Abstract
The master tumor suppressor p53 controls transcription of a wide-ranging gene network involved in apoptosis, cell cycle arrest, DNA damage repair, and senescence. Recent studies revealed pervasive binding of p53 to cis-regulatory elements (CREs), which are non-coding segments of DNA that spatially and temporally control transcription through the combinatorial binding of local transcription factors. Although the role of p53 as a strong trans-activator of gene expression is well known, the co-regulatory factors and local sequences acting at p53-bound CREs are comparatively understudied. We designed and executed a massively parallel reporter assay (MPRA) to investigate the effect of transcription factor binding motifs and local sequence context on p53-bound CRE activity. Our data indicate that p53-bound CREs are both positively and negatively affected by alterations in local sequence context and changes to co-regulatory TF motifs. Our data suggest p53 has the flexibility to cooperate with a variety of transcription factors in order to regulate CRE activity. By utilizing different sets of co-factors across CREs, we hypothesize that global p53 activity is guarded against loss of any one regulatory partner, allowing for dynamic and redundant control of p53-mediated transcription.
INTRODUCTION
The master tumor suppressor p53 is a transcription factor with key roles in preserving genome fidelity and cellular homeostasis. In support of these activities, p53 regulates a core transcriptional program involved in cellular processes like cell cycle arrest, apoptosis, DNA repair, and senescence (1–3). Loss of p53 activity is strongly linked to increased cancer risk and decreased life expectancy, and misregulation of p53 is associated with numerous other human disorders. Recent analyses suggest that p53 is mutated in greater than 30% of cancer cases and the majority of p53 variants are unable to bind DNA and enact a tumor suppressive gene expression program (4). The mechanisms by which tumorigenesis progresses in the presence of wild type p53 activity have not been well characterized. Recent evidence suggests that sequence variation within cis-regulatory elements (CREs) can influence p53 binding, transcriptional activity, and tumor suppressor function (5–7). The critical nature of the core p53 response element (p53RE) on p53 binding and CRE activity is well understood (8–10), but the influence of local sequence variation and the role of additional transcription factor motifs within a CRE on p53 activity remains an open and vital question.
Cis-regulatory elements, such as promoters and enhancers, govern gene expression through temporal, spatial, and quantitative control of transcription (11,12). While multiple models for CRE function have been proposed, the majority involve cooperative binding of transcription factors and cofactors acting locally to fine-tune gene expression (11–13). The presence and availability of transcription factors, repressors, and other cofactors vary across cell states such as development, stress, disease, and cell type (14–17). This variability provides a mechanism for differential CRE activity and downstream gene expression. Loss of transcription factor binding, through variation in DNA sequence or through changes in trans-factor availability, can strongly influence CRE activity and gene expression (11,12,18), with direct implications in numerous developmental and disease states (14,16).
While general transcription factors, like the TFIID complex (19,20), are involved in p53-dependent trans-activation at promoters, the requirement for other sequence-specific trans-factors at distally-acting CREs is unknown. A novel model was recently proposed whereby binding of a single transcription factor, in this case p53, was necessary and sufficient for CRE activity (9). This model was supported by another study suggesting that a canonical p53 response element (p53RE) is the only sequence-based determinant of p53-dependent CRE activity (10). However, multiple p53-dependent CREs have been reported to require other locally-acting transcription factors, in line with established CRE mechanisms like the enhanceosome and billboard models (18,21). For example, CRISPR/Cas9-based screening identified a CEBPβ-binding site within a CRE regulating CDKN1A/p21 required for p53-dependent senescence (22). Transcription factors such as those in the AP-1 family and SP1 have also been implicated in the activation of p53-dependent gene targets (23–25).
In order to directly address whether additional cofactors are required for p53-dependent transcriptional activity, we examined the effect of local sequence variation on putative p53 CREs using a massively parallel reporter assay (MPRA). Our results suggest that sequences flanking p53REs and transcription factors other than p53 are required for p53-dependent transcriptional activation. Consistent with previous reports, the p53RE is a strong determinant of p53-inducible activity. Loss of p53 occupancy through sequence manipulation or depletion of the protein strongly reduces CRE activity. We also identified sequences outside of the p53RE that positively or negatively regulate transcription. This includes a conserved SP1/KLF family binding site required for optimal transcription of the p53-dependent gene CCNG1 (cyclin G1). We also identified two distinct CREs with different local transcription factor requirements that are both necessary for p53-dependent transcriptional activation of GDF15, a gene recently identified as a key mediator of inflammation and metabolic function (26,27). Thus, p53-bound CREs do not depend on just one family of transcription factors for activity and can utilize multiple and different factors to regulate transcription. Thus, these data indicate p53’s flexibility to collaborate with different combinations of locally available transcription factors to regulate CRE activity and downstream gene activation involved in key organism-level traits like tumor suppression.
MATERIALS AND METHODS
Cell culture
HCT116 parental, TP53−/− and ATF3−/− lines were cultured in McCoy's 5A media with 10% fetal bovine serum. HCT116 ATF3−/− cell line was a kind gift of Chunhong Yan (Augusta University) (28). MCF10A cells were grown in HuMEC media (Gibco). Mouse embryonic fibroblasts (MEFs) were grown in DMEM with 10% FBS and were a kind gift of Jing Huang (National Cancer Institute, NIH). All cell lines were cultured at 37°C and 5% CO2 in a water-jacketed incubator.
Selection of candidate enhancers
Candidate enhancer regions were selected starting with regions of the hg19 genome assembly containing DNAse Hypersensitive Sites (DHS) (wgEncodeRegDnaseClusteredV3 downloaded from http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegDnaseClustered/). DHS were then filtered by the presence of a p53 family motif using gimmeMotifs (29). Ultimately, 296 DHS containing a p53 family motif and falling within 100 kb of a coding sequence transcriptional start site (TSS) were randomly selected for MPRA analysis. In parallel, 196 enhancers from the FANTOM Ubiquitous Enhancer group were selected as positive controls, with the central 100 bp segment of each enhancer used in the MPRA (http://enhancer.binf.ku.dk/presets/Ubiquitous_enhancers_S9.bed). Candidate p53-bound enhancers were shortened to 100 bp with the 20 bp p53 response element motif at the center with 40 bp of flanking genomic context on each side. Each candidate p53-bound enhancer was scrambled in 20 bp sections from 5′ to 3′ across the entire length producing. Nucleotide randomization preserved GC content and was performed using EMBOSS shuffleseq (30). As negative controls for regulatory activity, the entire 100 bp sequence for all candidate or ubiquitous enhancers was scrambled while preserving GC content.
Massively Parallel Reporter Assay (MPRA) oligo design
Our MPRA was designed using a previously published method (31). In brief, each candidate 100 bp regulatory sequence was coupled to five separate 12 nucleotide unique molecular identifier (UMI) sequences. Replicates of the test sequences plus controls totaled the library at 12 035 unique oligos in the orientation of: a 5′ Primer binding overlap, a 100 bp enhancer sequence, a spacer for restriction enzyme sites EcoRI and SbfI, a unique enhancer associated 12 bp barcode, and a 3′ Primer binding overlap. All sequences for the MPRA oligo library are found in Supplemental Table S1. The final 12 035 unique oligo pool was synthesized by CustomArray.
Two-step vector library cloning and verification
The MPRA lentiviral vector pLs-mP was a gift from Nadav Ahituv (Addgene plasmid # 81225; http://n2t.net/addgene:81225; RRID:Addgene_81225). pLs-mP was digested with the restriction enzymes EcoRI and SbfI yielding two fragments representing the plasmid backbone and the minimal promoter/eGFP. The candidate enhancer pool was PCR amplified using primers SL468 and SL469 in 3 separate reactions of 50 ng at 21 cycles each, gel purified, and combined (Supplemental Table S2, cloning_primers tab). The resulting PCR product was ligated to the EcoRI–SbfI digested pLs-mP backbone in three separate Gibson assembly reactions (HiFi DNA Assembly, NEB). 2 μl of each Gibson assembly reaction were transformed into Stbl4 electrocompetent Escherichia coli (Invitrogen) in three separate transformation reactions (1200 V, 200 Ω, BioRad). Transformation reactions were plated on 10 separate 15 cm LB-agar plates with 100 ug/ml ampicillin selection at 30°C for 48 h. Colonies were isolated directly from plates and plasmid DNA was individually prepped (ZymoPURE II Midi Plasmid Kit). The resulting DNA was combined to create the Step 1 library. This plasmid pool was then digested with EcoRI and SbfI in three separate reactions at 1 μg each and gel purified. The 780 bp fragment from the original EcoRI–SbfI reaction (containing the minimal promoter/EGFP fragment), was ligated into the digested Step 1 plasmid using T4 ligase. Ligation products were transformed as above and the resulting MPRA library was sequence verified by Illumina sequencing.
MPRA virus production and transduction
HEK293FT cells were used for virus preparation and were cultured in DMEM with 10% FBS without antibiotic. To make virus, 4 × 106 cells were seeded in 10 cm plates 24 h before transfection. Per 10 cm plate, cells were transfected with 8 ug of MPRA lentiviral backbone, 4 ug of M2G helper plasmid and 8ug of ps-PAX helper plasmid using JetPrime transfection reagent according to manufacturer's recommendations. Virus was collected from supernatant at 24 and 48 h, pooled, and filtered using 0.45 μm syringe filters. HCT116 TP53+/+ and TP53−/− cell lines were seeded 24 h before viral transduction in three 10 cm plates at a concentration of 2.0 × 106 cells/plate. Virus supernatant was combined with 8 μg/ml polybrene, added to the seeded cells, and incubated for 48 h. Cells were then treated for 6 h with DMSO (as a control) or 5 μM Nutlin-3A (45-SML0580, Millipore Sigma) to induce p53 activity. One plate was left untreated as the infection control plate for genomic DNA isolation. After 6 h of treatment, cells were collected in ice-cold 1× PBS, snap-frozen on liquid nitrogen, and stored at −80°C until analysis. Three biological replicates were performed for each condition.
Amplicon enrichment and RNA-seq library preparation
Total RNA was isolated from DMSO or Nutlin-3A treated cells (EZ RNA Kit, Omega Biotek) with on-column DNAseI treatment. 6 ug of resulting total RNA was then taken through an additional round of TurboDNase treatment (ThermoFisher) to ensure complete removal of contaminating genomic DNA. The resulting RNA was split into three first strand reverse transcription reactions each using custom barcoded primers to identify the cell line, treatment, and replicate number (Supplemental Table S2, Barcode_primers tab). All three cDNA reactions were combined and taken through a two-step PCR amplification process. In Round 1, each cDNA sample was amplified in 22 separate PCR reactions of three cycles each using barcoded primers (Supplemental Table S2). PCR reactions were then pooled and purified using 2.5× AMPure XP beads (Beckman-Coulter). The purified Round 1 PCR product went through a second round of PCR in eight reactions for 15 cycles each using barcoded primers as described in Supplemental Table S2. Step 2 PCR product was run on a 2% agarose gel and gel purified. Genomic DNA controls were prepared in a similar manner. 500 ng of genomic DNA was PCR amplified across 16 separate reactions of three cycles each using barcoded primers described in Supplemental Table S2. The pooled PCR product was combined and purified using 2.5× volume AMPure XP beads. The resulting purified DNA was then separated into 16 separate PCR reactions of 15 cycles each, pooled, and gel purified. After the two-step PCR reaction, DNA amplicons representing the expressed mRNA barcode and the genomic DNA infection control within each biological replicate were combined at equal molarity. An Illumina-compatible sequencing library was generated (NEBNext Ultra II DNA Library Kit, New England Biolabs) for each biological replicate and sequenced using the NextSeq 500 at the University at Albany Center for Functional Genomics.
