Abstract
Triple-negative breast cancer (TNBC) is a highly aggressive disease and of poor prognosis. It is very important to identify novel biomarkers to predict therapeutic response and outcome of TNBC. We investigated the association between polymorphisms in PARP1 gene and clinicopathological characteristics or survival of 272 patients with stage I-III primary TNBC treated with anthracycline/taxane based adjuvant chemotherapy. We found that after adjusted by age, grade, tumor size, lymph node status and vascular invasion, rs7531668 TA genotype carriers had significantly better DFS rate than TT genotype carriers, the 5 y DFS was 79.3% and 69.2% (P = 0.046, HR 0.526 95% CI 0.280–0.990). In lymph node negative subgroup, DFS of rs6664761 CC genotype carriers was much better than TT genotype carriers (P = 0.016, HR 0.261 95% CI 0.088–0.778) and DFS of rs7531668 AA genotype carriers was shorter than TT genotype carriers (P = 0.015, HR 3.361 95% CI 1.259–8.969). In subgroup of age ≤ 50, rs6664761 TC genotype predicted favorable DFS than TT genotype (P = 0.042, HR 0.405 95% CI 0.170–0.967). Polymorphisms in PARP1 gene had no influence on treatment toxicities. After multivariate analysis, tumor size (P = 0.037, HR = 2.829, 95% CI: 1.063–7.525) and lymph node status (P < 0.001, HR = 9.943, 95% CI: 2.974–33.243) were demonstrated to be independent prognostic factors. Our results suggested that polymorphisms in PARP1 gene might predict the DFS of TNBC patients treated with anthracycline/taxane based adjuvant chemotherapy.
Subject terms: Cancer therapy, Chemotherapy
Introduction
Breast cancer is one of the most common cancers and is the leading cause of cancer-related death in women around the world1. According to molecular profile, breast cancer was divided into several intrinsic subtypes: luminal-A, luminal-B, HER2-enriched, basal-like, and a normal breast-like group2. Triple negative breast cancer (TNBC) is defined as lacking expression of estrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2). It accounts for 15–20% of all breast cancers and is characterized by enhanced aggressiveness, young age of onset and poor prognosis3. All intrinsic subtypes can be found in TNBC, but 50%-75% of TNBC have basal phenotype4. Recently, four to six distinct subtypes have been defined within TNBC, such as basal-like and claudin-low5. Chemotherapy remains the main treatment for TNBC, but the overall survival for metastatic TNBC is only 13–18 months6. Though PARP inhibitors showed promising effect in BRCA mutation patients, their effectiveness in TNBC need to be further verified. So it is very important for us to explore novel biomarkers and potential therapeutic targets in TNBC patients7.
DNA damage caused by exogenous and endogenous factors plays an important role in carcinogenesis8. Multiple DNA repair pathways are vital for controlling DNA damage and maintaining genomic stability, such as base excision repair (BER) pathway9. Impaired DNA repair impacts upon carcinogenesis and response to DNA damaging radiotherapy and chemotherapeutics10. Poly[ADP-ribose] polymerase (PARP) is an abundant, highly conserved, cell signaling protein. The activation of PARP is essential for DNA single strand break (SSB) repair11, a sub-pathway related to BER. PARP-1, also known as ADPRT, is a major member of the PARP family, which is mainly responsible for the recognition of damaged bases and the recruitment of repaired proteins12.
It has been reported that PARP1 expression was correlated to clinicopathological variables and outcome of breast cancer patients13. Some investigators found that nuclear expression of PARP1 in invasive primary breast tumors is associated with chemotherapy sensitivity14. However, there is no systemic research about polymorphisms in PARP1 and prognosis of TNBC patients. In our study, we first demonstrated that polymorphisms in PARP1 gene were associated with survival of TNBC patients treated with anthracycline/taxane based adjuvant chemotherapy.
