Abstract
Intrahepatic macrophages influence the composition of the microenvironment, host immune response to liver injury, and development of fibrosis. Compared with stellate cells, the role of macrophages in the development of fibrosis remains unclear. Multispectral imaging allows detection of multiple markers in situ in human formalin‐fixed, paraffin‐embedded tissue. This cutting‐edge technology is ideal for analyzing human liver tissues, as it allows spectral unmixing of fluorophore signals, subtraction of auto‐fluorescence, and preservation of hepatic architecture. We analyzed five different antibodies commonly observed on macrophage populations (CD68, MAC387, CD163, CD14, and CD16). After optimization of the monoplex stains and development of a Spectral Library, we combined all of the antibodies into a multiplex protocol and used them to stain biopsies collected from representative patients with chronic liver diseases, including chronic hepatitis C, nonalcoholic steatohepatitis, and autoimmune hepatitis. Various imaging modalities were tested, including cell phenotyping, tissue segmentation, t‐distributed stochastic neighbor embedding plots, and phenotype matrices that facilitated comparison and visualization of the identified macrophage and other cellular profiles. We then tested the feasibility of this platform to analyze numerous regions of interest from liver biopsies with multiple patients per group, using batch analysis algorithms. Five populations showed significant differences between patients positive for hepatitis C virus with advanced fibrosis when compared with controls. Three of these were significantly increased in patients with advanced fibrosis when compared to controls, and these included CD163+CD16+, CD68+, and CD68+MAC387+. Conclusion: Spectral imaging microscopy is a powerful tool that enables in situ analysis of macrophages and other cells in human liver biopsies and may lead to more personalized therapeutic approaches in the future.
We optimized a spectral imaging microscopy platform to evaluate intrahepatic macrophages in liver biopsies from patients with chronic liver diseases (HCV, NASH, and AIH). We then compared differences in macrophage populations in patients with chronic viral hepatitis C and different stages of fibrosis (minimal and advanced), using batch analysis algorithms. Several macrophage phenotypes were significantly increased in patients with advanced fibrosis when compared to controls. Spectral imaging microscopy is a powerful tool that enables phenotypic characterization and quantification of intrahepatic macrophages, key players in hepatic fibrosis development that may be targets for future therapeutic intervention.

Abbreviations
- AIH
autoimmune hepatitis
- CPA
collagen proportionate area
- DAPI
4′,6‐diamidino‐2‐phenylindole
- FFPE
formalin‐fixed, paraffin‐embedded
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- IHC
immunohistochemical
- MHAI
modified hepatitis activity index
- NAS
nonalcoholic fatty liver disease activity score
- NASH
nonalcoholic steatohepatitis
- PASD
periodic acid–Schiff with diastase
- ROI
region of interest
- TBST
trishydroxymethylaminomethane‐buffered saline Tween 20
- TSA
tyramide signal amplification
- t‐SNE
t‐distributed stochastic neighbor embedding
Intrahepatic macrophages are of critical importance in progression of inflammation and fibrosis in nonalcoholic steatohepatitis (NASH). NASH develops by multiple hits stemming from lipid accumulation, metabolic disruption, and oxidative stress resulting in a pro‐inflammatory state with subsequent activation of Kupffer cells and recruitment of monocyte‐derived macrophages.1 Although less well understood, macrophages also play a role in development of autoimmune hepatitis (AIH) through their action as antigen‐presenting cells. Pro‐inflammatory (M1‐like) macrophages are increased in patients with AIH2 and are associated with increased fibrosis progression.3
Macrophages are difficult to isolate from human liver tissue and easily become activated, changing their phenotype when manipulated or cultured.4, 5 Although flow cytometry is able to analyze multiple antigens on suspensions of freshly isolated macrophages, it is unable to visualize them in the context of hepatic architecture,6 and fresh human tissue is not always available. Other innovative platforms such as single‐cell RNA sequencing or mass cytometry (e.g., CyTOF [Fluidigm Corp., South San Francisco, CA]) are able to analyze multiple markers on intrahepatic macrophages7, 8; however, these do not preserve hepatic architecture or allow the location of the identified cell populations to be determined. Moreover, mouse models of certain liver diseases, such as those induced by HCV infection or NASH, are poor surrogates, as they fail to replicate the chronicity observed in humans.9 Routine immunohistochemical (IHC) staining of human liver biopsies can identify macrophages in formalin‐fixed paraffin‐embedded (FFPE) tissues; however, there are many limitations, including the inability to stain multiple antigens on the same cellular compartment10 and dependence on the availability of primary antibodies raised in different species to prevent cross‐reactivity.6
A cutting‐edge technique has been developed that allows in situ characterization of human cells in FFPE tissues: the Vectra 3 quantitative pathology imaging system (Akoya Biosciences, Hopkinton, MA). This technology has been used primarily for evaluating tumor‐infiltrating lymphocytes.11 It allows spectral unmixing of fluorophore signals with subtraction of background auto‐fluorescence, producing a clean signal without interference from neighboring spectral wavelengths. For this study, we hypothesized that this platform would successfully quantify and phenotype intrahepatic macrophages in situ in patients with nonneoplastic liver disease.
Methods
Patient Biopsies
The University of Texas Medical Branch Institutional Review Board approved the protocol, and all studies were conducted on de‐identified, archived liver biopsies collected from 2006 to 2017. Biopsies were obtained as standard of care by licensed radiologists through the percutaneous route using an 18‐gauge core needle. First, we obtained representative liver biopsies from a control patient (n = 1) and from patients with chronic liver disease due to HCV (n = 1), NASH (n = 1), and AIH (n = 1), to optimize the multiplex staining and imaging analysis. Next, we collected liver biopsies from multiple patients per group and compared healthy controls (n = 8) to patients with clinically (by serology and molecular testing) and biopsy‐confirmed HCV with minimal fibrosis (n = 5) and advanced fibrosis (n = 6). Control biopsies were from patients without known liver disease who had normal transaminases and minimal histopathologic findings. The HCV+ and control patient groups were matched for age, sex, body mass index, human immunodeficiency virus (HIV), and genotype. At collection, tissue was immediately placed into 10% buffered formalin and processed using a TissueTekVIP tissue processor (Sakura Finetek, Torrance, CA), paraffin‐embedded, and sections were cut at 3 μm using Thermo‐Fisher CryoStar NX70 cryostats (Waltham, MA). Hematoxylin and eosin, periodic acid–Schiff with diastase (PASD), and Masson’s trichrome stains were conducted on a Ventana Ultra automated stainer (Roche Diagnostics, Tucson, AZ). All processing was performed in a College of American Pathologists–accredited laboratory by certified histotechnologists. Tissue blocks were stored at room temperature and sectioned immediately before multiplex staining, whenever possible. Slides that were unable to be stained within in 1 week were stored at 80°C in slide storage boxes wrapped with parafilm (to reduce moisture exposure).12
Fibrosis Quantitation
We used a quantitative method to determine the collagen proportionate area (CPA).13 Masson’s trichrome stains were digitally scanned on an Aperio ImageScope Digital slide scanner (Leica Biosystems, Wetzlar, Germany), and low‐power (×1.2) images of the entire surface area of each core was measured in number of pixels using Nikon’s NIS Elements‐BR Imaging Software (Tokyo, Japan). Next, blue‐stained areas from the trichrome stain were identified, and thresholds were set using the software. The fibrotic area was divided by the total biopsy surface area to obtain the percent fibrosis (% fibrosis = [∑fibrotic area in pixels/∑ total area in pixels] × 100). Liver capsules and large portal tracts (identified by presence of nerve bundles) were excluded.
