Abstract
Libraries of single-stranded oligodeoxynucleotides (ssODNs) can be enriched for sequences that specifically bind molecules on naïve complex biological samples like cells or tissues. Depending on the enrichment strategy, the ssODNs can identify molecules specifically associated with a defined biological condition, for example a pathological phenotype, and thus are potentially useful for biomarker discovery. We performed ADAPT, a variant of SELEX, on exosomes secreted by VCaP prostate cancer cells. A library of ∼1011 ssODNs was enriched for those that bind to VCaP exosomes and discriminate them from exosomes derived from LNCaP prostate cancer cells. Next-generation sequencing (NGS) identified the best discriminating ssODNs, nine of which were resynthesized and their discriminatory ability confirmed by qPCR. Affinity purification with one of the sequences (Sequence 7) combined with LC–MS/MS identified its molecular target complex, whereof most proteins are part of or associated with the multiprotein ESCRT complex participating in exosome biogenesis. Within this complex, YBX1 was identified as the directly-bound target protein. ADAPT thus is able to differentiate exosomes from cancer cell subtypes from the same lineage. The composition of ESCRT complexes in exosomes from VCaP versus LNCaP cells might constitute a discriminatory element between these prostate cancer subtypes.
INTRODUCTION
Extracellular vesicles (EVs), or exosomes, are small lipid vesicles ∼20–1500 nm in diameter that are secreted by most cell types into the extracellular milieu (1). Exosomes are highly abundant in most biological fluids such as blood plasma, saliva, cerebrospinal fluid, urine, etc. where up to 1012 exosome-particles/ml can be detected (2). The main biological function of exosomes is thought to be in intercellular communication. Exosomes carry a variety of bioactive molecules as cargo, from non-coding RNAs and proteins to metabolites and multi-molecular complexes, either in their interior or on their surface. Their molecular composition resembles that of the parent cell from which they are derived, which is thought to mirror the functional state of the cell, both in healthy and diseased states (2). Exosomes from tumor cells have been shown to be involved in fundamental aspects of cancer, including angiogenesis, immune evasion, and metastasis (3). Thus, exosomes have been described as a rich source for characterization of the tissue of origin and biomarker discovery due to the straightforward, minimally invasive access to tissue-derived samples by liquid biopsy, and because they reflect the dynamic alterations that can occur during tumor progression (4).
A method that has been used previously for deciphering proteins expressed by complex biological targets such as cells or tissues is ‘systematic evolution of ligands by exponential enrichment’ (SELEX) (5–7). The molecules derived from this process are single-stranded nucleic acid sequences, called aptamers, which can fold into complex 3D structures to bind huge varieties of target molecules ranging from small metabolites, other nucleic acids, proteins, post-translational modifications or multi-molecular complexes of biomolecules. Aptamers have been identified by SELEX on complex targets that interact with a malignant cell population or with malignant tissues, without a priori knowledge of the recognized target (8,9). The entire process of library enrichment can be controlled at virtually every stage because it occurs in vitro. This allows adjusting the specificity of a library for a given biological condition, by conducting the enrichment process so as to preferentially enrich sequences that associate with condition A (e.g. disease-related condition) and to disfavor the co-enrichment of sequences that associate with condition B (e.g. non disease-related condition) by including appropriate negative selection steps. In fact, the negative selection steps even allow the depletion of the library for sequences that bind targets common to both conditions, leaving in principle only those aptamers that bind to targets characteristic for the disease-related condition. The subsequent identification of these unknown targets by biophysical approaches such as mass spectrometry provides a potential avenue to biomarker discovery.
Recently, in a process called ‘adaptive dynamic artificial polyligand targeting’ (ADAPT), we described the enrichment of a complex library of ssODNs for sequences that preferentially associate with formalin-fixed paraffin embedded tissue of breast cancer patients and differentiate between patients that benefit from treatment with the Her2-antagonist trastuzumab from those that did not (10). In another study we employed ADAPT to enrich an ssODN library containing 1011 different sequences for binding to circulating exosomes isolated from blood plasma of women with or without breast cancer (11). A pool of synthetic versions of 2000 sequences contained in the enriched library was then used to profile an independent set of plasma exosomes from women with or without disease. The evaluation of healthy women and breast cancer-positive patients showed that ssODN polyligands distinguished between the two groups. By aptamer-mediated affinity purification and mass spectrometry we identified low-abundance exosome-associated proteins and protein complexes. We also showed that some of the ssODNs contained in the synthetic library associated with the C1Q-complex, a component of the serum complement system, in an aptameric binding mechanism.
In the present study, we employ ADAPT to an even more challenging problem, namely for enriching a ssODN-library for sequences that can distinguish between exosomes excreted by two subtypes of cancer cells originating from the same lineage. Differentiation between cancer subpopulations from related tissues requires the detection of differences in the expression levels of biomolecules that often reflect tumor heterogeneity. The two prostate cancer subtypes, ‘vertebral cancer of the prostate’ (VCaP) and ‘lymph node cancer of the prostate’ (LNCaP) comply with these premises: both subtypes express prostate specific antigen (PSA), prostatic acid phosphatase (PAP), p53 antigen and Rb protein (ATCC.org). Differential levels of proteins in VCaP versus LNCaP cell exosomes have been identified by mass spectrometry (12). VCaP cells also have increased androgen receptor (AR) gene copy number and show 5–10-fold higher levels of AR RNA and AR protein compared to LNCaP cells (13). VCaP cells also express the androgen receptor splice variant 7 (AR-V7) associated with increased resistance to androgen dependent therapy (ADT) whereas LNCaP cells have undetectable levels of the variant protein making it an ideal system to study AR-V7 splicing and its contribution to castration resistant prostate cancer (CRPC) progression (13) by ADAPT. Given the tumor heterogeneity of AR positive subtypes and their variability in response to ADT, we employ ADAPT to examine differential analysis of exosomes from these two PC sub-types with variable levels of AR and AR-V7.
MATERIALS AND METHODS
Exosome isolation
Cell lines were purchased from American Type Cell Culture. VCaP cells were grown in DMEM media and LNCaP cells were grown in RPMI media. Exosomes were isolated from VCaP and LNCaP cells similar to Théry et al. (14) with small modifications. Briefly, centrifugation of the exosomal supernatant at 400 × g and 2000 × g was performed to remove cell debris. The small vesicles were then collected using centrifugal filter units with 100 000 NMWL from Millipore. Applying the concentrated exosome suspension on a 30% sucrose cushion performed the final exosome purification by ultracentrifugation at 100 000 × g. Exosome purification was confirmed by transmission electron microscopy (TEM, Supplementary Figure S1).
Enrichment of aptamer library on exosomes derived from VCaP and LNCaP cells
Five rounds of SELEX were performed on VCaP and LNCaP exosomes derived from prostate cancer cell lines by ultracentrifugation. In order to block non-specific binding, exosomes were pre-incubated with 10 μl salmon sperm DNA (800 ng), 10 μl yeast tRNA (800 ng) in 160 μl ‘reaction buffer’ [1 × PBS, 3 mM MgCl2, 0.5% Pluronic® F127 (Sigma), 1 mg/ml human serum albumin (HSA)] for 20 min at 25°C with shaking at 500 rpm. In the first step of enrichment (round 1), 20 μl of a diverse library of 1011 ssODNs (5 ng) with a 35 nt random region was incubated in reaction buffer with 25 μg in 180 µl VCaP exosomes (positive selection: 25 μg pre-incubated exosomes for 30 min at 25°C with rotation (final volume: 200 μl). Exosomes were then precipitated with 6% PEG8000 (precipitation protocol: 200 μl 12% PEG8000 added to 200 μl ODN-bound exosomes, 30 min incubation on ice, centrifuge at 16 000 × g for 10 min at 4°C, remove supernatant, resuspend in 200 μl reaction buffer, centrifuge at 16 000 × g for 10 min at 4°C), and exosome-associated ssODNs were recovered by elution using 10 μl 0.25 M NaOH, incubation for 10 min at 50°C, shaking for 5–10 s at 550 rpm, addition of 10 μl 0.25 M HCl, centrifugation at 16 000 × g for 10 min. The supernatant was removed and the pellet was resuspend in 30 μl reaction buffer. This ssODN pool was taken straight into PCR after round 1 (11), but for subsequent rounds of enrichment (rounds 2 – 5), the eluted ssODNs from positive selection were further incubated with 25 μg LNCaP exosomes (negative selection). LNCaP exosomes were precipitated with 6% PEG8000 and then pelleted by centrifugation at 16 000 × g for 10 min. The pellet was discarded and the unbound ssODNs from the supernatant were collected and incubated with a fresh aliquot of 25 μg VCaP exosomes. Precipitation and two-step elution were again performed, with 6% PEG8000 and 0.25 M NaOH/HCl respectively. The eluted ssODN library was amplified by PCR, denatured to recover ssDNA and then purified. The amplified, enriched ssODN library was used as the starting material for the next round of enrichment at an input of 1011 sequences. Enrichment was monitored by next-generation sequencing. Libraries after each round of enrichment were amplified by PCR with unique indexing primers for multiplex analysis by NGS on an Illumina HiSeq2500 (Supplementary Figure S2).
Next-generation sequencing of ssODN-probed exosomes
Exosome probing was performed by incubating 25 μg VCaP and LNCaP exosomes, in duplicate, with 2 × 1010 copies (1 ng) enriched round 5 ssODN library for 30 min at 25°C. Exosomes were precipitated with 6% PEG8000 and centrifuged at 16 000 × g for 10 min. Supernatant was discarded and exosome pellets were resuspended in H2O. Exosome-associated ssODNs were amplified by PCR with unique indexing primers for multiplex analysis by NGS on an Illumina HiSeq2500. The nine sequences shown in Figure 2B were selected based on a combination of fold changes of at least 4.0 and normalized counts of at least 500 for probing on VCaP exosomes (positive samples). Normalized counts were obtained by dividing the raw counts by the total counts per sample and multiplying the result by the average sample count (Supplementary Figure S3).
