Abstract
The Japan Chronic Kidney Disease (CKD) Database (J-CKD-DB) is a large-scale, nation-wide registry based on electronic health record (EHR) data from participating university hospitals. Using a standardized exchangeable information storage, the J-CKD-DB succeeded to efficiently collect clinical data of CKD patients across hospitals despite their different EHR systems. CKD was defined as dipstick proteinuria ≥1+ and/or estimated glomerular filtration rate <60 mL/min/1.73 m2 base on both out- and inpatient laboratory data. As an initial analysis, we analyzed 39,121 CKD outpatients (median age was 71 years, 54.7% were men, median eGFR was 51.3 mL/min/1.73 m2) and observed that the number of patients with a CKD stage G1, G2, G3a, G3b, G4 and G5 were 1,001 (2.6%), 2,612 (6.7%), 23,333 (59.6%), 8,357 (21.4%), 2,710 (6.9%) and 1,108 (2.8%), respectively. According to the KDIGO risk classification, there were 30.1% and 25.5% of male and female patients with CKD at very high-risk, respectively. As the information from every clinical encounter from those participating hospitals will be continuously updated with an anonymized patient ID, the J-CKD-DB will be a dynamic registry of Japanese CKD patients by expanding and linking with other existing databases and a platform for a number of cross-sectional and prospective analyses to answer important clinical questions in CKD care.
Subject terms: Chronic kidney disease, Epidemiology
Introduction
Chronic kidney disease (CKD) is not only a precursor of end-stage renal disease (ESRD) but also a strong risk factor for various adverse outcomes such as cardiovascular disease (CVD) and dementia1–3. It is estimated that 10 to 12% (over 10 million people) of Japanese adults have CKD4–6, and there is a concern that the prevalence of CKD will be even higher in the future due to aging population. Therefore, it is urgently necessary to understand the patterns of CKD progression and investigate how to optimize preventive and therapeutic strategies through epidemiological and clinical research. To collect clinical data from CKD patients in Japan and address important clinical questions, the Committee for the Working Group for Renal Biopsy Database in the Japanese Society of Nephrology (JSN) established a nationwide, web-based, and prospective registry system in 2007. This led to the development of two registries — one with patients who underwent renal biopsy, called the Japan Renal Biopsy Registry (J-RBR)7, and the other with those who did not undergo renal biopsy, called the Japan Kidney Disease Registry (J-KDR)8. These registries have contributed to better understanding the epidemiology of biopsied and unbiopsied renal disease in Japan; however, these registries rely on manual online data entry and thus have a few important limitations with regard to scaling up (i.e., number of patients and types of variables) and data accuracy.
Recently, the application of Information and Communication Technology (ICT) to the medical field has established a foundation for big data analysis informing clinical management9,10. In medical institutions, enormous amount of electronic health record (EHR) data are accumulated daily11,12. However, there are several EHR systems that may not be able to communicate each other, precluding multicentre EHR-based research. In this context, the Ministry of Health, Labor and Welfare has developed a system, the Standardized Structured Medical Information eXchange (SS-MIX2)(http://www.ss-mix.org/consE/)13, which allows us to compile EHR data from different systems. Utilizing SS-MIX2, the JSN and the Japan Association for Medical Informatics have constructed a comprehensive clinical database of CKD patients called the Japan Chronic Kidney Disease Database (J-CKD-DB) through the collaboration with 21 university hospitals nationwide as of January 2018. The main aim of the J-CKD-DB registry is to develop a large-scale, nation-wide registry based on EHR data from the participating university hospitals to conduct investigations of the actual practice pattern of CKD in Japan, including cross-sectional and prospective studies. Here, we described the process of establishing J-CKD-DB, summarized characteristics of initial 39,121 CKD outpatients in J-CKD-DB, and discussed major research questions to be addressed in J-CKD-DB.
Results
Development of J-CKD-DB
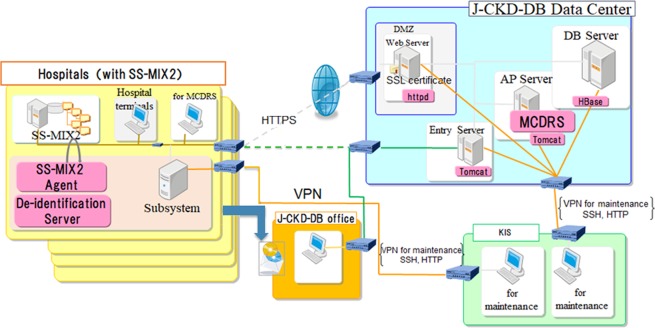
Figure 1 shows the overview of the J-CKD-DB system. The J-CKD-DB is a nationwide multicentre EHR-based database of CKD in Japan. The inclusion criteria of the J-CKD-DB were as follows: (1) age ≥ 18 years old and (2) proteinuria ≥ 1 + (dipstick test) and/or estimated glomerular filtration rate (eGFR) <60 mL/min/1.73 m2. All patients meeting these inclusion criteria based on outpatient or inpatient information were registered in the J-CKD-DB regardless of whether patients were under the care of nephrology or other specialties.
Figure 1.
