Abstract
Radiotherapy is one of the most common countermeasures for treating a wide range of tumors. However, the radioresistance of cancer cells is still a major limitation for radiotherapy applications. Efforts are continuously ongoing to explore sensitizing targets and develop radiosensitizers for improving the outcomes of radiotherapy. DNA double-strand breaks are the most lethal lesions induced by ionizing radiation and can trigger a series of cellular DNA damage responses (DDRs), including those helping cells recover from radiation injuries, such as the activation of DNA damage sensing and early transduction pathways, cell cycle arrest, and DNA repair. Obviously, these protective DDRs confer tumor radioresistance. Targeting DDR signaling pathways has become an attractive strategy for overcoming tumor radioresistance, and some important advances and breakthroughs have already been achieved in recent years. On the basis of comprehensively reviewing the DDR signal pathways, we provide an update on the novel and promising druggable targets emerging from DDR pathways that can be exploited for radiosensitization. We further discuss recent advances identified from preclinical studies, current clinical trials, and clinical application of chemical inhibitors targeting key DDR proteins, including DNA-PKcs (DNA-dependent protein kinase, catalytic subunit), ATM/ATR (ataxia–telangiectasia mutated and Rad3-related), the MRN (MRE11-RAD50-NBS1) complex, the PARP (poly[ADP-ribose] polymerase) family, MDC1, Wee1, LIG4 (ligase IV), CDK1, BRCA1 (BRCA1 C terminal), CHK1, and HIF-1 (hypoxia-inducible factor-1). Challenges for ionizing radiation-induced signal transduction and targeted therapy are also discussed based on recent achievements in the biological field of radiotherapy.
Subject terms: Cell biology, Genetics research
Introduction
The increasing prevalence of cancer worldwide is a major challenge to the improvement of quality and length of life. According to reports, 975,396 patients were newly diagnosed with cancer in 2012, and there were 358,392 deaths due to cancer in youth globally1. In 2015, the number of identified cancer cases increased to ~17.5 million, and the deaths from cancers increased to 8.7 million globally. Notably, from 2005 to 2015 (an 11-year span), the number of patients with cancer increased by 33%2. In 2016, ~9 million deaths were attributed to cancer, an increase of almost 18% over one decade3. Moreover, in the United States in 2017, there were 1,688,780 newly diagnosed cancer patients and almost 600,920 deaths due to cancer4. These numbers have increased rapidly annually, and in 2018, ~18.1 million newly diagnosed cancer patients and 9.6 million cancer deaths were reported worldwide5. In China, in 2014, there were 3.804 million newly diagnosed cancer patients and 2.296 million cancer deaths, and the statistical results showed that the crude incidence rate and the crude mortality rate were 278.07 per 100,000 people and 167.89 per 100,000 people, respectively6. Furthermore, it is estimated that by 2035, the number of annual cancer deaths will reach 14.5 million because worldwide cancer cases are expected to dramatically increase from 15 million at present to 24 million in the next 20 years3. Moreover, in parallel with the increasing rates of cancer diagnosis and death, the global burden of cancer has gradually increased over the past decade. Based on the Global Burden of Disease Cancer Collaboration announcement, 208.3 million disability-adjusted life-years (DALYs) were attributed to cancer globally in 2015. Lung cancer was the top cause of death among males, accounting for 25.9 million DALYs, while in females, breast cancer-attributable deaths were the top cause, accounting for 15.1 million DALYs2. Significant advances in the war against cancer have been achieved over the past decade. For instance, deaths from Hodgkin lymphoma declined significantly between 2005 and 2015 (−6.1%; 95% uncertainty interval: −10.6% to −1.3%), and other cancer deaths, such as deaths from esophageal cancer and stomach cancer, have also significantly decreased over the past decade2. Additionally, through large-action control of tobacco use and human papillomavirus vaccination in females, the burden of cancer in the female population has been substantially decreased in both economically developed and economically developing areas7. Although large-scale implementation of prevention and treatment methods has made these improvements possible, there is still a long way to go in the fight against cancer.
The management of cancer mainly involves surgery, radiotherapy, chemotherapy, and the rapidly evolving field immunotherapy8. The most commonly used cancer therapies over the past century include chemotherapy and radiotherapy methods9–12; among these, radiotherapy is widely and predominantly used prior to surgery and other treatment methods13,14. Radiotherapy is defined as the application of radiation for clinical cancer treatment, including external-beam radiation and local radioactive seed implants with the purpose of killing cancer cells or controlling cancer cell proliferation15,16. Radiotherapy is sometimes used alone, but at most times, it is applied in combination with other therapy strategies, such as surgery or oral medicine. Radiotherapy developed rapidly following the discoveries of X-rays by Roentgen, natural radioactivity by Becquerel, and radium by Curie over 125 years ago17. These three fundamental discoveries not only earned the discoverers Nobel Prizes but also founded the research field of radiology, as well as led to the establishment of radiotherapy techniques, such as external-beam radiotherapy with a long source surface distance and brachy therapy with a short-spacing surface space, which are commonly delivered through radium and X-rays18. Three countries, France, America, and Sweden, were the first to adopt radiation for gastric cancer and basal cell cancer treatment19,20. Over the past century, continuous technological improvements in radiotherapy have translated into better clinical practices, not only changing several fundamental concepts but also gradually changing clinical treatment guidelines. For instance, for a long time, radiation with a specific beam energy, such as telecobalt therapy, was applied in the clinic from 50–250 kV to 1.2 MeV, while the linear accelerator was between 6 and 20 MV; now, the computer revolution has made a three-dimensional (3D) approach in complex spaces a reality21. In addition, newly developed therapies based on high linear energy transfer (LET) particles, including protons and heavy ions such as carbon ions, are being used in cancer treatments22–25. However, with the increased usage of radiation not only in cancer treatment but also in medical examination globally, the adverse influence of radiation on the human body has attracted much attention in the public and scientific community, including the subsequent secondary cancer risks and damage to normal tissues after radiotherapy26. Indeed, advances in radiation and radiotherapy are contributing substantially to winning the battle against cancer27–29. Currently, ~50% of cancer patients are subjected to radiotherapy. The combination of radiotherapy, surgery, and other medical treatments has contributed to almost 50% of cancer patients having a long-term survival opportunity. A study performed in Australia reported that among newly diagnosed patients, almost 52% of patients were subjected to radiotherapy, and more than 23% of patients required multiple treatments for a better prognosis30. In China, more than 50% of clinically treated cancer patients have received radiotherapy, which contributed to cure in more than 40% of patients31,32. To standardize quality control, a set of basic guidelines for radiotherapy have been developed, in accordance with the relevant national laws and regulations and referring to relevant international guidelines; These guidelines were announced at the 14th National Congress of Radiation Oncology (CSTRO; available at https://cstro2017.medmeeting.org/cn) meeting32. Moreover, radiotherapy is a conservative treatment with the capacity to affect cancer without body image alteration. Most importantly, radiotherapy is very cost effective. The data released by the International Atomic Energy Agency suggest that radiotherapy is the most economical treatment measure overall, accounting for ~5% of the all-in expenses of patient care for cancers33. Most experts hold that treating cancer using radiation technology is essential and critical for not only saving thousands of cancer patient lives but also saving economic costs in cancer patients; thus, access to radiotherapy should be available globally in the near future 34.
However, radiotherapy is typically accompanied by the unavoidable development of cancer cell resistance to radiation exposure35,36. Radiotherapy resistance (RR), defined as a reduction in the effectiveness of antitumor therapy37, is a major obstacle in cancer treatment. RR either arises within cancer cells when cancer cell genes or phenotypes are altered in response to radiation exposure or is due to the cancer microenvironment protecting cancer cells against the treatment. The former is referred to as intrinsic resistance, while the latter is referred to as extrinsic resistance38. RR leads to cancer relapse, poor treatment response, poor prognosis, decreased quality of life, and increased disease treatment burden. Furthermore, RR induces damage to cancer-adjacent normal tissues, disrupting the physiological and biochemical functions of normal tissue, resulting in symptoms, including radiation-related diarrhea, rectal bleeding, and radiation dermatitis39–41, as well as an increased risk of subsequent secondary cancer26,42–44 or chronic noncommunicable diseases including type II diabetes45,46 or cardiovascular diseases47. Over the past century, to remove the barrier of RR, many studies have been carried out to investigate RR-related regulatory genes, molecules, and signaling pathways to uncover the underlying mechanisms of RR and to develop radiation sensitizers48,49. Currently, two large bottlenecks for successfully improving radiation resistance are as follows: (1) identifying the master regulator of the development and progression of RR and (2) determining how the master regulator can serve as a potential target to overcome RR. In this review, we discuss the promising targets of signaling pathways that can be proposed for cancer radiosensitization and may be translated into clinical radiotherapy targets.
DSBs are a major pattern of RIDD
There are many factors associated with increased RR in cancer cells. These factors include but are not limited to the following: the local cancer microenvironment, membrane signaling sensors, and the patient immune system, gut microbial community50, nutritional status51, and mental health status52. However, among the reported and discovered factors, DNA damage is a primary and intrinsic factor and the most crucial operator in the response to radiation exposure and the orchestration of the subsequent cascade of DNA repair response signaling pathways to control cancer cell cycle arrest and cell fate, i.e., death or survival53. In other words, the ability of radiation to control cancer predominantly depends on radiation-induced DNA damage (RIDD)53; as a result, the DNA damage response (DDR) of tumor cells and the ability of tumor cells to repair DNA damage are essential in determining the outcome of cancer cells.
The genomic integrity of cells is extremely important for cell growth as well as successful transmission of genetic information to the next generation54, However, many types of external or internal genotoxic insults challenge DNA integrity55, forcing the host to evolve and develop compensatory changes to combat DNA damage and maintain genomic integrity56 via several independent or complementary DNA repair pathways, allowing for a fail-safe mechanism whereby the disruption of one pathway will be compensated for by another pathway57. During cancer cell evolution, multiple comprehensive molecular signaling pathways have been developed to face the challenge of radiation stress, and this ability to evolve can contribute to increased cancer cell RR, leading to radiotherapy failure. Moreover, during the process of developing RR, a percentage of cells in tumor tissues not only acquire higher RR but also become more aggressive and are prone to lymph node and distant metastasis58. Thus, enhancement of the cancer response to radiation through DNA damage pathways has been a focus of radiotherapy studies for the past few decades 59–61.
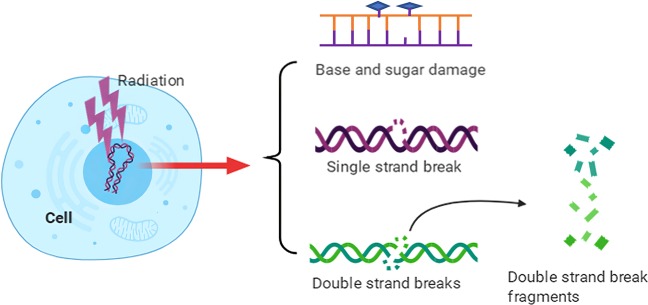
Typically, exposure to ionizing radiation (IR) is often suggested as a treatment for preventing cancer cell proliferation. There are various applications depending on the IR type, such as electromagnetic waves or particles. Currently, several techniques and standard values are well accepted for IR applications; for example, LET is usually performed at between 1 and 10 keV/µm using sources such as X-rays, γ-rays, protons, or carbon ions, while very high values, usually beyond >100 keV/µm, are found in some forms of space radiation62. As reported previously, once the radiation track deposits its energy in the DNA molecules of cancer cells, a fraction of the DNA damage sites will have two or more damages formed within one or two helical turns of DNA63,64. The common DNA damage pattern comprises base and sugar damage, crosslinks, single-strand breaks (SSBs) and double-strand breaks (DSBs), as shown in Fig. 1. Compared to those of SSBs, the patterns of DSBs are more intricate and include simple and complex types, as well as multiple physical characteristics not only referring to the break but also to the kinetics of the ability to repair the break65,66 In terms of simple DSBs, two-ended breaks of DNA may occur as a direct result of radiation, and these have rapid kinetic repair. Complex DNA DSBs, namely, “clustered DNA damage,” are a hallmark of IR and locally consist of more than two instances of oxidative base damage, basic sites, or SSBs around a DSB63,67,68. Compared to simple breaks, complex DSBs are more slowly and inefficiently repaired, resulting in genomic instability69–71. Indeed, the radiation exposure response to DNA damage may vary based on IR type. For instance, high-LET radiation, involving methods such as heavy ion and proton radiation, may preferentially induce clustered damage, a signature of IR; in contrast, isolated, endogenously induced lesions result in a homogeneous distribution67,72. Following irradiation with X-rays or γ-rays, clustered DNA lesions are often 3–4 times more abundant than single-strand damage67,73. Actually, the number of DSBs resulting from high-LET irradiation is much more than that resulting from low-LET irradiation. There is evidence that high-LET irradiation causes almost 500 DSBs/μm3 track volume74. Using 3D-structured illumination microscopy, Hagiwara et al.75 revealed that clustered DSBs could be formed in a space of 1 μm3 by high-LET irradiation; moreover, once these clustered DSBs occur, a higher risk of chromosomal rearrangement and lethality will subsequently develop. More efficient induction of complex clustered DNA damage is a major factor contributing to the higher relative biological effectiveness of heavy ions.
Fig. 1.
DNA damage induced by ionizing radiation. The major types of DNA damage induced by IR include base and sugar damage, single-strand breaks, double-strand breaks, clustered DNA damage, and covalent intrastrand or interstrand crosslinking
Moreover, once a complex DSB forms, the repair occurs slowly, and chromosomal aberrations can cause cell death or delayed mitosis without further repair73,76. Because IR induces genetic instability, RR is expected if cancer cells survive following treatment with IR. Hence, how cellular sensors respond to radiation and how early signal transducers work after IR will be reviewed in the next section based on recently reported evidence to understand the mechanisms of DNA damage signals more deeply and clearly.
Cellular DNA damage sensors and early signal transducers in response to IR
As a signal, DNA damage activates a series of biochemical reactions in response to IR insult, triggering a variety of cellular responses. Nevertheless, the key questions are how the DNA damage signal is sensed and recognized and how the cascade signaling of downstream biochemical reactions is triggered. DNA damage sensors and early signal transducers thus play essential roles in recognizing DNA damage77,78. The ideal DNA damage sensors are the first proteins to contact DNA damage sites, identifying damage signals and triggering cell signaling transduction79–81. Moreover, DNA damage sensors also have the ability to recruit DDR proteins to sites of DNA damage82,83. Signal transducers often play roles as functional partners of DNA damage sensors83,84. As DNA damage sensors and signal transducers usually coexist, it is difficult to classify them. However, signal transducers have kinase activity, transducing the DNA damage chemical signal to induce biochemical modification reactions and triggering the activity of downstream effectors 85.
The first DSB sensor identified from fission yeast was Rad24p by Ford et al. in 199486; this sensor is required for DNA damage checkpoint (DDC) activation and is essential for cell proliferation87. In Schizosaccharomyces pombe, Rad24p is required for some essential functions, as double deletion of its encoding gene is lethal87,88. As a DNA damage sensor, Rad24p is often considered to be the primary DNA damage responder, forming a complex with Ddc1p and Mec3p and triggering cell cycle arrest after DNA damage89. Furthermore, previous studies have revealed that Rad24p associates with Rfc2p-Rfc5p to form replication factor C, functioning in DNA replication or repair and DDC pathways90. In Rad24p-containing compounds, the Rad24p-Rfc2p or Rad24p-Rfc5p complexes can recruit the Rad24p-Ddc1p-Mec3p complex to create a “workshop” similar to the reaction machinery, triggering downstream kinases or effectors such as Rad53p89,91–93. The 14-3-3 isoforms are the mammalian homologs of fission yeast Rad24, functioning in DDC as well as in cell cycle control94. Following the discovery of Rad24p as the DSB sensor, Mec1p and Rad26p were subsequently identified as DNA damage sensors as well89,95. Mec1p has been considered to be the regulator of Ddc1p phosphorylation, a protein responsible for the yeast DDC89. Mec1p and Rad26p, from budding yeast and fission yeast, respectively96, have the characteristics of DNA damage sensors, activating phosphatidylinositol proteins97. The phosphorylation substrates of Mec1p and Rad26p include Ddc1p and Rad9p, another two DNA damage sensors98–100. In general, DNA response mechanisms have been extremely conserved during evolution within both yeast and mammalian cells. Although no effective or idealized DNA damage sensors have been confirmed in mammalian cells, several important molecules were recognized to be associated with DNA damage sensors and, importantly, to mediate and trigger IR-induced DSB signaling responses.
γH2AX
As reported by Siddiqui et al.101, H2AX can respond to DSBs in a phosphorylation pattern at a very early time. H2AX is a variant of the core histone protein H2A; upon DNA DSB occurrence, H2AX is phosphorylated at the S139 site, which forms γH2AX foci. Furthermore, in their review, Siddiqui et al.101 mentioned that γH2AX persisted after exposure to IR under treatment with various radiosensitizing drugs, indicating that this sensor could be used to monitor cancer therapy and to tailor cancer treatments. Using anti-γH2AX monoclonal antibodies and immunofluorescence hybridization techniques to visualize γH2AX localization at sites of DNA damage102,103, even with a very low dose of IR exposure, γH2AX foci can be visualized as well, but once the DNA damage is repaired, the foci are eliminated104,105. Kuo and Yang106 suggested that γH2AX foci represent DSBs in a 1:1 ratio and can be used as a biomarker for DNA damage106. Specifically, the disappearance of γH2AX typically occurs earlier than that of other IR exposure response proteins. Moreover, γH2AX can act as a platform to recruit other DNA repair proteins, such as BRCA1 (BRCA1 C terminal)107, 53BP1 (p53-binding protein 1),108, MDC1109, and Rad51110. Zhang et al.111 reported that glioma stem cells exhibited RR with increased γH2AX-positive cell rates after 6 Gy radiation due to upregulation of the long noncoding RNA (lncRNA) PCAT1. Katagi et al.112 evaluated the effects of histone demethylase inhibition on genes associated with DSB repair in diffuse intrinsic pontine glioma cells and found that the expression of DSB repair genes was significantly reduced, but the level of γH2AX increased and was sustained at a high level. Overall, γH2AX was considered to match the characteristics of a DNA damage sensor more closely than the expression of DSB repair genes. Currently, γH2AX is widely used as a marker to detect radiation-induced DSB repair by immunofluorescent staining of foci or immunocytochemistry113,114.
Nbs1/hMre11/hRad50 complex
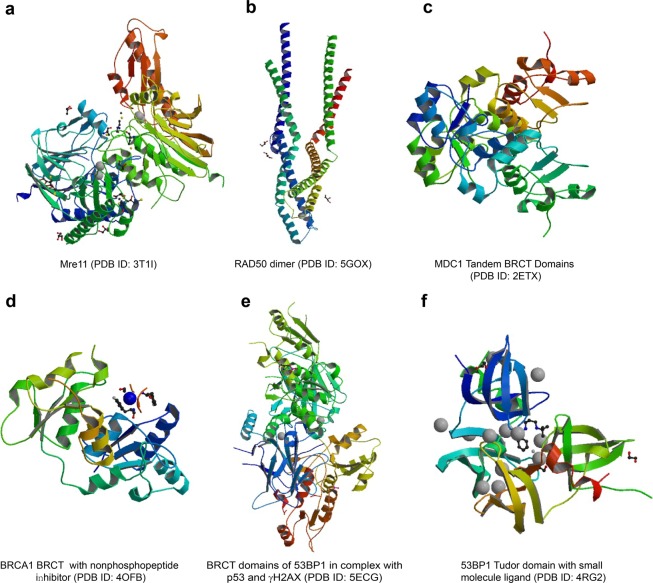
The MRN (Mre11-Rad50-Nbs1) complex, formed by Nbs1, hMre11, and hRad50, was first reported by Carney et al. in 1998115. According to this report, the MRN complex is responsible for linking DSB repair with cell cycle checkpoint functions. Habraken et al.116 suggested that the Nbs1/hMre11/hRad50 complex plays an important role in DNA DSB sensing and the signal transduction initiated by X-ray radiation. Similarly, a study by Kobayashi117 demonstrated that one of the roles of this Nbs1/hMre11/hRad50 complex is to recruit activated ataxia–telangiectasia mutated (ATM) to DNA damage locations, showing that it has the ability to recognize DNA damage initially. Moreover, according to Tauchi et al.118, the hMre11 binding region is necessary for both nuclear localization of the Nbs1/hMre11/hRad50 complex and for cellular radiation resistance; meanwhile, the fork head-associated domain of Nbs1 regulates nuclear foci formation of the multiple proteins in the complex. The hMre11 structure is comprised of an N-terminal core domain containing the nuclease and capping domains and a C-terminal domain containing the DNA-binding and glycine-arginine-rich motif (Fig. 2a). Deletion of the α2-β3 loop (AA84–119) in the hMre11 core structure prevented the formation of a stable hMre11 core dimer and inhibited Nbs1-binding activity119. The structure of the MRN complex contains two major dimerization interfaces that link Mre11, Rad50, and Nbs1 in DNA damage sensing and signaling. One dimerization interface is within the globular domain and involves Rad50 and Mre11. The second is the zinc hook situated distal to the globular domain separated by the antiparallel coiled-coil domains of Rad50. The two parallel coiled-coil domains of RAD50 proximal to the hook form a rod shape (Fig. 2b), which is crucial for stabilizing the interaction of Rad50 protomers within the dimeric assembly120. In the presence of Mn2+ ions, the hMre11 core shows exonuclease and endonuclease activities for 30–50 base spans. Structure and biochemistry analyses indicate that many tumorigenic mutations of hMrell are primarily associated with its Nbs1 binding and partly with its nuclease activities119. A study indicated that heterozygous p.l171V mutation in the Nbs1 gene was found in Korean patients with high-risk breast cancer121. In addition, a persistent increase in radiation-induced Nbs1 foci formation was accompanied by an increased frequency of spontaneous chromosome aberrations122. Another in vitro study indicated that heterozygosity of Nbn, the murine homolog of human Nbs1, contributes to mouse susceptibility to IR-induced tumorigenesis123. Della-Maria et al.124 further identified a novel mechanism in which the Nbs1/hMre11/hRad50 complex interacts with the DNA ligase III α/XRCC1 complex, which is linked with the nonhomologous end-joining (NHEJ) pathway in cancer cells following IR radiation. Ho et al.125 demonstrated that overexpression of the Nbs1/hMre11/hRad50 complex in rectal cancer was associated with RR and poor prognosis. Collectively, although Nbs1 is the phosphorylation target of ATM, the Nbs1/hMre11/hRad50 complex localizes upstream of ATM in the DDR, acting as a sensor.
Fig. 2.
Structures of major DNA damage signal sensors, their main functional domains and their interactions with their partners. The data are from the RCSB database (https://www.rcsb.org/)
Ku (Ku70/Ku80 heterodimer)
Once DSBs occur under IR stress, the primary repair pathways are triggered through two classical pathways, NHEJ and homologous recombination (HR). The NHEJ pathway is triggered via Ku, also known as the Ku70/Ku80 heterodimer, which is preferentially recognizes DSBs126. Depletion of the deubiquitylating enzyme UCHL3 resulted in the reduced chromatin-binding and IR-induced foci (IRIF) formation of Ku80 after DSB occurrence, moderately sensitizing cancer cells to IR127. A recent study by Pucci et al.128 reported nuclear localization of Ku in advanced rectal cancer patients with superior sensitivity to radiotherapy. However, in nonresponder patients, Ku70 was found to move from the nucleus to the cytoplasm, and strikingly, deregulation of Ku70/80 and the Ku70 partner clusterin was extensively associated with RR. Another study found that IR induces the accumulation of autophagosomes, and the radiosensitizing effect of autophagy-related BECN1 deficiency may result from the disruption of nuclear translocation and Ku protein activity, leading to the attenuation of DSB repair in malignant glioma. Similar to other DNA damage sensors, Ku includes a pocket structure. Once DNA damage occurs, Ku can bind to the DNA damage site and immediately embed the DNA break terminus into this pocket129. In a clinical study, the B cells of some B cell chronic lymphocytic leukemia patients were resistant to IR-induced apoptosis; when the B cell chronic lymphocytic leukemia cell subset was treated with radiation, the DNA end-binding ability of Ku was significantly increased by two- or three-fold in the radiation-resistant cell subset compared with that in the radiation-sensitive cell subset 130.