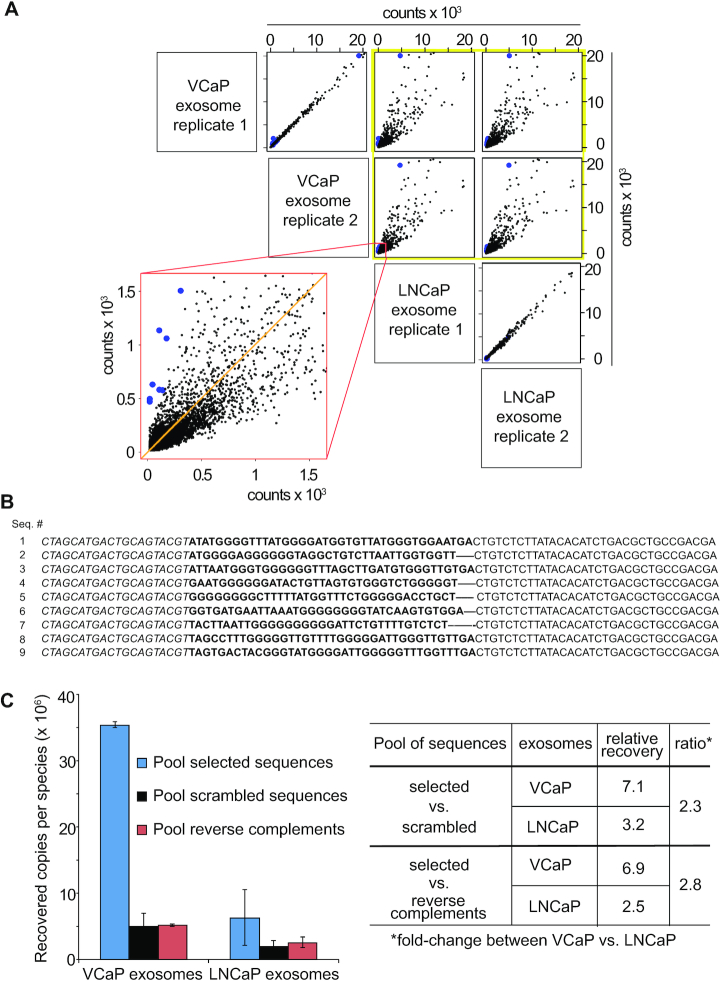
Figure 2.
Sequence identification and verification. (A) Library after five rounds of enrichment was used to probe exosomes from VCaP and LNCaP cells in order to identify individual ODNs that bound preferably to exosomes from VCaP cells (blue). Per exosome type two probing reactions (replicates 1 and 2) were performed and bound ODNs were identified by NGS; each dot represents one unique sequence with counts from different samples on both axes. Yellow frame: comparison of counts from VCaP exosome replicates 1 and 2 with LNCaP exosome replicates 1 and 2. Red frame: magnification of the VCaP exosome replicate 2 versus LNCaP exosome replicate 1 distribution of counts. A higher degree of scattering indicates the selection of sequences with a higher affinity to one or the other sample. For details on the NGS results see Supplementary Figure S3. (B) Sequences were resynthesized and binding of co-precipitated ODNs to VCaP exosomes was verified by qPCR. Sequences of the ssDNA clones 1–9. Italic: 5′-primer binding site; regular: 3′-primer binding site; bold: variable region. All nine sequences have the potential of forming G-quadruplex structures according to QGRS-mapper (68). For the related reverse complementary and scrambled versions of sequences 1–9 see Supplementary Table S1. (C) Left panel: Comparison of the averages across three lots of exosomes by binding of a pool of nine unique sequences (blue; represented as blue dots in A), a pool of their scrambled versions (black), and a pool of their reverse complements (red) by qPCR. Binding is expressed as the recovered copies per sequence from 2 μg of exosomes as determined by the BCA assay. Error bars: S.D.; n = 3. Right panel: Relative recovery across all exosome lots with selected sequences normalized to the indicated negative controls. Ratio: Fold-change between relative recoveries between VCaP versus LNCaP exosomes. Values of individual lots and the standards of pools in qPCR are shown in Supplementary Figure S5.
Quantitative PCR analysis of ssODN-probed exosomes
Exosome probing was performed by incubating 25 μg VCaP and LNCaP exosomes with 8 × 109 copies (0.4 ng) individual or pooled ssODN sequences for 30 min at 25°C. Exosomes were precipitated with 6% PEG8000 and centrifuged at 16 000 × g for 10 min. Supernatant was discarded and exosome pellets were resuspended in H2O. After determining the total protein recovery by the bicinchoninic acid (BCA; Pierce® BCA protein assay kit [ThermoFisher Scientific]) assay, from each precipitation reaction three times 2 μg of exosomes were used as the input for qPCR amplification (Accustart II Taq DNA polymerase). The total volume per reaction was 10 μl using 6 μl of SYBR Green I (Low ROX) Master Mix qPCR reagents (PerfeCTa from QuantaBio). qPCR was performed using MicroAmp Optical 384-well Reaction Plate w/ Barcode (Thermo/ABI/LifeTech) on the ViiA 7 (Applied Biosystems) instrument and consisted of a 95°C incubation for 30 s, followed by 40 cycles at 95°C for 10 s, 60°C for 1 min using the following primers (purchased from IDT-Integrated DNA Technologies, HPLC purified): 5′-CTA GCA TGA CTG CAG TAC GT-3′ (reverse) and 5′-TCG TCG GCA GCG TCA-3′ (forward). A standard curve consisting of the probing sequence(s) was run in duplicate, ranging from 5 × 104 to 5 × 108 copies. NTC (no template control) was run in duplicate. The Viia7 software (version 1.2.3) was used to analyze standard curves and quantify binding affinity of ssODNs to VCaP and LNCaP exosomes. Binding is expressed as the absolute recovered copies per sequence from 2 μg of exosomes.
Affinity purification
The ssODN sequence, herein referred to as Sequence 7, and the reverse complement of Sequence 7 (Sequence 7RC) were synthesized with a biotin affinity tag and 15 μg each of Sequence 7 and Sequence 7RC were immobilized to 10 µl at 10 mg/ml Dynabeads® MyOne™ Streptavidin C1 beads (ThermoFisher Scientific) by incubation in 100 µl for 30 min at room temperature in 50 mM HEPES pH 8.3, 3 mM MgCl2, 0.5 M NaCl, 0.2 mM EDTA, 0.1% Tween-20. In addition, a no ssODN control was included in the experiment. Beads were then washed and equilibrated into sample binding buffer, 1× PBS, 3 mM MgCl2. VCaP exosomes were prepared as described for library selection and 200 μg of resuspended exosomes were incubated with 10 μl of prepared beads or no aptamer control beads for 30 min at room temperature with intermittent mixing. Captured exosomes were then lysed on the bead with 1× PBS, 3 mM MgCl2, 0.5% Triton X-100 and washed two times with lysis buffer followed by three additional washes with 1× PBS, 3 mM MgCl2. All incubation and washing steps were carried out using a MagMAX™ Express-96 Magnetic Particle Processor (ThermoFisher Scientific). Samples were eluted in 0.3% TFA, 6 M urea for 10 min at 37°C. Eluted samples were run for 10 min on a NuPAGE™ Novex™ 4–12% gradient gel (Life Technologies) to remove any residual PEG. The gel was stained with Proteosilver® silver stain plus kit (Sigma). The entire lane (>10 kDa to the stack) was extracted and subjected to in-gel digestion with trypsin.
Liquid chromatography mass spectrometry
Samples containing tryptic peptides were analyzed by nanoflow reverse phase liquid chromatography using a Dionex Ultimate 3000 RSLCnano System (ThermoFisher Scientific) coupled in-line to a Q Exactive HF mass spectrometer (ThermoFisher Scientific). The nano LC system included an Acclaim PepMap 100 C18 5 μm 100A 300 μm × 5 mm trap column and an EASY-Spray C18 3 μm 100A 75 μm × 150 mm analytical column (ThermoFisher Scientific). Peptide samples were loaded onto the trap column and held for 5 min at a constant flow rate of 6 μl/min using running solvent A, where A consisted of 0.1% formic acid in water. Peptides were then eluted using a linear gradient of 2% to 40% B in 35 min, where B consisted of acetonitrile containing 0.1% formic acid. Blank samples consisting of 0.1% formic acid in water were injected between each sample and eluted with the same gradient profile as samples. The LC system was interfaced to the Q Exactive HF using an EASY-Spray electrospray ion source (ThermoFisher Scientific) and the samples were analyzed using positive ion spray voltage set to 2.4 kV, S-lens RF level at 55, and heated capillary at 300°C. The Q Exactive HF was operated in data-dependent acquisition mode selecting the top 10 most intense peaks for fragmentation. MS1 survey scans (m/z 375–1800) were acquired in the Orbitrap analyzer with a resolution of 60 000 at m/z 200, an accumulation target of 3 × 106, and maximum fill time of 50 ms. MS2 scans of the 10 most intense precursor ions were collected using a resolution of 15 000 at m/z 200, an accumulation target of 1 × 105, and maximum fill time of 120 ms, with an isolation window of 1.5 m/z, normalized collision energy of 27, and charged state recognition between 2 and 7. Dynamic exclusion was applied with exclusion duration of 5 s.
Data processing
Raw data files from the Q Exactive HF were analyzed using Sequest HT in the Proteome Discoverer 2.1.1.21 suite and searched against the SwissProt Homo sapiens fasta database (v2015-11-11, with 42 084 entries). Precursor selection was set to use MS1 precursor ions with a signal to noise threshold of 1.5. The search parameters for Sequest HT included full tryptic specificity with a maximum of 2 missed cleavages, minimum peptide length of 6, precursor mass tolerance of 10 ppm, and fragment mass tolerance of 0.02 Da. Set dynamic modifications searched were oxidation (15.9949 Da) of methionine, lysine, and proline; carbamidomethyl (57.0214 Da) of histidine, lysine and cysteine; acetylation (42.0105 Da) of lysine and N-terminus; phosphorylation of serine and threonine; and dimethylation (28.0532 Da) of arginine. Validation of peptide spectrum matches was performed using Percolator with a maximum delta Cn value of 0.05. False discovery rate (FDR) estimation was performed using a decoy database from the reverse sequences of the target database with a FDR setting of 0.01 for high confidence peptide matches, and validation base on q-value. High confidence proteins were accepted as having a q-value of 0.01 or lower as determine using a 0.01 target FDR threshold. Proteins detected with a fold-change ratio of the average precursor ion area intensity of Sequence 7/Sequence 7RC, or Sequence 7/no ssODN bead control >2.5 were considered as proteins likely to be enriched by Sequence 7.