Overview of the Japan Chronic Kidney Disease Database (J-CKD-DB) System. The SS-MIX2 (Standardized Structured Medical Information eXchange) leveraged recent progress made in healthcare information standards in Japan, including code standardization regarding laboratory data items and prescription data. The university hospitals participating in J-CKD-DB (left boxes) needed to have electronic health record systems that incorporated SS-MIX2 storage and a template-based structured-data entry function that could transfer the entered data to the SS-MIX2 storage. All data elements are extracted semi-automatically using SS-MIX2 storage and send to J-CKD-DB data centre through HTTPS (upper right box). MCDRS (Multi-purpose Clinical Data Repository System), a software system developed at the University of Tokyo, is adopted for designing and collecting the data elements. The administrative office of J-CKD-DB project is in Kawasaki Medical School (lower middle box). J-CKD-DB is maintained, and data cleaning is carried out at the office (lower right box). AP, application; DB, database; DMZ, demilitarized zone; HTTPS, hypertext transfer protocol secure; SSH, secure shell; SSL, secure sockets layer; VPN, virtual private network.
Among more than 80 university hospitals in Japan, 21 agreed to join the J-CKD-DB Registry (Supplementary Table S1). Table 1 summarizes information collected from the J-CKD-DB. The Multipurpose Clinical Data Registry System (MCDRS) for data extraction and registry has been developed through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program) by the Japan Society for Promotion of Science; this system allows the efficient collection of clinical data using the SS-MIX2 format11, which is application-independent, and current use includes community healthcare information systems, backup for disaster, multi-institutional database development, and clinical research.
Table 1.
Measures assessed at the baseline and during follow-up in the J-CKD-DB.
| Data elements | Description/unit | ||
|---|---|---|---|
| 1 | Hospital code | Issued by J-CKD-DB administration office | 4 digit code |
| 2 | Birth year & month | YYYYMM | |
| 3 | Gender | M: male, F: female | |
| 4 | Clinical department | 3 digit code | |
| 5 | Exceptional cases | 1: haemodialysis, 2: peritoneal dialysis, 3: renal transplantation, 99: others | |
| 6 | Start date of treatment | YYYYMMDD | |
| 7 | Patient | I: inpatient, O: outpatient | |
| 8 | Outcome | Repeat as necessary | 01: discharged, 05: referred to other hospitals, 20: deceased |
| 9 | Date of admission | Repeat as necessary | YYYYMMDD |
| 10 | Date of discharge | Repeat as necessary | YYYYMMDD |
| 11 | Date of laboratory test | Repeat as necessary | YYYYMMDD |
| 12 | Serum creatinine level |
Repeat as necessary -Laboratory test code (JLAC10 code) -Laboratory test value -Laboratory test unit -Date -Inpatient/outpatient |
mg/dL |
| 13 | Protein urine (qualitative) | -, ±, +, 2 + , 3 + , 4+ | |
| 14 | Urinary occult blood (qualitative) | -, ±, +, 2 + , 3 + , gross | |
| 15 | urine protein-to-creatinine ratio | g/gCre | |
| 16 | Protein urine (per day) | g/day | |
| 17 | Protein urine (quantitative) | mg/dL | |
| 18 | Urine protein-to-creatinine ratio | mg/gCre | |
| 19 | Urinary albumin (per day) | mg/day | |
| 20 | Urine creatinine (random urine) | mg/dL | |
| 21 | Urine creatinine (pooled urine) | mg/dL | |
| 22 | Urinary sodium (random urine) | mEq/L | |
| 23 | Urinary sodium (pooled urine) | mEq/L | |
| 24 | Urea nitrogen (random urine) | mg/dL | |
| 25 | Urea nitrogen (pooled urine) | mg/dL | |
| 26 | Urine Volume | mL/day | |
| 27 | Total serum protein | g/dL | |
| 28 | Serum albumin | g/dL | |
| 29 | Uric acid | mg/dL | |
| 30 | Urea-nitrogen | mg/dL | |
| 31 | Serum sodium |
Repeat as necessary -Laboratory test code (JLAC10 code) -laboratory test value -laboratory test unit -date -inpatient/outpatient |
mEq/L |
| 32 | Serum potassium | mEq/L | |
| 33 | Serum chloride | mEq/L | |
| 34 | Serum magnesium | mg/dL | |
| 35 | Total cholesterol | mg/dL | |
| 36 | HDL cholesterol | mg/dL | |
| 37 | LDL cholesterol | mg/dL | |
| 38 | Triglyceride | mg/dL | |
| 39 | HbA1c | % | |
| 40 | Glycated albumin | % | |
| 41 | Cystatin C | mg/L | |
| 42 | Serum calcium | mg/dL | |
| 43 | Serum phosphate | mg/dL | |
| 44 | Intact PTH | pg/mL | |
