi-colored rectangles) – that denotes a discrete terminus type and length. The length of the overhang is represented by the corresponding number of random bases (shown as Ns), with the exception of a blunt end (no additional bases). Template DNA is first phosphorylated in preparation for adapter ligation but not end-polished, leaving intact any natively sticky or blunt ends. The XACTLY adapter set is then hybridized and ligated to the template DNA. A second round of phosphorylation and ligation seals the nicks present due to the 5′ dephosphorylated adapters. Libraries are then ready for PCR amplification using Illumina-compatible indexing primers (P5 and P7).
During the initial step of XACTLY, we treat the template DNA with polynucleotide kinase to phosphorylate 5′ termini. Otherwise, we do not alter the template DNA termini. Next, we perform a two-step ligation. First, we ligate the 5′ phosphorylated template DNA to a pool of unphosphorylated, UEI-containing XACTLY adapters. This first ligation occurs between the 3′ end of the forward (P5) adapter and the 5′ end of the DNA template. Next, we purify to removes excess, unligated adapters. Finally, we phosphorylate the 5′ ends of the XACTLY adapters and perform a second ligation—this time between the 5′ end of the reverse (P7) adapter and the 3′ end of the DNA template—in order to complete the dsDNA library molecule. Fully formed XACTLY molecules are then indexed and amplified using a universal P5 primer (IS4) and a uniquely indexed P7 primer (7). Following Illumina paired-end sequencing, the UEI is used to classify sequence reads by the type, length and sequence of the overhang.
Assessing the identification of DNA termini
Accuracy and precision
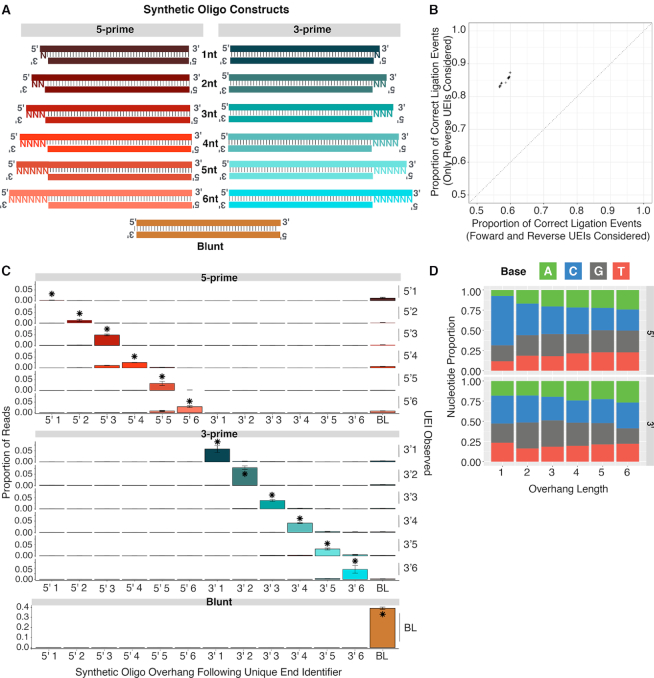
To determine the accuracy of the XACTLY assay we constructed a pool of 12 synthetic double-stranded control oligos, each designed to have a specific length and type (3′ or 5′) of single-stranded overhang. Each control oligo contained a unique 50 bp core and a common structure: blunt terminus on one side, and a 5′ or 3′ overhang of a specific length (1–6 nt) on the other side (Figure 2A). We generated XACTLY libraries using this pool of oligos as template. After sequencing the XACTLY libraries, we used the UEIs on the reverse read (P7) to quantify the assay's accuracy (see Limiting to Reverse Reads) by comparing how often the overhang indicated by the adapter UEI correctly matched the overhang engineered on the dsDNA control template. We limited analysis to reverse reads because the UEI present on the reverse adapter is more accurate in predicting the correct overhang than when we included the UEI present on both adapters (Figure 2B). Our model for explaining this phenomenon is provided in Supplementary Figure S1.
Figure 2.
Accuracy and precision using synthetic double-stranded control oligos. (A) Design schematic of control oligos – one blunt end, an identifiable 50 base pair UEI core, and an overhang of specific length and type on either end (purple rectangles). (B) Across replicate libraries, accuracy in overhang determination is highest when only overhanging UEIs are only considered when in reverse reads (includes blunt end UEIs from both reads), (C) Precision of UEI/control oligo combinations by UEI type. Columns are control oligo overhangs. Rows are UEI adapter overhangs. Asterisks show correct combinations; error bars show one standard deviation across libraries. (D) Base composition of overhangs by overhang type and length. Only overhangs for correct UEI/control oligo combinations are considered; proportions are means across libraries.