Results
Clinical characteristics and survival of TNBC patients
A cohort of 272 TNBC patients was enrolled in this study. The median age at diagnosis is 47 years old (range: 23–75). 166 (61.0%) patients were ≤ 50 years old. 203 (74.6%) patients were diagnosed with grade 3 tumors. Most patients were at stage II and III (189 patients, 69.5%). The 5-year OS and DFS rate were 86.9% and 72.2%, respectively. The survival rates for patients with different clinicopathological characteristics were listed in Table 1. Patients older than 50 years old had a significantly better 5 y DFS rate than those younger than 50 years old (79.4% vs. 68.0%, P = 0.044, HR = 0.572, 95%CI: 0.332–0.986). The Kaplan-Meier analysis revealed a significant higher DFS and OS for patients with tumor size ≤ 2 cm and negative lymph node metastasis. There was no significant association between grade, vascular invasion and TNBC survival. After multivariate analysis, tumor size (P = 0.037, HR = 2.829, 95%CI: 1.063–7.525) and lymph node status (P < 0.001, HR = 9.943, 95%CI: 2.974–33.243) were proved to be independent prognostic factors.
Table 1.
Clinicopathological characteristics and survival of TNBC.
| Variables | Patients (%) | 5yDFS(%) | HR(95%CI) | P | 5yOS(%) | HR(95%CI) | P |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| ≤50 | 166(61.0) | 68.0 | 1(Ref) | 86.9 | 1(Ref) | ||
| > 50 | 106(39.0) | 79.4 | 0.572(0.332–0.986) | 0.044 | 86.8 | 0.877(0.404–1.906) | 0.740 |
| Grade | |||||||
| 1–2 | 69(25.4) | 68.1 | 1(Ref) | 96.8 | 1(Ref) | ||
| 3 | 203(74.6) | 73.5 | 1.070(0.608–1.882) | 0.814 | 83.6 | 3.062(0.923–10.156) | 0.067 |
| Vascular invasion | |||||||
| negative | 254(93.4) | 72.9 | 1(Ref) | 87.0 | 1(Ref) | ||
| positive | 18(6.6) | 62.2 | 1.529(0.659–3.547) | 0.322 | 81.6 | 2.207(0.762–6.389) | 0.144 |
| Tumor size | |||||||
| ≤2 cm | 126(46.3) | 82.6 | 1(Ref) | 95.5 | 1(Ref) | ||
| >2 cm | 146(53.7) | 63.7 | 2.233(1.308–3.813) | 0.003 | 80.2 | 3.953(1.502–10.400) | 0.005 |
| Lymph node | |||||||
| negative | 160(58.8) | 80.0 | 1(Ref) | 98.0 | 1(Ref) | ||
| positive | 112(41.2) | 61.2 | 2.824(1.705–4.677) | <0.001 | 72.3 | 12.718(3.839–42.131) | <0.001 |
| TNM | |||||||
| I | 83(30.5) | 83.9 | 1(Ref) | 100.0 | 1(Ref) | ||
| II | 136(50.0) | 75.1 | 1.617(0.782–3.343) | 0.195 | 92.8 | 5.738(0.734–44.851) | 0.096 |
| III | 53(19.5) | 46.8 | 6.130(2.968–12.663) | <0.001 | 53.5 | 34.099(4.528–256.803) | 0.001 |
Abbreviations: DFS, disease free survival; OS, overall survival; HR, hazard ratios; CI, confidence interval; Ref, reference.
Polymorphisms in PARP1 and survival of TNBC patients
Tables 2, 3 listed the 5-year DFS and OS rate for patients with different genotypes. After adjusted by age, grade, tumor size, lymph node status and vascular invasion, rs7531668 TA genotype carriers had significantly better DFS rate than TT genotype carriers, the 5 y DFS was 79.3% and 69.2% (P = 0.046, HR 0.526 95% CI 0.280–0.990) (Fig. 1), respectively. There was no association between other polymorphisms in PARP1 and survival of TNBC patients.
Table 2.
PARP1 genotypes and disease-free survival.