Vectra 3 Platform
The Vectra 3 (Vectra AI, San Jose, CA) is an automated quantitative pathology imaging system that detects up to seven antibodies in the same tissue section. Vectra 3.0.3, inForm 2.4.0 (PerkinElmer, Waltham, MA), and Phenochart 1.0.4 (Akoya Biosciences) software programs come with the platform. For all experiments, we used Opal 7‐color manual IHC kits (50 slides, cat. #: NEL811001KT; Akoya Biosciences). This system acquires images from tissue sections labeled with Opal‐conjugated fluorophores combined with tyramide signal amplification (TSA), allowing for enhanced signal intensity and removal of antibodies with each antigen retrieval step. Images from monoplex and multiplex stained slides were acquired using the Vectra multispectral camera configured to capture discrete intervals (every 20 nm) across the spectrum from 420 nm to 720 nm.14, 15 The Vectra 3 uses a liquid crystal tunable filter, and each image generated has 35 wavelength λ channels, which allows extraction of spectral data from the images for analysis.
Spectral Library Development
Before developing a multiplex protocol for spectral imaging, refer to the detailed “Opal Multiplex Assay Development Guide” provided by Akoya Biosciences (https://www.akoyabio.com/application/files/5715/5510/3342/Akoya_Opal_Assay_Development_Guide.pdf). For all of the steps in this study, unstained sections from liver‐biopsy tissue blocks were used. We first assigned Opal fluorophores to each marker and performed titration of primary antibodies (Fig. 1). Fluorescence and simulated IHC views from inForm software and synthetic library spectra were used to evaluate proper cellular morphology and staining patterns (by a subspecialty‐trained, board‐certified pathologist [H.L.S.]). For the auto‐fluorescence inherently present in liver tissue to be subtracted from future analyses, an unstained control slide was incubated with primary antibody, without the addition of Opal TSA and 4′,6‐diamidino‐2‐phenylindole (DAPI). After the optimal staining condition for each antibody was obtained, Opal TSA concentrations were adjusted to achieve target intensities (5‐30 inForm normalized counts). Antigen‐retrieval buffers (AR6 [Biogenex, Fremont, CA] versus AR9 [Akoya Biosciences]) with the least‐nonspecific staining were selected (Fig. 1). Controls, including “monoplex” stains with each individual antibody/Opal fluorophore combination (without DAPI), DAPI alone, and an unstained liver biopsy tissue section were used to build the Spectral Library for subsequent spectral unmixing in the multiplex assay.14 Briefly, representative fields from the single color slides were imaged at 20× using the Vectra 3, and spectra were extracted from acquired images using inForm and saved to the Spectral Library. The quality of the Spectral Library was assessed by evaluating unmixed images to confirm the absence of spectral overlap or bleed‐over between channels, and by evaluating the values of unmixed peaks for each fluorophore (Fig. 2).
Figure 1.
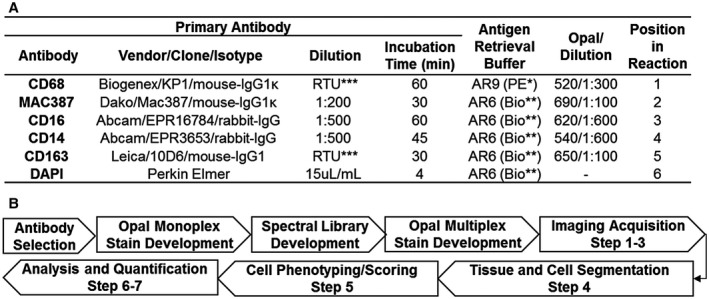
(A) Antibodies, optimized multiplex conditions, and steps used to identify intrahepatic macrophages in human FFPE liver biopsies. To optimize antibody concentrations, we first evaluated different dilutions of primary antibodies using the simulated bright field view in the inForm software program and chose the dilution with the best staining pattern (determined by a subspecialty‐trained, board‐certified pathologist). We then evaluated different dilutions of the Opal fluorophores until signal intensities were in the optimal range and well balanced between each of the monoplex slides. Once these steps were optimized, we then combined the antibodies together in the multiplex assay. *Akoya Biosciences, **Biogenex, ***Ready to Use (RTU). (B) Workflow for assay development and imaging analysis. The flow diagram highlights the optimization steps as well as the seven steps used for imaging acquisition and analysis. Step 1: Whole slides containing liver biopsy tissue were scanned with the Vectra 3 at low magnification (10×). Step 2: Vectra 3 was coupled with the Phenochart application, which allows whole‐slide navigation, annotation, and identification/selection of areas of interest for high‐resolution acquisition. For comparison of the initial single images, we selected representative 20× images that contained both portal tracts and lobules from either controls or patients with different types of liver disease (i.e., HCV, NASH, and AIH). For the experiments with multiple patients per group, the software randomly selected ROIs from 50% of the total surface area of each liver biopsy and captured these images using the 20× lens. Step 3: Images obtained were imported into inForm software to create a training project, which allows unmixing of signals and removal of tissue autofluorescence (using the previously optimized Spectral Library, Fig. 2). Step 4: Using inForm, we then optimized liver‐tissue segmentation (using both manual and trainable methods) into portal tracts and lobules, performed cell segmentation, and confirmed antibody staining within the proper compartment (i.e., nucleus, cytoplasm, and/or membrane). Step 5: Macrophages were phenotyped based on the labeling of CD68, CD163, MAC387, CD14, and CD16. The likelihood of co‐localization of different fluorophores was compensated during the phenotyping step using Visiopharm analysis, which uses two important parameters: signal intensity and percent coverage. Scoring of the macrophage markers was also possible after selecting the optimized marker threshold using inForm software, processing only two markers at a time. Step 6: Optimized settings and conditions were saved as an algorithm that was then used to analyze all of the images in “batch” (i.e., all images from all patients analyzed with the same setting conditions). Step 7: Data of individual cells were identified using the cell segmentation data feature of inForm.
Figure 2.
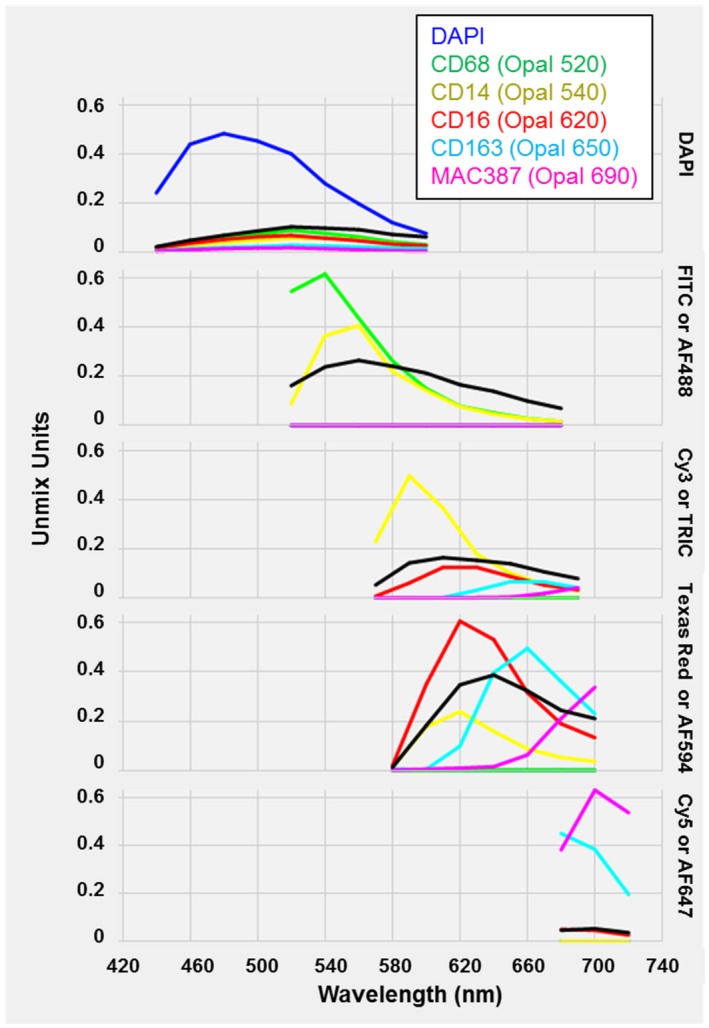
Spectral Library generated from optimized monoplex stains and an unstained liver biopsy section. A single slide from each antibody‐Opal fluorophore pair, a single slide with DAPI alone, and an unstained slide were analyzed using inForm software (included with the Vectra 3 platform) to create a “Spectral Library.” The Spectral Library is one of the main advantages of using the Vectra 3 platform, as it eliminates spectral overlap by unmixing the various fluorophore signals and allows subtraction of autofluorescence. Abbreviations: FITC, fluorescein isothiocyanate; TRITC, tetramethyl rhodamine isothiocyanate.
