Abstract
G-quadruplexes are four-stranded nucleic acid structures involved in multiple cellular pathways including DNA replication and telomere maintenance. Such structures are formed by G-rich DNA sequences typified by telomeric DNA repeats. Whilst there is evidence for proteins that bind and regulate G-quadruplex formation, the molecular basis for this remains poorly understood. The budding yeast telomeric protein Rap1, originally identified as a transcriptional regulator functioning by recognizing double-stranded DNA binding sites, was one of the first proteins to be discovered to also bind and promote G-quadruplex formation in vitro. Here, we present the 2.4 Å resolution crystal structure of the Rap1 DNA-binding domain in complex with a G-quadruplex. Our structure not only provides a detailed insight into the structural basis for G-quadruplex recognition by a protein, but also gives a mechanistic understanding of how the same DNA-binding domain adapts to specifically recognize different DNA structures. The key observation is the DNA-recognition helix functions in a bimodal manner: In double-stranded DNA recognition one helix face makes electrostatic interactions with the major groove of DNA, whereas in G-quadruplex recognition a different helix face is used to make primarily hydrophobic interactions with the planar face of a G-tetrad.
INTRODUCTION
G-quadruplexes (or G4s) are four-stranded nucleic acid structures know to form in nucleic acid sequences that contain runs of adjacent guanines (G-tracts). The building block of G-quadruplexes are four guanine nucleotides associated through Hoogsteen base-pairing into a cyclic arrangement forming a G-tetrad. The planar G-tetrads stack on top each other giving rise to a four-stranded helical structure (Figure 1A) (1,2). G-quadruplex formation is driven by monovalent cations such as potassium and sodium, and hence physiological buffer conditions favor their formation (1,2). Structural analyses have shown that G-quadruplexes are highly polymorphic and can be grouped into parallel, anti-parallel, or hybrid structures based on the relative orientation of the strands (3). Recent genome-wide sequence analyses have revealed that genomes are rich in sequences that have the potential to form G-quadruplexes (∼700 in Saccharomyces cerevisiae and 700 000 in man) and that their location is not random, correlating with functionally important genomic regions such as promoters and telomeres (4–7). Evidence is accumulating for a role of G-quadruplexes in the regulation of cellular pathways that cause double-helical DNA to be transiently single-stranded such as DNA replication, gene expression, and telomere maintenance (8). With their unique structure and their presence in regulatory DNA sequences, G-quadruplexes have emerged as molecular targets for anti-cancer drugs (9,10).
Figure 1.
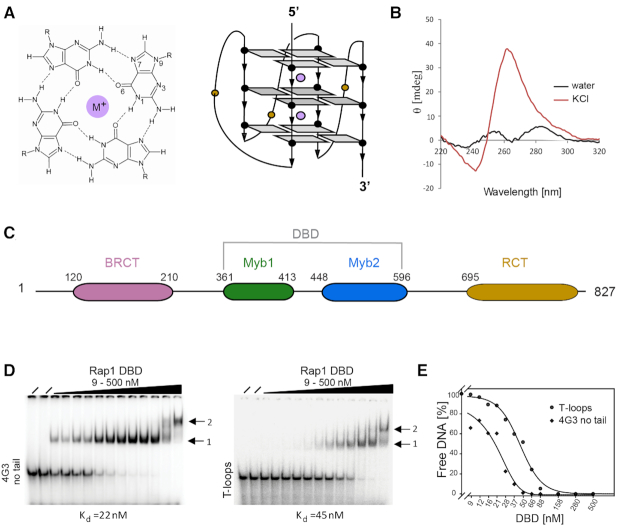
The DBD of yeast Rap1 is responsible for the interaction with G-quadruplex DNA. (A) Schematic representation of a G-tetrad and topology of a parallel G-quadruplex in which the loops linking the top and bottom G-tetrads cross the helical grooves. Purple spheres represent monovalent cations. (B) CD spectrum of the folded ‘4-G3-no tail’ oligonucleotide showing that it forms a parallel G-quadruplex. (C) Domain organization of the yeast Rap1 protein. The central DNA-binding domain consists of Myb1 and Myb2 homeodomains. (D) EMSA of the binding of the Rap1-DBD to the ‘4-G3 no tail’ 5′-TGGGTGTGTGGGTGTGTGGGTGTGTGGG-3′ G-quadruplex and the ‘T-loops’ 5′-TTGGGTGGGTGGGTGGGTT-3′. G-quadruplex. [32P] labeled DNA at 100 pM was incubated with increasing concentration of proteins (from 9 to 500 nM). (i) identifies the lower molecular weight complex consisting of one Rap1-DBD bound to one G-quadruplex and (ii) the larger ‘dimeric’ complex. (E) Binding curves for ‘T-loos’ and’ 4-G3-no tails’ G-quadruplexes: Apparent binding constants were calculated from protein concentrations at 50% binding.
Telomeres, the specific protein/DNA complexes found at the physical ends of eukaryotic linear chromosomes, contain the highest concentration of DNA sequences with the potential to form G-quadruplexes. The main function of telomeres is to protect chromosomes from inappropriate activation of DNA-damage pathways (11). Telomeric DNA sequences are highly conserved and consist of a tandem arrays of simple G-rich sequence repeats that typically contain tracts of three or four guanines such as TTAGGG in vertebrates and (TG)1–4G2–3 in S. cerevisiae (12). Telomeres consist of double-stranded repeats with the G-rich strand extending in the 3′ direction forming a single-stranded G-overhang (13). Soon after telomeres were first sequenced it was shown experimentally that telomeric G-rich strands spontaneously fold into G-quadruplex structures in physiological salt conditions (14–17). Significantly, for biological function, this was followed by the discovery in the mid-1990s that telomeric proteins such as the budding yeast Rap1 and the ciliate TEBPβ promote the formation and bind to G-quadruplex structures in vitro, suggesting that proteins could regulate their occurrence in cells (18–20). The breakthrough in obtaining in vivo evidence for the presence of G-quadruplexes came with the development of G-quadruplex structure-specific antibodies that allowed probing for G-quadruplexes directly in cells (21). The results from these immunostaining studies showed that G-quadruplex structures are present at the macronuclear telomeres in ciliates and that their formation is cell-cycle regulated through the phosphorylation of a telomere end-binding protein (TEBPβ). These observations suggested that G-quadruplexes may act as a capping mechanism for chromosome ends, and become unfolded during DNA replication (22).