Western blotting
Exosome concentrations were determined by BCA protein assay, which we found to reproduce better than Bradford assays. Exosomes samples (2–10 μg) were resolved by SDS-PAGE separation on 4–12% and 12% Bis–Tris gels (Invitrogen) and transferred to nitrocellulose membranes. Protein detection on the membrane was confirmed with Pierce™ Reversible Protein Stain Kit for Nitrocellulose Membranes (ThermoFisher Scientific). Immunoblotting was carried out using the iBind™ Western Blot System (Invitrogen) for the following primary antibodies: anti-CHMP1B (Santa Cruz), anti-CHMP2A (Santa Cruz), anti-CHMP2A (Abcam), anti-CHMP4B (ThermoFisher Scientific), anti-CHMP4C (ThermoFisher Scientific), anti-CPSF5/NUDT21 (ThermoFisher Scientific), anti-CSPF7 (Santa Cruz), anti-CXCL11 (ThermoFisher Scientific), anti-DNAJA2 (Abcam), anti-EEF1A2 (ThermoFisher Scientific), anti-HBA2 (ThermoFisher Scientific), anti-LAMA3 (ThermoFisher Scientific), anti-MVB12A/FAM125A (ThermoFisher Scientific), anti-PTBP1 (Abcam), anti-RBM3 (Abcam), anti-RPL30 (Abcam), anti-RPLP1 (ThermoFisher Scientific), anti-S100A8 (ThermoFisher Scientific), anti-SH3GL1 (ThermoFisher Scientific), anti-Syntenin-1 (Santa Cruz), anti-VPS28 (Santa Cruz) and anti-YBX1 (Cell Signaling). Membranes were then washed with water for 30 min. Bound antibodies were detected using the enhanced chemiluminescent SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) and images were captured using the Syngene PXi 4 imager (Syngene). Membranes were stripped and re-probed with anti-TSG101 (Santa Cruz) for the loading control, using Restore Western Blot Stripping Buffer (ThermoFisher Scientific). Intensities of bands were determined by densitometry. Intensities of bands were determined by densitometry.
ELONA and ELISA
High binding plates (Corning) were coated overnight, shaking at 4°C, with 10 nM of the following recombinant proteins of interest: CHMP1B (MyBioSource), CHMP2A (MyBioSource), CHMP4B (Origene), CHMP4C (Sigma-Aldrich), CPSF5 (Abcam), CSPF7 (LifeSpan BioScience), CXCL11 (Novus), DNAJA2 (Abnova), EEF1A2 (Abnova), HBA2 (Novus), LAMA3 (Origene), MVB12A (Novus), PTBP1 (Abcam), RBM3 (Abcam), RPL30 (Origene), RPLP1 (Novus), S100A8 (MyBioSource), SH3GL1 (Abnova), Syntenin-1 (Abcam), VPS28 (Abcam) and YBX1 (MyBioSource). Coating solution was removed the next day and washed two times. Plates were then blocked with 1× PBS, 3 mM MgCl2, 0.1 mg/ml salmon sperm DNA/yeast tRNA, 0.01× S1 and 3% BSA (ELONA) or 5% milk in TBS-T (ELISA), incubated at RT for 3 h. For the ELONA, the aptamer and SA-Poly HRP were pre-mixed for 15 min at 800 rpm, and then added to the plate for 1 h at RT. For the ELISA, the primary antibodies, same ones used in western blotting with the exception of: anti-CHMP4B (MyBioSource), anti-HBA2 (Santa Cruz), anti-S100A8 (R&D systems), anti-Syntenin-1 (ThermoFisher Scientific), were added for 1 h at RT, followed by 3× washes and addition of the secondary antibody for 1 h at RT. Plates were washed several times before adding the substrate solution (TMB; R&D Systems) for 5 min and stopped with stop solution (2 N H2SO4). Plates were read at 450/540 nm correction on a plate reader (Biotek).
RESULTS
In vitro selection of ssODN libraries targeting VCaP exosomes
Positive selection against isolated exosomes from VCaP cells was performed by incubation of the exosomes with a library of 1011 different ssODNs for 30 min. Unbound ssODNs were partitioned from the bound ones by precipitating exosomes with polyethylene glycol (PEG) as described previously for plasma exosomes (11). Bound ssODNs were eluted from precipitated exosomes by denaturation and amplified by asymmetric PCR to complete the first selection cycle (Figure 1A). To drive the selection pressure toward enrichment of ssODNs associated with molecular features specific for VCaP cell exosomes, each subsequent selection cycle consisted of the following steps: the ssODNs eluted from the precipitated VCaP cell exosomes were incubated for 30 min with exosomes purified from LNCaP cells in a negative selection step (Figure 1B), LNCaP exosomes were PEG-precipitated, and the supernatant was subjected to another positive selection step on exosomes from VCaP cells, after which the eluted VCaP exosome-bound ssODNs were PCR amplified (Figure 1). This procedure was repeated until enrichment round 5 was completed (Figure 1C). The eluted ssODNs from each selection round were amplified by PCR, and then subjected to next generation sequencing (NGS) to monitor enrichment (Supplementary Figure S2). As expected, an increase in copies per sequence species across the five subsequent rounds of enrichment was determined.
Figure 1.
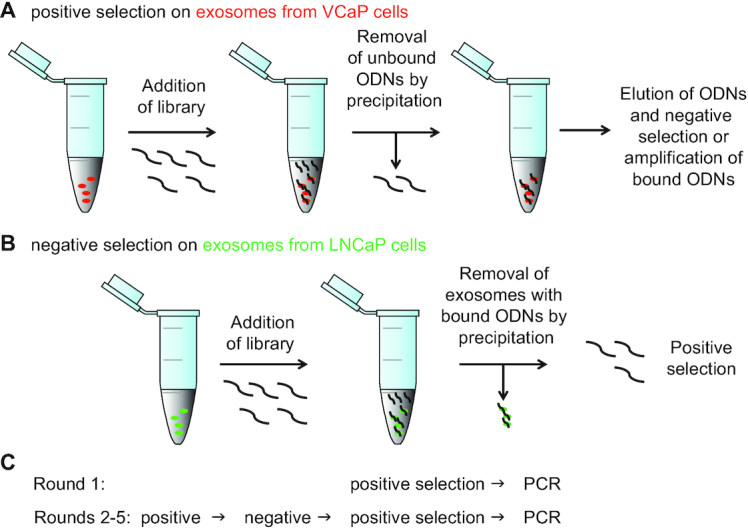
Selection scheme of ssODNs that bind to VCaP exosomes but not to LNCaP exosomes. (A) A highly diverse library of 1011 ssODNs was subjected to five rounds of positive selection. (B) Schematic for the negative selection cycle introduced at round 2 for the remaining selection. After mixing of exosomes with ODN library the unbound DNA was removed by precipitating exosomes and bound ssODNs by polymer. (C) Summary of the succession of selection rounds against exosomes from VCaP and LNCaP prostate cancer cell lines.
Identification and verification of VCaP exosome-binding ssODNs
To identify individual ssODN sequences that preferentially bound to VCaP exosomes, we used the enriched round 5 library to probe the binding of ssODNs to exosomes isolated from VCaP cells in comparison to exosomes isolated from LNCaP cells. We compared two replicates of the same exosome preparations from VCaP and from LNCaP cells, respectively, and identified exosome-bound sequences by NGS (Figure 2A). Supplementary Figure S3 shows the normalized read counts after probing of the round 5 library on VCaP and LNCaP exosomes that resulted in an averaged normalized count of at least 500 for sequences recovered from VCaP exosomes and at least a 4-fold higher recovery of sequences from VCaP exosomes compared to LNCaP exosomes. As expected, binding of ssODNs to replicates of the same exosome type resulted in a low degree of scattering of individual sequences, each of which is represented by a black dot (Figure 2A, upper left panel for intra VCaP, lowest panel for intra LNCaP). When counts of sequences bound to VCaP exosomes were compared to LNCaP exosomes, a high degree of scattering was observed, indicative of differential binding of ssODNs to different samples and various underlying sample profiles (Figure 2A, yellow frame). The lower left panel in Figure 2A (red frame) shows a magnification of the counts of sequences bound to VCaP exosome replicate 2 on the vertical axis, versus the counts of sequences bound to LNCaP exosome replicate 1 on the horizontal axis. We selected those sequences in which counts were most highly overrepresented (blue) compared to the orange line that indicates zero-deviation and three sequences that showed the strongest level of underrepresentation. These sequences should represent distinct ssODNs that identify biological differences between the two classes of exosomes being compared.
Nine of the exosome-bound synthetic sequences (Figure 2B), indicated as blue dots, were individually synthesized and subjected to further analysis. The randomized regions of sequences 1–9 appear to be lower in their C- and A-content than in their G- and T-content (Supplementary Figure S4A) whereas the distribution of the four bases after enrichment round 1 is more equal than after VCaP exosome probing using the round 5 library (Supplementary Figure S4B). Moreover, there is a shift in base-distribution from round 1 to round 5 toward a higher content of T and a low content of A, whereas the G/C-content remains more equally distributed after round 5. To test the reproducibility of VCaP exosome binding, we prepared three samples of exosomes from independent batches of VCaP cells and three samples of exosomes from independent batches of LNCaP cells. The nine VCaP-binding ssODNs shown in Figure 2B were resynthesized and pooled in equimolar quantities. After binding of the pooled sequences to the three batches of exosome preparations, removal of unbound, and recovery of bound sequences, the recovered ssODNs were subjected to qPCR analysis (Figure 2C). As negative controls, synthetic versions of the scrambled and the reverse complementary sequences (Supplementary Table S1), respectively, of each of the nine ssODNs were also pooled in equimolar quantities. The recovered copies per species were generally higher for the pool of selected sequences as compared to the pools of control sequences (Figure 2C, left panel; Supplementary Figure S5). However, the values of relative recovery obtained by normalization of the recovered copies per species of the pool of selected sequences versus either the pool of their scrambled or the pool of their reverse complement versions, respectively, was 2.3- or 2.8-fold higher for VCaP versus LNCaP exosomes, respectively (Figure 2C, right panel).