| Variables | Patients (%) | 5 y DFS(%) | Crude | Adjusted | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| Rs1136410 | ||||||
| TT | 94(34.6) | 69.4 | 1(Ref) | 1(Ref) | ||
| TC | 134(49.3) | 73.3 | 0.791(0.463–1.351) | 0.391 | 0.891 (0.516–1.537) | 0.678 |
| CC | 44(16.1) | 74.2 | 0.895(0.440–1.819) | 0.759 | 0.877(0.428–1.796) | 0.719 |
| rs11801168 | ||||||
| TT | 7(2.6) | 64.3 | 1(Ref) | 1(Ref) | ||
| TC | 68(25.0) | 76.7 | 0.885(0.202–3.881) | 0.872 | 1.070(0.240–4.772) | 0.929 |
| CC | 197(72.4) | 71.4 | 0.996(0.241–4.109) | 0.995 | 1.255(0.296–5.322) | 0.758 |
| rs12568297 | ||||||
| GG | 226(83.1) | 72.4 | 1(Ref) | 1(Ref) | ||
| GC | 41(15.1) | 75.1 | 1.085(0.552–2.133) | 0.814 | 1.020(0.513–2.027) | 0.956 |
| CC | 5(1.8) | 53.3 | 1.511(0.366–6.229) | 0.568 | 1.531(0.351–6.680) | 0.571 |
| Rs6664761 | ||||||
| TT | 33(12.1) | 68.3 | 1(Ref) | 1(Ref) | ||
| TC | 119(43.8) | 74.6 | 0.537(0.261–1.102) | 0.090 | 0.564(0.272–1.171) | 0.124 |
| CC | 120(44.1) | 70.5 | 0.712(0.358–1.417) | 0.333 | 0.755(0.376–1.516) | 0.429 |
| Rs7531668 | ||||||
| TT | 163(59.9) | 69.2 | 1(Ref) | 1(Ref) | ||
| TA | 78(28.7) | 79.3 | 0.594(0.319–1.108) | 0.102 | 0.526(0.280–0.990) | 0.046 |
| AA | 31(11.4) | 69.6 | 1.335(0.669–2.662) | 0.412 | 1.190(0.588–2.406) | 0.629 |
Abbreviations: DFS, disease free survival; OS, overall survival; HR, hazard ratios; CI, confidence interval; Ref, reference.
Table 3.
PARP1 genotypes and overall survival.
| Variables | 5 y OS(%) | Crude | Adjusted | ||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| rs1136410 | |||||
| TT | 87.3 | 1(Ref) | 1(Ref) | ||
| TC | 90.5 | 0.578(0.239–1.395) | 0.222 | 0.818(0.334–2.008) | 0.662 |
| CC | 77.1 | 1.382(0.555–3.439) | 0.487 | 1.572(0.607–4.073) | 0.352 |
| rs11801168 | |||||
| TT | 85.7 | 1(Ref) | 1(Ref) | ||
| TC | 91.5 | 0.344(0.066–1.788) | 0.204 | 0.663 (0.123–3.570) | 0.632 |
| CC | 85.3 | 0.486(0.113–2.090) | 0.332 | 0.990(0.214–4.577) | 0.990 |
| Rs12568297 | |||||
| GG | 85.5 | 1(Ref) | 1(Ref) | ||
| GC | 95.0 | 0.470(0.111–1.989) | 0.305 | 0.451(0.105–1.942) | 0.285 |
| CC | 80.0 | 2.823(0.658–12.110) | 0.162 | 1.678(0.343–8.214) | 0.523 |
| Rs6664761 | |||||
| TT | 85.2 | 1(Ref) | 1(Ref) | ||
| TC | 90.4 | 0.772(0.205–2.915) | 0.703 | 1.079(0.279–4.178) | 0.912 |
| CC | 84.4 | 1.522(0.446–5.196) | 0.503 | 1.974(0.563–6.919) | 0.288 |
| Rs7531668 | |||||
| TT | 84.3 | 1(Ref) | 1(Ref) | ||
| TA | 91.4 | 0.486 (0.183–1.289) | 0.147 | 0.483(0.180–1.292) | 0.147 |
| AA | 90.4 | 0.499(0.117–2.130) | 0.348 | 0.376(0.087–1.628) | 0.191 |
Abbreviations: DFS, disease free survival; OS, overall survival; HR, hazard ratios; CI, confidence interval; Ref, reference.
Figure 1.
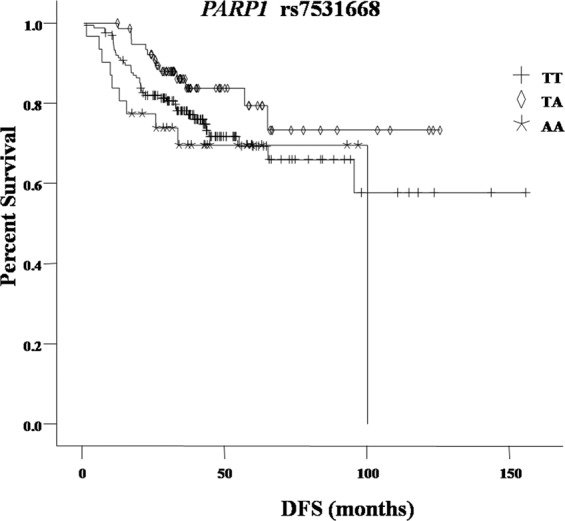
Kaplan-Meier curve of DFS for patients with different PARP1 rs7531668 genotypes.
