Abstract
Patients with advanced hepatocellular carcinoma (HCC) are often unable to tolerate chemotherapy due to liver dysfunction in the setting of cirrhosis. We investigate high‐density lipoprotein (HDL)‐mimicking peptide phospholipid scaffold (HPPS), which are nanoparticles that capitalize on normal lipoprotein metabolism and transport, as a solution for directed delivery of small interfering RNA (siRNA) cargo into HCC cells. Spalt‐like transcription factor 4 (SALL4), a fetal oncoprotein expressed in aggressive HCCs, is specifically targeted as a case study to evaluate the efficacy of HPPS carrying siRNA cargo. HPPS containing different formulations of siRNA therapy against SALL4 were generated specifically for HCC cells. These were investigated both in vitro and in vivo using fluorescence imaging. HPPS‐SALL4 effectively bound to scavenger receptor, class B type 1 (SR‐BI) and delivered the siRNA cargo into HCC cells, as seen in vitro. HPPS‐SALL4 effectively inhibited HCC tumor growth (P < 0.05) and induced a 3‐fold increase in apoptosis of the cancer cells in vivo compared to HPPS‐scramble. Additionally, there was no immunogenicity associated with HPPS‐SALL4 as measured by cytokine production. Conclusion: We have developed unique HDL‐like nanoparticles that directly deliver RNA interference (RNAi) therapy against SALL4 into the cytosol of HCC cells, effectively inhibiting HCC tumor growth without any systemic immunogenicity. This therapeutic modality avoids the need for hepatic metabolism in this cancer, which develops in the setting of cirrhosis and liver dysfunction. These natural lipoprotein‐like nanoparticles with RNAi therapy are a promising therapeutic strategy for HCC.
Patients with advanced HCC are often unable to tolerate chemotherapy due to liver dysfunction in the setting of cirrhosis. We herein investigate High‐density Lipoprotein (HDL)‐mimicking Peptide Phospholipid Scaffold (HPPS), nanoparticles that capitalize on normal lipoprotein metabolism and transport, as a solution for directed delivery of RNA interference (siRNA) cargo into HCC cells. Our HDL‐like nanoparticles effectively inhibit HCC tumor growth without any systemic immunogenicity and avoid the need for hepatic metabolism in this cancer that develops in the setting of cirrhosis and liver dysfunction. Therefore, these natural lipoprotein‐like nanoparticles with RNA interference therapy are a promising therapeutic strategy for HCC.

Abbreviations
- ApoA
apolipoprotein A
- chol
cholesterol
- DAB
3,3′‐diaminobenzidine
- FPLC
fast protein liquid chromatography
- HCC
hepatocellular carcinoma
- HDL
high‐density lipoprotein
- HPPS
high‐density lipoprotein‐mimicking peptide phospholipid scaffold
- IHC
immunohistochemistry
- PBS
phosphate‐buffered saline
- qRT‐PCR
quantitative reverse‐transcription polymerase chain reaction
- RNAi
RNA interference
- RT‐PCR
reverse‐transcription polymerase chain reaction
- SALL4
Spalt‐like transcription factor 4
- siRNA
small interfering RNA
- SR‐B1/SCARB1
scavenger receptor, class B type 1
- TUNEL
terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick‐end labeling
- UV
ultraviolet
- vol
volume
Hepatocellular carcinoma (HCC) continues to rise in incidence worldwide and is the third most common cause of cancer‐related death.( 1 , 2 ) The 5‐year survival rate is a dismal 12% for this high‐fatality cancer.( 2 , 3 ) Patients with HCC are often diagnosed at an advanced stage of disease and in the setting of cirrhosis with associated liver dysfunction where curative therapy is no longer an option.( 4 ) This means that treatment choices for HCC are dependent not only on the stage of cancer but also whether liver dysfunction is concurrently present.( 4 , 5 ) A meta‐analysis of observational studies has revealed that HCC treatment is underused; only 22% of patients receive curative treatments and 53% noncurative therapies.( 6 ) The multikinase inhibitor sorafenib has been used as the first‐line agent for advanced HCC.( 7 ) However, sorafenib can only be used in patients with adequate hepatic reserve and increases overall survival by only 3 months.( 7 , 8 , 9 ) Additionally, it is often compromised by systemic side effects, such as hand–foot skin reaction, diarrhea, nausea/vomiting, and weight loss in 81.9% of patients, requiring dose reduction or interruption of therapy.( 10 ) Effective titration of chemotherapy dosage while limiting side effects is difficult given dependence on drug‐metabolizing enzymes in the liver.( 9 ) There is, therefore, a dire need to develop therapies for advanced HCC that bypass hepatic metabolism and deliver therapy directly into the malignant hepatocytes.
High‐density lipoprotein (HDL)‐mimicking peptide phospholipid scaffold (HPPS) are natural lipoprotein‐like nanoparticles with multifunctional capabilities. These nanoparticles capitalize on normal lipoprotein metabolism and transport. Given that the liver serves as the principal site for HDL uptake, HPPS particles are a promising solution to delivery of better tolerated and more effective chemotherapy for HCC. HPPS as a biomimetic to HDL can directly deliver cargo (small interfering RNA [siRNA] or drug) into the cytosol of liver cells through specificity for the scavenger receptor, class B type 1 (SR‐BI, also known as SCARB1), resulting in the majority of these particles being taken up by the liver.( 11 , 12 )
In this study, we evaluate the potential of HPPS to effectively deliver therapeutics directly into HCC cells both in vitro and in vivo. As a case study to investigate this strategy, we use the RNA interference (RNAi) therapy of Spalt‐like transcription factor 4 (SALL4), an oncofetal gene reported as overexpressed in aggressive HCC.( 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 ) SALL4 encodes a transcription factor important for self‐renewal of embryonic stem cells and maintenance of their pluripotency, and therapeutic targeting of SALL4 through different approaches has been shown to effectively suppress tumor growth and progression.( 13 , 18 , 22 ) We examine the generation of HPPS, specifically for HCC cells, that contain different formulations of siRNA therapy against SALL4. Using HPPS to deliver RNAi therapy directly into HCC cells appears to be a viable therapeutic strategy and paves the way for further investigation into this modality for HCC.
Materials and Methods
Assessment of SR‐BI and SALL4 Expression in Human HCC Tumors
We determined the expression of SR‐BI by immunohistochemistry (IHC) in eight human HCC tumors versus adjacent liver parenchyma, using 4‐μm thick sections of formalin‐fixed paraffin‐embedded specimens from the University Health Network cancer biobank. After dewaxing procedures, antigen unmasking was performed with standard heat antigen retrieval (Tris‐ ethylene diamine tetraacetic acid buffer, pH 9.0). The dilution for SR‐BI antibody (ab52629; Abcam, Inc., Cambridge, MA) was 1:600 for 60 minutes. Endogenous peroxidase was blocked with 3% hydrogen peroxide. The detection system used was species specific by the ImmPRESS polymer system (anti‐rabbit immunoglobulin G, catalog number MP ‐7401; Vector Labs). After following the kit instructions, color development was performed with freshly prepared 3,3′‐diaminobenzidine (DAB; Dako Diagnostics). Finally, sections were counterstained lightly with Mayer’s hematoxylin, dehydrated in alcohols, cleared in xylene, and mounted with Permount mounting medium (catalog number SP15‐500; Fisher Scientific).
