Abstract
The genetic etiology of premature ovarian insufficiency (POI) has been well established to date, however, the role of long noncoding RNAs (lncRNAs) in POI is largely unknown. In this study, we identified a down-expressed lncRNA HCP5 in granulosa cells (GCs) from biochemical POI (bPOI) patients, which impaired DNA damage repair and promoted apoptosis of GCs. Mechanistically, we discovered that HCP5 stabilized the interaction between YB1 and its partner ILF2, which could mediate YB1 transferring into the nucleus of GCs. HCP5 silencing affected the localization of YB1 into nucleus and reduced the binding of YB1 to the promoter of MSH5 gene, thereby diminishing MSH5 expression. Taken together, we identified that the decreased expression of HCP5 in bPOI contributed to dysfunctional GCs by regulating MSH5 transcription and DNA damage repair via the interaction with YB1, providing a novel epigenetic mechanism for POI pathogenesis.
INTRODUCTION
Premature ovarian insufficiency (POI), defined as a cessation of menstruation prior to the age of 40 years, mainly manifests with irregular menstruation, elevated follicle-stimulating hormone (FSH >25 IU/l), and estrogen deficiency. POI is one of the most common reproductive endocrine disorders, which affects ∼1–2% of women of childbearing age (1,2). POI encompasses three stages in clinic, i.e. occult, biochemical and overt (formerly called premature ovarian failure) stage. Patients with biochemical POI (bPOI) typically have regular menstruation, but elevated FSH levels and reduced fertility (3). The etiology of POI is clinically challenging involving genetic, autoimmune, metabolic, and infectious factors. However, the pathogenesis remains to be elucidated in most cases. Of all the causes, genetic defects account for 20–25% of patients, including chromosomal abnormalities and gene mutations (4). Causative genes of POI have been extensively studied to date in coding regions with alterations to disrupt protein function (5,6). However, the protein-coding region only accounts for 1.5% of the whole human genome (7–9). The noncoding RNAs, including microRNAs, long noncoding RNAs (lncRNAs) and circRNAs, have recently begun to be explored in ovaries and human diseases.
LncRNAs are a group of noncoding RNAs which are longer than 200 nucleotides and poorly conserved among species. Large-scale transcriptome studies suggest that lncRNAs modulate the expression of protein-coding genes by altering chromatin modification, transcription, mRNA decay, protein subcellular localization and other processes (10,11). Also, lncRNAs have been proved to participate in the physiology and pathophysiology of neural, endocrine, and cardiovascular systems (12–14). Recently, 20 differentially expressed lncRNAs have been discovered in ovarian cortex from POI patients, which suggested that lncRNAs may be involved in the maintenance of ovarian function (15). However, the mechanism of lncRNAs contributing to human POI has yet to be determined.
Folliculogenesis is a delicate process regulated by a complex network. The cross-talk between oocyte and somatic cells plays a vital role during follicle development. Granulosa cells (GCs), as one group of ovarian somatic cells, provide essential nutrients, growth factors and secret steroids for oocyte development and maturation (16,17). GCs dysfunction would initiate follicle atresia and apoptosis, and eventually lead to oocyte loss (18,19). Therefore, exploring the role of lncRNAs in GCs will provide a comprehensive understanding of POI pathogenesis.
In the present study, a down-expressed lncRNA HCP5 was identified in GCs from bPOI patients through microarray analyses. Interestingly, its location is adjacent to the MSH5 gene, a known POI-causing gene. Functional experiments further revealed a regulatory role of HCP5 in MSH5 expression via YB1 to affect the DNA damage repair (DDR) progress of granulosa cells. Our findings highlighted HCP5 as a novel transcriptional activator of MSH5, and provided a new epigenetic mechanism for human POI.
MATERIALS AND METHODS
Patients
The study was approved by the Institutional Review Board of Reproductive Medicine of Shandong University, and informed consents were obtained from all participants. Thirty women with bPOI receiving in vitro fertilization or intracytoplasmic sperm injection and embryo transfer (IVF/ICSI-ET) at Reproductive Hospital affiliated to Shandong University were recruited. Inclusion criteria for bPOI consisted of (i) basal serum FSH ≥10 IU/l; (ii) <40 years of age; (iii) with regular menstruation (23–35 days) and (iv) unilateral ovarian antral follicle counts (AFC) <5. Women with known chromosomal abnormalities, history of ovarian surgery, chemotherapy, or radiotherapy were excluded. Thirty-two women with regular menstrual cycles and normal FSH level (<10 IU/l), who sought infertility treatment due to tubal obstruction or male factors were enrolled as controls. Clinical characteristics of all participants are summarized in Table 1.
Table 1.
Clinical characteristics of patients with bPOI and controls
| Variables | Control (n = 32) | bPOI (n = 30) | P value |
|---|---|---|---|
| Baseline characteristics | |||
| Age (years) | 28.97 ± 3.58 | 30.47 ± 3.70 | 0.111a |
| BMI (kg/m2) | 21.58 (19.90, 22.61) | 21.12 (19.30, 24.38) | 0.773b |
| Basal FSH (IU/l) | 6.09 ± 1.25 | 12.47 (11.87, 16.89) | <0.0001b |
| Basal LH (IU/l) | 5.14 ± 1.54 | 5.40 (3.72, 8.62) | 0.46b |
| Basal E2 (pg/ml) | 30.70 (24.38, 43.93) | 28.20 (14.58, 38.85) | 0.155b |
| AMH (ng/ml) | 2.98 (2.10, 4.86) | 0.43 (0.25, 0.61) | <0.0001b |
Values are presented as mean ± SD or median (interquartile range (IQR)) based on distribution. BMI, body mass index; LH, luteinizing hormone; E2, estradiol; AMH, anti-Mullerian hormone.
aStudent's t-test.
bMann–Whitney U-test.
Granulosa cells isolation
The follicular fluid of large follicles (>14 mm) was collected from each participant on the day of oocyte retrieval. After centrifugation, the supernatant was discarded. The precipitates were incubated with hyaluronidase (80 IU/ml) (Sigma, USA) for 30 min at 37°C, and then transferred into lymphocyte separation medium (Solarbio, CHN) and subjected to horizontal centrifugation at 1600 rpm for 10 min. GCs were isolated from the interlayer phase, then washed and resuspended in phosphate buffer saline (PBS). Finally, isolated GCs were snap-frozen and stored at −80°C until used.
Microarray analysis
Total RNA was extracted from granulosa cells of 10 bPOI and 10 control women using TRIzol Reagent (TaKaRa, China). RNA quantity and quality were measured by NanoDrop ND-1000, and RNA integrity was assessed by standard denaturing agarose gel electrophoresis. LncRNA expression profile was detected by Arraystar Human LncRNA Microarray V3.0 (GSE135697) (Arraystar, USA). Microarray scanning and data analyses were conducted by KangChen Bio-tech (China).