Polymorphisms inPARP1gene and survival of TNBC in different subgroups
Since there were only 7 and 5 patients with rs11801168 TT and rs12568287 CC genotypes, we only performed subgroup analysis for rs1136410, rs6664761 and rs7531668 (Table 4). In lymph node negative subgroup, DFS of rs6664761 CC genotype carriers was much better than TT genotype carriers (P = 0.016, HR 0.261 95% CI 0.088–0.778) and DFS of rs7531668 AA genotype carriers was shorter than TT genotype carriers (P = 0.015, HR 3.361 95% CI 1.259–8.969). In subgroup of age ≤50, rs6664761 TC genotype predicted favorable DFS than TT genotype (P = 0.042, HR 0.405 95% CI 0.170–0.967). No significant relationship was observed between polymorphisms and OS in any subgroups.
Table 4.
Subgroup analysis of polymorphisms and survival.
| subgroup | variants | DFS | |
|---|---|---|---|
| HR(95%CI) | P | ||
| Lymph node negative | rs1136410 | ||
| TT | 1(Ref) | ||
| TC | 0.937(0.404–2.170) | 0.879 | |
| CC | 0.219(0.027–1.716) | 0.148 | |
| Rs6664761 | |||
| TT | 1(Ref) | ||
| TC | 0.435(0.161–1.180) | 0.102 | |
| CC | 0.261(0.088–0.778) | 0.016 | |
| Rs7531668 | |||
| TT | 1(Ref) | ||
| TA | 1.083(0.406–2.890) | 0.873 | |
| AA | 3.361(1.259–8.969) | 0.015 | |
| Age ≤ 50 | rs1136410 | ||
| TT | 1(Ref) | ||
| TC | 0.683(0.353–1.319) | 0.256 | |
| CC | 1.073(0.498–2.315) | 0.857 | |
| Rs6664761 | |||
| TT | 1(Ref) | ||
| TC | 0.405(0.170–0.967) | 0.042 | |
| CC | 0.697(0.314–1.545) | 0.374 | |
| Rs7531668 | |||
| TT | 1(Ref) | ||
| TA | 0.530(0.243–1.154) | 0.110 | |
| AA | 1.711(0.785–3.731) | 0.177 | |
Abbreviations: DFS, disease free survival; HR, hazard ratios; CI, confidence interval; Ref, reference; UK, unknown.
Polymorphisms inPARP1gene and toxicities induced by anthracycline/taxane based chemotherapy
We further investigated the relationship between different genotypes and the risk of hematological (neutropenia, thrombocytopenia and anemia) and non-hematological toxicities (nausea, vomiting and neuropathy). We analyzed the distribution of genotypes between patients with or without chemotherapy induced toxicities. The results indicated that polymorphisms in PARP1 had no influence on either hematological or non-hematological toxicities (Tables 5, 6).
Table 5.
PARP1 genotypes and hematological toxicities.