SR‐BI expression was established using the Chen Liver data set( 23 ) available from Oncomine (https://www.oncomine.org),( 24 ) a public functional genomics data repository of high‐throughput array data. SR‐BI expression data, after the selection of P < 0.05 as threshold, were obtained comparing the HCC tumor samples (n = 104) to normal liver samples (n = 76) and plotted using Prism8 by GraphPad Software. This established the validity of examining HPPS as vehicles for delivery of cargo into HCC cells, given that SR‐BI is the receptor for these HDL‐mimicking nanoparticles, and the relevance of targeting SALL4 in HCC tumors.
Preparation and Optimization of HPPS‐SALL4
HPPS was prepared and quantified as described.( 25 ) Briefly, a lipid film consisting of 3 μmol of 1, 2‐dimyristoyl‐sn‐glycero‐3‐phosphocholine (DMPC) and 0.3 μmol cholesteryl oleate was prepared under a stream of nitrogen gas. The completely dried lipid film was hydrated with 1.0 mL phosphate‐buffered saline (PBS) buffer (150 mM, pH 7.5) and sonicated using Bioruptor at low frequency (30 kHz) for 30 cycles (30 seconds on/30 seconds off) at 40°C. The apolipoprotein A‐1 (ApoA‐1) mimetic reverse 4F peptide (Ac‐FAEKFKEAVKDYFAKFWD, 2.3 mg) in 1 mL PBS was added to the rehydrated solution, and the mixture was kept shaking at 4°C overnight. To optimize the HPPS‐SALL4 formulation, cholesterol (chol)‐si‐SALL4 and HPPS were mixed and incubated with various molar ratios of chol‐si‐SALL4:HPPS (1:1, 3:1, 5:1, 10:1). Chol‐modified SALL4 siRNAs were then mixed with the purified HPPS and incubated for 1 hour at room temperature. Following 1 hour incubation, the mixture was loaded onto a 1.5% agarose gel to check particle integrity by electrophoresis. The gel was run at 100 V for 45 minutes and subsequently imaged by ultraviolet (UV) transillumination. The HPPS‐SALL4 mixture with optimal ratio was then purified by fast protein liquid chromatography (FPLC) to remove free chol‐si‐SALL4. The purified HPPS‐SALL4 had a mean size of 13.8 ± 0.17 nm.
Morphology and Size Measurement
The morphology and size of HPPS‐SALL4 were determined by staining with 1% uranyl acetate and imaged by transmission electron microscopy using a modern Hitachi H‐7000 transmission electron microscope (Hitachi, Inc., Japan) equipped with a digital image acquisition system. Particle size distributions of HPPS‐SALL4 were measured by dynamic light scattering (Zetasizer Nano‐ZS90; Malvern Instruments, United Kingdom) using a 4.0 mW He‐Ne laser operating at 633 nm and a detector angle of 90 degrees.
Cell Culture
LdlA(mSR‐BI) and ldlA7 cell lines, with overexpression and normal expression of mouse SR‐BI (mSR‐BI), respectively, were gifts from Dr. Monty Krieger (Massachusetts Institute of Technology, Cambridge, MA). Human epidermoid carcinoma KB, fibrosarcoma HT1080, and HCC (HepG2, HEP3B, SNU398, and HUH‐7) cell lines were purchased from the American Type Culture Collection. LdlA7 cells were cultured in Hams F‐12 media with 2 mM l‐glutamine, 100 U/mL penicillin–streptomycin, and 5% fetal bovine serum (FBS). LdlA(mSR‐BI) cells were cultured under similar conditions with 300 μg/mL G418. The KB, HT1080, HepG2, HEP3B, SNU398, and Huh‐7 cells were cultured in modified Eagle’s medium supplemented with 2 mM l‐glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% FBS.
Confocal Study
Two Chinese hamster ovary (CHO) mutant clone cell lines (SR‐BI‐negative ldlA7 cells and ldlA[mSR‐BI] [ldlA7 with stably transfected mSR‐BI receptor]) were used as negative and positive controls for SR‐BI, respectively. These were compared to two human tumor cell lines (KB and HT1080). These cell lines were seeded onto eight‐well cover glass‐bottom chambers (Nunc Lab‐Tek; Sigma‐Aldrich) (2 × 104/well) and incubated for 24 hours at 37°C in an atmosphere of 5% CO2 in a humidified incubator. The Cy5.5‐chol‐siRNA‐loaded HPPS (HPPS‐Cy5.5‐siRNA) was added into chamber wells at a siRNA concentration of 200 nm. Following 3 hours of incubation, media was replaced with fresh cell culture media, and cells were imaged using Olympus FV1000 laser confocal scanning microscopy (Olympus, Tokyo, Japan) with an excitation wavelength of 633 nm for exciting Cy5.5 staining and 405 nm for exciting Hoechst 33258 staining.
Assessment by Western Blot of SR‐BI Expression in HCC Cell Lines
This experiment was performed in order to assess the baseline expression of SR‐BI, the receptor for our HDL‐mimicking nanoparticles (HPPS), in different cancer cell lines. The expression of SR‐BI on the cell surface is a requirement for uptake of HPPS. KB, HT1080, Hep3B, SNU398, and Huh7 HCC cell lines were seeded on a six‐well plate at a density of 2 × 105 cells per well and grown for 24 hours. Cell lysates were prepared with radio immunoprecipitation assay buffer plus complete protease inhibitors (Roche Diagnostics, Mannheim, Germany). Protein concentration was determined by bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL) and immunoblotted using antibodies specific for SCARB1 (anti‐SR‐BI antibody EP1556Y, 1:1,000; Abcam, Inc.). Immunoreactive proteins were detected using goat anti‐mouse horseradish peroxidase‐conjugated secondary antibody (GenScript, Pascataway, NJ) and Clarity Western enhanced chemiluminescence (Bio‐Rad Laboratories Ltd., Ontario, Canada). Membranes were stripped and immunoblotted with a mouse monoclonal antibody against beta‐actin (1:5,000; Sigma). Imaging was carried out using a Gel Logic 2200 Imaging System (Kodak, Rochester, NY).
