Abstract
The epithelial barrier in the gastrointestinal (GI) tract is a protective interface that endures constant exposure to the external environment while maintaining its close contact with the local immune system. Growing evidence has suggested that the intercellular crosstalk in the GI tract contributes to maintaining the homeostasis in coordination with the intestinal microbiome as well as the tissue-specific local immune elements. Thus, it is critical to map the complex crosstalks in the intestinal epithelial-microbiome-immune (EMI) axis to identify a pathological trigger in the development of intestinal inflammation, including inflammatory bowel disease. However, deciphering a specific contributor to the onset of pathophysiological cascades has been considerably hindered by the challenges in current in vivo and in vitro models. Here, we introduce various microphysiological engineering models of human immune responses in the EMI axis under the healthy conditions and gut inflammation. As a prospective model, we highlight how the human “gut inflammation-on-a-chip” can reconstitute the pathophysiological immune responses and contribute to understanding the independent role of inflammatory factors in the EMI axis on the initiation of immune responses under barrier dysfunction. We envision that the microengineered immune models can be useful to build a customizable patient's chip for the advance in precision medicine.
Keywords: Immune response, Microphysiological system, Gut inflammation-on-a-chip, Microbiome, Co-culture, Organoid
INTRODUCTION
The gastrointestinal (GI) epithelial barrier is a complex interface that endures constant exposure to the external environment (i.e., pathogen, microbiome, diets, or compounds) while also maintaining its close contact with the local immune system (1). Although epidemiological and clinical studies have identified unique profiles of the intestinal microbiome in health and disease (2,3) as well as shown the beneficial effects of probiotics (4,5) and fecal microbiome transplantation (6,7), the mechanistic processes involved in epithelial-microbiome-immune (EMI) axis are not fully understood.
Studying the human gut microbiome using in vitro models has been challenging because many commensal gut microbiome is oxygen-sensitive (8) and unculturable (9). The conventional static culture formats often cause the overgrowth of gut bacteria when co-cultured with epithelium, thus hampering the in vitro demonstration of host-microbiome crosstalk (10). Animal models harbor a vastly different composition of the microbiome population compared to that of humans, which compromises the translational value of experimental results (11). Moreover, although animal models have been used to induce disease-like symptoms and test for the validation of therapeutics, there have been notable limitations in the independent manipulation of the individual contributing factors (±immune element, ±gut microbiome), spatiotemporal modulation of the inflammatory triggers (e.g., before/after Ag presentation, directional introduction of the particular immune trigger), or in situ visualization at high resolution in real-time (e.g., time-lapse imaging of the fluorescently labeled cells under the controlled high-power magnification imaging). Thus, developing a translatable in vitro platform utilizing the gut-on-a-chip technology that allows the mechanistic investigation of the EMI axis is a promising technology in disease modeling (Fig. 1A and B). In this review, we aim to review the physiological role of the individual components in the EMI axis, unique techniques that have been developed to overcome the difficulty of mimicking such axis, and the future direction with the in vitro study of the EMI.
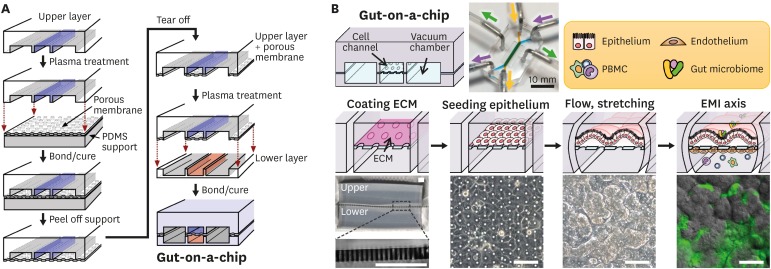
Figure 1. Microfabrication of a human gut-on-a-chip and microfluidic cultures of the EMI axis. (A) The gut-on-a-chip microfluidic device is fabricated using soft lithography method by placing each compartment in a layer-by-layer approach to build the central cell culture chamber with an upper (blue) and a lower (orange) microchannel (1 mm×1 cm×200 μm, width×length×height) and bi-lateral vacuum chambers (grey). (B) A schematic flowchart shows sequential steps to recreate intestinal EMI in a gut-on-a-chip by coating the central cell microchannels using ECM (“coating ECM”), followed by the attachment of epithelium (“seeding epithelium”), application of flow and mechanical stretching motions (“flow, stretching”), and maintenance of the steady-state physiological milieu including gut microbiome and immune cells (“EMI axis”). An inset photograph (top middle) shows a fully equipped gut-on-a-chip device linked to the silicone tubing that supplies culture medium (purple or yellow-green arrow heads) or cyclic vacuum suctions that induce peristalsis-like motions (green arrow heads). Schematics of the representative experimental steps (middle row) and corresponding micrographs of either the device or the cell morphology (bottom row). A zoomed-in snapshot in the left bottom shows the part (a light grey dashed box) of a PDMS porous membrane (25 µm in thickness). A phase contrast image in “seeding epithelium” shows the formation of an intestinal epithelial monolayer in the upper microchannel. An image in “flow, stretching” displays the villous growth under mechanically dynamic physiological conditions at ≤100 h since seeding. Finally, an overlaid image in the “EMI axis” shows the villus morphology (grey) and green fluorescent protein-labeled E. coli (green) after the co-culture for 24 h. Bars, 100 μm. Images were reprocessed from the references (65,66).
ECM, extracellular matrix.
EMULATING THE IMMUNE RESPONSE DURING THE INTESTINAL INTERCELLULAR CROSSTALK
The EMI axis
There has been substantial progress in developing a microfluidic system modeling the physiology and pathology of the GI tract (12,13,14,15,16). The human intestine is the most representative organ for the processes by which the tissue-specific immune system constitutively exerts homeostatic intercellular crosstalk with the host cells as well as the gut microbiome (17). Due to the spatial proximity (18) and biological interactions at the mucosal microenvironment (19), the major contributors involved in the multi-component interactions, including intestinal epithelium, gut microbiome, and the tissue-specific immune elements can be referred to as new terminology, “EMI axis.” These contributing cells should be involved in modeling the pathophysiology of inflammatory immune responses in a defined spatial structure. The prerequisites of each element to build inflammation models are summarized.