We measured the accuracy and precision of XACTLY using the control oligo pools, limiting the dataset to correctly formed library molecules (see Methods). We first calculated how often we captured the correct overhang type and length. As shown in Figure 2B, the overall UEI accuracy over each overhang type and length is 84.94 ± 0.72% (95% C.I.). Next, we measured the precision of each UEI by counting the times each UEI adapter was observed ligated to each type of synthetic oligo. For all overhang lengths and types tested, the most common ligation event is the correct one, with the exception of the 5′ 1-nt overhang (Figure 2C, asterisks indicate the correct ligation event). Overall, 3′ UEIs have higher fidelity than the 5′. UEI errors, when they occurred, often included ligation to the same type, but the wrong length of overhang, most commonly off by one nucleotide. Measurement of recall shows that for every overhang type and length considered, the proportion of control oligo ends ligated to the known correct UEI i.e. the expected UEI is the highest (Supplementary Figure S2).
Base composition
To determine if ligation accuracy or efficiency is influenced by the base composition of the overhangs, we used the sequence data and UEI data to determine the nucleotide sequence of each recovered single-stranded overhang. Due to the architecture of XACTLY libraries, the bases present in the 5′ overhangs derive from the insert template molecule, in this case the control oligo, whereas 3′ overhangs are derived from the DNA overhang of the adapter itself. We observe a uniform distribution of nucleotides for each overhang type and length except for 5′ 1-nt overhangs, where we observe an excess of cytosine (Figure 2D). To evaluate whether this cytosine bias is a product of the oligo synthesis process, we prepared standard end-repaired libraries (NEBNext® Ultra II) of our synthetic control oligos. The end repair step removes 3′ overhangs but fills-in 5′ overhangs via polymerase activity, allowing us to characterize the base composition of the synthetic DNA 5′ overhangs. Within the standard end-repaired libraries of the control oligos, we also observe an elevated read count of 5′ 1-nt cytosines. Therefore, our observations are likely to derive from biases in the custom oligo synthesis and not from biases introduced during ligation.
Detecting known overhangs at low concentrations
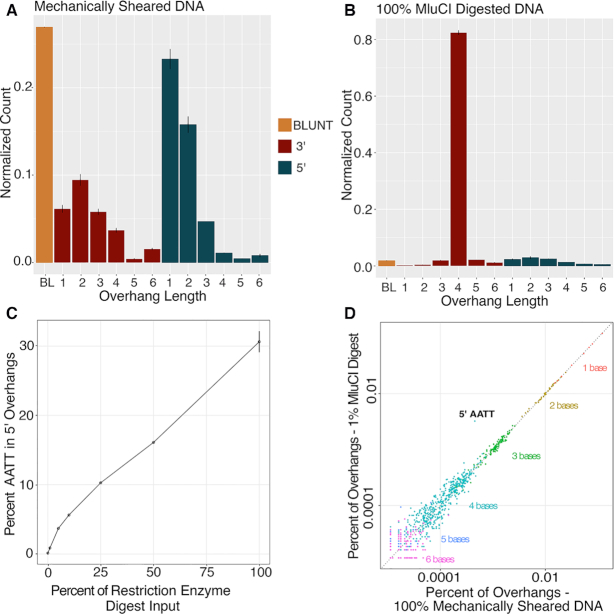
To evaluate the ability of XACTLY to detect the presence of a specific fraction of DNA molecules with a specific terminus type in a background of extraneous DNA molecules, we generated a dilution series by mixing DNA with a single known overhang sequence into a pool of DNA fragments with diverse overhangs. We created the pool of diverse termini by sonication (Diagenode Bioruptor) and size selection of NA12878 genomic DNA (gDNA). We created DNA with a single known overhang by digesting NA12878 gDNA with the restriction endonuclease MluCI, which creates 5′ 4-nt overhangs of the sequence AATT.