In Vitro Evaluation of Different siRNAs Against SALL4
This experiment was performed in order to assess the efficacy of different siRNA sequences against SALL4, enabling selection of the siRNA that best interferes with SALL4. SALL4 gene expression and down‐regulation by RNAi treatment were evaluated by quantitative reverse transcription PCR (RT‐qPCR). Hep3B, SNU398, and Huh7 HCC cells were seeded on a six‐well plate (1.2 × 105 cells/well). Various unmodified siRNA sequences, including SALL4‐sequence1, qiagen1, qiagen4, SALL4‐sequence 2, bioneer, and ambion, were added at a 50‐nM concentration by Lipofectamine 2000 delivery 24 hours after seeding. Details regarding the specific sequences for each siRNA are reported in the [Link], [Link], [Link], [Link]. The same cell lines without RNAi treatment were used as control. Following incubation for 48 hours, total RNA was isolated from each well using RNeasy Mini kit (Qiagen) and quantified by UV spectrophotometry. Complementary DNA (cDNA) was prepared from 2 μg total cellular RNA using RevertAid H Minus M‐MuLV Reverse Transcriptase (Fermentas, Canada) and oligo‐dT18 as primers, according to the manufacturer’s recommendations. The SALL4 gene was amplified in 12.5 μL PCR mix (2×), 9.5 μL water, 1 μL oligonucleotide primer, and 2 μL cDNA. Cycles of amplification were run as follows: denaturation at 94°C for 0.5 minutes, annealing at 57°C for 0.5 minutes, and extension at 72°C for 0.5 minutes. The primers were as follows: SALL4, sense GGTGGATGTCAAACCCAAAGA, and antisense AGTCCCAAAAACCTTGCTACAGTACT; beta 2‐microglobulin (B2M), sense 5′‐AGCAGAGAATGGAAAGTCAAA‐3′, antisense 5′‐TGTTGATGTTGGATAAGAGAA‐3′. Following amplification by PCR, samples were subjected to electrophoresis in 1% agarose gel containing 0.002% (volume [vol]/vol) Gelred. Gels were photographed with UV illumination.
Xenograft Models in Nude Mice
All animal studies were conducted in accordance with protocols approved by the Animal Care Committee at the University Health Network. Female nude mice (7‐8 weeks old) were inoculated subcutaneously with 2 × 106 KB cells (in 100 μL PBS) and HT1080 cells in the right and left flanks, respectively, to generate a dual tumor model for the SR‐BI targeting study. Female nude mice (7‐8 weeks old) were inoculated subcutaneously with 2 × 106 Hep3B cells on the right flank for the therapeutic RNAi study.
In Vivo and Ex Vivo Fluorescence Imaging
Dual xenograft tumor‐bearing mice (KB tumor on right and HT1080 tumor on left) were injected intravenously with HPPS‐Cy5.5‐siRNA at a Cy5.5‐chol‐siRNA dose of 1 mg/kg. Whole‐body fluorescence images were taken before injection and then at 5 minutes, 30 minutes, 3 hours, 6 hours, 9 hours, and 24 hours postinjection using the CRI Maestro imaging system with a red filter set (excitation filter, 615‐665 nm; emission filter, 700 nm long pass, signal collection from 680 to 950 nm in 10‐nm steps) and an exposure time of 1,000 milliseconds. After 24 hours of imaging, mice were killed, liver and tumor tissues were excised and washed with PBS, and tissues were imaged using the Maestro imaging system. Subsequently, all tissues were frozen in liquid nitrogen and then cut into 5‐µm thick slides using a Leica CM3050S cryostat. Slides were imaged by an Olympus FV1000 laser confocal scanning microscopy (Olympus) with an excitation wavelength of 633 nm.
In Vivo Therapeutic Efficacy of HPPS‐SALL4
Hep3B xenograft tumor‐bearing mice were used in the therapeutic study. Mice were subjected to treatment when the tumor volume reached 40‐50 mm3 on day 10 after the inoculation. They were randomly assigned to two treatment groups and were administered intravenously with HPPS‐SALL4 and HPPS‐scramble (n = 5), respectively. They were given three doses of chol‐siRNA at 8 mg/kg in 200 μL at days 0, 2, and 4. All mice were killed on day 6, and various organs and tumor tissues were excised.
In Vivo Antitumor Activity
Tumor dimensions were measured with vernier calipers, and volumes were calculated as follows: tumor volume (mm3) = width2 (mm2) × length (mm)/2. Tumor dimension was measured on day 0 before treatment and day 2, day 4, and day 6 after the initiation of treatment. All measurements were conducted in a blinded fashion.
Evaluation of Apoptosis by Terminal Deoxynucleotidyl Transferase–Mediated Deoxyuridine Triphosphate Nick‐End Labeling Assay
A terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick‐end labeling (TUNEL) assay was performed on frozen sections using the in‐situ end‐labeling technique for apoptosis as reported.( 26 ) Slides were fizzed with 1% paraformaldehyde in PBS, pH 7.4, preferably for 10 minutes at room temperature. After draining off excess liquid and washing twice with PBS (5 minutes each), slides were postfixed in precooled ethanol:acetic acid (2:1) for 5 minutes at −20°C. The ethanol:acetic acid mixture was removed, and slides were washed with two changes of PBS. Endogenous peroxidase activity was blocked using 3% aqueous hydrogen peroxide and endogenous biotin activity, using the avidin/biotin blocking kit (catalog number TA‐015‐BB; Lab Vision). Sections were treated with buffer A (50 mM TRIS‐HCl [pH 7.5], 50 mM MgCl2∙6H2O, 100 mM β‐mercaptoethanol, and 0.005% bovine serum albumin) for 5‐10 minutes and then were incubated with a biotin‐nucleotide cocktail in a water bath at 37°C for 1.5 hours. After washing with PBS, slides were labeled with Ultra Streptavidin Horseradish Peroxidase Labeling Reagent (catalog number BP2378ID; Labs Inc.) for 30 minutes at room temperature. Slides were then developed with freshly prepared DAB (Dako K3468), counterstained with Mayer’s hematoxylin, dehydrated, and mounted. Slides were scanned with an Aperio Whole Slide Scanner and analyzed using an Aperio ImageScope. Only DAB‐positive staining apoptosis cells with the morphology of cytoplasmic condensation, loss of cell to cell contact, and cell shrinkage were counted as TUNEL‐positive cells.
Evaluation of Immunogenicity of HPPS‐SALL4 In Vivo
To assess for the immunogenicity effects of HPPS‐SALL4 during treatment, healthy BALB/c female mice (6‐7 weeks old) received intravenous administration of saline (0.3 mL), HPPS (2,000 mg/kg), HPPS‐SALL4 (siRNA, 2,000 μg/kg; HPPS, 14 mg/kg), or chol‐si‐SALL4 (2,000 μg/kg) (n = 4 for each group). After injection, mouse blood samples (200 µL each) were collected from the saphenous vein for analysis of different cytokines at 3 hours and 7 days after injection.
Statistical Analysis
The two‐tailed Student t test was used to detect statistically significant differences at specific time points between HPPS‐SALL4 and HPPS‐scramble in the TUNEL and in vivo antitumor activity study. P < 0.05 was considered statistically significant.