Massively Parallel Reporter Assay (MPRA) data analysis
Sequencing reads for each individual experimental condition were flanked by a unique 5′ and 3′ amplicon barcode to allow separation from the pooled raw sequencing reads. Barcoded, experimental condition-specific reads were separated into individual files for further analysis using the FastX toolkit (fastx-barcode-splitter, http://hannonlab.cshl.edu). Barcodes for within-pool sequence identification can be found in Supplemental Table S2 (barcode_primers tab). The number of reads containing unique enhancer identifying sequences were then parsed and counted using fastX-collapser from the FastX-toolkit. Raw read counts for each enhancer sequence across cell lines, treatment conditions, and biological replicates be found in Supplemental Table S3. Differential enhancer activity across experimental conditions and cell lines was calculated from raw enhancer barcode read counts using DESeq2 (32). To account for differences in representation across the original viral enhancer sequence library, raw enhancer barcode read counts scaled to transcripts per million (TPM) and were then normalized to the read counts from genomic DNA (fold-change, RNA barcode/ DNA barcode). Normalized enhancer count values for mutant enhancer sequences were then compared to wild-type values using a one-way ANOVA with a post-hoc Tukey HSD test implemented in R (33).
In vivo CRISPR/Cas9 mutagenesis and amplicon ChIP-sequencing
HCT116 colon carcinoma cells were transduced with lentiCas9-Blast and cells stably expressing wild-type spCas9 were selected using 2 μg/ml blasticidin. LentiCas9-Blast was a gift from Feng Zhang (Addgene plasmid #52962; http://n2t.net/addgene:52962; RRID:Addgene_52962).
Streptomyces pyogenes (sp) guide RNA sequences (Supplemental Table S2) were cloned into the LentiGuide-Puro plasmid (plasmid #52963, Addgene). lentiGuide-Puro was a gift from Feng Zhang (Addgene plasmid # 52963; http://n2t.net/addgene:52963; RRID:Addgene_52963). Both LentiGuid-Puro and LentCas9-Blast were originally published in (34). Viral particles from LentiGuide-Puro were made by transfecting HEK293FT cells with the packaging plasmids psPax2 and pMD2.G and the respective guide RNA viral backbone cloned into pLS-mP. psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260). pMD2.G was a gift from Didier Trono (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259).
HCT116 cells stably expressing spCas9 were selected with 2 μg/ml puromycin 48 h after infection. The heterogenous cell pool was then treated for 6 h with either DMSO or 5 μM Nutlin-3A for either qPCR-mediated gene expression measurements or for ChIP. For ChIP, 10 million cells were crosslinked on plate with 1% formaldehyde for 10 min at room temperature until the reaction was quenched with 2.5% glycine. Crosslinked cells were processed using standard lysis procedures and chromatin was sonicated using a probe sonicator (Qsonica) at 25% amp for 10 pulses total; 10 seconds on, 50 s off for a total of 10 min at 4°C. Sheared chromatin was then used in a ChIP assay for p53 (clone DO1, BD Biosciences). 50 ng of purified DNA per experimental sample was used in a barcoding PCR reaction (Supplemental Table S2, barcode_primers tab), and amplicons were then used as template for created Illumina-compatible sequencing libraries (NEBNext Ultra II DNA Library Kit). Libraries were quantified by qPCR and run on an Illumina NextSeq 500 (150 bp single-end). Raw FastQ reads containing the amplicon primers (Supplemental Table S2, barcode_primers tab) were filtered (fastx-barcode-splitter, FastX-toolkit, http://hannonlab.cshl.edu) and used in subsequent analysis. The number of reads per DNA variant were quantified (fastX-collapser, FastX-toolkit) and the ChIP values were normalized to genomic DNA input. DNA variants were then sorted by the presence or absence of p53 or Sp1/KLF family motifs as determined by gimmeMotifs (29). Data tables for the amplicon ChIP-seq experiment can be found in Supplemental Table S4.
Quantitative real time PCR
Total RNA was isolated using the Omega E.Z RNA kit with an on-column treatment with 50 units of RNAse-free DNase I for 30 min. Single-stranded cDNA was generated with the High Capacity cDNA Reverse Transcription reagents (Applied Biosystems), and qPCR was performed on an Applied Biosystems 7900H with the relative standard curve method and iTaq Universal SYBR Green Supermix reagents (Biorad). qPCR primers are shown in Supplemental Table S2 (qPCR tab).
Chromatin immunoprecipitation for histone modifications
HCT116 colon carcinoma cells were treated for 6 h with 5uM Nutlin-3A and then crosslinked with methanol-free formaldehyde (1% final) at room temperature for 5 min. The reaction was then quenched with 2.5 M glycine for 5 min, followed by two washes with ice-cold PBS. Nuclear extraction and sonication were performed as previously described (35), with an average fragment size of 500 bp. Crosslinked material was then immunoprecipitated with 5 ug of either anti-H3K27ac (Active Motif 39133) or anti-H3K4me2 (Millipore 07-030) coupled to Protein A/G Dynabeads (Invitrogen) overnight at 4°C. Beads were then washed 3× with low-salt buffer, 1× with high-salt buffer, and 1× with LiCl buffer, followed by elution with 1% SDS and 500 mM NaCl at 65°C with shaking (35). Eluted DNA was then quantified and used for Illumina-compatible library preparation using the New England Biolabs NEBNext Ultra II DNA Library prep reagents. Libraries were sequenced on an Illumina NextSeq 500. Raw data were aligned to the human hg19 genome assembly using hisat2 (36). DNAse-seq, H3K4me1 and H3K4me3 histone modification data were obtained from the ENCODE Reference Epigenome Series for HCT116 colon carcinoma cell lines under accession ENCSR309SGV. eRNA data were obtained from GEO GSE53966 (2) and GEO GSE86165 (1). Normalized read counts within MPRA region coordinates were quantified using HOMER (37).
Luciferase plasmid cloning and expression assays
Candidate enhancer sequences were synthesized as dsDNA (IDT) and cloned into the pGL4.24 destabilized Luciferase reporter vector (Promega) using the HiFi DNA Assembly method (NEB). Enhancer variants were created using the inverse PCR method with Hot Start Q5 Polymerase (NEB) and primers available in Supplemental Table S2. Plasmids were reverse transfected according to manufacturer's recommendations (JetPrime, Polyplus Transfection) in triplicate in a 96-well plate. Plasmid DNA (0.2 μg) was transfected at a ratio of 9:1 for the candidate enhancer:constitutive promoter driving Renilla luciferase (pGL4.75, Promega). Luciferase activity was determined using the Promega Dual-Luciferase® Reporter Assay System according to manufacturer specifications on a Synergy HI plate reader (Bio-Tek).
Western blotting
Total protein was isolated using RIPA buffer, followed by a 15-min incubation on ice, and pelleting of and centrifugation to remove insoluble debris. Protein lysate samples were run at 180 V on NuPAGE 10% Bis–Tris protein gels from Invitrogen. Samples were blotted onto 0.2 μm nitrocellulose membrane and probed with p53 (clone DO1, BD Biosciences, #554923) or GAPDH (Cell signaling #5174S) antibodies. Proteins were visualized with HRP-conjugated secondary antibodies and SuperSignal Chemiluminescent reagents (Thermo Scientific) and imaged on a BioRad Chemidoc imager.
Protein expression and purification
Human p531–393 in pGEX-2TK (Ampicillin) coding for an in-frame N-terminal GST tag was a gift from Cheryl Arrowsmith (Addgene plasmid # 24860; http://n2t.net/addgene:24860; RRID:Addgene_24860 and (38)). p531–393 was expressed in Rosetta 2 DE3 cells (EMDMillipore) in 2YT media. Cells were grown at 37°C with shaking at 225 rpm until OD600 nm = 0.4–0.6, then shifted to 22°C and induced with 1 mM IPTG for 4 h. Cells were harvested by centrifugation at 4800 rpm for 15 min. Cell pellets were lysed in GST Buffer A (20 mM Tris–HCl pH 7.3 (RT), 300 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 5% glycerol, 10 mM Na-butyrate and supplemented with Complete protease inhibitor cocktail (Roche) and 0.1% NP40. Cells were lysed by sonication. Cell lysates were cleared by centrifugation at 18 000 rpm for 45 min at 4°C. Cell lysates were filtered with a 0.45 uM syringe filter and loaded on to an equilibrated 5 ml GSTrap HP column (GE Healthcare) using an Akta Purifier FPLC (GE Healthcare). The column was washed with GST Buffer A; bound protein was eluted using a linear gradient of 0–100% GST Buffer B (GST Buffer A + 10 mM glutathione (reduced)). Purity was determined by SDS PAGE using a 4–12% Bis–Tris gel run in 1× MOPS SDS buffer. Eluted fractions containing p531–393 constructs were pooled and concentrated using a Vivaspin 10 kDa MWCO centrifugal concentrator (GE healthcare). Concentrated protein was dialyzed overnight into GST-Buffer D (GST Buffer A + 20% glycerol). Protein concentration was determined using a nanodrop with calculated extinction co-efficient and molecular weight parameters. Purified protein was divided into single use aliquots, flash frozen in liquid nitrogen and stored at −80°C.
CCNG1 p53 electrophoretic mobility shift assay
Binding of p53 to CCNG1 enhancer sequences was tested by electrophoretic mobility shift assays (EMSAs). 60 bp HPLC purified DNA sequences (IDT, Supplemental Table S2, EMSA tab) were resuspended at 100 μM in 1× TE buffer. Equal volumes of complementary oligonucleotides were heated at 95°C for 5 min and annealed by cooling to 21°C for 30 min. Binding reactions were assembled in DNase/RNase free, low adhesion microcentrifuge tubes. Reactions contained 1× p53 binding buffer (50 mM Tris–HCl pH 7.3, 150 mM NaCl, 2 mM DTT, 0.1 mg/ml BSA). Unless otherwise stated, 50 nM of annealed DNA was added to each reaction. Binding reactions were started by adding the required concentration of p531–393 and allowed to proceed for 30 min at 21°C. Reactions were loaded immediately on a vertical 0.7% Agarose gel (Ultrapure agarose, Life Technologies) buffered with 1× TBE. Gels were run for 45 min at 60 V in 1× TBE buffer. Gels were stained for 10 min with 1× SYBR gold nucleic acid stain (ThermoFisher) in 1× TBE buffer and imaged on a G:box blue light transilluminator (Syngene). Densitometric analysis was carried out in Fiji (39); binding curves were fitted using the Hill equation with a two-site binding model (Hill coefficient = 2) using non-linear regression (nls) in R (33).
RNA sequencing
HCT116 parental, TP53−/− or ATF3−/− cells were treated at 80% confluency in a six-well plate with either DMSO or 5 μM Nutlin-3A for 6 h and total RNA was isolated (EZ RNA, Omega Biotek). PolyA+ RNA was purified using poly dT magnetic beads (Perkin Elmer) and fragmented at 90°C for 15 min. Fragmented RNA was used as the template for double-stranded cDNA production using random hexamers (first strand synthesis) and the dUTP method to preserve strandedness (second strand synthesis). The resulting double-stranded cDNA was then used to construct an Illumina-compatible sequencing library (BioO NextFlex RNA Library Kit, Perkin Elmer). Libraries were quantified using qPCR (NEBNext Library Quantification, New England Biolabs) and an Agilent Bioanalyzer and then pooled for sequencing on an Illumina NextSeq 500. Sequencing reads were aligned to the hg19/GRCh37 assembly and transcript counts were determined using quantMode in STAR (40). Differential gene expression and normalized gene counts were determined using DESeq2 (32).
Transcription factor peak intersections
Transcription factor peak files were obtained from the Cistrome database ((41), http://cistrome.org/db, accessed 1 July 2019). MPRA regions were converted into the hg38 genome assembly coordinates using liftOver from hg19 to hg38 (UCSC). Transcription factor peak summits were then intersected with MPRA regions using BedTools (intersectBed) (42). Peak intersection data were clustered by row (MPRA regions) and column (transcription factor) by One minus Pearson Correlation with complete linkage using Morpheus (https://software.broadinstitute.org/morpheus). Data underlying the cluster analysis can be found in Supplemental Table S5.