Epithelium
The intestinal epithelium is a primary cell type to form a physical tissue barrier that provides an interface between the lumen and the abluminal compartments. In this microenvironment, the EMI axis plays a key role in creating complex intercellular interactions, stimulations, and regulations (19). There are various intestinal epithelial cells, such as absorptive enterocyte, mucus-producing goblet cells, hormone-secreting enteroendocrine cell, anti-microbial peptide-releasing Paneth cells, Ag-permeable microfold (M) cells, and taste-sensing tuft cells (20,21,22). In general, absorptive and goblet cells are major contributors to recreate the epithelial barrier function by providing tight junction integrity and mucus layer, respectively. Hence, major cell lines such as Caco-2 (human colorectal adenocarcinoma, representing the absorptive enterocytes) (23) and HT-29 (human colorectal adenocarcinoma, representing the goblet cells) (24) have been predominantly used in biomedical researches or pharmaceutical tests. Anticipated physiological functions of intestinal epithelium include the expression of tight junction (e.g., zonula occludens 1, occludin, and claudin) (25) and adherence junction (e.g., E-cadherin) (26) and mucin (e.g., mucin 2) (27). However, immortalized cell lines are often derived from tumor cells (28) or been immortalized with a tumor Ag (29) with limited capability to recapitulate a coordinated function of multi-lineage cells (30). In recent days, biopsy-derived (31,32) or stem cell-derived (33) intestinal organoid culture method has been suggested to present diverse cell lineages compared to the immortalized single-lineage cell lines. The organoid cultures, however, have shown critical limitations in co-culturing living microbial cells due to the static culture nature and the enclosed lumen (34).
Immune components
Immune surveillance is critical to promote host defense as well as homeostasis (35,36,37), where tissue-specific immune elements control the primary immune responses in concert with systemic immunity. In the intestinal lamina propria, professional Ag-presenting cells (38,39) including dendritic cells (DCs), macrophages, and B lymphocytes as well as other local immune cells such as intraepithelial lymphocytes (40), innate lymphoid cells (41), and Th or Treg (42) closely communicate and interact to contribute to the intestinal homeostasis. In this unique spatial coordination, DCs (43) and macrophages (44) in the lamina propria collect exogenous Ags (e.g., infectious bacteria) (45) and share the epitopes of molecular components with B lymphocytes (46). In this event, the B cells that are homing from the mesenteric lymph node to the lamina propria transform into the IgA-producing plasma cells and produce secretory antimicrobial IgA (47,48). As given this example, immune cells continuously interact with endogenous or exogenous Ags as well as with other adjacent cells, then contribute to induce inflammatory responses. Thus, involving the right type of tissue-specific immune cells in intestinal inflammation models is an essential design step to demonstrate immune-mediated interactions. Recently, incorporation of immune system components into these microphysiological systems (MPS) was accomplished, and a major advance in such technological breakthrough includes the spatiotemporal induction of immune cells as well as living microbial cells in the model. Currently, most of in vitro models that demonstrate human immune reactions have relied on the PBMCs derived from a drawn blood sample (13). Alternatively, the differentiated immune cells (e.g., DCs from monocytes) (14) can be enriched in vitro and introduced into the MPS model to induce physiological immune tolerance in the gut (14,15). However, compared to animal models, it has been still nascent to recreate a tissue-specific immunity in an in vitro model, which remains a critical unmet in disease modeling.
Gut microbiome
The gut microbiome can contribute to maintaining, and perturbing, the EMI axis by producing short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate (49), and by releasing bacterial endotoxins (e.g., LPS) (50) or secreting polysaccharide A (51). SCFAs are the primary bacterial metabolites that are used as a major energy source for intestinal epithelium (52). The human commensal gut microbiome plays a pivotal role in degrading non-digestible fibers (i.e., prebiotics), including inulin (53), fructo-oligosaccharide (54), and galacto-oligosaccharides (55), and producing the SCFAs via symbiotic cross-feeding interactions in the colon (56). In addition, the intestinal microbiome can shape host immunity, where another major role of SCFAs is the orchestration of the balance of Th vs. Treg cells (49). Thus, stable maintenance of this syntrophic intercellular interaction of the commensal gut microbiome is important to sustain the physiological intestinal homeostasis in the EMI axis in a model (57). It is noted that the majority of intestinal bacteria are obligate anaerobic bacteria (58), and these anaerobic gut bacteria support intestinal homeostasis in the EMI in a mucosal anoxic-oxic interface (AOI) (59). The recent development of an AOI in a microfluidic gut-on-a-chip (60) enabled us to investigate the key physiological interactions of obligate anaerobic gut bacteria with the intestinal epithelium. We will further discuss how these technical challenges have been overcome in the in vitro recreation of AOI in the following section.
Prerequisites for modeling the EMI axis
Reverse engineering
Based on the reverse engineering approach (61), experimental demonstration of the EMI axis in vitro requires the “breakup” of individual components and their functionality from the complex microenvironment of the gut into the level of single components. Next, each component needs to be integrated independently, or collectively, to reform the sequence of interactions that induce intercellular crosstalk with a spatiotemporal resolution. As a reductionist's approach, simplified but physiologically relevant inter-connectivity between different cell types in the EMI axis can be established by adding the uncoupled elements (i.e., microbiome, epithelium, and immune cells) one-at-a-time or by removing a particular cell type in a defined culture format. More importantly, once a system is accessible to reconstitute a functional simulacrum in vitro, different types of cells from various sources (e.g., different patient donors, a defined pathobiont in the GI diseases, a distinct consortium of microbial communities, or differentiated immune cells after the clonal expansion) can be adapted to better mimic individual target tissues and organs.