Results
Clinical Relevance of Investigating HPPS as a Therapeutic Vehicle in HCC
SR‐BI is the receptor for HDL on the surface of hepatocytes. IHC staining for SR‐B1 was examined for 13 human cancer types, including eight human HCC tumor biopsy samples, and all HCC tumors were SR‐BI positive (Supporting Table S1). SR‐BI was more highly expressed in human HCCs compared to noncancerous adjacent liver tissue (Fig. 1A). Further confirmation was obtained through examination of SR‐B1 gene expression, which was 2.4‐fold increased (log2 fold change, 1.34; P = 3.38e‐05) on average in 104 HCC samples compared to normal liver tissue (n = 76) (Fig. 1B). These data suggest HPPS as a promising vehicle for therapeutic delivery of RNA interference into HCC cells.
Fig. 1.
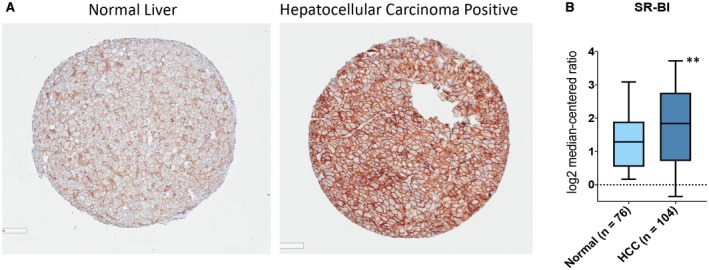
Clinical Relevance of investigating High‐density Lipoprotein (HDL) Peptide Phospholipid Scaffold (HPPS) as a therapeutic vehicle and SALL4 as a therapeutic target in Hepatocellular Carcinoma (HCC). (A) Expression of SR‐BI based on immunohistochemical staining in Human HCC tissue as compared to non‐cancerous adjacent liver. (B) Gene expression of SR‐BI. Box plots plotted using Prism8 (GraphPad Software) were derived from gene expression data in Oncomine, comparing expression of specific SR‐BI gene in in normal liver (left plot) and in HCC tissue (right plot). SB‐BI is higher (log2FC = 1.34, p‐value = 3.38e‐05) in 104 HCCs as compared to normal liver tissue (n = 76), making it therapeutically targetable. Graph shows median interquartile range (box), median (orizontal line), and outliers (whiskers). **Indicates statistical significance (T‐test P < 0.01).
Synthesis of HPPS‐SALL4
The HPPS‐siRNA nanoparticle has a hydrophobic cholesteryl oleate core enveloped by a phospholipid monolayer and intercalated with siRNA payloads; it is constrained by ApoA‐1 mimetic peptides to give an ultrasmall particle size (<20 nm), as illustrated in Fig. 2A.( 25 , 27 , 28 , 29 ) The steps in the synthesis of HPPS‐SALL4 are described in Materials and Methods and summarized in Fig. 2B. Briefly, cholesterol‐modified SALL4 siRNAs were mixed with the purified HPPS and incubated. The mixture was finally purified by fast FPLC to obtain HPPS‐SALL4, subjected to gel shift, and further purified by FPLC. The purified HPPS‐SALL4 had a mean size of 13.8 ± 0.17 nm (Supporting Fig. S1).
Fig. 2.
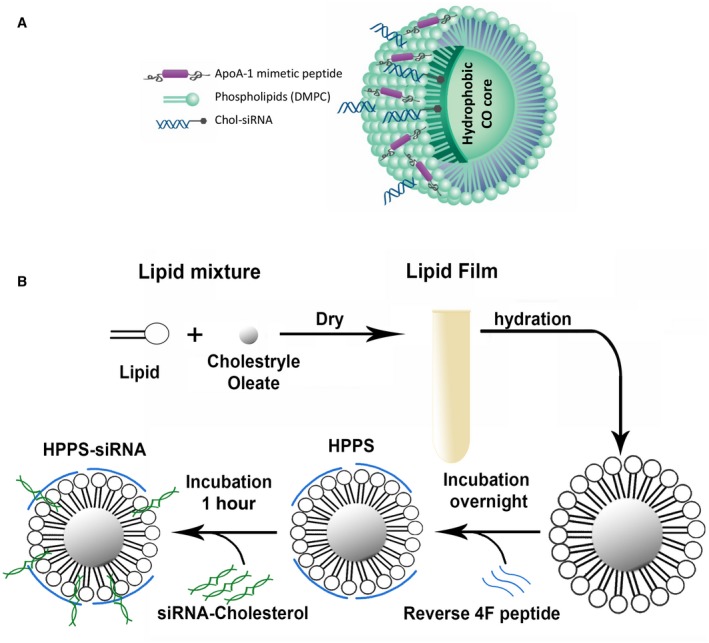
Schematic of HPPS‐siRNA and the synthesis procedure. (A) Schematic figure of HPPS containing siRNA against SALL4. (B) Steps in the synthesis of HPPS, starting with the lipid mixture. Abbreviations: CO, cholestryle oleate; DMPC, 1, 2‐dimyristoyl‐sn‐glycero‐3‐phosphocholine.
Specificity of the Delivery of HPPS‐SALL4 to HCC Cells
To investigate the specificity of HPPS‐SALL4, Cy5.5 fluorescent dye was conjugated to cholesterol‐labeled siRNA as a fluorescent mock siRNA (Cy5.5‐chol‐siRNA) or synthesized HPPS‐Cy5.5‐siRNA, and its delivery was investigated by fluorescence imaging. Confocal microscopy was used to assess intracellular uptake of Cy5.5‐siRNA into various cells with different levels of SR‐BI expression. Four cell lines were included. Two CHO mutant clones, (1) ldlA7 cells (SR‐BI‐negative cells) and (2) ldlA[mSR‐BI] (ldlA7 with stably transfected mSR‐BI receptor), were used to delineate the low and high SR‐BI receptor expression groups, respectively.( 29 ) KB and HT1080 cells, which are cancer cell lines with high and low SR‐BI expression, respectively, as evidenced by western blot analysis (Fig. 3A), were evaluated as well. HPPS selectively delivered Cy5.5‐siRNAs into SR‐BI‐positive cell lines versus negative cell lines (Fig. 3B). In addition, uptake of siRNA in KB cells was inhibited in the presence of excess native HDL (Fig. 3C). A 50‐fold increase of native HDL was able to block almost 68% of HPPS‐siRNA uptake in SR‐BI‐positive cancer cells (Fig. 3D). In vivo evaluation was further conducted on a dual tumor mouse model bearing KB (SR‐BI+) and HT1080 (SR‐BI−) tumors on the right and left flank, respectively. The results (Fig. 3E) revealed a significantly higher uptake of fluorescent siRNA by the KB tumor as opposed to the HT1080 tumors within 24 hours. In addition, ex vivo fluorescence imaging of the tissue resected at 24 hours postinjection showed dominantly higher siRNA signal in the KB tumor compared to normal liver tissue and SR‐BI‐negative HT1080 tumor (Supporting Fig. S2A). Fluorescence microscopic imaging of the tissue slices further confirmed significantly higher uptake of Cy5.5‐siRNA in KB tumor cells (Supporting Fig. S2B). These data support that HPPS enabled efficiently targeted delivery of siRNA into tumor cells by SR‐BI.