First, we sequenced XACTLY libraries generated from the sonicated template and from the MluCI-digested template DNA to characterize termini in both samples. The overhang length profile for the sonicated sample (Figure 3A) shows that sonication shearing of DNA creates a nonrandom profile characterized by a prevalence of blunt DSBs followed by breaks leaving 1- to 4-nt overhangs on both the 5′ and 3′ termini with an excess for 3′ 1-nt and 3′ 2-nt overhangs (Figure 3B). We also verify that neither sequencing depth nor the sequencer used affected observed end profiles (see downsampling data in Methods, Supplementary Figure S3 and Supplementary Table S4). Given our previous observations, 5′1 overhangs are likely underrepresented. As expected for MluCI, the length distribution for the MluCI-digested DNA shows predominately 5′ 4-nt overhangs (Figure 3B).
Figure 3.
Restriction digest experiments. (A) Overhang counts, divided by total per library, across two replicate libraries for 100% mechanically sheared DNA. (B) Overhang counts, divided by total per library, across two replicate libraries for 100% MluCI digested DNA. Values are means across two libraries; error bars show maximum and minimum value. (C) MluCI target sequence abundance with increasing concentration of MluCI. As the percentage of MluCI digested DNA increases (x-axis), so does the frequency of its target sequence (AATT) among 5′ overhang sequences (y-axis). (D) MluCI target sequence is identifiable even in 1% MluCI digested DNA. Points are counts of individual overhang sequences, divided by the sum of all such counts per library. Mean counts across two replicate libraries of only mechanically sheared DNA (x-axis) are shown against mean counts across two replicate libraries of 1% MluCI digested DNA (y-axis). The percent error of each count in 1% MluCI digested DNA was computed, using the count in mechanically sheared DNA as the expected value. All sequences for which this value, rounded to the thousandths place, fell at or above the 99.9th percentile of the distribution are shown labeled. The target sequence (AATT) has the highest percent error (6.2%) and is the only sequence with a value at or above the 99.9th percentile of the distribution (P < 0.001).
We mixed defined amounts of the MluCI-digested DNA to the sonicated DNA sample and generated XACTLY libraries from the pooled mixtures. The pools contained from 1% to 50% MluCI-digested DNA. We calculated the percentage of sequence reads in each library that were attributed to 5′ AATT overhangs (Figure 3C, Supplementary Table S5). Overall, there is concordance between the known MluCI fraction in the library pool and the proportion of termini correctly recovered within the sequenced data and reported as 5′ 4-nucleotide overhangs with the AATT overhang. Libraries with higher amounts of MluCI-digested DNA (100–10%) however show a lower than expected proportion of 5′ AATT overhangs. This is likely due to saturation of the available, compatible overhangs in the XACTLY pool for the specific overhang type, length, and sequence of this restriction product. Thus, for samples that are dominated by a specific species of overhang type, length, and sequence, the percentage detected by XACTLY may be an underestimate. The underestimate is also likely affected by MluCI products ligating to other, similar XACTLY overhangs. For example, 5′ AATTT is enriched in these MluCI spike in experiments. This may be due to mis-ligation or residual exonuclease activity acting on the other strand, creating longer overhangs.
Next, we estimated the concentration of MluCI-digested DNA at which we lose the 5′ AATT signal. We compared the sonicated libraries containing titrated amounts of MluCI-digested DNA to the control sonicated library that contained no spiked 5′ AATT overhangs. Even at the lowest dilution in the series (1% MluCI) we are able to detect an excess of 5′ AATT overhangs over all other overhangs, P < 0.001 (Figure 3D). This observation suggests that the XACTLY assay can detect and describe the presence of overhang motifs that make up <1% of a library.
Overhang profile of common DNA nucleases
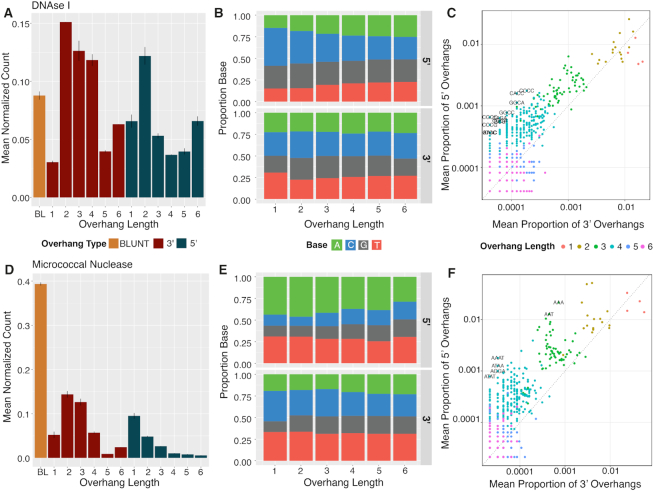
Nucleases are broad group of enzymes utilized in nearly every aspect of molecular biology, including cloning, gene editing, and fragmentation. General nucleases such as DNaseI or Micrococcal Nuclease (MNase) are used extensively in DNA footprinting and chromatin immunoprecipitation assays to interrogate DNA-protein interactions. To describe the digestion preferences of nucleases, researchers employ a variety of low throughput and time-consuming molecular biology, microarray, and sequenced-based methods (26–33). Because XACTLY libraries can assay DNA ends in an unbiased high-throughput manner, we used our assay to characterize with high resolution genomic DNA termini fragmented via nuclease digestion.