Multiplex Assay Development
Multiplex staining requires use of optimized conditions determined from the monoplex slides, as described previously. The detection of five macrophage markers in human‐liver FFPE tissue samples was performed using the Opal 7‐color manual IHC kits. Slides were heated at 60°C for 45‐60 minutes, residual paraffin was removed using xylene (Fisher, Fair Lawn, NJ) (3 × 10 minutes), and tissue was rehydrated in a graded series of histological grade ethanol solutions (100%: 1 × 10 minutes; 95%: 1 × 10 minutes; and rinsed in 70%), then rinsed with distilled water. Slides were fixed in 10% neutral‐buffered formalin (Fisher, Kalamazoo, MI) for 30‐45 minutes, rinsed in water, and placed in standardized antigen‐retrieval buffers (i.e., AR9 PE for CD68; Fig. 1). Slides were then microwaved at 95°C for 15 minutes using the EZ retriever system V3‐110V (Biogenex, Fremont, CA), which uses a temperature‐controlled antigen retrieval system. Slides were cooled to room temperature and then washed with water and tris‐buffered saline (trishydroxymethylaminomethane‐buffered saline Tween 20 [TBST], 0.1%]). A hydrophobic barrier pen (Vector Labs, Burlingame, CA) was used to surround tissue on the slide. Blocking was performed with 2‐3 drops of antibody diluent/blocking solution (included with the Opal kit) for 10 minutes at room temperature in a humidified chamber, followed by incubation with 150 µL of the first primary antibody (Fig. 1). Slides were incubated with two drops of secondary antibody mix (polymer horseradish peroxidase Ms + Rb) for 10 minutes. Slides were then incubated for 10 minutes with 150 µL of the standardized dilution of the assigned Opal selected for the antibody in position 1 (i.e., Opal 520 [1:300] for CD68). After primary antibody, secondary, and Opal incubations, slides were rinsed in TBST and washed in TBST (3 × 2 minutes). Slides were ready for another round of antigen retrieval after removal of the previously added antibodies, and this step was repeated until all targets of interest were detected using a different opal fluorophore for each marker. After each antigen‐retrieval step, the protocol restarts at the blocking step until all antibodies are used. AR6 buffer was always used before nuclear counterstaining with DAPI. Nuclei were stained with 150 µL of DAPI (Akoya Biosciences) diluted in TBST (15 µL/mL × 4 minutes). After nuclear staining, the slides were washed for 2 minutes in TBST and 2 minutes in water before mounting with Invitrogen ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, Grand Island, NY) and cover‐slipped. Samples were stored at 4°C until imaged.
Imaging Acquisition and Analysis
After we determined the absence of interference between each antibody signal and rebalanced specific signals to ensure that target intensities were in range, we proceeded to imaging acquisition and analysis. Detailed steps are shown in the flow diagram (Fig. 1). We first evaluated the macrophage multiplex panel and subsequent Vectra analysis using a single image from biopsies of patients with different liver diseases (HCV, NASH, and AIH) and then applied the platform to analyze biopsies from multiple patients per group using batch analysis. We used two different approaches for imaging analysis. First, for single‐image comparisons, multicomponent TIFF images were exported and provided to Visiopharm (Hoersholm, Denmark) for analysis. The heterogeneity and distribution of the various macrophage phenotypes in the study groups was determined using Visiopharm’s t‐distributed stochastic neighbor embedding (t‐SNE) and phenotypic matrix algorithms, which facilitated visualization and comparison of macrophage profiles in liver biopsies of different disease types. t‐SNE allows for visualization of high‐dimensional data in 2 or 3 dimensions, while preserving the innate structure and variance of the data. The likelihood of co‐localization of different fluorophores was compensated during the phenotyping step by using two important parameters: signal intensity and percent coverage. Second, for batch analysis, “cell_seg” files containing marker and location information of each cell in the image were exported and provided to the Bioinformatics group at the University of Michigan for thresholding and phenotype separation based on thresholds provided through InForm. Thresholds for specific markers were adjusted using images from control patients and then applied to all the images using a batch analysis approach, processing only two markers at a time. Mean normalized counts from the different macrophage markers were used in the analysis pipeline. The heterogeneity and distribution of these various cell phenotypes in the study groups were determined using t‐SNE, which allowed comparison of cellular profiles in liver biopsies at different stages of fibrosis. The t‐SNE was generated using all of the expression intensity values for the five biomarkers of interest: CD68, CD163, MAC387, CD14, and CD16. Using the raw intensity values in the “cell_seg” Excel file, principle component analysis (PCA) was performed to identify the 10 components with the highest variance, and the PCA‐embedded matrix was used to compute the t‐SNE plot. For the study, the expression of all 32 possible combinations of macrophage subpopulations under study were generated, and their proportions per patient and per group were also computed.
Statistical Analysis
The nonparametric Wilcoxon rank‐sum test (also called the Wilcoxon Man‐Whitney U test) was used to compare the relative abundance of the possible marker combinations among the three patient groups (control, minimal, and advanced fibrosis). Adjustment for multiple testing (due to the large number of possible marker combinations) was performed using the Benjamini‐Hochberg false discovery rate, with P < 0.05 chosen for significance. Analysis was performed using the image processing and statistics toolbox available in MATLAB 2019a (MathWorks, Natick, MA).
Results
Spectral Library Development
Standardization of the multiplex protocol requires evaluation of each macrophage marker singly in a “monoplex” assay, to optimize the antibody/Opal concentrations required to achieve the ideal morphologic staining pattern and proper target intensities (Fig. 1). AR6 buffer combined with use of the EZ retriever microwave system was effective in preventing nonspecific staining. Individual monoplex stains without DAPI were used to create the Spectral Library, which provided precise patterns for each fluorophore emission spectrum that was later required for spectral unmixing of multiplex stained slides (Fig. 2). The staining pattern of the macrophage markers, which was evaluated in an AIH liver biopsy by fluorescence (Fig. 3A) and simulated bright‐field view using inForm software (Fig. 3B), were restricted to specific cellular compartments, as previously reported.16, 17 CD14 was also expressed on liver sinusoidal endothelial cells as expected.18, 19 Although the distribution of the CD68+ and CD163+ macrophages looked similar, the CD163+ staining pattern was noticeably more intense and highlighted foci within the lobules that were devoid of hepatocytes and contained large aggregates of macrophages (Fig. 3A, PASD stain/inset). Individual monoplex images from a representative control patient are shown for comparison (Supporting Fig. S1).
Figure 3.