RNA isolation and RT-qPCR analysis
RNA isolation from GCs or cell lines was performed following standard protocols as described above. All mRNAs and lncRNAs were reverse transcribed using PrimeScript™ RT reagent Kit (TaKaRa, CHN) and followed by quantitative RT-PCR labeled by SYBR Green Master Mix (TaKaRa, CHN). The primers for qRT-PCR are listed in Supplementary Table S1.
Cell culture and transfection
The human granulosa-like tumor cell line KGN (20) was gifted from RIKEN BioResource Center in Japan. Another human granulosa-like tumor cell line COV434 was obtained from Prof. Ying Xu of Nanjing University in China. The luteinized granulosa cell line SVOG was a gift from Prof. Peter C.K. Leung of University of British Columbia in Canada to The Chinese University of Hong Kong-Shandong University (CUHK-SDU) Joint Laboratory on Reproductive Genetics. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM)/F-12 (KGN and SVOG) (HyClone, USA) or DMEM/High Glucose (COV434) supplemented with 10% fetal bovine serum (Biological Industries, ISV), and 1% penicillin-streptomycin (Invitrogen, USA) at 37°C in a humidified atmosphere of 5% CO2. Transfection of specific siRNAs for MSH5 or ILF2 and negative control (Genepharma, CHN) was performed using Lipofectamine 3000 Reagent (Invitrogen, USA) according to the manufacturer's protocol. After transfection, cells were incubated for 48 hours before further treatment. The sequences of all siRNAs are provided in Supplementary Table S2.
shRNA knockdown
The lentivirus vector carrying HCP5-specific shRNA with puromycin resistance gene (shRNA-HCP5) and negative control (shRNA-NC) lentivirus vector were synthesized and packaged into lentivirus particles by HanBio biotechnology (CHN). For infection, lentivirus was added to KGN, COV434 and SVOG cell culture medium, and positive cells were selected by 2 mg/ml puromycin for 7 days.
Western blot
Cells were washed twice with ice-cold PBS. After adding sodium dodecyl sulfonate (SDS) lysis buffer (Beyotime, CHN) including proteinase inhibitor cocktail (Cell Signaling Technology, USA), lysates were collected immediately according to the manufacturer's instruction. Approximately 30 μg proteins in each lane were separated by SDS polyacrylamide gels and subsequently transferred onto polyvinylidene fluoride membranes (Millipore, USA). Membranes were blocked in 5% non-fat milk for 1 h at room temperature and then incubated with specific primary antibodies following horseradish peroxidase-conjugated secondary antibodies (Proteintech Group, China). An ECL chemiluminescence kit (Millipore, USA) was used to detect immunoreactive protein bands by ChemiDoc MP Imaging System (BIO-RAD, USA). The details of antibodies are listed in Supplementary Table S3.
Cytoplasm/nucleus fractionation
The separation of cytoplasmic and nuclear RNA was carried out using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Canada) according to the manufacturer's instructions. RNA was purified and reverse-transcribed following standard protocols as described above from total and individual fractions, and then subjected to qPCR analysis. Expressions of HCP5 along with cytoplasmic and nuclear references in individual fractions were normalized to their expression levels in the input RNA, which was set as 100% (21). The proteins were extracted from each fraction using the PARIS™ Kit (Invitrogen, USA) according to the manufacturer's instructions.
Fluorescence in situ hybridization
A mix of probes targeting HCP5 was synthesized and labeled with Cy3 (RiboBio, China). The experiment was performed using Fluorescent in situ Hybridization Kit (RiboBio, China) according to the manufacturer's instructions and visualized under a laser confocal microscope (ANDOR, UK). The Cy3-labeled U6 and 18S probes were hybridized simultaneously as controls.
RNA pull-down assay
The plasmid pcDNA3.1 (Invitrogen, USA) containing sense or antisense full-length HCP5 cDNA was linearized with restriction enzyme XhoI and purified by phenol/chloroform extraction and ethanol precipitation. Biotin-labeled RNA was transcribed from linearized DNA templates in vitro using the MEGAscript™ T7 Transcription Kit (Invitrogen, USA) and purified using the MEGAclear™ Transcription Clean-Up Kit (Invitrogen, USA) according to the manufacturer's instructions. After incubating with the KGN cell lysates, proteins co-precipitated by the biotin-labeled transcripts were isolated with streptavidin beads and subjected to SDS-PAGE analysis. Then protein bands were visualized by silver staining followed by mass spectrometry (MS) or western blot.
Chromatin immunoprecipitation (ChIP)
The ChIP experiment was performed using the EZ-Magna ChIP™ A/G Kit (Millipore, USA) according to the manufacturer's instructions. For an individual ChIP assay, a total of 5 × 106 KGN cells were fixed in 1% formaldehyde at room temperature for 20 min and quenched by 0.125 M glycine for 5 min. Then the isolated chromatin DNA was sonicated and sheared to a length between 200 bp and 1000 bp by Bioruptor Pico sonication device (Diagenode, BEL). The sheared chromatin was immunoprecipitated at 4°C overnight with anti-YB1 (Abcam, USA) or anti-RNA polymerase II antibody (Abcam, USA). A homologous IgG was used as the isotype control. The co-precipitated DNA was purified by QIAquick Nucleotide Removal Kit (Qiagen, Germany) and then subjected to qPCR. The details of antibodies and specific primers for ChIP-qPCR are listed in Supplementary Table S3 and Supplementary Table S1, respectively.
RNA immunoprecipitation (RIP)
The RIP experiment was performed using the EZ-Magna RIP kit (Millipore, USA) according to the manufacturer's instructions. For each RIP assay, about 1 × 107 KGN, COV434 or SVOG cells were lysed in RIP lysis buffer supplemented with RNase inhibitor and protease inhibitor cocktail. After one freeze-thaw cycle, cell lysates were immunoprecipitated at 4°C overnight with anti-YB1 (Abcam, USA) or anti-ILF2 antibody (Abcam, USA). A homologous IgG was used as the isotype control. The co-precipitated RNA was extracted using TRIzol™ reagent according to the manufacturer's protocol. Then purified RNA was reverse-transcribed and subjected to qPCR analysis. Proteins isolated from the magnetic beads were detected using immunoblotting analysis. The details of antibodies are listed in Supplementary Table S3.
Immunofluorescence
The processed KGN cells were washed by PBS and fixed for 20 min in 4% paraformaldehyde at room temperature. After that, cells were permeabilized in 0.3% Triton X-100 and blocked by 5% bovine serum albumin in PBS for 1 h. Then samples were incubated with specific primary antibody at 4°C overnight and Alexa Fluor 568-conjugated goat anti-rabbit IgG secondary antibody for 1 h at room temperature. Nuclei were counterstained with DAPI. Images were visualized under a laser confocal microscope (ANDOR, UK). The details of antibodies are listed in Supplementary Table S3.