The S. cerevisiae budding yeast Rap1 (Repressor Activator Protein1) is an essential gene encoding a protein originally identified as a transcriptional regulator and only later discovered to be the major telomere-binding protein (23,24). Rap1 is a negative regulator of telomere length (23,25) and is involved in the silencing of genes located near telomeres as well as protecting from telomere–telomere fusions (26,27). Yeast Rap1 is a large protein (827aa) with a complex multidomain structure (Figure 1C): the N-terminus contains a BRCT domain that is conserved in other spices like human Rap1, but its function is not well understood; the C-terminal domain (RCT) is essential for the recruitment of Rif1/2 and Sir2/3/4, protein complexes necessary for the maintenance of telomere structure and function (26). The central section of Rap1 contains the DNA-binding domain (DBD) that is essential for yeast survival and is crucial for the interaction of Rap1 with double-stranded DNA binding sites located in promoters and at telomeres (28). Our earlier crystal structure of the Rap1-DBD in complex with a double-stranded telomeric DNA binding site revealed that Rap1 binds to two tandem telomeric repeats via two clearly defined Myb/homeodomains (Myb1 and Myb2) (29). DNA recognition is via the DNA-recognition helix of each domain entering the major groove of DNA making specific contacts with GG steps in adjacent telomeric repeats, whilst the N-terminal arm of each homeodomain binds in the minor groove—a DNA recognition mechanism conserved in the vertebrate telomeric proteins TRF1 and TRF2 (30). Over two decades ago, detailed DNA binding studies lead us to conclude that Rap1 can also bind specifically to a parallel DNA G-quadruplex and promote its formation, providing some of the first experimental evidence for the recognition of G-quadruplex structures by proteins (18,31).
The participation of G-quadruplexes in biology will likely require such structures to be recognized and the kinetics of their formation and resolution to be controlled by proteins (8). Therefore, an understanding of how proteins interact with G-quadruplex DNA (or RNA) structures is crucial for uncovering the biological roles of these non-canonical nucleic acid structures. Here, we address the basis for G-quadruplex DNA recognition by the budding yeast telomeric protein Rap1. To understand how Rap1 binds to G-quadruplex DNA, we defined the domain of Rap1 crucial for the interaction and determined the crystal structure of the Rap1-DBD bound to a parallel G-quadruplex. Our structural analysis provides an understanding at near-atomic resolution of how yeast Rap1 recognizes the structure of a G-quadruplex. Furthermore, comparison of this structure with our previous crystal structure of Rap1-DBD in complex with double-stranded telomeric DNA reveals how the same protein adapts its conformation and use of a DNA-recognition helix to recognize different DNA structures. The key observation is that different faces of the DNA-recognition helix are used for the specific recognition of double-stranded or G-quadruplex DNA: one face is used to make primarily hydrophobic interactions with the hydrophobic face of a G-tetrad, whereas the other face makes electrostatic interactions in the major groove of double-stranded DNA.
MATERIALS AND METHODS
Protein expression and purification
Gene fragments of the S. cerevisiae Rap1 (aa360–aa598) encompassing the DBD, the N-terminal BRCT domain (1–358) and the Rap1 C-terminal domain (600–827) were cloned into the pET30aTEV vector (Addgene). The constructs, which contain an N-terminus His6-tag followed by TEV protease cleavage site, were transformed and expressed in Escherichia coli BL21-CodonPlus (DE3)-RIPL competent cells. Cells were allowed to grow at 37°C until the OD600 reached 0.6–0.8, then cooled and protein expression induced with 0.5 mM IPTG at 18°C and cultures grown overnight at 18°C. Cell pellet were then re-suspended in lysis buffer (100 mM HEPES–NaOH pH 7.5, 0.5 M KCl, 10% glycerol, 5 mM β–mercaptoethanol, 5 mM MgCl2, 5 μg/ml DNase I, 1 mM PMSF, 10 mM Imidazole), sonicated and cleared by centrifugation to remove cell debris. The first purification step for all Rap1 constructs was by affinity chromatography. The supernatant was loaded onto a His-Trap HP (GE-Healthcare) column and proteins eluted with 500 mM Imidazole. Purified proteins were incubated overnight with TEV protease (in a molar ratio 1:50) at 4°C to cleave off the His6-Tag. Cleaved DBD as well as the other Rap1 domains were again loaded on a His-Trap HP column to remove the His-tag together with the TEV protease, and the flow-through fractions collected. The proteins were further fractionated by size-exclusion chromatography on a Superdex 75 prep grade 16/60 column and eluted with a buffer containing 20 mM MES–KOH pH 6.0, 400 mM KCl. The purified Rap1-DBD was concentrated to 2 mg/ml and stored at −80°C.
G-quadruplex DNA sample preparation
The G-rich DNA oligonucleotides were purchased from IDT. Desalted DNA pellets were dissolved in water at a concentration of 200 μM. Concentrations were confirmed by measuring the absorbance on a NanoDrop microvolume spectrophotometer (Thermo Fisher) using the extinction coefficients provided by the company. For G-quadruplex formation, DNA oligonucleotides were diluted to 50 μM in 10 mM Tris–HCl pH 7.5 and 150 mM KCl and G-quadruplexes were formed by annealing the DNA for 5 min at 95°C, followed by slow cooling to room temperature overnight. Oligonucleotides used in this study are listed in Table 1.
Table 1.
DNA oligonucleotide used in this study. rTelo is an RNA oligonucleotide. G-tracts are shown in bold
| Name | Sequence |
|---|---|
| 4-G3 | 5′-AATTCT6GGGTGTGT14GGGTGTGT22GGGTGTGT30GGGTGTGG-3′ |
| 4-G3 no tail | 5′-T1GGGTGTGT9GGGTGTGT17GGGTGTGT25GGG-3′ |
| T-loops | 5′-TT2GGGT6GGGT10GGGT14GGGTT-3′ |
| rTelo | 5′-U1GGGUGUGU9GGGUGUGU17GGGUGUGU25GGG-3′ |
Circular dichroism
Circular dichroism (CD) spectra were recorded at room temperature using a JASCO-815 spectropolarimeter with 1 cm path length cuvette. Spectra from 220 to 320 nm were recorded for 500 μl solutions containing 5 μM oligonucleotides in water, or folded into a G-quadruplex in the appropriate buffer. Each spectrum was derived from the average of three scans, the spectral contribution of the buffer subtracted and the ellipticity calculated using the provided program.