| 45 | Whole PTH | pg/mL | |
| 46 | Serum iron | μg/dL | |
| 47 | TIBC | μg/dL | |
| 48 | TSAT | % | |
| 49 | Ferritin | ng/mL | |
| 50 | Bicarbonate concentration | mEq/L | |
| 51 | BNP | pg/mL | |
| 52 | CRP | mg/dL | |
| 53 | WBC | /μL | |
| 54 | RBC | /μL | |
| 55 | Haemoglobin | g/dL | |
| 56 | Haematocrit | % | |
| 57 | PLT | /μL | |
| 58 | AST | U/L | |
| 59 | ALT | U/L | |
| 60 | Antinuclear antibody-FA | times | |
| 61 | Antinuclear antibody-EIA | ||
| 62 | MPO-ANCA(EIA) | U/mL | |
| 63 | MPO-ANCA (CLEIA) | times | |
| 64 | PR3-ANCA (EIA) | U/mL | |
| 65 | PR3-ANCA (CLEIA) | U/mL | |
| 66 | anti-GBM antibody (EIA) | U/mL | |
| 67 | anti-GBM antibody (CLEIA) | U/mL | |
| 68 | serum complement titer (CH50) | U/mL | |
| 69 | Prescriptions |
Repeat as necessary -Date -Pharmaceutical product code -Dosage -Rout of administration -Duration of administration |
Repeat for each pharmaceutical product |
| 70 | Diagnosis | Use ICD10 compliant Standard Master |
HDL, high-density lipoprotein; LDL, low-density lipoprotein; HbA1c, glycated haemoglobin; PTH, parathyroid hormone; TIBC, Total iron binding capacity; TSAT, Transferrin saturation; BNP, Brain natriuretic peptide; CRP, C-reactive protein; WBC, white blood cell count; RBC, red blood cell count; PLT, platelet count; AST, aspartate aminotransferase; MPO-ANCA, myeloperoxidase-antineutrophil cytoplasmic antibody; PR3-ANCA, Proteinase 3 antineutrophil cytoplasmic antibody; GBM, glomerular basement membrane; JLAC10, Japan Laboratory Code Version 10; ICD10, International Classifications of Disease, 10th revision.
Participant selection and baseline characteristics
Supplementary Fig. 1 shows the selection flowchart for an initial analysis which is focused on CKD outpatients. As of January 2018, over 100,000 CKD patients from 11 university hospitals (Phase 1 Database-Building hospitals) were registered in the database from 1 January 2014 to 31 December 2014. Four university hospitals had no information about admission history as of January 2018 and thus were not included. Finally, 39,121 outpatients without admission history were included from 7 university hospitals in the initial analysis.
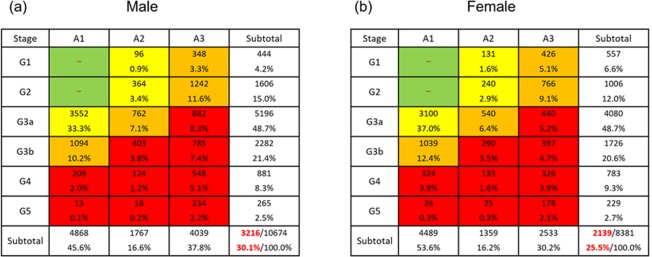
The key findings of the initial analysis are shown in Table 2. Among the 39,121 CKD outpatients (median age was 71 years, 54.7% were men, median eGFR was 51.3 mL/min/1.73 m2), we observed that the numbers of patients with CKD stage G1 (eGFR ≥ 90 mL/min/1.73 m2), G2 (60–89), G3a (45–59), G3b (30–44), G4 (15–30), and G5 ( < 15) were 1,001 (2.6%), 2,612 (6.7%), 23,333 (59.6%), 8,357 (21.4%), 2,710 (6.9%), and 1,108 (2.8%), respectively (Table 2), according to the CKD stages in the KDIGO guideline9. Among the 19,055 cases (48.7%) with available proteinuria data, the number of patients with CKD stage A1 (negative proteinuria), A2 (trace proteinuria [±]), and A3 (dipstick proteinuria ≥1+) were 9,357 (49.1%), 3,126 (16.4%), and 6,572 (34.5%), respectively (Table 2).
Table 2.
General characteristics of CKD outpatients in the J-CKD-DB at baseline.
| N | 39,121 |
| Age [IQI] (years) | 71 [62–79] |
| Age category | |
| 18–45 years | 2,424 (6.2%) |
| 45–64 years | 9,166 (23.4%) |
| 65–74 years | 12,303 (31.4%) |
| 75–84 years | 11,695 (9.9%) |
| ≥85 years | 3,533 (9.0%) |
| Gender: Male | 21,410 (54.7%) |
| eGFR [IQI] (mL/min/1.73 m2) | 51.3 [42.2–57.0] |
| eGFR stage | |
| G1 | 1,001 (2.6%) |
| G2 | 2,612 (6.7%) |
| G3a | 23,333 (59.6%) |
| G3b | 8,357 (21.4%) |
| G4 | 2,710 (6.9%) |
| G5 | 1,108 (2.8%) |
| Dipstick proteinuria | Overall: 19,055 |
| (−) | 9,357 (49.1%) |
| (±) | 3,126 (16.4%) |
| (1+) | 3,783 (19.9%) |
| (2+) | 1,919 (10.1%) |
| (3+) | 784 (4.1%) |
| (4+) | 86 (0.5%) |
| Haemoglobin (SD) (g/dL) | 13.2 (1.9) |
| Serum albumin (SD) (g/dL) | 4.1 (0.5) |
| Serum uric acid (SD) (mg/dL) | 5.9 (1.5) |
| Serum total cholesterol (mg/dL) | 186.4 (38.6) |
| Serum sodium (SD) (mEq/L) | 141.4 (2.8) |
| Serum potassium (SD) (mEq/L) | 4.3 (0.5) |
| Serum chloride (SD) (mEq/L) | 104.5 (3.3) |
| Serum calcium (SD) (mg/dL) | 9.1 (0.5) |
| Serum phosphate (SD) (mg/dL) | 3.4 (0.9) |
| Serum C-reactive protein [IQI] (mg/dL) | 0.10 [0.04–0.30] |
Continuous variables are described as median [inter-quartile interval, (IQI)] or mean (standard deviation, SD). Factors are described as n (%).