MDC1 and 53BP1
DDR alterations are a major cause of cancer cell resistance to radiotherapy131. Both proteins, one named mediator of MDC1 and another known as 53BP1, are also associated with the signaling of DNA DSBs131. MDC1 is a major modular phosphoprotein scaffold that plays an important role in the DDR process. The BRCT domains are crucial modules that mediate the protein–protein interaction in DNA damage sensing and DDR signaling and have been found in a number of DDR proteins, such as MDC1, 53BP1, BRCA1, and Nbs1. In addition to their conserved phosphopeptide recognition and binding functions, BRCT domains are also implicated in phosphorylation-independent protein interactions, poly(ADP-ribose) (PAR) binding and DNA binding132. The architecture of BRCT domains is variable, ranging from a single module to tandem BRCT repeats. Figure 2c–e displays the BRCT domain structures of MDC1, BRCA1, and 53BP1, respectively. Following induction of DNA DSBs, MDC1 is anchored to damaged sites through interaction of its BRCT repeat domain with the tail of γH2AX. Moreover, MDC1 often performs its roles in accompaniment with 53BP1; that is, it can make 53BP1 move to foci under the control of MDC1131,133. The function of 53BP1 in DDR is dependent on its recruitment to the damaged site through 53BP1 tandem Tudor domain-mediated recognition of methylated histone H4 (H4K20me2) and ubiquitinated histone H2A (H2AK15ub). 53BP1 binds with the DSB marker H2AX-pS139 through its BRCT2 domain in vitro and in cells (Fig. 2e), which is necessary for the recruitment of pATM to the damage site134. The Tudor domain of 53BP1 (Fig. 2f) also plays a critical role in the DDR through interactions with BRCT domains. As a 53BP1 regulator, the Tudor-interacting repair regulator (TIRR) directly binds to the 53BP1 Tudor domain and blocks the H4K20me2 binding surface. High-resolution structural analysis shows that the N-terminal region and the L8 loop of TIRR form an extensive binding interface with three loops of the 53BP1 Tudor domain135. TIRR masks the binding surface of H4K20me2. In colorectal cancer cells, following radiation, coimmunoprecipitation analyses showed that Ku70, γH2AX, and MDC1 were colocalized in nuclear foci136. Cairns et al.137 reported that MDC1 could be regulated by Bora. Another study showed that Bora could be phosphorylated by MDC1, leading to abolishment of irradiation-induced MDC1 foci formation, and downregulation of Bora increased the resistance to IR, likely due to a faster rate of DSB repair. A clinical study conducted by Cirauqui et al.138 showed that patients with head and neck cancer with low 53BP1 expression levels treated with radiotherapy had a higher complete response as well as a higher survival time than patients with high 53BP1. Generally, DNA damage induced by IR recruits MDC1 to sites of damage within ~1 min post irradiation, providing a γH2AX-dependent interaction platform for recruiting other DNA damage repair proteins, such as ATM and Nbs1, and the glycolytic enzyme PFKFB3.
BRCA1 and BRCA2
BRCA1 and BRCA2 are clinically correlated with hereditary breast and ovarian cancer. For BRCA1 mutation carriers, the relative risk of breast cancer is 1.19 (95% confidence interval [CI]: 1.02, 1.39), while for BRCA2 mutation carriers, the relative risk of breast cancer is 1.25 (95% CI: 1.01, 1.55)139. In prostate cancer patients with resistance to prostate-specific membrane antigen-targeting α-radiation therapy, BRCA1 and BRCA2 genes were deleted, and several variants of BRCA1 were detected140. BRCA1 consists of several domains, including an N-terminal region carrying the zinc-binding finger domain RING and two phosphopeptide-binding BRCT domains141,142. Similarly, there are also a few domains for BRCA2, that is, the transcriptional activation domain is located at the N terminus, and the DNA-binding domain is located close to the C-terminal region. Other regions include a conserved helical domain, three oligonucleotide binding folds, and a tower domain141,143. BRCA1 and BRCA2 play a crucial role in the repair of DSBs in the HR pathway144. After exposure to radiation, the BRCA1-RAP80-Abraxas complex binds ubiquitinated histone in response to DNA damage145–147. A recent report showed that BRCA1 could recruit CSB, a member of the SWI2 family, and MRN to form a complex at the late phase of S/G2. This interaction between BRCA1, CSB, and MRN is responsible for MRN-mediated DNA end resection148. In addition, the BRCA1-PALB2 interaction dictates the choice between HR and single-strand annealing149 and is associated with RR. These important roles of BRCA1/2 have suggested them as attractive, valuable, and sensitive diagnostic biomarkers in the prediction of radiotherapy outcomes 34.
The above discussion is associated with the progress of DDR-associated proteins; notably, with an increasing number of in-depth studies, some novel response proteins have been reported. A recent study found that a novel DDR was triggered by MT1-MMP (membrane-tethered matrix metalloproteinase)-integrin β1. This study indicated that suppression of MT1-MMP would improve breast cancer cell resistance to IR therapy 150.
In brief, it is well known that IR-induced DSBs are the most deleterious form of DNA damage, leading to cell death and viable chromosomal rearrangements. As a result, cells have evolved an efficient and rapid DDR to maintain genomic integrity. DNA damage sensors are response proteins that can detect DNA damage; sensor proteins can also recruit transducer proteins to provide signals to enzymes to respond to the break. To date, a series of DNA damage sensor proteins have been identified through numerous studies, including γH2AX, 53BP1, Nbs1, BRCA1/2, and Ku. These DNA damage sensors commonly have the following characteristics: (i) they localize to the sites of DSBs within a few seconds or minutes after IR exposure, forming microscopically visible nuclear domains referred to as IRIF; (ii) sensor proteins can modify the adjacent damage sites by methods such as phosphorylation of γH2AX; (iii) sensor proteins can recruit other proteins to sites of damage to form protein complexes such as the Nbs1/hMre11/hRad50 complex151; and (iv) these DNA damage sensors can also regulate each other. For instance, MDC1 expression was induced by radiation, and the overexpression of MDC1 could activate Nbs1 activity in the presence of DNA damage repair152. These sensors can also be regulated by upstream or downstream proteins. For instance, the human demethylase JMJD1C was stabilized by interaction with RNF8 and recruitment of RAP80-BRCA1, and MDC1 was demethylated at Lys45 through JMJD1C binding to RNF8 and MDC1, promoting cancer cell sensitivity to IR153. Reichert et al.154 reported that following exposure to radiation, a direct relationship between MDC1, γH2AX, and 53BP1 was identified, and higher amounts of DNA breaks were associated with an increased level of γH2AX/53BP1 foci post irradiation. Thus, it is suggested that identification of these sensors after the occurrence of DSBs under IR exposure may be a predictive biomarker in determining radiotherapy outcomes among patients with cancer. γH2AX is a typical example of a marker that has been translated from bench to bedside, and it has been employed in the clinic as a predictive biomarker for radiotherapy sensitivity in some kinds of cancers155. Table 1 presents several primary DNA damage sensors along with their roles and characteristics. Figure 2 illustrates the structures of these sensor proteins or their main functional domains and interacting partners. Figure 3 illustrates the regulation of DNA damage sensors following IR exposure in terms of the common DSB sensors and early signal transducers. Based on the above discussion of the roles and characteristics of DNA damage sensors, these sensors could be used as biomarkers to detect or evaluate DNA damage induced by IR in routine clinical use to determine optimal radiation dosing or as future targets for overcoming RR 156.
Table 1.
Summary of a few main DNA damage sensors induced by IR (human versions are shown)
| Length | Subcellular location | Interaction partners | Mutationsa | |
|---|---|---|---|---|
| γH2AX | 143 | Nucleus436, chromosome437 | Several other proteins436,438,439 | 141 (Q to N)440 |
| Nbs1 | 754 | Nucleus441, telomere442, chromosome441 | MCM9441; BRCA1, MSH2, MSH6, MLH1, ATM, BLM, RAD50, MRE11, and NBN443 | 28 (R to A); 45 (H to A); 136 (G to E)444 |
| Mre11 | 708 | Nucleus, telomere, chromosome441 | MCM9441,445 | 104 (S to C) in cancer446 |
| Rad50 | 1312 | Nucleus, telomere, chromosome441 | MCM8 and MCM9441; BRCA1447; MSH2, MSH6, MLH1, ATM, BLM, RAD50, MRE11, and NBN443 | 94 (I to L), 127 (V to I); 191 (T to I), 193 (R to W), 224 (R to H), 315 (V to L), 469 (G to A)448 |
| MDC1 | 2089 | Nucleus, chromosome449 | MRE11, RAD50, and NBN; CHEK2, the BRCA1-BARD1 complex, SMC1A and TP53BP1, ATM and FANCD2450,451 | 58 (R to A) and 1840 (K to R)451 |
| 53BP1 | 1972 | Nucleus452, chromosome453 | p53/TP53454; H2AFX438; CHEK2455; RIF1456; PAXIP1, IFI202A, and SHLD2457 | 6, 13, 25, 29, 105, and 166 (S to A) |
| BRCA1 | 1863 | Nucleus458, cytoplasm459 | BARD1, UIMC1/RAP80, ABRAXAS1, BRCC3/BRCC36, BABAM2, and BABAM1/NBA1460,461; RBBP8462; CHEK1, CHEK2, BAP1, BRCC3, AURKA, UBXN1, and PCLAF463; H2AFX436 | 26 (I to A)464; 308 (S to N)465; 1143 (S to A) and 1280 (S to A)466; 1692 (D to N) and 1749 (P to R)467 |
aAvailable from https://www.uniprot.org/
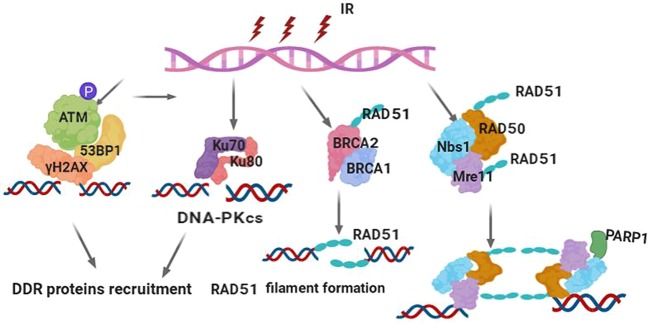
Fig. 3.
Damage sensors and their functional complexes in response to DNA double-strand breaks. (1) Upon DSB occurrence, the core histone protein variant H2AX is instantaneously phosphorylated on its S139 position to form γH2AX foci, which can be detected at the DSB site. γH2AX provides a platform to recruit DDR proteins, such as 53BP1, MDC1, and ATM, to DSBs to initiate DDR signal transduction. (2) DNA-dependent protein kinase (DNA-PK), composed of Ku70, Ku80, and the catalytic subunit DNA-PKcs, is a classical DSB-sensing and -binding complex. DSB binding by DNA-PK protects the broken DNA end from degradation by endogenous nucleases; on the other hand, it recruits and activates the downstream components in the NHEJ pathway of DSB repair. (3) BRCA1 and BRCA2 are key proteins involved in DSB binding and initiating the HR pathway and later repair processing. BRCA2 directly recruits RAD51 to sites of DNA damage through interaction with conserved BRCT motifs to stabilize the RAD51 nucleoprotein filament on the ssDNA end of DSBs. Following end resection of the DSBs, BRCA1 activates RAD51 to promote gene conversion of homologous recombination. (4) The MRN complex (Mre11-Rad50-Nbs1) is the primary sensor of DSBs and localizes to damage sites to initiate end resection and HR processing. The MRN complex also promotes the recruitment and activation of ATM and PARP-1. PARP-1 produces poly(ADP-ribose) polymers and extends DNA damage signaling
Signaling pathways of DDR and repair
IR-induced DNA damage repair
IR kills cancer cells via the induction of DSBs in cancer cell genomic DNA, resulting in genomic instability, apoptosis, cell cycle checkpoint alteration, or postmitotic death. During IR treatment, cancer cells evolve personalized DNA damage repair mechanisms against IR insults for survival66. The induction of DNA mechanisms required to realize the effects of IR has been referred to as “hormesis”157. It has been reported that three different primary pathways evolved to process DSB repair: the HR-based pathway, NHEJ, and alternative end joining (Fig. 4). The goal of these repair pathways is to handle different forms of DNA lesions, eventually achieving DSB removal and maintaining genomic integrity 158.
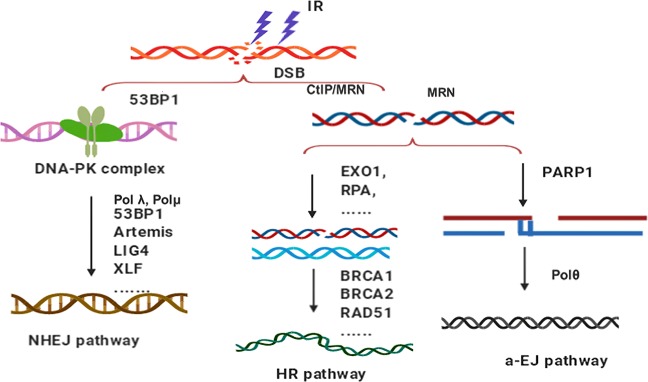
Fig. 4.
The pathways of DNA double-strand break repair. The nonhomologous end-joining (NHEJ) pathway is an error-prone repair pathway that functions through the cell cycle. The homologous recombination pathway is an error-free repair pathway that requires intact homologous DNA as a repair template and is active in the later S and G2 phases. The alternative end-joining (a-EJ) pathway, which repairs DNA double-strand breaks (DSBs), is initiated by end resection that generates 3′ single strand
Understanding the underlying mechanisms by which DNA damage is repaired in cancer cells post IR treatment would facilitate overcoming RR159. For instance, eurycomalactone, an active quassinoid isolated from Eurycoma longifolia Jack, markedly delayed the repair of radiation-induced DSBs in non-small-cell lung cancer (NSCLC) cells160. Koval et al.157 reported that a protective system could be activated among cancer cells in response to IR, leading to increased resistance to subsequent exposure to IR, and moreover, chronic exposure to γ-rays increased the expression of the mus210, mus219, and mus309 genes, even after 56 days, in Canton-S flies. Although the development of genome-wide sequencing techniques has allowed scientists to identify the molecular mechanisms of the radiation-induced adaptive response, including the Notch, tumor growth factor-β, mammalian target of rapamycin, and Wnt signaling pathways, the detailed mechanism of cancer cell defense in IR-induced hormesis remains unclear.
DNA DSB repair pathways
In the history of studying the DSB repair pathway, the HR pathway was the first to be discovered161. The HR pathway was named due to the close vicinity of homologous strands during mitosis. HR is specifically triggered in cells in the later S and G2/M phases162. In the 1980s, the second DSB repair pathway, the DNA end-joining pathway, was discovered. In contrast to HR, NHEJ is triggered in the G0/G1 phase as well as G2/M163. Nevertheless, NHEJ is supposed to be predominant in mammalian cells compared with microorganisms164,165. Since the term homologous was used previously in the HR pathway by radiobiological community, the second discovered pathway was defined as NHEJ166,167. However, some radiobiologists disagree with the naming approach and have suggested the existence of other DSB repair pathways because studies have revealed that in cancer cells with extreme radiotherapy sensitivity, both the HR and NHEJ pathways exist, suggesting that other repair pathways may also be functioning168.
Many studies have indicated that HR is essential for accessing the redundancy of genetic information that exists in the form of sister chromatids or homologous chromosomes when both strands of the DNA double helix are compromised169. When a chromosome is insulted by IR exposure, the DDR cooperates with cellular signaling pathways to maintain genomic stability and ensures cell survival170,171. As shown in Fig. 4, for the HR pathway, the DSB is resected from 5′ to 3′ on one strand of the DSB end, creating terminal 3′-OH single-strand DNA (ssDNA) tails169. In other words, during the process of HR repair, a homologous sequence is needed as the template172, aiming to restore HR accurately. Moreover, both one- or two-ended DSBs can be repaired through the HR pathway, and in particular, messy DNA breaks with covalently attached proteins can be repaired through HR169,173. Compared to NHEJ, HR is more complicated, involving numerous enzymes and proteins, but is more accurate and error free174. In summary, HR is slow, requires a template, is highly accurate, is only initiated at the later S and G2 phases, can repair both one- and two-ended breaks, and can repair protein-blocked ends175,176. The HR pathway for DSB repair has also been used as a genome editing tool. For instance, the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technique is now exploited in genome editing and is considered an incredible opportunity for curing genetic diseases177,178. Furthermore, the HR pathway is associated with RR. A recent study by Jin et al.179 found that Deinococcus shows high resistance to IR exposure due to a combination of passive and active defense mechanisms, such as self-repair of DNA damage through the HR pathway. Lopez Perez et al.180 reported that glioblastoma cancer cells exposed to carbon ions initiated the HR repair pathway with strong and long-lasting cell cycle delays, predominantly in G2, with a high rate of apoptosis. Many cancer cells aberrantly express the cancer/testes antigen HORMAD1. Knockout of HORMAD1 in cancer cells resulted in increased sensitivity to IR treatment, and the HR-mediated repair pathway targeting IR-induced DSBs was attenuated in HORMAD1-knockout cancer cells181. In addition, the cysteine protease cathepsin B contributes to RR by enhancing HR in glioblastoma182.
The basic mechanism of the CRISPR-Cas9 editing approach is to induce a site-specific DSB via bacteria, and the selection of the DSB repair pathway dictates the outcome of the editing183. That is, imprecise repair via the NHEJ pathway contributes to insertion or deletion mutations at the break sites; by contrast, repair via the HR pathway enables activation of the recombination machinery, which consequently deals with DNA segments or corrects pathogenic mutations184,185. The initiation of the HR pathway primarily occurs at the DNA break location, which function as ssDNA that can be used for locating a homologous dsDNA sequence. This sequence can then be used as a template for largely accurate repair. Meanwhile, extended DNA end resection contributes to nonligatable DNA breaks and hampers end joining; consequently, DNA end resection is dominant186–188. Hence, HR is initiated only when a repair template exists, which can limit the potential for illegitimate recombination. In general, misregulation of the HR repair pathway is critical for the generation of genome rearrangements in numerous cancers. It is important and necessary to further elucidate the details of HR, the roles of the relevant proteins, and how these proteins are regulated. We believe this will be an exciting direction in the future.
During the process of NHEJ, Ku first moves to and binds with DNA ends, and the binding shape is similar to a ring encircling the duplex DNA. This binding shape avoids DNA end degradation and recruits other proteins, such as DNA-dependent protein kinase, catalytic subunit (DNA-PKcs). After ligation of the broken DNA ends by the XRCC4-XLF complex and DNA ligase IV (LIG4), DNA-PKcs tethers with the Ku70/Ku80 ends172. NHEJ also plays an important role in cancer cell RR. Compared to HR, NHEJ generally has a rapid response, is template-independent, is often mutagenic, is cell cycle-independent, can only repair two-ended breaks, and cannot repair protein-blocked ends189,190. Bylicky et al.191 reported that increased expression of Ku70 may be a key factor for RR in normal human astrocyte cells; following X-ray radiation, the cells displayed a robust increase in the expression of NHEJ repair pathway-related enzymes within 15 min of radiation. Mu et al.192 reported that mangiferin induces sensitization of glioblastoma cells to radiotherapy via inhibition of the NHEJ repair pathway through regulation of various proteins, such as phosphorylation of ATM, 53BP1, and γH2AX. Wang et al.193 found that the lncRNA LINP1 facilitated DNA damage repair by decreasing the levels of cleaved caspase-3 and poly[ADP-ribose] polymerase (PARP) in response to IR and decreased the radiosensitivity of cervical cancer cells through the NHEJ pathway. These data show that the NHEJ repair pathway plays critical roles in controlling RR.
In cancer cells, both HR and NHEJ are important pathways for repairing DSBs caused by IR insult. For instance, the MEK1/2 (mitogen activated protein kinase kinase 1/2) inhibitor GSK212 mediates radiotherapy sensitivity by functionally repressing both HR and NHEJ, leading to delayed DNA repair and the persistent increased expression of γH2AX194. Thus, in clinical applications, upregulation of DNA repair pathways is recognized as a primary acquired mechanism through which cancer cells may become RR. Accordingly, radiotherapy sensitization strategies functioning via inhibition of IR-induced DNA repair and functional downregulation of the activity of both the HR and NHEJ pathways are expected to be applied clinically to control cancer. Figure 4 illustrates the repair pathways for DNA DSBs.
Activation of cell cycle checkpoints
The cell cycle is essential for cell growth, proliferation, and reproduction. The cell cycle allows cellular components to be replicated and delivered to the next generation of cells195. The cell cycle is a complex process that involves a large number of regulatory proteins, including cyclin family proteins, cyclin-dependent kinases (CDKs), CDK inhibitors (CKIs), including Ink4 family members (p15, p16, p18, and p19) and Cip/Kip family members (p21, p27, and p57), CDC25 isoforms, p53 family proteins, and MDM2196. Over the past few decades, studies of the cell cycle have attracted extensive attention in the scientific community. Generally, in eukaryotes, the cell cycle can be divided into four phases, termed G1 (the first gap period), S (synthesis, the phase in which DNA is replicated), G2 (the second gap period), and M (mitosis)197. Cell cycle checkpoints, which define the end product of a molecular regulatory pathway or signaling cascade, ensure an ordered succession of cell cycle events, and when perturbed, lead to cell cycle arrest198; these checkpoints are critical for protecting cells from progressing into the next phase of the cell cycle before prior molecular events, such as DNA damage and spindle structure disruption, have been resolved199. If premature entry into the next cell cycle phase occurs without checkpoint review, catastrophic consequences or cell death may occur200. Cell cycle arrest is caused by depletion of some key proteins regulating this process. The cell cycle can be thought of as similar to a wheel, while the cell cycle checkpoints are the spokes of the wheel. The running of the wheel normally can maintain the doubling of cellular components and their accurate segregation into the next generation of cells. The spokes of the wheel are the regulators of the cell cycle and play an essential role in the function of checkpoints.
Cell cycle checkpoints are classified as DNA structure checkpoints (DSCs, or DDCs) and spindle assembly checkpoints (SACs). IR-induced DNA damage is one of the major triggers for the activation of a number of DNA structure checkpoints, leading to cell cycle arrest at various points in G1, S, and G2/M201,202. The SAC functions in the mitotic phase. In summary, IR-induced DSBs are a key signal for activation of cell cycle arrest at several cell cycle stages: termed G1/S arrest, S-phase arrest, G2/M arrest, spindle checkpoint arrest, and M-phase arrest.
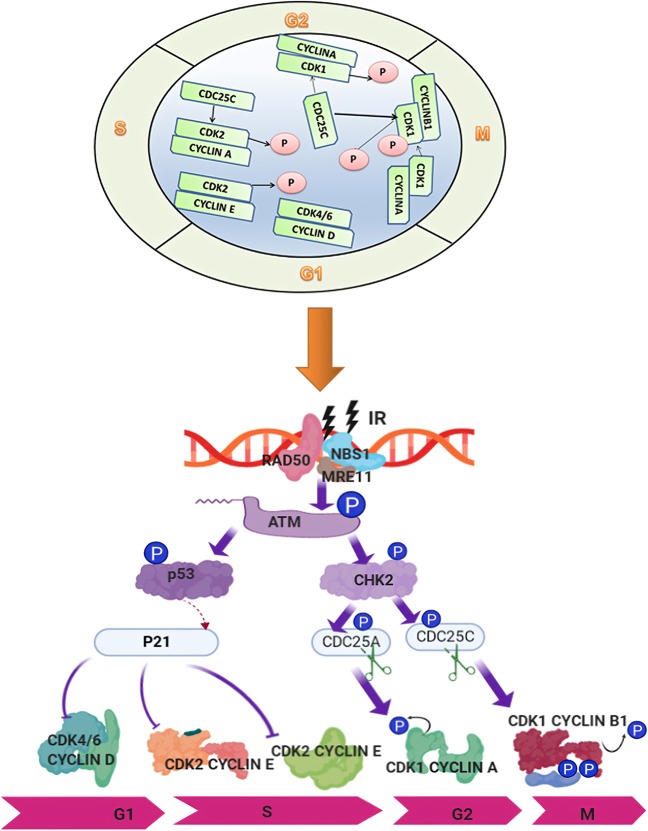
In G1/S arrest, cyclin D recruits CDK4 or CDK6 to form the cyclin D-CDK4/6 complex, which phosphorylates pRB, leading the transcription factor E2F to be released from pRB and to activation of cyclin E transcription. Cyclin E combines with CDK2 to form a complex, further phosphorylating pRB in a positive feedback loop and enhancing S-phase activities, promoting the transition from G1 to S phase203–206. However, exposure to IR may contribute to interruption of the G1/S transition, resulting in S-phase arrest. In theory, G1/S arrest would give cells with radiation exposure more time to perform DNA damage repair207–209. Previous studies have indicated that p53, a famous transcription factor, regulates the cell cycle210,211, especially by monitoring G1 and G2/M checkpoints195. G1 arrest has been reported to be associated with p53 status. Nagasawa et al.212 revealed the absence of G1/S arrest in cancer cells expressing normal p53 synchronized by mitotic selection following irradiation. Fabbro et al.213 found that p53 was phosphorylated and regulated by a series of proteins. First, BRCA1 is phosphorylated at two sites, Ser1423 and Ser1524, based on the regulation of ATM/ATR (ATM and Rad3-related), and then, ATM/ATR is activated by phosphorylation of BRCA1 to phosphorylate p53 at the Ser15 site. Consequently, phosphorylation of p53 serves to monitor G1/S arrest by inducing p21, which is reported to be a CDKi. Yoon et al.214 compared the G1 population difference post IR between colon cells with p53 (+/+) or without p53 (−/−) expression, and the results illustrated that the G1 population was significantly abolished in p53 (−/−) cancer cells compared with that in p53 (+/+) cancer cells post IR. They also found that KLF4 mediated p53 activation to control G1/S arrest following irradiation, indicating that p53 regulation in the IR response in cancer cells is complex and that p53 is a key factor in the process. Thus, recovery or activation of p53 could be a strategy for overcoming RR. Jiang and Wang215 showed that downregulation of mitochondrial transcription factor A increased the radiotherapy sensitivity of cancer cells through the p53 signaling pathway. It has now been confirmed that when DNA damage occurs in G1 cells, the G1/S checkpoint can be triggered via at least two signaling pathways, the ATM/p53/p21 and the ATM/CHK2/CDC25C pathways216. For instance, post irradiation, ATM is activated, and ATM phosphorylates p53 and MDM2, promoting dissociation of p53 from MDM2 and inhibiting p53 translocation from the nucleus to cytoplasm; on the other hand, CHK2 is activated, which phosphorylates and stabilizes p53, and the increased level of p53 triggers the transcription of downstream genes such as p21, contributing to G1/S arrest217–219. Compared to the ATM/p53/p21 pathway, the ATM/CHK2/CDC25C pathway induces rapid signaling in response to DNA damage220. G1/S arrest is induced when CDC25C is degraded via ATM/CHK2 after IR-induced DNA damage 221–223.