Electrophoretic mobility shift assay and native gel electrophoresis
Binding reactions were performed by incubating [32P]-labeled oligonucleotides at a constant concentration (100 pM) with increasing protein concentrations in a final volume of 12 μl. The binding buffer contained: 20 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM DTT, 6% glycerol, 100 μg/ml BSA and 100 mM KCl. After 30 min incubation at room temperature, 10 μl of each binding reaction was loaded on a pre-run 7% polyacrylamide native gel (acrylamide: bisacrylamide, 37.5:1 ratio). Electrophoresis was performed at 4°C for 1 h with constant voltage (200V) in 0.5× TB (89 mM Tris–borate, 89 mM boric acid, pH 8.3) electrophoresis buffer.
Crystallization of Rap1-DBD in complex with DNA G-quadruplexes
An aliquot of oligonucleotide pre-folded into G-quadruplex was mixed with the Rap1-DBD at 1:1 molecular ratio in binding buffer (20 mM MES–KOH pH 6.0, 400 mM KCl) and incubated at 25°C for 1 h. The complex was concentrated to 15 mg/ml using a Vivaspin 6 centrifugal concentrator with a 10 kDa cutoff. Crystals of Rap1-DBD in complex with the ‘T-loops’ G-quadruplex were grown by vapor-diffusion in sitting drops comprising equal volumes of complex (15 mg/ml) and precipitant solution (0.1 M Na citrate tribasic dihydrate pH 5.6, 10% isopropanol, 14% PEG 4000). Crystallization trials were carried at 25°C and crystals (∼200 μm in size) grew in about seven days. For data collection, crystals were transferred to a cryoprotectant buffer consisting of the precipitant solution supplemented with 25% glycerol, mounted in nylon loops and frozen in liquid nitrogen.
Crystallographic data collection and processing
Native data sets were collected at beamline X06DA at the Swiss Light Source (Switzerland). Data sets were indexed and integrated in MOSFLM (32). Further data analysis was performed using the CCP4 suite of programs (33). The structure of Rap1-DBD in complex with G-quadruplex was solved by molecular replacement with PHASER (34). Search models for the protein were created by using the two Myb domains in the original Rap1-DBD crystal structure in complex with double-stranded DNA entry (PDB ID: 1IGN (29)). The DNA model was created using the guanines from the NMR structure of a G-quadruplex with the same sequences as the one present in the crystals (PDB ID: 2lk7 (35)). Initial PHASER runs resulted in the solution for a single Myb domain. Therefore, the Rap1-DBD model was separated into two PDB files, each containing a separate Myb/homeodomains without the linker between them and molecular replacement calculations repeated. This separation allowed us to find a solution for both Myb domains. The model was further improved by manual adjustment in COOT and several cycles of refinement in Refmac (36). The quality of final model was evaluated by MOLPROBITY (37). The asymmetric unit of the crystal contains one protein molecule bound to one G-quadruplex and the final model consists of residues 361–483 and 507–573 of the Rap1-DBD. Due to poor electron density, the C- terminal (574–596) region as well as the loop between amino acid residues 484 and 506 in Myb2 are missing from the model. All of the ‘T-loops’ DNA sequence is also clearly interpretable. The data processing and refinement statistics for the final data set are shown in Supplementary Table S1. Coordinates and structure model have been deposited in the RCSB Protein Data Bank accession number 6LDM. Figures of the structure were drawn using PyMOL (38).
RESULTS
The DBD of budding yeast Rap1 is sufficient for G-quadruplex binding
Previous biochemical studies from our group had demonstrated that budding yeast Rap1 drives the formation, and forms a specific complex with a parallel G-quadruplex DNA (18,31). However, since it was unclear from these studies which of the domains of Rap1 was responsible for the interaction with G-quadruplex DNA, different Rap1 domains (BRCA, DBD and RCT) (Figure 1C) were expressed in E. coli, purified and analyzed for binding using the S. cerevisiae telomeric G-strand oligonucleotide ‘4-G3’ (Table 1), that folds into a parallel G-quadruplex in the presence of potassium chloride, as shown by the CD spectrum with a positive peak at 260 nm and a negative peak at 240 nm (Supplementary Figure S1A) (31).
The electrophoretic mobility shift assay (EMSA) demonstrates that the DNA binding domain of Rap1 (Rap1-DBD) is sufficient for specific binding to the ‘4-G3’ G-quadruplex structure (Supplementary Figure S1B) with an apparent Kd = 28 nM, which is comparable to that of full length Rap1 as previously reported by us (18). The BRCT and RCT domains show no binding activity (Supplementary Figure S1C and D). Therefore, we conclude that the Rap1-DBD, which is sufficient for sequence specific binding to double-stranded DNA sites (28), is also able to bind specifically to G-quadruplex DNA.