Fig. 3.
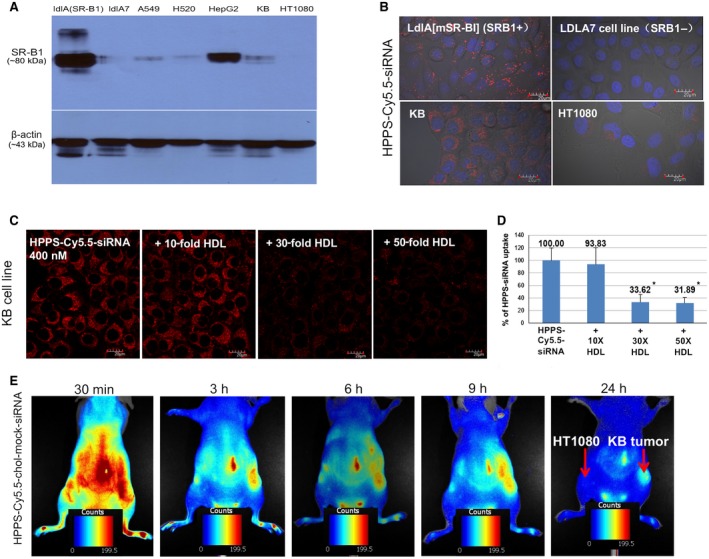
Demonstration of SR‐BI specificity in vitro and in vivo. (A) SR‐BI expression in different cell lines by western blot assay. (B) HPPS‐siRNA uptake into SR‐BI‐positive (ldlA[mSR‐BI] and KB) versus SR‐BI‐negative (ldlA7 and HT1080) cancer cells, magnification 60×. (C) Uptake into SR‐BI‐positive cancer cells (KB) is dependent on the concurrent presence of HDL; increasing HDL concentration results in decreased uptake of HPPS, magnification 60×. (D) Quantitative cellular uptake of HPPS‐siRNA in KB cells without or with the presence of HDL at different concentrations; data show mean ± SEM. (E) In vivo delivery in SR‐BI‐positive KB tumor versus less SR‐BI‐expressing HT1080 tumor, magnification 60×. *P < 0.01, t test; n = 3. Abbreviations: h, hour; min, minute.
Silencing Efficacy of SALL4 siRNAs
The different sequences of siRNA against SALL4 were evaluated in the three HCC cell lines (Hep3B, SNU‐398, and HuH‐7) that expressed high levels of SALL4( 30 ) by delivering unmodified siRNA at 50 nM by lipofectamine. Both sequence 1 and sequence 2 were able to induce significant down‐regulation of SALL4 in vitro in the examined cell lines (P < 0.05; Fig. 4A). To apply using HPPS delivery of siRNA for HCC application, we first checked expression of SR‐BI by quantitative RT‐PCR (qRT‐PCR) in different cancer cell lines, including three HCC cell lines (SNU398, Huh‐7, and Hep3B). The values were normalized against SR‐BI expression in SR‐BI+ KB cells (Fig. 4B). Hep3B showed higher SR‐BI expression when compared to other HCC cell lines (Huh7 or SNU398; P < 0.01) and were chosen as the targeted cell line for further RNAi evaluation by HPPS delivery in vitro and in vivo. To optimize loading of SALL4 siRNA onto HPPS, we selected siRNA sequence 2 for modification by conjugating with cholesterol and using 2’F modified phosphorothioate to improve HPPS loading and in vivo stability. We incubated chol‐si‐SALL4 with HPPS at various molar ratios of 1:1, 3:1, 5:1, and 10:1. The mixtures were then loaded onto a 2% agarose gel containing 0.002% (vol/vol) Gelred to monitor gel shift. The 3:1 molar ratio gave an integrity band gel migration with good siRNA recovery following FPLC purification (Fig. 4C). In addition, in vitro SALL4 gene down‐regulation in HepB3 cells by means of RT‐PCR following introduction of HPPS‐SALL4 also demonstrated that the 3:1 molar ratio gave the best therapeutic response as well as a batch‐independent RNAi therapeutic response (P < 0.05) (Fig. 4D). Therefore, the 3:1 molar ratio was found to be the optimal ratio for the HPPS‐SALL4 preparation and was selected for further in vivo evaluation.
Fig. 4.
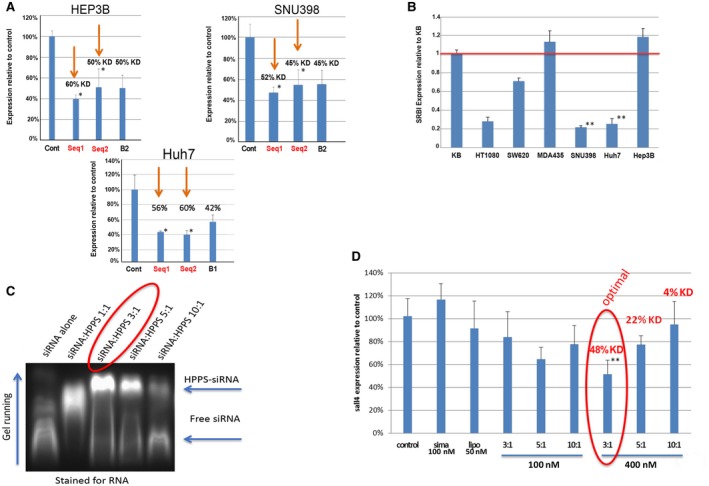
Evaluation RNAi efficacy. (A) RNAi efficacy in vitro was assessed by qRT‐PCR, with down‐regulation of SALL4 in Hep3B, SNU398, and Huh7 HCC cell lines by lipofectamine delivery of unmodified siRNA (50 nM). (B) Expression of SR‐BI by qRT‐PCR in different cancer cell lines, including three HCC cell lines (SNU398, Huh‐7, and Hep3B). Values are normalized against SR‐B1 expression in SR‐B1+ KB cells. (C) Optimization of ratio of siRNA to HPPS revealed that a 3:1 ratio resulted in the best loading, as observed by a gel with integrity band of gel migration and (D) best gene down‐regulation in SR‐BI highly expressed Hep3B cells. This gene knockdown was batch independent, Data show mean ± SEM. *P < 0.05, t test, n = 3; **P < 0.01), t test, n = 3. Abbreviations: Cont, control; KD, equilibrium dissociation constant; lipo, lipopolysaccharide; Seq, sequence; sirna, siRNA.