Transcription factor motif analysis
Three tools were used to identify motifs either present or enriched within the 100 bp regions from the MPRA p53-bound and p53-unbound peaks. Additionally, we analyzed transcription factor motif presence and enrichment within a set of p53 binding sites identified across multiple cell types in Verfaillie et al. (9). For the Verfaillie p53 set, we considered only the 1001/1149 (87%) p53 peaks found within DNAse Hypersensitive Clusters (UCSC GenomeBrowser, ENCODE Regulation Track (43,44)). Motif analysis was then performed on the entire DNAse Hypersensitive Cluster region. Known Motif enrichment using HOMER (37) was performed using the findMotifsGenome module (findMotifsGenome.pl -nomotif -size given) against the hg19 genome. Analysis for the presence of known motifs within MPRA regions and the Verfaillie p53 set was performed using the 2018 release of JASPAR Vertebrate Transcription Factor Motif Database (45) and both gimmeMotifs (29) and UCSC TableBrowser (46). Analysis of transcription factor motif enrichment using gimmeMotifs was run using the scan module (options -t -g hg19 -p JASPAR2018_vertebrates.pfm). The presence of hg19-based JASPAR vertebrate transcription factor motifs within p53-bound MPRA peaks and within the Verfaillie p53 set was performed using the UCSC TableBrowser and the JASPAR 2018 hg19 Track Hub. Only motifs with an enrichment score of 400 or higher (P < 0.0001) were considered for analysis. Histograms of motif enrichment within MPRA p53 or Verfaillie p53 peak sets were generated using bedTools (coverageBed -d option) and Morpheus.
Sequencing data availability
All sequencing data generated as part of this manuscript are available under Gene Expression Omnibus (GEO) Accession GSE137297. p53 ChIP data from Nutlin-3A-treated cells were obtained from GSE86222 (1). Conserved element data (bigWig) from PhyloP (47) and PhastCon (48) were obtained from the UCSC Genome Browser and plots were generated using deepTools (49).
RESULTS
Design and execution of a massively parallel reporter assay for determinants of p53-dependent CRE activity
We designed a barcoded and chromosomally integrated reporter system to assess the function of DNA sequences flanking the p53RE in p53-dependent CREs (Figure 1A). The lentivirus-based system expresses eGFP under the control of a minimal promoter and a putative p53-dependent CRE sequence (31). Each putative CRE was included in the library with five unique, 12 nucleotide barcodes encoded in the 3′UTR of eGFP to allow a molecular readout of transcriptional activity (Figure 1A). We selected 296 putative p53 binding locations based on the presence of a canonical p53 family binding motif (p53RE), distance from the nearest transcriptional start site (<100 kb), and localization within a DNAse hypersensitive site (DHS). The 20 bp p53 family motif was centered in a 100 bp fragment with 40 bp of flanking genomic context on each side. We also included 196 p53-independent and constitutively active CREs as determined by FANTOM Consortium CAGE data as positive controls of CRE activity (50). These sequences were cloned into the lentiviral plasmid backbone pLS-MP as described in the Methods and similar to previously described MPRA designs (31).
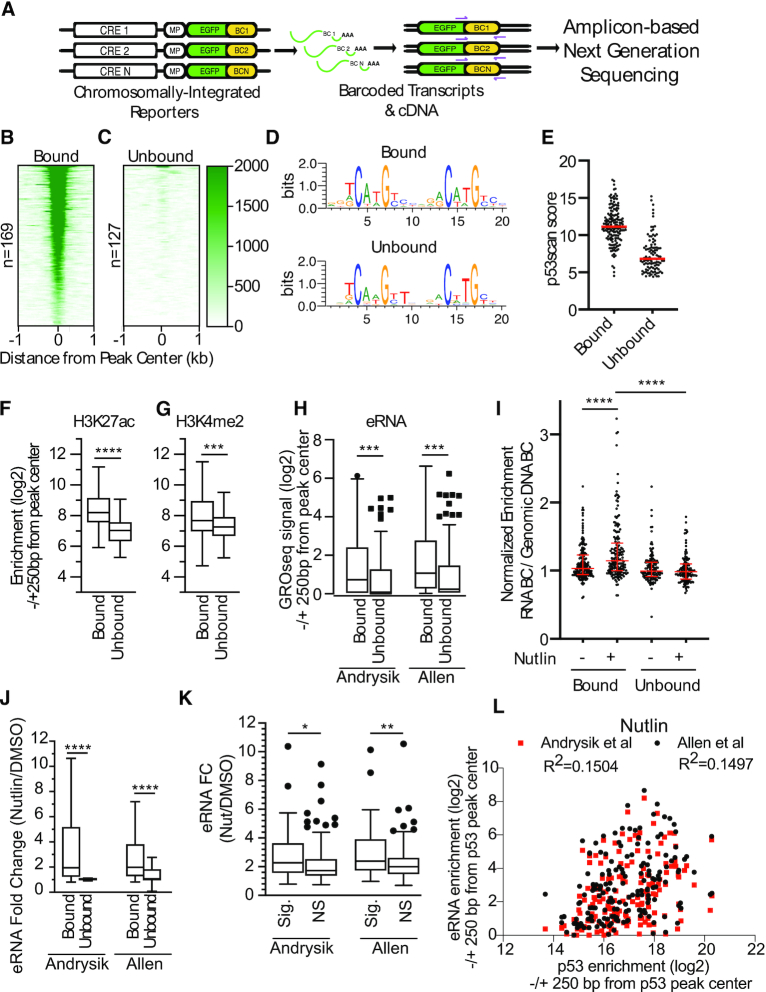
Figure 1.
(A) Schematic and workflow for a massively parallel reporter assay (MPRA) to study the effect of flanking sequence context on p53 transcriptional activity. Heatmaps of p53 ChIP-seq enrichment from Nutlin-3A-treated HCT116 colon carcinoma cell lines for (B) p53-bound or (C) p53-unbound regions found in the MPRA pool. (D) DNA sequence weight motifs for p53-bound (top) or unbound (bottom) regions from the MPRA pool. (E) Jitter plot of scores from p53scan depicting the adherence to a canonical p53 family motif sequence for the p53-bound and p53-unbound regions. Enrichment of (F) H3K27ac or (G) H3K4me2 in Nutlin-3A-treated HCT116 cells at p53-bound or unbound regions. Statistics represent an unpaired, two-tailed t-test (***P < 0.001, ****P < 0.0001). (H) eRNA enrichment measured by GRO-seq in p53-bound versus p53-unbound regions (±250 bp from p53 response element/p53RE). Statistics represent an unpaired Mann–Whitney U test (***P < 0.001). (I) Normalized transcriptional activity of p53-bound or unbound regions from the MPRA assay after 6 h of either DMSO or 10 uM Nutlin-3A treatment. Data are depicted as enrichment of the 3′UTR-encoded barcode linked to each putative regulatory region normalized to the enrichment of that same sequence integrated into the genome and represent the averages from three biological replicates of each condition (****P < 0.0001 by one-way ANOVA). (J) eRNA fold-change (Nutlin-3A versus DMSO conditions) as measured by GRO-seq at p53-bound versus p53-unbound regions. Statistics represent an unpaired Mann–Whitney U test (****P < 0.0001). (K) eRNA enrichment fold-change (Nutlin-3A versus DMSO conditions) as measured by GRO-seq Nutlin/DMSO at p53-bound regions that significantly increased (Sig.) in MPRA transcriptional activity upon Nutlin treatment versus p53-bound regions that did not significantly increase (NS) in MPRA transcriptional activity (*P < 0.05, **P < 0.01 by unpaired Mann–Whitney U test. (L) Spearman correlation analysis of eRNA enrichment (log2) at p53-bound CREs relative to enrichment of p53 (log2) in a region ± 250 bp from the p53RE between eRNA enrichment and p53-enrichment at MPRA regions.
We then examined the activity of our putative CREs using the model human colon carcinoma cell line HCT116 which is well-suited for studying p53-dependent transcriptional activity. Our 296 potential p53-dependent CREs clustered into two groups based on p53 occupancy using ChIP-seq data from HCT116 cells (1). 169 out of 296 regions were scored as p53 binding sites (peaks) by MACS2 (bound, Figure 1B, Model-based Analysis of ChIP-seq, (51)), whereas 127 regions lacked measurable p53 binding (unbound, Figure 1C). The average position weight matrix of the p53RE for each of the clusters is highly similar (Figure 1D), however, the consensus motif in p53-bound regions more closely resembles the optimal p53 consensus motif than do p53-unbound CREs (Figure 1E). CREs bound by p53 show higher enrichment of canonical enhancer-associated histone modifications H3K27ac (Figure 1F), H3K4me1 (Supplemental Figure S1A), and H3K4me2 (Figure 1G) than do those regions lacking p53 occupancy in HCT116 cells. The promoter-associated histone modification H3K4me3 is similarly enriched across both p53 bound and unbound CREs (Supplemental Figure S1B). p53-bound CREs are also found in regions with higher DNAse-accessibility (Supplemental Figure S1C) and are more enriched for the transcription initiation-associated RNA polymerase II C-terminal domain modification serine 5 phosphorylation (Supplemental Figure S1D). Consistent with the increased occupancy of transcriptionally associated features, enhancer RNA (eRNA) transcription is more prevalent at p53-bound CREs relative to those lacking p53 binding (Figure 1H) (1,2). These data suggest that CREs that are destined for binding by p53 have higher DNA accessibility, are more enriched for chromatin modifications associated with transcription, and produce more eRNA under basal/DMSO-treated conditions than those not bound by p53.
We then performed triplicate biological measurements of p53-dependent CRE activity in HCT116 using our MPRA approach. MPRA-transduced cells were treated for 6 h with either DMSO or the MDM2 inhibitor Nutlin-3A and expressed RNA barcodes were deep sequenced as described in Methods. Nutlin-3A leads to stabilization and activation of p53 at a similar level to what is seen with DNA damaging agents like etoposide (Figure 2A) (52). Importantly, enhancer activity measurements across biological replicates, treatment conditions, and cell lines were highly correlated (Supplemental Figure S2). As expected from our analysis of p53 occupancy, p53-bound CREs showed a bulk Nutlin-3A-dependent increase in activity compared to treatment with DMSO (Figure 1I, ****P < 0.0001, one-way ANOVA). Unbound CRE activity was not affected by Nutlin-3A treatment relative to DMSO and was substantially lower than p53-bound CREs (Figure 1I, ****P < 0.0001, one-way ANOVA). Activity of the ubiquitous CRE controls were unaffected by the induction of p53 by Nutlin-3A (Supplemental Figure S3A). eRNA transcription was more highly upregulated at p53-bound CREs upon Nutlin-3A treatment than at unbound CREs (Figure 1J), consistent with the observed Nutlin-3A-dependent increase in p53-bound CRE activity (Figure 1I). CREs with significantly increased MPRA activity upon Nutlin-3A-treatment had more robust eRNA induction (DMSO versus Nutlin-3A) than CREs with lower activity (Figure 1K). However, p53 enrichment is only weakly correlated with total eRNA abundance at a given CRE suggesting that p53 occupancy alone is not an indicator of eRNA transcription (Figure 1L for Nutlin-3A-treated conditions, Supplemental Figure S4 for DMSO-treated conditions), as has been previously reported (2,53,54). These data demonstrate that known markers of transcriptional activity, including histone modifications and eRNA transcription (54), distinguish p53-bound versus p53-unboud CREs, but that additional features contribute to the activity of p53-bound CREs.
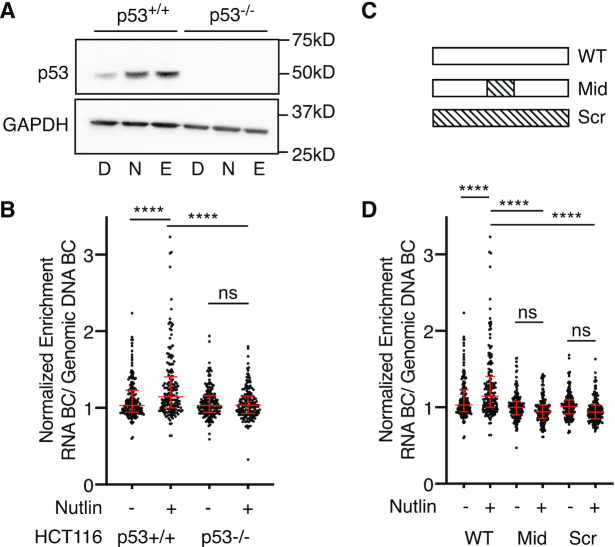
Figure 2.