Together, these data not only confirm the high reproducibility of the VCaP cell exosome-specific ssODNs within batches of VCaP cell exosomes that was already evident from the low scattering of NGS-data for two VCaP cell exosome probing replicates (Figure 2A, upper left panel), but also show that the recovery as analyzed by qPCR is specific for VCaP versus LNCaP cell exosomes. The sequences selected for binding to VCaP cell exosomes showed a considerably lower recovery rate when bound to and eluted from exosomes from LNCaP cells. Moreover, binding to VCaP cells is highly sequence-specific as indicated by the marginal recovery of the pool of reverse complementary sequences after incubation with exosomes from VCaP and LNCaP cells.
Having shown that the pooled nine sequences (Figure 2B) can discriminate between VCaP and LNCaP cell exosomes, we tested the ability of individual sequences to bind to VCaP exosomes by qPCR (Figure 3A). We recovered between 4 × 106 and 5 × 106 copies per sequence in case of sequences 1, 6, 8 and 9, between 7 × 106 and 1.5 × 107 for sequences 2–5, and up to >2 × 107 for Sequence 7. The reverse complement sequences were lower in that value, except for sequences 1, 6 and 9, where the RC versions gave higher values than the selected sequences. Except for sequence 9, all scrambled versions were lower than their respective selected sequences. Together, the data indicate that sequences 1–8 that exhibit high counts in VCaP versus LNCaP (Figure 1A, red frame, blue dots) do show specific interaction with VCaP cell derived exosomes, when compared to their scrambled versions.
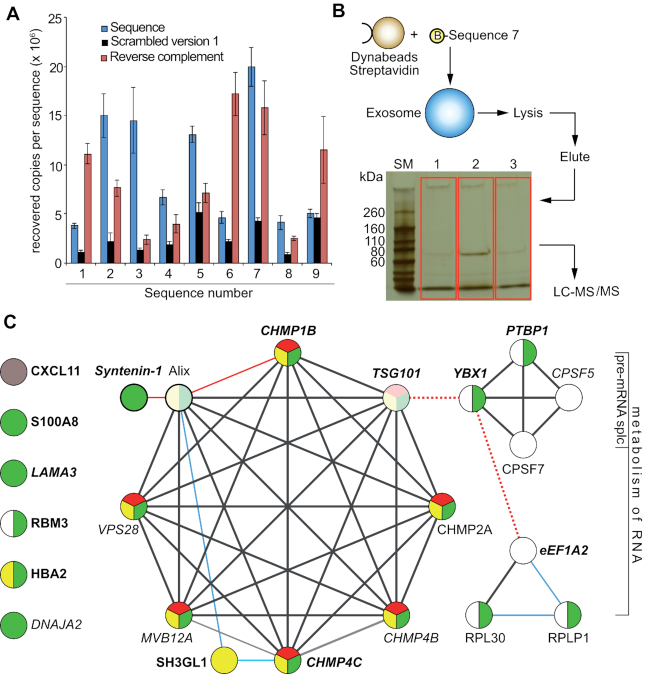
Figure 3.
Target ID. (A) Binding of nine unique sequences (represented as blue dots in 2A) to exosomes from VCaP cells by qPCR; blue bars represent the nine selected sequences. Sequence 7 shows the highest absolute recovery, and was used for target ID; black bars: scrambled versions 1 of the respective sequences; red bars: reverse complement versions of the respective sequences. Error bars: S.D.; n = 3. Binding is expressed as recovered copies per sequence from 2 μg of exosomes as determined by the BCA assay. (B) Affinity purification of target proteins bound to biotinylated Sequence 7 in combination with LC-MS/MS detection identified exosomal binding partners of the aptamer. The gel in red boxes was used for digestion and mass spec analysis. Lane 1: no DNA control; bare beads. Lane 2: pull-down with Sequence 7. Lane 3: pull-down with reverse complement of Sequence 7. (C) Reactome pathway enrichment and STRING 10.5 database analyses of the protein-protein networks of proteins that bound to Sequence 7 with ≥2.5-fold higher abundance than to the Sequence 7RC or to the no oligo bead only control. Black lines: direct or indirect protein-protein associations, edge confidence level high (0.7) to highest (0.9); blue lines: confidence level medium (0.4); red lines: confidence level low (0.15); red dotted lines: interaction not denoted by Reactome or STRING 10.5, but by references (21) for TSG101/YBX1 and (22) for YBX1/eEF1A2 mRNA. Circles represent proteins, protein isoforms, or PTMs produced by a single, protein-coding gene locus. Green circles: proteins associated with exosomes; yellow circles: enriched in the endocytic pathway; red circles: annotated by Reactome or STRING as part of the ESCRT complex. White circles: RNA binding proteins. Pale colored circles: connected through Alix (detected by MS but not necessarily enriched by Sequence 7). Proteins shown in boldface are associated with prostate cancer. CXCL11 (grey circle) is not part of any classification.
Target Identification
We next sought to identify binding partners of ssODNs bound to VCaP cell exosomes and chose Sequence 7 as a pull-down bait (15,16), as this sequence showed the highest level of recovered copies per sequence from the qPCR analysis (Figure 3A). These results are unlikely to be biased by the formation of primer-dimers since we have used the same primers for all sequences. If there were such a bias, it should affect all sequences in the same way. A biotinylated version of Sequence 7 was synthesized, immobilized on streptavidin-coated Dynabeads™ and incubated with VCaP exosomes. Unbound exosomes were removed by washing; the bound exosomes were lysed with detergent directly on the beads, followed by elution of the proteins (Figure 3B) from the magnetic beads. The eluted proteins were subjected to polyacrylamide gel electrophoresis (Figure 3B); lane 2, shows the proteins that bound to Sequence 7, whereas lane 1 is the negative control without DNA (no ssODN control), and lane 3 the negative control with the reverse complement Sequence 7 DNA (Sequence 7RC), which was chosen over the scrambled version as it provides a more stringent control. Lanes were excised from the gel, digested with trypsin, and subjected to LC-MS/MS analysis. Single replicates for each condition were performed.
The result of the mass-spectrometric analysis of protein binding partners of the VCaP cell exosome-specific ssODN Sequence 7 is listed in Table 1 and includes proteins that were either unique to Sequence 7 or showed higher abundance (>2.5-fold) when bound to Sequence 7 compared to the reverse complement of Sequence 7 or to the no ssODN control beads. Cellular component Gene Ontology (GO) enrichment analysis identified 16 of the 21 proteins as known to be associated with exosomes (Figure 3C, green circles). Protein pathway analysis was performed with the Reactome pathway knowledgebase (17,18). Reactome pathway enrichment analysis revealed eight proteins enriched in the ‘vesicle-mediated transport pathway’ (Figure 3C, yellow), six of which, CHMP2A, CHMP1B, CHMP4B, CHMP4C, VPS28, and MVB12A were specific to the ‘Endosomal Sorting Complex Required For Transport (ESCRT)’ pathway (Figure 3C, red). The ESCRT machinery is a high molecular weight multi-protein complex that participates in late endosome sorting and exosome biogenesis (19). Protein–protein interactions within these pathways were analyzed by the STRING database version 10.5 (string-db.org) that assesses direct (physical) and indirect (functional) associations (20). Several of the identified proteins, were also known or predicted to interact with the ESCRT complex (SH3GL1, Syntenin-1) (Figure 3C), connected through Alix (detected by MS but not necessarily enriched by Sequence 7), or through TSG101 (detected by MS but not differentially). Reactome pathway analysis also found proteins enriched in the ‘Metabolism of RNA’ pathway, including YBX1/YB-1, PTBP1, CPSF5, CPSF7, RPL30 and RPLP1. Two additional RNA binding proteins, RBM3 and eEF1A2, were also identified (white circles). YBX1/YB-1 was not annotated by the STRING database as interacting with TSG101, but it has been detected as bound to TSG101 through ubiquitin linkages on YBX1 leading to its increased secretion (21), potentially linking these networks together (Figure 3C, red dashed line). A functional link between YBX1 and eEF1A2 has been proposed based on the interaction of YBX1 and the mRNA of eEF1A2 (Figure 3C, red dashed line) (22). Many of the identified proteins have been reported to be associated with prostate cancer, including CHMP4C (41), Syntenin-1 (35), YBX1 (11), PTBP1/hnRNP-1 (11), RBM3 (23), eEF1A2 (24), SH3GL1 (25), S100A8 (26), HBA2 (27), CXCL11 (28), LAMA3 (29) and TSG101 (30) shown in bold in Figure 3C. In addition, CPSF5, a splicing factor critical for gene silencing in hepatocellular carcinoma (31) was identified, and DNAJA2, a protein that belongs to the heat shock protein family HSP40 and acts as a co-chaperone of HSP70 was also found.
Table 1.