To exclude the possibility that the single-stranded tails and loops present in the yeast telomeric ‘4-G3’ G-quadruplex (Table 1), and used above and in our previous studies (18,31), contribute to the observed binding, we analyzed the binding affinity of the same oligonucleotide lacking both single-stranded tails ‘4-G3 no tail’ (Table 1). The CD spectrum of ‘4-G3 no tail’ forms a parallel G-quadruplex in the presence of potassium chloride (Figure 1B), similarly to the longer ‘4-G3’ (Supplementary Figure S1A). The EMSA in Figure 1D shows that a complex is formed between the Rap1-DBD and the G-quadruplex, and that removing the single-stranded tails from ‘4-G3’ oligonucleotide does not result in a reduced binding affinity (apparent Kd = 28 nM versus = 22 nM). Subsequently, the role of the loops between G-tracts was investigated by analyzing the binding to the non-telomeric ‘T-loops’ oligonucleotide (Table 1) known to form a parallel G-quadruplex and chosen because like the yeast ‘4-G3’ contains four GGG tracts and hence forms a G-quadruplex containing a stack of three-G-tetrads, but is more stable (35). This G-quadruplex has single thymidines in the loops between the G-tracts, rather than the yeast ‘4-G3’ oligonucleotide that has longer TGTGT loops. Since the binding affinity for this G-quadruplex is approximately only 2-fold lower than for the yeast G-quadruplex (apparent Kd = 45 nM versus 22 nM) (Figure 1D and E), these results provide biochemical evidence that Rap1 specifically recognizes the DNA G-quadruplex structure itself. Interestingly, Rap1 does not discriminate between DNA and RNA G-quadruplexes, as it binds to a parallel, telomeric RNA G-quadruplexes ‘rTelo’ (Table 1) with the same affinity as to telomeric DNA G-quadruplexes (Supplementary Figure S1E and F).
G-quadruplex recognition by yeast Rap1
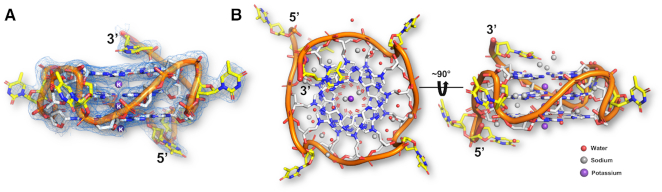
To establish the mechanism by which budding yeast Rap1 specifically recognizes G-quadruplex DNA, we determined the crystal structure at 2.4 Å resolution of the Rap1-DBD (residues 361–596) in complex with the parallel ‘T-loops’ G-quadruplex (Table 1, Figure 2). Despite a somewhat higher binding affinity, attempts to grow crystals of the Rap1-DBD in complex with the yeast ‘4-G3 no tail’ G-quadruplex or variations thereof failed. The asymmetric unit of the crystal contains one Rap1-DBD bound to one ‘T-loops’ G-quadruplex. In the crystal lattice, two Rap1-DBD/G-quadruplex complexes dimerize by stacking on top of each other through the planar and hydrophobic faces of the G-tetrads (Supplementary Figure S2). This ‘dimerization’, likely explains the larger molecular weight complex observed at higher protein concentrations in the EMSAs (Figure 1D). For the protein we observe clear density for amino acid residues 361–483 and 507–573 (Supplementary Figure S3) and no density for a loop consisting of residues 484–506, also not seen in the structure of Rap1-DBD complex with double-stranded DNA (29,39). The C-terminal 574–596 amino acid region involved in double-stranded DNA binding (29,39), is also not visible (Supplementary Figure S3). The overall architecture of the Rap1-DBD in the complex consists of two separate Myb/homeodomains linked by an ordered linker (Figure 2). As was previously observed in our crystal structure of the Rap1-DBD in complex with double-stranded DNA, the Myb1 domain consists a three-helix bundle whereas Myb2 consists of a four-helix bundle (29,39). For the G-quadruplex, there is clear density for the stack of three G-tetrads and the three short loops containing a single thymine cross the grooves of the parallel G-quadruplex helix joining the top and bottom G-tetrads in the stack (Figure 3A). The thymine bases in the loops point outwards permitting interactions with the protein. The density for the K+ ions, located between G-tetrad at the central axis of the G-quadruplex are also clearly interpretable (Figure 3A). Interestingly, the resolution of electron density map permits the assignment of a large number of ordered water molecules and Na+ ions, forming a water–sodium–water spine lining the grooves of the G-quadruplex helix (Figure 3B). The Na+ ions, present in both the protein and crystallization buffers, were tentatively assigned from contour levels in the electron-density map and their hydration geometry.
Figure 2.
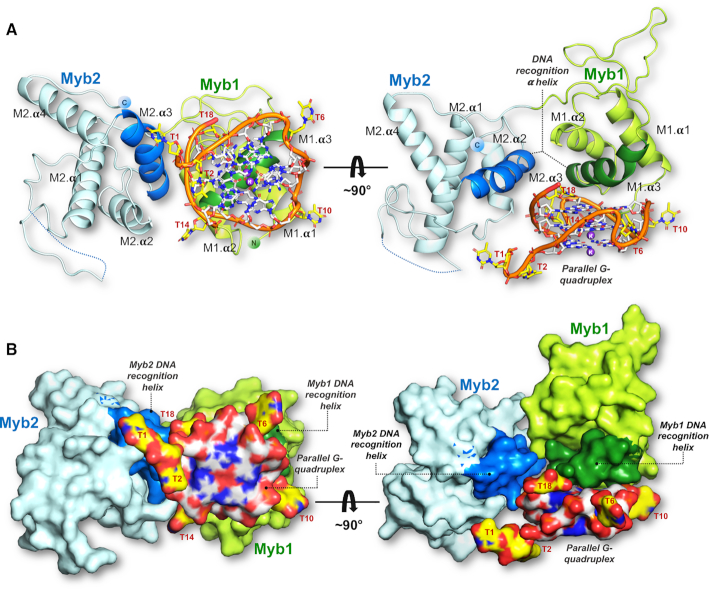
Global view of the structure of the complex between yeast Rap1-DBD and a parallel G-quadruplex. (A) Side and top view of the co-crystal structure where the Rap1-DBD is shown as a cartoon representation. (B) Surface representation of the complex shown in the same orientations as in (A). The two views are related by a 90° rotation. The structure is color coded: Myb1 domain (green), Myb2 (blue), and the DNA recognition α-helixes in the two Myb domains highlighted by a darker shade of the domain's color. The ribose–phosphate backbone of the G-quadruplex is shown in orange; light gray–blue represents guanines; yellow-blue thymines. The K+ ions at the central axis of the G-quadruplex are shown in purple.
Figure 3.