Co-immunoprecipitation
Co-immunoprecipitation assay for YB1 and ILF2 was performed by lysing KGN cells with NP-40 lysis buffer supplemented with protease inhibitor cocktails on ice for 30 min. Then lysates were sonicated three cycles (30 s on, 30 s off) to release nuclear proteins completely. A total of 0.5 mg of lysates was incubated overnight at 4°C with PureProteome™ Protein A/G Magnetic Beads (Millipore, USA) as well as anti-YB1 antibody (Abcam, USA). A rabbit IgG was used as negative control. After two times of washing using 0.05% Tween-20 in PBS, the immunoprecipitates were resuspended in loading buffer, degenerated and further analyzed by western blot.
Statistics
Statistical analysis was implemented using SPSS 13.0 (IBM, USA) and GraphPad Prism 8 (GraphPad Software, USA). Unless otherwise noted, statistical difference was analyzed by the two-tailed Student's t-test. The Mann–Whitney U-test was used when the data were not normally distributed. The Pearson correlation and linear regression analyses were used for the expression correlation between HCP5 and MSH5. Differences were considered statistically significant when P < 0.05.
RESULTS
The long noncoding RNA HCP5 is down-regulated in granulosa cells from bPOI patients
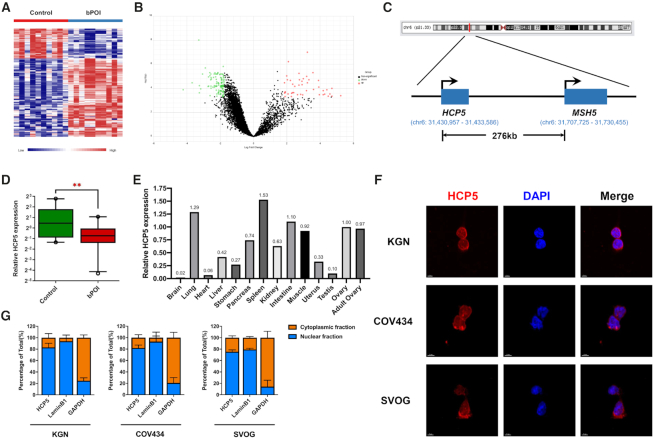
To explore aberrantly expressed lncRNAs involved in POI, we firstly conducted microarray analyses of granulosa cells from 10 patients with bPOI and 10 age-matched controls. According to the criteria by P < 0.0005 and fold change > 1.75, a total of 159 lncRNAs was discovered to be differentially expressed between patients and controls (Figure 1A, B). Among these candidates, we narrowed them down to consider only those lncRNAs which were co-localized and co-expressed with genes associated with POI. As a result, lncRNA HCP5 (RefSeq accession number NR_040662), which is from 276kb upstream of the MSH5 gene on chromosome 6 (Figure 1C), was the only POI-causing gene co-localized lncRNA identified to be differentially expressed in granulosa cells from bPOI patients compared with controls. Furthermore, quantitative RT-PCR analysis confirmed that HCP5 was differentially expressed in GCs from an independent cohort of 20 bPOI patients and 22 control women (Figure 1D). HCP5 is associated with autoimmune diseases, infections, and cancers in human (22); however, nothing is known about its role in ovary.
Figure 1.
LncRNA HCP5 is down-regulated in GCs from patients with bPOI. (A) Microarray heatmap of differentially expressed lncRNAs between GCs from controls (n = 10) and bPOI patients (n = 10). (B) Volcano plot of differentially expressed lncRNAs between GCs from controls and bPOI patients. (C) Schematic representation of location of HCP5 and MSH5 in chromosome. (D) The expression level of HCP5 was validated by qRT-PCR in GCs from an independent cohort of controls (n = 22) and bPOI patients (n = 20). Ct values were normalized to GAPDH. Data are presented as the median ± interquartile range. **P < 0.01.Two-tailed Mann–Whitney U-test. (E) The mRNA level of HCP5 in 21-week fetal tissues was validated by qRT-PCR. The expression level in ovary was set as ‘1’. GAPDH was used as internal reference. (F) Subcellular localization of HCP5 was detected by RNA FISH assay in KGN, COV434 and SVOG cells. (G) Relative HCP5 expression levels in the cytoplasmic and nuclear fractions of KGN, COV434 and SVOG cells by qRT-PCR. Lamin B1 was used as nuclear control. GAPDH was used as cytoplasmic control.
Our results showed that HCP5 was expressed ubiquitously in multiple tissues of 21-week human fetus, relatively high in the ovary (Figure 1E). Meanwhile, we assessed the coding potential of HCP5 using bioinformatic tools, including Coding Potential Calculator (CPC2) (23) and Coding-Potential Assessment Tool (CPAT) (24), and confirmed the annotation of HCP5 as a long noncoding RNA (Supplementary Figure S1A, B). Then fluorescence in situ hybridization (FISH) was performed to detect the subcellular localization of HCP5. As shown by the result, HCP5 mainly located in the nucleus, and particularly around the nuclear membrane of KGN, COV434 and SVOG cells (Figure 1F), which was further verified by cell fractionation combined with qPCR assay (Figure 1G). Together, the decreased expression of lncRNA HCP5 was firstly identified in GCs from bPOI patients.
HCP5 regulates MSH5 expression in granulosa cells
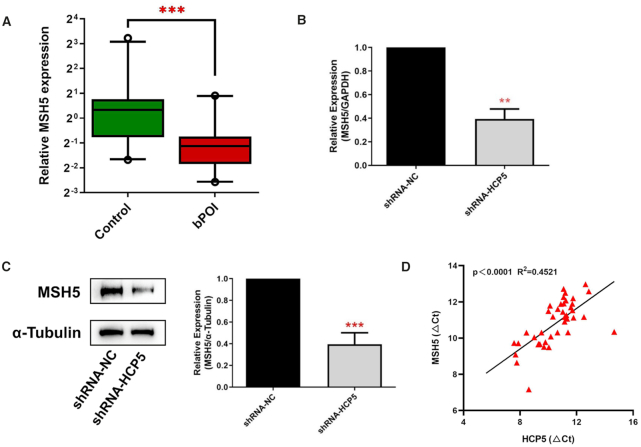
LncRNAs have been reported to play a cis-regulatory role in their neighboring genes (25,26). Therefore, given the adjacent localization between HCP5 and MSH5, we proposed that HCP5 may participate in the regulation of MSH5 expression. RT-qPCR results showed that MSH5 was downregulated in the GCs from bPOI women compared with controls (Figure 2A), which was consistent with the expression of HCP5. Moreover, knockdown of HCP5 by shRNA in KGN cells resulted in an obviously decreased expression of MSH5 in both mRNA and protein levels (Figure 2B, C).
