Abstract
In this study, 19 endophytic fungi were isolated from Lafoensia pacari, Guazuma ulmifolia, Campomanesia xanthocarpa and Siparuna guianensis. Seventeen strains were molecularly identified as belonging to the genera Colletotrichum, Diaporthe, Bjerkandera, Talaromyces, Cochliobolus, Phaeophlebiopsis, Curvularia, and Xylaraceae. Assays for detecting antioxidant activity were performed by free radical scavenging activity using the DDPH and ABTS + methods. Based on the results with DPPH, two strains were selected to evaluate the presence of flavonoids and anti-inflammatory activity. A strong positive correlation was found between these activities and the presence of flavonoids. The anti-inflammatory activity of endophytic fungi is under explored; however, the Talaromyces obtained the best result of 87.33% protection of erythrocytes and Colletotrichium of 60.71%. This study demonstrated that endophytic fungi associated with selected plants are potential sources of novel antioxidant products.
Keywords: Anti-inflammatories, Antioxidants, Bioactive compounds, Endophytic, Medicinal plants
Introduction
Endophytic microorganisms live inside plant tissues without damaging the host plant. Associations with tissues may be symbiotic or mutualistic and promote benefits for the plant such as disease resistance, tolerance to stress conditions such as herbivorous attack, greater availability of nutrients and, finally an increase in biodiversity (Golinska et al. 2015).
It is believed that all plants have endophytic microorganisms and also that throughout the evolutionary process microorganisms have incorporated some metabolic pathways of plants so that fungi and endophytic bacteria can produce a similar type of phytochemical compounds (Strobel et al., 2004). Endophytic fungi have played an important role as a source of pharmacological active metabolites (Debbab et al. 2013). Recently, the number of studies on bioactive secondary fungal metabolites have progressively increased for use in chronic diseases (Chen et al. 2017). These metabolites produced by endophytes associated with medicinal plants can be explored in an attempt to cure various diseases (Tejesvi et al. 2007).
Other aspects that arouse interest in these microorganisms are the diversity of biological molecules produced and the low risk of these molecules being cytotoxic as symbiotic and mutualistic relations do not cause damage or death to the host plant, which is of great importance for drug manufacturing (Strobel and Daisy 2003; Alvin et al. 2014).
The diversity of microorganism species and the compounds produced by them are severely influenced by the environment. Thus, some strategies can be adapted to search for new compounds, such as selecting plants with a known ethnobotanical history, for instance medicinal plants and selecting environments with great biodiversity (Golinska et al. 2015). Endophytic bioprospecting promotes discovery of natural products with therapeutic value (Kusari and Spiteller (2011).
Medicinal plants have been used to directly isolate and characterize bioactive metabolites. However, the discovery of fungal endophytes within these plants, capable of producing the same compounds, has changed the focus of new drug sources, and fungi stands out. Bioactive natural products of endophytic fungi isolated from different plant species are attracting researchers' attention (Nisa et al. 2015).
In this paper, filamentous fungi were isolated with reports of medicinal properties in the Brazilian savannah of the following species: Lafoensia pacari, Campomanesia xanthocarpa, Guazuma ulmifolia and Siparuna guianensis. Plant species are described in traditional medicine to treat stomach diseases, inflammations, diarrhoea, fever and diabetes (Valentini et al. 2010; Alarcon-Aguilara et al. 1998). To the best of our knowledge, studies on the microbiota of endophytic fungi of these plants have not yet been performed. It can be observed that these plants contain fungi that can produce substances with antioxidant and anti-inflammatory properties.
Materials and methods
Plant samples
Stem samples and leaves with a healthy appearance of Lafoensia pacari (23°35′08.2″S 47°31′06.7″W), Guazuma ulmofia (23°35′08.6″S47°31′08.1″W), Campomanesia xanthocarpa (23°35′09.1″S 47°31′08.9″W) and Siparuna guianensis (23°35′06.2″S 47°31′25.4″W) were collected from forest fragments in the Brazilian savanna (Cerrado biome) in the state of São Paulo in June, 2016. Samples were treated in 24 h.
Isolation of the endophytic fungus
The samples were disinfected with 70% alcohol immersion for 1 min, sodium hypochlorite 2% for 5 min, 70% alcohol for 30 s and sterile distilled water twice. To confirm the efficiency of this disinfection process, a 0.1 mL aliquot of the distilled water used in the last wash was inoculated into malt agar and incubated at 28 °C for 5 days (adapted from Zhao et al. 2012). The samples were aseptically dissected into 25 mm2 pieces of the area and placed on malt agar at 28 °C. The growth of fungal colonies was monitored daily (Santos et al. 2015). The isolated fungi were stored using the Castellani method (1939).