Clinical features of CKD outpatients in japan
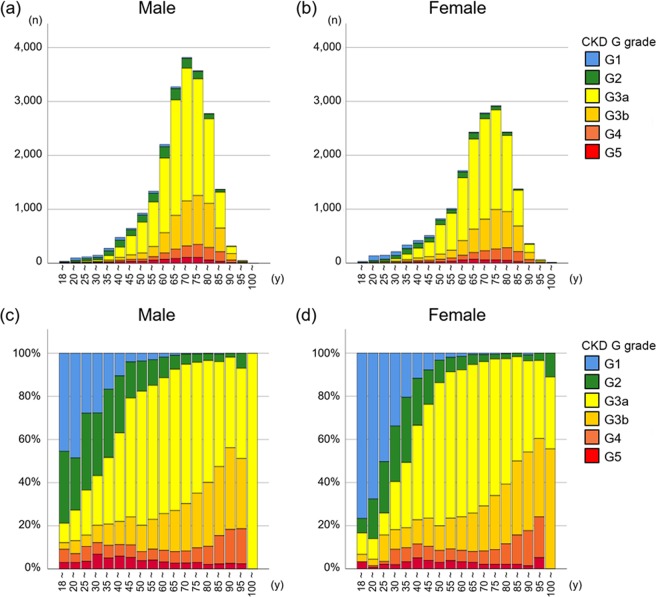
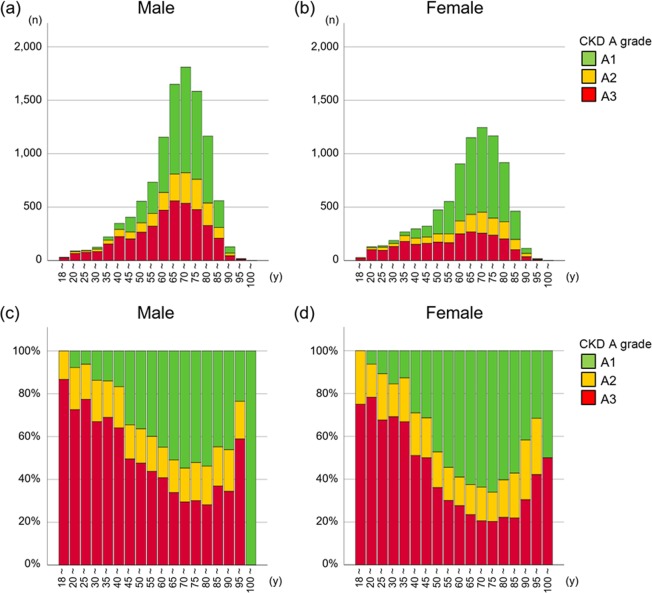
As anticipated, majority of CKD patients were older than 65 years old, with the most prevalent age category of 70–79 years in both sexes (Fig. 2a,b). The most prevalent G category after the age of 40 was G3 in both sexes. The proportion of G3b and G4 increased gradually along with age in both sexes (Fig. 2c,d, Supplementary Table S2). The proportion of CKD stage G5 was higher in younger patients (Fig. 2c,d, Supplementary Table S2). In all age groups, proteinuria was more predominant in men than in women (Fig. 3a,b, Supplementary Table S3). The proportion of those with CKD stage A3 was higher among younger patients (60%–80%) than among older ones (20%–40%) (Fig. 3c,d), suggesting that younger patients with advanced CKD are more likely to be referred to and managed in university hospitals than are older patients.
Figure 2.
Age distribution (a,b) and proportion (c,d) of CKD G stage by sex in the J-CKD-DB according to the KDIGO criteria.
Figure 3.
Age distribution (a,b) and proportion (c,d) of CKD A stage by sex in the J-CKD-DB according to the KDIGO criteria.