Figure 2.
HCP5 regulates MSH5 expression in granulosa cells. (A) The mRNA levels of MSH5 were validated by qRT-PCR in GCs from controls (n = 22) and bPOI patients (n = 20). Ct values were normalized to GAPDH. Data are presented as the median ± interquartile range. ***P < 0.001. Two-tailed Mann–Whitney U-test. (B, C) qRT-PCR and western blot indicated that MSH5 mRNA (B) and protein (C) levels were significantly reduced by shRNA-mediated knockdown of HCP5 in KGN cells. Protein quantification was analyzed using ImageJ software (right). ***P < 0.001. Data shown represent three independent experiments. (D) The correlation between HCP5 transcript levels and MSH5 mRNA levels was measured in the same cohort of GCs as in (A). The Ct values normalized to GAPDH were subjected to Pearson correlation analysis.
Since lncRNA HCP5 could regulate MSH5 expression at transcriptional level, whether HCP5 was co-expressed with MSH5 mRNA in GCs was further examined. We measured the expression of HCP5 and MSH5 in GCs from 20 cases and 22 controls. As shown in Figure 2D, lncRNA HCP5 was significantly correlated with MSH5 mRNA level. Conversely, HCP5 expression was not affected by siRNA-mediated MSH5 knockdown (Supplementary Figure S1C). These results indicated a predominant role of HCP5 in modulating MSH5 expression.
MSH5 is the target of HCP5 during DNA double-strand breaks (DSBs) repair
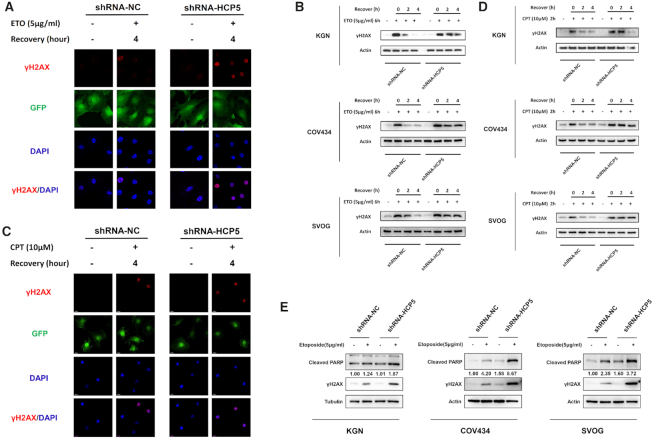
MSH5 principally participates in mismatch repair, and the MSH4-MSH5 heterodimers play an essential role in homologous recombination (HR) repair for DSBs (27). To assess whether silencing HCP5 can adversely affect DNA repair process, three independent specific shRNAs were transfected into KGN cells to silence HCP5. A qPCR assay was performed to test the knockdown efficiency (Supplementary Figure S1D). The most efficient shRNA-HCP5#1 was used to decrease the expression of HCP5 in vitro (Supplementary Figure S1E).
KGN cells were stably transfected with HCP5-specific shRNA#1 and negative control, following incubation with 5 μg/ml etoposide (ETO) for 1 h to induce DSBs. The γH2AX foci, a sensitive marker of DSBs, were observed by immunofluorescence, and gradually disappeared along with the repair processing (Figure 3A). Then protein levels of γH2AX were detected by western blot. Results showed that knockdown of HCP5 distinctly extended the time for DSB repair induced by ETO in KGN, COV434 and SVOG cells (Figure 3B). Given the essential role of MSH5 in HR pathway upon DSBs, we therefore examined the effect of HCP5 knockdown on the repair of camptothecin (CPT)-induced DNA damage, which could be only repaired by HR but not NHEJ pathway. Briefly, KGN cells were treated with 10 μM CPT for 2 h. Consistent with ETO treatment, γH2AX foci were observed and faded away with the recovery as well (Figure 3C). Also, silencing HCP5 distinctly prolonged the time for CPT-induced DSB repair in KGN, COV434 and SVOG cells (Figure 3D). This indicated the HR repair of DSBs was delayed in HCP5-deficiency cells. Moreover, knockdown of HCP5 markedly enhanced ETO-induced apoptosis as evidenced by cleaved poly (ADP-ribose) polymerase 1 (PARP1) (Figure 3E).
Figure 3.
Knockdown of HCP5 impairs DSBs HR repair and promotes apoptosis of GCs through MSH5. (A) Immunofluorescence showed the γH2AX foci formation in HCP5-silencing and negative control KGN cells suffered from ETO treatment. (B) After exposed to ETO for 6 h, the γH2AX levels were detected by western blot in HCP5-silencing and negative control KGN, COV434 and SVOG cells. Data shown represent three independent experiments. (C) Immunofluorescence showed the γH2AX foci formation in HCP5-silencing and negative control KGN cells suffered from CPT treatment. (D) After exposed to CPT for 2 h, the γH2AX levels were detected by western blot in HCP5-silencing and negative control KGN, COV434 and SVOG cells. Data shown represent three independent experiments. (E) Knockdown of HCP5 enhanced the cleavage of PARP and formation of γH2AX caused by treatment with Etoposide in KGN, COV434 and SVOG cells. Data shown represent three independent experiments.
Given the role of HCP5 in the regulation of MSH5 expression, we further explored the functional effect of MSH5 downregulation. The siRNA-mediated MSH5 knockdown was validated effective in KGN cells (Supplementary Figure S1F, G). Similarly, the impaired DNA damage repair and increased apoptosis after MSH5 knockdown were observed, which recapitulated the phenomena of HCP5-knockdown (Supplementary Figure S1H, I, J). Collectively, these results indicated that HCP5 participated in DNA damage repair and apoptosis of GCs through regulating MSH5 expression.