HPPS‐SALL4 Effectively Decreases HCC In Vivo
We further investigated HPPS‐SALL4 for in vivo RNAi therapy against SALL4. A higher gene expression level of SALL4 has been previously shown to portend a poor prognosis,( 16 ) which we confirmed by analysis of the The Cancer Genome Atlas data (Supporting Fig. S3). Hep3B tumor‐bearing mice were treated with HPPS‐SALL4 following the time line detailed in Fig. 5A. Briefly, HPPS‐SALL4 or HPPS‐scramble (HPPS loaded with scramble SALL4 siRNA) was administered once every 2 days by intravenous injection at days 0, 2, and 4. Tumor volume was monitored before treatment and 6 days after the first dose; tumors were then excised to determine therapeutic response. HPPS‐SALL4 treatment resulted in a dramatic reduction in HCC tumor volume when compared with the HPPS‐scramble group (Fig. 5B,C). As silencing of SALL4 will inhibit cell proliferation and induce apoptosis,( 20 , 31 ) we next examined the apoptosis of tumor tissue by TUNEL assay. HPPS‐SALL4 induced over 3‐fold increase in apoptosis when compared with HPPS‐scramble (P < 0.05) (Fig. 5D,E). Collectively, these in vivo antitumor studies provide convincing evidence that HPPS is not only able to efficiently deliver siRNA to the HCC tumor but is also capable of facilitating gene knockdown of siRNA in the tumor and improving antitumor efficacy.
Fig. 5.
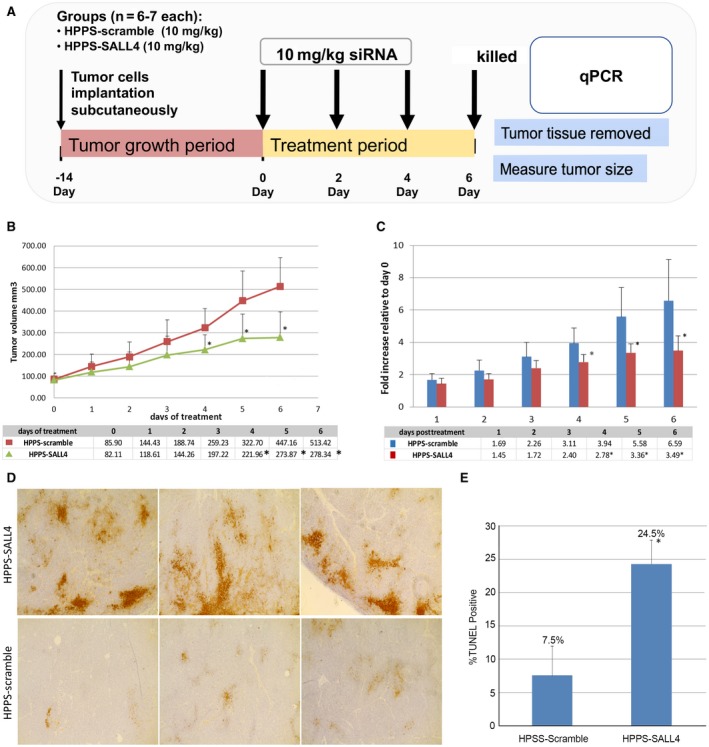
HPPS‐SALL4 versus HPPS‐scramble. (A) Time line of HEP3B HCC model treated with HPPS‐SALL4 versus HPPS‐scramble. (B) HPPS‐SALL4 seq2 induces significant tumor growth inhibition by tumor volume. (C) HPPS‐SALL4 seq2 induces significant inhibition by fold change in tumor growth. (D) TUNEL staining of HEP3B tumors treated with either HPPS‐SALL4 or HPPS‐scramble siRNA, with HPPS‐SALL4 inducing an over 3‐fold increase in apoptosis, magnification 20×. (E) Percentage apoptosis of HCC tumors treated by HPPS‐SALL4 versus HPPS‐scramble (control) by TUNEL assay. Data show mean ± SD. *P < 0.05, t test; n = 5. Abbreviation: seq, sequence.
Furthermore, the immunogenicity of HPPS‐SALL4 was evaluated on healthy BALB/c mice by injecting the same amount of HPPS‐SALL4 as the RNAi treatment group. The concentration of different cytokines was measured at 3 hours and 7 days after injection. Mice injected with saline (0.3 mL), HPPS alone, and siRNA alone were used as controls. HPPS‐SALL4 did not trigger any significant immune response (Fig. 6).
Fig. 6.
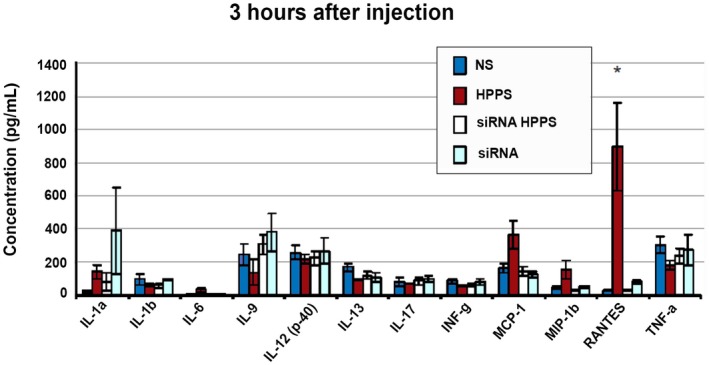
Evaluation of HPPS immunogenicity in vivo as evidenced by concentrations of different cytokines 3 hours and 7 days after injection with different concentrations of HPPS and siRNA. Mice were injected in the tail vein with saline (0.3 mL), HPPS (2,000 mg/kg), siRNA HPPS (siRNA 2,000 μg/kg, HPPS 14 mg/kg), or siRNA (2,000 μg/kg). Data show mean ± SEM. *P < 0.05, t test, n = 4. Abbreviations: IL, interleukin; INF, interferon; MCP, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; NS, saline; RANTES, regulated upon activation, normal T cell expressed, and secreted; TNF, tumor necrosis factor.
Discussion
HCC is a high‐fatality cancer often diagnosed at an advanced stage and most commonly in patients with cirrhosis and associated liver dysfunction.( 5 ) Many patients are unable to tolerate chemotherapy due to impaired hepatic metabolism and systemic side effects. The presence of underlying cirrhosis and hepatic dysfunction are considerations that are unique to HCC as a cancer. Lipoprotein‐like nanoparticles are particularly appropriate to consider and investigate in HCC because these particles are able to deliver cargo directly into malignant hepatocytes and systemic side effects can be avoided in susceptible patients.
Our study demonstrates for the first time that siRNA delivered by HDL‐like HPPS particles is a viable therapeutic strategy to inhibit HCC tumor growth. We optimized the formulation of cholesterol‐conjugated siRNA loaded onto HPPS at a ratio of 3:1 and discovered that it was able to mediate a significant reduction in HCC tumor growth. Additionally, HPPS‐SALL4 induced a significant increase in tumor apoptosis without any evidence of immunogenicity.