G-quadruplex structure, hydration and metal binding. (A) Side-view of the parallel ‘T-loops’ G-quadruplex shown within its well-defined 2Fo – Fc electron density map countered at 1.2 σ. (B) Two views related by a 90° rotation along the G-quadruplex helix axis, illustrating the hydration and metal ion binding of the G-quadruplex. The 3′ and 5′ ends of the G-quadruplex are indicated. Water molecules are shown in red, Na+ ions in grey and K+ ions in purple. The T-loops G-quadruplex is formed by the oligonucleotide sequence: 5′- -3′. Guanines are color-coded in the sequence to indicate the guanine number forming each G-tetrad:
-3′. Guanines are color-coded in the sequence to indicate the guanine number forming each G-tetrad:  , 5′G-tetrad,
, 5′G-tetrad,  , middle G-tetrad and
, middle G-tetrad and  , 3′G-tetrad.
, 3′G-tetrad.
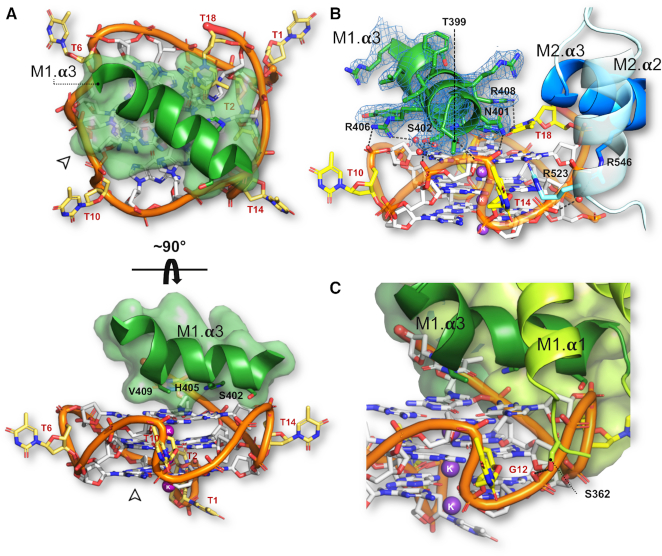
However, contrary to Rap1’s mode of recognition of double-stranded DNA in which the two Myb/homeodomains domains make essentially equivalent sequence-specific DNA interactions, only the Myb1 domain interacts specifically with the G-quadruplex (Figure 2). Recognition is via the third α-helix of the Myb domain - the DNA-recognition helix -docking onto the planar surface of the bottom (3′) G-tetrad (Figure 2A, Supplementary Figure S4). The parallel topology of the G-quadruplex results in the top and bottom faces of the G-tetrads in the stack to be accessible for protein binding, providing an explanation for the promotion of a parallel G-quadruplex by Rap1 when incubated with yeast telomeric oligonucleotides (31). The DNA-recognition helix of Myb1 (spanning residues Gly-400 to Tyr-410) sits diagonally across the G-tetrad almost entirely covering the exposed hydrophobic surface of the guanine bases (Figure 4A). It is positioned so that the imidazole ring of His-405 stacks on the pyrimidine–imidazole ring of G13 and Val-409 packs against G8 through van der Waal interactions. These amino acid side chains together with Ser-402 form a planar, primarily hydrophobic patch on one face of the DNA-recognition helix that specifies the interaction with the G-tetrad (Figure 4B). This mode of interaction was also observed for the G-quadruplex binding α-helix of the G-quadruplex helicase DHX36/RHAU (Figure 6C, discussed below) (40,41), and also resembles the mode of binding used by planar G-quadruplex ligands (42).
Figure 4.
Interaction of Rap1-DBD with the G-quadruplex: Only one Myb domain interacts specifically. (A) Top and side view of the docking of the Myb1 DNA-recognition helix onto the planar face the 3′ G-tetrad of the parallel G-quadruplex. The two views are related by a 90° rotation along the G-quadruplex helix axis. Transparent spheres represent van der Waals radii. The bottom panel shows Ser402, His405, Val409 located on adjacent turns of the α-helix making contacts with the planar hydrophobic surface of the G-tetrad. (B) View showing direct and water-mediated contacts by Myb1 DNA recognition helix (dark green) and Myb2 DNA-recognition helix and helix 2 to the G-quadruplex. The 2Fo-Fc map countered at 1.2σ for the Myb1 DNA-recognition helix is shown (C) View showing the N-terminal arm of Myb1 interacting with a groove of the G-quadruplex.
Figure 6.
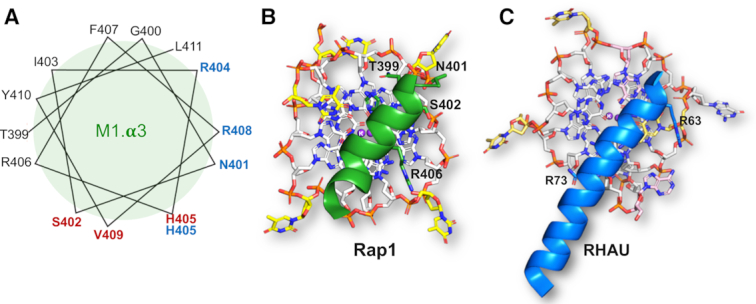
The Myb1 domain uses different faces of the DNA-recognition helix to recognize G-quadruplex and double-helical DNA. (A) Helical wheel representation of Myb1 DNA-recognition helix. Amino acids side chains forming specific contacts with double-helical DNA are shown in blue and those contacting the G-tetrad in in the G-quadruplex are shown in red. (B) View of the interaction of the Myb1 DNA-recognition helix with the G-tetrad looking down the G-quadruplex helix axis. (C) View of the Interaction of the RSM helix from RHAU with the G-tetrad looking down the G-quadruplex helix axis (taken from (40)). In both (B) and (C), the view is from the top showing the orientation of the α-helices onto the planar face of the G-tetrad. In both (A) and (B), only charged interactions are labelled as the hydrophobic ones are hidden in this view at the G-tetrad/DNA-recognition helix interphase.
In addition to the polar/hydrophobic interactions, the DNA-recognition helix of Myb1 is anchored to the face of the G-tetrad by a network of direct and water-mediated hydrogen bonds: Thr-399 and Asn-401 located at the N-terminal end of the DNA recognition helix contact the phosphate group of T14; Ser-402 makes a water-mediated contact to G9 as well as to the phosphate group of G13; Arg-406 at the C-terminus of the helix contacts the phosphate group of T10 as well as making a water mediated contact to G9 (Figure 4B, interaction map Figure 5C). The interaction of the Myb1 domain with the G-quadruplex is further stabilized through a short N-terminal arm (typical of homeodomains) entering a groove of the G-quadruplex helix, placing Ser-362 within hydrogen bonding distance to the phosphate group of G12 (Figure 4C).