Recreating the tissue interface
Since these multiple cell types within the EMI axis are localized in the three-dimensional (3D) microenvironmental structure, recreating the lumen-capillary or lumen-lymphatic tissue interface is of great importance when working to demonstrate a polarized tissue organization and directional stimulation of microbial (i.e., luminal) or immune cells (i.e., capillary or lymphatic) in the model. To recreate an organ-level, tissue-specific microenvironment of the human living gut, the establishment of an intact intestinal epithelial barrier is a prerequisite to building the lumen-capillary tissue interface (25,26). The intestinal epithelial layer needs to possess similar compositions of differentiated lineages, including absorptive, mucus-secretory, enteroendocrine, and paneth cells (27,62,63,64). As previously mentioned, mucus production, expression of tight junction proteins, and production of antimicrobial peptides are major functional prerequisites in an epithelial layer. Also, the histogenesis of the 3D villus microarchitecture that displays the crypt-villus characteristics is also important for illustrating the effect of 3D topology on microbial niche formation, stem cell regeneration, and immune-microbiome interactions. Another key requirement in the intestinal modeling is the demonstration of physical deformations that mimics the biomechanics of bowel movement, so-called peristalsis (12,65). The macroscopic mechanical deformations and the microscopic villus motility are necessary, not only for showing the mixing and propulsion of bolus (66) but also for reflecting the villous morphogenesis (12), regenerative cellular signaling (67), and epithelial differentiation (68).
Microbial co-culture
Typically, microbial co-culture can be performed by adding an inoculum of microbial cells on the apical compartment of the epithelial monolayer that has reasonable barrier integrity for a limited period within a day (69). As previously discussed, this limited culture period is mainly due to the overgrowth of microbial cells, where an accumulation of bacterial organic wastes rapidly diminishes the pH of culture medium, followed by the damage of epithelial barrier. Thus, the proposed model should maintain a controlled microbial population without undesirable bacterial overgrowth (66). To study on a longitudinal host-microbiome interaction and monitor how the immune cells respond to the microbiome and their products in the EMI axis, it is necessary to recreate an intact epithelial layer that provides a physical barrier and prevents aberrant transmigration of microbial cells from the lumen to the capillary side. This physical compartmentalization also contributes to control the infiltration of immune cells from the vascular side to the lumen side. In this experimental setup, it is critical to flow culture medium and exert mechanical deformations to suppress bacterial overgrowth (60,66,68). Moreover, the majority of the gut microbiome comprises obligate anaerobic bacteria. Thus, it is also critical to create a local oxygen gradient and establish an in vitro AOI in the model. An AOI can be accomplished by several approaches. The easiest method is to perfuse anoxic and oxic culture media into the upper and lower microchannels, respectively, of a two-channel microfluidic device (e.g., gut-on-a-chip) (60). This method has the advantage to circumvent using complex equipment, including an anaerobic glove box or an oxygen controller (70). Alternatively, the “human-microbial crosstalk (HuMiX)” model employs the perfusion of anoxic and oxic culture media by continuously infusing the nitrogen gas into the adjacent microchannel. The oxygen level can be detected by an oxygen sensor (71) and controlled, in which a facultative and an obligate anaerobic bacteria were co-cultured in the HuMiX model to assess the effect on the epithelial cells. Another method to grow anaerobic gut bacteria is to fabricate the anaerobic chamber while using a calibrated optical probe system to verify the hypoxic conditions within the microfluidic culture system (70). An AOI can lead to better stability in the maintenance of diverse composition of the fecal gut microbiome, which may allow us to investigate the host-microbiome crosstalk to the homeostasis of the GI tract of the intestinal disease (60). Such an AOI together with physiological flow and mechanical motions in an organ-on-a-chip may overcome the limited longitudinal host-microbiome crosstalk observation in the conventional static in vitro cultures (60,70,71).
Modeling the pathophysiology of inflammatory immune responses
MPS to model the physiology of an EMI axis
The MPS refers to a microfluidics-based experimental microcircuit that provides an in vivo-relevant and accessible microenvironment driven by a reverse engineering approach with high modularity to engineer each comprising element on-demand (61). The human gut-on-a-chip is a representative MPS that models the intestinal physiology (66,68) and the pathophysiology that occurs in the EMI axis of various GI diseases (12,13). In the following sections, we will focus on the introduction and application of the gut-on-a-chip microsystem in host-microbe co-cultures and inflammatory immune responses. The gut-on-a-chip is made of polydimethylsiloxane (PDMS), a transparent, elastic, and gas-permeable silicone material, and has 2 superposed microchannels separated by a porous, flexible, extracellular matrix-coated PDMS membrane (Fig. 1A). The human intestinal epithelium can be cultivated on the porous membrane in the upper microchannel, forming an intact intestinal epithelial barrier and thereby recreating the luminal microenvironment of the gut. On the other side of the membrane, either lymphatic or capillary endothelium can be grown, thus creating a counter microenvironment in the lower microchannel that represents the lymphatic or capillary vasculature. Epithelial cells in the gut-on-a-chip can form a 3D microstructure (72) reminiscent of in vivo intestinal villi, where gut microbiome and immune cells can be co-cultured in the upper and lower channels, respectively, to recreate the EMI axis on-chip (12,13) (Fig. 1B). Optionally, it is possible to grow anaerobic gut bacteria once after the AOI condition is established and stabilized (60). Finally, PBMCs as immune elements can be introduced into the lower microchannel to emulate the recruitment of immune cells from the nearby capillary vasculature (12,13). Recent studies demonstrated the introduction of tissue-specific differentiated immune cells such as DCs or macrophages (14), which may lead to more physiological biomimicry. However, as previously mentioned, the preparation of the various immune elements for the inflammatory modeling of the human intestine has been nascent, where aggressive collaborations between biomedical engineers and immunologists have been appreciated.