List of proteins identified in VCaP cell exosomes after pull-downs with Sequence 7a
| Protein | Description | Significance | WB verified | Reference |
|---|---|---|---|---|
| CHMP1B | Charged multivesicular body protein 1b | Part of ESCRT machinery that plays role in exosome biogenesis. Identified in exosomes from expressed prostatic secretions | Yes | (60–64) |
| CHMP2A | No | |||
| CHMP4B | Charged multivesicular body protein 4b | Yes | ||
| CHMP4C | Charged multivesicular body protein 4c | Yes | ||
| MVB12A | Multivesicular body subunit 12A | Yes | ||
| SH3GL1 | Endophilin-A2 | No | ||
| VPS28 | Vacuolar protein sorting-associated protein 28 homolog | Yes | ||
| Syntenin-1 | Syntenin-1 | Associated to ESCRT machinery that plays role in exosome and prostasome biogenesis | Yes | (60,62,63,65) |
| YBX1 | Nuclease-sensitive element-binding protein 1 | Connected to ESCRT-1 complex by interacting with TSG101 Marker in CRPC and role in resistance to enzalutamide | Yes | (46) |
| RPLP1 | 60S acidic ribosomal protein P1 | No | ||
| RBM3 | RNA-binding protein 3 | No | ||
| RPL30 | 60S ribosomal protein L30 | No | ||
| HBA2 | Hemoglobin subunit alpha | No | ||
| S100A8 | Protein S100-A8 | No | ||
| CXCL11 | C-X-C motif chemokine 11 | Increased gene expression in prostate cancer | No | (28) |
| DNAJA2 | DnaJ heat shock protein family (Hsp40) member A2 | Upregulated in spermatocytes; close homolog of DNAJA1, which is involved in spermatogenesis and AR signaling | Yes | (48,49) |
| PTBP1 | Polypyrimidine tract-binding protein 1, hnRNP-1 | Cancer associated splicing factor. Known association with breast tumorigenesis; required for tumor cell growth | Yes | (66) |
| eEF1A2 | Elongation factor 1-alpha 2 | Promotes proliferation and inhibits apoptosis in prostate cancer | Yes | (67) |
| LAMA3 | Laminin subunit alpha-3 | Yes | ||
| CPSF5/ NUDT21 | Cleavage and polyadenylation specificity factor subunit 5 | Yes | ||
| CPSF7 | Cleavage and polyadenylation specificity factor subunit 7 | No |
aAdditional MS data are listed in Supplementary Table S2.
Verification and characterization of targets affinity purified with Sequence 7
To further verify the identified targets and to test for different protein levels in exosomes isolated from VCaP versus LNCaP cells, we compared their relative abundances in exosomes from VCaP cells with those from LNCaP cells by western blotting (Figure 4A) and quantified their relative abundance in these two types of exosomes (Figure 4B). Band intensities on the blots suggested that proteins comprising the ESCRT-complex, or being associated with it, namely CHMP1B, CHMP4B, CHMP4C, Syntenin-1, and YBX1 showed considerably higher abundance in exosomes from VCaP cells than in exosomes from LNCaP cells. TSG101 is a component of the ESCRT machinery and an established exosomal marker in prostate cancer (32). We consistently found TSG101 to be present at similar levels in exosomes from both cell lines and thus used this marker for quantification by stripping each respective western blot and re-blotting them with a TSG101 specific antibody (Figure 4A). Similar levels of TSG101 were observed for all analyzed blots, thus confirming equal protein loading and the elevated levels of abundance of the tested proteins in exosomes from VCaP cells versus LNCaP cells. Since TSG101 is a component of the ESCRT machinery, equal protein loading was also confirmed by staining with MemCode® Reversible Protein Stain (ThermoFisher Scientific) of each blot (data not shown). Among the 21 proteins listed in Table 1, nine were disqualified due to unreliable protein detection by the respective antibodies, precluding reliable quantitative evaluation.
Figure 4.
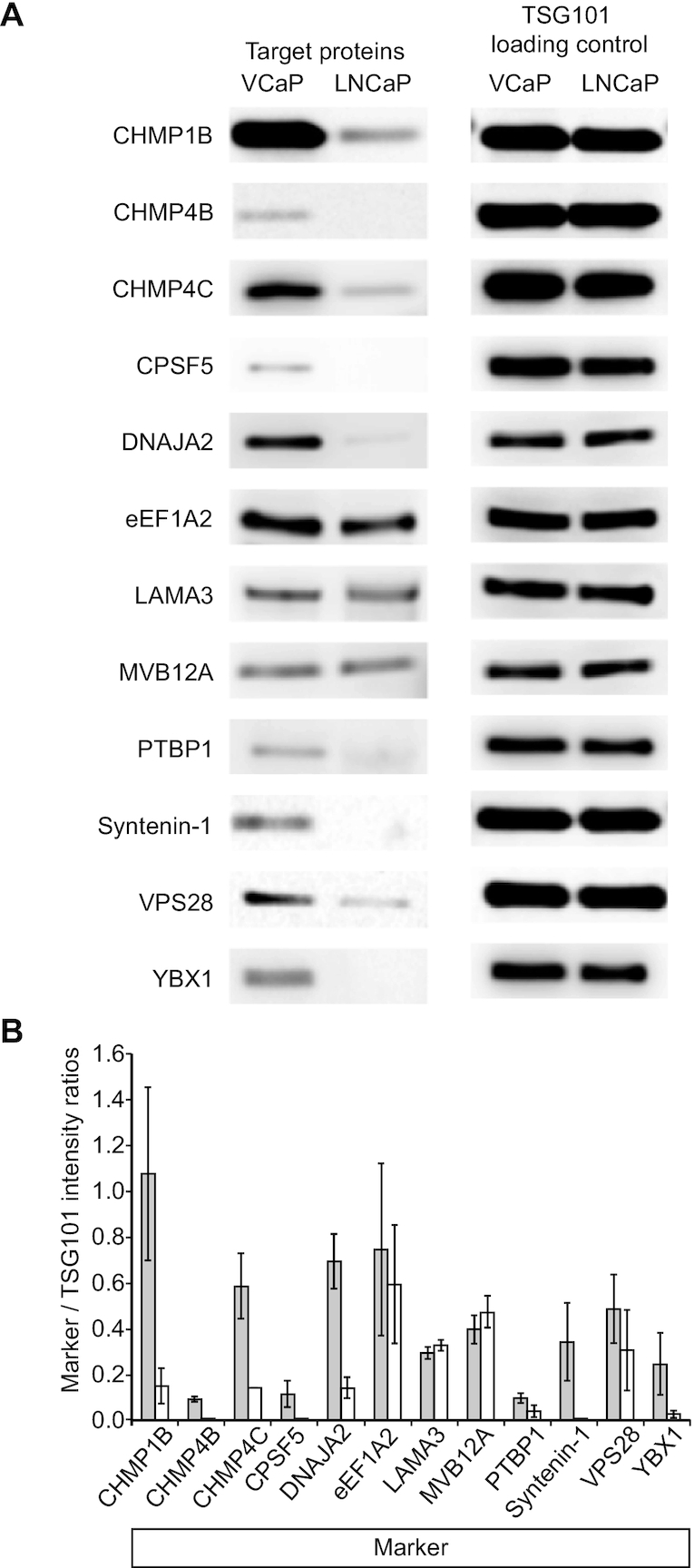
Differential detection of identified proteins in VCaP vs LNCaP exosomes by western blotting. (A) Of the 12 proteins tested (CHMP1B, CHMP4B, CHMP4C, CPSF5, DNAJA2, PTBP1, Syntenin-1 and YBX1 all exhibit higher expression in exosomes from VCaP cells (VCaP) compared to exosomes from LNCaP cells (LNCaP) as quantified by the intensity ratios of targets to the respective TSG101 loading and transfer controls. Only eEF1A2, LAMA3 and MVB12A showed similar expression levels in VCaP and LNCaP exosomes. (B) Relative quantification of intensities from several western blot experiments (VCaP exosomes: dark bars; LNCaP exosomes: light bars) in relation to the respective TSG101 intensities. CHMP1B: n = 4; CHMP4B: n = 2; CHMP4C: n = 2; CPSF5: n = 2; DNAJA2: n = 3; EEF1A2: n = 2; LAMA3: n = 2; MVB12A: n = 2; PTBP1: n = 2; Syntenin-1: n = 2; VPS28: n = 4; YBX1: n = 2. Error bars: mean ± S.D. All proteins verified by western blotting are shown in italic in Figure 3C. We tested all proteins listed in Table 1 and Figure 3C, respectively, but obtained ambiguous results due to insufficient antibody specificity (data are not shown).
We next determined the intensity ratios between the twelve marker proteins and their respective TSG101 loading controls from multiple experiments (n ≥ 2) by densitometry. This quantification revealed that all proteins, except MVB12A, eEF1A2, LAMA3 and VPS28 are present at elevated levels in VCaP cells as compared to LNCaP cells. Similar to TSG101, the levels of MVB12A, a TSG101 interacting protein and also a member of the ESCRT-I complex (34), LAMA3, a protein not known to be associated with the ESCRT complex, and eEF1A2, were approximately equal (Figure 4B) in both cell line exosomes. The most marked difference between levels was found for Syntenin-1 (>10-fold). DNAJA2, CPSF5, YBX1, CHMP1B and CHMP4B showed a 6–7-fold higher level in VCaP cell exosomes than in exosomes from LNCaP cells. A 4–5-fold higher level in exosomes from VCaP cells compared to LNCaP cells was observed for CHMP4C. Between 2- and 3-fold higher levels in VCaP versus LNCaP exosomes was measured for PTBP1. VPS28 exhibited rather high fluctuation in detection levels between experimental replicates, resulting in rather high experimental error, making it unclear as to whether there are differences in its detection levels between exosomes from VCaP versus LNCaP cells.
Sequence 7 binds specifically to YBX1
To determine the direct binding partner among the proteins that were affinity purified with Sequence 7 we performed enzyme-linked oligonucleotide assays (ELONAs). Recombinant purified candidate target proteins were immobilized on plates, incubated with biotinylated Sequence 7 premixed with streptavidin-poly HRP (SA-poly HRP), and quantified by measuring the absorbance at 450–540 nm (Figure 5A). The reverse complementary ssODN of Sequence 7 (Sequence 7RC) served as a negative control. For comparison and to ensure protein integrity, each of the immobilized targets was independently confirmed by an enzyme-linked immunosorbent assay (ELISA), using the respective target-specific antibody (Supplementary Figure S6).