During the IR-induced cell injury response, S-phase arrest is activated to inhibit DNA synthesis224. Various patterns, including DSBs, DNA crosslinks, and DNA adducts, can trigger S-phase arrest195,225. Deficiencies in S-phase checkpoints accelerate DNA synthesis under the condition of DSBs, an abnormal phenomenon that can occur in cells from patients with ataxia–telangiectasia (A–T), Nijmegen breakage syndrome, or other chromosome syndromes225,226. According to previous studies, ATM mediates S-phase checkpoints via three parallel signaling pathways: ATM/CHK2/CDC25a/CDK2, ATM/MRN/SMC1, and ATM/MRN/RPA225,227. In addition, post IR, ATM phosphorylates both Nbs1 and CHK2, leading to S-phase checkpoint activation. Ultimately, the distinct steps of DNA replication are suppressed227. Regulation of the S-phase checkpoint is complex, involving multiple pathways; thus, determining whether cancer cells are dependent on one, both, or neither of these intra-S-phase checkpoints in response to IR is necessary.
G2/M arrest prevents cells from entering the M (mitosis) phase in the presence of DSBs224. G2/M arrest often occurs at 0.5 to 4 h post exposure to IR in mammalian cells and then resolves228. The higher the IR dose, the more obvious the G2/M arrest, and the more delayed the recovery effect; sometimes recovery is impossible, and cell death occurs as a result of mechanisms such as mitotic catastrophe. However, it should be noted that the level of cell cycle arrest and recovery time differ in different cell lines. In addition, a deficiency in some genes involved in regulating G2/M arrest, including PLK1, ATM, and CHK1, alters the cell cycle response to IR-induced DNA damage229,230. G2/M arrest is also associated with RR. Gogineni et al.231 found that when meningioma cells were subjected to IR, G2/M arrest could be triggered quickly, and the key event in the underlying mechanism was the phosphorylation of CHK2, CDC25C, and CDC2; this phosphorylation interferes with CHK2 activation and the cyclin B1/CDC2 interaction, resulting in permanent arrest followed by apoptosis. Aninditha et al.232 compared the effects of heavy ions and photons on malignant melanoma cell G2/M arrest and found that heavy ions caused a greater increase in G2/M arrest than photons, showing that heavy ions have better properties for improving RR for malignant melanoma cells than photons. A study conducted by Wang et al.233 showed that IR increased colorectal cancer cell sensitivity to the melatonin by triggering G2/M arrest as well as downregulating the expression of ATM, a key mediator in DSB repair. Peng et al.228 showed that in response to IR, radioresistant cells exhibited a recoverable G2/M phase during a prolonged cell cycle. These data validate the potential of targeting G2/M-related proteins involved in the response to IR to control RR in cancer patients. Figure 5 illustrates the regulation of cell cycle checkpoint-related proteins in the response to IR.
Fig. 5.
Functional complexes of cyclins and cyclin-dependent kinases (CDKs) and the signaling pathways involved in the regulation of cell cycle checkpoints in response to IR-induced DNA damage. CDK4/6/cyclin D promotes progression through the G1 phase. In late G1, the active CDK2/cyclin E complex promotes the G1/S transition. The CDK2/cyclin A complex promotes progression through S phase. The CDK1/cyclin A complex regulates progression through the G2 phase in preparation for mitosis. The G2/M-phase transition is initiated and promoted by the CDK1/cyclin B complex. The activity of CDK1/cyclin B is tightly maintained by the CDC25C phosphatase. Following DSB induction by IR, ATM is activated by the MRN complex, which then phosphorylates p53. Activated p53 transactivates the expression of p21Cip1, which inhibits CDK2, consequently inducing G1/S arrest. On the other hand, ATM phosphorylates and activates CHK2, which in turn phosphorylates and inactivates CDC25C; the latter is then cytoplasmically sequestered by 14-3-3 proteins. Consequently, the inhibitory phosphorylation of CDK1 by Wee1 and Myt1 on Tyr15 and Thr14 is maintained, and G2/M arrest is induced
Targeting DNA damage repair to sensitize cancer cells to radiation
DSBs generated by radiotherapy are the most efficient molecular events damaging and killing cancer cells; however, the inherent DNA damage repair efficiency of cancer cells may cause cellular resistance and weaken the therapeutic outcome. Genes and proteins involved in DSB repair are targets for cancer therapy since their alteration, interaction, translocation, and regulation can impact the repair process, making cancer cells more resistant or more sensitive to radiotherapy. Thus, targeting DNA damage repair as a method to sensitize cancer cells to radiotherapy is a promising therapeutic strategy for the precise and effective treatment of cancer patients. In this section, recently reported literature regarding some of the most important DDR-associated proteins involved in RR is reviewed and discussed.
Targeting DNA-PKcs
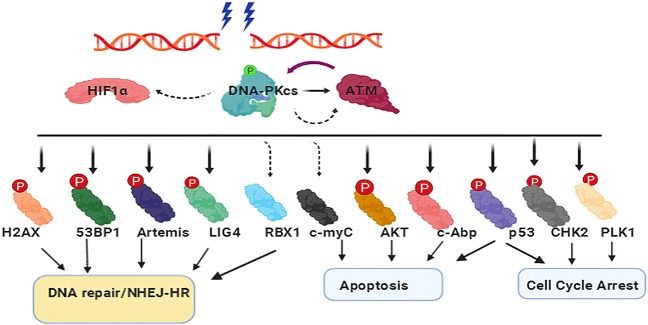
DNA-dependent protein kinase (DNA-PK) is first introduced in this section due to its importance and because it has been intensively studied previously in the NHEJ pathway.234. It has been confirmed that DNA-PKcs can identify DSBs post-IR insult and facilitate “messy” broken end processing and DNA ligation by recruiting the proteins responsible for DNA damage repair processing and ultimately ligating the broken DNA ends235,236. DNA-PKcs was first discovered due to the observation that dsDNA can modulate the phosphorylation of a series of proteins237. In early published reports, DNA-PKcs was associated with repairing DSBs through the NHEJ pathway; however, with further study, it was illustrated that DNA-PKcs has multiple functions, including selection of NHEJ and HR repair pathways238–241, regulation of cell cycle checkpoints242–245, and maintenance of telomeres245–247. As one of the largest family members of the PIKK (phosphatidylinositol 3-kinase-related kinase) family, DNA-PKcs consists of 4128 amino acids248. During the past few decades, extensive studies have been conducted to reveal how DNA-PKcs works in DDR pathways71,249. Briefly, DNA-PKcs and other proteins, including Artemis and XLF, are recruited by Ku to form a DNA-PK functional complex250. Then, DNA-PKcs phosphorylates components of the NHEJ machinery, and autophosphorylation or ATM-catalyzed phosphorylation at Thr2609, Ser2056, and Thr2647 allows for DNA end processing251–253. Cells harboring decreased levels of DNA-PKcs showed increased sensitivity to IR compared to control cells254. In the HR pathway, replication protein A coupled with the phosphorylation of p53 affected HR in a mechanism mediated by DNA-PKcs 255.
Our research team has focused on studying DNA-PKcs for almost three decades. We have reported multiple essential DNA-PKcs functions and mechanisms in the IR-induced DDR. For instance, cyclin B1 ubiquitination was activated and its protein stability was affected by DNA-PKcs via the CDH1-APC pathway256. We also found that radioresistance may be a result of the effect of c-Myc on ATM phosphorylation and DNA-PKcs kinase activity257. Furthermore, DNA-PKcs associates with PLK1 and contributes to chromosome segregation and cytokinesis242. We demonstrated the effects of anti-DPK3-scFv on radiosensitization by targeting DNA-PKcs258. Another discovery was that Ku could recruit DNA-PKcs and CHK2245, and a dominant role for DNA-PKcs in regulating H2AX phosphorylation in the post-IR-induced DDR was identified259. Moreover, suppression of DNA-PKcs changes multiple signal transduction-associated genes at the transcriptional level and eventually affects cell proliferation and differentiation260. Our studies further identified that DNA-PKcs is a critical component of DNA damage repair pathways.
To date, DNA-PKcs is the best known regulator/mediator of the IR-induced DDR; furthermore, it has been implicated as an emerging intervention target in cancer therapy, particularly in radiotherapy or genotoxic chemotherapy261. More recently, the targeting of DNA-PKcs has been used in cancer radiotherapy. According to Liu et al.262, suppression of DNA-PKcs was markedly abrogated by the IR-induced transcription factor hypoxia-inducible factor-1α (HIF-1α), leading to IR-induced decreases in migration and invasion and enhanced radiotherapy sensitivity in glioblastoma. Mamo et al.263 reported that inhibition of DNA-PKcs sensitized human osteosarcoma cells in response to IR. Targeting DNA-PKcs with various inhibitors has been reported to be effective for potentiating radiotherapy and has been proposed as an effective strategy to improve cancer patient outcomes264. In recent decades, significant progress has been made in the development of large amounts of DNA-PKcs inhibitors from basic experiments, and many treatments have already been tested in clinical trials or applied in clinical therapy. Wortmannin was the first identified DNA-PKcs inhibitor265, but it lacks specificity, and its in vivo toxicity makes it difficult to use in clinical applications. Another nonselective inhibitor is LY294002, which has a similar structure to wortmannin266. Davidson et al.266 summarized a series of DNA-PKcs inhibitors in their review, which included LY294002, NU7026, NU7441, IC86621, IC87102, IC87361, OK-1035, SU11752, vanillin, NK314, and IC486241. LY294002 is a competitive inhibitor that binds reversibly to the kinase domain of DNA-PK267. Compared to nonselective inhibitors, NU7026 is more selective for DNA-PKcs268. NU7441 is a strong inhibitor of DNA-PKcs. It could improve RR in liver cancer cells by participating in DDR pathways and activating cell cycle arrest269. IC86621, IC87102, and IC87361 are inhibitors based on the LY294002 structure. These inhibitors promoted increased sensitization to IR and decreased repair of spontaneous and IR-induced DSBs270. Vanillin, derived from vanilla pods, showed selective inhibition of DNA-PK and specifically affects NHEJ271. In our laboratory, we identified a vanillin derivative, BVAN08, as a DNA-PKcs inhibitor that can efficiently induce autophagic cell death and mitotic catastrophe in radioresistant cancer cells. In addition to DNA repair inhibition, cancer cell killing by BVAN08 is related to destruction of the c-Myc oncoprotein and G2/M-phase function272,273. In addition to these inhibitors, several novel inhibitors have been recently discovered and published. M3814, an oral DNA-PK inhibitor, showed preclinical activity274. Sun et al.275 demonstrated that M3814 effectively blocked IR-induced DSB repair. AZD7648 is reported to be a potent and selective DNA-PKcs inhibitor, enhancing radiation sensitivity276. VX-984, a novel drug that was developed as a selective inhibitor of DNA-PKcs, enhanced cell death during radiotherapy277. Doxycycline, the first US Food and Drug Administration (FDA)-approved DNA-PK inhibitor, reduced DNA-PKcs protein expression by ~15-fold and functioned as a radiosensitizer in breast cancer cells278. A recent study showed that phosphorylation of H2AX and KAP1 could be facilitated by DNA-PKcs, resulting in chromatin decondensation and quickly recruiting the DDR complex to DNA damage sites 279.
Targeting ATM/ATR
ATM was discovered during a clinical case observation; that is, in 1967, Gotoff et al.280 reported that a patient with a rare inherited autosomal-recessive genetic A–T condition exhibited immunodeficiency. A previous study showed that patients with A–T were more sensitive to radiotherapy than patients without A–T281. Subsequently, a study indicated that not only G1/S arrest but also G2/M arrest could not be activated in A–T cells after IR exposure282. Later work identified the ATM gene, with a transcript size of 12 kb283. ATR, originally discovered in budding yeast, has been found to exhibit S and G2 checkpoint deficiency284. Later work identified the C-terminal phosphoinositide 3-kinase (PI3K)-like kinase domain while cloning the MEC1 gene because the sequence was similar to Mec1/Esr/Sad3 and fission yeast Rad3285. In 1996, Bentley et al.286 identified that ATR could functionally enhance esr1-1 radiation sensitivity in Saccharomyces cerevisiae. With the discoveries of ATM and ATR, multiple lines of scientific enquiry converged.
Both ATM and ATR are large polypeptides, and their domain organizations are similar, but their structural features are generally different126. One of the main roles of ATM and ATR is that they can phosphorylate serine or threonine residues according to some biochemical reports287. ATM and ATR share certain substrates and have some overlapping functions. ATM is activated and recruited to DSB sites by the MRN complex, which serves as a DNA damage sensor, while ATR is activated and recruited to DSB sites with its stable binding partner ATRIP (ATR-interacting protein)288. Previous studies have demonstrated that ATM is a master regulator of the cellular response to DSBs289. Indeed, the major ATM function is that it can initiate a cascade for the DSB signaling response, resulting in the phosphorylation of almost hundreds of substrates when cells are undergoing the DDR290. For instance, ATM activates CHK2 kinase and phosphorylates multiple sites, triggering apoptosis, and cell cycle arrest, and deficiency of ATM-mediated signaling reactions causes sensitization of cells to IR291. A recent study demonstrated that resting peripheral blood lymphocytes were more sensitive to radiotherapy when ATM was inhibited than when it functioned normally. Meanwhile, it was found post IR that ATM phosphorylation activity was decreased through stimulation of CD3/CD28292. Moreover, the synergistic relationships among three key proteins, ATM, ATR, and DNA-PKcs, have been found to present different functions with low- or high-dose IR. When subjected to low-dose IR, the G2 checkpoint is tightly regulated by ATM and ATR mainly through interactions with another cell cycle mediator, CHK1. However, when subjected to high-dose IR, the ATM and ATR complex becomes relaxed. Both ATM and ATR can affect the G2 checkpoint independently, resulting in DSB end resection293. Thus, some experts, such as Blackford and Jackson126, suggest that ATM, ATR, and DNA-PKcs react with each other, forming a complex and mediating DDR.
As key mediators of the DDR, the ATM and ATR kinases have been suggested to have extreme potential for improving radiotherapy outcomes because of their abilities to promote DDR and mediate cell cycle arrest294. To date, reported ATM inhibitors include caffeine, wortmannin, CP-466722, KU-55933, KU-60019, and KU-559403. Caffeine was first reported to sensitize cells to IR in 1995, and an increased radiosensitizing effect was observed in cells with p53 deficiency295. Wortmannin targets both ATM and DNA-PKcs to increase cell radiosensitivity. KU-55933 is potentially a selective ATM inhibitor. It confers marked sensitization to IR296. KU-60019 is an analog of KU-55933, inhibiting ATM downstream signaling and sensitizing cells to IR in vitro297. Glioblastoma-initiating cell-driven cancers with low p53 expression and high PI3K expression might be effectively radiosensitized by KU-60019298. Reported ATR inhibitors include schisandrin B, NU6027, NVP-BEZ235, VE-821, VE-822, AZ20, and AZD6738. Schisandrin B, reported in 2009, is a natural extract from the medicinal herb Schisandra chinensis. In human lung cancer cells treated with ultraviolet light, it could inhibit the activity of ATR phosphorylation substrates and abrogate G2/M cell cycle checkpoints299. NU6027, a nonselective inhibitor, has been shown in a few cancer cell lines to have potential for improving IR-induced RR. NVP-BEZ235, another reported potential and effective inhibitor, caused marked radiosensitivity in Ras-overexpressing cancers. VE-821 has been shown to be a potent ATR inhibitor by inhibiting phosphorylation of the ATR downstream target CHK1 at Ser345 in the colorectal cancer cell line HCT116. As a key inhibitor, VE-822 has been found to have potential because of its ability to increase persistent DNA damage. VE-822 was also reported to decrease HR for cancer cells post IR300, suggesting that this inhibitor is promising for overcoming RR in patients with advanced pancreatic ductal adenocarcinoma. AZ20 is another inhibitor of ATR301. A phase I study of AZD6738 was conducted to analyze the tolerability, safety, and biological effects of palliative radiotherapy in cancer patients in the United Kingdom in 2019302. These ATM and ATR inhibitors were identified based on a broad range of preclinical studies and extensive literature; however, the potential for increased normal tissue toxicity is likely to be an important concern, and identifying their selective effects in concert with radiotherapy will require further investigation.
Targeting DNA LIG4
DNA LIG4 is an essential DNA repair component in the radiation-induced NHEJ pathway303. Commonly, LIG4, XRCC4, and Cernunos-XLF are recruited to the breakage site and temporarily attach to the ends of the DNA to ensure ligation of the DSB304. LIG4 deficiency leads to a rare primary immunodeficiency called LIG4 syndrome305. Patients who have been diagnosed with LIG4 syndrome present increased sensitivity to radiotherapy, but they also have an increased risk of neurological abnormalities and bone marrow failure, as well as increased susceptibility to cancer306. The symptoms of LIG4 syndrome show that LIG4 is vital in the DDR. Numerous studies have reported that mutations in LIG4 confer clinical radiosensitization. Riballo et al.307 indicated that mutation of LIG4 impaired the formation of an adenylate complex in addition to reducing the rejoining activity. Furthermore, healthy participants with the rs1805388 polymorphism of LIG4 were more sensitive to radiation based on γH2AX foci analysis than healthy participants without this polymorphism308. Nevertheless, a Lig4−/−p53−/− cell line had a higher sensitivity to high-LET radiation than a Lig4+/+p53−/− cell line, suggesting that LET-induced DNA damage is partially repaired through LIG4309 McKay et al.310 screened tissues from a unique bank of samples from radiosensitive cancer patients for expression defects in major DSB proteins such as LIG4. LIG4 and RCC4 proteins showed reduced expression in addition to a corresponding reduction in both gene products at the mRNA level. The impact of LIG4 on RR may be due to the following reported molecular mechanisms. The activity of LIG4 is regulated by other proteins, such as XRCC4, which is the key contributor to the stabilization of LIG4311. In addition, DNA-binding protein-1 negatively regulates DNA repair processes by downregulating the expression of LIG4312. Indeed, a LIG4 peptide was shown to be a substrate of DNA-PK in vitro; a phosphorylation site for DNA-PKcs is present at Thr650 in human LIG4, and LIG stability is regulated by multiple factors, including negative regulation by DNA-PK 313.
Screening for inhibitors of LIG4 offers a chance for target-based drug discovery to design RR drugs. Tseng et al.314 conducted a screen of 5280 compounds and found that rabeprazole and U73122 could specifically block the adenylate transfer step and DNA rejoining to inhibit IR-induced DNA damage repair by targeting LIG4. SCR7, identified by Srivastava et al.315, blocks LIG4-mediated joining by interfering with its DNA binding. NU7026 affects the radiosensitivity of wild-type LIG4 mouse embryonic fibroblasts316. Although inhibitors of LIG4 are considered potential anticancer drugs, they are likely not effective in cancer cells with mutations, which may affect radiotherapy outcomes. Thus, in the future, more investigations regarding LIG4 functions in the IR-induced DDR need to be conducted.
Targeting PARP-1
PARP-1 is the most extensively studied nuclear enzyme of the PARP superfamily317. As reported previously, PARP-1 has been suggested to be a key regulator of DNA damage repair318. In the response to DNA damage, poly (ADP-ribosyl)ation (PARylation) of proteins is an initial reaction319. For instance, in the case of DNA damage, PARP-1 recognizes NAD+ as a major substrate320. PARylation of proteins post translation is suggested to provide a local signal of DNA damage because of the existence of poly(ADP-ribose)-binding domains, and DDR factors can regulate the functions of relevant proteins321. Furthermore, PARP-1 has been extensively studied, as it is a widely known regulator of DNA damage repair, particularly DSB repair 322.
Mechanistically, PARylation targets include nuclear DDR proteins, such as DNA-PKcs323 and PARP-1 itself. Since PARylated proteins can associate with negatively charged PAR, PARylated proteins can interact with DNA as well as regulate DNA damage as a signal324,325. Moreover, recent evidence has shown that PARP-1 has the potential for catalyzing the heterodimer formed with XPC-RAD23B and free PAR, suggesting the critical role of PARP-1 in IR-induced DNA damage 319.
Based on research indicating that inhibition of PARP-1 might sensitize cancer cells to radiotherapy326,327, the function of PARP-1 in DNA repair has been utilized in targeted radiotherapy. Currently, targeted radionuclide therapy with PARP-1 is a novel approach for cancer therapy328. Jannetti et al.329 reported that 131I-labeled PARP-1 therapy showed high potential for treating mice with glioblastoma, as the mice showed significantly longer survival than mice that received control vehicle in a subcutaneous model. Inhibitors of PARP-1 have also been developed to enhance the sensitivity of cancer cells to radiotherapy. The first identified inhibitor was nicotinamide, ~30 years ago330. After that, several generations of PARP-1 inhibitors were identified. Many of these inhibitors have been shown to enhance the anticancer efficacy of DNA-damaging agents such as IR331,332. For inhibitors of PARP (PARPi), their structures are similar to nicotinamide. PARPi mainly perform the following two functions. One is to inhibit PARP-1 catalytic activity, and the other is restrict PARP-1. The aims are typically to either inhibit PARylation or suppress PARP-1 release333,334. However, some older PARP-1 inhibitors showed limitations, such as nonselectivity and nonspecificity, in clinical radiotherapy331. Recently, some more specific and effective novel inhibitors have been developed. Ryu et al.335 suggested that the PARP-1 inhibitor KJ-28d might enhance the sensitivity of NSCLC to radiotherapy. Olaparib was the first PARP inhibitor approved (in December 2014) for cancer therapy by the FDA (https://www.astrazeneca.com/media-center/press-releases/2014/lynparza-approved-us-fda-brca-mutated-ovarian-cancer-treatment-19122014.html#) and by the European Union (https://www.astrazeneca.com/media-center/press-releases/2014/lynparza-approved-european-union-brca-mutated-ovarian-cancer-treatment-18122014.html#). It was approved for use in patients with advanced ovarian cancer and BRCA mutations. In January 2016, the US FDA further granted the Breakthrough Therapy Designation to olaparib for treating patients with metastatic-castration-resistant prostate cancer carrying BRCA1/2 or ATM mutations (https://www.astrazeneca.com/media-center/press-releases/2016/Lynparza-Olaparib-granted-Breakthrough-Therapy-Designation-by-US-FDA-for-treatment-of-BRCA1-2-or-ATM-gene-mutated-metastatic-Castration-Resistant-Prostate-Cancer-28012016.html#). Generally, cancer cells lacking either of the tumor suppressors BRCA1 and BRCA2, which are key components in the HR pathway of DSB repair, are selectively sensitive to PARP family inhibitors. Mechanistically, SSBs are primarily repaired by PARP-1. However, when DNA damage is caused by PARPi, inhibiting PARP-1 may not be lethal because there are still other repair pathways that function in the DDR, such as HR. However, the absence of BRCA1/2 results in a deficiency of HR activity, and cytotoxicity is present because DNA lesions caused by PARPi cannot be repaired due to the lack of HR activation336. Bourton et al.337 demonstrated that compared to normal BRCA cells, BRCA1+/− lymphoblastoid cells treated with olaparib, followed by IR exhibited decreased BRCA1 protein levels and increased apoptosis, resulting in radiation hypersensitivity; these results suggest that the combination of a PARP-1 inhibitor with radiotherapy has clinical relevance in treating BRCA1-associated cancers. AZD2281 is also an effective radiosensitizer for carbon-ion radiotherapy, indicating that PARP-1 has a wide therapeutic range in combination with LET radiation by blocking the DNA damage repair response327. The PARP-1 inhibitor ABT-888 enhanced radiosensitizing effects in hepatocellular carcinoma338. In cell experiments, Mk-4827, a PARP-1/2 inhibitor, promoted lung and breast cancer cell sensitivity to radiation339. Some inhibitors have been implicated to improve therapy sensitivity or inhibit cancer recurrence in cancer patients in the clinic. For example, niraparib is used in patients with ovarian cancer340. Niraparib is also found to inhibit the DDR in cancer cells, leading to an initial sensitization of cancer cells to radiotherapy341. Collectively, although PARP-1 inhibitors have been identified and tested after clinical trials and their function in enhancing the response of cancers to IR has been documented, the underlying mechanisms of radiotherapy sensitization by these inhibitors remain to be fully elucidated 342.