Demonstration of immune-mediated inflammatory responses
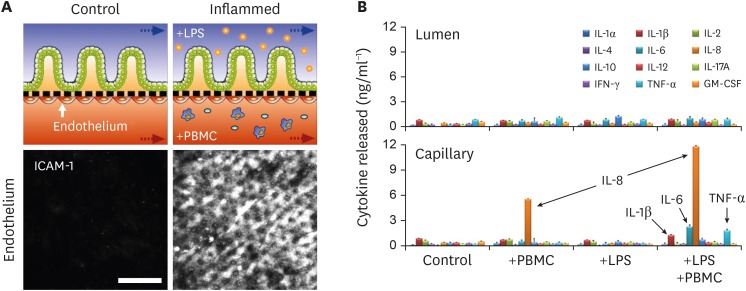
Notably, the microengineered models of the human intestinal EMI axis enable us to recapitulate the pathophysiology of immune-associated inflammatory responses by leveraging the modularity and accessbility (12,13,60,66). For example, a different strain of bacteria or a bacterial LPS was introduced to the luminal upper microchannel lined by an intact epithelium (50) to interrogate how the specific luminal perturbation provokes the epithelial damage and inflammatory reactions. LPS produced by gram-negative bacteria is a potent ligand of the TLR4 (50). An excessive amount of LPS (15 µg/mL) introduced to the gut-on-a-chip that emulates an outgrowth or infection of Gram-negative bacteria resulted in the disruption of the villous epithelial morphology as well as the barrier function (12). Interestingly, in vitro immune responses occurred when the microbial and immune cells co-stimulated the epithelial layer, by which the recruitment of PBMCs, activation of intercellular adhesion molecule 1 molecules on the endothelial surface (Fig. 2A), disruption of the villous microarchitecture, expression of TLR4 on the villous epithelium, and the directional secretion of pro-inflammatory cytokines (Fig. 2B). On the contrary, an independent stimulation of each cell type in the EMI axis did not show any inflammatory responses (12).
Figure 2. Demonstration of the pathophysiological immune responses in the gut-on-a-chip challenged to LPS and PBMCs. (A) The schematics in the upper layer show the experimental design of the healthy (control) versus stimulated microenvironment (inflamed) of the intestinal tissue interface composed of the intestinal villus epithelium and vascular endothelium. In this setup, inflammatory responses are induced by introducing LPS and PBMC at the upper and the lower microchannels, respectively, for 48 h. Confocal micrographs confirm the strong activation of ICAM-1 on the apical surface of the capillary endothelium when the EMI axis is perturbed and inflamed. Bar, 50 μm.(B) The basolateral secretion of proinflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) illustrates the mucosal inflammation in response to the co-stimulation of LPS or E. coli cells and PBMCs. It is noted that a directional release of the proinflammatory cytokines demonstrates possible immune cell recruitment from the lamina propria in vitro. Data were reorganized from the references (12).
ICAM-1, intercellular adhesion molecule 1.
The pathomimetic gut inflammation-on-a-chip
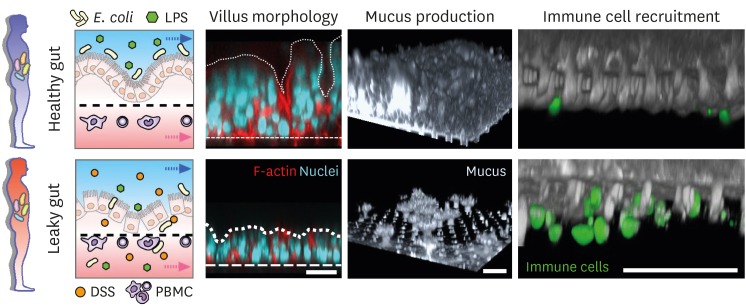
Since gut inflammation involves multifactorial crosstalk followed by simultaneous pathological outcomes, it is challenging to unravel the onset trigger of the complex inflammation process. Indeed, it is noted that experimental animal models are impossible to independently uncouple the inflammatory factors in the EMI axis and identify a specific inflammatory trigger that develops the disease cascade. To surmount this caveat, MPS models may be a compelling alternative to dissect the mechanistic contribution of individual inflammatory factors in various combinations. The “gut-inflammation-on-a-chip” for instance allowed us to match the cells in the EMI axis one-at-a-time and assess inflammatory readouts to identify which factor is a critical trigger that initiates the entire inflammatory cascade (13). Briefly, an intact intestinal epithelial barrier is necessary and sufficient to sustain the physiological homeostatic tolerance in the co-presence of both luminal (e.g., LPS or gut bacteria) and immune elements (e.g., PBMCs) (13). Under the complex intercellular crosstalk in the EMI axis, spatiotemporal inflammatory responses such as oxidative stress, inflammatory epithelial injury, secretion of inflammatory cytokine, immune cell recruitment or infiltration, and microbial vascular invasion were observed during the dysregulated epithelial barrier, suggesting that the maintenance of a good barrier function is pertinent to the intestinal homeostasis (Fig. 3).
Figure 3. A demonstration of the inflammatory crosstalk and experimental results in patients with healthy versus leaky gut. In the EMI axis, an intact epithelial layer (healthy gut) provides 3D villous morphology, high mucus production, and limited immune cell recruitment that maintains the “homeostatic tolerance” in response to the continuous exposure to the luminal (e.g., E. coli, LPS) and submucosal stimulation (e.g., PBMCs). On the contrary, a compromised epithelial barrier due to the external stimulation (here, DSS treatment) results in refractory villous destruction, loss of mucus, and substantial recruitment of activated immune cells (leaky gut). Arrows in the left schematic show the direction of fluid flow in each microchannel. Dotted and dashed lines in “villus morphology” pinpoint the contour of villi and the location of the basement membrane, respectively. Bar, 50 µm. Images were reprocessed from reference (13).
DSS, dextran sulfate sodium.