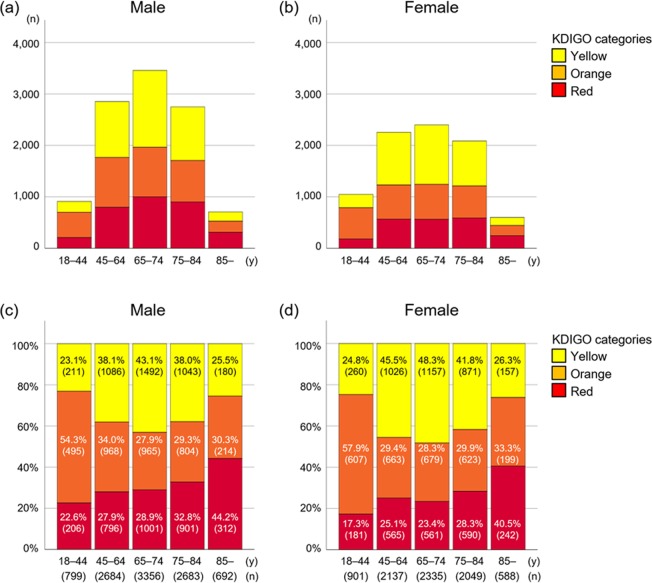
According to the KDIGO risk classification, very high-risk (red zone in Fig. 4a,b) cases accounted for 30.1% and 25.5% of all cases of male and female patients, respectively. Through all age strata, the very high-risk (red zone) cases were higher in males than in female (Fig. 5a,b). Among young patients 18–44 years, the most prevalence risk category was high-risk (orange zone), accounting for 54.3% in male and 57.9% in female (Fig. 5c,d). The prevalence of very high-risk (red zone) CKD increased gradually along with age in both sexes (i.e., 44.2% in male and 40.5% in female among patients over 85 years) (Fig. 5c,d).
Figure 4.
The KDIGO risk classification by age and sex in the J-CKD-DB. According to the KDIGO risk classification, very high-risk (red zone) cases accounted for 30.1% and 25.5% of all cases of male (a) and female (b) patients, respectively.
Figure 5.
Age distribution (a,b) and proportion (c,d) of patients by KDIGO risk categories in the J-CKD-DB. The highest proportion of very high-risk (red zone) cases in both sexes is among patients over 85 years of age (c,d).
Discussion
In the present study, we have established the largest registry of CKD patients in Japan, called the J-CKD-DB based on advanced EHR systems to automatically extract data. As an initial analysis, we analyzed 39,121 CKD outpatients and observed that majority of CKD patients were older than 65 years old, with the most prevalent age category of 70–79 years in both sexes. Younger CKD patients in J-CKD-DB tended to be at more advanced A stages than older patients.
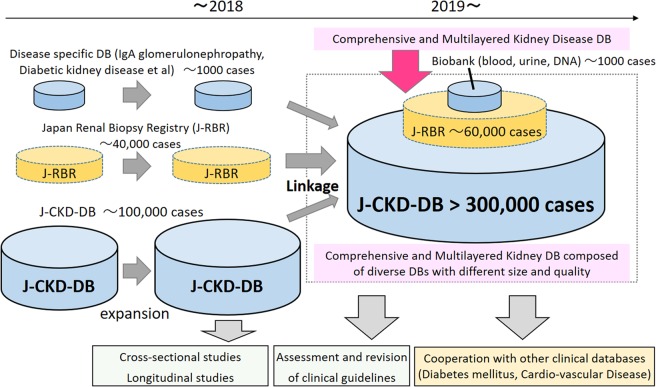
A major strength of the J-CKD-DB is the largest registry of CKD patients in Japan. Since the all data elements are extracted automatically using SS-MIX2 storage as determined from EHR data11,12, data collection and analysis will be facilitated by the J-CKD-DB in several ways. First, it is possible to build a multi-layered database by linking with an existing database, such as J-RBR/J-KDR7,8 for complementary purposes (Fig. 6). Furthermore, we are also planning to connect the J-CKD-DB to biological samples (blood, serum and urine) and genomic information after informed consent and establish a multi-layered database that will comprise the J-RBR/J-KDR at the middle layer and a biological sample database at the top layer and the J-CKD-DB at the bottom layer (Fig. 6). Although there are some systems to build up the database of CKD automatically in the world14–17, the J-CKD-DB has an advantage of expanding and linking with these multi-layer databases over the other systems. Second, it is possible to conduct various studies by publicly inviting research questions, which will provide evidence on CKD management. Third, the database can be used to analyse guideline-based quality indicators, compliance rates, and the heterogeneity of medical quality across institutions. By iterating the above processes, the J-CKD-DB can develop evidence-based strategies, informing future clinical guidelines and improving the quality of medical treatment for CKD patients.
Figure 6.
The plan for the J-CKD-DB project. The J-CKD-DB (bottom layer) will be integrated into other databases including J-RBR/J-KDR (Japan Renal Biopsy Registry and Japan Kidney Disease Registry) (middle layer). It is also planning to connect J-CKD-DB to biological samples and genomic information after informed consent (top layer) and establishing a multi-layered database that will comprise the J-RBR. Numbers are number of patients/measurements.
In the process of establishing the J-CKD-DB, we noticed a few issues to be addressed. Although laboratory and prescription data have generally been described according to the standardized code system, SS-MIX2, throughout Japan, we found variations in those coding systems across the participating hospitals11. We found it challenging to convert the local code of medical care and tests to the standardized code by referencing mapping tables. One solution is to consider local variations, which will contribute to the construction of additional clinical effect databases in the future.