We generated XACTLY libraries using the two widely used endonucleases, DNaseI and MNase. The overhang length profile for DNaseI-digested naked DNA (Figure 4A) shows a prevalence of 3′ 2-nt overhangs, as well as 5′ 2-nt to 4-nt overhangs. Of the latter, overhangs of three or more nucleotides are GC-rich (Supplemental Figure 3A). XACTLY adapters used in this study extend only to 6-nt, however the relative abundance of 6-nt overhangs in the overhang length distribution plot suggests that DNaseI creates overhangs greater than 6-nt in length. The overhang base composition profile of DNaseI-digested DNA (Figure 4B) shows a decreasing preference for cytosine in the 5′ overhang as the length of the overhang increases, and a slight preference for 3′1-nt thymine. The base composition of nucleotides upstream of DNaseI cut sites (Supplementary Figure S4A) shows a preference for cutting DNA at A/T sites in the -1 position of 5′ overhangs. Sequences of overhangs created by DNaseI digestion are primarily 5′ GC-rich overhangs (Figure 4C).
Figure 4.
. Overhangs created by enzymatic DNA shearing. Overhang counts, base composition of overhangs, and overhang sequences in a gDNA pool treated with the DNaseI (A–C) and Micrococcal nuclease (D–F). Input DNA for all libraries is human genomic DNA extracted from GM12878 cells. All results are the average of two independent libraries; error bars on the overhang abundance plots show maximum and minimum value. In c and f, we computed the difference between the normalized 3′ and 5′ count of each overhang sequence, divided by the mean of the two counts. Shown in text in C and F are sequences for which this value was significant (above the upper 99th percentile of the distribution).
Conversely, the overhang length plot for MNase-digested DNA (Figure 4D) shows that MNase has a strong preference for the creation of blunt DNA termini (39.5% of the overhang data), with longer overhangs becoming decreasingly likely. When an overhang is produced, MNase shows an overall preference for creating A/T rich 5′ overhangs (Figure 4E, F). The base composition of nucleotides upstream (5′) of MNase cut sites corroborates early studies that observed that MNase is more likely to cut 5′ to A or T than C or G (29,33). As expected, we see a preference for cleaving on the 5′ side of A or T whether the termini were blunt or overhanging. Nucleotides downstream (3′) tend to also preference A/T but following a pyrimidine on the 3′ side of the cut (Supplementary Figure S4B). (Supplementary Table S6).
Recovery of DNA termini generated by in vivo nuclease activity in whole blood
Recently, circulating cell-free DNA (cfDNA) profiling has garnered considerable attention for use in non-invasive prenatal testing and cancer diagnostics (34–36). Obtaining high quality DNA from blood plasma begins with blood collection itself. Blood coagulation or clotting is a process associated with increased nuclease activity (37–40) and the type of blood collection tube (BCT) has been shown to affect the quantity and quality of a cfDNA extract (41). Here, we assayed how common BCTs maintain cfDNA integrity by constructing XACTLY libraries of cfDNA extracted from various BCTs spiked with known control oligos (described above). We included BCTs containing commonly used anticoagulants (Supplementary Table S2), and a control tube without anticoagulants (red top tube; RTT).