Targeting HIF-1
The 2019 Nobel Prize in Physiology or Medicine was awarded to three scientists, William Kaelin, Gregg Semenza, and Peter Ratcliffe, for their work on the discovery of HIF-1 as the gene switch controlling the cellular identification of and response to changed oxygen status343. HIF-1 is formed by HIF-1α and HIF-1β344, belonging to the basic helix–loop–helix PER-ARNT‐SIM (bHLH‐PAS) protein family345. HIF-1α becomes stable in response to hypoxia, but is degraded under normal conditions, while HIF-1β is constitutively expressed. Since HIF-1α expression changes under hypoxic conditions, most studies have focused on the effect of HIF-1α on the cellular identification of and response to oxygen levels. There are multiple functional domains in HIF-1α; in particular, the bHLH and PAS domains are linked to dimerization, DNA binding, and oxygen-dependent degradation domains that make HIF-1α more susceptible to proteasomal degradation under normal conditions and transactivation domains that are responsible for the activation of HIF-1 target genes346,347. HIF-1 activation mainly occurs due to hypoxia caused by vascular damage under radiotherapy conditions348, as well as due to reactive oxygen species (ROS) produced from IR insult349. Moreover, IR exposure upregulates glucose availability, promoting HIF-1α translation350. In radioresistant NSCLC cells, PAI-1 was secreted post IR through upregulation of HIF-1α, leading to the radioresistance of adjacent cells in a paracrine manner351. In addition, increased ROS production stabilizes HIF-1α, enhancing cancer-related fibroblasts through glycolysis, leading to the Warburg effect and the secretion of lactate to feed nearby cancer cells, ultimately inducing RR352. As metabolic changes occur in radioresistant cancer cells, several signaling pathways are activated by IR with altered HIF-1 expression353. Presently, investigation of the metabolic mechanisms is ongoing. HIF-1 signaling can also be activated by IR through damage to endothelial cells, producing hypoxia and regulating VEGF (vascular endothelial growth factor) and CXCL12 (C-X-C motif chemokine 12)354. HIF-1 is involved in the mitochondria-mediated RR of cancer stem cells355. In fact, HIF-1 has the potential to improve RR and increase radiotherapy outcomes.
In 2007, in a review by Brown356, it was noted that in cancer therapy, it is important to consider that cancer cells exist under a wide range of oxygen concentrations, which include those concentrations induced by hypoxia and necrosis, and one therapeutic strategy is to selectively induce HIF-1 under hypoxic condition. To this end, a series of HIF-1 inhibitors have been developed in the past decade. Typically, HIF-1-associated inhibitors can be divided into the following categories: adamantyl-based inhibitors, boron-based inhibitors, sulfonamides, moracins, manassantin A and its analogs, chalcones, ring-truncated deguelin derivatives, YC-1-related derivatives, and heterocycles. Among these reported inhibitors, some have been associated with RR. LW6 improved resistance to radiotherapy in hypoxic lung cancer cells by inhibiting the accumulation of HIF-1α357 via inhibition of the mitochondrial respiratory chain. Another HIF-1 inhibitor, PX-478, could enhance pancreatic cancer cell sensitivity to radiotherapy. The potential mechanism may be related to inhibiting cancer cell proliferation and, in particular, activating HIF-1 proangiogenic signaling to reverse RR in hypoxic cancer cells358. YC-1, another potential HIF-1 inhibitor, could reverse RR in A549 lung cancer cells by changing the optical redox (OR) status of cancer cells358. Topotecan was found to alleviate IR-induced brain necrosis in a mouse model359. However, most HIF-1 inhibitors are nonselective and do not directly inhibit HIF-1 protein. Furthermore, the HIF-1 signaling pathway is very complex. More than 1% of genes present high sensitivity to hypoxia. Thus, in future studies, we need to address the following questions: What are the underlying mechanisms of potential selective HIF-1 inhibitors? How do they influence RR: Notably, it should be mentioned that the effects of most HIF-1 inhibitors may be off-target, indicating that other signaling pathways may also be affected by these inhibitors. Hence, during the discovery process of novel selective inhibitors, knowledge of the molecular mechanism of action is essential.
Targeting HDACs
One of the roles of histone deacetylases (HDACs) is to remove acetyl groups from the amino-terminal lysine residues by catalyzing reactions on histone proteins360. HDAC expression is broad, but the highest expression is mainly in the brain, heart, muscle, and testis. According to investigations of the role of HDACs in cancer RR, HDACs act as guardians of IR-induced DNA damage and promote RR361. For instance, HDAC6 knockdown predisposes glioblastoma cancer cells to radiotherapy-induced apoptosis 361.
Most HDAC inhibitors (HDACis) were discovered recently. They can target HDACs, improving RR. Various biological processes can be affected by these inhibitors. Cell growth, apoptosis, DNA repair, and terminal differentiation are the main processes influenced. HDACis have been found to suppress many proteins important in the DDR by downregulating proteins in the HR and NHEJ repair pathways in vitro362. HDAC induces the acetylation of histone and nonhistone proteins and modulates the acetylation of proteins related to DSB repair363. Several HDACis have been approved through clinical trials. They are short-chain fatty acids, benzamides, hydroxamic acids, and cyclic tetrapeptides364. Their approval information can be reviewed on clinical trial websites such as https://www.who.int/ictrp/ and https://clinicaltrials.gov/. Their molecular mechanisms involve enhancing radiation exposure by targeting multiple RR pathway molecules, including EGFR (epidermal growth factor receptor) and AKT365,366. Furthermore, HDACis overcome RR through physical modification of chromatin structure. HDACis may also acetylate heat-shock protein 90 (HSP90), promoting receptor degradation. Most importantly, HDACis are involved in DSB repair through prolonged expression of γH2AX and therefore downmodulate RAD51 and DNA-PK expression, eventually sensitizing cancer cells to IR367,368. In the past decade, several novel HDACis have been studied in cancer cell and animal models. Wang et al.369 reported that curcumin-mediated HDAC inhibition suppressed the radiation-induced DDR and led to elevated DNA damage sensitivity, indicating that curcumin is important for DSB repair. A study by Frame et al.370 demonstrated that trichostatin A conferred radiosensitivity to prostate stem-like cells. In addition, suberoylanilide hydroxamic acid (SAHA) enhanced radiotherapy sensitivity and suppressed lung metastasis in breast cancer, indicating that SAHA may serve as a potential sensitizer for radiotherapy371. Robert et al.363 found that HDACis, such as trichostatin A, resulted in both physical and functional alteration of PARP-1 binding at DSBs, potentially avoiding NHEJ processes in cancer cells and eventually decreasing NHEJ repair.
Targeting CDK1
CDK1 plays an essential role in cell cycle regulation through modulation of the centrosome cycle and mitotic onset, enhancement of the G2/M transition, and regulation of G1 progression and the G1/S transition in combination with different cyclins372. CDK1 activity can be switched off by WEE1- or PKMYT1-mediated phosphorylation373. Once DNA damage occurs by IR, CDK1 is inactivated to stop the cell cycle at the G2 checkpoint to facilitate DSB repair374. CDK1 activation occurs by a series of steps. In brief, it first binds to cyclin B, and then, its phosphorylation of Thr161 is induced by the CAK/CDK7 complex; at the same time, its phosphorylation on Thr14 and Tyr15 sites is inhibited by CDC25, after which the CDK1/cyclin B complex is activated to redirect cyclin-kinase complexes from the cytoplasm to the nucleus375. In fact, because cancer cells usually promote CDK1 activity, this property can be useful for targeted cancer therapy376. Indeed, cells in S phase at the time of radiation exhibit a slowing of DNA synthesis mediated by the ATM/NBS1 and ATM/CHK2/CDC25A/CDK2 pathways377,378. Thus, the regulatory proteins that control cell cycle processes through CDK1 inhibition may be beneficial in radiotherapy.
Raghavan et al.379 tested a new-generation CDK1 inhibitor, AZD5438, in combination with radiotherapy in lung cancer lines, such as A549, H1299, and H460. The results showed that AZD5438 significantly enhanced the radiation sensitivity of lung cancer cells. RO-3306, a CDK1-targeting inhibitor, could promote activation of Bax and induce mitochondrial apoptosis, indicating that inhibiting CDK1 may be effective in acute myeloid leukemia cells380 by enhancing the downstream p53 signaling pathway. Yang and co-workers381 investigated the potential of taurine as an inhibitor of CDK1 and demonstrated that taurine significantly inhibited the IR-induced downregulation of CDK1. Notably, JNJ-7706621, an inhibitor that can inhibit both CDK1 and AURKA/B kinase concomitantly, has been shown to be able to revert radioimmunotherapy resistance in lymphoma cancer cells. This has an intriguing clinical implication: in some TP53-deficient cancers, the potential application of dual protein inhibitors may be a strategy to overcome IR-induced resistance. MEK162 is a nonspecific CDK1 inhibitor that downregulates and dephosphorylates multiple cell cycle checkpoint proteins, including CDK1, CDK2, and Wee1382, leading to a prolonged DNA damage signal in response to IR exposure in glioblastoma cells. In fact, regulation of the cell cycle in cancer cells is complex, and CDK1 can be upregulated or downregulated by multiple cell cycle checkpoint proteins. The regulation of cell cycle proteins in response to IR exposure can be thought to resemble a spider web. Exposure to IR is similar to the moth flying into the spider web; multiple proteins located in this web react immediately, regulating the interaction and mediating the integrity of the web. Thus, compounds that inhibit CDK1-related proteins may also inhibit CDK1 activity. Satyanarayana et al.383 showed that p21, which is upregulated by CDK1, is activated in response to γ-irradiation, resulting in the inhibition of CDK1 for further DNA damage repair. As cancer cells require specific interphase CDKs for proliferation, CDK1 inhibitors may provide a therapeutic benefit against RR.
Targeting Wee1-like protein kinase (Wee1)
Wee1 serves as a negative regulator of cell cycle entry into mitosis at the G2 to M transition. Wee1 could mediate the phosphorylation of CDK1 at Tyr15. Wee1 could prevent the cyclin B1/CDK1 complex from relocating to the nucleus from the cytoplasm prior to the onset of mitosis384,385. Indeed, Wee1 phosphorylates and inactivates the cyclin B1/CDK1 complex in a specific way386. Wee1 activity increases during S and G2 and decreases in M phase when it is hyperphosphorylated387–390. Inhibition of Wee1 activity has been considered a potential avenue for cancer radiotherapy391.
To date, several Wee1 inhibitors have been investigated, some of which are concomitant CDK1 inhibitors, suggesting that Weel is an effective target for sensitizing various types of cancers to radiotherapy392–394. Experimental evidence has demonstrated that Wee1 inhibitor AZD1775 significantly inhibits cancer growth and impairs RAD51 focus formation in response to radiotherapy394. The mechanisms underlying how inhibition of the Wee1 activity by AZD1775, resulting in promotion of the G2/M transition, is to be considered395. In this context, cancer cells, which harbor G1 checkpoint aberrations, could enter premature mitosis; ultimately, mitotic catastrophe would occur because of this unsuccessful DDR388. To date, on the clinical study website (https://clinicaltrials.gov/), there are ~50 clinical studies regarding AZD1775, of which 12 have been completed, 16 trials are recruiting, and 9 have been terminated or withdrawn. Most of these clinical trials have been conducted in various cancers and feature combinations with chemotherapy or radiotherapy. MK 1775, a selective WEE1 inhibitor, sensitizes lung cancer cells to RIDD and apoptosis through suppression of Sirt1396. Human leukemic T cells pretreated with 681641, a Wee1 kinase II inhibitor, have significantly increased sensitivity to radiation. If pretreated with both inhibitors, 681641 and R1-1, a Rad51 inhibitor, further enhanced apoptosis was seen in cancer cells397. PD0166285, another reported WEE1 inhibitor, is a major drug for inducing radiotherapy sensitivity because it can lead cancer cells to fail to perform an IR-induced DDR398. Because of the important role of Wee1 in the G2/M checkpoint along with CHK1/2, using Wee1 inhibitors such as AZD1775 or MK 1775, can relieve G2 arrest, sensitizing cancer cells to radiotherapy.
Targeting serine/threonine-protein kinase (CHK1)
Similar to Wee1 and CDK1, CHK1 is also required for checkpoint-mediated cell cycle arrest and activation of DNA repair in response to DNA damage. The mechanisms of CHK1 regulation are complex and involve multiple steps399. Briefly, during HR, CHK1 is activated by its interaction with RAD51, promoting the intra-S and G2/M cell cycle checkpoints and modulating the cellular response to replication stress400. Increasing evidence illustrates that cancer cell survival and proliferation can be targeted as potential strategies for sensitizing cells to radiotherapy because cell survival and proliferation are significantly affected by CHK1401. In 1996, UCN-01 became the first reported CHK1 inhibitor; it has broad-spectrum efficacy against the protein kinase C family402, but due to its nonspecificity and long half-life, its clinical usage has been limited. According to the clinical trials website (https://clinicaltrials.gov), 12 clinical trials on CHK1 inhibitors have been completed, including trials for LY2606368, PF-00477736, and SRA737. In the trial data, these CHK1 inhibitors showed promising anticancer combination effects with other drugs that could generate replication-dependent DNA damage403,404. LY2606368 was the first selective CHK1 inhibitor. King et al.405 reported that inhibition of CHK1 by LY2603618 contributed to reduced DNA synthesis and elevated H2AX phosphorylation, indicative of DNA damage and premature entry into mitosis. PF-00477736, a selective inhibitor, was evaluated for its potential for radiosensitization in several cancer cell lines. The results showed that PF-00477736 contributes to substantial radiosensitization406. SRA737, another inhibitor, was also found to have the potential to suppress cell growth when used in combination with niraparib407. However, some CHK1 inhibitors developed at an early stage have been found to induce serious adverse events; for example, a clinical study showed that 14.29% of participants treated with SCH900776, a CHK1 inhibitor, were at risk of cardiac disorders (https://clinicaltrials.gov/). During the past decade, several new inhibitors have been discovered. In 2017, Suzuki et al.408 reported that a novel CHK1 inhibitor, MK8776, promoted IR-induced cell death via enhancement of aberrant mitosis and exacerbated mitotic catastrophe at a minimally toxic concentration without influencing DNA damage repair. Another novel CHK1 inhibitor, CCT244747, the first orally bioavailable CHK1 inhibitor, sensitized bladder and head and neck cancer cells to IR through modulation of G2/M checkpoint control, suggesting that CCT244747 may be suitable for oral administration409,410. Notably, combined treatment of the CHK1 inhibitor AZD6738 with the WEE1 inhibitor AZD1775 exerted a significant synergistic cytotoxic effect in mantle cell lymphoma and diffuse large B cell lymphoma cancer cells with a marked S-phase delay and increased DNA damage, indicating that combination treatment may provide a promising therapeutic avenue for B cell lymphoma patients411. Another study also indicated that serious adverse cytotoxic effects were found when cancer cells were treated with three different targeted inhibitors, LY2606368, cisplatin, and talazoparib, together412. However, although some inhibitors alone or in combination with other inhibitors may induce certain adverse effects, these inhibitors show great potential for cancer therapy. Therefore, during the development of inhibitors, seeking low cytotoxicity and high efficacy agents targeting CHK1 is encouraged. The combination of CHK1 inhibitors with inhibitors of other targets may translate into effective clinical applications in the future.
Indeed, increasing our knowledge of the underlying mechanisms of IR-induced DDR as well as clarifying the precise roles of key genes and proteins and their interaction partners involved in DDR signaling pathways is critical to the clinical discovery of novel intervention targets and will aid in the eventual development of effective strategies against cancer. In fact, numerous studies have focused on DNA damage repair genes and proteins and their regulation, suggesting essential roles in sensitization to radiotherapy. These observations provide insights for more radiotherapy sensitization approaches to kill cancer cells selectively and specifically.
Previous investigations of targets and inhibitors involved in radiotherapy-induced DNA damage repair have improved radiotherapy treatments targeting signaling pathways for various cancers. In the future, given the roles of these targeted signaling pathways associated with the IR-induced DDR, the relevant molecular mechanisms will require further elucidation to develop novel effective inhibitors and to decrease the toxicity of currently available inhibitors. Several current inhibitors have not achieved success in clinical trials, indicating the need for a better understanding of how these inhibitors participate in radiation-mediated DNA damage. Table 2 lists the targeted proteins involved in the response to radiotherapy. Table 3 lists the inhibitors of these targeted proteins and their functions in radiotherapy. Figure 6 displays the major downstream targets of ATM- and DNA-PK-mediated signaling pathways and their involvement in DDR.
Table 2.
Targetable proteins involved in DNA damage repair in response to radiation
| Length | Activity regulation | Molecular function | Main biological processesa | |
|---|---|---|---|---|
| DNA-PKcs | 4128 | Autophosphorylation or phosphorylation by ATM | ATP binding, DNA-dependent protein kinase activity, double-stranded DNA binding, protein domain-specific binding | DSB repair, negative regulation of the response to γ-radiation |
| ATM | 3056 | ATM dimers are normally inactivated but are activated with DNA in the presence of MRN; inhibited by wortmannin | ATP binding, DNA binding, DNA-dependent protein kinase activity, identical protein binding, protein serine/threonine kinase activity | Cell cycle arrest, cellular response to γ-radiation and X-ray radiation, mitotic telomere clustering, DSB repair via HR and NHEJ, DNA replication, DNA damage checkpoint |
| ATR | 2644 | Stimulated by TOPBP1468, activated by ETAA469, inhibited by caffeine, wortmannin, and LY294002 | ATP binding, DNA binding, MutSα and MutLα complex binding, protein kinase activity | Cellular response to γ-radiation and UV light, DNA damage checkpoint, DNA repair, positive regulation of DDR, signal transduction, regulation of signal transduction by p53 class mediator, negative regulation of DNA replication |
| CHK1 | 476 | Activated through phosphorylation | ATP binding, histone kinase activity, kinase activity, protein domain-specific binding, protein kinase activity | Cell cycle arrest, cellular response to γ-radiation and X-ray radiation, mitotic telomere clustering, DSB repair via HR and NHEJ, DNA replication, DNA damage checkpoint |
| LIG4 | 911 | A direct target of β-catenin | ATP binding, DNA binding, DNA ligase activity, DNA ligase activity, metal ion binding, protein C terminus binding | Cell cycle, cell division, cell proliferation, cellular response to IR, DNA biosynthetic process, DNA ligation, DNA ligation involved in DNA repair, DSB repair via HR and NHEJ, response to γ-radiation and X-ray radiation, single-strand break repair |
| PARP-1 | 1014 | NAD+ as a substrate and automodification of PARP‐1 itself470 | DNA binding, enzyme binding, estrogen receptor binding, histone deacetylase binding, NAD binding, protein kinase binding | Apoptotic process, cellular response to DNA damage-stimulated DNA repair, DSB by HE, mitochondrial DNA repair, response to γ-radiation |
| WEE1 | 646 | Activated through phosphorylation471 | ATP binding, kinase activity, protein kinase activity, protein tyrosine kinase activity | Cell division, establishment of cell polarity, G2/M transition of the cell cycle, negative regulation of G1/S transition in the cell cycle |
| CDK1 | 297 | Activated through phosphorylation | ATP binding, chromatin binding, cyclin binding, histone kinase activity, HSP70 protein kinase activity, protein serine/threonine kinase activity | Apoptotic process, mitotic G2 DNA damage checkpoint, positive regulation of G2/M transition in the cell cycle and gene expression and protein import into the nucleus, protein phosphorylation |
aAvailable from https://www.uniprot.org/
Table 3.
DDR inhibitors and their association with radiotherapy resistance
| Target | Inhibitor | Cancer type | Result | Mechanism | Refs. |
|---|---|---|---|---|---|
| DNA-PKcs | VX-984 | Glioblastomas | Concentration-dependent inhibition of radiation-induced DNA-PKcs phosphorylation | VX-984-induced radiosensitization is mediated by inhibition of DNA repair | 472 |
| NU5455 | Human orthotopic lung cancer | Enhanced the activity of doxorubicin | N/A | 264 | |
| KU60648 | Osteosarcoma | Enhanced cell cycle distribution and DNA damage | Increased cell accumulation at the G2/M transition point and increased percentage of cells with γH2AX foci | 263 | |
| NU7441 | Cervical and breast cancers | Enhanced radiosensitivity | Lowered cancer cell survival rates, increased apoptosis, and a G2-phase arrest | 473 | |
| NU7026 | Non-small-cell lung cancer | Enhanced radiosensitivity and increased the levels of ATM and ATR | Promoted apoptosis and G2/M arrest | 474 | |
| Wortmannin | Bladder cancer | Enhanced radiation-induced apoptosis with defective p53 activity (bladder cancer cell line, RT112 cells) | Inhibited DNA-PK, resulting in the inhibition of DSB repair | 475 | |
| ATM | AZD1390 | Glioma | Resulted in tumor cell hypersensitivity to IR | Induced G2 cell cycle phase accumulation, micronuclei, and apoptosis | 476 |
| AZ32 | Glioma | Blocked the DDR | Interacted with mutant p53 | 477 | |
| GSK635416A | Head and neck squamous cell carcinoma | Enhanced radiosensitization with cisplatin and cetuximab | Increased DSBs | 478 | |
| KU-55933 | Bladder cancer | Enhanced radiosensitivity | Elevated ATM expression and S-phase cell distribution and DSB repair kinetics | 296 | |
| ATR | AZD6738 | Kras-mutant cancer | Attenuated radiation-induced CD8+ T cell exhaustion and potentiated CD8+ T cell activity. | Suppressed PD-L1 upregulation on tumor cells and decreased the number of tumor-infiltrating Tregs | 479 |
| VE-821 | HeLa cells | Enhanced radiosensitivity | Abrogated the G2/M checkpoint | 480 | |
| HIF-1 | BAY 87-2243 | Non-small-cell lung cancer | Enhanced radiosensitivity | Inhibited mitochondrial complex I activity | 344,481,482 |
| Saikosaponin-d | Hepatoma | Enhanced radiosensitivity | Upregulated p53 and Bax, downregulated Bcl-2 by attenuating HIF-1α expression under hypoxic condition | 483 | |
| BAY-84-7296 | Squamous cell carcinomas | Enhanced radiosensitivity | N/A | 484 | |
| HDAC | Trichostatin A (TSA) | Human cervical carcinoma | Enhanced radiosensitivity | Downregulated the expression of HIF-1α and VEGF proteins | 485 |
| Vorinostat/SAHA | Enhanced radiosensitivity | Stimulated caspase activation consistent with apoptotic cell death | 486 | ||
| ITF2357 | Glioma | Enhanced radiosensitivity | Induced G1/S growth arrest | 487 | |
| LBH589 | Undifferentiated pleomorphic sarcoma | Suppressed tumor growth | Induced apoptosis and G2/M cell cycle arrest | ||
| WEE1 | AZD1775 | Hepatocellular carcinoma | Enhanced radiosensitivity | Delayed resolution of γH2AX foci and the induction of pan-nuclear γH2AX staining | 488 |
| MK 1775 | Lung cancer | Enhanced radiosensitivity | Suppressed Cdk1 phosphorylation | 489 | |
| PD0166285 | Osteosarcoma (OS) | Enhanced radiosensitivity | Abrogated G2 arrest | 398 | |
| CDK1 | AZD5438 | Lung cancer | Enhanced radiosensitivity | Inhibited Cdk1, prolonged G2/M arrest | 379 |
| CHK1 | CCT244747 | Bladder and head and neck cancer | Enhanced radiosensitivity | Abrogated G2 arrest | 409 |
| MK8776 | Human triple-negative breast cancer | Enhanced radiosensitivity | Suppressed autophagy | 490 |
Fig. 6.