DNA extraction, PCR amplification and molecular identification
Genomic DNA was extracted based on the protocol by Silva et al. (2012). Samples of fungal mycelium (0.1–0.2 g) were incubated in 500 μl of lysis buffer (Tris 50 mM, EDTA 50 mM, NaCl 250 mM, SDS 1.0%, pH 8.0 and 0.2 g glass beads) at 90 °C for 5 min and cooled at − 18 °C until freezing. The samples were mixed by vortex for 30 min, centrifuged for 5 min at 10,000g. The supernatants were extracted successfully in organic solvents (500 μL) and collected after centrifugation for 15 min at 10,000g. The first extraction was in phenol–chloroform (1:1), and the second and third in chloroform.
The supernatants were added to 500 μL of isopropanol and incubated at − 18 °C for 16 h and centrifuged at 20,000g at 4 °C for 20 min. The aqueous phase was removed and the precipitates were collected. The precipitates were washed in 200 μL of ethanol (70%) and centrifuged for 15 min at 4 °C. The precipitates containing the genomic DNA were suspended in 50 μl aliquots of ultra-purified water and stored at − 18 °C.
The ITS region of the rDNA was amplified and sequenced with the universal primers ITS1 (5′‐TCC GTA GGTGAA CCT GCG G‐3′) and ITS4 (5′‐TCC TCC GCT TAT TGA TAT GC‐3´). The polymerase chain reaction—PCR (25 µL total volume) contained 1 μL of template, 1.25 μL of each primer (5 μM each), 0.10 μL of Taq PCR, 0.20 μL dNTP (0.8 mM), 1 μL of MgCl2 (1 mM) and 15.20 μL of ddH2O. The PCR reaction was performed using the method described by Cui et al. (2015).
The PCR products were purified using the Wizard® Genomic DNA-Promega kit, the sequencing was performed by the "Júlio de Mesquita" Sao Paulo State University (UNESP), Faculty of Agrarian and Veterinary Sciences (FCAV). The sequences were edited and verified manually using the Bioedit Sequence Alignment Editor software, Hall (1999). The sequences were submitted to the National Center for Biotechnology Information (NCBI) and compared to sequences of the database using the BLAST tool to identify strains.
Construction of the dendrogram
The ITS sequence from the isolates was aligned using the ClustalW (Thompson et al., 1994) inside the BioEdit 7.2.6 program (Hall 1999). Genetic distance analyses were conducted using the MEGA 10.0 program (Tamura et al. 2013). The distance matrix was calculated with the Kimura (1980) and the construction of the dendrogram from the genetic distances was done by the Neighbor-Joining method (Saitou and Nei 1987), with bootstrap values calculated from 1000 re-samples, using the software included in the MEGA 10.0 program.
Preparation of endophytic fungal extract
The isolates were reactivated in malt agar for 5 days at 28 °C. Each fungus was inoculated into 250 mL Erlenmeyer flasks containing 100 mL of Czapek-Dox medium composed by: NaNO3 (3.0 gL−1); K2HPO4 (1.0 gL−1); MgSO4 7H2O (0.5 gL−1); KCl (0.5 gL−1); FeSO4 (0.01 gL−1); and sucrose (30 gL−1). The fungi were cultivated for 20 days at 150 rpm at 30 °C, adapted from the Cui et al. (2015). Each fungal extract was separated from the mycelium by centrifugation at 7500 rpm for 10 min at 4 °C and then filtered on filter paper (modified from Arora and Chandra 2011).
Determination of antioxidant activity by the DPPH method
The radical-scavenging assay 2. 2-diphenyl-1-picrylhydrazyl (DPPH) was performed according to Zhao et al. (2012) with alterations. Aliquots of 1 mL of each sample of the extract were added to 2 mL (50 µg mL−1) of MeOH solution of DPPH. The solution was vibrated and then incubated in the absence of light for 30 min at 35 °C and the absorbance was measured at 517 nm in a spectrophotometer.
The negative control was the culture medium used (Czapek Dox), the positive control with 1000 μg ascorbic acid ml−1 and the blank sample with ethyl alcohol without the presence of DPPH.
Determination of antioxidant activity by the ABTS+ method
ABTS (2.2 azino bis (3-ethylbenzene thiazoline 6 sulfonic acid) radical was prepared by the reaction between ABTS (7 mMol) and potassium persulfate (2.45 mMol) at a ratio of 1:1 (v/v) in water. The mixture was then incubated in the dark at room temperature for 16 h before use. ABTS solution stock was diluted in ethanol (95%) to obtain an absorbance of 0.700 at 734 nm. Then, 0.5 mL of each fungal extract were mixed with 4.5 mL of ABTS solution. The samples were incubated for 30 min at room temperature. The negative control was the culture medium (Czapek Dox), the positive control with ascorbic acid (1000 μg mL−1) and the blank with distilled water. The radical scavenging activity was calculated according to the equation , where: A0 = absorbance of the control reaction (containing all reagents except the sample), A1 = absorbance of the sample or positive substance) (Li et al. 2017).