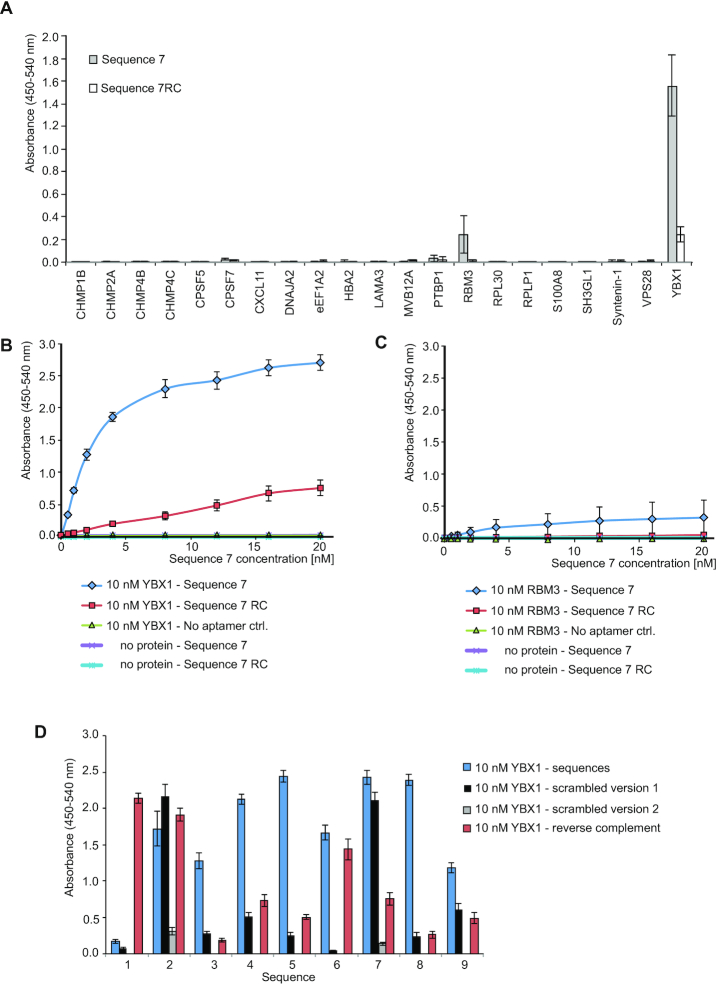
Figure 5.
Direct ELONA experiments to identify the Sequence 7-bound target protein among a set of recombinant proteins. (A) Absorbance levels of horseradish peroxidase (HRP) activity using 10 nM of the indicated recombinant proteins detected by 20 nM of biotinylated Sequence 7 or Sequence 7RC premixed with streptavidin-poly-HRP. Error bars: S.D.; n = 3 for all proteins except for YBX1 (n = 6). (B) Titration of 10 nM YBX1 with increasing concentrations of Sequence 7 (blue diamonds) versus Sequence 7RC (red squares), and no ssODN (green triangles). The no protein/Sequence 7 (purple cross) and no protein/Sequence 7RC (blue star) controls did not change with concentration. (C) The same experiment carried out using 10 nM RBM3 instead of YBX1. Assuming the curve to be fully saturated at 3.0 absorbance with half-maximal 1.5 absorbance, a KD of approximately 3.0 nM can be estimated. (D) ELONA performed for each of the nine sequences and their respective scrambled version 1 on 10 nM YBX1. Sequences 2 and 7 were also tested with their respective scrambled versions 2. Error bars: S.D. n = 6 for selected sequences 2 and 7, n = 3 for all other sequences.
Among all 21 proteins tested, a strong signal specific for Sequence 7 was obtained only with recombinant YBX1. Sequence 7RC gave a considerably weaker signal with YBX1. In the case of RBM3, Sequence 7 yielded a signal that was comparative to that obtained for the combination YBX1/Sequence 7RC, whereas the combination RBM3/Sequence 7RC was negative (Figure 5A). These results point to YBX1 as the direct Sequence 7 target. To quantify these initial binding data, we performed an ELONA titration using a fixed concentration of 10 nM YBX1 and increasing concentrations of Sequence 7 (Figure 5B). Assuming the curve to be fully saturated at 3.0 absorbance with half-maximal 1.5 absorbance, a KD of approximately 3.0 nM can be estimated. For comparison, the same experiment was also carried out using RBM3 at 10 nM fixed concentration. Sequence 7RC and no aptamer, respectively, served as negative controls for both proteins. Only with YBX1 a saturating binding curve was obtained with Sequence 7. A weak, but not saturating signal with flat slope was obtained with the combination YBX1/Sequence 7RC, whereas an even flatter slope was observed for the combination RBM3/Sequence 7. No changes in absorbance were seen for all other negative control experiments even at the highest ssODN concentrations of 20 nM. Taken together, these data confirm YBX1 as the direct target of Sequence 7.
To investigate the binding behavior of sequences 1–6, 8 and 9 in comparison to Sequence 7 toward YBX1 we carried out an ELONA. As controls the respective reverse complements, and the respective scrambled versions of sequences 1–9 were also tested (Figure 5D). Except for sequence 1, all other sequences exhibited high levels of absorbance in the ELONA, with sequences 5, 7 and 8 yielding the highest absolute values. The scrambled version 1 of sequences 3–6 and 8 all showed considerably lower binding to YBX1, in contrast to those of sequences 2 and 7, which were equal to the selected sequences. We therefore designed another scrambled version for these two sequences and found the scrambled version 2 no longer bind to YBX1. The reverse complement versions exhibited reduced or low-level binding, except for sequences 1, 2 and 6. Together, the ELONA study indicates that most of the nine sequences do interact with YBX1, at different levels and with different selectivity with respect to their control sequences.
Sequence 7 specifically captures exosomes from VCaP but not LNCaP cell culture media
To test whether Sequence 7 is capable of specifically capturing VCaP but not LNCaP cell-derived exosomes, pull-down experiments were carried out (Supplementary Data Set S7). We grew VCaP or LNCaP cells separately in exosome-depleted media. To ensure that equal amounts of exosomes were present in the same volume of cell culture media supernatant from both VCaP and LNCaP cells, we performed a classic immunoprecipitation (IP) followed by western blot capturing exosomes with an anti-CD9 biotinylated antibody and probing for TSG101 using an anti-TSG101 antibody. Aptamer pull-downs (AP) were done from the same volume of cell culture media supernatant capturing with Sequence 7, Sequence 7scr, Sequence 7RC, and beads-only and probing for TSG101 with an anti-TSG101 antibody. Three independent experiments were carried out, the results of which are described and discussed in detail in the Supplementary Data Set S7. These data show that VCaP exosomes and not LNCaP exosomes can be pulled down specifically with Sequence 7. The data set also shows that Sequence 7 can pull down more exosomes from VCaP supernatant than its control sequences 7RC and 7scr, but the absolute levels of these pulldowns are variable in the three independent experiments, and so are the differences in the levels between Sequence 7 and its negative control sequences. Because the aptamers were selected on purified exosomes not in cell culture media, the variability between Sequence 7 and its negative control sequences seen in the three independent experiments shown in Supplementary Data Set 7 is not too surprising given that aptamers tend to work best under the conditions under which they were selected.
DISCUSSION
We carried out an in vitro selection using a ssODN-library to target exosomes from the prostate cancer cell line VCaP, which led to a set of enriched sequences that discriminate between exosomes from VCaP and LNCaP cells –subtypes of androgen receptor positive prostate cancer cells. VCaP was first described in 2001 as a prostate cancer cell line that was derived from a vertebral metastatic lesion to a lumbar vertebral body of a patient with hormone-refractory prostate cancer (33). VCaP cells differ morphologically from LNCaP cells with more pronounced dendritic extensions whereas LNCaP are adherent epithelial cells growing in aggregates and as single cells (34). The employed selection scheme is related to our recently described molecular profiling technology called ‘Adaptive dynamic artificial polyligand targeting’ (ADAPT), a platform that combines aptamer selections with next generation sequencing and target identification (11). ADAPT can be deployed against multiple cancer types in various biological matrices, and offers broad potential applications in biomarker discovery. Here we show that ADAPT could not only discriminate between cancer types but between subtypes of a specific lineage.
Among the nine enriched sequences, we found Sequence 7 to be the sequence with the highest relative recovery in qPCR, and used this sequence for target identification by affinity purification and mass spectrometry. Most of the identified proteins associated with Sequence 7 are known to be involved in the formation of the endosomal sorting complexes required for transport (ESCRT) machinery (Figure 6). The ESCRT machinery is a large multi-component assembly of proteins and consists of the sub-complexes I, II and III (19). Its main biological function involves cellular cargo transportation; including remodeling of early endosome membranes for ubiquitinylated receptor sorting (35), membrane bending, scission and repair (36,37), endosomal sorting and intraluminal vesicle formation (38), and cell-to-cell communication by ecto- and exosomal secretion (19,39,40). These activities imply the ESCRT complex as a central gatekeeper of cargo flow toward degradation (41). Moreover, several ESCRT proteins have been implicated in playing a role in cancer (40), and recently in prostate cancer (42). Specifically, the latter study showed that the farnesyltransferase inhibitor manumycin A suppresses exosome biogenesis and secretion in the castration-resistant prostate cancer cell line C4–2B. This effect was partly mediated by ERK-dependent inhibition of the oncogenic splicing factor hnRNP-H1. Another study (43) identified hnRNP-L as a potential hub protein for prostate cancer, a known interactor of PTBP1/hnRNP-1 (44) that we found here to be 2- to 3-fold upregulated in VCaP versus LNCaP cells. Decreased PTBP1/hnRNP-1 protein recruitment to the AR gene was previously observed in VCaP cells under ADT conditions induced by treatment with the non-steroidal anti-androgen Enzalutamide, suggesting a role in AR and AR-V7 splicing (13). The transcriptional regulator Y-box binding protein-1 YBX1 was also found associated with core splicing components in exosomes from Ras-transformed MDCK cells following epithelial-mesenchymal transition (EMT) (45); we detected 6- to 7-fold higher YBX1 levels in exosomes from VCaP versus LNCaP cells. YBX1 associates with the ESCRT-1 complex by interacting with TSG101 when ubiquitinylated (21), and was recently shown to be required for developing resistance against Enzalutamide treatment in prostate cancers expressing AR-V7 (46). Thus, YBX1 was suggested as a target for inhibition to prevent the development of castration resistance where splice variants are expressed that render AR inhibitors such as Enzalutamide ineffective. It was also suggested that YBX1 might be a predictive biomarker for ADT in metastatic prostate cancer (46).