Interregulation of the PIKK family members DNA-PKcs and ATM and their downstream substrates in the DDR pathway activated following DSB induction by IR. The dotted arrow represents the regulation at the transcription level. The solid arrow indicates the kinase activity-dependent regulation at the post-translational level
Challenges for IR-induced signal transduction and targeted therapy
As discussed above and in previous studies, radiation induces genomic DNA damage, challenging the integrity of the genome, particularly by inducing DSBs and causing various types of chromosomal aberrations in cancer cells413. From bacterial to mammalian cells, DNA damage signaling kinases play a central role in coordinating the DDR414. Once exposed to IR, DNA damage signaling kinases mediate hallmark responses, including cell cycle arrest, initiation of apoptosis, and induction of transcription415,416. In addition, these DNA damage signaling kinases are also regulated in an integrated network. In their review, Lanz et al.414 noted that DNA damage signaling kinases target dozens, if not hundreds, of DNA repair proteins, thereby modulating repair pathways. Indeed, the DNA damage signaling kinases and their targets are regulated by a complex network, the underlying mechanisms of which remain to be completely elucidated. Below, we list a number of key scientific issues for a holistic view of the IR-induced DNA damage signal transduction network that may be of concern over the next decade:
As described above, phosphorylation events mediated by DNA damage signaling kinases in response to IR are critical for the control of DNA damage repair pathways. However, the substrates and underlying molecular mechanisms remain incompletely clear. For instance, inhibition of ATM and DNA-PKcs strongly impairs the NHEJ pathway, but we do not know how the previous phosphorylation events occur in this process or which key signal transductions are essential for NHEJ. With the development of techniques such as mass spectrometry and gene sequencing technology, the ability to quantitatively detect phosphorylation events in the process of IR-induced DNA damage repair can provide new insight for the identification of signaling kinase substrates and molecular mechanisms. Phosphoproteomics has been used not only to identify the targets of DNA damage signaling kinases in mammalian cell lines but also to identify phosphorylation events catalyzed by DNA damage signaling kinases417–419. Looking ahead, given the large datasets arising from mass spectrometry and high-throughput gene sequencing, statistical analysis methods must be developed for more efficient prediction and analysis.
How do DNA damage signaling kinases function differently when subjected to high-, low-, and very low-dose IR exposure? Similarly, at what dose of radiation do phenotypic alterations in DNA occur? While DDR and repair signaling pathways under conditions of high-dose IR exposure, such as the levels provided by clinical radiotherapy, have been intensively investigated, the effects of low-dose radiation, particularly environmental radiation exposure, such as that generated from pediatric computed tomography or chest examination by X-ray, on the DNA damage repair signaling pathways have been ignored. Kim et al.420 reported on how exposure to low-dose radiation affects the human body, showing that low-dose radiation stress causes DNA damage and induces DNA damage-related signaling pathways, including apoptosis, cell cycle arrest, and DNA repair. Nagle et al.421 reported that low-dose irradiation of high-density murine and human glandular stem cells represented a dose threshold for DNA damage repair activation, leading to low-dose hyperradiosensitivity. Gaetani et al.422 investigated the effects of low-dose IR on DNA damage in circulating cells of occupationally exposed workers; the results showed that among workers with low-dose IR exposure, DNA repair activity was increased, and moreover, workers with cancer cases in their family history showed significantly reduced 8-oxoguanine glycosylase 1-dependent DNA repair activity compared with those workers without any family history of cancer. Although the effects of low-dose radiation on DNA have attracted increasing attention over the past decade, the molecular mechanisms, biomarkers, and possible clinical applicability require further investigation.
What is the crosstalk between DSB repair pathways in response to radiotherapy? Current studies have focused on the roles of HR and NHEJ, the two main pathways involved in regulating IR-induced DSB repair, and cell cycle checkpoints. However, there may be other unidentified repair pathways that have yet to be discovered, and furthermore, as different repair pathways exist, there may also be crosstalk. Limpose et al.423 showed that base excision repair, which was suggested to function independently in a previous perspective, was proven to directly interact with other DDR pathways rather than operating in isolation. Sunada et al.424 demonstrated that breast cancer cells with BRCA1/2 mutation treated with a PARP inhibitor had improved susceptibility via the crosstalk of DSB repair pathways. Several novel regulatory mechanisms have been discovered using crosstalk assays. For instance, Yuan et al.425 conducted cell experiments and revealed a feedback loop of Yes-associated protein (YAP) and p53 protein, as well as SIRT1-mediated regulation of YAP-p53 feedback loop-related deacetylation in certain residues. The authors suggested that this regulatory route was a new mechanism related to lung tumorigenesis. Exploring the interplay between repair pathways is important for our understanding of fundamental processes relevant to improving radiotherapy outcomes. Currently, omics technology and high-throughput assays to simultaneously survey the crosstalk of multiple DNA repair pathways are being developed69,426. For instance, the CRISPR/Cas9 system has been widely used and is being improved rapidly.
Recently, increasing numbers of studies have shown that combining radiation therapy with immunotherapy triggers a series of cell responses, including inducing cell death. This area is drawing increasing attention in the clinical and scientific communities427. As a result, the identification of inhibitors with potential to improve radioimmunotherapy resistance should consider both the regulation of radiation-related signal transduction and immunotherapy signal transduction428. Furthermore, it is necessary to uncover the mechanisms underlying why some patients experience durable responses, while others develop therapy resistance when treated with the combination of radiotherapy and immune therapy. Importantly, a better mechanistic understanding of the combination of radiotherapy and immune therapy is needed to benefit more patients in clinical applications. It is worth considering whether and how the translocation of DNA fragments from the nucleus to the cytoplasm can be recognized and used to trigger a response that can be translated into a novel strategy of improving cancer cell killing at the cellular and whole body levels. It is well documented that the tumor microenvironment can be altered. For instance, post IR, some inflammatory cytokines and immune cells are altered significantly. Harding et al.429 revealed that cell cycle progression through mitosis following DSBs leads to the formation of micronuclei. Furthermore, the pattern-recognition receptor cyclic GMP-AMP synthase (cGAS) is a cytosolic DNA sensor that activates innate immunity prior to activation of inflammatory signaling. Low expression of cGAS in patients with lung cancer is associated with poor survival, likely because cGAS deletion abrogates the senescence-associated secretory phenotype430. Obviously, targeted therapies to activate the cGAS-STING pathway in cancer cells can mediate cellular senescence and activate antitumor immunity, which could be another promising strategy for providing significant therapeutic benefits for cancer patients.
Is enough data available to assess inhibitor toxicity, safety, and carcinogenicity for normal tissues? As discussed above, inhibition of DNA damage repair can improve radiotherapy sensitivity in a wide range of cancers. However, most inhibitors lack relevant toxicity assessments, safety assays, and investigations of the risk of carcinogenesis prior to clinical application. Although several studies have been conducted to indicate that inhibition of cyclooxygenase-2 or nuclear factor-κB sensitizes cancer cells without causing normal cell/tissue toxicity431–434, the underlying molecular mechanisms should be determined to improve our understanding of the toxicity. Likewise, it is also important to provide more selective, low toxicity, and higher efficiency radiotherapy treatments at a low cost for cancer patients. A newly published review by Pilie et al.71 pointed out that the current strategies for treating cancers target DDR signaling pathways; hence, when the current DNA damage repair inhibitors are under development or in clinical trials prior to use in the larger population, attention should be paid to minimizing overlapping toxicities.
Are there enough early and sensitive biomarkers for predicting, preventing, or controlling RR? Although current studies have provided multiple DNA damage sensors and DNA damage repair regulatory proteins as possible targets to improve radiotherapy sensitivity, the challenge ahead is how to translate basic radiobiology studies into clinical applications. Moreover, any innovation and progress in radiotherapy depend on insights realized by basic radiobiological research435.
Clinical applications are generated from the discoveries of radiobiological studies, including DNA damage repair signal transduction pathways and specific IR-induced DDR proteins, such as DNA-PKcs, ATM, and ATR. We would like to use a fairy tale to explain the key points of this review. RR is similar to a mermaid, whose scales can be considered similar to the different signaling transduction-related pathways and proteins. The mermaid wants to shed her scales and become human; however, without an understanding of from where each scale originates and how they are regulated by internal or environmental factors, the mermaid scales are difficult to remove. Once the underlying mechanisms have been elucidated, the mermaid can shed her scales and become a human being.
Indeed, studying signaling transduction pathways in the field of radiobiology will dramatically contribute to the development of targeted radiotherapies. More than one century of study in this field has borne plenty of fruit, and the quality of radiotherapy has gradually improved. Further studies should continue to uncover the underlying mechanisms of IR-induced DNA damage repair, cancer metabolism, cancer stem cells, and the cancer microenvironment, ensuring that our knowledge of radiobiology increases to improve the outcomes of cancer treatments.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant Nos. U1803124, 81530085, and 31870847) and the Natural Science Foundation of Hunan Province (Grant No. 2019JJ40396).
Author contributions
P.-K.Z. conceived and designed the study. R.-X.H. searched and reviewed the literature and drafted the illustrations and initial manuscript. P.-K.Z. critically reviewed and revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Fidler MM, et al. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: a population-based study. Lancet Oncol. 2017;18:1579–1589. doi: 10.1016/S1470-2045(17)30677-0. [DOI] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer, C. et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaborators, G. B. D. C. O. D. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 5.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, et al. Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torre LA, et al. Global cancer in women: burden and trends. Cancer Epidemiol. Biomark. Prev. 2017;26:444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg MS. Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer. 2019;19:587–602. doi: 10.1038/s41568-019-0186-9. [DOI] [PubMed] [Google Scholar]
- 9.Shaked Y. The pro-tumorigenic host response to cancer therapies. Nat. Rev. Cancer. 2019;19:667–685. doi: 10.1038/s41568-019-0209-6. [DOI] [PubMed] [Google Scholar]
- 10.Falkson CB, et al. Radiotherapy with curative intent in patients with early-stage, medically inoperable, non-small-cell lung cancer: a systematic review. Clin. Lung Cancer. 2017;18:105–121 e105. doi: 10.1016/j.cllc.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Tam SY, Wu VWC. A review on the special radiotherapy techniques of colorectal cancer. Front. Oncol. 2019;9:208. doi: 10.3389/fonc.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taito M, et al. Voice rehabilitation for laryngeal cancer after radiotherapy: a systematic review and meta-analysis. Eur. Arch. Oto-Rhino-laryngol. 2019;276:1573–1583. doi: 10.1007/s00405-019-05452-2. [DOI] [PubMed] [Google Scholar]
- 13.Robinson, S. D. et al. Radical accelerated radiotherapy for non-small cell lung cancer (NSCLC): a 5-year retrospective review of two dose fractionation schedules. Radiother. Oncol.143, 37–43 (2019). [DOI] [PubMed]
- 14.Razvi Y, et al. A review of the rapid response radiotherapy program in patients with advanced cancer referred for palliative radiotherapy over two decades. Support. Care Cancer. 2019;27:2131–2134. doi: 10.1007/s00520-018-4474-9. [DOI] [PubMed] [Google Scholar]
- 15.Ghotra VP, Geldof AA, Danen EH. Targeted radiosensitization in prostate cancer. Curr. Pharm. Des. 2013;19:2819–2828. doi: 10.2174/1381612811319150017. [DOI] [PubMed] [Google Scholar]
- 16.Ma L, et al. A current review of dose-escalated radiotherapy in locally advanced non-small cell lung cancer. Radiol. Oncol. 2019;53:6–14. doi: 10.2478/raon-2019-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar S, Singh RK, Meena R. Emerging targets for radioprotection and radiosensitization in radiotherapy. Tumour Biol. 2016;37:11589–11609. doi: 10.1007/s13277-016-5117-8. [DOI] [PubMed] [Google Scholar]
- 18.Allison RR, Patel RM, McLawhorn RA. Radiation oncology: physics advances that minimize morbidity. Fut. Oncol. 2014;10:2329–2344. doi: 10.2217/fon.14.176. [DOI] [PubMed] [Google Scholar]
- 19.Roland CG. Priority of clinical x-ray reports: a classic dethroned? Can. J. Surg. 1962;5:247–251. [PubMed] [Google Scholar]
- 20.Thariat J, et al. Past, present, and future of radiotherapy for the benefit of patients. Nat. Rev. Clin. Oncol. 2013;10:52–60. doi: 10.1038/nrclinonc.2012.203. [DOI] [PubMed] [Google Scholar]
- 21.Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int. J. Med. Sci. 2012;9:193–199. doi: 10.7150/ijms.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Royce TJ, Efstathiou JA. Proton therapy for prostate cancer: a review of the rationale, evidence, and current state. Urol. Oncol. 2019;37:628–636. doi: 10.1016/j.urolonc.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Lorenz E, et al. Radiotherapy for childhood cancer and subsequent thyroid cancer risk: a systematic review. Eur. J. Epidemiol. 2018;33:1139–1162. doi: 10.1007/s10654-018-0467-8. [DOI] [PubMed] [Google Scholar]
- 24.Gebauer J, et al. Long-term endocrine and metabolic consequences of cancer treatment: a systematic review. Endocr. Rev. 2019;40:711–767. doi: 10.1210/er.2018-00092. [DOI] [PubMed] [Google Scholar]
- 25.Mattke M, et al. High control rates of proton- and carbon-ion-beam treatment with intensity-modulated active raster scanning in 101 patients with skull base chondrosarcoma at the Heidelberg Ion Beam Therapy Center. Cancer. 2018;124:2036–2044. doi: 10.1002/cncr.31298. [DOI] [PubMed] [Google Scholar]
- 26.Huang R, et al. Radiotherapy exposure in cancer patients and subsequent risk of stroke: a systematic review and meta-analysis. Front. Neurol. 2019;10:233. doi: 10.3389/fneur.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turek M, Krzyczmonik M, Balczewski P. New hopes in cancer battle—a review of new molecules and treatment strategies. Med. Chem. 2016;12:700–719. doi: 10.2174/1573406412666160502153700. [DOI] [PubMed] [Google Scholar]
- 28.Killock D. Brain cancer: chemoradiotherapy for low-grade glioma: battle won, but the war goes on. Nat. Rev. Clin. Oncol. 2016;13:328–329. doi: 10.1038/nrclinonc.2016.61. [DOI] [PubMed] [Google Scholar]
- 29.Dabaja BS, Mikhaeel NG. In the battle between protons and photons for hematologic malignancies, the patient must win. Int. J. Radiat. Oncol. Biol. Phys. 2016;95:43–45. doi: 10.1016/j.ijrobp.2015.09.043. [DOI] [PubMed] [Google Scholar]
- 30.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. 2005;104:1129–1137. doi: 10.1002/cncr.21324. [DOI] [PubMed] [Google Scholar]
- 31.Xian S, Zhang S. New advances in clinical treatment of radiation oncology. Sci. Technol. Rev. 2014;32:37–41. [Google Scholar]
- 32.He J, et al. Basic guidelines of quality control for radiotherapy. Chin. J. Radiat. Oncol. 2018;27:335–342. [Google Scholar]
- 33.Baskar R, Itahana K. Radiation therapy and cancer control in developing countries: can we save more lives? Int. J. Med. Sci. 2017;14:13–17. doi: 10.7150/ijms.17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atun R, et al. Expanding global access to radiotherapy. Lancet Oncol. 2015;16:1153–1186. doi: 10.1016/S1470-2045(15)00222-3. [DOI] [PubMed] [Google Scholar]
- 35.Seo YS, et al. Radiation-induced changes in tumor vessels and microenvironment contribute to therapeutic resistance in glioblastoma. Front. Oncol. 2019;9:1259. doi: 10.3389/fonc.2019.01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranawat P, Rawat S. Radiation resistance in thermophiles: mechanisms and applications. World J. Microbiol. Biotechnol. 2017;33:112. doi: 10.1007/s11274-017-2279-5. [DOI] [PubMed] [Google Scholar]
- 37.Steinbichler TB, et al. Therapy resistance mediated by exosomes. Mol. Cancer. 2019;18:58. doi: 10.1186/s12943-019-0970-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat. Rev. Cancer. 2009;9:665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 39.Hegedus F, Mathew LM, Schwartz RA. Radiation dermatitis: an overview. Int. J. Dermatol. 2017;56:909–914. doi: 10.1111/ijd.13371. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Abulizi A, Li M. Protective effect of Ganoderma (Lingzhi) on radiation and chemotherapy. Adv. Exp. Med. Biol. 2019;1182:119–142. doi: 10.1007/978-981-32-9421-9_4. [DOI] [PubMed] [Google Scholar]
- 41.Lam, E. et al. A systematic review and meta-analysis of clinician-reported versus patient-reported outcomes of radiation dermatitis. Breast. 50, 125–134 (2019). [DOI] [PMC free article] [PubMed]
- 42.Facoetti A, Barcellini A, Valvo F, Pullia M. The role of particle therapy in the risk of radio-induced second tumors: a review of the literature. Anticancer Res. 2019;39:4613–4617. doi: 10.21873/anticanres.13641. [DOI] [PubMed] [Google Scholar]
- 43.Pearce MS, et al. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet. 2012;380:499–505. doi: 10.1016/S0140-6736(12)60815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JW, Wernicke AG. Risk and survival outcomes of radiation-induced CNS tumors. J. Neuro-Oncol. 2016;129:15–22. doi: 10.1007/s11060-016-2148-3. [DOI] [PubMed] [Google Scholar]
- 45.Friedman, D. N. et al. Radiation dose and volume to the pancreas and subsequent risk of diabetes mellitus. J. Natl Cancer Inst. (2019). 10.1093/jnci/djz152. [DOI] [PMC free article] [PubMed]
- 46.Groot HJ, et al. Risk of diabetes after para-aortic radiation for testicular cancer. Br. J. cancer. 2018;119:901–907. doi: 10.1038/s41416-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Nimwegen FA, et al. Radiation dose–response relationship for risk of coronary heart disease in survivors of Hodgkin lymphoma. J. Clin. Oncol. 2016;34:235–243. doi: 10.1200/JCO.2015.63.4444. [DOI] [PubMed] [Google Scholar]
- 48.Luo D, et al. Targeted gold nanocluster-enhanced radiotherapy of prostate cancer. Small. 2019;15:e1900968. doi: 10.1002/smll.201900968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khoo AM, et al. Radiosensitization of prostate cancers in vitro and in vivo to erbium-filtered orthovoltage X-rays using actively targeted gold nanoparticles. Sci. Rep. 2017;7:18044. doi: 10.1038/s41598-017-18304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat. Rev. Cancer. 2017;17:271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 51.Castro-Eguiluz D, et al. Nutrient recommendations for cancer patients treated with pelvic radiotherapy, with or without comorbidities. Rev. Invest. Clin. 2018;70:130–135. doi: 10.24875/RIC.18002526. [DOI] [PubMed] [Google Scholar]
- 52.Aghajanzadeh, S., Karlsson, T., Tuomi, L. & Finizia, C. The effect of jaw exercises on anxiety and depression in patients with head and neck cancer receiving radiotherapy: prospective 2-year follow-up study. Head Neck. 42, 330–335 (2020). [DOI] [PubMed]
- 53.Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid. redox Signal. 2014;21:251–259. doi: 10.1089/ars.2013.5668. [DOI] [PubMed] [Google Scholar]
- 54.Anand SK, Sharma A, Singh N, Kakkar P. Entrenching role of cell cycle checkpoints and autophagy for maintenance of genomic integrity. DNA Rep. 2019;86:102748. doi: 10.1016/j.dnarep.2019.102748. [DOI] [PubMed] [Google Scholar]
- 55.Huang R, Zhou P. Double-edged effects of noncoding RNAs in responses to environmental genotoxic insults: perspectives with regards to molecule-ecology network. Environ. Pollut. 2019;247:64–71. doi: 10.1016/j.envpol.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 56.Ferreira S, Dutreix M. DNA repair inhibitors to enhance radiotherapy: progresses and limitations. Cancer Radiother. 2019;23:883–890. doi: 10.1016/j.canrad.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 57.Shukla V, et al. HMCES functions in the alternative end-joining pathway of the DNA DSB repair during class switch recombination in B cells. Mol. Cell. 2020;77:384–394. doi: 10.1016/j.molcel.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong H, et al. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell. Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 59.Ward JF. Mechanisms of DNA repair and their potential modification for radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 1986;12:1027–1032. doi: 10.1016/0360-3016(86)90220-8. [DOI] [PubMed] [Google Scholar]
- 60.Deycmar, S. et al. The relative biological effectiveness of proton irradiation in dependence of DNA damage repair. Br. J. Radiol. 20190494 (2019). [DOI] [PMC free article] [PubMed]
- 61.Kim, W. et al. Cellular stress responses in radiotherapy. Cells8, 1105 (2019). [DOI] [PMC free article] [PubMed]
- 62.Mavragani, I. V., Nikitaki, Z., Kalospyros, S. A. & Georgakilas, A. G. Ionizing radiation and complex DNA damage: from prediction to detection challenges and biological significance. Cancers11, 1789 (2019). [DOI] [PMC free article] [PubMed]
- 63.Lomax ME, Folkes LK, O’Neill P. Biological consequences of radiation-induced DNA damage: relevance to radiotherapy. Clin. Oncol. 2013;25:578–585. doi: 10.1016/j.clon.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Hei TK, et al. Radiation induced non-targeted response: mechanism and potential clinical implications. Curr. Mol. Pharmacol. 2011;4:96–105. doi: 10.2174/1874467211104020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Groth P, et al. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–6594. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mladenov E, Magin S, Soni A, Iliakis G. DNA double-strand break repair as determinant of cellular radiosensitivity to killing and target in radiation therapy. Front. Oncol. 2013;3:113. doi: 10.3389/fonc.2013.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sage E, Shikazono N. Radiation-induced clustered DNA lesions: Repair and mutagenesis. Free Radic. Biol. Med. 2017;107:125–135. doi: 10.1016/j.freeradbiomed.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 68.Nikjoo H, et al. Radiation track, DNA damage and response-a review. Rep. Prog. Phys. Phys. Soc. 2016;79:116601. doi: 10.1088/0034-4885/79/11/116601. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Rep. 2016;38:94–101. doi: 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 71.Pilie PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2019;16:81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ou HL, Schumacher B. DNA damage responses and p53 in the aging process. Blood. 2018;131:488–495. doi: 10.1182/blood-2017-07-746396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikitaki Z, et al. Measurement of complex DNA damage induction and repair in human cellular systems after exposure to ionizing radiations of varying linear energy transfer (LET) Free Radic. Res. 2016;50:S64–S78. doi: 10.1080/10715762.2016.1232484. [DOI] [PubMed] [Google Scholar]
- 74.Lorat Y, et al. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy—the heavy burden to repair. DNA Rep. 2015;28:93–106. doi: 10.1016/j.dnarep.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Hagiwara Y, et al. 3D-structured illumination microscopy reveals clustered DNA double-strand break formation in widespread gammaH2AX foci after high LET heavy-ion particle radiation. Oncotarget. 2017;8:109370–109381. doi: 10.18632/oncotarget.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart RD. Induction of DNA damage by light ions relative to (60)Co gamma-rays. Int. J. Part. Ther. 2018;5:25–39. doi: 10.14338/IJPT-18-00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018;215:1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maremonti, E. et al. In vivo assessment of reactive oxygen species production and oxidative stress effects induced by chronic exposure to gamma radiation in Caenorhabditis elegans. Free Radic. Biol. Med. (2019) 10.1016/j.freeradbiomed.2019.11.037. [DOI] [PubMed]
- 79.Schuch AP, Garcia CC, Makita K, Menck CF. DNA damage as a biological sensor for environmental sunlight. Photochem. Photobiol. Sci. 2013;12:1259–1272. doi: 10.1039/c3pp00004d. [DOI] [PubMed] [Google Scholar]
- 80.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 81.Kouranti I, Peyroche A. Protein degradation in DNA damage response. Semin. Cell Dev. Biol. 2012;23:538–545. doi: 10.1016/j.semcdb.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 82.Hau PM, et al. Role of ATM in the formation of the replication compartment during lytic replication of Epstein–Barr virus in nasopharyngeal epithelial cells. J. Virol. 2015;89:652–668. doi: 10.1128/JVI.01437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heijink AM, Krajewska M, van Vugt MA. The DNA damage response during mitosis. Mutat. Res. 2013;750:45–55. doi: 10.1016/j.mrfmmm.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Hau, P. M. & Tsao, S. W. Epstein–Barr virus hijacks DNA damage response transducers to orchestrate its life cycle. Viruses. 9, 341 (2017). [DOI] [PMC free article] [PubMed]