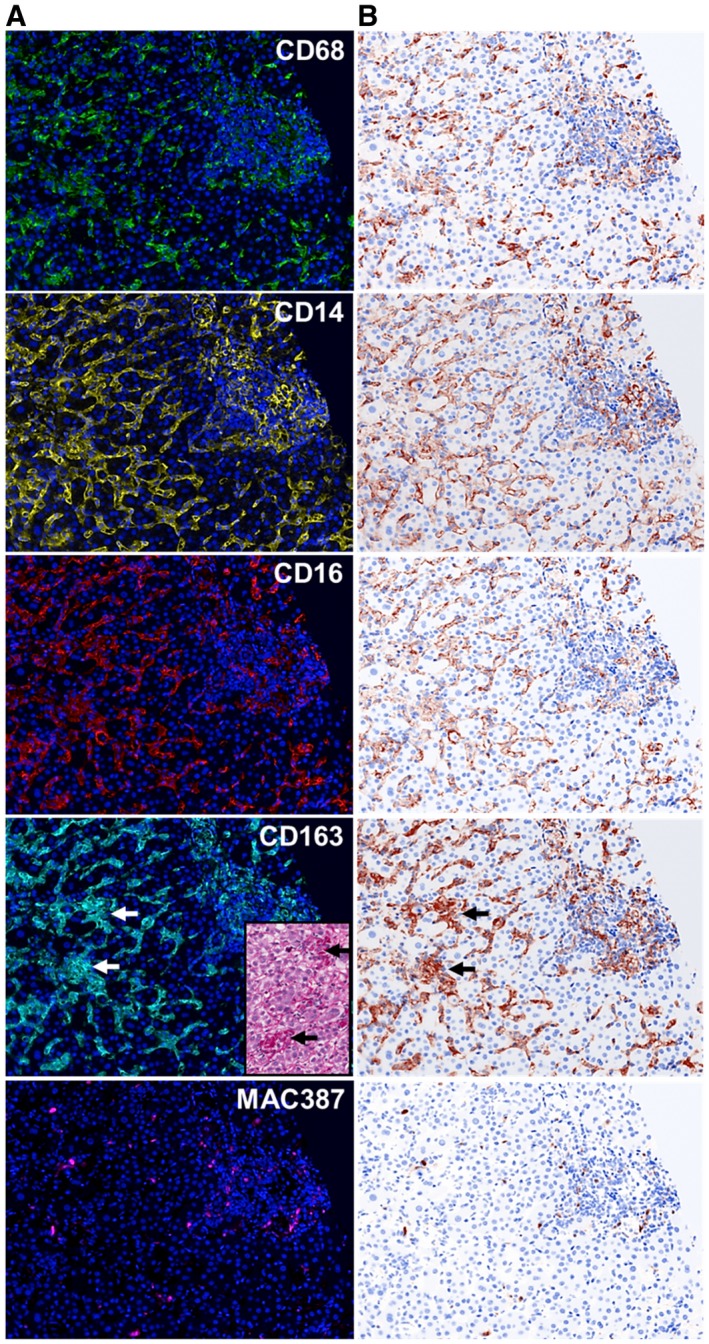
Visualization of each fluorophore channel acquired from multiplex‐staining a liver biopsy from a patient with AIH. (A) We stained a representative unstained liver biopsy slide from a patient with AIH with the multiplex panel. A single 20× fluorescence image that included both portal tract and lobular regions was obtained after spectral unmixing was applied. These steps allowed the expression of each individual macrophage marker to be visualized in the context of hepatic architecture: CD68 (green‐Opal 520), CD14 (yellow‐Opal 540), CD16 (red‐ Opal 620), CD163 (cyan‐Opal 650), MAC387 (magenta‐Opal 690), and nuclear stain (Blue‐DAPI). (B) Simulated brightfield images were generated to recreate the staining pattern that would be observed by conventional IHC using chromogenic methods. Large aggregates of macrophages were observed in the lobules, and these showed high expression of CD163; these cells correlated with the large aggregates of macrophages that were observed by light microscopy (inset; PASD stain). The patient with AIH had active chronic hepatitis with a modified hepatitis activity index of 13/18 and a fibrosis stage of 2/6.(30) For comparison, images from a representative control patient are shown in Supporting Fig. S1.
Biopsies From Patients With Chronic Liver Diseases Showed Expansion of Cellular Phenotypes When Compared With Controls
Next we combined the optimized antibodies from Figs. 1, 2, 3 to create a multiplex panel. We first used it to stain unstained slides from a control (n = 1) or patients with HCV (n = 1), NASH (n = 1), and AIH (n = 1), and selected a representative multiplex image from each patient that included both portal and lobular areas (Fig. 4). Figure 4 shows these representative images (Fig. 4A) and examples of the tissue segmentation feature of the software (Fig. 4B). Each disease displayed a unique pattern of portal and lobular populations (Fig. 4A). When compared with controls, biopsies from patients with chronic liver diseases showed increased staining in portal tracts and lobules (Fig. 4A,B). The liver biopsy obtained from the patient with active AIH (modified hepatitis activity index [MHAI]: 13/18; Ishak fibrosis stage: 2/6) showed the most prominent staining within both the portal tracts and lobules, particularly when compared with controls (Fig. 4A). This patient had increased percentages of cells expressing many of the markers used in this study when compared with the other disease types and controls (Supporting Fig. S2). All types of chronic liver disease showed increased expression of the pan‐macrophage marker CD68 and MAC387+ monocyte‐derived macrophages in the portal tracts when compared with controls; however, numbers were markedly increased in the patients with NASH and AIH (Supporting Fig. S2). Visiopharm analysis of the multicomponent TIFF files acquired from the representative images shown in Fig. 4A determined the differences in cellular phenotypes (Fig. 5A) and cluster analyses (Fig. 5B). In Fig. 5A,B, each color represents a different phenotype of cell, including macrophages. t‐SNE plots highlighted expansion and increased complexity of cellular phenotypes in diseased livers when compared with the representative control patient in whom the cells were more tightly clustered and similar (Fig. 5B). Figure 6 highlights the unique cellular phenotypes identified in each of the representative multiplex images from Figs. 4 and 5. Visiopharm phenotype matrix algorithms were used to determine the phenotype of each colored dot/cell shown in Fig. 5A. Gray dots indicate cells that were negative for all markers in the multiplex assay. The most commonly identified phenotypes in each of the disease types can be determined by use of the scale on the right‐hand side of the figure, in which dark green indicates the highest prevalence of that particular phenotype (Fig. 6). Even though these phenotypes were identified from single representative images, the control patient image showed increased expression of CD14+ cells, and the image from the patient with active AIH showed a high prevalence of two main phenotypes of macrophages: CD14+CD163+ and CD68+CD14+CD163+.
Figure 4.
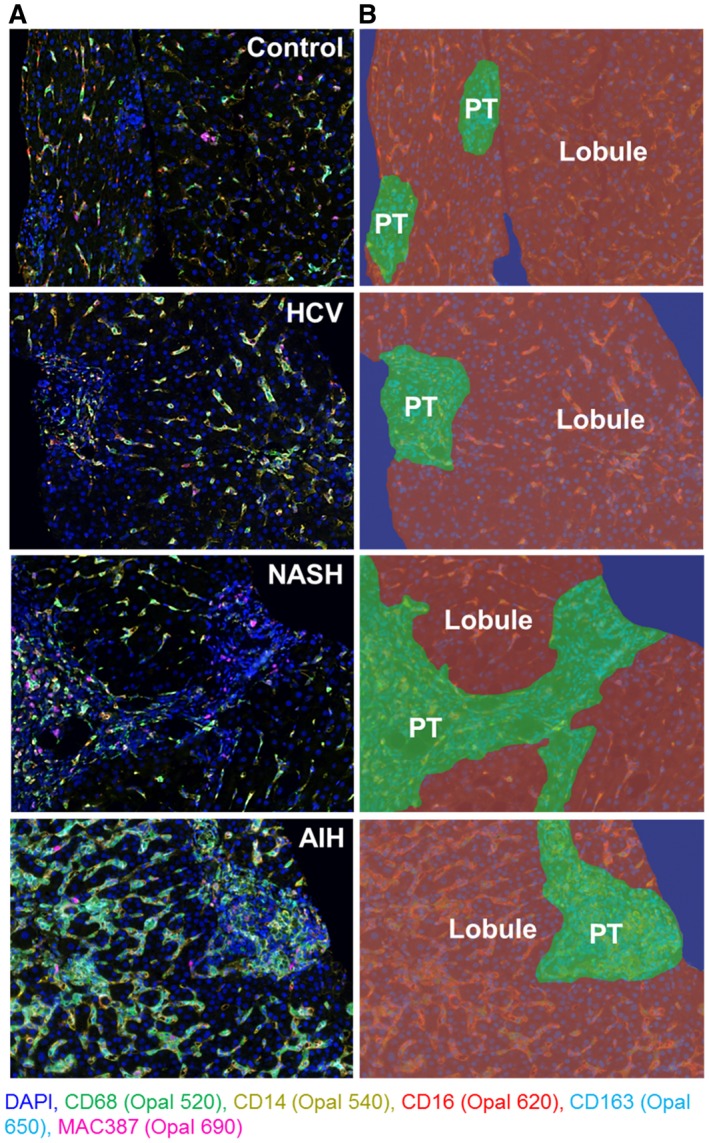
Spectral imaging analysis of patients with different chronic liver diseases (HCV, NASH, and AIH) highlights intrahepatic macrophages. We tested the optimized multiplex panel on unstained slides obtained from liver biopsies collected from representative patients with different types of chronic liver disease (n = 1 of each disease type) that had similar fibrosis stages and compared them with a control patient who did not have liver disease (i.e., no histopathologic changes and normal liver enzymes at the time of biopsy) (n = 1). (A) Representative unmixed multispectral images obtained after staining unstained slides with the macrophage multiplex panel. From each biopsy, we chose single images that contained both portal tracts and lobules. Images were acquired at 20× using inForm software. (B) Representative images after using the tissue‐segmentation feature of inForm on the multiplex images from (A). The 20× images were divided into portal tract (green), lobular (brown), and nontissue (blue) areas. HCV: MHAI, 4/18; fibrosis stage, 3/6. AIH: MHAI, 13/18; fibrosis stage, 2/6. NASH: nonalcoholic fatty liver disease activity score (NAS), 4/8; fibrosis stage, 2/4. Abbreviation: PT, portal tract.