HPPS is a promising modality for treatment delivery directly into liver cancer cells in patients with cirrhosis and liver dysfunction,( 12 , 27 , 32 , 33 , 34 , 35 ) offering a number of advantages over other nanoparticles. It is biocompatible and largely avoids detection by mononuclear phagocytic cells in the body’s defense system.( 25 ) Its long blood circulation characteristics allow a favorable systemic drug delivery without need for PEGylation.( 36 ) However, HPPS is large enough to avoid rapid elimination by kidneys.( 37 ) It is a powerful vehicle for direct cytosolic delivery of drugs, including siRNA by SR‐BI, thereby bypassing the endosomal trapping seen with most nanoparticles.( 25 , 27 ) There is no concern for the long‐term toxicity associated with gold or iron oxide nanoparticles.( 38 ) In fact, we have previously demonstrated that HPPS containing siRNA, even at very high doses of 2,000 mg/kg, was not associated with any liver or systemic toxicity( 27 , 28 ); transaminases did not increase. We have additionally shown in the current study that there was no associated immunogenicity. The use of HPPS is advantageous when compared with approaches such as siRNA (unstable, quickly degraded( 39 )) and adenoviral vectors (previously associated with safety concerns).( 40 ) Finally, the receptor for HPPS is more highly expressed in HCC cells compared to noncancerous liver cells, making HPPS more selective for HCC tumors.
Delivery of RNAi therapy avoids the need for hepatic drug metabolism, which is significantly compromised in patients with cirrhosis. Using HPPS as a vector prevents rapid degradation of the siRNAs and allows for optimized delivery into liver cells. We targeted SALL4 by siRNA because it has been reported to be overexpressed in 56% of HCCs and has a strong prognostic value for aggressive HCC.( 30 ) SALL4 is a biomarker for a progenitor subclass of HCC and has a unique expression pattern in human livers at various stages. This transcription factor is initially activated in fetal liver tissue, inhibited in adult liver tissue, and then re‐expressed as an oncofetal protein in HCC.( 41 ) Therefore, targeting of SALL4 would not affect normal, noncancerous, adult liver cells. The viability of SALL4 as a therapeutic target in HCC has been demonstrated in mouse models in which inhibition of SALL4 was shown to effectively inhibit HCC growth and progression.( 17 , 41 ) Knockdown of SALL4 by short hairpin RNA in vitro has also been shown to induce apoptosis.( 30 ) HPPS‐mediated delivery of siRNAs targeting other genes could reasonably be envisioned for therapeutics of HCC.
Image‐guided treatment planning was not used in this study but would be able to further define the appropriate dosage and optimal conditions for delivery.( 32 ) An additional limitation of our study was the use of a xenograft model in nude mice as these mice are immune deficient. The siRNA was loaded onto the particle surface rather than directly into the particle, which would have been optimal in order to minimize any systemic impact. The siRNA sequence could be further optimized in our future study for better therapeutic efficacy. Nonetheless, this approach selectively and efficiently delivered RNAi therapy to the liver through the SR‐BI receptor and did not induce immunogenicity, at least in the short term. With respect to immunogenicity, we recognize that the quantification at 7 days may not fully describe the adaptive immune response. We will plan to perform longer term follow‐up of adaptive immunity in the future with evaluation of HPPS immunogenicity in vivo beyond a month.
In conclusion, we have identified the HPPS lipoprotein‐like nanoparticle as a viable vector for siRNA therapeutic targeting of HCC tumors. This is a promising therapeutic tool that enables cytosolic siRNA delivery into HCC cells without systemic toxicity or dependence on hepatic metabolism. The summary of the findings is illustrated in Fig. 7. Given that patients with HCC often have underlying cirrhosis and hepatic dysfunction that limit therapeutic options and ability to tolerate medications, this targeted delivery is a particularly relevant and interesting therapeutic strategy to help patients diagnosed with this high‐fatality cancer.
Fig. 7.
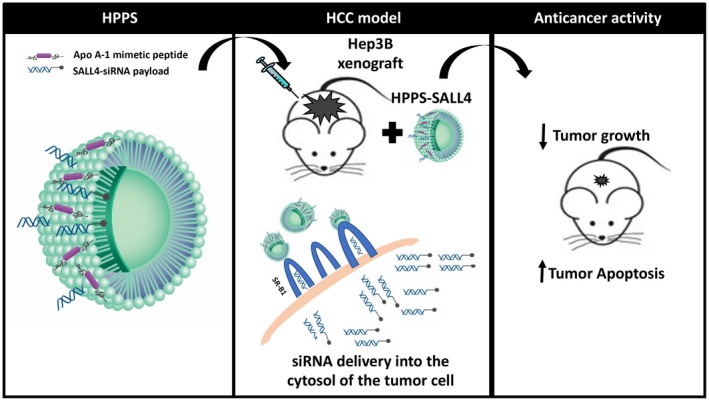
Summary schematic figure.
Supporting information
FigS1
FigS2
FigS3
TableS1
Supported by the Canadian Institute of Health Research, Canadian Cancer Society Research Institute (GZ), Canada Foundation for Innovation (GZ), Natural Sciences and Engineering Research Council of Canada (GZ), Princess Margaret Cancer Foundation (GZ), Canada Research Chairs Program (GZ), and DLVR Therapeutics Inc. (GZ).
The raw data required to reproduce our findings are available upon request.
Potential conflict of interest: Dr. Zheng consults for, received grants from, and owns intellectual property rights in DLVR. The other authors have nothing to report.
Contributor Information
Gang Zheng, Email: Gang.zheng@uhnresearch.ca.
Juan Chen, Email: Juan.chen@uhnresearch.ca.
Mamatha Bhat, Email: Mamatha.bhat@uhn.ca.