Figure 5.
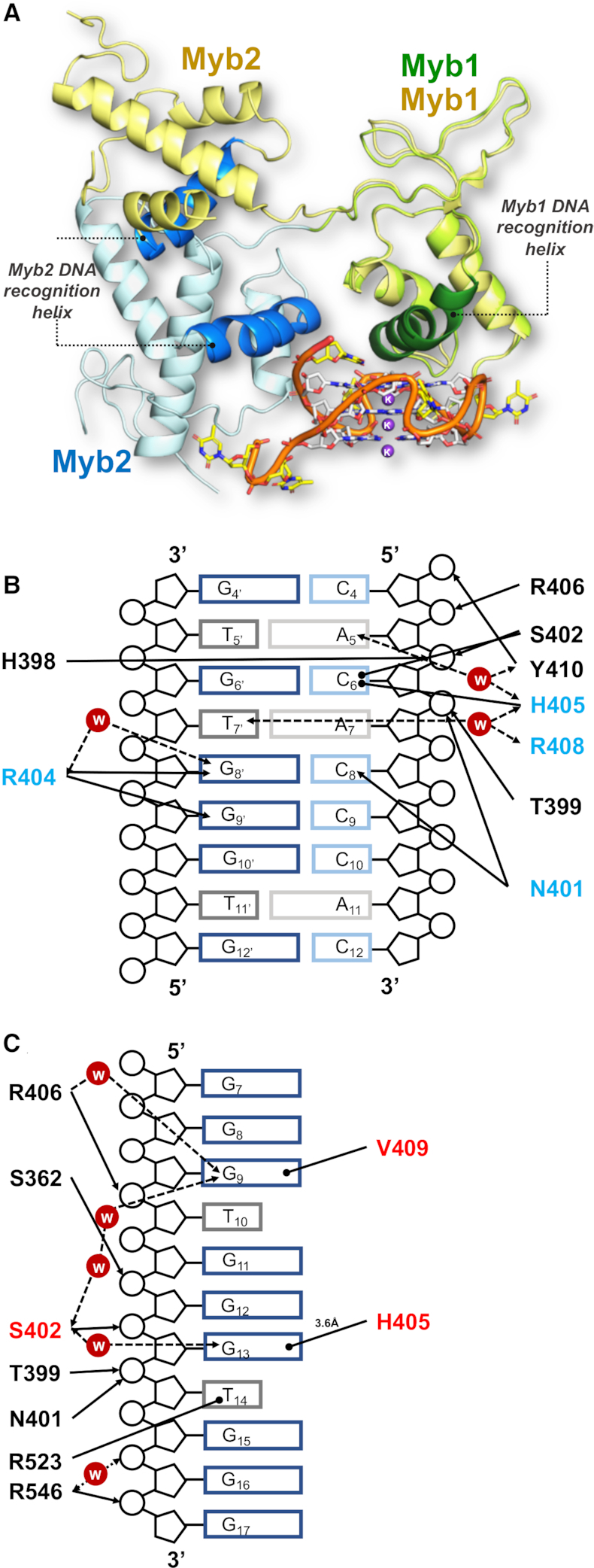
The Rap1-DBD undergoes a structural rearrangement on G-quadruplex binding (A) Superimposition of the Rap1-DBD in complex with double-helical DNA (yellow, PDB: 1IGN (29)) and with the G-quadruplex colour-coded as in previous figures: Myb1 in green and Myb2 in blue. The Myb 1 DNA-recognition helix is shown in dark green and that of Myb 2 in mid blue. The two structures were superimposed on the Myb1 domain structure. (B) Schematic map of the interactions of the Myb1 domain with its double-stranded telomeric DNA site (taken from (25)). Amino-acid side-chains involved in base-specific contacts are highlighted in blue. (C) Schematic map of the interactions of the Myb1 domain with the G-quadruplex DNA. Amino-acid side-chains involved in base-specific contacts are highlighted in blue. In both contact maps (B) and (C): arrows indicate direct and water-mediated hydrogen-bonds between protein and DNA; arrows with round heads indicate hydrophobic/stacking interactions. The distance and angular cut-off are 3.0 Å and 20°, respectively.
The Myb2 domain embraces the G-quadruplex by packing against the side of the G-quadruplex (Figure 2) through the interaction of Arg-546 with the phosphate groups of both G16 via a water molecule, and G17 directly. The Myb2 interaction is further stabilized by Arg-523 stacking on T14 located in the third loop of the G-quadruplex (Figures 4B and 5C).
We then asked why does only the Myb1 domain of the Rap1-DBD interact specifically with the G-quadruplex DNA and not Myb2? Inspection of the amino acid sequence alignment of the DNA recognition helices of Myb1 and Myb2 reveals significant differences (Supplementary Figure S5A). In Myb2, in place of His-405 and Val-409 important for the specific interactions with the face of the G-tetrad by Myb1, there are the larger and charged amino acid side chains aspartic acid (Asp-543) and lysine (Lys-547), respectively. Moreover, Ser-402 in Myb1 is swapped to an alanine (Ala-540) in Myb2. Consequently, Myb2 cannot make equivalent interactions. To test the effect of the amino acid differences in the two Myb domains, peptides encompassing the DNA-recognition helices of Myb1 and Myb2 were analyzed for G-quadruplex binding. The EMSA (Supplementary Figure S5B) shows that although both isolated DNA-recognition helices can interact with the G-quadruplex, albeit with a 40–200-fold lower affinity than the Rap1-DBD (Kd = 20–30 nM versus 1.2–5.4 μM respectively), the apparent binding affinity of the Myb2 peptide is 4- to 5-fold lower than that of Myb1 (Supplementary Figure S5B), providing experimental support for why the Myb1 domain of the Rap1-DBD preferentially interacts with the G-tetrad.