Other models for mimicking inflammatory responses in vitro
In a similar approach, there have been a number of MPS models that mimicked the intestinal EMI axis to build intercellular inflammatory interactions in vitro. Using a sequentially connected porous inserts (15) or a microfluidic device (14), differentiated subsets of immune cells, including tissue-specific macrophages or DCs, were added to the MPS device into either luminal or vascular compartment to induce immune responses in the intestine (14). For example, 2 microbial cells, Lactobacillus rhamnosus and Candida albicans, were co-cultured in aerobic condition on the formed intestinal epithelial layer. Under the formed EMI, probiotic effects of L. rhamnosus against C. albicans-induced tissue injury and inflammatory responses was tested. However, no mechanical deformations to emulate the bowel movement existed, which can affect bacterial growth rate and less physiological. Another MPS model combined an ex vivo tissue explant that secures the intestinal EMI axis to an engineered circuit (16). This study employed resected large intestine segments of mice and directly connected this resected “tube” to the device to maintain the original intestinal architecture and the homeostasis created by flowing the culture medium in the luminal side. In this integrative in vitro-ex vivo configuration, the differentiated intestinal epithelium, immune cells, and enteric neuronal cells were successfully co-cultured to investigate the role of the gut bacteria in activation of Th17 or Treg cells as well as the enteric nervous system (16). However, a limited longevity of the system due to the use of ex vivo tissue hampers a long-term study of the EMI crosstalk, and human intestinal EMI cannot be applied to the devised platform.
FUTURE PERSPECTIVES
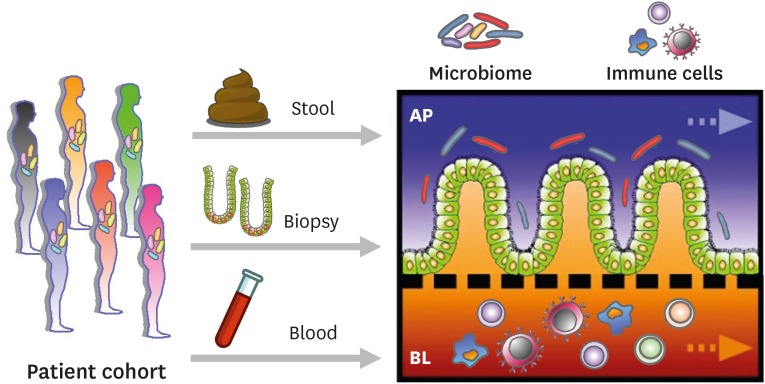
The impact of a human MPS platform has been outlined in various contexts in biomedical and clinical applications. By leveraging the modular accessibility, a simple microfluidic device can grow different types of cells for mimicking the pathophysiological immune responses of diverse organs and tissues, by which the tissue-specific immunological crosstalk can be uncoupled and recoupled in a defined space and time. By the phenomenal advances in stem cell- or tissue-derived organoid cultures, a modular MPS system allows for the collection of patient-specific organoids, immune cells, and other surrounding cells (e.g., fecal microbiome, tissue-derived mesenchymal stromal cells) (70,73) to build a better model that recapitulates a disease-specific milieu in a patient's “avatar model” (Fig. 4). There has been substantial progress in developing microfluidic chip systems modeling the physiology of various organ systems, including the heart (74), liver (75), lung (76), kidney (77), and GI tract (12,13,14,15,16).
Figure 4. Reconstitution of the patient-specific EMI axis in the gut-on-a-chip MPS. The gut-on-a-chip that conveys donor-specific primary intestinal cells from patient cohorts with different GI diseases may reconstitute the patient's intestinal microenvironment in a modular way. Epithelium, gut microbiome, and immune cells isolated from the biopsy, stool, and blood samples can be used to recreate a patient-specific EMI axis under a physiologically active microenvironment.
AP, apical; BL, basolateral.
There is a notable limitation of the 3D intestinal organoid system, which is that it prevents access to the apical side of the epithelium for studying the interactions with dietary constituents, microorganisms, and pharmaceutical compounds transported through epithelial cells (34,78). However, organoid-derived epithelial layers have been shown to grow while maintaining the accessible apical surface on MPS systems (31,32,72). This approach may allow for a more physiologic reflection of genetic variants and inter/intra-heterogeneity of human chronic diseases such as cancers (79), lead to the discovery of the independent effect of disease triggers at various immune microenvironments (13,80,81), and result in validation of the pharmacological responses of immunotherapeutic drugs in different race/ethnicity backgrounds (82,83). It may also be possible to precisely evaluate the independent contribution of immune elements on the intestinal tissue homeostasis and regeneration using an organ chip model. For instance, germ-free animals often show abnormal or non-physiological immune activations regardless of a lack of exogenous perturbations, where the hyper-reactivity in immune responses resulted in false-positive or negative outcomes (84). We envision that the application of the human organ-on-a-chip can be a discerning strategy to preclude the possible limitation of germ-free models and to concordantly understand the role of immune components in diseases.
Regardless of technological advances and progress, there are several challenges to be further delineated and discussed. A notable limitation includes the restricted resource of human immune cells, suggesting that substantial collaborations between biomedical engineers and immunologists to explore a robust protocol for the enrichment of patient-specific immune cell subtypes (e.g., induced pluripotent stem cell-derived immune cells) (85). A considerable collaboration with experts in the biobanking communities (86) and scientists who have explored liquid biopsy technologies (87) is a crucial component for securing precious and valuable bioresources. There may be an alternative avenue for immunologists to contemplate using the MPS as a comparative model to build a rare immunological disease that has been poorly established using mouse models (88). The transdisciplinary approach may contribute to bridge the gap between in vivo, ex vivo, in vitro, and in silico models and reinforce to discover the breakthrough to explore disease mechanism and test new therapeutics.