References
- 1. International Agency for Research on Cancer. World Health Organization . Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. http://www.globocan.iarc.fr/Pages/fact_sheets_population.aspx [Google Scholar]
- 2. National Cancer Institute . Surveillance, Epidemiology, and End Results (SEER) Program . SEER*Stat Database: Incidence. SEER 9 Regs Research Data, November 2009 Sub (1973‐2007). Bethesda, MD: National Cancer Institute; 2010. [Google Scholar]
- 3. Mittal S, El‐Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol 2013;47(Suppl.):S2‐S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan ST, Poon RT, Yeung C, Lam CM, Lo CM, Yuen WK, et al. Outcome after partial hepatectomy for hepatocellular cancer within the Milan criteria. Br J Surg 2011;98:1292‐1300. [DOI] [PubMed] [Google Scholar]
- 5. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358‐380. [DOI] [PubMed] [Google Scholar]
- 6. Tan D, Yopp A, Beg MS, Gopal P, Singal AG. Meta‐analysis: underutilisation and disparities of treatment among patients with hepatocellular carcinoma in the United States. Aliment Pharmacol Ther 2013;38:703‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al.; SHARP Investigators Study Group . Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378‐390. [DOI] [PubMed] [Google Scholar]
- 8. Palatini P, De Martin S. Pharmacokinetic drug interactions in liver disease: an update. World J Gastroenterol 2016;22:1260‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diep U, Chudow M, Sunjic KM. Pharmacokinetic changes in liver failure and impact on drug therapy. AACN Adv Crit Care 2017;28:93‐101. [DOI] [PubMed] [Google Scholar]
- 10. McNamara MG, Slagter AE, Nuttall C, Frizziero M, Pihlak R, Lamarca A, et al. Sorafenib as first‐line therapy in patients with advanced Child‐Pugh B hepatocellular carcinoma‐a meta‐analysis. Eur J Cancer 2018;105:1‐9. [DOI] [PubMed] [Google Scholar]
- 11. Lovell JF, Jin CS, Huynh E, Jin H, Kim C, Rubinstein JL, et al. Porphysome nanovesicles generated by porphyrin bilayers for use as multimodal biophotonic contrast agents. Nat Mater 2011;10:324‐332. [DOI] [PubMed] [Google Scholar]
- 12. Lanza GM, Moonen C, Baker JR Jr, Chang E, Cheng Z, Grodzinski P, et al. Assessing the barriers to image‐guided drug delivery. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2014;6:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S, et al. MicroRNA‐98 acts as a tumor suppressor in hepatocellular carcinoma via targeting SALL4. Oncotarget 2016;7:74059‐74073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, et al. The transcription factor SALL4 regulates stemness of EpCAM‐positive hepatocellular carcinoma. J Hepatol 2014;60:127‐134. [DOI] [PubMed] [Google Scholar]
- 15. Yin F, Han X, Yao SK, Wang XL, Yang HC. Importance of SALL4 in the development and prognosis of hepatocellular carcinoma. World J Gastroenterol 2016;22:2837‐2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park H, Lee H, Seo AN, Cho JY, Choi YR, Yoon Y‐S, et al. SALL4 expression in hepatocellular carcinomas is associated with EpCAM‐positivity and a poor prognosis. J Pathol Transl Med 2015;49:373‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, et al. Sal‐like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology 2013;57:1469‐1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu BH, Jobichen C, Chia CSB, Chan THM, Tang JP, Chung TXY, et al. Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide. Proc Natl Acad Sci U S A 2018;115:E7119‐E7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu TC, Vachharajani N, Chapman WC, Brunt EM. SALL4 immunoreactivity predicts prognosis in Western hepatocellular carcinoma patients but is a rare event: a study of 236 cases. Am J Surg Pathol 2014;38:966‐972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han SX, Wang JL, Guo XJ, He CC, Ying X, Ma JL, et al. Serum SALL4 is a novel prognosis biomarker with tumor recurrence and poor survival of patients in hepatocellular carcinoma. J Immunol Res 2014;2014:262385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan H, Cui Z, Zhang H, Mani SK, Diab A, Lefrancois L, et al. DNA demethylation induces SALL4 gene re‐expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection. Oncogene 2017;36:2435‐2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian Q, Xiao Y, Wu Y, Liu Y, Song Z, Gao W, et al. MicroRNA‐33b suppresses the proliferation and metastasis of hepatocellular carcinoma cells through the inhibition of Sal‐like protein 4 expression. Int J Mol Med 2016;38:1587‐1595. [DOI] [PubMed] [Google Scholar]
- 23. Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929‐1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data‐mining platform. Neoplasia 2004;6:1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Z, Cao W, Jin H, Lovell JF, Yang M, Ding L, et al. Biomimetic nanocarrier for direct cytosolic drug delivery. Angew Chem Int Ed Engl 2009;48:9171‐9175. [DOI] [PubMed] [Google Scholar]
- 26. Lozano G, Bejarano I, Espino J, González‐Flores D, Ortiz A, García JF, et al. Density gradient capacitation is the most suitable method to improve fertilization and to reduce DNA fragmentation positive spermatozoa of infertile men. Anatol J Obstet Ginecol 2009;3:1‐7. [Google Scholar]
- 27. Lin Q, Chen J, Jin H, Ng KK, Yang M, Cao W, et al. Efficient systemic delivery of siRNA by using high‐density lipoprotein‐mimicking peptide lipid nanoparticles. Nanomedicine (Lond) 2012;7:1813‐1825. [DOI] [PubMed] [Google Scholar]
- 28. Yang M, Chen J, Cao W, Ding L, Ng KK, Jin H, et al. Attenuation of nontargeted cell‐kill using a high‐density lipoprotein‐mimicking peptide–phospholipid nanoscaffold. Nanomedicine (Lond) 2011;6:631‐641. [DOI] [PubMed] [Google Scholar]
- 29. Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M. Identification of scavenger receptor SR‐BI as a high density lipoprotein receptor. Science 1996;271:518‐520. [DOI] [PubMed] [Google Scholar]
- 30. Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med 2013;368:2266‐2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood 2008;112:805‐813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin Q, Jin CS, Huang H, Ding L, Zhang Z, Chen J, et al. Nanoparticle‐enabled, image‐guided treatment planning of target specific RNAi therapeutics in an orthotopic prostate cancer model. Small 2014;10:3072‐3082. [DOI] [PubMed] [Google Scholar]
- 33. Lin Q, Huang H, Chen J, Zheng G. Using fluorescence imaging to track drug delivery and guide treatment planning in vivo. Methods Mol Biol 2016;1444:153‐166. [DOI] [PubMed] [Google Scholar]
- 34. Lin Q, Chen J, Zhang Z, Zheng G. Lipid‐based nanoparticles in the systemic delivery of siRNA. Nanomedicine (Lond) 2014;9:105‐120. [DOI] [PubMed] [Google Scholar]
- 35. Lin Q, Chen J, Ng KK, Cao W, Zhang Z, Zheng G. Imaging the cytosolic drug delivery mechanism of HDL‐like nanoparticles. Pharm Res 2014;31:1438‐1449. [DOI] [PubMed] [Google Scholar]
- 36. Pluen A, Boucher Y, Ramanujan S, McKee TD, Gohongi T, di Tomaso E, et al. Role of tumor‐host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc Natl Acad Sci U S A 2001;98:4628‐4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng G, Chen J, Li H, Glickson JD. Rerouting lipoprotein nanoparticles to selected alternate receptors for the targeted delivery of cancer diagnostic and therapeutic agents. Proc Natl Acad Sci U S A 2005;102:17757‐17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lovell JF, Jin CS, Huynh E, MacDonald TD, Cao W, Zheng G. Enzymatic regioselection for the synthesis and biodegradation of porphysome nanovesicles. Angew Chem Int Ed Engl 2012;51:2429‐2433. [DOI] [PubMed] [Google Scholar]
- 39. Chen SH, Zhaori G. Potential clinical applications of siRNA technique: benefits and limitations. Eur J Clin Invest 2011;41:221‐232. [DOI] [PubMed] [Google Scholar]
- 40. Lee CS, Bishop ES, Zhang R, Yu X, Farina EM, Yan S, et al. Adenovirus‐mediated gene delivery: potential applications for gene and cell‐based therapies in the new era of personalized medicine. Genes Dis 2017;4:43‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009;136:1000‐1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FigS1
FigS2
FigS3
TableS1