Determination of total flavonoids
The total flavonoid content was estimated using the colorimetric method according to Cui et al. (2015), with some modifications. Five milliliters of each extract was mixed with 0.3 mL of NaNO3 (5%). After 5 min, 0.3 mL of AlCl3 (10%) was added and after 5 min, 2 mL of NaOH (1 mol L−1) was added. The final solution was obtained by adding distilled water to the volume of 10 ml, followed by rigorous stirring. After 5 min, the absorbances were measured at 510 nm. The blank of the samples consisted of all reagents except for the fungal extract and the positive control consisted of a 50 μg solution of flavonoid rutin.
Determination of anti-inflammatory activity
The method used was human red blood cell (HRBC) membrane stability after inducting hemolysis by heating. Blood samples were collected from healthy adult volunteers who had not taken any medication or steroids at least 2 weeks before the experiment (Ananthi and Chitra 2013). The experiment was authorized by the Human Ethics Committee—Plataforma Brasil—consultation number 5357917.0.0000.5504. The venipuncture was collected into vacuum collection tubes of 3.6 ml with buffered sodium citrate solution 0.109 M. The samples were centrifuged at 560 g for 10 min at room temperature and the supernatant (plasma) was discarded. The precipitated cells were washed in saline solution (0.85%) and centrifuged again until the supernatant was cleared. The final solution was obtained with 10% v/v (Félix-silva et al. 2014).
The test reaction consisted of 2 mL of each fungal extract solution 1 mg mL−1 (lyophilized fungal extract in normal 0.85% saline solution) mixed with 1 mL of erythrocyte solution (HRBC). The reaction was homogenized manually, incubated at 56 °C for 30 min, cooled to room temperature and centrifuged at 560 g for 5 min at 25 °C, the supernatant was collected and the absorbance was measured in a spectrophotometer at 560 nm. The blank of the reaction consisted of distilled water, the positive control with the drug Ibuprofen 400 mg and the negative control with 0.85% saline solution. The membrane protection percentage (based on the amount of hemoglobin present in the suspension) was calculated according to Rang et al. (2005).
Spectroscopy analysis in the Fourier transform infrared (FTIR)
200 mL samples of each extract were frozen at − 80 °C for 24 h and then lyophilized for 48 h. Dry samples (0.2–0.5 g) were homogenized in KBr -Potassium Bromide and the readings were measured in the region of 400–4000 cm−1, in 4 cm−1 resolution transmission with 64 scans.
Statistical analysis
Significance of the data was determined using one-way ANOVA. The Tukey test was applied to compare mean values whenever the data were significant. All statistical analyses were performed using the InStat for Windows software program (GraphPad Software, San Diego, CA, USA). The significance levels were set at p < 0.05.
Results
Identification of endophytic fungi
Twenty morphologically distinct endophytic fungi were obtained. Fungi identification followed the proposed criteria by Cui et al. (2015). For the same species, the sequence similarity was established as ≥ 99%, the same genera ≥ 95% and the same family < 95%, Table 1.
Table 1.
Identification of endophytic fungi from the comparison of their DNA sequences (based on ITS) to sequences deposited on the NCBI platform
| Fungal isolation | Isolation source | Plant parts | Speciesa | Accession numbers for submitted sequence |
|---|---|---|---|---|
| SF2F | Leaf | Diaporthe sp. | MK757988 | |
| SF5F | Leaf | Diaporthe sp. | MK757989 | |
| SF6F | Leaf | Colletotrichum sp. (orchidearum complex) | MK782046 | |
| SF8F | Siparuna guianensis | Leaf | Colletotrichum sp. (gigasporum complex) | MK757990 |
| SF10F | Leaf | Bjerkandera sp. | MK757991 | |
| SF4C | Stem | Diaporthe sp. | MK757992 | |
| SF7C | Stem | Fungal endophyte | MK757993 | |
| LF2F | Leaf | Phaeophlebiopsis sp. | MK757994 | |
| LF1C |
Lafoensia Pacari |
Stem | Diaporthe sp. | MK757995 |
| LF3C | Caule | Talaromyces sp. | MK757996 | |
| CF1F | Leaf | Curvularia sp. | MK764885 | |
| CF6F | Campomanesia xanthocarpa | Leaf | Bjerkandera sp. | MK764886 |
| CF7F | Leaf | Diaporthe sp. | MK764887 | |
| GF1F | Leaf | Bipolaris sp. | MK764888 | |
| GF2F | Leaf | Xylariaceae sp. | MK764890 | |
| GF3F | Leaf | Colletotrichum sp. (gloeosporioides complex) | MK771151 | |
| GF5F | Guazuma | Leaf | Gnomoniaceae | MK764891 |
| GF13F | ulmifolia | Leaf | Diaporthe sp. | MK764892 |
| GF16F | Leaf | Colletotrichum sp. (boninense complex) | MK764893 |
aIdentified according to consensus phylogram available in the support material
17 isolates were identified, in which a total of 7 genera were found: Colletotrichum, Diaporthe, Bjerkandera, Talaromyces, Bipolaris, Phaeophlebiopsis and Curvularia. The genus Diaporthe was found in all plants and the species. Curvularia and Talaromyces funiculosus was not isolated only in C. xanthocarpa. Colletrichium was found in S. guianensis and G. ulmifolia. Bjerkandera was found in S. guianensis and C. xanthocarpa.