As a concluding remark, advances in human organs-on-chips and their integrations can potentially contribute to unraveling the immunological contributions in health and disease. We envision that a pertinent gut inflammation-on-a-chip model will be a cornerstone in the development of new disease models that include tissue-specific or systemic immune cells as well as the disease-specific immune milieu.
ACKNOWLEDGEMENTS
We thank Erin N. Simpson for assistance with editing this paper. This work was supported partially by Bio & Medical Technology Development Program of the National Research Foundation funded by the Ministry of Science and ICT (2018M3A9H3025030 to H.J.K.), Cancer Research Institute (UTA18-000889 to H.J.K.), NIH/NCI/IMAT (1R21CA236690-01 to H.J.K.), Leona M. & Harry B. Helmsley Charitable Trust (1912-03604 to H.J.K.), Alternative Research and Development Foundation (UTA18-001198 to H.J.K.), F99/K00 Predoctoral to Postdoctoral Transition Award (1F99CA245801-01 to W.S.), Asan Foundation Biomedical Science Scholarship (to W.S.), the ACVIM Advanced Research Training Fellowship (771066 to Y.M.A.), and Burroughs Wellcome Fund Collaborative Research Travel Grant (BWF1019990.01 to Y.M.A.).
Abbreviations
- 3D
three-dimensional
- AOI
anoxic-oxic interface
- DC
dendritic cell
- EMI
epithelial-microbiome-immune
- GI
gastrointestinal
- HuMiX
human-microbial crosstalk
- MPS
microphysiological system
- PDMS
polydimethylsiloxane
- SCFA
short-chain fatty acid
Footnotes
Conflicts of Interest: H.J.K. is a founder of 3D Health Solutions Inc. and holds an equity interest in the company.
- Data curation: Shin W, Kim HJ.
- Funding acquisition: Ambrosini YM, Shin W, Kim HJ.
- Project administration: Kim HJ.
- Supervision: Kim HJ.
- Writing - original draft: Ambrosini YM, Shin W.
- Writing - review & editing: Ambrosini YM, Shin W, Min S, Kim HJ.
References
- 1.Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, Picu A, Petcu L, Chifiriuc MC. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magro DO, Santos A, Guadagnini D, de Godoy FM, Silva SH, Lemos WJ, Vitulo N, Torriani S, Pinheiro LV, Martinez CA, et al. Remission in Crohn's disease is accompanied by alterations in the gut microbiota and mucins production. Sci Rep. 2019;9:13263. doi: 10.1038/s41598-019-49893-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow SA, Schröder H. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:1–9. doi: 10.1155/2018/4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basson AR, Lam M, Cominelli F. Complementary and alternative medicine strategies for therapeutic gut microbiota modulation in inflammatory bowel disease and their next-generation approaches. Gastroenterol Clin North Am. 2017;46:689–729. doi: 10.1016/j.gtc.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Javanmard A, Ashtari S, Sabet B, Davoodi SH, Rostami-Nejad M, Esmaeil Akbari M, Niaz A, Mortazavian AM. Probiotics and their role in gastrointestinal cancers prevention and treatment; an overview. Gastroenterol Hepatol Bed Bench. 2018;11:284–295. [PMC free article] [PubMed] [Google Scholar]
- 6.Bruno G, Gagliardi A, Oliva A, Trancassini M, Macone A, Cicerone C, D'Abramo A, Iebba V, Auria S, Bonfiglio G, et al. Fecal Microbial Transplantation impact on gut microbiota composition and metabolome, microbial translocation and T-lymphocyte immune activation in recurrent Clostridium difficile infection patients. New Microbiol. 2019;42:221–224. [PubMed] [Google Scholar]
- 7.Vaughn BP, Vatanen T, Allegretti JR, Bai A, Xavier RJ, Korzenik J, Gevers D, Ting A, Robson SC, Moss AC. Increased intestinal microbial diversity following fecal microbiota transplant for active Crohn's disease. Inflamm Bowel Dis. 2016;22:2182–2190. doi: 10.1097/MIB.0000000000000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012;28:63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014;22:267–274. doi: 10.1016/j.tim.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Park GS, Park MH, Shin W, Zhao C, Sheikh S, Oh SJ, Kim HJ. Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro . Stem Cell Rev Rep. 2017;13:321–334. doi: 10.1007/s12015-017-9739-z. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin W, Kim HJ. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci U S A. 2018;115:E10539–E10547. doi: 10.1073/pnas.1810819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurer M, Gresnigt MS, Last A, Wollny T, Berlinghof F, Pospich R, Cseresnyes Z, Medyukhina A, Graf K, Gröger M, et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials. 2019;220:119396. doi: 10.1016/j.biomaterials.2019.119396. [DOI] [PubMed] [Google Scholar]
- 15.Ramadan Q, Jafarpoorchekab H, Huang C, Silacci P, Carrara S, Koklü G, Ghaye J, Ramsden J, Ruffert C, Vergeres G, et al. NutriChip: nutrition analysis meets microfluidics. Lab Chip. 2013;13:196–203. doi: 10.1039/c2lc40845g. [DOI] [PubMed] [Google Scholar]
- 16.Yissachar N, Zhou Y, Ung L, Lai NY, Mohan JF, Ehrlicher A, Weitz DA, Kasper DL, Chiu IM, Mathis D, et al. An Intestinal organ culture system uncovers a role for the nervous system in microbe-immune crosstalk. Cell. 2017;168:1135–1148.e12. doi: 10.1016/j.cell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose M, Mukherjee P. Role of microbiome in modulating immune responses in cancer. Mediators Inflamm. 2019;2019:4107917. doi: 10.1155/2019/4107917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 21.Schneider C, O'Leary CE, Locksley RM. Regulation of immune responses by tuft cells. Nat Rev Immunol. 2019;19:584–593. doi: 10.1038/s41577-019-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillon A, Lo DD. M cells: intelligent engineering of mucosal immune surveillance. Front Immunol. 2019;10:1499. doi: 10.3389/fimmu.2019.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol. 2005;21:1–26. doi: 10.1007/s10565-005-0085-6. [DOI] [PubMed] [Google Scholar]