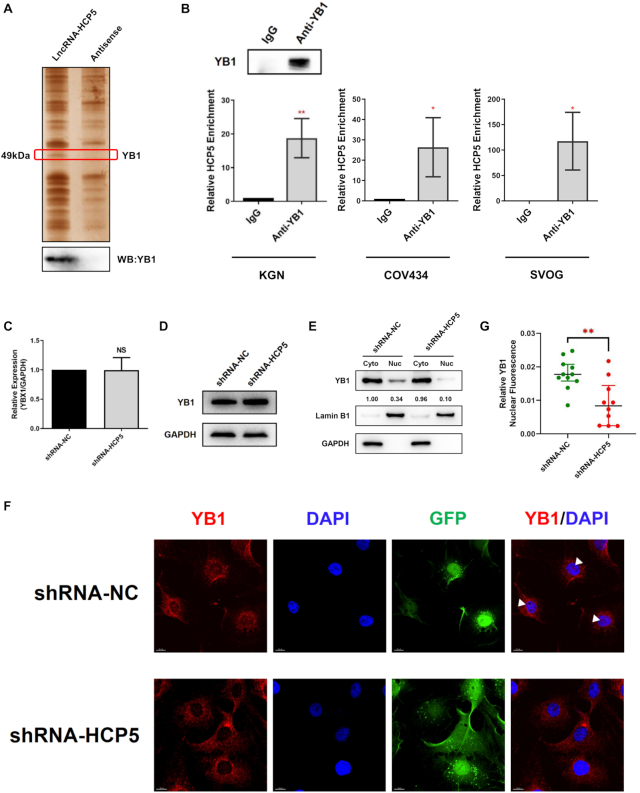
HCP5 directly binds to YB1
To investigate the protein partner of HCP5, RNA pull-down assay combined with MS was conducted with biotin-labeled HCP5 full-length transcript and its antisense transcript. As shown in Figure 4A, a differential HCP5-binding protein band was visualized at ∼50 kDa after silver staining. The specific band was cut off, digested by trypsin, and subjected to MS. As a result, Y-box binding protein 1 (YB1), which is a member of the family of DNA- and RNA-binding proteins with an evolutionarily ancient and conserved cold shock domain (28), was identified as a specific HCP5-interacting protein (Supplementary Figure S2A). Western blot confirmed that YB1 was enriched in the HCP5 pull-down proteins (Figure 4A). In addition, the potential binding fragments of HCP5 to YB1 were predicted by the catRAPID algorithm (29) (Supplementary Figure S2B).
Figure 4.
HCP5 directly binds to YB1 and is essential its nuclear localization. (A) Detection of HCP5-binding proteins by RNA pull-down assays. A specific band of ∼50 kDa (red box) was specific pulled down by HCP5 and subsequently identified as YB1 using mass spectrometry (Upper). Western blot showing the specific association between YB1 and HCP5 in the samples obtained from RNA pull-down (lower). The antisense transcript of HCP5 was used as a negative control. (B) Confirmation of the interaction between YB1 and HCP5 by RNA-binding protein immunoprecipitation (RIP) using YB1 antibody in KGN, COV434 and SVOG cells. Results are expressed as the mean ± SD (n = 3). *P < 0.05 and **P < 0.01. Two-tailed Student's t-test. (C) YB1 mRNA levels were analyzed by qRT-PCR after HCP5 silencing by shRNA in KGN cells. Values of qRT-PCR were obtained from triplicates and expressed as the mean ± SD (n = 3). Two-tailed Student's t-test. (D) YB1 protein levels were analyzed by western blot after HCP5 knockdown in KGN cells. Data shown represent three independent experiments. (E) Subcellular localization of YB1 protein was detected by western blot after HCP5 knockdown in KGN cells. Lamin B1 was used as nuclear control. GAPDH was used as cytoplasmic control. (F) Subcellular localization of YB1 protein was confirmed by immunofluorescence assay after HCP5 silencing. White triangles indicate representative YB1 expression in the nucleus. (G) Quantification of nuclear immunofluorescence intensity (mean gray value) in HCP5- knockdown (n = 10) and negative control (n = 11) KGN cells. Data are presented as the median ± interquartile range. **P < 0.01. Two-tailed Mann–Whitney U-test.
Since RNA pull-down assay uncovered HCP5-YB1 interaction in vitro, we performed RNA-binding protein immunoprecipitation (RIP) to detect the endogenous association between HCP5 and YB1. RIP assay using anti-YB1 antibody, followed by qRT-PCR, confirmed a direct interaction between HCP5 and YB1 in KGN, COV434 and SVOG cells (Figure 4B). Taken together, YB1 protein was a direct binding partner of lncRNA HCP5.
HCP5 is essential for YB1 localization to nucleus by acting as an RNA scaffold for ILF2 and YB1
We then explored the regulatory role of HCP5 on its binding protein YB1. Knockdown of HCP5 did not alter YB1 expression in either mRNA or protein level (Figure 4C, D), indicating that HCP5 did not affect the overall expression of YB1 in KGN cells. Considering that YB1 activity was mainly determined by its shuttling between nucleus and cytoplasm (28), whether HCP5 participated in the cellular localization of YB1 was further examined. We detected YB1 expression in the nuclear and cytoplasmic fraction of KGN cells after HCP5 silencing. Western blot showed that YB1 was present in both nuclear and cytoplasmic compartments of KGN cells; however, HCP5 silencing obviously reduced YB1 residing in the nucleus (Figure 4E). Intriguingly, immunofluorescence assay confirmed the reduced nuclear staining of YB1 protein and excluded nucleolar localization of YB1 when HCP5 was knocked down (Figure 4F, G and Supplementary Figure S2C). These results indicated that HCP5 silencing prevented YB1 from transporting into the nucleus.
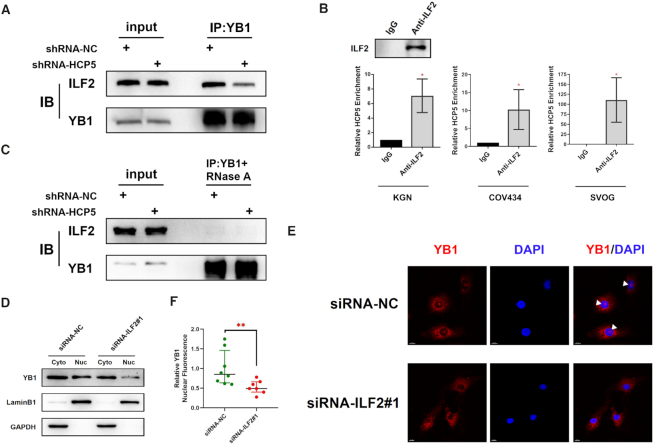
Mechanistically, it is still unclear how HCP5 induced YB1 localization into the nucleus. It was previously proposed that activated Akt could bind to and phosphorylate cold shock domain at Ser102 of YB1 and then promoted its nuclear translocation (30). However, YB1 phosphorylation status was not altered after HCP5 silencing (Supplementary Figure S2D). Given that Interleukin Enhancer Binding Factor 2 (ILF2) could interact with YB1 and modulate its nuclear localization (31,32), a co-immunoprecipitation assay was conducted to assess the impact of HCP5 on YB1–ILF2 complex. Notably, knockdown of HCP5 decreased the relative amount of ILF2 that co-precipitated by YB1 antibody (Figure 5A). Considering HCP5 localization in the nucleus, especially the nuclear membrane, we hypothesized that HCP5 might serve as a modular scaffold between YB1 and ILF2. Therefore, we performed RIP assay to confirm the interaction between HCP5 and ILF2. As expected, anti-ILF2 antibody co-precipitated HCP5 in KGN, COV434 and SVOG cells (Figure 5B), whereas knockdown of HCP5 did not affect subcellular localization of ILF2 (Supplementary Figure S2E, F). In addition, we observed diminished association between YB1 and ILF2 upon RNase A treatment, which further validated the essential role of an RNA scaffold in YB1-ILF2 interaction (Figure 5C). Finally, siRNA-mediated silencing of ILF2 resulted in a marked reduction of nuclear localization of YB1, which was in accordance with the results when HCP5 knockdown (Figure 5D–F and Supplementary Figure S2G, H). Thus, it was concluded that HCP5 served as an RNA scaffold for the interaction between YB1 and ILF2 and could be responsible for the nuclear localization of YB1.