Antioxidant activity screening
Two types of assays with synthetic-free radicals were performed: for the DPPH assay, the antioxidant ranged from 0 to 87.33 ± 0.09% and for the ABTS method the variation was from 5.09 ± 0.68 to 99.00 ± 0.32. Table 2 shows the results obtained using ascorbic acid as control.
Table 2.
Screening of antioxidant activity of endophytic fungi through DPPH and ABTS assays
| Fungal isolation | Antioxidant activity in DPPH (%) | Antioxidant activity in ABTS (%) |
|---|---|---|
| SF2F—Diaporthe | 24.48 ± 0.16 | 91.44 ± 0.60 |
| SF5F- Diaporthe | 29.49 ± 0.38 | 92.01 ± 2.22 |
| SF6F- Colletotrichium | 27.98 ± 1.71 | 86.59 ± 4.57 |
| SF8F- Colletotrichium | 35.42 ± 0.77 | 97.21 ± 1.81 |
| SF10F- Bjerkandera | 30.96 ± 0.20 | 25.84 ± 6.47 |
| SF4C- Diaporthe | 22.71 ± 1.27 | 50.37 ± 7.40 |
| LF2F- Phaeophlebiopsis | 9.41 ± 0.04 | 5.09 ± 0.68 |
| LF1C—Diaporthe | 0 | 95.56 ± 3.26 |
| LF3C—Talaromyces | 87.33 ± 0.09 | 90.23 ± 0.65 |
| CF1F—Curvularia | 5.89 ± 0.26 | 70.1 ± 0 |
| CF6F—Bjerkandera | 1.34 ± 0.05 | 6.73 ± 0 |
| CF7F- Diaporthe | 52.71 ± 0.47 | 93.65 ± 1.09 |
| GF1F—Bipolaris | 0.65 ± 0.81 | 99.00 ± 0.32 |
| GF3F—Colletotrichium | 0 | 70.90 ± 0 |
| GF5F—Gnomoniaceae | 36.82 ± 1.94 | 96.36 ± 0 |
| GF13F—Diaporthe | 39.30 ± 1.88 | 89.94 ± 1,50 |
| GF16F—Colletotrichium | 60.71 ± 0.61 | 96.36 ± 0 |
Based on the results of the two methods, divergence was observed between the responses obtained with ABTS and DPPH. The radical scavenging capacity was stronger for ABTS and this free radical possibly has a more sensitive response compared to DPPH. Finally, the LF3C (Talaromyces) strain showed greater activity in DPPH, and was statistically better than the other isolates (Table 3). The second isolate to present the highest DPPH activity was GF16F (Colletotrichium) with 60.71%.
Table 3.
Results showed statistical importance compared between GF16F and LF3C
| DPPH | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungal isolation (%) | CF1F | CF6F | CF7F | GF1F | GF3F | GF5F | GF13F | GF16F | LF2F | LF1C | LF3C | SF5F | F8F | SF10F | SF4C |
| GF16F (60.71 ± 0.61) | *** ↓ | *** | *** | *** | *** | *** | *** | na | *** | *** | *** ↑ | *** | *** | *** | *** |
| LF3C (87.33 ± 0.09) | *** ↓ | *** | *** | *** | *** | *** | *** | *** | *** | *** | na | *** | *** | *** | *** |
| ABTS | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CF1F | CF6F | CF7F | GF1F | GF3F | GF5F | GF13F | GF16F | LF2F | LF1C | LF3C | SF5F | SF8F | SF10F | SF4C | ||||||||
| GF16F (96 ± 0.00) | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | |||||||
| LF3C (90.23 ± 0.65) | ns | *** | ns | ns | *** | ns | ns | ns | *** | ns | na | ns | ns | *** | *** | |||||||
n = 3 per each data fungal isolation
ns = not significant, na not analyzed
↑↓Increased or decreased in relation to the reference
(*p < 0.05, **p < 0.01 and ***p < 0.001)
The results of antioxidant activity and growth stability in the malt agar medium were used as criteria to select LF3C for complementary tests. Several studies have demonstrated the ability of endophytic fungi to produce antioxidant compounds. Due to the structural differences of antioxidants, more than one method of analysis is recommended (Pan et al. 2017).