| Variables | Neutropenia | Thrombocytopenia | Anemia | |||
|---|---|---|---|---|---|---|
| Rs1136410 | P | HR (95%CI) | P | HR (95%CI) | P | HR (95%CI) |
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.635 | 0.880 (0.519–1.492) | 0.212 | 0.521 (0.187–1.451) | 0.832 | 0.895 (0.321–2.494) |
| CC | 0.550 | 0.803 (0.392–1.646) | 0.477 | 1.491 (0.496–4.487) | 0.067 | 2.762 (0.932–8.183) |
| rs11801168 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.991 | 1.009 (0.209–4.859) | 0.845 | 0.800 (0.085–7.529) | 0.640 | 0.581 (0.060–5.659) |
| CC | 0.644 | 0.699 (0.152–3.203) | 0.443 | 0.424 (0.047–3.789) | 0.609 | 0.567 (0.064–4.986) |
| Rs12568297 | ||||||
| GG | 1(Ref) | 1(Ref) | 1(Ref) | |||
| GC | 0.442 | 1.301 (0.666–2.541) | 0.269 | 1.823 (0.629–5.286) | 0.378 | 1.605 (0.560–4.596) |
| CC | 0.646 | 1.527 (0.250–9.312) | 0.301 | 3.281 (0.346–31.116) | 0.354 | 2.889 (0.306–27.234) |
| Rs6664761 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.370 | 1.427 (0.655–3.108) | 0.313 | 2.936 (0.362–23.808) | 0.313 | 2.936 (0.362–23.808) |
| CC | 0.307 | 1.500 (0.689–3.265) | 0.270 | 3.229 (0.402–25.971) | 0.199 | 3.888 (0.490–30.870) |
| Rs7531668 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TA | 0.900 | 0.966 (0.563–1.658) | 0.698 | 0.822 (0.306–2,208) | 0.500 | 0.716 (0.271–1.893) |
| AA | 0.299 | 0.663 (0.305–1.441) | 0.290 | 0.329 (0.042–2.585) | 0.233 | 0.286 (0.037–2.234) |
Abbreviations: HR, hazard ratios; CI, confidence interval; Ref, reference.
Table 6.
PARP1 genotypes and non-hematological toxicities.
| Variables | Nausea | Vomiting | Neuropathy | |||
|---|---|---|---|---|---|---|
| Rs1136410 | P | HR (95%CI) | P | HR (95%CI) | P | HR (95%CI) |
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.848 | 1.054 (0.614–1.811) | 0.883 | 1.060 (0.484–2.321) | 0.978 | 1.008 (0.573–1.774) |
| CC | 0.296 | 1.471 (0.714–3.032) | 0.303 | 0.500 (0.134–1.871) | 0.779 | 0.895 (0.410–1.950) |
| rs11801168 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.634 | 0.681 (0.141–3.305) | 0.845 | 0.800 (0.085–7.529) | 0.779 | 1.278 (0.230–7.101) |
| CC | 0.991 | 0.991 (0.216–4.547) | 0.868 | 0.832 (0.096–7.215) | 0.893 | 1.121 (0.212–5.941) |
| Rs12568297 | ||||||
| GG | 1(Ref) | 1(Ref) | 1(Ref) | |||
| GC | 0.057 | 0.488 (0.233–1.021) | 0.324 | 0.536 (0.155–1.850) | 0.985 | 0.993 (0.486–2.030) |
| CC | 0.896 | 0.887 (0.145–5.409) | 0.641 | 1.698 (0.183–15.726) | 0.579 | 0.535 (0.059–4.870) |
| Rs6664761 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TC | 0.481 | 0.756 (0.347–1.647) | 0.900 | 0.933 (0.317–2.752) | 0.438 | 1.402 (0.597–3.294) |
| CC | 0.634 | 0.828 (0.381–1.799) | 0.325 | 0.565 (0.182–1.760) | 0.761 | 1.143 (0.484–2.700) |
| Rs7531668 | ||||||
| TT | 1(Ref) | 1(Ref) | 1(Ref) | |||
| TA | 0.439 | 0.802 (0.459–1.402) | 0.979 | 0.989 (0.425–2.298) | 0.606 | 1.163 (0.656–2.060) |
| AA | 0.453 | 1.343 (0.622–2.902) | 0.490 | 1.457 (0.500–4.249) | 0.544 | 0.764 (0.320–1.823) |
Abbreviations: HR, hazard ratios; CI, confidence interval; Ref, reference.
Discussion
PARP1 is the major member of PARP family. It was identified by Chambon et al. in 1963 as a protein whose enzymatic activity allows it to generate ADP-ribose polymers15. DNA double-strand breaks (DSBs) can lead to fragmentation, loss or rearrangement of chromosomes16. DSBs are repaired through two pathways: homologous recombination (HR) and error-prone non-homologous end joining (NHEJ), which repair DSBs generated during the S-phase and outside the S-phase of the cell cycle respectively17. PARP1 is involved in both HR and NHEJ pathways18,19. In our study, we systemically investigated the association between polymorphisms in PARP1 and prognosis of TNBC, we first demonstrated that rs7531668 was related to DFS of all patients and lymph node negative patients and rs6664761 genotype predicted DFS in lymph node negative and age ≤ 50 subgroups. We also found that polymorphisms in PARP1 gene had no influence on treatment toxicities.