Rap1 undergoes conformational changes to recognize different DNA structure
To obtain a mechanistic understanding of how the DNA-binding domain of Rap1 is able to recognize and bind both to double-stranded and G-quadruplex DNA, we compared the crystal structures of Rap1-DBD in complex with double-stranded DNA (2.25 Å resolution) (29), with the structure presented here. The three-dimensional structures of the two complexes were superimposed by aligning on the Myb1 domain (Figure 5A). Although both Myb domains have the same 3D folds whether bound to double-helical or G-quadruplex DNA, their relative spatial orientation in the two complexes is dramatically different (Figure 5A). In the structure of Rap1-DBD in complex with the G-quadruplex, the Myb2 domain is twisted by 117° and displaced by 36 Å in respect to its position when bound to double-stranded DNA. From the structure of the complex with double-stranded DNA, it could be deduced that the relative orientation of the two tandemly arranged Myb domains in the Rap1-DBD is primarily determined by binding in the major grooves of two telomeric sequence repeats (GGTGT) spaced eight base pairs apart, and that the amino acid stretch linking the two domains becomes structured upon binding in the DNA minor groove (29). In other words, the linker between the two domains is likely to be unstructured in solution, permitting the different spatial orientation of the two Myb domains when bound to the G-quadruplex. Indeed, in the structure with the G-quadruplex where the linker region is not involved in DNA interactions but is instead structured through interactions with the closely spaced Myb domains, the orientation of the Myb1 domain is determined by the binding of its DNA-recognition helix to the surface of the G-tetrad and that of Myb2 by interactions from its DNA-recognition helix with the ribose-phosphate backbone of the G-quadruplex (Figure 4B)
Furthermore, although the DNA-recognition helix of Myb1 is used to recognize and bind both to double stranded and to G-quadruplex DNA, the location and chemical properties of the interacting amino acid side chains show crucial differences (Figure 5B and C). Firstly, the specific recognition of the double-stranded telomeric DNA sequence is primarily through highly basic amino acid side-chains making hydrogen bonds to the conserved G–G steps in adjacent major grooves in the double-stranded telomeric DNA (29), whereas that to G-quadruplex DNA is primarily through polar/hydrophobic side-chains recognizing the planar hydrophobic surface of the 3′ G-tetrad in the G-quadruplex (Figure 5B and C). A detailed analysis of the role of amino acid side-chains shows that in both structures, Thr-399 and Asn-401 and Arg-406 contact the ribose-phosphate backbone. His-405, that is crucial for the interaction with the G-tetrad by stacking on G13, in the complex with double-stranded DNA makes an unusual interaction with the cytosine that base pairs with G6, permitted by a protein induced distortion of the ribose-phosphate backbone at this point (29). The interaction with the planar face of the G-quartet is additionally increased by Val-409 stacking on G9, a residue not involved in interactions with double-stranded DNA (Figure 5A and B). Significantly, we note that the residues primarily involved in G-tetrad recognition (Ser-402, His-405, Val-409) are located on adjacent helical turns on one face of the DNA-recognition helix, whereas residues making base-specific hydrogen bonds to double-stranded DNA (Asn-401, Arg-404, Arg-408, Ser-410) are on a different face, representing a rotation of the α-helix by ∼120° (Figure 6A). In conclusion, comparison of the crystal structures of the two complexes reveals that the mechanism for the recognition of double-helical and G-quadruplex DNA structures is bimodal by using different faces of the DNA recognition helix.
DISCUSSION
The participation of G-quadruplexes in biology will require such structures to be recognized and the kinetics of their formation and resolution to be controlled by proteins (1). The crystal structure of Rap1-DBD in complex with a parallel G-quadruplex presented here provides an understanding at near-atomic resolution of how a Myb/homeo DNA-binding domain specifically recognizes the structure ofG-quadruplex DNA. Significantly, it also reveals, for the first time, how a conserved DNA-binding domain generally used to recognize double-helical DNA has a dual role through adapting its overall conformation and use of its DNA-recognition helix to also specifically bind to G-quadruplex DNA.
The crystal structure of the Rap1-DBD/G-quadruplex complex contains a non-telomeric G-quadruplex in which the three loops linking the G-tetrads (‘T-loops’) consist of single thymidines, whereas the cognate S. cerevisiae sequence ‘4-G3’ has longer TGTGT loops. Although we were unable to obtain crystals with the yeast G-quadruplex, or the ‘T-loops’ G-quadruplex containing the yeast loop sequence inserted in different loop positions (not shown), the topology and core of the G-quadruplex consisting of a stack of three G-tetrads is the same. Firstly, since the apparent binding affinity of the Rap1-DBD for the yeast ‘4-G3’ and ‘T-loops’ G-quadruplexes is very similar (Kd = 22 versus 45nM), it seems unlikely that the loops in the G-quadruplex contribute to G-quadruplex recognition and binding. Secondly, from the structure of the complex we can conclude that the primary and specific recognition by the protein is via the binding of a DNA-recognition helix to the flat surface of a G-tetrad. Thirdly, the structure of the complex shows that only the third loop (T14) of the ‘T-loops’ G-quadruplex makes a significant interaction with the Rap1-DBD located between Myb1 and Myb2 domains, and at this position in the structure there is space to accommodate a longer loop (Supplementary Figure S4).
Although the available structural information is limited, comparison of the structure presented here with the crystal structure of the DEAH/RHA helicase DHX36/RHAU in complex with a parallel G-quadruplex (41,43), suggests that the mode of interaction by an α-helix with a G-tetrad is conserved. Similarly to the DNA-recognition helix of the Myb1 domain of Rap1 (Figure 6B), the interaction of the RHAU G-quadruplex-binding α-helix, consisting of the RHAU-specific motif RSM shown experimentally to be essential for G-quadruplex recognition (40), to the flat face of a G-tetrad is determined by a non-polar/hydrophobic surface on the helix (Ile-65, Trp-68, Tyr-69 and Ala-70) making van der Waals interactions (Figure 6C). In this case also, the interaction is further stabilized by basic residues from the α-helix hydrogen bonding to the ribose-phosphate backbone (Figure 6B and C). It is these electrostatic interactions that lock the DNA-recognition helix on to the surface of the G-tetrad, and also explain the relatively high nanomolar binding affinities (20–40 nM) we observe for the Rap1-DBD for G-quadruplex DNA.