Presence of flavonoids
The total flavonoid content was expressed in µg quercetin mL−1. The total flavonoids were 72.68 ± 3.09 of LF3C. The determination by the colorimetric method with aluminum chloride has a positive result when the reaction solution turns yellow. In the presence of aluminum chloride occurs the complexation of Al+3 ions with hydroxyl groups of positions 3 and 5 of the structure of the flavonoid. The greater the presence of flavonoids, the more the solution will tend to yellow (Deng et al. 1998). The presence of flavonoids in the extract may be related to antioxidant action (Cui et al. 2015).
Anti-inflammatory activity
The extract of the LF3C strain presented anti-inflammatory activity of 94.82% ± 1.43. Using HRBC solution is widely found in research with medicinal plant extracts, however, it has not yet been applied to extracts of fungal origin. Some results obtained with plant extracts can be cited, such as those of Parvin et al. (2015) and Kumar et al. (2012), where they obtained inhibition of hemolysis between 40 and 68.40% with the medicinal plants Skimmia anquetilia and Crescentia cujete. Therefore, the results obtained with the LF3C strain can be considered promising.
The HRBC stabilization method was selected, because it is easy to evaluate possible anti-inflammatory compounds in vitro as the erythrocyte membrane behavior is analogous to the lysosomal membrane (Parvin et al. 2015). Lysosomes are responsible for the release of proteolytic enzymes and lysosomal constituents of neutrophils that cause inflammatory processes and tissue damage after being released into the extracellular environment. When the lysosome membrane is stabilized by anti-inflammatories, the release of these enzymes becomes reduced (Guyton and Hall 2006; Saleem et al. 2011). The anti-inflammatory action is detected by the method when the erythrocyte membranes are stabilized preventing the disruption and release of hemoglobin.
Structural analysis of the extract by FTIR
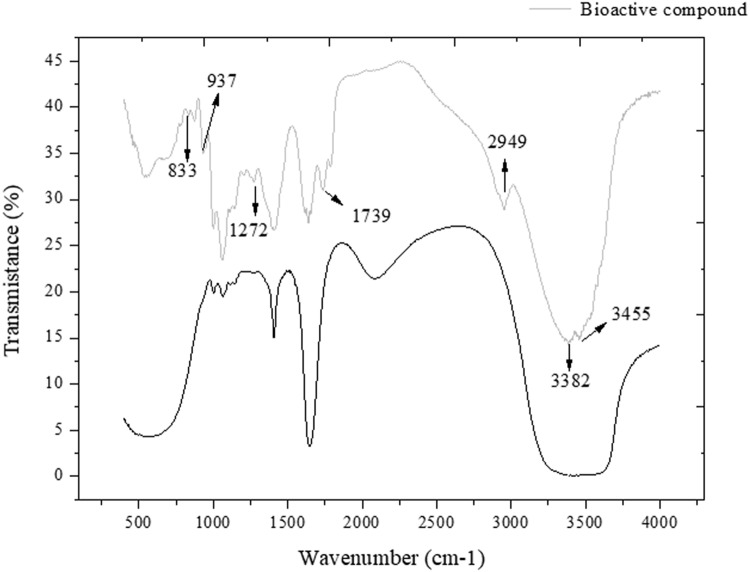
The FTIR analyses were performed to partially characterize the possible functional groups of the fungal extract of the LF3C strain. The FTIR analyses were performed to partially characterize the possible functional groups of the fungal extract of the LF3C strain (Fig. 1). Vibration spectroscopy techniques are used to demonstrate hydroxyl and carbonyl groups present in flavonoids (Pinheiro and Justino 2012).
Fig. 1.
FTIR bands related to the extract of Talaromyces (LF3C)
The peaks found were compared to studies on antioxidants and flavonoids. A spectrum of the Czapek Dox culture medium was compared to the LF3C extract. The peaks that were common between the two were not considered in the analyses as they may only be remnants of the culture medium. Table 4 shows the functional groups found.
Table 4.