PARP1 gene resides on the long arm of chromosome 1. It spans 23 exons and 22 introns13. Till now, there is no report about polymorphisms investigated in our study and prognosis of breast cancer. However, the expression level of PARP1 has been reported to be associated with the survival of breast cancer patients, but the results are inconsistent. Rojo et al. found that nuclear PARP-1 is overexpressed during the malignant transformation of the breast, particularly in triple-negative tumors, and independently predicts poor prognosis in operable invasive breast cancer20. Donizy et al. reported that nuclear-cytoplasm expression (NCE) of PARP-1 was associated with unfavorable prognosis in lymph node negative early breast cancer21. While in Aiad’s study, the authors demonstrated that PARP-1 immunohistochemical expression is a marker of good prognosis in locally advanced breast cancer22. A meta-analysis included 3506 patients from eight studies, the results indicated that higher PARP expression indicated a worse clinical outcome in early stage breast cancer, with a HR of 3.08 (95% CI, 1.14 ± 8.29, P = 0.03) for disease-free survival and a HR of 1.82 (95% CI, 1.20 ± 2.76; P = 0.005) for overall survival23. But in locally advanced breast cancer, the authors observed no association between PARP expression level and survival23. The results from above studies suggested that PARP1 protein might be a stronger prognostic marker in early stage patients. In our study, two polymorphisms were associated with DFS in lymph node negative patients but not in lymph node positive patients, which supported the results from above researches.
PARP1 was also related to sensitivity of some chemotherapeutic agents. Minckwitz et al. found that high cPARP expression predicted high sensitivity to neoadjuvant taxane/anthracycline-based chemotherapy24. Zhai et al. found that higher nuclear PARP1 expression correlated with increased in vitro chemosensitivity against docetaxel and epirubicin but not cisplatin and vinorelbine14. In their study, patients with high nPARP1 expression were more sensitive to anthracycline/taxane based neoadjuvant chemotherapy and with higher pathologic responses14. Results from Egyptian researchers also demonstrated that PARP1 immunohistochemical expression is predictive of response to anthracycline/taxane based neoadjuvant chemotherapy in locally advanced breast cancer patients22. As PARP1 involves in repairing DNA double-strand breaks, it is easy for us to understand its influence on sensitivity of agents which cause DNA damage, such as anthracyclines. While PARP1 may also lead to an intrinsic cell death program (PARP1-dependent cell death)25, which might explain its effect on the sensitivity of other drugs, such as taxanes.
Polymorphisms in PARP1 have been reported to be associated with the risk of several kinds of cancers. In one meta-analysis, the authors found that rs1136410 may be involved in cancer development at least in some ethnic groups (Asian) or some specific cancer types (gastric, cervical, and lung cancers, and glioma)26. Alanazi et al. confirmed that rs1136410 was associated with risk of breast cancer in Saudi population27. Rs1136410 is the mostly investigated polymorphism in PARP1 which locates at codon 762 in exon 17. Rs1136410 leads to a valineto-alanine substitution in the catalytic domain and then reduces the activities of PARP128. In vitro enzymatic analysis of PARP1-Ala762 and PARP1-Val762 demonstrated that PARP1-Ala762 displayed 57.2% of the activity of PARP1-Val762 for auto-poly(ADP-ribosyl)ation and 61.9% of the activity of PARP1-Val762 for trans-poly(ADP-ribosyl)ation of histone H128. As expression level of PARP1 protein was proved to predict prognosis and chemotherapy sensitivity in breast cancer patients20,21, we assumed that rs1136410 genotypes might relate to prognosis of TNBC. But in our study, no significant association was observed. There are some reasons for this result. Firstly, the prognostic value might vary between different ethnic groups. Secondly, the complicated interactions with other polymorphisms could also affect the results. Thirdly, all patients in our study received taxane/anthracycline-based adjuvant chemotherapy, which might compromise the prognostic value of PARP1, since higher PARP1 expression level has been found to predict higher sensitivity of taxanes and anthracyclines24. The underlying mechanisms of other SNPs on survival of TNBC are not yet clear and are going to be investigated in our further studies.