Figure 5.
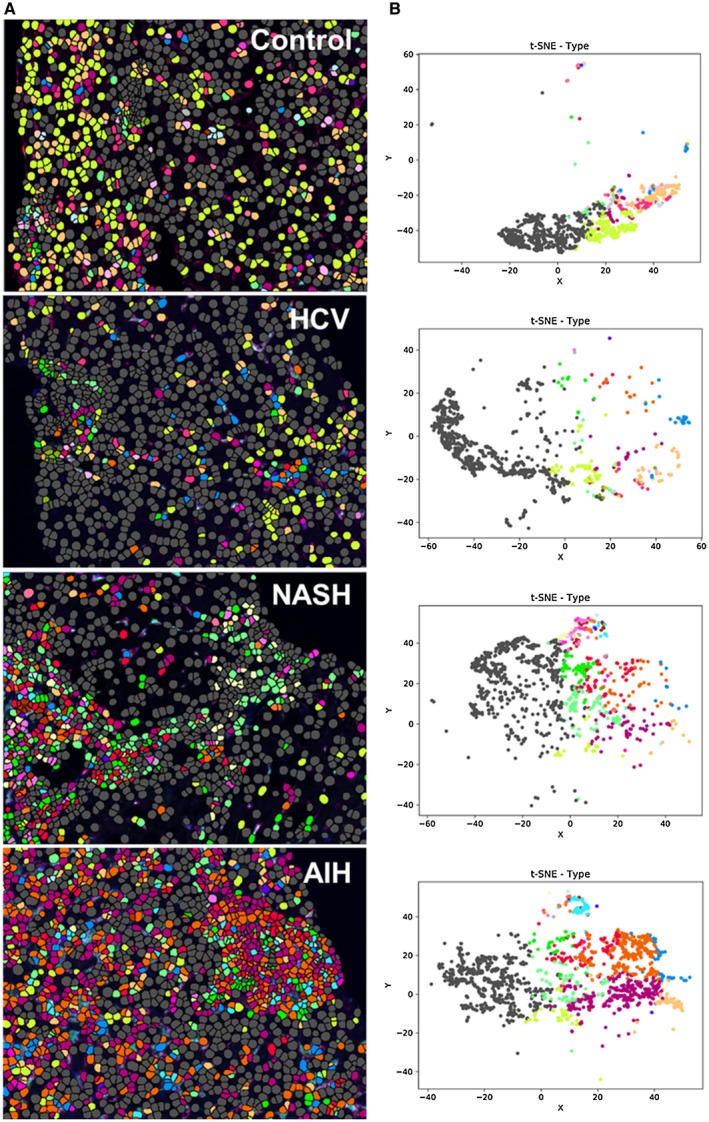
Spectral images analyzed with Visiopharm applications highlight cellular expansion and increased complexity in diseased livers when compared with controls. (A) Single multiplex images from Fig. 4A were used to export multicomponent TIFF files for Visiopharm analysis. A phenotyping application was used to determine the number of different cellular phenotypes present in each image after staining with the 6‐color (including DAPI) multiplex panel. Each colored dot represents a unique cellular phenotype. Gray dots represent cells that were negative for all of the markers in the multiplex panel (i.e., CD68, CD14, CD16, CD163, and MAC387). (B) t‐SNE plots highlight the unique patterns of concatenated cellular markers that are present in the representative liver images from patients with chronic liver diseases when compared with a control patient. This algorithm uses nonlinear dimensional reduction to allow visualization of high dimensional data sets. Cells with similar properties appear closer together and those that are dissimilar appear farther apart in the 2‐dimensional map. The control patient showed much less diversity in the types of macrophages and other cellular phenotypes identified when compared to patients with chronic liver disease. AIH: MHAI, 13/18; fibrosis stage, 2/6. HCV: MHAI, 4/18; fibrosis stage, 3/6. NASH: NAS, 4/8; fibrosis stage, 2/4.
Figure 6.
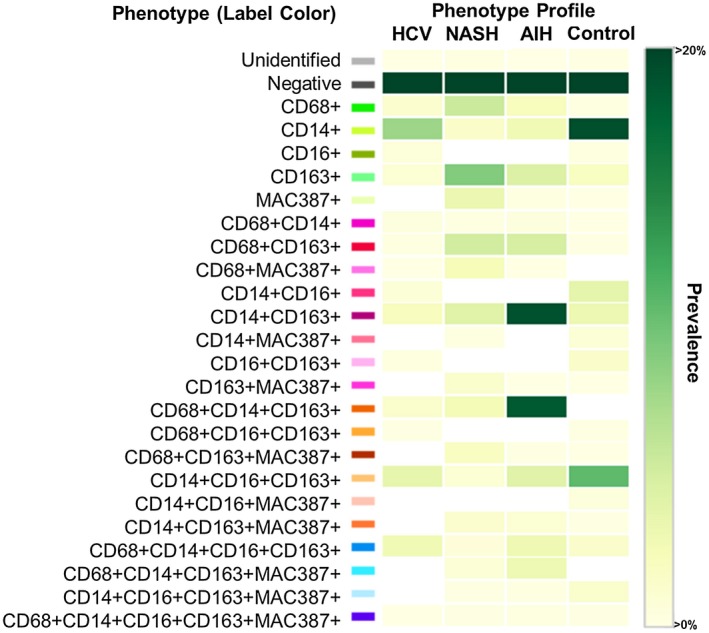
Phenotypic matrix algorithms were used to identify macrophage and other cellular phenotypes present in liver biopsies obtained from patients with chronic liver disease when compared with a control. Using the same exported multicomponent TIFF files from Fig. 5, we used Visiopharm phenotype matrix algorithms to determine the different cellular phenotypes present in each of the 20× multiplex images. The colors shown for each of the different phenotypes correlate with the various colored dots (i.e., individual cells) shown in the multiplex images from Fig. 5. Dark green and white boxes indicate the populations with the highest and lowest prevalence, respectively. Approximately 20 different macrophage populations (i.e., those that are positive for CD68, CD163, or MAC387) were identified using the five markers in the multiplex staining assay. The patient with AIH showed two prominent populations (CD14+CD163+ and CD68+CD14+CD163+), and the control patient had the highest prevalence of CD14+ expression alone (in the absence of other markers). AIH: MHAI, 13/18; fibrosis stage, 2/6. HCV: MHAI, 4/18; fibrosis stage, 3/6. NASH: NAS, 4/8; fibrosis stage, 2/4.