- 24.Phillips TE, Huet C, Bilbo PR, Podolsky DK, Louvard D, Neutra MR. Human intestinal goblet cells in monolayer culture: characterization of a mucus-secreting subclone derived from the HT29 colon adenocarcinoma cell line. Gastroenterology. 1988;94:1390–1403. doi: 10.1016/0016-5085(88)90678-6. [DOI] [PubMed] [Google Scholar]
- 25.Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res. 2015;13:11–18. doi: 10.5217/ir.2015.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JE, Jensen PJ, Johnson KR, Wheelock MJ. E-cadherin mediates adherens junction organization through protein kinase C. J Cell Sci. 1994;107:3615–3621. doi: 10.1242/jcs.107.12.3615. [DOI] [PubMed] [Google Scholar]
- 27.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scherer WF, Syverton JT, Gey GO. Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 1953;97:695–710. doi: 10.1084/jem.97.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng XH, Beyenbach KW, Quaroni A. Cultured monolayers of the dog jejunum with the structural and functional properties resembling the normal epithelium. Am J Physiol Gastrointest Liver Physiol. 2005;288:G705–G717. doi: 10.1152/ajpgi.00518.2003. [DOI] [PubMed] [Google Scholar]
- 30.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–458. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasendra M, Luc R, Yin J, Manatakis DV, Kulkarni G, Lucchesi C, Sliz J, Apostolou A, Sunuwar L, Obrigewitch J, et al. Duodenum intestine-chip for preclinical drug assessment in a human relevant model. Elife. 2020;9:e50135. doi: 10.7554/eLife.50135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Ahmad AA, Sims CE, Magness ST, Allbritton NL. In vitro generation of colonic epithelium from primary cells guided by microstructures. Lab Chip. 2014;14:1622–1631. doi: 10.1039/c3lc51353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 34.Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, Carroll I, Azcarate-Peril MA, Rawls JF, Allbritton NL, et al. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell Mol Gastroenterol Hepatol. 2018;6:301–319. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelsall BL, Leon F. Involvement of intestinal dendritic cells in oral tolerance, immunity to pathogens, and inflammatory bowel disease. Immunol Rev. 2005;206:132–148. doi: 10.1111/j.0105-2896.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 36.Stephen-Victor E, Chatila TA. Regulation of oral immune tolerance by the microbiome in food allergy. Curr Opin Immunol. 2019;60:141–147. doi: 10.1016/j.coi.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alpan O. Oral tolerance and gut-oriented immune response to dietary proteins. Curr Allergy Asthma Rep. 2001;1:572–577. doi: 10.1007/s11882-001-0067-6. [DOI] [PubMed] [Google Scholar]
- 38.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung C, Hugot JP, Barreau F. Peyer's patches: the immune sensors of the intestine. Int J Inflamm. 2010;2010:823710. doi: 10.4061/2010/823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Kaer L, Olivares-Villagómez D. Development, homeostasis, and functions of intestinal intraepithelial lymphocytes. J Immunol. 2018;200:2235–2244. doi: 10.4049/jimmunol.1701704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geremia A, Arancibia-Cárcamo CV. Innate lymphoid cells in intestinal inflammation. Front Immunol. 2017;8:1296. doi: 10.3389/fimmu.2017.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, Mucida D. Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science. 2016;352:1581–1586. doi: 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stagg AJ. Intestinal dendritic cells in health and gut inflammation. Front Immunol. 2018;9:2883. doi: 10.3389/fimmu.2018.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bain CC, Schridde A. Origin, differentiation, and function of intestinal macrophages. Front Immunol. 2018;9:2733. doi: 10.3389/fimmu.2018.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Wu J, Wang J, Zhang W, Xu B, Xu X, Zong L. Targeted delivery of antigen to intestinal dendritic cells induces oral tolerance and prevents autoimmune diabetes in NOD mice. Diabetologia. 2018;61:1384–1396. doi: 10.1007/s00125-018-4593-3. [DOI] [PubMed] [Google Scholar]
- 46.Regner EH, Ohri N, Stahly A, Gerich ME, Fennimore BP, Ir D, Jubair WK, Görg C, Siebert J, Robertson CE, et al. Functional intraepithelial lymphocyte changes in inflammatory bowel disease and spondyloarthritis have disease specific correlations with intestinal microbiota. Arthritis Res Ther. 2018;20:149. doi: 10.1186/s13075-018-1639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 48.Brandtzaeg P. Function of mucosa-associated lymphoid tissue in antibody formation. Immunol Invest. 2010;39:303–355. doi: 10.3109/08820131003680369. [DOI] [PubMed] [Google Scholar]
- 49.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.d'Hennezel E, Abubucker S, Murphy LO, Cullen TW. Total lipopolysaccharide from the human gut microbiome silences toll-like receptor signaling. mSystems. 2017;2:e00046-17. doi: 10.1128/mSystems.00046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter NT, Martens EC. The critical roles of polysaccharides in gut microbial ecology and physiology. Annu Rev Microbiol. 2017;71:349–369. doi: 10.1146/annurev-micro-102215-095316. [DOI] [PubMed] [Google Scholar]
- 52.Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brehm CU, Pfefferle PI. Inulin, the gut microbiome and the deeper breath of asthma patients - novel pathways in asthma treatment. EBioMedicine. 2019;46:15–16. doi: 10.1016/j.ebiom.2019.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao B, Gu J, Li D, Cui S, Zhao J, Zhang H, Chen W. Effects of different doses of fructooligosaccharides (FOS) on the composition of mice fecal microbiota, especially the bifidobacterium composition. Nutrients. 2018;10:1105. doi: 10.3390/nu10081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng W, Lu J, Lin W, Wei X, Li H, Zhao X, Jiang A, Yuan J. Effects of a galacto-oligosaccharide-rich diet on fecal microbiota and metabolite profiles in mice. Food Funct. 2018;9:1612–1620. doi: 10.1039/c7fo01720k. [DOI] [PubMed] [Google Scholar]