In conclusion, our results first demonstrated that polymorphisms in PARP1 were associated with survival of TNBC patients receiving anthracycline/taxane based adjuvant chemotherapy especially in lymph node negative and age ≤ 50 subgroups. This study supported the findings from previous researches of PARP1 protein. We found no association between these polymorphisms and toxicities induced by chemotherapy. Since other polymorphisms have never been reported except rs1136410, further investigations are needed to verify the results.
Materials and methods
Patients
In our study, a total of 272 patients with stage I-III primary TNBC treated with anthracycline/taxane based adjuvant chemotherapy were enrolled between January 2004 and December 2014. Stage was determined according to American Join Committee on Cancer 2010 classification29. TNBC was defined according to guidelines issued by the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) in 201030,31. This investigation was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Hospital and Jiangxi Cancer Hospital. It was conducted in accordance with the ethical standards of the Declaration of Helsinki and following the national and international guidelines. All patients have consented to their blood and clinical information being used in this study. Clinical and pathological data were collected. Patients were followed until December 2018 to collect data on recurrence and death.
Single nucleotide polymorphism selection and genotyping
Genotype data from PARP1 gene regions encompassing 10 kb of upstream and 10 kb of downstream flanking sequences were extracted from the HapMap We used Chinese Han population. Haploview 4.2 software was to identify Tag SNPs. The inclusion criteria were: 1) SNPs known in ethnic Han Chinese population; 2) a minor allele frequency (MAF) > 0.05 and r2 > 0.8. A total of 5 candidate SNPs were selected for genotyping (Table 7).
Table 7.
Information for the SNPs genotyped in this study.
| SNPs | Position | Location | Alleles | MAF |
|---|---|---|---|---|
| rs1136410 | 1:226367601 | coding sequence variant | C/T | 0.1969 |
| rs11801168 | 1:226357130 | downstream transcript variant | T/C | 0.4036 |
| rs12568297 | 1:226356905 | downstream transcript variant | G/C | 0.2400 |
| rs6664761 | 1:226362490 | intron variant | T/C | 0.2392 |
| rs7531668 | 1:226408318 | upstream transcript variant | T/A | 0.2632 |
Abbreviations: SNP, single nucleotide polymorphism; MAF, minor allele frequency.
Genomic DNA was extracted from the peripheral blood samples of each patient and was isolated by the routine phenol–chloroform method. Primers and probes were designed by MassARRAY Typer 4.0 software. MassARRAY MALDI-TOF System (Sequenom Inc., San Diego, CA, USA)32,33 was used for genotyping by the method described in the Sequenom Genotyping Protocol.
Statistical analyses
SPSS 18.0 statistical software (SPSS Inc, Chicago, IL, USA) was used for analysis. 5-year DFS and OS rates of patients with different genotypes were estimated by Kaplan–Meier product limit method and compared by the log-rank test. Hazard ratios of recurrence/metastasis and death with 95% confidence intervals (CI) were estimated by Cox regression model. The multivariate analysis was adjusted for age, histological grade, tumor size, lymph node status and vascular invasion. The distribution of genotypes in patients with or without toxicities were compared by two-sided Pearson’s Chi-square tests, odds ratios (ORs) and 95% confidence intervals (CI) were calculated by logistic regression. All statistical tests were two-sided, and P < 0.05 was considered significant.
Ethical Standards
This article does not contain any studies with human or animal subjects performed by any of the authors.
Acknowledgements
This study was funded by Natural Science Foundation of Jiangxi Province of China (No. 20151BAB205043).
Author contributions
All authors contributed significantly to this work. Y.F. conceived and designed the present study. Y.Q.L. and Y.L.L. collected the samples and performed the research study. Y.Q.L. and J.L. wrote the paper. Y.F. and J.P.X. reviewed and revised the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Thses authors contributed equally: Yuqian Liao and Yulu Liao.