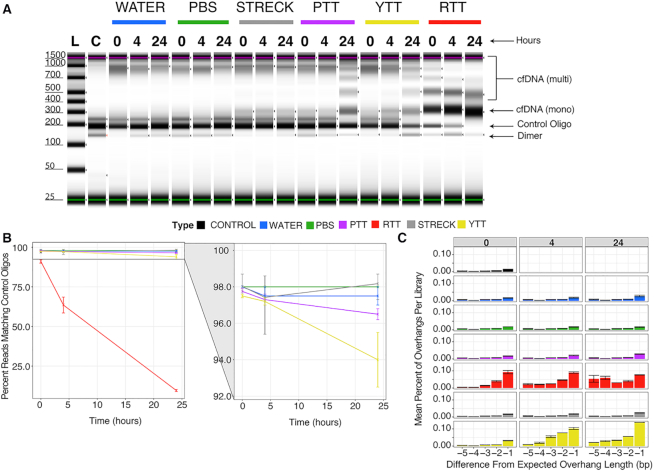
Before extracting plasma (or serum) and isolating cfDNA (see Materials and Methods), we spiked control oligos into each of four tube types. Mixtures of control oligos and cfDNA were extracted at 0, 4 and 24 h following the oligo spike-in and converted into XACTLY libraries. In Streck® BCTs (SBCTs), which contain additives that inhibit nuclease activity and cell lysis, the human cfDNA fragment length profile or abundance does not change over time. Conversely, multi-nucleosome fragments appears in YTTs (anticoagulant – citrate), and PTTs (anticoagulant – potassium EDTA) at 24 h, reminiscent of apoptotic cellular gDNA (42) (Figure 5A). In the RTTs, multi-nucleosome fragments are seen as early as 0 h suggesting that apoptotic processes may be initiated during blood coagulation that are associated with higher nucleolytic activity.
Figure 5.
Effect of blood collection tubes on human cfDNA length and overhang profiles of control oligos. Various blood collection tubes containing whole blood were spiked with synthetic control oligos and incubated for a duration of 0, 4 or 24 h. DNA extracted from each tube contained in a mixture of human cfDNA and synthetic oligos, and was used as template for XACTLY libraries. To control for the effect of time and extraction protocol on DNA termini, the same procedure was performed with control oligos spiked into tubes of water and PBS. (A) Tapestation electrophoresis trace showing the size distribution of XACTLY libraries generated from each tube and timepoint. Control oligo – bands corresponding to control oligos library size ∼200 bp. cfDNA (mono) – mononucleosome band at ∼300 bp; cfDNA (multi) – multinucleosomal bands > 400 bp, demonstrating a ladder-like pattern of DNA fragmentation. cfDNA (multi) appear from 0 to 24 ho in RTT, and at 24 h in YTT and PTT; no multi-nucleosome bands are seen in the Streck tubes; Dimer – small amount of adapter dimers seen in the library. (B) Percentage of reads matching known control oligo sequences at different time points in different collection tube types. Control oligos disappear from RTT and YTT more quickly than control tubes, with the greatest reduction in RTT. Control oligo levels remain stable in Streck tubes, water and PBS, and to a lesser extent in PTT. (C) Differences between expected and observed control oligo overhang lengths demonstrate loss in overhang length in RTT by 4 h and by 24 h in YTT. Shown are differences from expected length between –1 (i.e. chewed back by one base) and –5 (the 99th percentile of the distribution); error bars show maximum and minimum values.
The incubation of whole blood containing control oligos prior to cfDNA extraction allowed us to quantify the amount of loss or change of known DNA ends due to nuclease(s) that remained active following the blood draw. It also provided us with an opportunity to assess the artefactual effect of sample preparation on native DNA breaks by evaluating control oligos spiked into water and PBS negative controls that underwent incubation and extraction. For each time-point and blood collection tube, including negative controls, we evaluated the XACTLY UEIs ligated to the control oligos and compared them with the overhang profile obtained from control oligos libraries that had not undergone incubation or extraction. In this way, we can detect if the original termini type of the control oligo has been altered by nuclease activity present in each sample. The overhang profiles of libraries generated from control oligos spiked into water resemble those of the native control oligo libraries.
We observe little loss or change to the control DNA end profiles in the SBCTs, the PTTs, or the negative controls (Figure 5A–C). In YTTs, which do not contain any known nuclease inhibitors, we observe changes in both 3′ and 5′ overhang profiles. These changes indicate the presence of one or more active circulating exonucleases (Figure 5C). By 24 h, the 3′ overhang signal of control oligos is significantly diminished in YTTs, suggesting that the 3′ to 5′ exonuclease(s) may be more processive than the 5′ to 3′ exonuclease(s). In the RTTs, we observe the complete loss of 3′ overhang counts within 4 h, as well as depletion of true blunt-ends, identified by the generation of new overhangs on formerly blunt-ended molecules. By 24 h, the control oligos are no longer visible in RTTs (Figure 5A, B), as is expected in an environment with high nuclease activity. In sum, these observations show that the XACTLY method discerns changes in overhang patterns of cfDNA and can be used to investigate the effect(s) of circulating nucleases in the blood.