Spectral Imaging Microscopy Facilitated Analysis of Intrahepatic Macrophages in Multiple Patients per Group
We then showed that this platform could be used in a more high‐throughput manner by using batch analysis to obtain many regions of interest (ROIs) from each liver biopsy and included multiple patients per group. We obtained liver biopsies from controls (n = 8) and patients with chronic HCV infection and either minimal (n = 5) or advanced fibrosis (n = 6). Demographic and clinical information for the patients who were HCV‐positive are given in Supporting Table S1. Ishak criteria were used for scoring the liver biopsies.20 Supporting Fig. S3 compares the liver biopsies from representative patients with minimal fibrosis (Supporting Fig. S3A,B, MHAI: 4/18; fibrosis stage: 1/6) and advanced fibrosis (Supporting Fig. S3C,D, MHAI: 7/18; fibrosis stage: 6/6). CPA13 was used to confirm that the patients who were HCV‐positive with advanced fibrosis had significantly higher amounts of fibrosis in their liver biopsies when compared to those with minimal fibrosis (Supporting Fig. S4). Patients with advanced fibrosis had a median CPA of 24.4% (range: 15.5‐54.865), and those with minimal fibrosis had a median of CPA of 5.299% (range: 1.613‐8.021) (P < 0.01). Vectra 3 Phenochart software was used to scan the entire liver biopsy tissue (at 10×) from all patients in each group. Phenochart was used to randomly select ROIs that totaled at least 50% of the entire biopsy tissue, which resulted in 50‐70 images (at 20×) acquired from each patient (Fig. 7A‐C).
Figure 7.
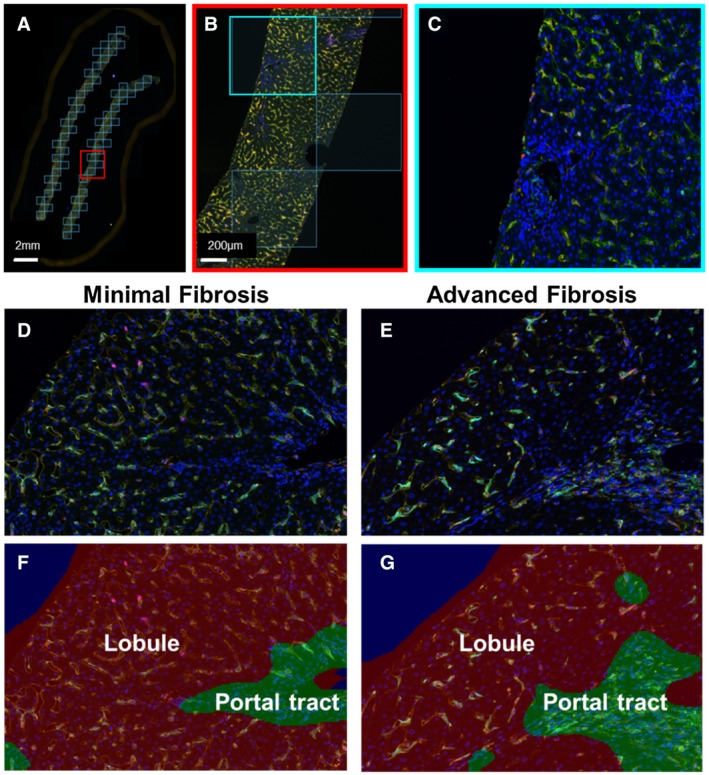
Example of batch analysis and collection of ROIs in each patient’s liver biopsy. We tested the feasibility of the platform for collecting multiple images per liver biopsy and added more patients per group. We used the multiplex panel to stain biopsies from control patients (n = 8) and patients with chronic hepatitis C and either minimal (n = 5) or advanced fibrosis (n = 6). Clinical information pertaining to these patients is provided in Supporting Table S1, and representative examples of liver biopsies collected from patients with minimal or advanced fibrosis are shown in Supporting Fig. S3 and S4. (A) The Phenochart application of inForm software was used to randomly stamp multiple ROIs in each liver biopsy (totaling at least 50% of the tissue). These corresponded to 50‐70 images per patient, depending on the size of the liver biopsy. (B,C) Each of the ROIs were then acquired at 20×. (D,E) Representative multiplex images from patients who were HCV‐positive with minimal or advanced fibrosis. (F,G) Examples of the tissue segmentation feature of the software that allowed separation of the parenchyma into portal tracts and lobules. MHAI scores and fibrosis stages for all patients in Figs. 7 and 8 are provided in Supporting Table S1.
Patients Who Are HCV‐Positive With Advanced Fibrosis Had Significantly Increased Intrahepatic Macrophage Populations When Compared With Controls
Representative multiplex images from patients with minimal (Fig. 7D,F) and advanced fibrosis (Fig. 7E,G) are shown for comparison. The mean normalized data were analyzed using t‐SNE plots of all the markers (Fig. 8A). We identified five unique macrophage clusters and overlaid them onto their corresponding t‐SNE plots (Fig. 8B), which showed significantly different expression patterns between controls and patients with advanced fibrosis. Specifically, the proportions of CD68+CD14+CD163+ macrophages and the overall expression of CD14+ clusters were significantly increased in control livers when compared to patients with advanced fibrosis (Fig. 8C, #1 and #2, respectively). Although the proportion of CD14+ cells in the patients with minimal fibrosis was not significantly different when compared with the controls or patients with advanced fibrosis, it was also the population with the highest frequency in this group. Liver biopsies from patients with advanced fibrosis revealed three distinct macrophage subpopulations that had significantly increased numbers when compared with controls: (1) CD68‐CD14‐CD163+CD16+MAC387‐, (2) CD68+CD14‐CD163‐CD16‐MAC387‐, and (3) CD68+CD14‐CD163‐CD16‐MAC387+ (Fig. 8C, #3, #4 and #5, respectively). Supporting Fig. S5 shows a t‐SNE plot that includes all patients from each group. The plot was separated for each patient and provides a low‐dimensional representation of the distribution of biomarker expression values. There was high mixing among all groups, but each shows several distinct regions.
Figure 8.
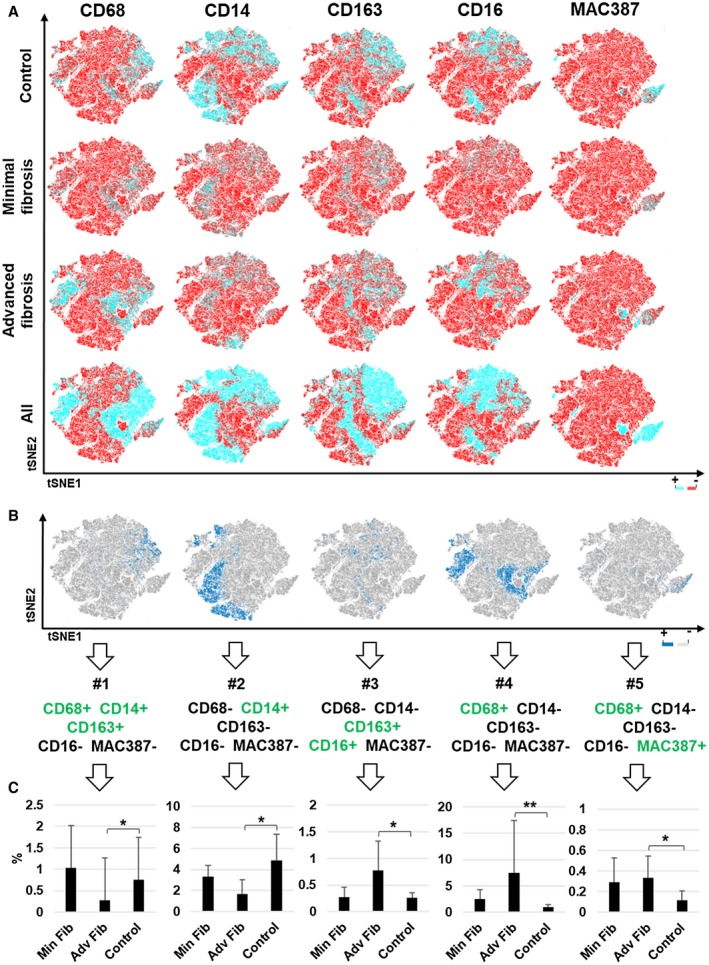
t‐SNE analysis highlighted significantly different cellular populations between patients with advanced fibrosis due to chronic hepatitis C when compared with controls. We analyzed the multiple images collected from the same patients shown in Fig. 7 (controls, n = 8; minimal fibrosis, n = 5; and advanced fibrosis, n = 6) using expression heat maps from t‐SNE and phenotype cluster analyses. (A) Heat maps showing differences in expression of each individual macrophage marker (CD68, CD14, CD163, CD16, and MAC387) within the control, minimal fibrosis, and advanced fibrosis groups. (B,C) Five clusters that were significantly increased between control and patients who were HCV‐positive with advanced fibrosis were overlaid individually on the concatenated t‐SNE plot from Fig. 8A. Clusters of cells expressing the different markers, positive (blue) or negative (gray), are shown. These five subpopulations showed significant differences between the control and advanced fibrosis groups. All data are presented as median with significance calculated using the Wilcoxon rank sum test (*P < 0.05; ***P < 0.01).