The present study demonstrated that clinical features of 39,121 CKD outpatients in university hospitals in Japan. As anticipated, the prevalence of proteinuria increased as GFR decreased. Male patients in J-CKD-DB tended to have higher levels of proteinuria than female patients. Accordingly, the prevalence of CKD with very high-risk (according to the KDIGO risk classification) was higher in male than in female patients across all age strata. When we looked the proportion of CKD risk stages within each age category, the highest proportion of high-risk (orange zone) CKD was seen among younger patients aged 18–44 years, with high proportion of CKD stage G5 and A3 in both sexes. In the field of adolescent and young adult patients with childhood-onset CKD, progressive CKD due to various diseases including childhood-onset nephrotic syndrome, chronic glomerulonephritis such as IgA nephropathy, congenital malformations of the kidney and urinary tract (CAKUT), persist after patients become adults18. Especially, the median age at which CAKUT progresses to ESKD was reported to be around 35 years19. Based on these reports, younger patients with advanced CKD are more likely to be referred to and managed in university hospitals. Overall, the prevalence of proteinuria was higher in more severe stages of eGFR, although only 48.7% of CKD patients in J-CKD-DB had data on proteinuria. Further, we will present the results of cross-sectional analyses in more detail and collecting more patients to conduct longitudinal follow-up study for patients in the J-CKD-DB as a J-CKD-DB extension (J-CKD-DB-Ex).
Weaknesses of the J-CKD-DB are as follows: First, EHR data may be inferior to observational epidemiological studies in some information, e.g. the cause of CKD, body mass index, blood pressure levels, social status, patient-reported experience measures, and incidence of CVDs because these elements are not included in SS-MIX2. Hence, some research questions related to these variables were not answered. Second, eGFR values based on a single measurement of serum creatinine are prone to causing misclassification, specifically in CKD G3a without proteinuria, thus not meeting the chronicity criterion. However, this approach has been used often in CKD research20,21.
In conclusion, we launched the J-CKD-DB based on advanced EHR systems to automatically extract data literally and literally eliminate manual input. As the information from every clinical encounter will be updated with an anonymized patient ID, the J-CKD-DB will be used for both cross-sectional and prospective investigations for patients with CKD in Japan to answer important clinical research questions and contribute to the improvement in the quality of CKD care.
Materials and Methods
Study design
This was an observational retrospective study using electronic health records.
Setting and data source
The J-CKD-DB has been designed as a nationwide multicentre EHR-based database of patients with CKD in collaboration with the JSN and the JAMI. It was initiated in June 2016 as a comprehensive database of the clinical effective information of the Ministry of Health, Labor and Welfare (UMIN trial number, UMIN000026272). To ensure the smooth implementation and maintenance of the systems essential for J-CKD-DB; the facilities participating in J-CKD-DB needed to have EHR systems that incorporated SS-MIX2 storage (Fig. 1)11. The ethical committee of Kawasaki Medical School and JSN comprehensively approved the study (JSN no. 28), and a local committee of participating university hospitals individually approved the study. Because this is a retrospective study and the data analyzed were anonymized, informed consent from participants was obtained through an opt-out method on the web-site of each participating university hospitals in accordance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects in Japan. The administrative office of J-CKD-DB project is in Kawasaki Medical School and is in charge of data cleansing and maintenance.
In the future, the J-CKD-DB will be linked to other databases. Firstly, linkage to J-RBR/J-KDR7,8, which has over 40,000 participants, is planned in late 2020 (Fig. 2). Therefore, to enhance future analysis, physicians in each university hospitals have manually flagged special registrations in the database as follows: (1) haemodialysis cases, (2) peritoneal dialysis cases, (3) kidney transplantation cases, (4) cases in which kidney biopsy is being performed, and (5) cases registered in the J-RBR.
Participants
Inclusion criteria of J-CKD-DB are as follows: 1) Age ≥ 18 years old, 2) Proteinuria ≥ 1 + (dipstick test) according to the previous studies4,5 and/or estimated glomerular filtration rate (eGFR) <60 mL/min/1.73 m2 where estimated GFR is calculated using Japanese equation of eGFR: eGFR (mL/min/1.73 m2) = 194 × serum creatinine value −1.094 × age−0.287 (female × 0.739)22. The registry includes all patients with CKD (both out- and inpatients) in the facilities participating in J-CKD-DB.
Data collection
Baseline data abstraction and compilation were done from 1 January 2014 to 31 December 2014. To avoid input error and burden on physicians, all data elements are extracted automatically using SS-MIX2 storage and send to J-CKD-DB data centre (Fig. 1). Fundamental standards adopted in SS-MIX2 are Health Level Seven (HL7) V2.5 (ISO 27931)23 data format for patient profile, prescriptions (HOT reference code24), laboratory test results (Japan Laboratory Code Version 10 [JLAC10] code25), diagnoses (International Classifications of Disease, 10th revision [ICD10]26), incidences of major outcomes (such as death, hospital admission, discharge and transfer to other hospitals), etc.