(A) Immunoblotting for p53 (top) or GAPDH (bottom) expression in HCT116 p53+/+ or p53−/− colon carcinoma cells after 6 hours of treatment with DMSO (D), 10 uM Nutlin-3A (N) or 100 uM etoposide (E). (B) Normalized transcriptional activity of the wild-type p53-bound regions in either HCT116 p53+/+ or p53−/− cells after a 6-h treatment of either DMSO or 10 uM Nutlin-3A. Data are depicted as enrichment of the 3′UTR-encoded barcode linked to each putative regulatory region normalized to the enrichment of that same sequence integrated into the genome and represent the averages from three biological replicates of each condition (****P < 0.0001 using an ordinary one-way ANOVA). (C) Sequences within the wild-type p53-bound regions from the MPRA (WT) were shuffled (while preserving GC content) to alter either the 20 bp p53 binding site (Mid) or the entire 100 bp MPRA sequence. (D) Normalized transcriptional activity of p53-bound sequences for the wild-type (WT), p53-binding site scramble (Mid), or the full scramble (Scr) regions in HCT116 p53+/+ cells after a 6-h treatment of either DMSO or 10 uM Nutlin-3A. Data are depicted as enrichment of the 3′UTR-encoded barcode linked to each putative regulatory region normalized to the enrichment of that same sequence integrated into the genome and represent the averages from three biological replicates of each condition (****P < 0.0001 by one-way ANOVA).
p53-bound CREs require direct-binding of p53 for Nutlin-3A-induced activity
In order to determine if Nutlin-3A-induced activity of p53-bound CREs is p53-dependent, we assessed transcriptional activity in matched HCT116 TP53+/+ and TP53−/− cell lines (Figure 2A). Nutlin-3A-induced activity of p53-bound CREs was diminished in HCT116 TP53−/− cells suggesting these enhancers are dependent on wild-type p53 (Figure 2B). As expected, ubiquitously expressed CRE controls were unaffected by the loss of p53 expression (Supplemental Figure S3A). To test whether CRE activity is direct or indirectly dependent on p53, we compared wild-type CRE sequences to those with either the 20 bp p53RE (Mid) or the entire CRE sequence (Scr) randomized (Figure 2C). As a control for randomization, we fully scrambled the 196 ubiquitous CRE control sequences while preserving GC content, leading to a loss of activity (P < 0.0001, one ANOVA, Supplemental Figure S3B). Scrambling either the p53RE or the entire CRE sequence abrogates Nutlin-3A-dependent enhancer activity (Figure 2D, ****P < 0.0001, one-way ANOVA). In aggregate, wild-type CREs are more highly active than Mid or Scr CREs, suggesting that p53 strongly influences overall CRE activity (Figure 2D). Taken together, these data suggest that Nutlin-3A-induced activity of CREs requires direct binding of p53, in agreement with previous observations (9,10).
Variation in flanking sequence context alters p53-dependent CRE activity
Previous work suggests that p53-dependent CREs uniquely work in a single-factor mechanism in which only the presence of p53 is required for activation of transcription (9). Thus, this model implies that other transcription factors are required for CRE activity. Similarly, analysis of DNA sequences flanking functional p53REs revealed no consistent enrichment or requirement for other transcription factor binding motifs or sequences outside of the p53RE (9,10). Because this model represents a potential novel mechanism for enhancer function and diverges from canonical CRE models, we sought to directly test whether p53-dependent CRE activity requires sequences or transcription factor motifs outside of the p53RE. We systematically scrambled non-overlapping 20 bp regions of each CRE starting at the 5′ end (Figure 3A). As previously discussed, we also included controls where the p53RE and the entire CRE sequence were randomized (Figure 3A). We then asked whether individual CRE variants had significantly different transcriptional activity than their wild-type counterpart in Nutlin-3A-treated, wild-type HCT116 cells. Both the Mid and Scr variant CREs displayed significantly reduced activity relative to wild-type CREs (Figure 3B, P < 0.001, one-way ANOVA), consistent with a loss of p53 binding and p53-dependent transactivation. Overall, 93 Mid or Scr CRE variants had statistically significant differential activity relative to the wild-type CRE, with 88 (95%) displaying reduced activity when sequences were randomized (Figure 3C, one-way ANOVA with Tukey HSD, P < 0.05). Only 1.5% of Mid or Scr variants (5/338) had statistically increased activity relative to the wild-type sequence (Figure 3C), with two-fifth of those more active variants representing scrambled CRE sequences that generated a novel canonical p53 binding site. The remaining three variants possessed unique combinations of TF motifs with increased activity relative to the wild-type, p53RE-containing sequences. These data suggest that, overall, disruption of the p53RE is sufficient to decrease CRE activity.
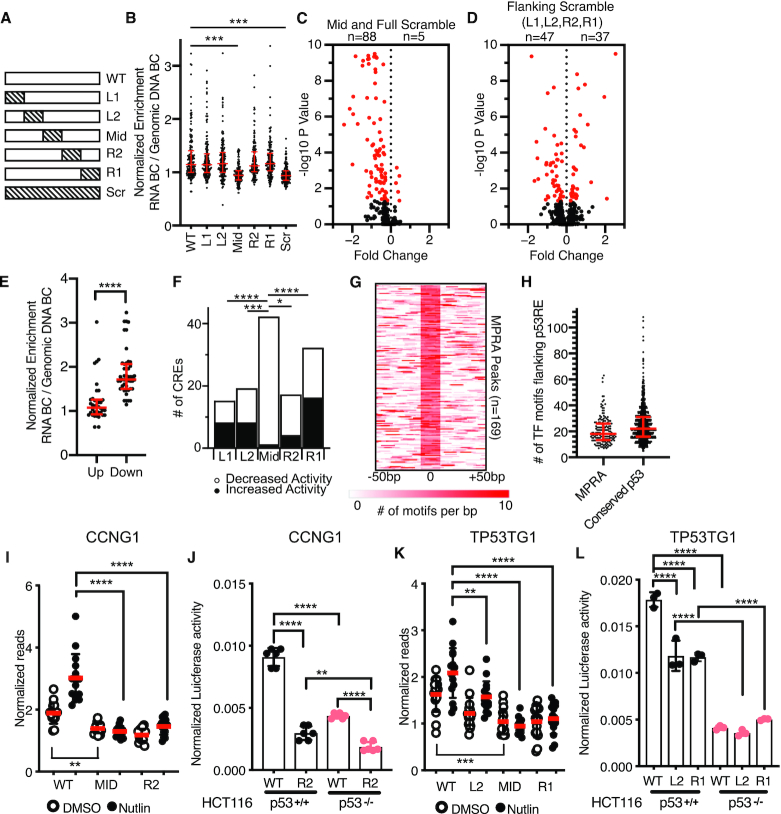
Figure 3.
(A) Schematic depicting scrambling of 20 bp sequences within 100 bp p53-bound MPRA regions. Sequence scrambling was performed to preserve total GC content within the 20 bp scrambled region relative to the wild-type sequence. (B) Normalized transcriptional activity of the MPRA sequences depicted in (A) in HCT116 p53+/+ cells after a 6-h treatment with 10 uM Nutlin-3A. Data are depicted as enrichment of the 3′UTR-encoded barcode linked to each putative regulatory region normalized to the enrichment of that same sequence integrated into the genome and represent the averages from three biological replicates of each condition. (***P < 0.001 using an ordinary one-way ANOVA). (C) Volcano plot of wild-type MPRA sequence activity compared to Mid or Full scramble. Data are plotted as Fold Change (WT over scramble) versus –log10P-value (from a one-way ANOVA with Tukey post-hoc test). (D) Volcano plot of wild-type MPRA sequence activity compared to flanking scramble (L1, L2, R2 or R1). Data are plotted as Fold Change (WT over scramble) versus –log10P-value (from a one-way ANOVA with Tukey post-hoc test). (E) Jitter plot depicting CRE activity values for wild-type p53-bound CREs (in the Nutlin-3A-treated condition) for those variants with increased (up) or decreased (down) activity relative to wild-type. P-value represents the result of an unpaired Mann–Whitney U test (****P < 0.0001). (F) Number of CRE variants per position that significantly increase or decrease in MPRA transcriptional activity compared to WT sequences (*P < 0.05, ***P < 0.001, ****P < 0.0001 by Fisher Exact Test). (G) Heatmap of transcription factor motif enrichment across 100 bp of the 169 p53-bound MPRA regions in the current study. Data represent a per base pair score for the presence of JASPAR-derived transcription factor motifs (P < 0.0001, corresponding to a JASPAR score of 400 or greater). Regions are centered on the putative p53 family response element. (G) Median values and the interquartile range are depicted in red. (H) Jitter plot representing the number of JASPAR-derived transcription factor motifs flanking the p53RE of the p53-bound CRE (P < 0.0001, corresponding to a JASPAR score of 400 or greater). Regions are centered on the putative p53 family response element (p53RE) and are extended –250 bp and +250 bp upstream. (I) Normalized transcriptional activity of the WT, Mid or R2 version of Region 36/CCNG1 from the MPRA in HCT116 p53+/+ cells after 6 h of either DMSO or 10 uM Nutlin-3A treatment. (****P < 0.0001 using an ordinary one-way ANOVA). (J) Normalized Luciferase activity of either wild-type or R2-scrambled Region 36/CCNG1 sequence cloned upstream of a minimal promoter and driving expression of firefly luciferase in either HCT116 p53+/+ or p53−/− colon carcinoma cells. Firefly luciferase values were normalized to those of Renilla luciferase driven by a CMV promoter and co-transfected with the candidate Firefly plasmid (**P < 0.01, ****P < 0.0001 using an ordinary one-way ANOVA) (K) Normalized transcriptional activity of the WT, L2, Mid or R1 version of the TP53TG1 CRE from the MPRA in HCT116 p53+/+ cells after 6 h of either DMSO or 10 uM Nutlin-3A treatment (**P < 0.01, ****P < 0.0001 using an ordinary one-way ANOVA). (L) Normalized Luciferase activity of either wild-type, L2, or R1-scrambled TP53TG1 CRE sequence cloned upstream of a minimal promoter and driving expression of firefly luciferase in either HCT116 p53+/+ or p53−/− colon carcinoma cells. Firefly luciferase values were normalized to those of Renilla luciferase driven by a CMV promoter and co-transfected with the candidate Firefly plasmid (****P < 0.0001 by one-way ANOVA).
Scrambling DNA sequence flanking a p53RE does not affect transcriptional activity in aggregate (Figure 3B, P > 0.05, one-way ANOVA). However, individual CREs have significantly altered transcriptional outputs when context-specific regions are scrambled (Figure 3D, one-way ANOVA with Tukey HSD, P < 0.05). Variants with decreased p53-dependent CRE activity have wild-type counterparts with higher expression values than compared to those variants with increased activity (Figure 3E, P < 0.0001, Mann–Whitney U test). We then wanted to determine whether the position of the scrambled sequence relative to the p53RE influenced the change in CRE activity by examining the proportion of significantly upregulated and downregulated CREs at each position. The Mid position, containing a p53RE, was more strongly associated with decreased CRE activity upon scrambling relative to any other position (Figure 3F, Fisher's Exact Test), consistent with a loss of p53-mediated activation. The R1 position (20–40 bp 3′ of the p53 RE) had a larger number of variants with displaying significantly different activity than wild-type than any of the other flanking region variants (Fisher's Exact Test, P < 0.05 for L1, L2 and R2), although the underlying mechanism for this observation is unclear. These data suggest that DNA elements important for CRE function can be found in any position relative to the p53RE and that local context and DNA sequence content may play key roles in p53-bound CRE activity.