The percentages of the individual phenotypes present in each patient’s liver biopsy are shown in Supporting Fig. S6. As expected, some of the patients with minimal fibrosis (e.g., patient 5 in the minimal fibrosis group) had higher percentages of both protective and pathogenic populations in their livers, whereas some of the patients with advanced fibrosis had lower percentages of potentially pathogenic phenotypes (e.g., patient 5 in the advanced fibrosis group).
Discussion
Studying macrophages in situ in human liver biopsy tissue, where they naturally reside, is an ideal approach to increase understanding of their role in the complex hepatic microenvironment. Advantages of spectral imaging include (1) detection of multiple antigens simultaneously, even on the same cell or cellular compartment; (2) elimination of spectral overlap and auto‐fluorescence inherently present in liver tissue by applying an optimized “Spectral Library (Fig. 2);” (3) evaluation of FFPE tissues, a common biospecimen source; and (4) characterization of cells in situ within preserved hepatic architecture and microenvironment. These attributes are of great importance when studying cells such as macrophages that exhibit marked plasticity21, 22 and become activated when manipulated.4, 5
A critical step for successful multiplex staining with this platform was proper preservation of FFPE material. We recommend using freshly cut unstained slides stored at room temperature if used within 1 week, or alternatively stored at −80°C and protected from moisture. For initial monoplex stains, optimal staining was determined empirically by evaluating different antibody concentrations followed by adjusting Opal‐TSA concentrations to obtain target intensities. The staining sequence for the antibodies in the multiplex reaction and use of different buffers (i.e., AR6 or AR9) proved to be critical steps in optimization (Fig. 1).
Using just five markers, the results of this initial study further support that categorizing macrophages into “M1” and “M2” phenotypes is too simplistic. A single multiplex image acquired from patients with HCV, AIH, or NASH showed that compared with controls, patients with diseased livers have increased numbers of macrophages and cellular complexity (Figs. 4, 5, 6, Supporting Fig. S2). Involvement of hepatic macrophages in fibrosis development is most widely accepted in NASH, where it is known that Kupffer cells become activated to a pro‐inflammatory phenotype that enhances recruitment of monocyte‐derived macrophages into portal tracts and lobules, leading to subsequent activation of stellate cells.23, 24 Among the three chronic diseases compared in this study, liver biopsies obtained from patients with AIH showed the most intense staining with the multiplex panel, where large aggregates of cells were observed in both the portal tracts and lobules (Figs. 3 and 4). However, this patient had more active hepatitis (i.e., MHAI: 13/18) when compared with the other patients.
Visiopharm applications, including cellular phenotyping, t‐SNE plots, and phenotype matrix algorithms, were able to highlight unique cellular populations by analysis of multiplex images and are powerful tools that facilitate comparison among different disease states. Before any definite conclusions can be made regarding the various phenotypes identified by analysis of the single images acquired from individual patients in this initial optimization study, larger experiments with more patients per group are needed. We were, however, able to gain initial insight regarding macrophage phenotypes present in situ within the liver using this panel. Figure 6 shows that some populations of macrophages express both CD163 and CD68, whereas others appear to have one or the other. Similar to previous reports,25 diseased livers, especially those examples from patients with NASH and AIH, showed increased MAC387+ macrophages in the portal tracts (Figs. 4 and 5, Supporting Fig. S2) and more cell populations expressing this marker, as shown by the phenotype matrix plots (Fig. 6).
Liaskou et al. concluded that intermediate CD14++CD16+ macrophages accumulate in diseased livers, are recruited from blood, differentiated from local CD14++CD16‐ monocytes, and are critical players in fibrosis development.17 These cells had macrophage and dendritic cell–like features and most expressed CD163 and CD68, had phagocytic capability, secreted inflammatory cytokines (e.g., interleukin [IL]‐1, IL‐6, and tumor necrosis factor alpha), and profibrogenic chemokines (e.g., chemokine [C‐C motif] ligand [CCL] 2 and CCL5). Phenotype matrix plots (Fig. 6) showed that several of the identified phenotypes co‐expressed CD14 and CD16, and some CD14+CD16+ cells expressed CD163, CD68, MAC387, or low levels of all of these markers.
Because intrahepatic macrophages play pathogenic roles during chronic hepatitis C infection,16, 26 and we had plenty of archived material, we used this disease to optimize batch analysis using spectral imaging (Fig. 7). Multiplex images showed distinct differences between cell populations identified in controls, and patients with minimal or advanced fibrosis (Fig. 8). Imaging analysis revealed significant increases in two cell clusters from patients without known liver disease, specifically a population of CD68+CD14+ macrophages and, importantly, overall increased expression of CD14. Interestingly, the representative control patient shown in Figs. 4, 5, 6 also showed increased expression of CD14 alone, in the absence of the other markers. The increase in CD14 expression in control patients when compared to those with advanced fibrosis or inflammatory liver disease is likely due to higher expression of CD14 not only on macrophages, but also on liver sinusoidal endothelial cells. These cells are known to express CD14, which decreases following activation, as observed with HCV infection.18 Identification of an apparently protective CD68+CD14+ cell population (Fig. 8) is similar to other studies of tolerogenic livers and a recent study of human intrahepatic macrophages that analyzed populations using single‐cell RNA sequencing, where they had immunoregulatory and patrolling type functions.7, 27 Three main macrophage clusters were significantly increased in patients who were HCV‐positive with advanced fibrosis when compared to controls, and included CD163+CD16+, CD68+, and CD68+MAC387+. Results using spectral imaging confirm previous observations that suggest a predominance of CD14+ cells during homeostasis and increased accumulation of CD163+, CD68+, and MAC387+ macrophages in human chronic liver diseases.17, 26, 28, 29
Characterization of the CD163+ macrophage function has varied depending on the study; some reports suggest they confer a characteristic M2‐like phenotype that is increased in portal tracts and lobules of patients with chronic HCV.26, 29, 30 Results of this study showed CD163+ populations that expressed either CD14 or CD16 or both (Fig. 6). In addition, patients who were HCV‐positive with advanced fibrosis when compared to controls had increased percentages of CD163+CD16+ macrophages, whereas controls had increased CD163+CD14+ macrophages (Fig. 8). The intermediate pro‐inflammatory monocyte subset CD14++CD16+, which accumulates in the chronically inflamed human liver, also expresses CD163, and this phenotype is thought to represent a pro‐fibrogenic macrophage.17, 31 Many patients in this study were co‐infected with HIV (73%) (Supporting Table S1), and these patients have been shown to have significantly higher expression of CD163 on CD14++CD16+ monocytes when compared with healthy individuals.32 We recently showed that HIV‐infected macrophages did not influence fibrogenic gene expression of stellate cells, but rather enhanced HCV‐mediated activation of stellate cells.33
This initial study had several limitations. We evaluated only five antibodies that have been identified on macrophages: CD68, CD14, CD16, CD163, and MAC387. Several others will be used in future studies, including MARCO,7 CD206,34 Tim4,35 and chemokine receptors such as CCR2 and CCR5.23 Another challenge is acquiring adequate experience in analyzing large amounts of imaging data, especially when many ROIs/images (approximately 50‐70 per patient in this study) are acquired. Although we were able to conduct initial analyses using the software programs that are included with the Vectra 3 platform (i.e., Phenochart, inForm, and phenoptr), the multiplex panel stained multiple markers in the same compartment of the cell, which exceeded the capabilities of these programs. Therefore, we sought out imaging analysis experts from Visiopharm and the University of Michigan, co‐authors in this manuscript, to assist in these analyses. Even though five protective and pathogenic phenotypes were shown to be significantly different between the controls and patients with advanced fibrosis, individual analysis of the patients in each group showed variability that likely contributed to the lack of significance between the patients with minimal and advanced fibrosis.