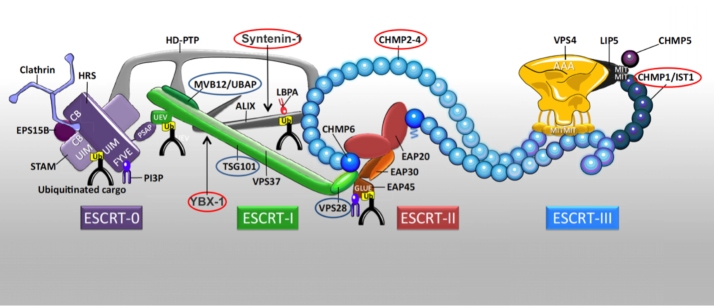
Figure 6.
Composition of the ESCRT Machinery. Proteins in red circles were found to be present at higher abundance in exosomes from VCaP cells compared to exosomes from LNCaP cells as part of this study. Proteins in blue circles were found to be present in similar quantities in both types of exosomes. Adapted with permission from (19). Copyright 2017 Elsevier.
Sequence 7 is not the only aptamer identified in this study that binds to YBX1. Sequences 2–6, 8 and, to a lower extent sequence 9 also bind to YBX1, whereas sequence 1 does not. This finding is not unexpected since all of the nine sequences share a related G-rich motif (Supplementary Figure S4A). YBX1 was found to also bind to tiRNAs that are able to assemble G-quadruplex-like structures at oligoguanine-motifs they usually contain at their 5′-ends (47). The differences of each individual sequence in their YBX1 binding as determined by an ELONA (Figure 5D) may be the result of differences in the G-rich motif. On the other hand, one may hypothesize that in the context of the ESCRT complex each sequence binds to YBX1 as its primary target, but might also sense slight differences in the chemical environment of YBX1 resulting from the proteins in the complex that are in close proximity to YBX1.
DNAJA2 was identified as another highly-upregulated protein in exosomes from VCaP cells compared to LNCaP exosomes. This protein belongs to the HSP40-homologous class of co-chaperones and was found to be upregulated in spermatocytes (48). It shares high homology with DNAJA1, which is involved in spermatogenesis and AR signaling (49). We did not find any reports suggesting that DNAJA2 directly interacts with the ESCRT machinery, or any members of the ESCRT complexes in VCaP. However, LNCaP95 is a prostate cancer cell line that also expresses AR-V7 associated with increased resistance to ADT, like VCaP (but unlike LNCaP). In LNCaP95 cells, an anti-HSP70 antibody immunoprecipitated DNAJA3 along with YBX1 and CPSF5 (50). DNAJA3 is highly homologous to DNAJA2, and YBX1 and CPSF5 were both also found in our study. Moreover, DNAJA2 is involved in Ago2-RNA silencing complex assembly (Ago2-RISC) (51) which has been shown to modulate the splicing of HIV-1 viral transcripts independent of their miRNA binding function, as well as having a role in coupling of chromatin silencing and alternative splicing (52).
Syntenin-1 and the CHMP-proteins 1B, 4B and 4C also showed considerably higher levels in exosomes from VCaP cells compared to those from LNCaP cells. These proteins are well-documented members of the ESCRT machinery (Figure 6). Interestingly, Chiasserini et al. (53) recently identified two populations of prostasomes, extracellular vesicles that are secreted by epithelial cells of the prostate (2). In one of the prostasome populations the ESCRT proteins Alix, Syntenin-1, and TSG101 were overrepresented ∼3-fold, which is within the order of magnitude of the differential levels of abundance of Syntenin-1 in VCaP cell exosomes that we observe in this study when using Sequence 7 pull-downs compared to the Sequence 7 negative control sequences. TSG101, VPS28 and CHMP4B were also among the proteins that were recently found to be essentially required for restricting the constitutive signaling of NF-κB in HEK293 cells (41).
Most of the proteins that were affinity purified with Sequence 7 constitute known members of the ESCRT complex, with the direct target of Sequence 7 being YBX1, presumably bound by an aptameric binding mechanism. To our knowledge, aptamers that selectively target the ESCRT complex on exosomes associated with a specific prostate cancer cell line, or individual members thereof, have not been described. The only example that relates aptamer technology to prostate cancer is the anti-PSMA aptamer that binds the human prostate-specific membrane antigen PSMA, and was shown to specifically target prostate tumor cells (54). When this aptamer was conjugated to the toxin gelonin, the aptamer ‘escorted’ the toxin to tumor cells that are characterized by PSMA overexpression (55,56). Others have used this aptamer for cell-type specific delivery of siRNAs (56), cis-Pt(IV)-prodrug nanoparticles (57), or quantum dots (58). Because these aptamers can be used to target drugs to specific cell types they are commonly designated as ‘escort’-aptamers, but they are completely unrelated to targeting the ESCRT-machinery.
This study demonstrates that the ADAPT platform can lead to the identification of differences in protein expression patterns between exosomes from two related prostate cancer cell lines VCaP and LNCaP. Sequence 7 is likely to bind the ESCRT complex by a direct aptameric interaction with YBX1 associated with a multicomponent protein complex. There is no biomarker-guided strategy for advanced prostate cancer treatment except up-front ADT.
Taken together, our study shows that ADAPT is a powerful method that allows for the enrichment of polyligands that can distinguish even between different subpopulations of the same disease. Our analysis and characterization revealed differences in the composition of ESCRT and associated complexes between exosomes derived from VCaP and LNCaP cells. Several of the ESCRT-comprising proteins appear to be present at similar or equal levels, while others clearly show elevated levels in VCaP cell exosomes. Whether these differences qualify the respective proteins as novel biomarkers for these different prostate cancers, will be the subject of further investigation.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (59) with the dataset identifier PXD011496.
NGS data of libraries after enrichment and probing have been deposited at the Sequence Read Archive (SRA), SRA identifier: SRP191574; Accession number: PRJNA531690; study unique name: Libraries of single-stranded oligodeoxynucleotides enriched for binding to exosomes from VCaP prostate cancer cells.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Caris Life Sciences.
Conflict of interest statement. The authors declare competing financial interest. M.F. and G.M. are scientific consultants for Caris Life Sciences. T.H., H.A.O’N., S.C.L., K.M.F., J.E.D., M.R., A.S.B., T.T., V.M., J.Z., Z.Z., M.N.R., X.W., M.R.M. and D.S. are or were full-time employees of Caris Life Sciences.
REFERENCES
- 1. El Andaloussi S., Mager I., Breakefield X.O., Wood M.J.. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013; 12:347–357. [DOI] [PubMed] [Google Scholar]
- 2. Raposo G., Stoorvogel W.. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013; 200:373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whiteside T.L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016; 74:103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perakis S., Speicher M.R.. Emerging concepts in liquid biopsies. BMC Med. 2017; 15:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brody E.N., Gold L.. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000; 74:5–13. [DOI] [PubMed] [Google Scholar]
- 6. Famulok M., Mayer G.. Aptamer modules as sensors and detectors. Acc. Chem. Res. 2011; 44:1349–1358. [DOI] [PubMed] [Google Scholar]
- 7. Zhou J., Rossi J.. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017; 16:181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mi J., Liu Y., Rabbani Z.N., Yang Z., Urban J.H., Sullenger B.A., Clary B.M.. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010; 6:22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lyu Y., Chen G., Shangguan D., Zhang L., Wan S., Wu Y., Zhang H., Duan L., Liu C., You M. et al.. Generating cell targeting aptamers for nanotheranostics using cell-SELEX. Theranostics. 2016; 6:1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Domenyuk V., Gatalica Z., Santhanam R., Wei X., Stark A., Kennedy P., Toussaint B., Levenberg S., Wang J., Xiao N. et al.. Poly-ligand profiling differentiates trastuzumab-treated breast cancer patients according to their outcomes. Nat. Commun. 2018; 9:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Domenyuk V., Zhong Z., Stark A., Xiao N., O’Neill H.A., Wei X., Wang J., Tinder T.T., Tonapi S., Duncan J. et al.. Plasma exosome profiling of cancer patients by a next generation systems biology approach. Sci. Rep. 2017; 7:42741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hosseini-Beheshti E., Pham S., Adomat H., Li N., Tomlinson Guns E.S.. Exosomes as biomarker enriched microvesicles: characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol. Cell Proteomics. 2012; 11:863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu L., Xie N, S. S., Plymate S., Mostaghel E., Dong X.. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014; 33:3140–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thery C., Amigorena S., Raposo G., Clayton A.. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006; doi:10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 15. Vinkenborg J.L., Mayer G., Famulok M.. Aptamer-based affinity labeling of proteins. Angew. Chem. Int. Ed. Engl. 2012; 51:9176–9180. [DOI] [PubMed] [Google Scholar]
- 16. Tonapi S.S., Pannu V., Duncan J.E., Rosenow M., Helmstetter A., Magee D., Zhang Q., Tinder T.T., Richards M., Halbert D.D. et al.. Translocation of a cell surface spliceosomal complex induces alternative splicing events and lymphoma cell necrosis. Cell Chem. Biol. 2019; 26:756–764. [DOI] [PubMed] [Google Scholar]
- 17. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. et al.. The reactome pathway knowledgebase. Nucleic Acids Res. 2018; 46:D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. et al.. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016; 44:D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christ L., Raiborg C., Wenzel E.M., Campsteijn C., Stenmark H.. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 2017; 42:42–56. [DOI] [PubMed] [Google Scholar]
- 20. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. et al.. STRING v10: protein-protein interaction networks, integrated over the tree of life. NucleicAcids Res. 2015; 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palicharla V.R., Maddika S.. HACE1 mediated K27 ubiquitin linkage leads to YB-1 protein secretion. Cell Signal. 2015; 27:2355–2362. [DOI] [PubMed] [Google Scholar]