Functional clusters found in the extract of the LF3C strain by FTIR analyses
| Funcional class | Peaks | References |
|---|---|---|
| C–H | 833 e 937 | Agatonovic-kustrin et al. (2013) |
| − C–OH | 1272 | Heneczkowski et al. (2001) |
| C=O | 1390 | Heneczkowski et al. (2001) |
| C–H | 1739 | Kumar et al. (2015) |
| OH | 2949 | Agatonovic-kustrin et al. (2013); Heneczkowski et al. (2001) |
| OH | 3382 | Ahmad et al. (2016) |
| OH | 3455 | Ahmad et al. (2016) |
Peaks 833, 937 and 1739 cm−1 potentially represent out-of-plane deformation caused by the presence of aromatic C–H groups. The values of the peaks indicate the presence of C–OH and C=O, which are close to those found for the FTIR spectra of flavonoid quercetin-5′-sulfonated acid. The presence of OH indicated by the peaks 3382 cm−1 and 3455 cm−1 is associated with alcohols and phenols (Table 4). Finally, the OH peak grouping 2947 cm−1 was not relevant for the interpretation, because it usually indicates the presence of water. There are several types of flavonoids, but all share the general structure of C6–C3–C6 phenyl benzopyran, consisting of aromatic rings. The clusters found corroborate the presence of aromatic rings that may be linked to the phenylbenzopyran structure.
Discussion
Endophytic fungi have shown great diversity as further studies are conducted, as it is estimated that all plants have fungi within their tissues (Rodriguez et al. 2009). Plants with ethnobotanical history and differentiated biology as medicinal plants are pointed out as sources of new bioactive compounds (Strobel et al. 2004).
Specific conditions such as humidity, temperature, and nutrients in the soil in which the host plant is located can determine the species of colonizer fungi, including the type of metabolites produced (Wu et al. 2013; Jia et al. 2016). The structure of the fungal populations changes drastically according to the environment; however, plants belonging to the same regions may present a high degree of similarity of taxa and species (D'Amico et al. 2008). The genus talaromyces is widespread in plants, foods, soil and spawn (Zhai et al. 2016). This fungus can produce a wide range of secondary metabolites (Bara et al. 2013).
In this study, 17 different fungi were isolated corroborating the medicinal plants as a source of endophytic microorganisms. The selected plants come from the Cerrado biome (Brazilian savannah) and are used in traditional medicine. Among the genera found, Diaporthe and Talaromyces were predominant, present in three of the plant species used, reinforcing the environmental condition as an important factor in the distribution of endophytic fungi. In the present study, fungi were investigated for their antioxidant properties and afterwards their anti-inflammatory capacity. Most of the isolated fungi presented antioxidant activity. The CF7F and LF3C strains obtained strong activity, especially the species Talaromyces (LF3C). Diaporthe (CF3F) was only found in G. ulmifolia and Phaeophlebiopsis was observed in L. pacari. The plants were collected in the same region in nearby forest fragments, which may have influenced the similarity of the distribution of the species of the genera found.
Antioxidant compounds produced by endophytes can help host plants against the action of species of reactive oxygen that in excess cause cellular damage, also promoting protection against environmental stresses (Prasad et al. 2013).
The presence of flavonoids in plants is essential. Flavonoids act on pigmentation of flowers and fruits, the visual attractiveness of pollinators and protection against ultraviolet light (Ferreyra et al. 2012). Flavonoids also have isolated endophytic fungi and other compounds such as polyphenols and saponins (Pan et al. 2017).
The mechanisms of anti-inflammatory action of flavonoids are not fully elucidated. The inhibition of phospholipase A2 is proposed generating enzymes, cyclooxygenase, and lipoxygenases, which decrease the protein concentration of the inflammatory process, inhibition of the release of histamine, phosphodiesterase and protein kinases (Rathee et al. 2009).
The crude extract from the LF3C strain was selected to present the most promising results for an FTIR analysis. The peaks presented are similar to the presence of aromatic structures, typical of flavonoids. The anti-inflammatory activity of flavonoids produced by fungi has not yet been reported.
The interaction relationships between endophytic fungi, plant cells, metabolites and other diverse environmental and biological factors may offer a wide range of bioactive compounds (Pan et al. 2017; Surveswaran et al. 2007). Thus, the endophytic fungi present in the plants studied here may be a source of new resources for obtaining compounds of interest in biotechnological applications or studies concerning ecological relationships.