DISCUSSION
Most NGS library preparation methods to date end-repair DNA in preparation for sequencing, obliterating precise signals of dsDNA termini degradation and fragmentation. We have developed a novel NGS assay, XACTLY, to interrogate the termini of fragmented DNA. When applied to a population of extracted dsDNA fragments, XACTLY recovers, tags, and prepares overhanging and blunt-ended molecules for sequencing. Our bioinformatics pipeline then generates a DNA termini profile, reporting the relative abundance of all lengths and types (5′ and 3′) of single-stranded overhangs, if present, on each DNA fragment with an overall accuracy between 80 and 90%. The method simultaneously obtains the nucleotide sequence of each available single-stranded overhang. This feature allows for direct observation of overhang sequences that follow cleavage events as well as the precise genomic location of each fragment end.
Within one day, the XACTLY assay can be performed with as little as 1 nanogram of fragmented DNA template. The analyses described here required low depth of coverage, i.e. fewer than 200 000 read pairs, making XACTLY significantly cost effective as a DNA termini exploration tool when compared to NGS assays that require multi-fold coverage. Together these attributes make XACTLY a more accessible NGS-based assay.
XACTLY can be applied as a novel NGS research and discovery tool to characterize in vitro DNA fragmentation or, in the unique context of cell-free DNA, the products of in vivo genomic DNA fragmentation. The results presented here demonstrate that the XACTLY method corroborates several previous observations about the types of fragmentation products generated by endonucleases. Using XACTLY we characterize the overhangs at dsDNA breaks generated via mechanical or enzymatic cleavage in vitro. We show that XACTLY can identify restriction products even at our lowest titration of 1%.
For exploring broad biological signals on DNA fragment ends, the assay leverages the distinct benefits of high-throughput shotgun sequencing, providing a snapshot of a diverse population of molecules whose end profiles remain consistent even at extremely low depths of sequencing coverage. In contrast, NGS-based assays that involve rare variant detection require orders of magnitude of sequencing data, amounting to hundreds of millions of shotgun reads to achieve 30X coverage of human genome. For applications that want to examine specific allelic variants along with the DNA overhang characteristics, higher DNA inputs and sequencing depth or targeted enrichment is always recommended.
Using XACTLY we show that the degradation of cfDNA ends varies depending on the blood collection tube/anticoagulant used. While the importance of blood collection tubes in cfDNA recovery has been described previously (43–45), XACTLY reveals precisely how best practice sample collection enables recovery of cfDNA fragments that remain unaltered from the time of blood draw. This observation may be of particular interest in clinical and research settings where use of techniques to assay rare tumor-derived cfDNA fragments has surged in recent years.
Although not presented, the XACTLY NGS assay protocol can be easily modified for applications with high molecular weight DNA input such as genomic DNA and chromatin-bound DNA to examine global in situ double-stranded DNA breaks and to characterize effects of altered nuclease activity in response to experimental intervention. With the growing use of CRISPR-Cas9 and other gene-editing tools, XACTLY may also prove useful in the characterization of cleavage products produced specifically by various Cas9 enzymes or other programmable and/or novel nucleases.
Enrichment or depletion strategies are also feasible with minimal modification to the described method. By synthesizing specific overhang adapters, or by modifying the pool to omit or include overhangs of interest, the method can be transformed into a targeted tool for isolating known or suspected DNA termini, whether by type, length and/or sequence.
To date no high-throughput assay exists for identifying the termini of extracted fragmented DNA or single-stranded overhang sequences generated by nuclease activity or other cleavage events. We hope future research applications of the XACTLY assay provide an avenue for understanding the sequence preferences and cleavage products of apoptotic and other human endogenous nucleases, for example, in liquid biopsy and cell-free DNA research. In sum, XACTLY is a new high-throughput, hypothesis-free method for characterizing fragmented DNA termini, adding new information about molecular history for analysis of cell-free and other fragmented DNA molecules.
DATA AVAILABILITY
Raw sequencing data generated from XACTLY libraries of sheared gDNA (NA12878) are available in Bioproject PRJNA579700.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr David Haussler for his advisory role during the development of the method and analysis of data, as well as Jessica Morgan Garcia for her support and assistance performing labwork, and graphic design for figure generation.