- 22. Dong J., Akcakanat A., Stivers D.N., Zhang J., Kim D., Meric-Bernstam F.. RNA-binding specificity of Y-box protein 1. RNA Biol. 2009; 6:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grupp K., Wilking J., Prien K., Hube-Magg C., Sirma H., Simon R., Steurer S., Budaus L., Haese A., Izbicki J. et al.. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur. J. Cancer. 2014; 50:852–861. [DOI] [PubMed] [Google Scholar]
- 24. Worst T.S., Waldbillig F., Abdelhadi A., Weis C.A., Gottschalt M., Steidler A., von Hardenberg J., Michel M.S., Erben P.. The EEF1A2 gene expression as risk predictor in localized prostate cancer. BMC Urol. 2017; 17:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kharaziha P., Chioureas D., Rutishauser D., Baltatzis G., Lennartsson L., Fonseca P., Azimi A., Hultenby K., Zubarev R., Ullen A. et al.. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget. 2015; 6:21740–21754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yun S.J., Yan C., Jeong P., Kang H.W., Kim Y.H., Kim E.A., Lee O.J., Kim W.T., Moon S.K., Kim I.Y. et al.. Comparison of mRNA, protein, and urinary nucleic acid levels of S100A8 and S100A9 between prostate cancer and BPH. Ann. Surg. Oncol. 2015; 22:2439–2445. [DOI] [PubMed] [Google Scholar]
- 27. Gužvić M., Braun B., Ganzer R., Burger M., Nerlich M., Winkler S., Werner-Klein M., Czyż Z.T., Polzer B., Klein C.A.. Combined genome and transcriptome analysis of single disseminated cancer cells from bone marrow of prostate cancer patients reveals unexpected transcriptomes. Cancer Res. 2014; 74:7383–7394. [DOI] [PubMed] [Google Scholar]
- 28. Hsiao J.J., Ng B.H., Smits M.M., Wang J., Jasavala R.J., Martinez H.D., Lee J., Alston J.J., Misonou H., Trimmer J.S. et al.. Androgen receptor and chemokine receptors 4 and 7 form a signaling axis to regulate CXCL12-dependent cellular motility. BMC Cancer. 2015; 15:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sathyanarayana U.G., Padar A., Suzuki M., Maruyama R., Shigematsu H., Hsieh J.T., Frenkel E.P., Gazdar A.F.. Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin. Cancer Res. 2003; 9:6395–6400. [PubMed] [Google Scholar]
- 30. Sun Z., Pan J., Bubley G., Balk S.P.. Frequent abnormalities of TSG101 transcripts in human prostate cancer. Oncogene. 1997; 15:3121–3125. [DOI] [PubMed] [Google Scholar]
- 31. Sun M., Ding J., Li D., Yang G., Cheng Z., Zhu Q.. NUDT21 regulates 3′-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017; 410:158–168. [DOI] [PubMed] [Google Scholar]
- 32. Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtiö J, El Andaloussi S et al.. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016; 6:22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korenchuk S., Lehr J.E., L M.C., Lee Y.G., Whitney S., Vessella R., Lin D.L., Pienta K.J.. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001; 15:163–168. [PubMed] [Google Scholar]
- 34. Horoszewicz J.S., Leong S.S., Kawinski E., Karr J.P., Rosenthal H., Chu T.M., Mirand E.A., Murphy G.P.. LNCaP model of human prostatic carcinoma. Cancer Res. 1983; 43:1809–1818. [PubMed] [Google Scholar]
- 35. Saksena S., Sun J., Chu T., Emr S.D.. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 2007; 32:561–573. [DOI] [PubMed] [Google Scholar]
- 36. Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., Emr S.D.. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002; 3:271–282. [DOI] [PubMed] [Google Scholar]
- 37. Jimenez A.J., Maiuri P., Lafaurie-Janvore J., Divoux S., Piel M., Perez F.. ESCRT machinery is required for plasma membrane repair. Science. 2014; 343:1247136. [DOI] [PubMed] [Google Scholar]
- 38. Stoorvogel W. Resolving sorting mechanisms into exosomes. Cell Res. 2015; 25:531–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams R.L., Urbe S.. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007; 8:355–368. [DOI] [PubMed] [Google Scholar]
- 40. Gingras M.C., Kazan J.M., Pause A.. Role of ESCRT component HD-PTP/PTPN23 in cancer. Biochem. Soc. Trans. 2017; 45:845–854. [DOI] [PubMed] [Google Scholar]
- 41. Maminska A., Bartosik A., Banach-Orlowska M., Pilecka I., Jastrzebski K., Zdzalik-Bielecka D., Castanon I., Poulain M., Neyen C., Wolinska-Niziol L. et al.. ESCRT proteins restrict constitutive NF-kappaB signaling by trafficking cytokine receptors. Sci Signal. 2016; 9:ra8. [DOI] [PubMed] [Google Scholar]
- 42. Datta A., Kim H., Lal M., McGee L., Johnson A., Moustafa A.A., Jones J.C., Mondal D., Ferrer M., Abdel-Mageed A.B.. Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells. Cancer Lett. 2017; 408:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang F.N., He H.C., Zhang Y.Q., Yang D.L., Huang J.H., Zhu Y.X., Mo R.J., Chen G., Yang S.B., Chen Y.R. et al.. An integrative proteomics and interaction network-based classifier for prostate cancer diagnosis. PLoS One. 2013; 8:e63941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim J.H., Hahm B., Kim Y.K., Choi M., Jang S.K.. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 2000; 298:395–405. [DOI] [PubMed] [Google Scholar]
- 45. Tauro B.J., Mathias R.A., Greening D.W., Gopal S.K., Ji H., Kapp E.A., Coleman B.M., Hill A.F., Kusebauch U., Hallows J.L. et al.. Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol. Cell Proteomics. 2013; 12:2148–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shiota M., Fujimoto N., Imada K., Yokomizo A., Itsumi M., Takeuchi A., Kuruma H., Inokuchi J., Tatsugami K., Uchiumi T. et al.. Potential role for YB-1 in castration-resistant prostate cancer and resistance to enzalutamide through the androgen receptor V7. J. Natl. Cancer Inst. 2016; doi:10.1093/jnci/djw005. [DOI] [PubMed] [Google Scholar]
- 47. Lyons S.M., Achorn C., Kedersha N.L., Anderson P.J., Ivanov P.. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016; 44:6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abdul K.M., Terada K., Gotoh T., Hafizur R.M., Mori M.. Characterization and functional analysis of a heart-enriched DnaJ/ Hsp40 homolog dj4/DjA4. Cell Stress Chaperones. 2002; 7:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Terada K., Yomogida K., Imai T., Kiyonari H., Takeda N., Kadomatsu T., Yano M., Aizawa S., Mori M.. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO J. 2005; 24:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kita K., Shiota M., Tanaka M., Otsuka A., Matsumoto M., Kato M., Tamada S., Iwao H., Miura K., Nakatani T. et al.. Heat shock protein 70 inhibitors suppress androgen receptor expression in LNCaP95 prostate cancer cells. Cancer Sci. 2017; 108:1820–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naruse K., Matsuura-Suzuki E., Watanabe M., Iwasaki S., Tomari Y.. In vitro reconstitution of chaperone-mediated human RISC assembly. RNA. 2018; 24:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.C. et al.. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012; 19:998–1004. [DOI] [PubMed] [Google Scholar]
- 53. Chiasserini D., Mazzoni M., Bordi F., Sennato S., Susta F., Orvietani P.L., Binaglia L., Palmerini C.A.. Identification and partial characterization of two populations of prostasomes by a combination of dynamic light scattering and proteomic analysis. J. Membr. Biol. 2015; 248:991–1004. [DOI] [PubMed] [Google Scholar]
- 54. Lupold S.E., Hicke B.J., Lin Y., Coffey D.S.. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002; 62:4029–4033. [PubMed] [Google Scholar]
- 55. Chu T.C., Marks J.W. 3rd, Lavery L.A., Faulkner S., Rosenblum M.G., Ellington A.D., Levy M.. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006; 66:5989–5992. [DOI] [PubMed] [Google Scholar]
- 56. McNamara J.O. 2nd, Andrechek E.R., Wang Y., Viles K.D., Rempel R.E., Gilboa E., Sullenger B.A., Giangrande P.H.. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006; 24:1005–1015. [DOI] [PubMed] [Google Scholar]
- 57. Dhar S., Gu F.X., Langer R., Farokhzad O.C., Lippard S.J.. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17356–17361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bagalkot V., Zhang L., Levy-Nissenbaum E., Jon S., Kantoff P.W., Langer R., Farokhzad O.C.. Quantum dot-aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007; 7:3065–3070. [DOI] [PubMed] [Google Scholar]
- 59. Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al.. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Principe S., Jones E.E., Kim Y., Sinha A., Nyalwidhe J.O., Brooks J., Semmes O.J., Troyer D.A., Lance R.S., Kislinger T. et al.. In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics. 2013; 13:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fujita K., Kume H., Matsuzaki K., Kawashima A., Ujike T., Nagahara A., Uemura M., Miyagawa Y., Tomonaga T., Nonomura N.. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017; 7:42961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kashyap R., Roucourt B., Lembo F., Fares J., Carcavilla A.M., Restouin A., Zimmermann P., Ghossoub R.. Syntenin controls migration, growth, proliferation, and cell cycle progression in cancer cells. Front. Pharmacol. 2015; 6:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qian X.L., Li Y.Q., Yu B., Gu F., Liu F.F., Li W.D., Zhang X.M., Fu L.. Syndecan binding protein (SDCBP) is overexpressed in estrogen receptor negative breast cancers, and is a potential promoter for tumor proliferation. PLoS One. 2013; 8:e60046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Raiborg C., Stenmark H.. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009; 458:445–452. [DOI] [PubMed] [Google Scholar]
- 65. Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E. et al.. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012; 14:677–685. [DOI] [PubMed] [Google Scholar]
- 66. He X., Arslan A.D., Ho T.T., Yuan C., Stampfer M.R., Beck W.T.. Involvement of polypyrimidine tract-binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. 2014; 3:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun Y., Du C., Wang B., Zhang Y., Liu X., Ren G.. Up-regulation of eEF1A2 promotes proliferation and inhibits apoptosis in prostate cancer. Biochem. Biophys. Res. Commun. 2014; 450:1–6. [DOI] [PubMed] [Google Scholar]
- 68. Kikin O., D’Antonio L., Bagga P.S.. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006; 34:W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (59) with the dataset identifier PXD011496.
NGS data of libraries after enrichment and probing have been deposited at the Sequence Read Archive (SRA), SRA identifier: SRP191574; Accession number: PRJNA531690; study unique name: Libraries of single-stranded oligodeoxynucleotides enriched for binding to exosomes from VCaP prostate cancer cells.