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Ananthi T, Chitra M. Screening of invitro anti-inflammatory activity of Michelia champaca linn. Flowers Asian J Pharm ClinRes. 2013;6:71–72. [Google Scholar]
- Alvin A, Miller KI, Neilan BA. Exploring the potential of endophytes from medicinal plants as sources of antimycobacterial compounds. Microbiol Res. 2014;169:483–495. doi: 10.1016/j.micres.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agatonovic-kustrin S, Morton DW, Yusof SP. The use of fourier transform infrared (FTIR) spectroscopy and artificial neural networks (ANNs) to assess wine quality. Modern Chem AppLs. 2013;1:1–8. [Google Scholar]
- Ahmad S, Abdel-salam NM, Ullah R. In vitro antimicrobial bioassays, dpph radical scavenging activity, and FTIR spectroscopy analysis of heliotropium bacciferum. Biomed Res Int. 2016;2016:1–12. doi: 10.1155/2016/3818945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon-Aguilara FJ, Roman-Ramos R, Perez-Gutierrez S, Aguilar-Contreras A, Contreras-Weber CC, Flores-Saenz JL. Study of the antihyperglycemic effect of plants used as antidiabetics. J Ethnopharmacol. 1998;61:101–110. doi: 10.1016/s0378-8741(98)00020-8. [DOI] [PubMed] [Google Scholar]
- Arora DS, Chandra P. Antioxidant activity of Aspergillus fumigatus. ISRN Pharmacol. 2011;2011:1–11. doi: 10.5402/2011/619395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bara R, Aly AH, Pretsch A, Wray V, Wang B, Proksch P, Debbab A. Antibiotically active metabolites from Talaromyces wortmannii, an endophyte of Aloe vera. J Antibiot. 2013;66:491–493. doi: 10.1038/ja.2013.28. [DOI] [PubMed] [Google Scholar]
- Berg G. Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84:11–18. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- Chen S, He L, Chen D, Cai R, Long Y, Lu Y, She Z. Talaramide A, anunusual alkaloid from the mangrove endophytic fungus Talaromyces sp. (HZYX1) as an inhibitor of mycobacterial PknG. New J Chem. 2017;41:4273–4276. [Google Scholar]
- Cui J, Guo T, Ren Z, Zhang N, Wang M. Diversity and antioxidant activity of culturable endophytic fungi from alpine plants of Rhodiola crenulata, R. angusta and R. sachalinensis. Plos One. 2015;13:1–16. doi: 10.1371/journal.pone.0118204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico M, Frisullo S, Cirulli M. Endophytic fungi occurring in Fennel, lettuce, chicory and celery comercial cropsin Southern Italy. Mycol Res. 2008;112:100–107. doi: 10.1016/j.mycres.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Debbab A, Aly AH, Proksch P. Mangrove derived fungal endophytesea chemical and biological perception. Fungal Divers. 2013;61:1–27. [Google Scholar]
- Deng H, Gary J, Van berkel GJ. Electrospray mass spectrometry and uv/visible spectrophotometry studies of aluminum (III) –flavonoid complexes. J mass spectrom. 1998;33:1080–1087. [Google Scholar]
- Ferreyra MLF, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;28:222. doi: 10.3389/fpls.2012.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix-Silva J, Souza T, Menezes Y, Cabral B, Câmara RBG, Silva-junior AA, Rocha HAO, Rebecchi IMM, Zucolotto SM, Fernandes-pedrosa MF. Aqueous leaf extract of Jatropha gossypiifolia L. (Euphorbiaceae) inhibits enzymatic and biological actions of bothrops jararaca snake venom. PLoS ONE. 2014;8:e104952. doi: 10.1371/journal.pone.0104952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, Rai M. Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Antonie Van Leeuwenhoek. 2015;108:267–289. doi: 10.1007/s10482-015-0502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyton AC, Hall JE, editors. Tratado de Fisiologia Médica. Rio de Janeiro: Elsevier; 2006. [Google Scholar]
- Hall TA. Bioedit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/nt. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hammer Ø, Dat H, Ryan PD. PAST. Paleontological statistics software package for education and data analysis. Paleontol Electron. 2001;4:9. [Google Scholar]
- Heneczkowski M, Kopacz M, Nowak D, Kuziar A. Infrared spectrum analysis of some flavonoids. Acta Pol Pharm. 2001;58:415–420. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar V, Ali bhat Z, KumarKhanChashoo DNI. Evaluation of antiinflammatory potential of leaf extracts of Skimmia anquetilia. Asian Pac J Trop Med. 2012;2:627–630. doi: 10.1016/S2221-1691(12)60109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SS, Manoj P, Giridhar P. Fourier transform infrared spectroscopy (FTIR) analysis, chlorophyll content and antioxidant properties of native and defatted foliage of green leafy vegetables. J Food Sci Technol. 2015;52:9131–9139. doi: 10.1007/s13197-015-1959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari S, Spiteller M. Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat Prod Rep. 2011;28:1203–1207. doi: 10.1039/c1np00030f. [DOI] [PubMed] [Google Scholar]