Previous analyses of local sequence context at p53-bound CREs suggested a lack of enrichment of transcription factor motifs, besides the p53 RE, that might influence CRE activity (10). Our MPRA approach demonstrates that sequences flanking p53RE can contribute to CRE activity (Figure 3D), but the context and content of DNA sequences influencing p53-bound CRE activity is unclear. Therefore, to determine whether the scrambling of flanking sequences might disrupt specific DNA-encoded information, like transcription factor motifs, we undertook a series of motif enrichment analyses on wild-type and scrambled sequences. First, we examined the genome-wide enrichment of known transcription factor motifs across p53-bound CREs using HOMER (37). Expectedly, p53 family motifs were highly enriched in the MPRA regions relative to size and GC-content matched genomic regions (Supplemental Table S6). We also observed statistically significant (Bonferroni q value < 0.05) enrichment of other known transcription factor motifs, including those in the AP-1, GATA and ETS families (Supplemental Table S6). The observed enrichment of transcription factor motifs near p53RE is similar in a group of 1149 consensus p53 binding sites (9). Of note, a wider range of transcription factor families are represented in this consensus set of p53 binding sites, likely due to the increase in number and length of surveyed regions included in the analysis (±250 bp from p53RE) (Supplemental Table S7, Supplemental Figure S5, Figure 3H). In order to account for potential bias in a single motif enrichment strategy, we also used gimmeMotifs (29) and the JASPAR vertebrate transcription factor database (45) to identify the position of transcription factor motifs relative to the p53RE. p53-bound CREs are enriched for flanking transcription factor motifs (Figure 3G and H) with a median of 18 distinct motifs found per p53-bound CRE (Figure 3H). Examination of transcription factor chromatin immunoprecipitation (ChIP) data from the Cistrome Browser (41) suggests that numerous transcription factors are likely to occupy our group of p53-bound CREs (Supplemental Figure S6). Thus, our data suggests that regions proximal to p53REs are enriched for transcription factor binding motifs that may be involved in p53-dependent CRE activity.
We then focused exclusively on CRE variants that displayed reduced activity relative to the wild-type CRE sequence to better understand how flanking sequence context might affect p53-dependent CRE activity. Our rationale was to identify potential transcription factors or functional DNA elements that facilitate p53-dependent transcriptional activity. The 47 p53-bound CREs with reduced activity (Figure 3D) had a median loss of two JASPAR-defined TF motifs relative to the wild-type sequence (Supplemental Figure S7A). TF motif enrichment demonstrates that the AP-1 family motif is enriched in WT sequences and depleted from scrambled CREs (Supplemental Table S8), being lost from 17% of CREs with reduced activity. AP-1 family factors have wide-ranging roles in transcription regulation, including mediating chromatin accessibility at CREs (55–57). Overall, motifs for 95 different TFs are lost within the 47 scrambled REs with reduced activity, including AP-1, GATA, SP1 and ETS family members (Supplemental Table S9). Loss of activity is not correlated with loss of canonical transcription initiation sequences, like the INR element (Supplemental Figure S7B) (58–61). These data, taken together, suggest that loss of a broad set of transcription factor motifs flanking p53RE can influence p53-dependent CRE activity.
We then moved to validate our observation that altering TF motifs flanking p53RE could reduce p53-dependent CRE activity. We first examined a CRE localized within the first intron of the CCNG1 gene that is induced upon Nutlin-3A treatment in a p53-dependent manner (Figure 3I). This activity is abrogated when either the p53RE or the 3′ adjacent 20bp region are scrambled (position R2, Figure 3I). We observe similar results for a putative CRE localized within the second intron of TP53TG1 where scrambling either the 5′ adjacent 20 or 40 bp downstream of the p53RE (Figure 3K) leads to diminished p53-dependent CRE activity. We then sought to validate these MPRA results by utilizing a standard Luciferase reporter-based assay of CRE activity. In contrast to the 100bp sequence tested in the MPRA, we assessed the activity of a larger sequence encompassing an entire region of DNase hypersensitivity (DHS) as determined by ENCODE. DHS are putative regulatory regions often possessing transcriptional activity (43). Both the CCNG1 and TP53TG1 wild-type CREs are dependent on p53 for full activity (Figure 3J, L, p53+/+), consistent with the MPRA data. We confirmed that flanking region variants with loss of activity in the MPRA displayed a similar reduction of activity in traditional luciferase enhancer assays (Figure 3J, L), suggesting our MPRA results are not an artifact of the restricted sequence size (100 bp) or differences in assay conditions. Under DMSO treated conditions, CCNG1 and TP53TG1 CRE activity is reduced in either p53-depleted cells (Figure 3J,L) or in the p53RE mutant (mid) relative to the wild-type sequence (Figure 3I, K). These results are likely indicative of either basal p53 activity and genomic occupancy in unstimulated cells (10) or that a population of cells is experiencing intrinsic stress, such as DNA damage during S phase (62). For the CCNG1 enhancer, loss of activity in the R2 variant is further reduced in the absence of p53 (Figure 3J, P < 0.01, one-way ANOVA), suggesting potential combinatorial activity of p53 and the wild-type R2 sequence within the CCNG1 CRE. This combinatorial activity was not observed for TP53TG1, as the wild-type CRE and the L2 and R1 variants have similar activity in cells lacking p53 (Figure 3L). data suggest that p53-dependent CREs require different sequences and motifs, and potentially TFs, flanking the p53RE for optimal activity.
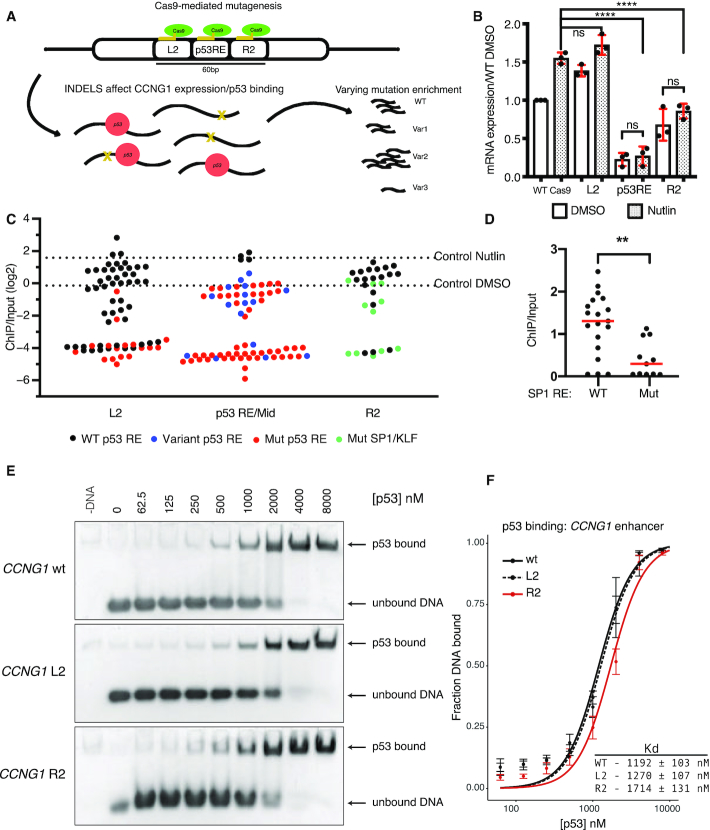
An SP1/KLF family motif is required for p53-dependent activity of the CCNG1 CRE
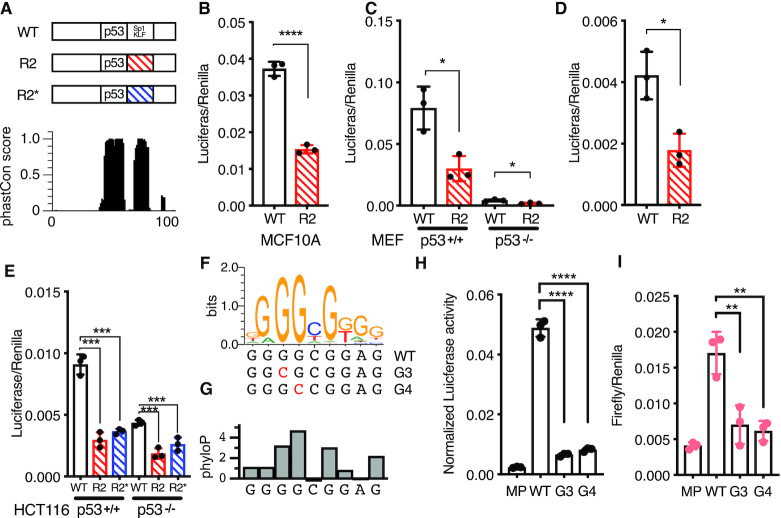
We continued investigating the role of flanking DNA sequences on p53-dependent CRE activity by further examining the CCNG1 CRE. Both the DNA sequence within the R2 position and the p53RE are highly conserved across vertebrates suggesting that this region may have a conserved functional regulatory role (Figure 4A). The p53-dependence of the CCNG1 CRE is similar across human cell types, as the R2 variant leads to a similar reduction in enhancer activity relative to the wild-type sequence when assayed in the non-transformed human cell line MCF10A (Figure 4B). We then assessed the activity of the wild-type and R2 variant CCNG1 CREs in Trp53+/+ and Trp53−/− mouse embryonic fibroblasts (MEFs). The R2 variant has reduced activity compared to the wild-type CCNG1 CRE in both wild-type and p53-deficient MEFs consistent with our observations in human cell lines (Figure 4C, D). In order to determine if the decrease in CCNG1 CRE activity via R2 scrambling is due to a loss of the wild-type sequence or a gain of function from the scrambled sequence, we created a second R2 variant (R2*) that preserves GC content but is further randomized from the wild-type sequence (Figure 4A). Both R2 and R2* variants lead to loss of CCNG1 CRE activity relative to wild-type and are further reduced in the absence of p53 (Figure 4E). These results suggest that 20 bp flanking the p53RE within CCNG1 are required for p53-dependent CRE activity. The 20 bp immediately downstream of the p53RE within CCNG1 enhancer contains several known transcription factor motifs based on analysis from JASPAR (45), suggesting that the loss of activity may be due to a loss of TF binding. The highest-scoring transcription factor motif from JASPAR are for members of the SP1/KLF family, and this G-rich motif is highly conserved across vertebrates (Figure 4F-G). We therefore made single base-pair mutations in the most conserved G3 and G4 residues of the SP1/KLF motif (Figure 4F, G) and asked whether loss of these residues was sufficient to reduce CRE activity (63). Consistent with scrambling the entire 20 bp R2 region, mutation of either the G3 or G4 position within the SP1/KLF motif severely diminishes transcriptional output in both wild-type and p53-deficient cell lines (Figure 4H, I). The additional reduction in CRE activity in p53-deficient cells seen in SP1/KLF motif variants suggests this region is functionally important independent of p53. Taken together, these data suggest that CCNG1 CRE activity requires both p53 and a regulatory motif belonging to the SP1/KLF family.
Figure 4.
(A) (top) Schematic of the 100 bp Region 36/CCNG1 enhancer. R2 position is annotated with the predicted motif ‘SP1/KLF’ 20 bp 3′ adjacent to the p53RE and (bottom) per-basepair vertebrate phastCon score. (B) Normalized Luciferase activity (test sequence Firefly versus constitutive Renilla) for WT Region36/CCNG and the R2 version in the MCF10A mammary epithelial cell line (****P < 0.0001 paired t-test). (C) Normalized Luciferase activity (test sequence Firefly versus constitutive Renilla) for WT Region36/CCNG and the R2 version in either p53+/+ or p53−/− mouse embryonic fibroblasts (*P < 0.05, paired t-test). (D) Rescaled view of p53−/− reporter assay data from (C) depicting normalized Luciferase data for the wild-type or R2 version of the CCNG1 enhancer in p53−/− mouse embryonic fibroblasts (*P < 0.05, paired t-test). (E) Normalized luciferase activity for either the wild-type, R2 or R2* version of the CCNG1 enhancer in HCT116 p53+/+ or p53−/− cells. (***P < 0.001 by one-way ANOVA). (F) The canonical Sp1/KLF family motif sequence in the CCNG1 enhancer as a transcription factor logo compared to the wild-type, G3, or G4 variants. This motif is located within the R2 site of the CCNG1 CRE, between 0 and 20 bp from the p53 response element. (G) phyloP vertebrate conservation of the Sp1/KLF family motif within the wild-type CCNG1 enhancer sequence showing high conservation at the G3 and G4 positions. Normalized enhancer activity for the minimal promoter only (MP), WT, G2 or G3 CCNG1 variants in (H) HCT116 p53+/+ or (I) HCT116 p53−/− cells (**P < 0.01, ****P < 0.0001, by one-way ANOVA).