Figure 5.
HCP5 is essential for YB1 locating to nucleus by acting as a scaffold for ILF2 and YB1. (A) The association between YB1 and ILF2 was shown in co-immunoprecipitation assays after silencing HCP5 in KGN cells. Data shown represent three independent experiments. (B) Confirmation of the interaction between ILF2 and HCP5 by RIP using ILF2 antibody in KGN, COV434 and SVOG cells. Results are expressed as the mean ± SD (n = 3). *P < 0.05. Two-tailed Student's t-test. (C) The association between YB1 and ILF2 was shown in co-immunoprecipitation assays after silencing HCP5 in KGN cells upon RNase A treatment. Data shown represent three independent experiments. (D) Subcellular localization of YB1 protein was detected by western blot after ILF2 silencing by siRNA in KGN cells. Lamin B1 was used as nuclear control. GAPDH was used as cytoplasmic control. (E) Subcellular localization of YB1 protein was confirmed by immunofluorescence assay after ILF2 silencing. White triangles indicate representative YB1 expression in the nucleus. (F) Quantification of nuclear immunofluorescence intensity (mean gray value) in ILF2-knockdown (n = 7) and negative control (n = 8) KGN cells. Data are presented as the median ± interquartile range. **P < 0.01. Two-tailed Mann–Whitney U-test.
Knockdown of HCP5 reduced YB1 binding to MSH5 promoter and inhibited the transcriptional activation of MSH5
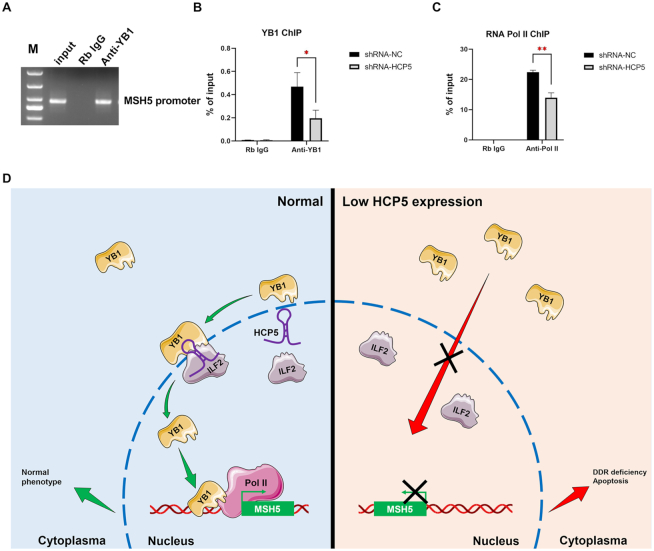
Notably, nuclear YB1 could locate to the promoters or other key regulatory regions of target genes, where the transcription is initiated (33). We further explored whether HCP5 knockdown affected YB1 occupancy to the MSH5 promoter. Chromatin immunoprecipitation (ChIP) assay followed by qPCR demonstrated significant enrichment of YB1 on MSH5 promoter (Figure 6A). Furthermore, knockdown of HCP5 significantly reduced the binding of YB1 to the promoter of MSH5 (Figure 6B).
Figure 6.
Knockdown of HCP5 reduced YB1 binding to MSH5 promoter region and inhibited the transcriptional activation of MSH5. (A) ChIP analyses of KGN cells were performed with IgG or YB1 antibody. ChIP products were amplified by PCR with specific primers of the MSH5 promoter region and subjected to electrophoresis analyses. (B) ChIP analyses of KGN cells were performed with IgG or YB1 antibody after HCP5 silencing, followed by qPCR in the MSH5 promoter region. Values of qPCR were obtained from triplicates and expressed as the mean ± SD (n = 3). *P < 0.05. Two-tailed Student's t-test. (C) ChIP analyses of KGN cells were performed with IgG or RNA pol II antibody after HCP5 silencing, followed by qPCR in the MSH5 promoter region. Values of qPCR were obtained from triplicates and expressed as the mean ± SD (n = 3). **P < 0.01. Two-tailed Student's t-test. (D) Schematic summary of the critical role of HCP5 in MSH5 expression and GCs function through direct binding and modulating the subcellular localization of YB1.
It was previously reported that recruitment of YB1 to the specific binding site on promoter is crucial for RNA polymerase II (Pol II) loading, and activating gene transcription (34). Therefore, we further investigated the influence of HCP5 knockdown on RNA Pol II binding to MSH5 promoter. As shown in Figure 6C, shRNA-mediated HCP5 silencing led to a reduced binding between RNA Pol II and MSH5 promoter, which was consistent with the inhibition of HCP5 shRNA on MSH5 mRNA level. Collectively, it could conclude that HCP5 regulated the transcription of MSH5 by facilitating the recruitment of YB1 and RNA Pol II to MSH5 promoter.
DISCUSSION
Before exploring noncoding RNAs responsible for POI, the identification of causative variations for POI is limited within protein-coding genes (4–6). However, the causing-gene mutations could only explain 10–15% of POI cases (35,36). Except for small noncoding RNAs (37,38), studies on the roles of lncRNAs played in human POI are lacking. To our knowledge, this is the first finding of a lncRNA involved in GCs DNA damage repair process and responsible for POI.
The ovarian reserve is principally determined by the quantity and quality of oocytes. Either initially inadequate or accelerated exhaustion of primordial follicle pool will lead to ovarian insufficiency. In mammals, the role of GCs determining the fate of follicles has been well documented (39,40). Moreover, diminished GCs induced follicle atresia and finally resulted in POI in vivo (18,19). Several lncRNAs expressed in granulosa cells have been discovered to be specifically associated with polycystic ovary syndrome, endometriosis and ovarian hyperstimulation syndrome (41–44). Herein, we explored the lncRNAs expression profile in GCs from patients with bPOI and identified down-regulated lncRNA HCP5. It has been recognized that increased DNA damage and repair deficiency in granulosa cells facilitate follicle atresia and ovarian aging (45,46). Consistently, our results indicated that HCP5-knockdown disrupted the DSBs repair activity of GCs, and thereby led to GCs apoptosis in vitro.