- Jia M, Chen L, Xin H, Zheng C, Rahman K, Han T, Qin L. A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front Microbiol. 2016;9:906. doi: 10.3389/fmicb.2016.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xue F, Yu XGC-MS. FTIR and Raman analysis of antioxidant components of red pigments from Stemphylium lycopersici. Curr Microbiol. 2017;74:532–539. doi: 10.1007/s00284-017-1220-3. [DOI] [PubMed] [Google Scholar]
- Nisa H, Kamili AN, Nawchoo IA, Shafi S, Shameem N, Bandh SA. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: a review. Microb Pathog. 2015;82:50–59. doi: 10.1016/j.micpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Pan F, Su T, Cai S, Wu W. Fungal endophyte-derived Fritillaria unibracteata var. wabuensis: diversity, antioxidant capacities in vitro and relations to phenolic, flavonoid or saponin compounds. Sci Rep. 2017;7:42008. doi: 10.1038/srep42008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin S, Das N, Jahan N, Akhter A, Nahar L, Islam E. Evaluation of in vitro anti-inflammatory and antibacterial potential of Crescentia cujete leaves and stem bark. BMC Res Notes. 2015;8:412. doi: 10.1186/s13104-015-1384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro PF, Justino GC (2012) Structural analysis of flavonoids and related compounds—a review of spectroscopic applications, phytochemicals—a global perspective of their role in nutrition and Healthvenketeshwer Rao (Ed.) InTech: 33–56.
- Prasad R, Kamal S, Sharma PK, Oelmuller R, Varma A. Root endophyte Piriformospora indica DSM 11827 alters plant morphology, enhances biomass and antioxidant activity of medicinal plant Bacopa monniera. J Basic Microbiol. 2013;53:1016–1024. doi: 10.1002/jobm.201200367. [DOI] [PubMed] [Google Scholar]
- Rang HP, Dale MM, Ritter JM, Moore PK. Pharmacology. 5. India: Elsevier; 2005. pp. 27–29. [Google Scholar]
- Rathee P, Chaudhary H, Rathee S, Rathee D, Kumar V, Kohli K. Mechanism of action of flavonoids as anti-inflammatory agents: a review. Inflamm Allergy - Drug Targets. 2009;8:229–235. doi: 10.2174/187152809788681029. [DOI] [PubMed] [Google Scholar]
- Rodriguez R, White J, Jr, Arnold A, Redman R. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saleem TKM, Azeem AK, Dilip C, Sankar C, Prasanth NV, Duraisa R. Anti-inflammatory activity of the leaf extacts of Gendarussa vulgaris Nees. Asian Pac J Trop Med. 2011;1:147–149. doi: 10.1016/S2221-1691(11)60014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos IP, Silva LCN, Silva MV, Araújo JM, Cavalcanti MS, Lima VLM. Antibacterial activity of endophytic fungi from leaves of Indigofera suffruticosa Miller (Fabaceae) Front Microbiol. 2015;6:1–6. doi: 10.3389/fmicb.2015.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GA, Bernardi TL, Schaker PDC, Menegotto M, Rapid VP, Yeast DNA. Extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Brazilian Arch Bio Tech. 2012;55:319–327. [Google Scholar]
- Stierle A, Strobel G, Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- Strobel GA, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Pro. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- Surveswaran S, Cai YZ, Corke H, Sun M. Systematic evaluation of natural phenolic antioxidants from 133 Indian medicinal plants. Food Chem. 2007;102:938–953. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejesvi MV, Nalini MS, Mahesh B, et al. New hopes from endophytic fungal secondary metabolite. Bol Soc Quim Mex. 2007;1:19–26. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Han T, Li W, Jia M, Xue L, Rahman K, Qin L. Geographic and tissue influences on endophytic fungal communities of Taxus chinensis var. mairei in China. Curr Microbiol. 2013;66:40–48. doi: 10.1007/s00284-012-0235-z. [DOI] [PubMed] [Google Scholar]
- Valentini CMA, Rodríguez-Ortíz CE, Coelho MFB. Siparuna guianensis Aublet (negramina): uma revisão. Brazilian J med plants. 2010;12:96–104. [Google Scholar]
- Zhai MM, Li J, Jiang CX, Shi YP, Di DL, Crews P, Wu QX. The bioactive secondary metabolites from Talaromyces species. Nat Prod Bioprospect. 2016;6:1–24. doi: 10.1007/s13659-015-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xu L, Jiang C. Methods for the study of endophytic microorganisms from traditional chinese medicine. Methods Enzymol. 2012;517:3–21. doi: 10.1016/B978-0-12-404634-4.00001-2. [DOI] [PubMed] [Google Scholar]