References
- 1.Siegel RL, Miller KD. Cancer statistics. 2019. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Haffty BG, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:5652–5657. doi: 10.1200/jco.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 4.Perou CM. Molecular stratification of triple-negative breast cancers. The oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 5.Udyavar, A. R. et al. Genomic Alterations Associated with Recurrence and TNBC Subtype in High-risk Early Breast Cancers. 10.1158/1541-7786.mcr-18-0619 (2018). [DOI] [PubMed]
- 6.Andre F, Zielinski CC. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Annals of oncology: official journal of the European Society for Medical Oncology. 2012;23(Suppl 6):vi46–51. doi: 10.1093/annonc/mds195. [DOI] [PubMed] [Google Scholar]
- 7.Burstein MD, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:1688–1698. doi: 10.1158/1078-0432.ccr-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 9.Carter RJ, Parsons JL. Base Excision Repair, a Pathway Regulated by Posttranslational Modifications. Molecular and cellular biology. 2016;36:1426–1437. doi: 10.1128/mcb.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 11.Ame JC, Spenlehauer C. & de Murcia, G. The PARP superfamily. BioEssays: news and reviews in molecular, cellular and developmental biology. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 12.Abd Elmageed ZY, Naura AS, Errami Y, Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): new roles in intracellular transport. Cellular signalling. 2012;24:1–8. doi: 10.1016/j.cellsig.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Green AR, et al. Biological and clinical significance of PARP1 protein expression in breast cancer. Breast cancer research and treatment. 2015;149:353–362. doi: 10.1007/s10549-014-3230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai L, et al. The nuclear expression of poly (ADP-ribose) polymerase-1 (PARP1) in invasive primary breast tumors is associated with chemotherapy sensitivity. Pathology, research and practice. 2015;211:130–137. doi: 10.1016/j.prp.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochemical and biophysical research communications. 1963;11:39–43. doi: 10.1016/0006-291X(63)90024-X. [DOI] [PubMed] [Google Scholar]
- 16.Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA repair. 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiewer, M. J. et al. PARP-1 regulates DNA repair factor availability. 10, 10.15252/emmm.201708816 (2018). [DOI] [PMC free article] [PubMed]
- 19.Lee WP, et al. Helicobacter pylori-induced chronic inflammation causes telomere shortening of gastric mucosa by promoting PARP-1-mediated non-homologous end joining of DNA. Archives of biochemistry and biophysics. 2016;606:90–98. doi: 10.1016/j.abb.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Rojo F, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology. 2012;23:1156–1164. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- 21.Donizy P, et al. Nuclear-cytoplasmic PARP-1 expression as an unfavorable prognostic marker in lymph nodenegative early breast cancer: 15-year follow-up. Oncology reports. 2014;31:1777–1787. doi: 10.3892/or.2014.3024. [DOI] [PubMed] [Google Scholar]
- 22.Aiad HA, et al. The prognostic and predictive significance of PARP-1 in locally advanced breast cancer of Egyptian patients receiving neoadjuvant chemotherapy. Applied immunohistochemistry & molecular morphology: AIMM. 2015;23:571–579. doi: 10.1097/pai.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 23.Qiao W, Pan L, Kou C, Li K, Yang M. Prognostic and clinicopathological value of poly (adenosine diphosphate-ribose) polymerase expression in breast cancer: A meta-analysis. PloS one. 2017;12:e0172413. doi: 10.1371/journal.pone.0172413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Minckwitz G, et al. Cytoplasmic poly(adenosine diphosphate-ribose) polymerase expression is predictive and prognostic in patients with breast cancer treated with neoadjuvant chemotherapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:2150–2157. doi: 10.1200/jco.2010.31.9079. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Science signaling. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Q, et al. PARP-1 Val762Ala polymorphism and risk of cancer: a meta-analysis based on 39 case-control studies. PloS one. 2014;9:e98022. doi: 10.1371/journal.pone.0098022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alanazi M, et al. Association between PARP-1 V762A polymorphism and breast cancer susceptibility in Saudi population. PloS one. 2013;8:e85541. doi: 10.1371/journal.pone.0085541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochemical and biophysical research communications. 2007;354:122–126. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- 29.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 30.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:3997–4013. doi: 10.1200/jco.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 31.Hammond ME, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:2784–2795. doi: 10.1200/jco.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiu L, Zhang C, Wu Z, Peng J. Establishment and Application of a Universal Coronavirus Screening Method Using MALDI-TOF Mass Spectrometry. Frontiers in microbiology. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung KW, et al. Rapid and Simultaneous Detection of Major Drug Resistance Mutations in Reverse Transcriptase Gene for HIV-1 CRF01_AE, CRF07_BC and Subtype B in China Using Sequenom MassARRAY(R) System. PloS one. 2016;11:e0153641. doi: 10.1371/journal.pone.0153641. [DOI] [PMC free article] [PubMed] [Google Scholar]