CKD G staging was classified according to the Kidney Disease: Improving Global Outcomes (KDIGO) guideline27, with an eGFR ≥90, 60–89, 45–59, 30–44, 15–29, and <15 mL/min/1.73 m2 classified as G1, G2, G3a, G3b, G4, and G5, respectively. Dipstick proteinuria was classified into 3 categories: KDIGO A1 category of negative proteinuria (−); A2 category of trace proteinuria (±); and A3 category of ≥1 + 3,28. In the initial analysis, the patients with eGFR <5 ml/min/1.73m2 were excluded to eliminate the possibility of including patients with ESRD, because we could not distinguish those on hemodialysis, peritoneal dialysis, or those after kidney transplantation as of conducting the initial analysis. Furthermore, patients with eGFR ≥ 200 mL/min/1.73 m2 were excluded as clinically implausible29. Patients with eGFR ≥ 60 mL/min/1.73 m2 without trace proteinuria who did not meet the definition of CKD were excluded.
Statistical analyses
Values are presented as median with interquartile interval (IQI), mean (SD), or count with percentage, as appropriate. Distributions of the variables were evaluated by histogram, quantile-quantile plot, and the Kolmogorov-Smirnov test. Clinical parameters were compared among the eGFR or proteinuria categories using the Kruskal–Wallis nonparametric test. All data were statistically analyzed using IBM SPSS Advanced Statistical Version 26.0 (SPSS, Chicago, IL, USA), and p < 0.05 was considered to indicate a significant difference.
Supplementary information
Acknowledgements
This work was supported in part by a grant for Development of Comprehensive Database of Chronic Kidney Disease, Research on Policy Planning and Evaluation from the Ministry of Health, Labour and Welfare of Japan (H28-ICT-ippan-002) and was partially conducted by means of the Multi-purpose Clinical Data Repository System (MCDRS, http://mcdrs.jp). In addition to the authors, the following investigators and institutions participated in this study: Asahikawa Medical University Hospital, Yuji Tani and Hiroyuki Hirokawa; Kagawa University Hospital, Hideto Yokoi; University of Tokyo Graduate School of Medicine, Tetsuhiro Tanaka and Yosuke Hirakawa; Nagoya University Hospital, Yoshimune Shiratori; Kanazawa University Hospital, Shinsuke Yamaoka and Keisuke Nagase; Kochi University Hospital, Yutaka Hatakeyama and Yoshiyasu Okuhara; University of Tsukuba Hospital, Rina Kagawa and Makoto Ohara; Niigata University Hospital, Kohei Akazawa; Kyoto University Hospital, Shusuke Hiragi and Tomohiro Kuroda; Okayama University Hospital, Hiroshi Morinaga, Hideo Gobara; Wakayama Medical University Hospital, Masayuki Irie and Toru Mima; Shimane University Hospital, Shusaku Tsumoto; Yokohama City University Hospital, Akinobu Nemoto; Osaka University Hospital, Yasushi Matsumura; Kyushu University, Hisako Yoshida. We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
N. Nakagawa, T.S., E.K., H.N., M.N., S.M., T.W., Y.T., K.Y., I.N., M.Y., H.S., T.Sh., T.I., K.T., Y.I., H.O., K.Ts., H.Y. and N. Nakashima., H.K. and M.O. performed the data collection and extraction. M.N., S.M., T.W., K.Y., H.S., H.Y., N. Nakashima., K.O., M.O. and N.K. participated in the study design. N. Nakagawa, T.S, E.K., H.K. and M.O. performed statistical analysis. N. Nakagawa, T.S., E.K., H.N., K.M., M.O. and N.K. wrote the main manuscript text and prepared Figs. 1–6. All authors reviewed the manuscript.
Data availability
The datasets used and/or analysed during the current study are not an open-access. However, we would encourage external investigators to consider applying to use the data for secondary analyses, to maximize the scientific output from the data. Further details on the J-CKD-DB, including information for joining and a full list of publications, are available from the website: http://j-ckd-db.jp/english/index.html.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64123-z.