Loss of the SP1/KLF motif leads to reduced CCNG1 transcription and reduced p53 binding
Thus far, our data indicate that the p53-dependent CCNG1 CRE also requires key DNA sequences flanking the p53RE and that these DNA sequences likely represents a binding motif for the SP1/KLF family. Therefore, we assessed the role of the p53RE and the R2 position sequence in their native genomic contexts using CRISPR/Cas9-mediated mutagenesis (Figure 5A). Guide RNA sequences were targeted to either the p53RE, the R2 position, or the L2 position, which is 5′ adjacent to the p53RE, generating a population of indel mutations (Figure 5A, Supplemental Table S4). Targeting of the p53RE substantially reduced endogenous CCNG1 mRNA abundance relative to non-targeted Cas9 cells (Figure 5B, P < 0.0001). Consistent with in vitro observations of CCNG1 enhancer activity, mutations within the R2 position reduce CCNG1 mRNA levels, albeit not as severely as mutations in the p53RE (Figure 5B, P < 0.0001). As a control for sequence variants proximal to the p53RE, we generated indel mutations in the L2 position which is found in the 20 bp immediately preceding the p53RE. Targeting the L2 position did not reduce endogenous CCNG1 mRNA levels suggesting that proximal DNA mutations are not sufficient to decrease transcriptional output (Figure 5B). Additionally, the L2 control suggests that the act of targeting Cas9 to the CCNG1 intron does not affect CCNG1 transcription on its own. These data further suggest that the p53RE and sequences immediately downstream of the p53RE are required for endogenous expression of the CCNG1 mRNA.
Figure 5.
(A) Schematic depicting strategy to introduce insertion-deletion (indels) mutations into the native CCNG1 locus in HCT116 cell lines. Pools of indel-containing cells were then used for measurement of CCNG1 mRNA expression and for analysis of p53 binding by ChIP-sequencing. (B) qRT-PCR data of CCNG1 mRNA expression in control (WT Cas9), L2, p53RE or R2-targeted Cas9 experiments after 6 h of DMSO or 10uM Nutlin-3A treatment in HCT116 p53+/+ cell lines (****P < 0.0001, by one-way ANOVA). (C) Jitter plot of p53 ChIP-enrichment for each Cas9-induced mutation at the native CCNG1 enhancer. Black dots represent genetic variants that retain a wild-type p53 response element motif, while red dots are genetic variants with a mutated p53 response-element. Green dots represent sequences with a mutated or missing ‘SP1/KLF’ motif. Dashed line represents p53 ChIP/Input enrichment (log2) for the control cell line after 6 h of Nutlin-3A treatment. (D) Comparison of p53 ChIP enrichment from variants with either a canonical or mutated Sp1/KLF family motif from the R2-targeted Cas9 experiment (red line is the median, **P < 0.01, unpaired t-test). (E) Electrophoretic Mobility Shift Assay (EMSA) for recombinant p53 bound to the wild type, L2 or R2 variant of the CCNG1 cis-regulatory element DNA sequence. Images are representative of four independent biological replicates. (F) Quantification of EMSA experiment in (E) performed as described in Materials and Methods. Fitted curves were fitted using a two-site binding model (Hill equation; Hill coefficient = 2) using a non-linear regression.
We generated a pool of indel mutations at three locations within the CCNG1 CRE, with mutations near the p53RE and the R2 position leading to a reduction in endogenous CCNG1 mRNA expression (Figure 5A, B). We therefore coupled chromatin immunoprecipitation of p53 to amplicon sequencing to simultaneously determine indel mutations and their potential effect on p53 occupancy at the CCNG1 CRE. As a control, we performed p53 ChIP under DMSO and Nutlin-3A-treated conditions on HCT116 p53+/+ cells expressing wild-type Cas9 without a gRNA to target it to DNA. We then amplified a 150 bp region in the CCNG1 CRE from the p53-immunoprecipitated and input samples and sequenced them using Illumina approaches. We observed a 2.98-fold enrichment of the amplified sequence in the Nutlin-3A-induced samples relative to DMSO (Fig 5C, dotted lines), suggesting our amplicon ChIP approach was valid.
We next performed ChIP experiments from DMSO and Nutlin-3A-treated cell lines with Cas9 targeted to the p53RE, L2 or R2 positions. Sixty six unique DNA variants were identified within our pool when targeting Cas9 to the p53RE within the CCNG1 CRE (Supplemental Table S4, Figure 5C). As expected, the wild-type CCNG1 CRE sequence and three variants with an intact p53RE were enriched near the levels seen in the Cas9 control (Figure 3C, p53RE, black dots). Enrichment of p53 is below or at DMSO levels when a p53RE is present but varies from the wild-type version (Figure 5C, blue dots). As expected, CCNG1 variants lacking a p53RE show a strong reduction in p53 binding. Variants proximal to the p53RE generally reduced p53 binding to the level of DMSO treatment (no enrichment), but many variants were depleted to the level seen when the p53RE was mutated (Figure 5C). These data are consistent with data showing that Cas9-induced mutations proximal to GATA1 binding sites alter GATA1 occupancy (64). None of the R2 mutations identified contained variants within the p53RE; however, when the conserved SP1/KLF motif was mutated or lost, p53 occupancy was reduced relative to the presence of an intact motif (Figure 5C (green dots), D, P < 0.01). Our ChIP-based approach suggested that sequence variation flanking a p53RE can alter in vivo p53 binding in a context-dependent manner. We also performed electrophoretic mobility shift assays (EMSA) to better understand the effect of flanking sequence variation on p53 binding to p53RE. Increasing concentrations of recombinant p53 were combined with 60 bp double-stranded DNA fragments representing the wild-type CCNG1 p53RE and either the L2 or R2 variants from the original MPRA experiment (Figure 5E). The R2 variant had a modest, but statistically significant, increase in Kd relative to either the wild-type or L2 variants (Figure 5F). The reduction in p53 binding observed in the R2 variant by EMSA is consistent with our results from the in vivo variant ChIP experiment as well as the reduced p53-dependent transcription of endogenous CCNG1. Surprisingly, although a number of L2 variants had reduced p53 binding in vivo, we observed similar p53 binding affinities for both the wild-type and L2 sequences by EMSA (Figure 5F). The fully randomized L2 variant does not affect p53-dependent CRE activity or affect p53 binding in vitro, suggesting that loss of p53 binding observed in vivo for specific L2 variants may be context-specific. These data, along with our examination of endogenous CCNG1 mRNA expression, indicate that the specific sequences proximal to the p53RE in the CCNG1 enhancer, which includes an SP1/KLF family motif, leads to increased p53 occupancy and higher CCNG1 mRNA expression.
p53-dependent transcription of GDF15 requires regulatory factors at two separate distal CREs
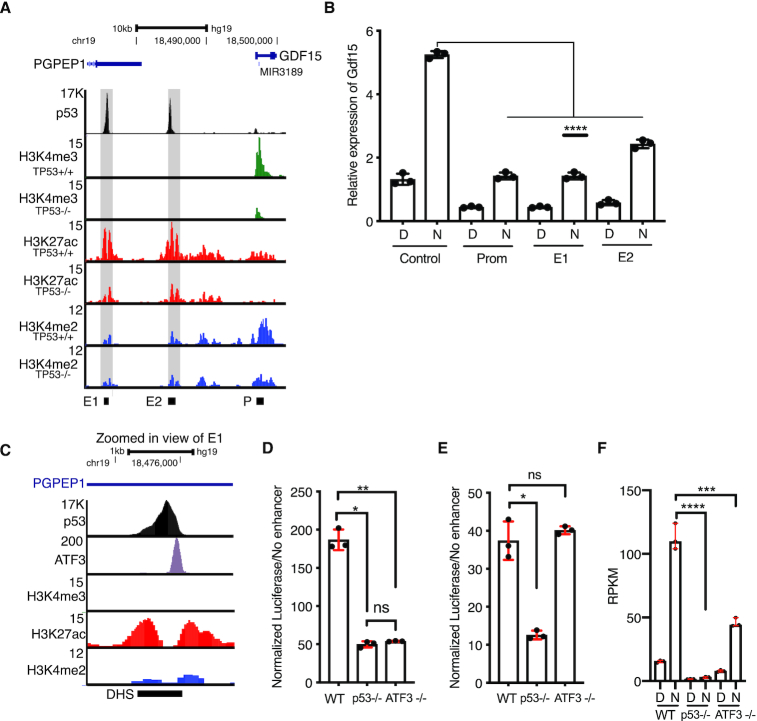
Data from our MPRA approach and follow-up experiments demonstrate that p53-dependent transcriptional activity at CREs is altered when sequences flanking the p53RE are perturbed. The CREs regulating CCNG1 (Figure 3I, J) and TP53TG1 (Figure 3K, L) are intragenic or proximal to the gene promoter similar to the majority of known p53-bound CREs directly controlling downstream gene expression (1,65). In order to better understand the regulatory potential and mechanisms of gene distal p53 binding events, we searched our MPRA dataset for p53-dependent CREs distal to the promoters of known p53 target genes. One such region is approximately 11kb upstream of GDF15, which is well-characterized p53 target gene (2,66). As predicted, basal and induced GDF15 expression is strongly dependent on p53 (Figure 6F). GDF15 has also recently been identified as a key modulator of inflammatory and metabolic responses (26,27), but the CREs key to regulation after p53 activation are known. This putative CRE (called E2) is enriched with the histone modifications H3K27ac and H3K4me2 and depleted for H3K4me3, a pattern strongly associated with transcriptional CREs. p53 is strongly bound to this region in vivo (Figure 6A). While examining the genomic context of this putative enhancer, we identified a second putative p53-bound CRE approximately 20kb upstream of GDF15 in the 3′UTR of the PGPEP1 gene, which we define as E1. Both p53-bound regions are enriched for H3K27ac and H3K4me2 in the absence of p53 (Figure 6A) suggesting potential CRE activity independent of p53. This observation is consistent with recent reports of basal enhancer RNA transcription and histone modification enrichment at potential CREs in the absence of p53 (2,35,53). We therefore wanted to determine whether these p53-bound regions act as CREs for the endogenous expression of GDF15. We took advantage of a recently described approach to inactivate enhancers (67). In this approach, a catalytically inactive form of Cas9 (dCas9) was fused to the transcriptional repressor domain KRAB, and this strong repressor was targeted to either a control region (an enhancer for an unrelated gene, FGF2), the two p53-bound regions, or the GDF15 promoter. Compared to the non-targeting control, targeting dCas9-KRAB to either p53-bound distal region (E1 or E2) reduced expression of endogenous GDF15 mRNA in both basal (DMSO) or p53-activated (Nutlin-3A) conditions (Figure 6B). Repression of GDF15 mRNA levels when targeting either E1 or E2 was similar to that of targeting the GDF15 promoter region (Figure 6B). These results provide evidence that the E1 and E2 regions, bound by p53, are likely CREs regulating the expression of GDF15.
Figure 6.
(A) Genome browser view of the GDF15 locus showing p53, H3K4me3, H3K27ac, and H3K4me2 enrichment in HCT116 p53+/+ or HCT116 p53−/− cell lines. E1 = enhancer 1, E2 = enhancer 2, and P = GDF15 promoter. Grey shaded boxes are placed over the E1 and E2 regions. (B) qRT-PCR relative expression of GDF15 mRNA in control, promoter, E1, or E2-targeted dCas-KRAB-expressing HCT116 p53+/+ cells treated with either DMSO (D) or 10uM Nutlin-3A (N) for 6 hours. (****P < 0.0001, one-way ANOVA). (C) Genome browser view of the GDF15 E1 region showing ChIP-seq enrichment of p53, ATF3, H3K4me3, H3K27ac or H3K4me2. DHS = DNAse hypersensitive site. Normalized Luciferse activity (relative to minimal promoter only) of the GDF15 E1 enhancer (D) or the GDF15 E2 enhancer (E) in wild-type, p53−/− or ATF3−/− HCT116 colon carcinoma cells (*P < 0.05, **P < 0.01, one-way ANOVA). (F) Reads per kilobase per million (RPKM) expression value of GDF15 mRNA from three replicates of polyA+ RNAseq of wild-type, p53−/− or ATF3−/− HCT116 treated for 6 h with either DMSO or 10 uM Nutlin-3A (***P < 0.001, ****P < 0.0001, one-way ANOVA).