In this study, MSH5, which has been nominated as POF13 by Online Mendelian Inheritance in Man (OMIM) was proved to be the executor of HCP5. MSH5, as a member of the MutS protein family, forms a heterodimeric complex with MSH4, plays an essential role during homologous recombination repair for DSBs (27). Causative mutations of MSH5 gene have been reported in women with POI (47,48). In the present study, the adjacent location of HCP5 to MSH5 gene indicated that it may act as a cis-regulatory factor of MSH5. Furthermore, HCP5 silencing inhibited MSH5 expression. MSH5 silencing also led to a longer time for DSBs repair and promoted ETO-induced apoptosis in GCs. Combined with the similar loss-of-function influence of HCP5 and MSH5 on GCs, we elucidated that MSH5 was the effector of HCP5 during DNA damage repair, which further enriched the understanding of MSH5 on ovarian function, not only during meiotic recombination in oocyte, but also protecting GCs from DNA damage.
It has been proved that lncRNAs exert their biological functions mainly through interacting with RNA-binding proteins (49,50). The biotin-labeled RNA pull-down/MS and RIP assay discovered that HCP5 bound with YB1. YB1 is a pleiotropic protein which participates in DNA transcription, mRNA splicing, mRNA stabilization, and mRNA translation by shuttling between the nucleus and the cytoplasm (28). Although HCP5 did not affect the overall expression of YB1, strikingly, the nuclear enrichment of YB1 was dramatically reduced after HCP5 silencing. In the nucleus, YB1 usually serves as a transcriptional activator (51). It was further demonstrated that silencing HCP5 decreased the binding of YB1 to the promoter of MSH5. Also, the co-precipitated MSH5 promoter level by RNA Pol II was reduced after silencing HCP5, which validated a pro-transcription regulation of HCP5 on MSH5 gene. These findings elaborated that HCP5 regulated MSH5 transcription and expression, by controlling the binding between YB1 and MSH5 promoter. Of importance, given that proteins are synthesized in cytoplasm where away from the transcription start site, HCP5 is timely suited to regulate MSH5 in situ once DNA damage occurs in GCs.
Different approaches by which YB1 was transported into the nucleus have been investigated. Interestingly, ILF2 was proved to modulate YB1 nuclear localization by directly interacting with the protein itself, which was further supported by a positive correlation between ILF2 expression and YB1 nuclear enrichment (31). Our results demonstrated that YB1 and ILF2 could both co-precipitate with HCP5. When HCP5 was silenced, the binding of YB1 with ILF2 showed significantly diminished. More strikingly, ILF2 knockdown resulted in obvious reduction of YB1 nuclear localization. Thus, HCP5 served as an RNA scaffold for the interaction between YB1 and ILF2 and induced YB1 shuttling to the nucleus in GCs. Combined with the interaction between YB1 and ILF2 reported previously (32), we revealed a direct connection between lncRNA HCP5 and YB1-ILF2 heterodimerization during DNA damage repair process in GCs.
In conclusion, we elucidated the mechanism of LncRNA HCP5 responsible for human POI, i.e. HCP5 regulated MSH5 expression and GCs function through direct binding with YB1 and modulating its subcellular localization (Figure 6D). This study discovered a novel lncRNA HCP5 contributed to dysfunctional GCs by transcriptionally regulating MSH5 and DNA damage repair via YB1, providing a novel epigenetic mechanism for POI pathogenesis.
DATA AVAILABILITY
GEO accession ID for the microarray data is GSE135697.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research & Developmental Program of China [2017YFC1001100]; Science Foundation for Distinguished Young Scholars of Shandong [JQ201720]; National Natural Science Foundation of China [81522018, 81471509, 81771541]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Coulam C.B., Adamson S.C., Annegers J.F.. Incidence of premature ovarian failure. Obstet. Gynecol. 1986; 67:604–606. [PubMed] [Google Scholar]
- 2. Wu X., Cai H., Kallianpur A., Li H., Yang G., Gao J., Xiang Y.B., Ji B.T., Yu T., Zheng W. et al.. Impact of premature ovarian failure on mortality and morbidity among Chinese women. PLoS One. 2014; 9:e89597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Welt C.K. Primary ovarian insufficiency: a more accurate term for premature ovarian failure. Clin. Endocrinol. (Oxf.). 2008; 68:499–509. [DOI] [PubMed] [Google Scholar]
- 4. Qin Y., Jiao X., Simpson J.L., Chen Z.J.. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update. 2015; 21:787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiao X., Ke H., Qin Y., Chen Z.J.. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol. Metab. 2018; 29:795–807. [DOI] [PubMed] [Google Scholar]
- 6. Huhtaniemi I., Hovatta O., La Marca A., Livera G., Monniaux D., Persani L., Heddar A., Jarzabek K., Laisk-Podar T., Salumets A. et al.. Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol. Metab. 2018; 29:400–419. [DOI] [PubMed] [Google Scholar]
- 7. Elgar G., Vavouri T.. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008; 24:344–352. [DOI] [PubMed] [Google Scholar]
- 8. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al.. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 10. Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012; 338:1435–1439. [DOI] [PubMed] [Google Scholar]
- 11. Kim T.K., Hemberg M., Gray J.M.. Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015; 7:a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ng S.Y., Bogu G.K., Soh B.S., Stanton L.W.. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013; 51:349–359. [DOI] [PubMed] [Google Scholar]
- 13. Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. et al.. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013; 152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morán I., Akerman İ., van de Bunt M., Xie R., Benazra M., Nammo T., Arnes L., Nakić N., García-Hurtado J., Rodríguez-Seguí S. et al.. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012; 16:435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yao G., He J., Kong Y., Zhai J., Xu Y., Yang G., Kong D., Dong F., Shi S., Yang Q. et al.. Transcriptional profiling of long noncoding RNAs and their target transcripts in ovarian cortical tissues from women with normal menstrual cycles and primary ovarian insufficiency. Mol. Reprod. Dev. 2019; 86:847–861. [DOI] [PubMed] [Google Scholar]
- 16. Dumesic D.A., Meldrum D.R., Katz-Jaffe M.G., Krisher R.L., Schoolcraft W.B.. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015; 103:303–316. [DOI] [PubMed] [Google Scholar]
- 17. Li R., Albertini D.F.. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 2013; 14:141–152. [DOI] [PubMed] [Google Scholar]