References
- 1.Matsushita K, et al. Cohort profile: the chronic kidney disease prognosis consortium. Int. J. Epidemiol. 2013;42:1660–1668. doi: 10.1093/ije/dys173. [DOI] [PubMed] [Google Scholar]
- 2.Levin A, et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet. 2017;390:1888–1917. doi: 10.1016/S0140-6736(17)30788-2. [DOI] [PubMed] [Google Scholar]
- 3.Fukui A, Yokoo T, Nangaku M, Kashihara N. New measures against chronic kidney diseases in Japan since 2018. Clin. Exp. Nephrol. 2019;23:1263–1271. doi: 10.1007/s10157-019-01786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai E, et al. Prevalence of chronic kidney disease in the Japanese general population. Clin. Exp. Nephrol. 2009;13:621–630. doi: 10.1007/s10157-009-0199-x. [DOI] [PubMed] [Google Scholar]
- 5.Nagata M, et al. Trends in the prevalence of chronic kidney disease and its risk factors in a general Japanese population: the Hisayama Study. Nephrol. Dial. Transplant. 2010;25:2557–2564. doi: 10.1093/ndt/gfq062. [DOI] [PubMed] [Google Scholar]
- 6.Hill NR, et al. Global Prevalence of Chronic Kidney Disease - A Systematic Review and Meta-Analysis. PLoS One. 2016;11:e0158765. doi: 10.1371/journal.pone.0158765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugiyama H, et al. Japan Renal Biopsy Registry: the first nationwide, web-based, and prospective registry system of renal biopsies in Japan. Clin. Exp. Nephrol. 2011;15:493–503. doi: 10.1007/s10157-011-0430-4. [DOI] [PubMed] [Google Scholar]
- 8.Sugiyama H, et al. Japan Renal Biopsy Registry and Japan Kidney Disease Registry: Committee Report for 2009 and 2010. Clin. Exp. Nephrol. 2013;17:155–173. doi: 10.1007/s10157-012-0746-8. [DOI] [PubMed] [Google Scholar]
- 9.Drawz PE, et al. CKD as a Model for Improving Chronic Disease Care through Electronic Health Records. Clin. J. Am. Soc. Nephrol. 2015;10:1488–1499. doi: 10.2215/CJN.00940115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, et al. Learning from Longitudinal Data in Electronic Health Record and Genetic Data to Improve Cardiovascular Event Prediction. Sci. Rep. 2019;9:717. doi: 10.1038/s41598-018-36745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugiyama T, et al. Design of and rationale for the Japan Diabetes compREhensive database project based on an Advanced electronic Medical record System (J-DREAMS) Diabetology Int. 2017;8:375–382. doi: 10.1007/s13340-017-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su G, et al. Association of Kidney Function with Infections by Multidrug-Resistant Organisms: An Electronic Medical Record Analysis. Sci. Rep. 2018;8:13372. doi: 10.1038/s41598-018-31612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura M, et al. SS-MIX: a ministry project to promote standardized healthcare information exchange. Methods Inf. Med. 2011;50:131–139. doi: 10.3414/ME11-06-0005. [DOI] [PubMed] [Google Scholar]
- 14.Navaneethan SD, et al. Development and validation of an electronic health record-based chronic kidney disease registry. Clin. J. Am. Soc. Nephrol. 2011;6:40–49. doi: 10.2215/CJN.04230510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparini A, et al. Prevalence and recognition of chronic kidney disease in Stockholm healthcare. Nephrol. Dial. Transplant. 2016;31:2086–2094. doi: 10.1093/ndt/gfw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendu ML, et al. Development of an electronic health record-based chronic kidney disease registry to promote population health management. BMC Nephrol. 2019;20:72. doi: 10.1186/s12882-019-1260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bello AK, et al. Quality of Chronic Kidney Disease Management in Canadian Primary Care. JAMA Netw. Open. 2019;2:e1910704. doi: 10.1001/jamanetworkopen.2019.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubota W, et al. A consensus statement on health-care transition of patients with childhood-onset chronic kidney diseases: providing adequate medical care in adolescence and young adulthood. Clin. Exp. Nephrol. 2018;22:743–751. doi: 10.1007/s10157-018-1589-8. [DOI] [PubMed] [Google Scholar]
- 19.Wuhl E, et al. Timing and outcome of renal replacement therapy in patients with congenital malformations of the kidney and urinary tract. Clin. J. Am. Soc. Nephrol. 2013;8:67–74. doi: 10.2215/CJN.03310412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coresh J, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet. 2012;379:815–822. doi: 10.1016/S0140-6736(12)60033-6. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo S, et al. Revised equations for estimated GFR from serum creatinine in Japan. Am. J. Kidney Dis. 2009;53:982–992. doi: 10.1053/j.ajkd.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 23.Rajeev D, et al. Development of an electronic public health case report using HL7 v2.5 to meet public health needs. J. Am. Med. Inform. Assoc. 2010;17:34–41. doi: 10.1197/jamia.M3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawazoe Y, Imai T, Ohe K. A Querying Method over RDF-ized Health Level Seven v2.5 Messages Using Life Science Knowledge Resources. JMIR Med. Inf. 2016;4:e12. doi: 10.2196/medinform.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kume N, et al. Development of Unified Lab Test Result Master for Multiple Facilities. Stud. Health Technol. Inform. 2015;216:1050. [PubMed] [Google Scholar]
- 26.Hohl CM, et al. ICD-10 codes used to identify adverse drug events in administrative data: a systematic review. J. Am. Med. Inform. Assoc. 2014;21:547–557. doi: 10.1136/amiajnl-2013-002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013;3:1–150. doi: 10.1038/kisup.2012.73. [DOI] [Google Scholar]
- 28.Park JI, Baek H, Kim BR, Jung HH. Comparison of urine dipstick and albumin:creatinine ratio for chronic kidney disease screening: A population-based study. PLoS One. 2017;12:e0171106. doi: 10.1371/journal.pone.0171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohta M, Babazono T, Uchigata Y, Iwamoto Y. Comparison of the prevalence of chronic kidney disease in Japanese patients with Type 1 and Type 2 diabetes. Diabet. Med. 2010;27:1017–1023. doi: 10.1111/j.1464-5491.2010.03049.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are not an open-access. However, we would encourage external investigators to consider applying to use the data for secondary analyses, to maximize the scientific output from the data. Further details on the J-CKD-DB, including information for joining and a full list of publications, are available from the website: http://j-ckd-db.jp/english/index.html.