We next wanted to determine whether potential sequences or transcription factors might regulate the activity of these p53-bound CREs for GDF15. We sought to use the breadth of publicly available ChIP-seq datasets to identify potential transcription factors bound the GDF15 E1 CRE. Using information from the CISTROME database (41), we found that ATF3, a member of the AP-1 family of transcription factors, strongly binds to the GDF15 E1 CRE in HCT116 cells (Figure 6C). The summit of the ATF3 binding event coincides with an ATF3 DNA motif, ∼125 bp downstream of the p53RE (Figure 6C). We therefore focused on ATF3 because of its previous association as a positive regulator of p53 activity and its well-known role as a modulator of the inflammatory response (28,68–70), of which GDF15 is a central regulator (27). Using a luciferase reporter approach, we assessed the activity of the GDF15 E1 enhancer in wild-type, p53-deficient, or ATF3-deficient HCT116 cells. Loss of either p53 or ATF3 leads to a substantial reduction of E1 enhancer activity (Figure 6D). As expected by the lack of binding in vivo, the activity of the GDF15 E2 enhancer was unaffected by the loss of ATF3, whereas it is strongly dependent on p53 for activity (Figure 6D, E). Interestingly, although GDF15 E2 contains multiple AP-1 family motifs, it is not bound by or regulated by AP-1 member ATF3. Expression of endogenous GDF15 mRNA as determined by polyA+ RNAseq is reduced in either p53 or ATF3-deficient HCT116 cells consistent with in vivo binding and activity of ATF3 at the GDF15 E1 enhancer (Figure 6F). Taken together, our data indicate combinatorial activity of both p53 and ATF3 is required for activity of the GDF15 E1 enhancer and that both the E1 and E2 enhancer directly regulate expression of GDF15.
DISCUSSION
A number of high throughput analyses of p53 genomic occupancy revealed that p53 predominantly binds to cis-regulatory regions (CRE) like enhancers and promoters (34,35,39). The ability of p53 to activate transcription is well established, however, specific molecular mechanisms underlying p53 activity at CRE are less well studied. Here, we describe the use of a massively parallel reporter assay (MPRA) to characterize the effect of local sequence variation and transcription factor motifs on p53-dependent CRE activity. Our results support the canonical model of p53 activity where p53 primarily functions as a strong transcriptional activator at cis-regulatory elements (CRE) (73). Further, we confirmed that the p53 response element (p53RE) is a strong predictor of p53-dependent transcriptional regulation as has been observed across multiple experimental systems (8–10). We then examined the contribution of local sequence context on enhancer activity by systematically altering sequences flanking the p53RE (Figure 3A). Our results indicate that sequences outside of the core p53 binding site are required for optimal transcriptional activation (Figure 3C). These functional sequences include both putative and confirmed transcription factor binding motifs, suggesting that p53 requires additional DNA-bound factors for its ability to activate transcription through CREs.
Two recent MPRA studies proposed a novel ‘single-factor’ model for CRE regulation by p53 (9,10). In this model, p53 is solely responsible for transcriptional output of an individual p53-bound CRE and does not require other transcription factors. Our data suggest that loss of p53 through either genetic depletion or through alteration of a p53RE sequence severely diminishes p53-dependent CRE activity (Figure 2B, D). Therefore, our data support the necessity of p53 for stimulus-dependent activation of p53-bound enhancers in agreement with the single-factor model (9,10). Conversely, our results also demonstrate that additional transcription factors can co-regulate p53-dependent CRE activity. Our screening approach identified numerous motifs that positively or negatively affect CRE activity in a context-dependent manner (Figure 3C), including those directly bound by other transcription factors. ATF3 binding to the GDF15 E1 enhancer is required for p53-dependent CRE activity and endogenous GDF15 mRNA expression (Figure 6D-E). Additionally, a CRE regulating CCNG1 transcription requires both a p53RE and an adjacent SP1/KLF family motif for activity (Figure 4E, F). Loss of either motif diminishes native CCNG1 transcription driven by the CRE (Figure 5A). p53 occupancy at the CCNG1 enhancer is reduced when sequences flanking the p53RE diverge from the wild type sequence as observed in our in vivo ChIP experiments (Figure 5C). In vitro EMSA experiments suggest that only variation in the 3′ adjacent sequence, which contains the SP1/KLF motif, alters p53 binding affinity (Figure 5E-F). Similarly, only mutations within the SP1/KLF motif affected endogenous CCNG1 mRNA levels (Figure 5B). CRE activity is reduced when evolutionarily conserved residues in the SP1/KLF motif are altered in both wild type and p53-deficient cells. These data suggest the CCNG1 CRE requires both p53 and an additional factor bound to the SP1/KLF family motif for optimal activity.
The requirement for other transcription factors in the co-regulation of p53-dependent transcriptional activity at CREs has not been characterized on a broad scale. Certainly, individual p53 CREs have been previously demonstrated to require co-regulatory factors in reporter assays, including the requirement for the p53RE and an AP-1 element bound by JunD for DNA damage-dependent activation of GADD45A transcription (74), amongst others. A recent in vivo CRISPR/Cas9-screening approach identified CEBPβ binding within a p53-dependent CRE required for optimal transcription of CDKN1A and initiation of senescence (22). Single nucleotide polymorphisms associated with lung cancer risk found within a p53-regulated CRE disrupt canonical transcription factor motifs, reduce p53 binding, and alter expression of TNFRSF19 (7). In this study, we identified both ATF3 and a likely member of the SP1/KLF family as co-regulatory transcription factors required for p53-dependent CRE activity. Of note, we have not identified specific transcription factors that bind to the CCNG1 or TP53TG1 CREs whose p53-dependent activity is altered upon variation in flanking sequence. In the case of CCNG1, the SP1/KLF family motif can be bound by over 12 family members, most of which are expressed in the cell type (HCT116 colon carcinoma) used in this study. Identification of specific transcription factors binding to individual DNA elements is often challenging, but updated approaches like the enhanced yeast 1-hybrid or quantitative mass spectrometry methods are now possible (75,76). Importantly, we cannot rule out potential transcription factor-independent roles for these motifs in regulating p53-dependent CRE activity, such as the possibility that DNA shape or nucleosome positioning changes in vivo might be affected by changes in CRE sequence.
MPRAs are powerful tools for rapidly dissecting how sequence variation and context contributes to the activity of CREs. Despite their power, specific assay design choices and the non-native genomic context of the MPRA approach might help to explain some of the discrepancies between our work and previous reports (9,10). First, we used a random, lentiviral-based genomic integration strategy to deliver our MPRA constructs, whereas previous p53-based approaches have used transient, plasmid-based delivery (9,10). Genomic integration presumably allows for the greater influence of chromatin and higher-order genomic structure which directly influence transcription factor binding and activity (31). In direct comparison with episomal DNA, the activity of integrated massively parallel reporter constructs was more reproducible and more closely aligned with expected activity based on CRE-associated biochemical patterns like histone modification and accessible chromatin in previous MPRA analyses (31). Second, our assay was specifically designed to test the effect of sequence variation on p53-dependent CRE activity while post hoc computational approaches were previously used to identify sequence features defining CRE activity. In both the primary MPRA screen and in traditional plasmid-based enhancer assays, we observed that variation in transcription factor motif sequences flanking the p53RE could alter CRE activity. Altering p53RE-adjacent TF motif sequences did not always alter CRE activity, suggesting sequence and context-dependent effects (Figure 3D). Our MPRA approach was limited to assaying the activity of short segments (100 bp) of p53-bound CREs during the early phase of p53 activity and in only one cell type. Given that CREs regulate can transcription in space and time, in addition to abundance, additional cell types and contexts may ultimately reveal additional TF requirements for p53-bound CREs. Further, CREs are typically larger than 100 bp in length suggesting additional transcription factors are likely involved in their regulation that were not directly tested in this study.
Our data suggest that p53 activity at CREs often requires additional transcription factors. However, we cannot rule out that the recently proposed single factor model explains p53 activity at other locations, such as at the hundreds of p53 binding sites that lack evidence of CRE-associated histone modifications or features like eRNA transcription (10,53,54). The most likely scenario is that p53-bound CREs exist in a spectrum and have variable co-factor requirements, such that both the single factor and multi-factor model underlie the activity of different sets of p53-bound CREs. Ultimately, additional work is needed to dissect the specific motif features, transcription factor requirements, and context-dependence of p53-bound CREs and how they function to enact the broad tumor suppressor activity of p53.
Previous reports could not identify sequence-based features beyond the p53RE that predicted p53-bound CRE activity using machine learning and traditional motif enrichment approaches (9,10). While accurate to say that there are no other transcription factor motifs or sequence-features that are as enriched as the p53RE (Supplemental Tables S6 and S7), we find that other transcription factor motifs are well represented in the CREs we studied and within a previously identified core set of p53-bound CREs (Supplemental Tables S6 and S7). This includes overrepresentation of the motif for the stress-dependent transcription factor ATF3 (Supplemental Table S6), which binds to a number of p53-dependent CREs and regulates activity of a CRE for GDF15 (Figure 6D, E). ATF3 is a well-studied regulator of p53-dependent transcription through control of p53 stability and co-factor recruitment (28,68,69). Previous reports clearly demonstrate that ATF3 can directly alter p53 stability and modulate p53 activity through interactions with histone modifying enzymes (68,69,77). Our work uniquely identifies a direct role for ATF3 DNA binding within a p53-bound CRE and demonstrates a positive effect on p53-dependent transcriptional activity. Given that ATF3 binds to numerous p53-bound regions of the genome (28) and their previously reported relationship, further examination into the local interplay between p53 and ATF3 at DNA is warranted.
p53 is a pioneer transcription factor and can mediate context-dependent chromatin remodeling at CREs (10,35,53). Despite this activity, the large majority of p53 genomic binding events occur within regions that are accessible before p53 engagement (10,35,53,71), similar to what is observed for glucocorticoid receptor binding (78). These regions also contain chromatin modifications associated with active CRE, including H3K27ac and H3K4me1/2 before p53 binding (1,35,53,71,72,72). Further, p53 depletion does not alter basal CRE-associated chromatin modifications or chromatin structure at the large majority of CRE (35,53) suggesting that other transcription factors mediate chromatin accessibility at most p53-bound CREs. Consistent with this model, we observe enrichment of enhancer-associated histone modifications H3K27ac and H3K4me2 at GDF15 E1 and E2 enhancers in the absence of p53 (Figure 6A.) Enhancer-derived RNA (eRNA) has also been identified as a strong predictor of transcription factor binding and CRE activity (54). Our data suggest that those CREs with eRNA transcription are more likely to be bound by p53 and are more likely to see p53-dependent gains in enhancer activity (Figure 1H, J, K). In further support of a multi-factor model, eRNA are transcribed from p53-regulated CREs in the absence of p53 (2,53) suggesting other transcription factors are bound and active as previously suggested (54). These data suggest that other factors are likely responsible for establishing and maintaining chromatin structure and basal activity at the majority of p53-bound CREs. Recent reports suggest that p53 binding and activity is strongly influenced by cell type-specific chromatin accessibility (1,2,35), which itself is controlled by differential DNA binding transcription factor activity (11,12,79–81). Indeed, we recently identified p63, a p53 family member, as a factor required for chromatin accessibility and activity of certain p53-bound enhancers in epithelial cell types (35). How p53 functions across various cell and tissue contexts is still a vital and open question, but recent reports suggested that p53 binding and activity can be influenced by cell type suggest significant work remains to address the broad scope of p53-dependent transcription (2,35,82).
We propose that the lack of a core set of commonly enriched transcription factors within p53-dependent CREs is a potentially important regulatory feature of the p53 network. By utilizing different sets of transcription factor co-regulators, we hypothesize that global p53 transcriptional activity is buffered against loss of any one regulatory partner. This hypothesis for p53 is strongly reminiscent of the ‘billboard’ model seen at many Drosophila developmental enhancers (11,18), whereby different combinations of factors can bind to a CRE and produce similar transcriptional outputs. The flexible billboard model for p53-bound enhancers is also consistent with a recently proposed ‘distributed p53 network’ model whereby p53 transcriptionally controls many genes, but that any one p53 target gene is dispensable for tumor suppression (1).