Author contributions: R.E.G., K.H., B.S. conceived the investigation; R.E.G., D.H., K.H., C.T., V.R. designed experiments and interpreted results; N.K.S. and V.R. performed data analyses and visualization; C.N., C.T., V.R., K.H., J.K. performed laboratory work; K.H., N.K.S., C.T., V.R. wrote the manuscript with contributions from all co-authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Gordon and Betty Moore Foundation [GBMF 3804 to B.S. and R.E.G.]; NIH SBIR Award [R43CA232935]. Funding for open access charge: Claret Bioscience LLC.
Conflict of interest statement. R.E.G., B.S., K.H., D.H., C.T., V.R., C.N., N.K.S. are cofounders, shareholders, advisors and/or officers/consultants of Claret Bioscience LLC, a genomics company that commercializes sequencing and analysis tools for cfDNA and other nucleic acid sources. The described methods are the subject of active patent applications.
REFERENCES
- 1. Didenko V.V., Hornsby P.J.. Presence of double-strand breaks with single-base 3′ overhangs in cells undergoing apoptosis but not necrosis. J. Cell Biol. 1996; 135:1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Didenko V.V., Ngo H., Baskin D.S.. Early necrotic DNA degradation: presence of blunt-ended DNA breaks, 3′ and 5′ overhangs in apoptosis, but only 5′ overhangs in early necrosis. Am. J. Pathol. 2003; 162:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Widlak P., Li P., Wang X., Garrard W.T.. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 2000; 275:8226–8232. [DOI] [PubMed] [Google Scholar]
- 4. Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S.. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998; 391:43–50. [DOI] [PubMed] [Google Scholar]
- 5. Nagata S., Nagase H., Kawane K., Mukae N., Fukuyama H.. Degradation of chromosomal DNA during apoptosis. Cell Death Differ. 2003; 10:108–116. [DOI] [PubMed] [Google Scholar]
- 6. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer M., Kircher M.. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010; 2010:doi:10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 8. Adey A., Morrison H.G., Asan, Xun X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X. et al.. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010; 11:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins A.R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004; 26:249–261. [DOI] [PubMed] [Google Scholar]
- 10. Singh N.P., McCoy M.T., Tice R.R., Schneider E.L.. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988; 175:184–191. [DOI] [PubMed] [Google Scholar]
- 11. Ansari B., Coates P.J., Greenstein B.D., Hall P.A.. In situ end-labelling detects DNA strand breaks in apoptosis and other physiological and pathological states. J. Pathol. 1993; 170:1–8. [DOI] [PubMed] [Google Scholar]
- 12. Ostling O., Johanson K.J.. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 1984; 123:291–298. [DOI] [PubMed] [Google Scholar]
- 13. Tice R.R., Agurell E., Anderson D., Burlinson B., Hartmann A., Kobayashi H., Miyamae Y., Rojas E., Ryu J.C., Sasaki Y.F.. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 2000; 35:206–221. [DOI] [PubMed] [Google Scholar]
- 14. Gavrieli Y., Sherman Y., Ben-Sasson S.A.. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992; 119:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crosetto N., Mitra A., Silva M.J., Bienko M., Dojer N., Wang Q., Karaca E., Chiarle R., Skrzypczak M., Ginalski K. et al.. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013; 10:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Canela A., Sridharan S., Sciascia N., Tubbs A., Meltzer P., Sleckman B.P., Nussenzweig A.. DNA breaks and end resection measured genome-wide by end sequencing. Mol. Cell. 2016; 63:898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan K.C., Jiang P., Sun K., Cheng Y.K., Tong Y.K., Cheng S.H., Wong A.I., Hudecova I., Leung T.Y., Chiu R.W. et al.. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. PNAS. 2016; 113:E8159–E8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan W.X., Mirzazadeh R., Garnerone S., Scott D., Schneider M.W., Kallas T., Custodio J., Wernersson E., Li Y., Gao L. et al.. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017; 8:15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kircher M. Analysis of high-throughput ancient DNA sequencing data. Methods Mol. Biol. 2012; 840:197–228. [DOI] [PubMed] [Google Scholar]