- 18. Sun Z., Zhang H., Wang X., Wang Q.C., Zhang C., Wang J.Q., Wang Y.H., An C.Q., Yang K.Y., Wang Y. et al.. TMCO1 is essential for ovarian follicle development by regulating ER Ca store of granulosa cells. Cell Death Differ. 2018; 25:1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yeung C.K., Wang G., Yao Y., Liang J., Tenny Chung C.Y., Chuai M., Lee K.K., Yang X.. BRE modulates granulosa cell death to affect ovarian follicle development and atresia in the mouse. Cell Death. Dis. 2017; 8:e2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishi Y., Yanase T., Mu Y., Oba K., Ichino I., Saito M., Nomura M., Mukasa C., Okabe T., Goto K. et al.. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001; 142:437–445. [DOI] [PubMed] [Google Scholar]
- 21. Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J. et al.. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016; 165:1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang W., Lou W., Ding B., Yang B., Lu H., Kong Q., Fan W.. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY). 2019; 11:2610–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G.. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017; 45:W12–W16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W.. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maamar H., Cabili M.N., Rinn J., Raj A.. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013; 27:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Q., Chao T.C., Patil V.S., Qin Y., Tiwari S.K., Chiou J., Dobin A., Tsai C.M., Li Z., Dang J. et al.. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019; 38:e100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snowden T., Acharya S., Butz C., Berardini M., Fishel R.. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell. 2004; 15:437–451. [DOI] [PubMed] [Google Scholar]
- 28. Lyabin D.N., Eliseeva I.A., Ovchinnikov L.P.. YB-1 protein: functions and regulation. Wiley Interdiscip. Rev. RNA. 2014; 5:95–110. [DOI] [PubMed] [Google Scholar]
- 29. Bellucci M., Agostini F., Masin M., Tartaglia G.G.. Predicting protein associations with long noncoding RNAs. Nat. Methods. 2011; 8:444–445. [DOI] [PubMed] [Google Scholar]
- 30. Sutherland B.W., Kucab J., Wu J., Lee C., Cheang M.C., Yorida E., Turbin D., Dedhar S., Nelson C., Pollak M. et al.. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005; 24:4281–4292. [DOI] [PubMed] [Google Scholar]
- 31. Marchesini M., Ogoti Y., Fiorini E., Aktas Samur A., Nezi L., D’Anca M., Storti P., Samur M.K., Ganan-Gomez I., Fulciniti M.T. et al.. ILF2 is a regulator of RNA splicing and DNA damage response in 1q21-amplified multiple myeloma. Cancer Cell. 2017; 32:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chatel-Chaix L., Germain M.A., Motorina A., Bonneil E., Thibault P., Baril M., Lamarre D.. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J. Virol. 2013; 87:11704–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dolfini D., Mantovani R.. Targeting the Y/CCAAT box in cancer: YB-1 (YBX1) or NF-Y. Cell Death Differ. 2013; 20:676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sengupta S., Mantha A.K., Mitra S., Bhakat K.K.. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene. 2011; 30:482–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dixit H., Rao L., Padmalatha V., Raseswari T., Kapu A.K., Panda B., Murthy K., Tosh D., Nallari P., Deenadayal M. et al.. Genes governing premature ovarian failure. Reprod. Biomed. Online. 2010; 20:724–740. [DOI] [PubMed] [Google Scholar]
- 36. van Kasteren Y.M., Hundscheid R.D., Smits A.P., Cremers F.P., van Zonneveld P., Braat D.D.. Familial idiopathic premature ovarian failure: an overrated and underestimated genetic disease. Hum. Reprod. 1999; 14:2455–2459. [DOI] [PubMed] [Google Scholar]
- 37. Dang Y., Wang X., Hao Y., Zhang X., Zhao S., Ma J., Qin Y., Chen Z.J.. MicroRNA-379-5p is associate with biochemical premature ovarian insufficiency through PARP1 and XRCC6. Cell Death. Dis. 2018; 9:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo Y., Sun J., Lai D.. Role of microRNAs in premature ovarian insufficiency. Reprod. Biol. Endocrinol. 2017; 15:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boone D.L., Carnegie J.A., Rippstein P.U., Tsang B.K.. Induction of apoptosis in equine chorionic gonadotropin (eCG)-primed rat ovaries by anti-eCG antibody. Biol. Reprod. 1997; 57:420–427. [DOI] [PubMed] [Google Scholar]
- 40. Gao F., Zhang J., Wang X., Yang J., Chen D., Huff V., Liu Y.X.. Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Hum. Mol. Genet. 2014; 23:333–341. [DOI] [PubMed] [Google Scholar]
- 41. Liu Y.-d., Li Y., Feng S.-x., Ye D.-s., Chen X., Zhou X.-y., Chen S.-l.. Long noncoding RNAs: potential regulators involved in the pathogenesis of polycystic ovary syndrome. Endocrinology. 2017; 158:3890–3899. [DOI] [PubMed] [Google Scholar]
- 42. Zhao J., Xu J., Wang W., Zhao H., Liu H., Liu X., Liu J., Sun Y., Dunaif A., Du Y. et al.. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine. 2018; 36:526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y., Liu Y.D., Chen S.L., Chen X., Ye D.S., Zhou X.Y., Zhe J., Zhang J.. Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway. Mol. Hum. Reprod. 2019; 25:17–29. [DOI] [PubMed] [Google Scholar]
- 44. Lin H., Li Y., Xing W., Qiu Q., Wang W., Zhang Q.. Genome-wide screening differential long non-coding RNAs expression profiles discloses its roles involved in OHSS development. J. Assist. Reprod. Genet. 2018; 35:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang D., Zhang X., Zeng M., Yuan J., Liu M., Yin Y., Wu X., Keefe D.L., Liu L.. Increased DNA damage and repair deficiency in granulosa cells are associated with ovarian aging in rhesus monkey. J. Assist. Reprod. Genet. 2015; 32:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu J., Yang M., Jing L., Ren L., Wei J., Zhang J., Zhang F., Duan J., Zhou X., Sun Z.. Silica nanoparticle exposure inducing granulosa cell apoptosis and follicular atresia in female Balb/c mice. Environ. Sci. Pollut. Res. Int. 2018; 25:3423–3434. [DOI] [PubMed] [Google Scholar]
- 47. Mandon-Pepin B., Touraine P., Kuttenn F., Derbois C., Rouxel A., Matsuda F., Nicolas A., Cotinot C., Fellous M.. Genetic investigation of four meiotic genes in women with premature ovarian failure. Eur. J. Endocrinol. 2008; 158:107–115. [DOI] [PubMed] [Google Scholar]
- 48. Guo T., Zhao S., Zhao S., Chen M., Li G., Jiao X., Wang Z., Zhao Y., Qin Y., Gao F. et al.. Mutations in MSH5 in primary ovarian insufficiency. Hum. Mol. Genet. 2017; 26:1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun M., Kraus W.L.. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 2015; 36:25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wapinski O., Chang H.Y.. Long noncoding RNAs and human disease. Trends Cell Biol. 2011; 21:354–361. [DOI] [PubMed] [Google Scholar]
- 51. Wu J., Stratford A.L., Astanehe A., Dunn S.E.. YB-1 is a transcription/translation factor that orchestrates the oncogenome by hardwiring signal transduction to gene expression. Transl Oncogenomics. 2007; 2:49–65. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEO accession ID for the microarray data is GSE135697.