DATA AVAILABILITY
All sequencing data generated as part of this study are available under Gene Expression Omnibus (GEO) Accession GSE137297.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the Center for Functional Genomics at the University at Albany for sequencing support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) NIGMS [R15 GM128049 to M.A.S]; University at Albany (to M.A.S.); Wellcome Trust [213501/Z/18/Z to D.B.]. Funding for open access charge: University at Albany.
Conflict of interest statement. None declared.
REFERENCES
- 1. Andrysik Z., Galbraith M.D., Guarnieri A.L., Zaccara S., Sullivan K.D., Pandey A., MacBeth M., Inga A., Espinosa J.M.. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res. 2017; 27:1645–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen M.A., Andrysik Z., Dengler V.L., Mellert H.S., Guarnieri A., Freeman J.A., Sullivan K.D., Galbraith M.D., Luo X., Kraus W.L. et al.. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014; 3:e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017; 36:3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donehower L.A., Soussi T., Korkut A., Liu Y., Schultz A., Cardenas M., Li X., Babur O., Hsu T.-K., Lichtarge O. et al.. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019; 28:1370–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeron-Medina J., Wang X., Repapi E., Campbell M.R., Su D., Castro-Giner F., Davies B., Peterse E.F., Sacilotto N., Walker G.J. et al.. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell. 2013; 155:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Menendez D., Snipe J., Marzec J., Innes C.L., Polack F.P., Caballero M., Schurman S.H., Kleeberger S.R., Resnick M.A.. p53-responsive TLR8 SNP enhances human innate immune response to respiratory syncytial virus. J. Clin. Invest. 2019; 129:4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shao L., Zuo X., Yang Y., Zhang Y., Yang N., Shen B., Wang J., Wang X., Li R., Jin G. et al.. The inherited variations of a p53-responsive enhancer in 13q12.12 confer lung cancer risk by attenuating TNFRSF19 expression. Genome Biol. 2019; 20:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B.. Definition of a consensus binding site for p53. Nat. Genet. 1992; 1:45–49. [DOI] [PubMed] [Google Scholar]
- 9. Verfaillie A., Svetlichnyy D., Imrichova H., Davie K., Fiers M., Kalender Atak Z., Hulselmans G., Christiaens V., Aerts S.. Multiplex enhancer-reporter assays uncover unsophisticated TP53 enhancer logic. Genome Res. 2016; 26:882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Younger S.T., Rinn J.L.. p53 regulates enhancer accessibility and activity in response to DNA damage. Nucleic Acids Res. 2017; 45:9889–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spitz F., Furlong E.E.M.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 12. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 13. Schoenfelder S., Fraser P.. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019; 20:437–455. [DOI] [PubMed] [Google Scholar]
- 14. Long H.K., Prescott S.L., Wysocka J.. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016; 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herz H.M., Hu D., Shilatifard A.. Enhancer malfunction in cancer. Mol. Cell. 2014; 53:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rickels R., Shilatifard A.. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 2018; 28:608–630. [DOI] [PubMed] [Google Scholar]
- 17. Smith E., Shilatifard A.. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014; 21:210–219. [DOI] [PubMed] [Google Scholar]
- 18. Kulkarni M.M., Arnosti D.N.. Information display by transcriptional enhancers. Development. 2003; 130:6569–6575. [DOI] [PubMed] [Google Scholar]
- 19. Seto E., Usheva A., Zambetti G.P., Momand J., Horikoshi N., Weinmann R., Levine A.J., Shenk T.. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:12028–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X., Farmer G., Zhu H., Prywes R., Prives C.. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993; 7:1837–1849. [DOI] [PubMed] [Google Scholar]
- 21. Thanos D., Maniatis T.. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995; 83:1091–1100. [DOI] [PubMed] [Google Scholar]
- 22. Korkmaz G., Lopes R., Ugalde A.P., Nevedomskaya E., Han R., Myacheva K., Zwart W., Elkon R., Agami R.. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016; 34:192–198. [DOI] [PubMed] [Google Scholar]
- 23. Koutsodontis G., Tentes I., Papakosta P., Moustakas A., Kardassis D.. Sp1 Plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21 WAF1/Cip1 gene by the p53 tumor suppressor protein. J. Biol. Chem. 2001; 276:29116–29125. [DOI] [PubMed] [Google Scholar]
- 24. Nikulenkov F., Spinnler C., Li H., Tonelli C., Shi Y., Turunen M., Kivioja T., Ignatiev I., Kel A., Taipale J. et al.. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012; 19:1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Zhang Y., Ströse A., Tedesco D., Gurova K., Selivanova G.. Integrated high-throughput analysis identifies Sp1 as a crucial determinant of p53-mediated apoptosis. Cell Death Differ. 2014; 21:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patel S., Alvarez-Guaita A., Melvin A., Rimmington D., Dattilo A., Miedzybrodzka E.L., Cimino I., Maurin A.-C., Roberts G.P., Meek C.L. et al.. GDF15 Provides an endocrine signal of nutritional stress in mice and humans. Cell Metab. 2019; 29:707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luan H.H., Wang A., Hilliard B.K., Carvalho F., Rosen C.E., Ahasic A.M., Herzog E.L., Kang I., Pisani M.A., Yu S. et al.. GDF15 Is an inflammation-induced central mediator of tissue tolerance. Cell. 2019; 178:1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao J., Li X., Guo M., Yu J., Yan C.. The common stress responsive transcription factor ATF3 binds genomic sites enriched with p300 and H3K27ac for transcriptional regulation. BMC Genomics. 2016; 17:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Heeringen S.J., Veenstra G.J.C.. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinforma. Oxf. Engl. 2011; 27:270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rice P., Longden I., Bleasby A.. EMBOSS: the European molecular biology open software suite. Trends Genet. TIG. 2000; 16:276–277. [DOI] [PubMed] [Google Scholar]
- 31. Inoue F., Kircher M., Martin B., Cooper G.M., Witten D.M., McManus M.T., Ahituv N., Shendure J.. A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 2017; 27:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. R Core Team R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing. 2018; Vienna. [Google Scholar]
- 34. Sanjana N.E., Shalem O., Zhang F.. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014; 11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karsli Uzunbas G., Ahmed F., Sammons M.A.. Control of p53-dependent transcription and enhancer activity by the p53 family member p63. J. Biol. Chem. 2019; 294:10720–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ayed A., Mulder F.A., Yi G.S., Lu Y., Kay L.E., Arrowsmith C.H.. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001; 8:756–760. [DOI] [PubMed] [Google Scholar]
- 39. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf. Engl. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L. et al.. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017; 45:D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al.. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G. et al.. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018; 46:D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. The FANTOM Consortium Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C. et al.. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C. et al.. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004; 303:844–848. [DOI] [PubMed] [Google Scholar]
- 53. Sammons M.A., Zhu J., Drake A.M., Berger S.L.. TP53 engagement with the genome occurs in distinct local chromatin environments via pioneer factor activity. Genome Res. 2015; 25:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Azofeifa J.G., Allen M.A., Hendrix J.R., Read T., Rubin J.D., Dowell R.D.. Enhancer RNA profiling predicts transcription factor activity. Genome Res. 2018; 28:334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vierbuchen T., Ling E., Cowley C.J., Couch C.H., Wang X., Harmin D.A., Roberts C.W.M., Greenberg M.E.. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell. 2017; 68:1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Biddie S.C., John S., Sabo P.J., Thurman R.E., Johnson T.A., Schiltz R.L., Miranda T.B., Sung M.-H., Trump S., Lightman S.L. et al.. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011; 43:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Phanstiel D.H., Van Bortle K., Spacek D., Hess G.T., Shamim M.S., Machol I., Love M.I., Aiden E.L., Bassik M.C., Snyder M.P.. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol. Cell. 2017; 67:1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vo Ngoc L., Cassidy C.J., Huang C.Y., Duttke S.H.C., Kadonaga J.T.. The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters. Genes Dev. 2017; 31:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C. et al.. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006; 38:626–635. [DOI] [PubMed] [Google Scholar]
- 60. Frith M.C., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Hayshizaki Y., Sandelin A.. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006; 16:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lo K., Smale S.T.. Generality of a functional initiator consensus sequence. Gene. 1996; 182:13–22. [DOI] [PubMed] [Google Scholar]
- 62. Loewer A., Batchelor E., Gaglia G., Lahav G.. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010; 142:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. et al.. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 64. Behera V., Evans P., Face C.J., Hamagami N., Sankaranarayanan L., Keller C.A., Giardine B., Tan K., Hardison R.C., Shi J. et al.. Exploiting genetic variation to uncover rules of transcription factor binding and chromatin accessibility. Nat. Commun. 2018; 9:782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wei C.-L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z. et al.. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006; 124:207–219. [DOI] [PubMed] [Google Scholar]
- 66. Kannan K., Amariglio N., Rechavi G., Givol D.. Profile of gene expression regulated by induced p53: connection to the TGF-beta family. FEBS Lett. 2000; 470:77–82. [DOI] [PubMed] [Google Scholar]
- 67. Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A.. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015; 12:1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cui H., Li X., Han C., Wang Q.-E., Wang H., Ding H.-F., Zhang J., Yan C.. The stress-responsive gene ATF3 mediates dichotomous UV responses by regulating the Tip60 and p53 proteins. J. Biol. Chem. 2016; 291:10847–10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Taketani K., Kawauchi J., Tanaka-Okamoto M., Ishizaki H., Tanaka Y., Sakai T., Miyoshi J., Maehara Y., Kitajima S.. Key role of ATF3 in p53-dependent DR5 induction upon DNA damage of human colon cancer cells. Oncogene. 2012; 31:2210–2221. [DOI] [PubMed] [Google Scholar]
- 70. Gilchrist M., Thorsson V., Li B., Rust A.G., Korb M., Roach J.C., Kennedy K., Hai T., Bolouri H., Aderem A.. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006; 441:173–178. [DOI] [PubMed] [Google Scholar]
- 71. Su D., Wang X., Campbell M.R., Song L., Safi A., Crawford G.E., Bell D.A.. Interactions of chromatin context, binding site sequence content, and sequence evolution in Stress-Induced p53 occupancy and transactivation. PLoS Genet. 2015; 11:e1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Younger S.T., Kenzelmann-Broz D., Jung H., Attardi L.D., Rinn J.L.. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res. 2015; 43:4447–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fischer M., Steiner L., Engeland K.. The transcription factor p53: not a repressor, solely an activator. Cell Cycle Georget. Tex. 2014; 13:3037–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Daino K., Ichimura S., Nenoi M.. Both the basal transcriptional activity of the GADD45A gene and its enhancement after ionizing irradiation are mediated by AP-1 element. Biochim. Biophys. Acta BBA - Gene Struct. Expr. 2006; 1759:458–469. [DOI] [PubMed] [Google Scholar]
- 75. Fuxman Bass J.I., Sahni N., Shrestha S., Garcia-Gonzalez A., Mori A., Bhat N., Yi S., Hill D.E., Vidal M., Walhout A.J.M.. Human gene-centered transcription factor networks for enhancers and disease variants. Cell. 2015; 161:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reece-Hoyes J.S., Diallo A., Lajoie B., Kent A., Shrestha S., Kadreppa S., Pesyna C., Dekker J., Myers C.L., Walhout A.J.M.. Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat. Methods. 2011; 8:1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yan C., Lu D., Hai T., Boyd D.D.. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2005; 24:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McDowell I.C., Barrera A., D’Ippolito A.M., Vockley C.M., Hong L.K., Leichter S.M., Bartelt L.C., Majoros W.H., Song L., Safi A. et al.. Glucocorticoid receptor recruits to enhancers and drives activation by motif-directed binding. Genome Res. 2018; 28:1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Reiter F., Wienerroither S., Stark A.. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017; 43:73–81. [DOI] [PubMed] [Google Scholar]
- 80. Zaret K.S., Carroll J.S.. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011; 25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zaret K.S., Mango S.E.. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 2016; 37:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nguyen T.-A.T., Grimm S.A., Bushel P.R., Li J., Li Y., Bennett B.D., Lavender C.A., Ward J.M., Fargo D.C., Anderson C.W. et al.. Revealing a human p53 universe. Nucleic Acids Res. 2018; 46:8153–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated as part of this study are available under Gene Expression Omnibus (GEO) Accession GSE137297.