- 21. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:16 March 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 22. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Šošić M., Šikić M.. Edlib: a C/C ++ library for fast, exact sequence alignment using edit distance. Bioinformatics. 2017; 33:1394–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crawford G.E., Davis S., Scacheri P.C., Renaud G., Halawi M.J., Erdos M.R., Green R., Meltzer P.S., Wolfsberg T.G., Collins F.S.. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006; 3:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sabo P.J., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A. et al.. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006; 3:511–518. [DOI] [PubMed] [Google Scholar]
- 28. Koohy H., Down T.A., Hubbard T.J.. Chromatin accessibility data sets show bias due to sequence specificity of the DNase I enzyme. PLoS One. 2013; 8:e69853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dingwall C., Lomonossoff G.P., Laskey R.A.. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981; 9:2659–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rushizky G.W., Knight C.A., Roberts W.K., Dekker C.A.. A map of the products resulting from the action of micrococcal nuclease on thymus deoxyribonucleic acid and its use as a guide to specificity. Biochem. Biophys. Res. Commun. 1960; 2:153–158. [Google Scholar]
- 31. Dekker C.A. Nucleic acids selected topics related to their enzymology and chemistry. Annu. Rev. Biochem. 1960; 29:453–474. [DOI] [PubMed] [Google Scholar]
- 32. Sulkowski E., Laskowski M. Sr. Mechanism of action of micrococcal nuclease on deoxyribonucleic acid. J. Biol. Chem. 1962; 237:2620–2625. [PubMed] [Google Scholar]
- 33. Drew H.R. Structural specificities of five commonly used DNA nucleases. J. Mol. Biol. 1984; 176:535–557. [DOI] [PubMed] [Google Scholar]
- 34. Aravanis A.M., Lee M., Klausner R.D.. Next-Generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017; 168:571–574. [DOI] [PubMed] [Google Scholar]
- 35. Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A. et al.. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008; 14:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan H.C., Blumenfeld Y.J., Chitkara U., Hudgins L., Quake S.R.. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. PNAS. 2008; 105:16266–16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chitrabamrung S., Rubin R.L., Tan E.M.. Serum deoxyribonuclease I and clinical activity in systemic lupus erythematosus. Rheumatol. Int. 1981; 1:55–60. [DOI] [PubMed] [Google Scholar]
- 38. Ershova E., Sergeeva V., Klimenko M., Avetisova K., Klimenko P., Kostyuk E., Veiko N., Veiko R., Izevskaya V., Kutsev S. et al.. Circulating cell-free DNA concentration and DNase I activity of peripheral blood plasma change in case of pregnancy with intrauterine growth restriction compared to normal pregnancy. Biomed Rep. 2017; 7:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamkovich S.N., Cherepanova A.V., Kolesnikova E.V., Rykova E.Y., Pyshnyi D.V., Vlassov V.V., Laktionov P.P.. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006; 1075:191–196. [DOI] [PubMed] [Google Scholar]
- 40. Patel P.S., Patel B.P., Rawal R.M., Raval G.N., Patel M.M., Patel J.B., Jha F.P., Patel D.D.. Evaluation of serum alkaline DNase activity in treatment monitoring of head and neck cancer patients. Tumour Biol. 2000; 21:82–89. [DOI] [PubMed] [Google Scholar]
- 41. Barra G.B., Santa Rita T.H., de Almeida Vasques J., Chianca C.F., Nery L.F., Santana Soares Costa S.. EDTA-mediated inhibition of DNases protects circulating cell-free DNA from ex vivo degradation in blood samples. Clin. Biochem. 2015; 48:976–981. [DOI] [PubMed] [Google Scholar]
- 42. Wylie A.H. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980; 284:555–556. [DOI] [PubMed] [Google Scholar]
- 43. Kang Q., Henry N.L., Paoletti C., Jiang H., Vats P., Chinnaiyan A.M., Hayes D.F., Merajver S.D., Rae J.M., Tewari M.. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin. Biochem. 2016; 49:1354–1360. [DOI] [PubMed] [Google Scholar]
- 44. Wong D., Moturi S., Angkachatchai V., Mueller R., DeSantis G., van den Boom D., Ehrich M.. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin. Biochem. 2013; 46:1099–1104. [DOI] [PubMed] [Google Scholar]
- 45. Lahiri D.K., Schnabel B.. DNA isolation by a rapid method from human blood samples: effects of MgCl2, EDTA, storage time, and temperature on DNA yield and quality. Biochem. Genet. 1993; 31:321–328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data generated from XACTLY libraries of sheared gDNA (NA12878) are available in Bioproject PRJNA579700.