Significantly, comparison of the use of the DNA-recognition helix of the Rap1 Myb1 domain in double-helical DNA and G-quadruplex recognition suggests the mechanism by which the bimodal DNA recognition is made possible is through the DNA-recognition helix having two different faces: hydrophobic/polar amino acids located on one face and basic residues on the other, related by approximately 120° (Figure 6A). These observations suggest that the Myb1 domain might have evolved to enable Rap1 to have this dual role, and imparted on it the ability to switch between binding to double-helical and G-quadruplex DNA. Intriguingly, our observation that Rap1-DBD binds with the same high affinity to a telomeric RNA oligonucleotide folded into a parallel G-quadruplex (Supplementary Figure S1E and F), raises the possibility that Rap1 may exploit its G-quadruplex binding activity to interact with TERRA, the G-rich RNA transcript of telomeric DNA (44).
Finally, our high-resolution insight into how a protein interacts with a parallel G-quadruplex may aid the design of small-molecule ligands with better target selectivity and affinity for G-quadruplexes, and hence increase the potential of G-quadruplexes as therapeutic targets (9).
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 6LDM.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues from the NTU Institute of Structural Biology (NISB) for providing insights and expertise that greatly assisted this research. We acknowledge the Swiss Light Source (SLS), Villigen PSI, Switzerland for providing X-ray synchrotron beamtime at the X06DA (PXIII) beamline. We are grateful to Simon Lattmann for stimulating discussions and suggestions throughout the project. We are also indebted to Rafael Giraldo for discovering a quarter of a century ago that S. cerevisiae Rap1 binds to telomeric DNA G-quadruplexes.
Notes
Present address: Anna Traczyk, Genome Institute of Singapore, A*Star, 60 Biopolis St, Singapore 138672, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education Academic Research Fund (AcRF) Tier 3 [MOE2012-T3-1-001]. Funding for open access charge: Singapore Ministry of Education Academic Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Todd A.K. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huppert J.L. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 7. Capra J.A., Paeschke K., Singh M., Zakian V.A.. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010; 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lipps H.J., Rhodes D.. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009; 19:414–422. [DOI] [PubMed] [Google Scholar]
- 9. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 10. Islam M.K., Jackson P.J., Rahman K.M., Thurston D.E.. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016; 8:1259–1290. [DOI] [PubMed] [Google Scholar]
- 11. Maciejowski J., De Lange T.. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017; 18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wellinger R.J., Zakian V.A.. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012; 191:1073–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blackburn E.H. Telomeres. Elsevier Science. 1991; 16:378–381. [DOI] [PubMed] [Google Scholar]
- 14. Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989; 342:825–829. [DOI] [PubMed] [Google Scholar]
- 15. Williamson J.R., Raghuraman M.K., Cech T.R.. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell. 1989; 59:871–880. [DOI] [PubMed] [Google Scholar]
- 16. Sen D., Gilbert W.. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990; 344:410–414. [DOI] [PubMed] [Google Scholar]
- 17. Szostak J.W., Blackburn E.H.. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982; 29:245–255. [DOI] [PubMed] [Google Scholar]
- 18. Giraldo R., Rhodes D.. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994; 13:2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang G., Cech T.R.. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993; 74:875–885. [DOI] [PubMed] [Google Scholar]
- 20. Fang G., Cech T.R.. Characterization of a G-quartet formation reaction promoted by the β-subunit of the oxytricha telomere-binding protein. Biochemistry. 1993; 32:11646–11657. [DOI] [PubMed] [Google Scholar]
- 21. Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Plückthun A.. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paeschke K., Simonsson T., Postberg J., Rhodes D., Lipps H.J.. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005; 12:847–854. [DOI] [PubMed] [Google Scholar]
- 23. Lustig A.J., Kurtz S., Shore D.. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990; 250:549–553. [DOI] [PubMed] [Google Scholar]
- 24. Shore D., Nasmyth K.. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987; 51:721–732. [DOI] [PubMed] [Google Scholar]
- 25. Conrad M.N., Wright J.H., Wolf A.J., Zakian V.A.. RAP1 protein interacts with yeast telomeres in vivo: Overproduction alters telomere structure and decreases chromosome stability. Cell. 1990; 63:739–750. [DOI] [PubMed] [Google Scholar]
- 26. Lue N.F. Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 2010; 35:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kyrion G., Liu K., Liu C., Lustig A.J.. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993; 7:1146–1159. [DOI] [PubMed] [Google Scholar]
- 28. Graham I.R., Haw R.A., Spink K.G., Halden K.A., Chambers A.. In vivo analysis of functional regions within yeast Rap1p. Mol. Cell Biol. 1999; 19:7481–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konig P., Giraldo R., Chapman L., Rhodes D.. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996; 85:125–136. [DOI] [PubMed] [Google Scholar]
- 30. Court R., Chapman L., Fairall L., Rhodes D.. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005; 6:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giraldo R., Suzuki M., Chapman L., Rhodes D.. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:7658–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W.. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. et al.. Overview of the CCP 4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Do N.Q., Phan A.T.. Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded g-quadruplexes: NMR structural study. Chem. - A Eur. J. 2012; 18:14752–14759. [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., Snoeyink J., Richardson J.S. et al.. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007; 35:375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schrödinger L. The PyMOL Molecular Graphics System, Version 2.3. 2015;
- 39. Taylor H.O., O’Reilly M., Leslie A.G., Rhodes D.. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol. 2000; 303:693–707. [DOI] [PubMed] [Google Scholar]
- 40. Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y.. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010; 38:6219–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen M.C., Tippana R., Demeshkina N.A., Murat P., Balasubramanian S., Myong S., Ferré-D’Amaré A.R.. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018; 558:465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohnmacht S.A., Neidle S.. Small-molecule quadruplex-targeted drug discovery. Bioorganic Med. Chem. Lett. 2014; 24:2602–2612. [DOI] [PubMed] [Google Scholar]
- 43. Heddi B., Cheong V.V., Martadinata H., Phan A.T.. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: solution structure of a peptide–quadruplex complex. Proc. Natl. Acad. Sci. U.S.A. 2015; 637371:201422605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 6LDM.