- 86.Ford JC, et al. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science. 1994;265:533–535. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- 87.Voicu PM, et al. In Schizosaccharomyces pombe the 14-3-3 protein Rad24p is involved in negative control of pho1 gene expression. Yeast. 2007;24:121–127. doi: 10.1002/yea.1433. [DOI] [PubMed] [Google Scholar]
- 88.Brown AL, et al. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc. Natl Acad. Sci. USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 2003;23:2999–3007. doi: 10.1128/MCB.23.8.2999-3007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang H, et al. Characterization of DNA damage-stimulated self-interaction of Saccharomyces cerevisiae checkpoint protein Rad17p. J. Biol. Chem. 2001;276:26715–26723. doi: 10.1074/jbc.M103682200. [DOI] [PubMed] [Google Scholar]
- 92.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/S1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 93.Maradeo ME, Garg A, Skibbens RV. Rfc5p regulates alternate RFC complex functions in sister chromatid pairing reactions in budding yeast. Cell Cycle. 2010;9:4370–4378. doi: 10.4161/cc.9.21.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kasahara K, et al. 14-3-3gamma mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage. EMBO J. 2010;29:2802–2812. doi: 10.1038/emboj.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol. 1999;9:963–966. doi: 10.1016/S0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- 96.McCulley JL, Petes TD. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc. Natl Acad. Sci. USA. 2010;107:11465–11470. doi: 10.1073/pnas.1006281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. The contribution of the S-phase checkpoint genes MEC1 and SGS1 to genome stability maintenance in Candida albicans. Fungal Genet. Biol. 2011;48:823–830. doi: 10.1016/j.fgb.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Foss EJ. Is Rad9p upstream or downstream from Mec1p? Cold Spring Harb. Symp. Quant. Biol. 2000;65:347–351. doi: 10.1101/sqb.2000.65.347. [DOI] [PubMed] [Google Scholar]
- 99.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell. 2002;9:857–869. doi: 10.1016/S1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 100.Rouse J, Jackson SP. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siddiqui MS, Francois M, Fenech MF, Leifert WR. Persistent gammaH2AX: a promising molecular marker of DNA damage and aging. Mutat. Res. Rev. Mutat. Res. 2015;766:1–19. doi: 10.1016/j.mrrev.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 102.Ku A, Facca VJ, Cai Z, Reilly RM. Auger electrons for cancer therapy— review. EJNMMI Radiopharm. Chem. 2019;4:27. doi: 10.1186/s41181-019-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Motoyama S, et al. Advantages of evaluating gammaH2AX induction in non-clinical drug development. Genes Environ. 2018;40:10. doi: 10.1186/s41021-018-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Campillo-Marcos I, Lazo PA. Implication of the VRK1 chromatin kinase in the signaling responses to DNA damage: a therapeutic target? Cell. Mol. Life Sci. 2018;75:2375–2388. doi: 10.1007/s00018-018-2811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Popp HD, et al. Leukocyte DNA damage after reduced and conventional absorbed radiation doses using 3rd generation dual-source CT technology. Eur. J. Radiol. Open. 2016;3:134–137. doi: 10.1016/j.ejro.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuo LJ, Yang LX. Gamma-H2AX—a novel biomarker for DNA double-strand breaks. In vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 107.Liu H, et al. Brca1 is involved in tolerance to adefovir dipivoxilinduced DNA damage. Int. J. Mol. Med. 2019;43:2491–2498. doi: 10.3892/ijmm.2019.4164. [DOI] [PubMed] [Google Scholar]
- 108.Kitabatake K, Yoshida E, Kaji T, Tsukimoto M. Involvement of adenosine A2B receptor in radiation-induced translocation of epidermal growth factor receptor and DNA damage response leading to radioresistance in human lung cancer cells. Biochim. Biophys. Acta Gen. Subj. 2020;1864:129457. doi: 10.1016/j.bbagen.2019.129457. [DOI] [PubMed] [Google Scholar]
- 109.Gustafsson NMS, et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat. Commun. 2018;9:3872. doi: 10.1038/s41467-018-06287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Im J, Lawrence J, Seelig D, Nho RS. FoxM1-dependent RAD51 and BRCA2 signaling protects idiopathic pulmonary fibrosis fibroblasts from radiation-induced cell death. Cell Death Dis. 2018;9:584. doi: 10.1038/s41419-018-0652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang P, et al. Knockdown of long non-coding RNA PCAT1 in glioma stem cells promotes radiation sensitivity. Med. Mol. Morphol. 2019;52:114–122. doi: 10.1007/s00795-018-0209-8. [DOI] [PubMed] [Google Scholar]
- 112.Katagi H, et al. Radiosensitization by histone H3 demethylase inhibition in diffuse intrinsic pontine glioma. Clin. Cancer Res. 2019;25:5572–5583. doi: 10.1158/1078-0432.CCR-18-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rothkamm K, et al. DNA damage foci: meaning and significance. Environ. Mol. Mutagen. 2015;56:491–504. doi: 10.1002/em.21944. [DOI] [PubMed] [Google Scholar]
- 114.Sharma A, Singh K, Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 115.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/S0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 116.Habraken Y, Jolois O, Piette J. Differential involvement of the hMRE11/hRAD50/NBS1 complex, BRCA1 and MLH1 in NF-kappaB activation by camptothecin and X-ray. Oncogene. 2003;22:6090–6099. doi: 10.1038/sj.onc.1206893. [DOI] [PubMed] [Google Scholar]
- 117.Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J. Radiat. Res. 2004;45:473–478. doi: 10.1269/jrr.45.473. [DOI] [PubMed] [Google Scholar]
- 118.Tauchi H, et al. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J. Biol. Chem. 2001;276:12–15. doi: 10.1074/jbc.C000578200. [DOI] [PubMed] [Google Scholar]
- 119.Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 120.Park YB, et al. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 2017;24:248–257. doi: 10.1038/nsmb.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim H, et al. Heterozygous germline mutations in NBS1 among Korean patients with high-risk breast cancer negative for BRCA1/2 mutation. Fam. Cancer. 2015;14:365–371. doi: 10.1007/s10689-015-9789-9. [DOI] [PubMed] [Google Scholar]
- 122.Someya M, et al. Association of ionizing radiation-induced foci of NBS1 with chromosomal instability and breast cancer susceptibility. Radiat. Res. 2006;166:575–582. doi: 10.1667/RR0638.1. [DOI] [PubMed] [Google Scholar]
- 123.Dumon-Jones V, et al. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263–7269. [PubMed] [Google Scholar]
- 124.Della-Maria J, et al. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. J. Biol. Chem. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ho V, et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer. 2018;18:869. doi: 10.1186/s12885-018-4776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 127.Nishi R, et al. The deubiquitylating enzyme UCHL3 regulates Ku80 retention at sites of DNA damage. Sci. Rep. 2018;8:17891. doi: 10.1038/s41598-018-36235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pucci S, et al. Ku70, Ku80, and sClusterin: a cluster of predicting factors for response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2017;97:381–388. doi: 10.1016/j.ijrobp.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 129.Koike M, Yutoku Y, Koike A. Cloning, localization and focus formation at DNA damage sites of canine Ku70. J. Vet. Med. Sci. 2017;79:554–561. doi: 10.1292/jvms.16-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deriano L, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105:4776–4783. doi: 10.1182/blood-2004-07-2888. [DOI] [PubMed] [Google Scholar]
- 131.Gou Q, et al. Downregulation of MDC1 and 53BP1 by short hairpin RNA enhances radiosensitivity in laryngeal carcinoma cells. Oncol. Rep. 2015;34:251–257. doi: 10.3892/or.2015.3980. [DOI] [PubMed] [Google Scholar]
- 132.Gerloff DL, Woods NT, Farago AA, Monteiro AN. BRCT domains: a little more than kin, and less than kind. FEBS Lett. 2012;586:2711–2716. doi: 10.1016/j.febslet.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen Y, et al. And-1 coordinates with CtIP for efficient homologous recombination and DNA damage checkpoint maintenance. Nucleic Acids Res. 2017;45:2516–2530. doi: 10.1093/nar/gkw1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baldock RA, et al. ATM localization and heterochromatin repair depend on direct interaction of the 53BP1-BRCT2 domain with gammaH2AX. Cell Rep. 2015;13:2081–2089. doi: 10.1016/j.celrep.2015.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dai Y, et al. Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR. Nat. Commun. 2018;9:2123. doi: 10.1038/s41467-018-04557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Capalbo G, et al. Radiation-induced survivin nuclear accumulation is linked to DNA damage repair. Int. J. Radiat. Oncol. Biol. Phys. 2010;77:226–234. doi: 10.1016/j.ijrobp.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 137.Cairns J, et al. Bora downregulation results in radioresistance by promoting repair of double strand breaks. PLoS ONE. 2015;10:e0119208. doi: 10.1371/journal.pone.0119208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cirauqui B, et al. DNA repair pathways to regulate response to chemoradiotherapy in patients with locally advanced head and neck cancer. Tumour Biol. 2016;37:13435–13443. doi: 10.1007/s13277-016-5149-0. [DOI] [PubMed] [Google Scholar]
- 139.Li, H. et al. Alcohol consumption, cigarette smoking, and risk of breast cancer for BRCA1 and BRCA2 mutation carriers: results from the BRCA1 and BRCA2 Cohort Consortium. Cancer Epidemiol. Biomark. Prev.29, 368–378 (2020). [DOI] [PMC free article] [PubMed]
- 140.Kratochwil, C. et al. Patients resistant against PSMA-targeting alpha-radiation therapy often harbor mutations in DNA-repair associated genes. J. Nucl. Med. (2019). 10.2967/jnumed.119.234559. [DOI] [PubMed]
- 141.Gorodetska I, Kozeretska I, Dubrovska A. BRCA genes: the role in genome stability, cancer stemness and therapy resistance. J. Cancer. 2019;10:2109–2127. doi: 10.7150/jca.30410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Atipairin A, Canyuk B, Ratanaphan A. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by the platinum-based anticancer drugs. Breast Cancer Res. Treat. 2011;126:203–209. doi: 10.1007/s10549-010-1182-7. [DOI] [PubMed] [Google Scholar]
- 143.Nishikawa H, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 2009;69:111–119. doi: 10.1158/0008-5472.CAN-08-3355. [DOI] [PubMed] [Google Scholar]
- 144.Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- 145.Li Y, et al. USP13 regulates the RAP80-BRCA1 complex dependent DNA damage response. Nat. Commun. 2017;8:15752. doi: 10.1038/ncomms15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Her J, Soo Lee N, Kim Y, Kim H. Factors forming the BRCA1-A complex orchestrate BRCA1 recruitment to the sites of DNA damage. Acta Biochim. Biophys. Sin. 2016;48:658–664. doi: 10.1093/abbs/gmw047. [DOI] [PubMed] [Google Scholar]
- 147.Ling H, et al. Genetic evaluation of BRCA1 associated a complex genes with triple-negative breast cancer susceptibility in Chinese women. Oncotarget. 2016;7:9759–9772. doi: 10.18632/oncotarget.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Batenburg NL, et al. CSB interacts with BRCA1 in late S/G2 to promote MRN- and CtIP-mediated DNA end resection. Nucleic Acids Res. 2019;47:10678–10692. doi: 10.1093/nar/gkz784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Anantha, R. W. et al. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. eLife. 6, e21350 (2017). [DOI] [PMC free article] [PubMed]
- 150.Thakur V, et al. The membrane tethered matrix metalloproteinase MT1-MMP triggers an outside-in DNA damage response that impacts chemo- and radiotherapy responses of breast cancer. Cancer Lett. 2019;443:115–124. doi: 10.1016/j.canlet.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Costes SV, et al. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat. Res. 2010;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu J, et al. Silencing of MBD1 reverses pancreatic cancer therapy resistance through inhibition of DNA damage repair. Int. J. Oncol. 2013;42:2046–2052. doi: 10.3892/ijo.2013.1901. [DOI] [PubMed] [Google Scholar]
- 153.Watanabe S, et al. JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 2013;20:1425–1433. doi: 10.1038/nsmb.2702. [DOI] [PubMed] [Google Scholar]
- 154.Reichert S, et al. Survivin inhibition and DNA double-strand break repair: a molecular mechanism to overcome radioresistance in glioblastoma. Radiother. Oncol. 2011;101:51–58. doi: 10.1016/j.radonc.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 155.Zhao J, et al. pATM and gammaH2AX are effective radiation biomarkers in assessing the radiosensitivity of (12)C(6+) in human tumor cells. Cancer Cell Int. 2017;17:49. doi: 10.1186/s12935-017-0419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pajic M, et al. miR-139-5p modulates radiotherapy resistance in breast cancer by repressing multiple gene networks of DNA repair and ROS defense. Cancer Res. 2018;78:501–515. doi: 10.1158/0008-5472.CAN-16-3105. [DOI] [PubMed] [Google Scholar]
- 157.Koval, L., Proshkina, E., Shaposhnikov, M. & Moskalev, A. The role of DNA repair genes in radiation-induced adaptive response in Drosophila melanogaster is differential and conditional. Biogerontology21, 45–56 (2020). [DOI] [PubMed]
- 158.Iliakis, G., Mladenov, E. & Mladenova, V. Necessities in the processing of DNA double strand breaks and their effects on genomic instability and cancer. Cancers11, 1671 (2019). [DOI] [PMC free article] [PubMed]
- 159.Karanam, N. K., Ding, L., Aroumougame, A. & Story, M. D. Tumor treating fields cause replication stress and interfere with DNA replication fork maintenance: Implications for cancer therapy. Transl. Res.217, 33–46 (2020). [DOI] [PubMed]
- 160.Dukaew, N. et al. Enhancement of radiosensitivity by eurycomalactone in human NSCLC cells through G2/M cell cycle arrest and delayed DNA double-strand break repair. Oncol. Res.28, 161–175 (2020). [DOI] [PMC free article] [PubMed]
- 161.Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J. Theor. Biol. 1976;59:97–106. doi: 10.1016/S0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 162.Lodovichi, S. et al. Effect of BRCA1 missense variants on gene reversion in DNA double-strand break repair mutants and cell cycle-arrested cells of Saccharomyces cerevisiae. Mutagenesis35, 189–195 (2020). [DOI] [PubMed]
- 163.Roth DB, Wilson JH. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc. Natl Acad. Sci. USA. 1985;82:3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Roth DB, Porter TN, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/MCB.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mukherjee K, et al. Systematic analysis of linker histone PTM hotspots reveals phosphorylation sites that modulate homologous recombination and DSB repair. DNA Rep. 2019;86:102763. doi: 10.1016/j.dnarep.2019.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Berthel, E. et al. What does the history of research on the repair of DNA double-strand breaks tell us?—A comprehensive review of human radiosensitivity. Int. J. Mol. Sci.20, 5339 (2020). [DOI] [PMC free article] [PubMed]
- 167.Foray N, Bourguignon M, Hamada N. Individual response to ionizing radiation. Mutat. Res. 2016;770:369–386. doi: 10.1016/j.mrrev.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 168.Joubert A, Foray N. Intrinsic radiosensitivity and DNA double-strand breaks in human cells. Cancer Radiother. 2007;11:129–142. doi: 10.1016/j.canrad.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 169.Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018;293:10524–10535. doi: 10.1074/jbc.TM118.000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Toulany, M. Targeting DNA double-strand break repair pathways to improve radiotherapy response. Genes10, 25 (2019). [DOI] [PMC free article] [PubMed]
- 171.Klein HL, et al. Guidelines for DNA recombination and repair studies: cellular assays of DNA repair pathways. Microb. Cell. 2019;6:1–64. doi: 10.15698/mic2019.01.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ranjha L, Howard SM, Cejka P. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018;127:187–214. doi: 10.1007/s00412-017-0658-1. [DOI] [PubMed] [Google Scholar]
- 173.Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J. Natl Cancer Inst. 2018;110:704–713. doi: 10.1093/jnci/djy085. [DOI] [PubMed] [Google Scholar]
- 174.Mazur, A. K., Nguyen, T. S. & Gladyshev, E. Direct homologous dsDNA–dsDNA pairing: How, where, and why? J. Mol. Biol.432, 737–744 (2020). [DOI] [PubMed]
- 175.Turan, V. & Oktay, K. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum. Reprod. Update26, 43–57 (2020). [DOI] [PMC free article] [PubMed]
- 176.Brito, D. V. C. et al. Mimicking age-associated Gadd45gamma dysregulation results in memory impairments in young adult mice. J. Neurosci.40, 1197–1210 (2020). [DOI] [PMC free article] [PubMed]
- 177.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fellmann C, et al. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017;16:89–100. doi: 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jin M, et al. The diversity and commonalities of the radiation-resistance mechanisms of Deinococcus and its up-to-date applications. AMB Express. 2019;9:138. doi: 10.1186/s13568-019-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lopez Perez R, et al. DNA damage response of clinical carbon ion versus photon radiation in human glioblastoma cells. Radiother. Oncol. 2019;133:77–86. doi: 10.1016/j.radonc.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 181.Gao Y, et al. The cancer/testes (CT) antigen HORMAD1 promotes homologous recombinational DNA repair and radioresistance in lung adenocarcinoma cells. Sci. Rep. 2018;8:15304. doi: 10.1038/s41598-018-33601-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang X, et al. Cathepsin B contributes to radioresistance by enhancing homologous recombination in glioblastoma. Biomed. Pharmacother. 2018;107:390–396. doi: 10.1016/j.biopha.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 183.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ipoutcha T, et al. Multiple origins and specific evolution of CRISPR/Cas9 systems in minimal bacteria (Mollicutes) Front. Microbiol. 2019;10:2701. doi: 10.3389/fmicb.2019.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Buyukyoruk M, Wiedenheft B. Type I-F CRISPR-Cas provides protection from DNA, but not RNA phages. Cell Discov. 2019;5:54. doi: 10.1038/s41421-019-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dave, A. et al. Homologous recombination repair intermediates promote efficient de novo telomere addition at DNA double-strand breaks. Nucleic Acids Res.48, 1271–1284 (2020). [DOI] [PMC free article] [PubMed]
- 187.Gibbs, D. R. & Dhakal, S. Homologous recombination under the single-molecule fluorescence microscope. Int. J. Mol. Sci.20, 6102 (2019). [DOI] [PMC free article] [PubMed]
- 188.Sun, Y., McCorvie, T. J., Yates, L. A. & Zhang, X. Structural basis of homologous recombination. Cell. Mol. Life Sci.77, 3–18 (2020). [DOI] [PMC free article] [PubMed]
- 189.Nogueira, A., Fernandes, M., Catarino, R. & Medeiros, R. RAD52 functions in homologous recombination and its importance on genomic integrity maintenance and cancer therapy. Cancers11 (2019). [DOI] [PMC free article] [PubMed]
- 190.Biau J, Chautard E, Verrelle P, Dutreix M. Altering DNA repair to improve radiation therapy: specific and multiple pathway targeting. Front. Oncol. 2019;9:1009. doi: 10.3389/fonc.2019.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bylicky MA, Mueller GP, Day RM. Radiation resistance of normal human astrocytes: the role of non-homologous end joining DNA repair activity. J. Radiat. Res. 2019;60:37–50. doi: 10.1093/jrr/rry084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mu F, et al. Mangiferin induces radiosensitization in glioblastoma cells by inhibiting nonhomologous end joining. Oncol. Rep. 2018;40:3663–3673. doi: 10.3892/or.2018.6756. [DOI] [PubMed] [Google Scholar]
- 193.Wang X, et al. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle. 2018;17:439–447. doi: 10.1080/15384101.2018.1442625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Estrada-Bernal A, et al. MEK inhibitor GSK1120212-mediated radiosensitization of pancreatic cancer cells involves inhibition of DNA double-strand break repair pathways. Cell Cycle. 2015;14:3713–3724. doi: 10.1080/15384101.2015.1104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Barnum KJ, O’Connell MJ. Cell cycle regulation by checkpoints. Methods Mol. Biol. 2014;1170:29–40. doi: 10.1007/978-1-4939-0888-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schafer KA. The cell cycle: a review. Vet. Pathol. 1998;35:461–478. doi: 10.1177/030098589803500601. [DOI] [PubMed] [Google Scholar]
- 197.Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 2013;23:345–356. doi: 10.1016/j.tcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 198.Ghelli Luserna di Rora A, Iacobucci I, Martinelli G. The cell cycle checkpoint inhibitors in the treatment of leukemias. J. Hematol. Oncol. 2017;10:77. doi: 10.1186/s13045-017-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang H, Zhang X, Teng L, Legerski RJ. DNA damage checkpoint recovery and cancer development. Exp. Cell Res. 2015;334:350–358. doi: 10.1016/j.yexcr.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 200.Panzica MT, McNally FJ. Mechanisms that prevent catastrophic interactions between paternal chromosomes and the oocyte meiotic spindle. Cell Cycle. 2018;17:529–534. doi: 10.1080/15384101.2018.1431495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ikeda, H. et al. Expression profile of cell cycle-related genes in human fibroblasts exposed simultaneously to radiation and simulated microgravity. Int. J. Mol. Sci20, 4791 (2019). [DOI] [PMC free article] [PubMed]
- 202.Ismael, M. et al. The targeting of rna polymerase I transcription using CX-5461 in combination with radiation enhances tumour cell killing effects in human solid cancers. Cancers. 11, 1429 (2019). [DOI] [PMC free article] [PubMed]
- 203.Peng G, et al. Alterations of cell cycle control proteins SHP1/2, p16, CDK4 and cyclin D1 in radioresistant nasopharyngeal carcinoma cells. Mol. Med. Rep. 2014;10:1709–1716. doi: 10.3892/mmr.2014.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Finkielstein CV, Chen LG, Maller JL. A role for G1/S cyclin-dependent protein kinases in the apoptotic response to ionizing radiation. J. Biol. Chem. 2002;277:38476–38485. doi: 10.1074/jbc.M206184200. [DOI] [PubMed] [Google Scholar]
- 205.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/S0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 206.Poon RY, Toyoshima H, Hunter T. Redistribution of the CDK inhibitor p27 between different cyclin.CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol. Biol. cell. 1995;6:1197–1213. doi: 10.1091/mbc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lv, L. et al. SOX6 suppresses the development of lung adenocarcinoma by regulating expression of p53, p21(CIPI), cyclin D1 and beta-catenin. FEBS Open Biol.10, 135–146 (2020). [DOI] [PMC free article] [PubMed]
- 208.Tanoue Y, et al. Differential roles of Rad18 and Chk2 in genome maintenance and skin carcinogenesis following UV exposure. J. Invest. Dermatol. 2018;138:2550–2557. doi: 10.1016/j.jid.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 209.Luo L, et al. Study on the mechanism of cell cycle checkpoint kinase 2 (CHEK2) gene dysfunction in chemotherapeutic drug resistance of triple negative breast cancer cells. Med. Sci. Monit. 2018;24:3176–3183. doi: 10.12659/MSM.907256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Andrysik Z, Kim J, Tan AC, Espinosa JM. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013;3:1346–1354. doi: 10.1016/j.celrep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kottemann MC, Bale AE. Characterization of DNA damage-dependent cell cycle checkpoints in a menin-deficient model. DNA Rep. 2009;8:944–952. doi: 10.1016/j.dnarep.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nagasawa H, et al. Absence of a radiation-induced first-cycle G1–S arrest in p53+ human tumor cells synchronized by mitotic selection. Cancer Res. 1998;58:2036–2041. [PubMed] [Google Scholar]
- 213.Fabbro M, et al. BRCA1-BARD1 complexes are required for p53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J. Biol. Chem. 2004;279:31251–31258. doi: 10.1074/jbc.M405372200. [DOI] [PubMed] [Google Scholar]
- 214.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J. Biol. Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jiang X, Wang J. Down-regulation of TFAM increases the sensitivity of tumour cells to radiation via p53/TIGAR signalling pathway. J. Cell. Mol. Med. 2019;23:4545–4558. doi: 10.1111/jcmm.14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Li SJ, et al. Low-dose irradiation promotes proliferation of the human breast cancer MDA-MB-231 cells through accumulation of mutant P53. Int. J. Oncol. 2017;50:290–296. doi: 10.3892/ijo.2016.3795. [DOI] [PubMed] [Google Scholar]
- 217.Chen, S. et al. Inhibition of MELK produces potential anti-tumour effects in bladder cancer by inducing G1/S cell cycle arrest via the ATM/CHK2/p53 pathway. J. Cell. Mol. Med.24, 1804–1821 (2020). [DOI] [PMC free article] [PubMed]
- 218.Lashgari A, Fauteux M, Marechal A, Gaudreau L. Cellular Depletion of BRD8 causes p53-dependent apoptosis and induces a DNA damage response in non-stressed cells. Sci. Rep. 2018;8:14089. doi: 10.1038/s41598-018-32323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gomez V, et al. Regulation of DNA damage responses and cell cycle progression by hMOB2. Cell. Signal. 2015;27:326–339. doi: 10.1016/j.cellsig.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Luo Q, et al. Sodium fluoride arrests renal G2/M phase cell-cycle progression by activating ATM-Chk2-P53/Cdc25C signaling pathway in mice. Cell. Physiol. Biochem. 2018;51:2421–2433. doi: 10.1159/000495899. [DOI] [PubMed] [Google Scholar]