- 56.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 57.Okumura R, Takeda K. Maintenance of intestinal homeostasis by mucosal barriers. Inflamm Regen. 2018;38:5. doi: 10.1186/s41232-018-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh CJ, Guinane CM, O'Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett. 2014;588:4120–4130. doi: 10.1016/j.febslet.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 59.Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med. 2013;55:130–140. doi: 10.1016/j.freeradbiomed.2012.10.554. [DOI] [PubMed] [Google Scholar]
- 60.Shin W, Wu A, Massidda MW, Foster C, Thomas N, Lee DW, Koh H, Ju Y, Kim J, Kim HJ. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front Bioeng Biotechnol. 2019;7:13. doi: 10.3389/fbioe.2019.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell. 2016;164:1105–1109. doi: 10.1016/j.cell.2016.02.049. [DOI] [PubMed] [Google Scholar]
- 62.Lock JY, Carlson TL, Carrier RL. Mucus models to evaluate the diffusion of drugs and particles. Adv Drug Deliv Rev. 2018;124:34–49. doi: 10.1016/j.addr.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Worthington JJ, Reimann F, Gribble FM. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2018;11:3–20. doi: 10.1038/mi.2017.73. [DOI] [PubMed] [Google Scholar]
- 65.Shin W, Kim HJ. Pathomimetic modeling of human intestinal diseases and underlying host-gut microbiome interactions in a gut-on-a-chip. Methods Cell Biol. 2018;146:135–148. doi: 10.1016/bs.mcb.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 66.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 67.Basson MD, Wang Q, Chaturvedi LS, More S, Vomhof-DeKrey EE, Al-Marsoummi S, Sun K, Kuhn LA, Kovalenko P, Kiupel M. Schlafen 12 interaction with SerpinB12 and deubiquitylases drives human enterocyte differentiation. Cell Physiol Biochem. 2018;48:1274–1290. doi: 10.1159/000492019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 69.Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol. 2018;5:659–668. doi: 10.1016/j.jcmgh.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin W, Hinojosa CD, Ingber DE, Kim HJ. Human intestinal morphogenesis controlled by transepithelial morphogen gradient and flow-dependent physical cues in a microengineered gut-on-a-chip. iScience. 2019;15:391–406. doi: 10.1016/j.isci.2019.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt L, Mellado-Gomez E, Attar M, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–386.e17. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grosberg A, Alford PW, McCain ML, Parker KK. Ensembles of engineered cardiac tissues for physiological and pharmacological study: heart on a chip. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma LD, Wang YT, Wang JR, Wu JL, Meng XS, Hu P, Mu X, Liang QL, Luo GA. Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip. 2018;18:2547–2562. doi: 10.1039/c8lc00333e. [DOI] [PubMed] [Google Scholar]
- 76.Bovard D, Sandoz A, Luettich K, Frentzel S, Iskandar A, Marescotti D, Trivedi K, Guedj E, Dutertre Q, Peitsch MC, et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip. 2018;18:3814–3829. doi: 10.1039/c8lc01029c. [DOI] [PubMed] [Google Scholar]
- 77.Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY, Ingber DE. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol. 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- 78.Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8:352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Miao Z, Yang Q, Wang Y, Zhang J. The dynamic roles of mesenchymal stem cells in colon cancer. Can J Gastroenterol Hepatol. 2018;2018:7628763. doi: 10.1155/2018/7628763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sunuwar L, Yin J, Kasendra M, Karalis K, Kaper J, Fleckenstein J, Donowitz M. Mechanical stimuli affect Escherichia coli heat-stable enterotoxin-cyclic GMP signaling in a human enteroid intestine-chip model. Infect Immun. 2019;88:e00866-19. doi: 10.1128/IAI.00866-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3:e00159-12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shum E, Su C, Zhu C, Gucalp RA, Haigentz M, Packer SH, Baker C, Eng Y, Ravera E, Browne R, et al. PD-L1 expression profile and immunotherapy (IO) experience in African American (AA) and Hispanic (H) lung cancer (LC) patients: Addressing disparities at a minority-based academic cancer center. J Clin Oncol. 2017;35(15_suppl):e18073 [Google Scholar]
- 83.Sontheimer-Phelps A, Chou DB, Tovaglieri A, Ferrante TC, Duckworth T, Fadel C, Frismantas V, Sutherland AD, Jalili-Firoozinezhad S, Kasendra M, et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol Gastroenterol Hepatol. 2019;9:507–526. doi: 10.1016/j.jcmgh.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Packey CD, Shanahan MT, Manick S, Bower MA, Ellermann M, Tonkonogy SL, Carroll IM, Sartor RB. Molecular detection of bacterial contamination in gnotobiotic rodent units. Gut Microbes. 2013;4:361–370. doi: 10.4161/gmic.25824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bernareggi D, Pouyanfard S, Kaufman DS. Development of innate immune cells from human pluripotent stem cells. Exp Hematol. 2019;71:13–23. doi: 10.1016/j.exphem.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barbazan J, Dunkel Y, Li H, Nitsche U, Janssen KP, Messer K, Ghosh P. Prognostic impact of modulators of G proteins in circulating tumor cells from patients with metastatic colorectal cancer. Sci Rep. 2016;6:22112. doi: 10.1038/srep22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reimers N, Pantel K. Liquid biopsy: novel technologies and clinical applications. Clin Chem Lab Med. 2019;57:312–316. doi: 10.1515/cclm-2018-0610. [DOI] [PubMed] [Google Scholar]
- 88.Wagar LE, DiFazio RM, Davis MM. Advanced model systems and tools for basic and translational human immunology. Genome Med. 2018;10:73. doi: 10.1186/s13073-018-0584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]