- 221.Thanasoula M, Escandell JM, Suwaki N, Tarsounas M. ATM/ATR checkpoint activation downregulates CDC25C to prevent mitotic entry with uncapped telomeres. EMBO J. 2012;31:3398–3410. doi: 10.1038/emboj.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bahassi el M, et al. Priming phosphorylation of Chk2 by polo-like kinase 3 (Plk3) mediates its full activation by ATM and a downstream checkpoint in response to DNA damage. Mutat. Res. 2006;596:166–176. doi: 10.1016/j.mrfmmm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 223.Li J, Stern DF. DNA damage regulates Chk2 association with chromatin. J. Biol. Chem. 2005;280:37948–37956. doi: 10.1074/jbc.M509299200. [DOI] [PubMed] [Google Scholar]
- 224.Wang B. Analyzing cell cycle checkpoints in response to ionizing radiation in mammalian cells. Methods Mol. Biol. 2014;1170:313–320. doi: 10.1007/978-1-4939-0888-2_15. [DOI] [PubMed] [Google Scholar]
- 225.Deckbar D, Jeggo PA, Lobrich M. Understanding the limitations of radiation-induced cell cycle checkpoints. Crit. Rev. Biochem. Mol. Biol. 2011;46:271–283. doi: 10.3109/10409238.2011.575764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Warren NJH, Eastman A. Inhibition of checkpoint kinase 1 following gemcitabine-mediated S phase arrest results in CDC7- and CDK2-dependent replication catastrophe. J. Biol. Chem. 2019;294:1763–1778. doi: 10.1074/jbc.RA118.005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Falck J, et al. The DNA damage-dependent intra-S phase checkpoint is regulated by parallel pathways. Nat. Genet. 2002;30:290–294. doi: 10.1038/ng845. [DOI] [PubMed] [Google Scholar]
- 228.Peng Y, et al. Glutamine synthetase facilitates cancer cells to recover from irradiation-induced G2/M arrest. Cancer Biol. Ther. 2020;21:43–51. doi: 10.1080/15384047.2019.1665394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Reda M, et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett. 2019;467:9–18. doi: 10.1016/j.canlet.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhou, X. et al. EBV encoded miRNA BART8-3p promotes radioresistance in nasopharyngeal carcinoma by regulating ATM/ATR signaling pathway. Biosci. Rep.39, BSR20190415 (2019). [DOI] [PMC free article] [PubMed]
- 231.Gogineni VR, et al. Chk2-mediated G2/M cell cycle arrest maintains radiation resistance in malignant meningioma cells. Cancer Lett. 2011;313:64–75. doi: 10.1016/j.canlet.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Aninditha KP, et al. In vitro sensitivity of malignant melanoma cells lines to photon and heavy ion radiation. Clin. Transl. Radiat. Oncol. 2019;17:51–56. doi: 10.1016/j.ctro.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wang, Q. et al. Melatonin sensitizes human colorectal cancer cells to gamma-ray ionizing radiation in vitro and in vivo. Int. J. Mol. Sci. 19, 3974 (2018). [DOI] [PMC free article] [PubMed]
- 234.Dolman ME, et al. DNA-dependent protein kinase as molecular target for radiosensitization of neuroblastoma cells. PLoS ONE. 2015;10:e0145744. doi: 10.1371/journal.pone.0145744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Innes, C. L. et al. DNA damage responses in murine pre-B cells with genetic deficiencies in damage response genes. Cell Cycle. 19, 67–83 (2020). [DOI] [PMC free article] [PubMed]
- 236.Lu H, et al. DNA-PKcs promotes chromatin decondensation to facilitate initiation of the DNA damage response. Nucleic Acids Res. 2019;47:9467–9479. doi: 10.1093/nar/gkz694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Walker AI, Hunt T, Jackson RJ, Anderson CW. Double-stranded DNA induces the phosphorylation of several proteins including the 90 000 mol. wt. heat-shock protein in animal cell extracts. EMBO J. 1985;4:139–145. doi: 10.1002/j.1460-2075.1985.tb02328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xie, Y. et al. RBX1 prompts degradation of EXO1 to limit the homologous recombination pathway of DNA double-strand break repair in G1 phase. Cell Death Differ.27, 1383–1397 (2020). [DOI] [PMC free article] [PubMed]
- 239.Davis AJ, et al. BRCA1 modulates the autophosphorylation status of DNA-PKcs in S phase of the cell cycle. Nucleic Acids Res. 2014;42:11487–11501. doi: 10.1093/nar/gku824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhou Y, Paull TT. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM) J. Biol. Chem. 2013;288:37112–37125. doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Neal JA, et al. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol. Cell. Biol. 2011;31:1719–1733. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Huang B, et al. DNA-PKcs associates with PLK1 and is involved in proper chromosome segregation and cytokinesis. J. Cell. Biochem. 2014;115:1077–1088. doi: 10.1002/jcb.24703. [DOI] [PubMed] [Google Scholar]
- 243.Tu WZ, et al. GammaH2AX foci formation in the absence of DNA damage: mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 2013;587:3437–3443. doi: 10.1016/j.febslet.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 244.Lee KJ, et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J. Biol. Chem. 2011;286:12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shang ZF, et al. Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 phosphorylation in response to DNA damage. Cancer Res. 2010;70:3657–3666. doi: 10.1158/0008-5472.CAN-09-3362. [DOI] [PubMed] [Google Scholar]
- 246.Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol. Cell. Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li Y, et al. Transient introduction of human telomerase mRNA improves hallmarks of progeria cells. Aging Cell. 2019;18:e12979. doi: 10.1111/acel.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jeggo PA. DNA-PK: at the cross-roads of biochemistry and genetics. Mutat. Res. 1997;384:1–14. doi: 10.1016/S0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 249.Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yan Q, et al. Cleavage of Ku80 by caspase-2 promotes non-homologous end joining-mediated DNA repair. DNA Rep. 2017;60:18–28. doi: 10.1016/j.dnarep.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 251.Cui X, et al. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang S, et al. Spontaneous tumor development in bone marrow-rescued DNA-PKcs(3A/3A) mice due to dysfunction of telomere leading strand deprotection. Oncogene. 2016;35:3909–3918. doi: 10.1038/onc.2015.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Lee BS, et al. Functional intersection of ATM and DNA-dependent protein kinase catalytic subunit in coding end joining during V(D)J recombination. Mol. Cell. Biol. 2013;33:3568–3579. doi: 10.1128/MCB.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Douglas P, et al. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Rep. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 255.Serrano MA, et al. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene. 2013;32:2452–2462. doi: 10.1038/onc.2012.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Shang ZF, et al. DNA-PKcs negatively regulates cyclin B1 protein stability through facilitating its ubiquitination mediated by Cdh1-APC/C pathway. Int. J. Biol. Sci. 2015;11:1026–1035. doi: 10.7150/ijbs.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Cui F, et al. The involvement of c-Myc in the DNA double-strand break repair via regulating radiation-induced phosphorylation of ATM and DNA-PKcs activity. Mol. Cell. Biochem. 2015;406:43–51. doi: 10.1007/s11010-015-2422-2. [DOI] [PubMed] [Google Scholar]
- 258.Du L, et al. Radiosensitization and growth inhibition of cancer cells mediated by an scFv antibody gene against DNA-PKcs in vitro and in vivo. Radiat. Oncol. 2010;5:70. doi: 10.1186/1748-717X-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.An J, et al. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol. Biol. 2010;11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.An J, et al. Silencing of DNA-PKcs alters the transcriptional profile of certain signal transduction genes related to proliferation and differentiation in HeLa cells. Int. J. Mol. Med. 2005;16:455–462. [PubMed] [Google Scholar]
- 261.Mohiuddin IS, Kang MH. DNA-PK as an emerging therapeutic target in cancer. Front. Oncol. 2019;9:635. doi: 10.3389/fonc.2019.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liu Y, et al. DNA-PKcs deficiency inhibits glioblastoma cell-derived angiogenesis after ionizing radiation. J. Cell. Physiol. 2015;230:1094–1103. doi: 10.1002/jcp.24841. [DOI] [PubMed] [Google Scholar]
- 263.Mamo T, et al. Inhibiting DNA-PKCS radiosensitizes human osteosarcoma cells. Biochem. Biophys. Res. Commun. 2017;486:307–313. doi: 10.1016/j.bbrc.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Willoughby, C. E. et al. Selective DNA-PKcs inhibition extends the therapeutic index of localized radiotherapy and chemotherapy. J. Clin. Invest.130, 258–271 (2020). [DOI] [PMC free article] [PubMed]
- 265.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/S0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 266.Davidson, D., Amrein, L., Panasci, L. & Aloyz, R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 4, 5 (2013). [DOI] [PMC free article] [PubMed]
- 267.Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 268.Gurung RL, et al. Targeting DNA-PKcs and telomerase in brain tumour cells. Mol. Cancer. 2014;13:232. doi: 10.1186/1476-4598-13-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yang C, et al. NU7441 enhances the radiosensitivity of liver cancer cells. Cell. Physiol. Biochem. 2016;38:1897–1905. doi: 10.1159/000445551. [DOI] [PubMed] [Google Scholar]
- 270.Shinohara ET, et al. DNA-dependent protein kinase is a molecular target for the development of noncytotoxic radiation-sensitizing drugs. Cancer Res. 2005;65:4987–4992. doi: 10.1158/0008-5472.CAN-04-4250. [DOI] [PubMed] [Google Scholar]
- 271.Durant S, Karran P. Vanillins—a novel family of DNA-PK inhibitors. Nucleic Acids Res. 2003;31:5501–5512. doi: 10.1093/nar/gkg753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang B, et al. Proteomic profiling revealed the functional networks associated with mitotic catastrophe of HepG2 hepatoma cells induced by 6-bromine-5-hydroxy-4-methoxybenzaldehyde. Toxicol. Appl. Pharmacol. 2011;252:307–317. doi: 10.1016/j.taap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 273.Yan YQ, et al. Induction of apoptosis and autophagic cell death by the vanillin derivative 6-bromine-5-hydroxy-4-methoxybenzaldehyde is accompanied by the cleavage of DNA-PKcs and rapid destruction of c-Myc oncoprotein in HepG2 cells. Cancer Lett. 2007;252:280–289. doi: 10.1016/j.canlet.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 274.Wise HC, et al. Activity of M3814, an oral DNA-PK inhibitor, in combination with topoisomerase II inhibitors in ovarian cancer models. Sci. Rep. 2019;9:18882. doi: 10.1038/s41598-019-54796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sun Q, et al. Therapeutic implications of p53 status on cancer cell fate following exposure to ionizing radiation and the DNA-PK inhibitor M3814. Mol. Cancer Res. 2019;17:2457–2468. doi: 10.1158/1541-7786.MCR-19-0362. [DOI] [PubMed] [Google Scholar]
- 276.Fok JHL, et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat. Commun. 2019;10:5065. doi: 10.1038/s41467-019-12836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Khan AJ, et al. VX-984 is a selective inhibitor of non-homologous end joining, with possible preferential activity in transformed cells. Oncotarget. 2018;9:25833–25841. doi: 10.18632/oncotarget.25383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lamb R, et al. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget. 2015;6:14005–14025. doi: 10.18632/oncotarget.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Oike T, et al. Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 2012;84:815–821. doi: 10.1016/j.ijrobp.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 280.Gotoff SP, Amirmokri E, Liebner EJ. Ataxia telangiectasia. Neoplasia, untoward response to x-irradiation, and tuberous sclerosis. Am. J. Dis. Child. 1967;114:617–625. doi: 10.1001/archpedi.1967.02090270073006. [DOI] [PubMed] [Google Scholar]
- 281.Taylor AM, et al. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 282.Imray FP, Kidson C. Perturbations of cell-cycle progression in gamma-irradiated ataxia telangiectasia and Huntington’s disease cells detected by DNA flow cytometric analysis. Mutat. Res. 1983;112:369–382. doi: 10.1016/0167-8817(83)90030-5. [DOI] [PubMed] [Google Scholar]
- 283.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 284.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 285.al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bentley NJ, et al. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. doi: 10.1002/j.1460-2075.1996.tb01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 288.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 289.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell. Biol. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 290.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 291.Hirao A, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Heylmann D, et al. Sensitivity of CD3/CD28-stimulated versus non-stimulated lymphocytes to ionizing radiation and genotoxic anticancer drugs: key role of ATM in the differential radiation response. Cell Death Dis. 2018;9:1053. doi: 10.1038/s41419-018-1095-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mladenov E, et al. Radiation-dose-dependent functional synergisms between ATM, ATR and DNA-PKcs in checkpoint control and resection in G2-phase. Sci. Rep. 2019;9:8255. doi: 10.1038/s41598-019-44771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 295.Powell SN, et al. Differential sensitivity of p53(−) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995;55:1643–1648. [PubMed] [Google Scholar]
- 296.Zhang T, et al. The ATM inhibitor KU55933 sensitizes radioresistant bladder cancer cells with DAB2IP gene defect. Int. J. Radiat. Biol. 2015;91:368–378. doi: 10.3109/09553002.2015.1001531. [DOI] [PubMed] [Google Scholar]
- 297.Vecchio D, et al. Pharmacokinetics, pharmacodynamics and efficacy on pediatric tumors of the glioma radiosensitizer KU60019. Int. J. Cancer. 2015;136:1445–1457. doi: 10.1002/ijc.29121. [DOI] [PubMed] [Google Scholar]
- 298.Vecchio D, et al. Predictability, efficacy and safety of radiosensitization of glioblastoma-initiating cells by the ATM inhibitor KU-60019. Int. J. Cancer. 2014;135:479–491. doi: 10.1002/ijc.28680. [DOI] [PubMed] [Google Scholar]
- 299.Nishida H, et al. Inhibition of ATR protein kinase activity by schisandrin B in DNA damage response. Nucleic Acids Res. 2009;37:5678–5689. doi: 10.1093/nar/gkp593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Fokas E, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Foote, K. M. et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-y l}-1H-indole (AZ20): a potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J. Med. Chem. 56, 2125–2138 (2013). [DOI] [PubMed]
- 302.Dillon MT, et al. PATRIOT: a phase I study to assess the tolerability, safety and biological effects of a specific ataxia telangiectasia and Rad3-related (ATR) inhibitor (AZD6738) as a single agent and in combination with palliative radiation therapy in patients with solid tumours. Clin. Transl. Radiat. Oncol. 2018;12:16–20. doi: 10.1016/j.ctro.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mo LJ, et al. Exosome-packaged miR-1246 contributes to bystander DNA damage by targeting LIG4. Br. J. Cancer. 2018;119:492–502. doi: 10.1038/s41416-018-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Williams GJ, et al. Structural insights into NHEJ: building up an integrated picture of the dynamic DSB repair super complex, one component and interaction at a time. DNA Rep. 2014;17:110–120. doi: 10.1016/j.dnarep.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Altmann T, Gennery AR. DNA ligase IV syndrome; a review. Orphanet J. Rare Dis. 2016;11:137. doi: 10.1186/s13023-016-0520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.O’Driscoll M, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/S1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 307.Riballo E, et al. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J. Biol. Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]
- 308.Mumbrekar KD, Goutham HV, Vadhiraja BM, Bola Sadashiva SR. Polymorphisms in double strand break repair related genes influence radiosensitivity phenotype in lymphocytes from healthy individuals. DNA Rep. 2016;40:27–34. doi: 10.1016/j.dnarep.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 309.Kondo N, et al. DNA damage induced by boron neutron capture therapy is partially repaired by DNA ligase IV. Radiat. Environ. Biophys. 2016;55:89–94. doi: 10.1007/s00411-015-0625-2. [DOI] [PubMed] [Google Scholar]
- 310.McKay MJ, et al. Non-homologous end-joining protein expression screen from radiosensitive cancer patients yields a novel DNA double strand break repair phenotype. Ann. Transl. Med. 2017;5:96. doi: 10.21037/atm.2017.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Koike M, Yutoku Y, Koike A. Establishment of hamster cell lines with EGFP-tagged human XRCC4 and protection from low-dose X-ray radiation. J. Vet. Med. Sci. 2012;74:1269–1275. doi: 10.1292/jvms.12-0112. [DOI] [PubMed] [Google Scholar]
- 312.Guo Q, et al. ID1 affects the efficacy of radiotherapy in glioblastoma through inhibition of DNA repair pathways. Med. Oncol. 2013;30:325. doi: 10.1007/s12032-012-0325-6. [DOI] [PubMed] [Google Scholar]
- 313.Wang YG, et al. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J. Biol. Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 314.Tseng HM, et al. A high-throughput scintillation proximity-based assay for human DNA ligase IV. Assay Drug Dev. Technol. 2012;10:235–249. doi: 10.1089/adt.2011.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Srivastava M, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 316.Ma H, et al. Combining carbon ion irradiation and non-homologous end-joining repair inhibitor NU7026 efficiently kills cancer cells. Radiat. Oncol. 2015;10:225. doi: 10.1186/s13014-015-0536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Kim DS, et al. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell. 2019;75:1270–1285 e1214. doi: 10.1016/j.molcel.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Swiatecka G, Smagon H. Primary heart amyloidosis. Kardiol. Pol. 1976;19:67–73. [PubMed] [Google Scholar]
- 319.Maltseva EA, Rechkunova NI, Sukhanova MV, Lavrik OI. Poly(ADP-ribose) polymerase 1 modulates interaction of the nucleotide excision repair factor XPC-RAD23B with DNA via poly(ADP-ribosyl)ation. J. Biol. Chem. 2015;290:21811–21820. doi: 10.1074/jbc.M115.646638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Durante M, et al. Effects of PARP-1 deficiency and histamine H4 receptor inhibition in an inflammatory model of lung fibrosis in mice. Front. Pharmacol. 2019;10:525. doi: 10.3389/fphar.2019.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sodhi RK, Singh N, Jaggi AS. Poly(ADP-ribose) polymerase-1 (PARP-1) and its therapeutic implications. Vasc. Pharmacol. 2010;53:77–87. doi: 10.1016/j.vph.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 322.Kai M. Roles of RNA-binding proteins in DNA damage response. Int. J. Mol. Sci. 2016;17:310. doi: 10.3390/ijms17030310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Han Y, et al. DNAPKcs PARylation regulates DNAPK kinase activity in the DNA damage response. Mol. Med. Rep. 2019;20:3609–3616. doi: 10.3892/mmr.2019.10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.de Murcia JM, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Soni A, et al. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014;42:6380–6392. doi: 10.1093/nar/gku298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Vormoor B, et al. Sensitizing Ewing sarcoma to chemo- and radiotherapy by inhibition of the DNA-repair enzymes DNA protein kinase (DNA-PK) and poly-ADP-ribose polymerase (PARP) 1/2. Oncotarget. 2017;8:113418–113430. doi: 10.18632/oncotarget.21300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Hirai T, et al. Radiosensitization by PARP inhibition to proton beam irradiation in cancer cells. Biochem. Biophys. Res. Commun. 2016;478:234–240. doi: 10.1016/j.bbrc.2016.07.062. [DOI] [PubMed] [Google Scholar]
- 328.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer. 2011;11:239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 329.Jannetti SA, et al. PARP-1-targeted radiotherapy in mouse models of glioblastoma. J. Nucl. Med. 2018;59:1225–1233. doi: 10.2967/jnumed.117.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ieraci A, Herrera DG. Nicotinamide inhibits ethanol-induced caspase-3 and PARP-1 Over-activation and subsequent neurodegeneration in the developing mouse cerebellum. Cerebellum. 2018;17:326–335. doi: 10.1007/s12311-017-0916-z. [DOI] [PubMed] [Google Scholar]
- 331.Calabrese CR, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 332.Noel G, et al. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1,8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA synthesis. Mol. Cancer Ther. 2006;5:564–574. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 333.Mateo J, et al. A decade of clinical development of PARP inhibitors in perspective. Ann. Oncol. 2019;30:1437–1447. doi: 10.1093/annonc/mdz192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Murai J, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ryu, H. et al. A small compound KJ-28d enhances the sensitivity of non-small cell lung cancer to radio- and chemotherapy. Int. J. Mol. Sci.20, 6026 (2019). [DOI] [PMC free article] [PubMed]
- 336.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 337.Bourton EC, et al. The PARP-1 inhibitor Olaparib suppresses BRCA1 protein levels, increases apoptosis and causes radiation hypersensitivity in BRCA1(+/−) lymphoblastoid cells. J. Cancer. 2017;8:4048–4056. doi: 10.7150/jca.21338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Guillot C, et al. PARP inhibition and the radiosensitizing effects of the PARP inhibitor ABT-888 in in vitro hepatocellular carcinoma models. BMC Cancer. 2014;14:603. doi: 10.1186/1471-2407-14-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Wang L, et al. MK-4827, a PARP-1/-2 inhibitor, strongly enhances response of human lung and breast cancer xenografts to radiation. Invest. N. Drugs. 2012;30:2113–2120. doi: 10.1007/s10637-011-9770-x. [DOI] [PubMed] [Google Scholar]
- 340.Mirza MR, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 341.Alotaibi M, et al. Radiosensitization by PARP Inhibition in DNA repair proficient and deficient tumor cells: proliferative recovery in senescent cells. Radiat. Res. 2016;185:229–245. doi: 10.1667/RR14202.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Pernin V, et al. PARP inhibitors and radiotherapy: rational and prospects for a clinical use. Cancer Radiother. 2014;18:790–798. doi: 10.1016/j.canrad.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 343.Ledford H, Callaway E. Biologists who decoded how cells sense oxygen win medicine Nobel. Nature. 2019;574:161–162. doi: 10.1038/d41586-019-02963-0. [DOI] [PubMed] [Google Scholar]
- 344.Bhattarai D, Xu X, Lee K. Hypoxia-inducible factor-1 (HIF-1) inhibitors from the last decade (2007 to 2016): a “structure–activity relationship” perspective. Med. Res. Rev. 2018;38:1404–1442. doi: 10.1002/med.21477. [DOI] [PubMed] [Google Scholar]
- 345.Ban HS, Uto Y, Won M, Nakamura H. Hypoxia-inducible factor (HIF) inhibitors: a patent survey (2011–2015) Expert Opin. Ther. Pat. 2016;26:309–322. doi: 10.1517/13543776.2016.1146252. [DOI] [PubMed] [Google Scholar]
- 346.Vaupel P, Multhoff G. Hypoxia-/HIF-1alpha-driven factors of the tumor microenvironment impeding antitumor immune responses and promoting malignant progression. Adv. Exp. Med. Biol. 2018;1072:171–175. doi: 10.1007/978-3-319-91287-5_27. [DOI] [PubMed] [Google Scholar]
- 347.Cui W, Wu F, Ma L. Hypoxia associated biomarkers in lung cancer—an update. Eur. Rev. Med. Pharmacol. Sci. 2017;21:43–46. [PubMed] [Google Scholar]
- 348.Nenu I, Gafencu GA, Popescu T, Kacso G. Lactate—a new frontier in the immunology and therapy of prostate cancer. J. Cancer Res. Ther. 2017;13:406–411. doi: 10.4103/0973-1482.163692. [DOI] [PubMed] [Google Scholar]
- 349.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/S1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 350.Harada H, et al. The Akt/mTOR pathway assures the synthesis of HIF-1alpha protein in a glucose- and reoxygenation-dependent manner in irradiated tumors. J. Biol. Chem. 2009;284:5332–5342. doi: 10.1074/jbc.M806653200. [DOI] [PubMed] [Google Scholar]
- 351.Kang J, et al. Plasminogen activator inhibitor-1 enhances radioresistance and aggressiveness of non-small cell lung cancer cells. Oncotarget. 2016;7:23961–23974. doi: 10.18632/oncotarget.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Lee M, Yoon JH. Metabolic interplay between glycolysis and mitochondrial oxidation: the reverse Warburg effect and its therapeutic implication. World J. Biol. Chem. 2015;6:148–161. doi: 10.4331/wjbc.v6.i3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Dittmann K, et al. Nuclear EGFR renders cells radio-resistant by binding mRNA species and triggering a metabolic switch to increase lactate production. Radiother. Oncol. 2015;116:431–437. doi: 10.1016/j.radonc.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 354.Sun X, et al. Combination with stereotactic body radiotherapy offers a promising strategy to overcome resistance to immunotherapy in advanced renal cell cancer. J. Oncol. 2019;2019:1483406. doi: 10.1155/2019/1483406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Lyakhovich A, Lleonart ME. Bypassing mechanisms of mitochondria-mediated cancer stem cells resistance to chemo- and radiotherapy. Oxid. Med. Cell. Longev. 2016;2016:1716341. doi: 10.1155/2016/1716341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Brown JM. Tumor hypoxia in cancer therapy. Methods Enzymol. 2007;435:297–321. doi: 10.1016/S0076-6879(07)35015-5. [DOI] [PubMed] [Google Scholar]
- 357.Sato M, et al. LW6, a hypoxia-inducible factor 1 inhibitor, selectively induces apoptosis in hypoxic cells through depolarization of mitochondria in A549 human lung cancer cells. Mol. Med. Rep. 2015;12:3462–3468. doi: 10.3892/mmr.2015.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Schwartz DL, et al. Radiosensitization and stromal imaging response correlates for the HIF-1 inhibitor PX-478 given with or without chemotherapy in pancreatic cancer. Mol. Cancer Ther. 2010;9:2057–2067. doi: 10.1158/1535-7163.MCT-09-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yang R, et al. Inhibitors of HIF-1alpha and CXCR4 mitigate the development of radiation necrosis in mouse brain. Int. J. Radiat. Oncol. Biol. Phys. 2018;100:1016–1025. doi: 10.1016/j.ijrobp.2017.12.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Paap B, Wilson DM, 3rd, Sutherland BM. Human abasic endonuclease action on multilesion abasic clusters: implications for radiation-induced biological damage. Nucleic Acids Res. 2008;36:2717–2727. doi: 10.1093/nar/gkn118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Marampon F, et al. HDAC4 and HDAC6 sustain DNA double strand break repair and stem-like phenotype by promoting radioresistance in glioblastoma cells. Cancer Lett. 2017;397:1–11. doi: 10.1016/j.canlet.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 362.Zhu L, Wu K, Ma S, Zhang S. HDAC inhibitors: a new radiosensitizer for non-small-cell lung cancer. Tumori. 2015;101:257–262. doi: 10.5301/tj.5000347. [DOI] [PubMed] [Google Scholar]
- 363.Robert C, et al. Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin. Leuk. Res. 2016;45:14–23. doi: 10.1016/j.leukres.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Chinnaiyan P, Allen GW, Harari PM. Radiation and new molecular agents, part II: targeting HDAC, HSP90, IGF-1R, PI3K, and Ras. Semin. Radiat. Oncol. 2006;16:59–64. doi: 10.1016/j.semradonc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 365.Shi XY, et al. Histone deacetylase (HDAC) inhibitor, suberoylanilide hydroxamic acid (SAHA), induces apoptosis in prostate cancer cell lines via the Akt/FOXO3a signaling pathway. Med. Sci. Monit. 2017;23:5793–5802. doi: 10.12659/MSM.904597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Wang G, et al. HDAC inhibition prevents white matter injury by modulating microglia/macrophage polarization through the GSK3beta/PTEN/Akt axis. Proc. Natl Acad. Sci. USA. 2015;112:2853–2858. doi: 10.1073/pnas.1501441112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Mehndiratta S, et al. N-alkyl-hydroxybenzoyl anilide hydroxamates as dual inhibitors of HDAC and HSP90, downregulating IFN-gamma induced PD-L1 expression. Eur. J. Med. Chem. 2020;185:111725. doi: 10.1016/j.ejmech.2019.111725. [DOI] [PubMed] [Google Scholar]
- 368.Hernandez A, Lopez-Lluch G, Navas P, Pintor-Toro JA. HDAC and Hsp90 inhibitors down-regulate PTTG1/securin but do not induce aneuploidy. Genes Chromosomes Cancer. 2009;48:194–201. doi: 10.1002/gcc.20630. [DOI] [PubMed] [Google Scholar]
- 369.Wang SH, et al. Curcumin-mediated HDAC inhibition suppresses the DNA damage response and contributes to increased DNA damage sensitivity. PLoS ONE. 2015;10:e0134110. doi: 10.1371/journal.pone.0134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Frame FM, et al. HDAC inhibitor confers radiosensitivity to prostate stem-like cells. Br. J. Cancer. 2013;109:3023–3033. doi: 10.1038/bjc.2013.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Chiu HW, et al. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, enhances radiosensitivity and suppresses lung metastasis in breast cancer in vitro and in vivo. PLoS ONE. 2013;8:e76340. doi: 10.1371/journal.pone.0076340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Wang Y, et al. Radiosensitization by irinotecan is attributed to G2/M phase arrest, followed by enhanced apoptosis, probably through the ATM/Chk/Cdc25C/Cdc2 pathway in p53-mutant colorectal cancer cells. Int. J. Oncol. 2018;53:1667–1680. doi: 10.3892/ijo.2018.4514. [DOI] [PubMed] [Google Scholar]
- 373.Wang J, et al. Tetrandrine enhances radiosensitivity through the CDC25C/CDK1/cyclin B1 pathway in nasopharyngeal carcinoma cells. Cell Cycle. 2018;17:671–680. doi: 10.1080/15384101.2017.1415679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Wang Z, et al. Radiosensitization of metformin in pancreatic cancer cells via abrogating the G2 checkpoint and inhibiting DNA damage repair. Cancer Lett. 2015;369:192–201. doi: 10.1016/j.canlet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 375.Yu TY, Garcia VE, Symington LS. CDK and Mec1/Tel1-catalyzed phosphorylation of Sae2 regulate different responses to DNA damage. Nucleic Acids Res. 2019;47:11238–11249. doi: 10.1093/nar/gkz814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Prevo R, et al. CDK1 inhibition sensitizes normal cells to DNA damage in a cell cycle dependent manner. Cell Cycle. 2018;17:1513–1523. doi: 10.1080/15384101.2018.1491236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lee, M. S., Joo, J. W., Kang, H. A. & Kim, K. Mec1 modulates interhomolog crossover and interplays with Tel1 at post double-strand break stages. J. Microbiol. Biotechnol.30, 469–475 (2020). [DOI] [PMC free article] [PubMed]
- 378.Mladenov, E. et al. Strong suppression of gene conversion with increasing DNA double-strand break load delimited by 53BP1 and RAD52. Nucleic Acids Res.48, 1905–1924 (2020). [DOI] [PMC free article] [PubMed]
- 379.Raghavan P, et al. AZD5438, an inhibitor of Cdk1, 2, and 9, enhances the radiosensitivity of non-small cell lung carcinoma cells. Int. J. Radiat. Oncol. Biol. Phys. 2012;84:e507–e514. doi: 10.1016/j.ijrobp.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Kojima K, et al. Cyclin-dependent kinase 1 inhibitor RO-3306 enhances p53-mediated Bax activation and mitochondrial apoptosis in AML. Cancer Sci. 2009;100:1128–1136. doi: 10.1111/j.1349-7006.2009.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rodland GE, et al. The dual cell cycle kinase inhibitor JNJ-7706621 reverses resistance to CD37-targeted radioimmunotherapy in activated B cell like diffuse large B cell lymphoma cell lines. Front. Oncol. 2019;9:1301. doi: 10.3389/fonc.2019.01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Narayan RS, et al. Identification of MEK162 as a radiosensitizer for the treatment of glioblastoma. Mol. Cancer Ther. 2018;17:347–354. doi: 10.1158/1535-7163.MCT-17-0480. [DOI] [PubMed] [Google Scholar]
- 383.Satyanarayana A, Hilton MB, Kaldis P. p21 inhibits Cdk1 in the absence of Cdk2 to maintain the G1/S phase DNA damage checkpoint. Mol. Biol. Cell. 2008;19:65–77. doi: 10.1091/mbc.e07-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Sun L, et al. WEE1 kinase inhibition reverses G2/M cell cycle checkpoint activation to sensitize cancer cells to immunotherapy. Oncoimmunology. 2018;7:e1488359. doi: 10.1080/2162402X.2018.1488359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Friedman J, et al. Inhibition of WEE1 kinase and cell cycle checkpoint activation sensitizes head and neck cancers to natural killer cell therapies. J. Immunother. Cancer. 2018;6:59. doi: 10.1186/s40425-018-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Qiu L, Wang JJ, Ying SH, Feng MG. Wee1 and Cdc25 control morphogenesis, virulence and multistress tolerance of Beauveria bassiana by balancing cell cycle-required cyclin-dependent kinase 1 activity. Environ. Microbiol. 2015;17:1119–1133. doi: 10.1111/1462-2920.12530. [DOI] [PubMed] [Google Scholar]
- 387.Nojima H, et al. Differential properties of mitosis-associated events following CHK1 and WEE1 inhibitor treatments in human tongue carcinoma cells. Exp. Cell Res. 2020;386:111720. doi: 10.1016/j.yexcr.2019.111720. [DOI] [PubMed] [Google Scholar]
- 388.Fu S, et al. Strategic development of AZD1775, a Wee1 kinase inhibitor, for cancer therapy. Expert Opin. Invest. Drugs. 2018;27:741–751. doi: 10.1080/13543784.2018.1511700. [DOI] [PubMed] [Google Scholar]
- 389.Forment JV, O’Connor MJ. Targeting the replication stress response in cancer. Pharmacol. Ther. 2018;188:155–167. doi: 10.1016/j.pharmthera.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 390.Schmidt, M. et al. Regulation of G2/M transition by inhibition of WEE1 and PKMYT1 kinases. Molecules22, 2045 (2017). [DOI] [PMC free article] [PubMed]
- 391.Lee YY, et al. Anti-tumor effects of Wee1 kinase inhibitor with radiotherapy in human cervical cancer. Sci. Rep. 2019;9:15394. doi: 10.1038/s41598-019-51959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Lescarbeau RS, et al. Quantitative phosphoproteomics reveals Wee1 kinase as a therapeutic target in a model of proneural glioblastoma. Mol. Cancer Ther. 2016;15:1332–1343. doi: 10.1158/1535-7163.MCT-15-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Matheson CJ, et al. A WEE1 inhibitor analog of AZD1775 maintains synergy with cisplatin and demonstrates reduced single-agent cytotoxicity in medulloblastoma cells. ACS Chem. Biol. 2016;11:921–930. doi: 10.1021/acschembio.5b00725. [DOI] [PubMed] [Google Scholar]
- 394.Kausar T, et al. Sensitization of pancreatic cancers to gemcitabine chemoradiation by WEE1 kinase inhibition depends on homologous recombination repair. Neoplasia. 2015;17:757–766. doi: 10.1016/j.neo.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Yang, L. et al. Wee1 kinase inhibitor AZD1775 effectively sensitizes esophageal cancer to radiotherapy. Clin. Cancer Res. (2020). 10.1158/1078-0432.CCR-19-3373. [DOI] [PMC free article] [PubMed]
- 396.Chen G, et al. Suppression of Sirt1 sensitizes lung cancer cells to WEE1 inhibitor MK-1775-induced DNA damage and apoptosis. Oncogene. 2017;36:6863–6872. doi: 10.1038/onc.2017.297. [DOI] [PubMed] [Google Scholar]
- 397.Havelek R, et al. Specific inhibition of Wee1 kinase and Rad51 recombinase: a strategy to enhance the sensitivity of leukemic T-cells to ionizing radiation-induced DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2014;453:569–575. doi: 10.1016/j.bbrc.2014.09.123. [DOI] [PubMed] [Google Scholar]
- 398.PosthumaDeBoer J, et al. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer. 2011;11:156. doi: 10.1186/1471-2407-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 400.Gonzalez Besteiro MA, Gottifredi V. The fork and the kinase: a DNA replication tale from a CHK1 perspective. Mutat. Res. Rev. Mutat. Res. 2015;763:168–180. doi: 10.1016/j.mrrev.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Chen Z, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int. J. Cancer. 2006;119:2784–2794. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 402.Bunch RT, Eastman A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin. Cancer Res. 1996;2:791–797. [PubMed] [Google Scholar]
- 403.Patties I, et al. The Chk1 inhibitor SAR-020106 sensitizes human glioblastoma cells to irradiation, to temozolomide, and to decitabine treatment. J. Exp. Clin. Cancer Res. 2019;38:420. doi: 10.1186/s13046-019-1434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Parmar K, et al. The CHK1 inhibitor prexasertib exhibits monotherapy activity in high-grade serous ovarian cancer models and sensitizes to PARP inhibition. Clin. Cancer Res. 2019;25:6127–6140. doi: 10.1158/1078-0432.CCR-19-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.King C, et al. Characterization and preclinical development of LY2603618: a selective and potent Chk1 inhibitor. Invest. N. Drugs. 2014;32:213–226. doi: 10.1007/s10637-013-0036-7. [DOI] [PubMed] [Google Scholar]
- 406.Guster JD, et al. The inhibition of PARP but not EGFR results in the radiosensitization of HPV/p16-positive HNSCC cell lines. Radiother. Oncol. 2014;113:345–351. doi: 10.1016/j.radonc.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 407.Booth L, Roberts J, Poklepovic A, Dent P. The CHK1 inhibitor SRA737 synergizes with PARP1 inhibitors to kill carcinoma cells. Cancer Biol. Ther. 2018;19:786–796. doi: 10.1080/15384047.2018.1472189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Suzuki M, et al. MK-8776, a novel Chk1 inhibitor, exhibits an improved radiosensitizing effect compared to UCN-01 by exacerbating radiation-induced aberrant mitosis. Transl. Oncol. 2017;10:491–500. doi: 10.1016/j.tranon.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Patel R, et al. An orally bioavailable Chk1 inhibitor, CCT244747, sensitizes bladder and head and neck cancer cell lines to radiation. Radiother. Oncol. 2017;122:470–475. doi: 10.1016/j.radonc.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Barker HE, et al. CHK1 inhibition radiosensitizes head and neck cancers to paclitaxel-based chemoradiotherapy. Mol. Cancer Ther. 2016;15:2042–2054. doi: 10.1158/1535-7163.MCT-15-0998. [DOI] [PubMed] [Google Scholar]
- 411.Restelli V, et al. DNA damage response inhibitor combinations exert synergistic antitumor activity in aggressive B-cell lymphomas. Mol. Cancer Ther. 2019;18:1255–1264. doi: 10.1158/1535-7163.MCT-18-0919. [DOI] [PubMed] [Google Scholar]
- 412.Heidler, C. L. et al. Prexasertib (LY2606368) reduces clonogenic survival by inducing apoptosis in primary patient-derived osteosarcoma cells and synergizes with cisplatin and talazoparib. Int. J Cancer. (2019). 10.1002/ijc.32814. [DOI] [PMC free article] [PubMed]
- 413.Sung P. Introduction to the Thematic Minireview Series: DNA double-strand break repair and pathway choice. J. Biol. Chem. 2018;293:10500–10501. doi: 10.1074/jbc.TM118.003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38:e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Tadesse, S. et al. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov. Today. 25, 406–413 (2020). [DOI] [PubMed]
- 416.Pillon MC, Lo YH, Stanley RE. IT’S 2 for the price of 1: multifaceted ITS2 processing machines in RNA and DNA maintenance. DNA Rep. 2019;81:102653. doi: 10.1016/j.dnarep.2019.102653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Katsogiannou M, et al. Integrative proteomic and phosphoproteomic profiling of prostate cell lines. PLoS ONE. 2019;14:e0224148. doi: 10.1371/journal.pone.0224148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Waterworth WM, et al. Phosphoproteomic analysis reveals plant DNA damage signalling pathways with a functional role for histone H2AX phosphorylation in plant growth under genotoxic stress. Plant J. 2019;100:1007–1021. doi: 10.1111/tpj.14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Owusu M, et al. Mapping the human kinome in response to DNA damage. Cell Rep. 2019;26:555–563 e556. doi: 10.1016/j.celrep.2018.12.087. [DOI] [PubMed] [Google Scholar]
- 420.Kim DC, et al. Effects of low dose ionizing radiation on DNA damage-caused pathways by reverse-phase protein array and Bayesian networks. J. Bioinform. Comput. Biol. 2017;15:1750006. doi: 10.1142/S0219720017500068. [DOI] [PubMed] [Google Scholar]
- 421.Nagle PW, et al. Lack of DNA damage response at low radiation doses in adult stem cells contributes to organ dysfunction. Clin. Cancer Res. 2018;24:6583–6593. doi: 10.1158/1078-0432.CCR-18-0533. [DOI] [PubMed] [Google Scholar]
- 422.Gaetani S, et al. DNA damage response in workers exposed to low-dose ionising radiation. Occup. Environ. Med. 2018;75:724–729. doi: 10.1136/oemed-2018-105094. [DOI] [PubMed] [Google Scholar]
- 423.Limpose KL, Corbett AH, Doetsch PW. BERing the burden of damage: Pathway crosstalk and posttranslational modification of base excision repair proteins regulate DNA damage management. DNA Rep. 2017;56:51–64. doi: 10.1016/j.dnarep.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Sunada S, Nakanishi A, Miki Y. Crosstalk of DNA double-strand break repair pathways in poly(ADP-ribose) polymerase inhibitor treatment of breast cancer susceptibility gene 1/2-mutated cancer. Cancer Sci. 2018;109:893–899. doi: 10.1111/cas.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Yuan F, et al. A new regulatory mechanism between P53 And YAP crosstalk By SIRT1 mediated deacetylation to regulate cell cycle and apoptosis in A549 cell lines. Cancer Manag. Res. 2019;11:8619–8633. doi: 10.2147/CMAR.S214826. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 426.Raghunandan, M. et al. Functional crosstalk between the Fanconi anemia and ATRX/DAXX histone chaperone pathways promotes replication fork recovery. Hum. Mol. Genet. (2019). 10.1093/hmg/ddz250. [DOI] [PMC free article] [PubMed]
- 427.Darragh LB, Oweida AJ, Karam SD. Overcoming resistance to combination radiation-immunotherapy: a focus on contributing pathways within the tumor microenvironment. Front. Immunol. 2018;9:3154. doi: 10.3389/fimmu.2018.03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Schaue D, Ratikan JA, Iwamoto KS, McBride WH. Maximizing tumor immunity with fractionated radiation. Int. J. Radiat. Oncol. Biol. Phys. 2012;83:1306–1310. doi: 10.1016/j.ijrobp.2011.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Harding SM, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Yang H, et al. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Cryer B. A COX-2-specific inhibitor plus a proton-pump inhibitor: is this a reasonable approach to reduction in NSAIDs’ GI toxicity? Am. J. Gastroenterol. 2006;101:711–713. doi: 10.1111/j.1572-0241.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- 432.Chen L, et al. Selective COX-2 inhibitor celecoxib combined with EGFR-TKI ZD1839 on non-small cell lung cancer cell lines: in vitro toxicity and mechanism study. Med. Oncol. 2008;25:161–171. doi: 10.1007/s12032-007-9015-1. [DOI] [PubMed] [Google Scholar]
- 433.Marton A, et al. Vanillin analogues o-vanillin and 2,4,6-trihydroxybenzaldehyde inhibit NFkB activation and suppress growth of A375 human melanoma. Anticancer Res. 2016;36:5743–5750. doi: 10.21873/anticanres.11157. [DOI] [PubMed] [Google Scholar]
- 434.Shanmuganathan S, Angayarkanni N. Chebulagic acid chebulinic acid and gallic acid, the active principles of triphala, inhibit TNFalpha induced pro-angiogenic and pro-inflammatory activities in retinal capillary endothelial cells by inhibiting p38, ERK and NFkB phosphorylation. Vasc. Pharmacol. 2018;108:23–35. doi: 10.1016/j.vph.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 435.Kirsch DG, et al. The future of radiobiology. J. Natl Cancer Inst. 2018;110:329–340. doi: 10.1093/jnci/djx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Kobayashi J, et al. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 2002;12:1846–1851. doi: 10.1016/S0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 437.Mischo HE, Hemmerich P, Grosse F, Zhang S. Actinomycin D induces histone gamma-H2AX foci and complex formation of gamma-H2AX with Ku70 and nuclear DNA helicase II. J. Biol. Chem. 2005;280:9586–9594. doi: 10.1074/jbc.M411444200. [DOI] [PubMed] [Google Scholar]
- 438.Stewart GS, et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 439.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Rogakou EP, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 441.Lee KY, et al. MCM8-9 complex promotes resection of double-strand break ends by MRE11-RAD50-NBS1 complex. Nat. Commun. 2015;6:7744. doi: 10.1038/ncomms8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Zhu XD, et al. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 443.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 444.Gatei M, et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 445.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 446.Fukuda T, et al. Alterations of the double-strand break repair gene MRE11 in cancer. Cancer Res. 2001;61:23–26. [PubMed] [Google Scholar]
- 447.Zhong Q, et al. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 448.Heikkinen K, et al. Mutation screening of Mre11 complex genes: indication of RAD50 involvement in breast and ovarian cancer susceptibility. J. Med. Genet. 2003;40:e131. doi: 10.1136/jmg.40.12.e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Becherel OJ, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Peng A, Chen PL. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 2003;278:8873–8876. doi: 10.1074/jbc.C300001200. [DOI] [PubMed] [Google Scholar]
- 451.Luo K, et al. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 2012;31:3008–3019. doi: 10.1038/emboj.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Drane P, et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature. 2017;543:211–216. doi: 10.1038/nature21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 454.Derbyshire DJ, et al. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 456.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Tomida, J. et al. FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 37, e99543 (2018). [DOI] [PMC free article] [PubMed]
- 458.Wu Q, et al. Structure of BRCA1-BRCT/Abraxas complex reveals phosphorylation-dependent BRCT dimerization at DNA damage sites. Mol. cell. 2016;61:434–448. doi: 10.1016/j.molcel.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Hiraike H, et al. Identification of DBC1 as a transcriptional repressor for BRCA1. Br. J. Cancer. 2010;102:1061–1067. doi: 10.1038/sj.bjc.6605577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Shao G, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Yu X, et al. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Kais Z, et al. KIAA0101 interacts with BRCA1 and regulates centrosome number. Mol. Cancer Res. 2011;9:1091–1099. doi: 10.1158/1541-7786.MCR-10-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Wu-Baer F, Ludwig T, Baer R. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol. Cell. Biol. 2010;30:2787–2798. doi: 10.1128/MCB.01056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Ouchi M, et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J. Biol. Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 466.Tibbetts RS, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Gough CA, Gojobori T, Imanishi T. Cancer-related mutations in BRCA1-BRCT cause long-range structural changes in protein–protein binding sites: a molecular dynamics study. Proteins. 2007;66:69–86. doi: 10.1002/prot.21188. [DOI] [PubMed] [Google Scholar]
- 468.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 469.Bass TE, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016;18:1185–1195. doi: 10.1038/ncb3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Schiewer, M. J. et al. PARP-1 regulates DNA repair factor availability. EMBO Mol. Med. 10, e8816 (2018). [DOI] [PMC free article] [PubMed]
- 471.Kratassiouk G, et al. The WEE1 regulators CPEB1 and miR-15b switch from inhibitor to activators at G2/M. Cell Cycle. 2016;15:667–677. doi: 10.1080/15384101.2016.1147631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Timme CR, et al. The DNA-PK inhibitor VX-984 enhances the radiosensitivity of glioblastoma cells grown in vitro and as orthotopic xenografts. Mol. Cancer Ther. 2018;17:1207–1216. doi: 10.1158/1535-7163.MCT-17-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.van Oorschot B, et al. Targeting DNA double strand break repair with hyperthermia and DNA-PKcs inhibition to enhance the effect of radiation treatment. Oncotarget. 2016;7:65504–65513. doi: 10.18632/oncotarget.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Yang L, et al. Inhibition of DNA-PKcs enhances radiosensitivity and increases the levels of ATM and ATR in NSCLC cells exposed to carbon ion irradiation. Oncol. Lett. 2015;10:2856–2864. doi: 10.3892/ol.2015.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Ortiz T, et al. Enhanced induction of apoptosis in a radio-resistant bladder tumor cell line by combined treatments with X-rays and wortmannin. Radiat. Environ. Biophys. 2008;47:445–452. doi: 10.1007/s00411-008-0188-6. [DOI] [PubMed] [Google Scholar]
- 476.Durant ST, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018;4:eaat1719. doi: 10.1126/sciadv.aat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Karlin J, et al. Orally bioavailable and blood–brain barrier-penetrating ATM inhibitor (AZ32) radiosensitizes intracranial gliomas in mice. Mol. Cancer Ther. 2018;17:1637–1647. doi: 10.1158/1535-7163.MCT-17-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Dohmen AJC, et al. Identification of a novel ATM inhibitor with cancer cell specific radiosensitization activity. Oncotarget. 2017;8:73925–73937. doi: 10.18632/oncotarget.18034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Vendetti FP, et al. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J. Clin. Invest. 2018;128:3926–3940. doi: 10.1172/JCI96519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Fujisawa H, et al. VE-821, an ATR inhibitor, causes radiosensitization in human tumor cells irradiated with high LET radiation. Radiat. Oncol. 2015;10:175. doi: 10.1186/s13014-015-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Xia Y, Choi HK, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur. J. Med. Chem. 2012;49:24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 482.Ellinghaus P, et al. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has antitumor activities by inhibition of mitochondrial complex I. Cancer Med. 2013;2:611–624. doi: 10.1002/cam4.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Wang BF, et al. Saikosaponin-D enhances radiosensitivity of hepatoma cells under hypoxic conditions by inhibiting hypoxia-inducible factor-1alpha. Cell. Physiol. Biochem. 2014;33:37–51. doi: 10.1159/000356648. [DOI] [PubMed] [Google Scholar]
- 484.Helbig L, et al. Hypoxia-inducible factor pathway inhibition resolves tumor hypoxia and improves local tumor control after single-dose irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2014;88:159–166. doi: 10.1016/j.ijrobp.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 485.Yu J, et al. Regulation of radiosensitivity by HDAC inhibitor trichostatin A in the human cervical carcinoma cell line Hela. Eur. J. Gynaecol. Oncol. 2012;33:285–290. [PubMed] [Google Scholar]
- 486.Miles MA, Harris MA, Hawkins CJ. Proteasome inhibitors trigger mutations via activation of caspases and CAD, but mutagenesis provoked by the HDAC inhibitors vorinostat and romidepsin is caspase/CAD-independent. Apoptosis Int. J. Program. Cell Death. 2019;24:404–413. doi: 10.1007/s10495-019-01543-x. [DOI] [PubMed] [Google Scholar]
- 487.Marampon F, et al. Histone deacetylase inhibitor ITF2357 (givinostat) reverts transformed phenotype and counteracts stemness in in vitro and in vivo models of human glioblastoma. J. Cancer Res. Clin. Oncol. 2019;145:393–409. doi: 10.1007/s00432-018-2800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Cuneo KC, et al. Wee1 kinase inhibitor AZD1775 radiosensitizes hepatocellular carcinoma regardless of TP53 mutational status through induction of replication stress. Int. J. Radiat. Oncol. 2016;95:782–790. doi: 10.1016/j.ijrobp.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Ma H, et al. Targeting of carbon ion-induced G2 checkpoint activation in lung cancer cells using Wee-1 inhibitor MK-1775. Radiat. Res. 2015;184:660–669. doi: 10.1667/RR14171.1. [DOI] [PubMed] [Google Scholar]
- 490.Zhou ZR, et al. The Chk1 inhibitor MK-8776 increases the radiosensitivity of human triple-negative breast cancer by inhibiting autophagy. Acta Pharmacol. Sin. 2017;38:513–523. doi: 10.1038/aps.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]