One of the main advantages of this platform when compared to other recently developed cutting‐edge approaches such as single‐cell RNA sequencing and Fluidigm CyTOF, is the preservation of tissue architecture. Because the hepatic architecture is left intact, spatial and neighboring analyses can be performed using the spectral imaging platform. However, we found this to be challenging in the current study, in which multiple markers were present on the same compartment of the same cell. Determining the different zones in which the identified populations are located is critical when evaluating macrophage phenotypes in chronic liver disease, as those present in the portal tracts and lobules are not the same and are known to have unique phenotypes and functional characteristics.30 We attempted to separate the hepatic architecture into portal tracts and lobules using the tissue segmentation feature in inForm. We easily trained the software to recognize portal tracts as tumor‐like regions and lobules as non‐tumor‐like regions in patients with moderate/advanced fibrosis. However, the same training algorithms could not be applied to control patients, where portal stroma was subtle in appearance and challenging for the software to recognize. We did have better results when we trained the software to analyze each group of patients separately (controls, minimal, and advanced fibrosis). We were able to analyze the expression of the individual markers in the portal tracts and lobules of the representative images from Fig. 4 using Akoya Biosciences free online software, phenoptr (Supporting Fig. S2). The free online software is also able to perform neighboring analyses, which we tested using the representative multiplex images from Fig. 4 (Supporting Fig. S7). Visiopharm has also developed a neighborhood matrix (similar to Fig. 6, but for intercellular distances) that plots all possible phenotypes on the x/y axes and then color codes them from red to blue based on the significance of the distance between two objects. We are currently in the process of developing additional algorithms for spatial analysis of intrahepatic macrophages with collaborators at Visiopharm and the University of Michigan.
Patients undergo invasive procedures to obtain liver biopsies. Instead of merely providing a diagnosis, as clinicians and pathologists we should glean as much information as possible from this precious material. Several treatments targeting macrophages are Food and Drug Administration–approved,36 and the dual CCR2/CCR5 inhibitor that decreases monocyte recruitment to the liver is now in phase 3 clinical trials.23, 37 Results of multiplex imaging platforms may allow us to personalize treatment depending on cellular phenotypes present, similar to approaches that determine phenotypic expression on tumor‐infiltrating lymphocytes in patients with cancer.11, 38, 39, 40 The biggest challenge is developing more rapid and efficient big‐data analytics that can cope with the surplus of data generated. In summary, use of multispectral imaging has the potential to not only change our understanding of intrahepatic macrophages, but also the way in which patient liver biopsies are evaluated in the future.
Supporting information
Acknowledgment
We would like to thank Judy Pham for her assistance with fibrosis quantification of liver biopsies. We also thank Kevin Mottershead and Grady Carlson from Akoya Biosciences for their support and guidance in using the Vectra Platform. We extend sincere gratitude to Jeffrey East, physician assistant, and Shana White, study coordinator, for their assistance in acquiring and organizing the clinical samples. Sam Diaz de Leon and Blanca Flores provided secretarial support and assistance with obtaining the archived liver biopsy tissue blocks. Drs. David Walker and Cornelius Elferink provided a critical review of the manuscript.
H.L.S. and O.A.S. were supported by a Moody Endowment Grant (2014‐07) and a National Center for Advancing Translational Services Clinical and Translational Science Awards Grant (KL2 Scholars Program; KL2TR001441). The Vectra 3 microscope was purchased with funds from the University of Texas Systems Faculty Science and Technology Acquisition and Retention Program. A.R. and S.K. were supported by a gift from Agilent Technologies, institutional startup funds from the University of Michigan, and a Research Scholar Grant from the American Cancer Society (RSG‐16‐005‐01).
Potential conflict of interest: Dr. Rao consults for Voxel Analytics. He received grants from ACS, Agilent Technologies, and CHI. Dr. Freiberg is employed by Visiopharm.
References
Author names in bold designate shared co‐first authorship.
- 1. Li P, He K, Li J, Liu Z, Gong J. The role of Kupffer cells in hepatic diseases. Mol Immunol 2017;85:222‐229. [DOI] [PubMed] [Google Scholar]
- 2. Guo LP, Zhou L, Li HX, Zhang J, Wang BM. The study of liver macrophages polarization in patients with autoimmune hepatitis. Zhonghua Nei Ke Za Zhi 2017;56:763‐765. [DOI] [PubMed] [Google Scholar]
- 3. Lin R, Zhang J, Zhou L, Wang B. Altered function of monocytes/macrophages in patients with autoimmune hepatitis. Mol Med Rep 2016;13:3874‐3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malorny U, Neumann C, Sorg C. Influence of various detachment procedures on the functional state of cultured murine macrophages. Immunobiology 1981;159:327‐336. [DOI] [PubMed] [Google Scholar]
- 5. Chen S, So EC, Strome SE, Zhang X. Impact of detachment methods on M2 macrophage phenotype and function. J Immunol Methods 2015;426:56‐61. [DOI] [PubMed] [Google Scholar]
- 6. Gorris MAJ, Halilovic A, Rabold K, van Duffelen A, Wickramasinghe IN, Verweij D, et al. Eight‐color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J Immunol 2018;200:347‐354. [DOI] [PubMed] [Google Scholar]
- 7. MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morrell ED, Wiedeman A, Long SA, Gharib SA, West TE, Skerrett SJ, et al. Cytometry TOF identifies alveolar macrophage subtypes in acute respiratory distress syndrome. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wermuth PJ, Jimenez SA. The significance of macrophage polarization subtypes for animal models of tissue fibrosis and human fibrotic diseases. Clin Transl Med 2015;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mezheyeuski A, Bergsland CH, Backman M, Djureinovic D, Sjöblom T, Bruun J, et al. Multispectral imaging for quantitative and compartment‐specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients. J Pathol 2018;244:421‐431. [DOI] [PubMed] [Google Scholar]
- 11. Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol 2013;44:29‐38. [DOI] [PubMed] [Google Scholar]
- 12. Xie R, Chung JY, Ylaya K, Williams RL, Guerrero N, Nakatsuka N, et al. Factors influencing the degradation of archival formalin‐fixed paraffin‐embedded tissue sections. J Histochem Cytochem 2011;59:356‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang Y, de Boer WB, Adams LA, MacQuillan G, Bulsara MK, Jeffrey GP. Image analysis of liver biopsy samples measures fibrosis and predicts clinical outcome. J Hepatol 2014;61:22‐27. [DOI] [PubMed] [Google Scholar]
- 14. Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014;70:46‐58. [DOI] [PubMed] [Google Scholar]
- 15. Faget L, Hnasko TS. Tyramide signal amplification for immunofluorescent enhancement. Methods Mol Biol 2015;1318:161‐172. [DOI] [PubMed] [Google Scholar]
- 16. McGuinness PH, Painter D, Davies S, McCaughan GW. Increases in intrahepatic CD68 positive cells, MAC387 positive cells, and proinflammatory cytokines (particularly interleukin 18) in chronic hepatitis C infection. Gut 2000;46:260‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liaskou E, Zimmermann HW, Li KK, Oo YH, Suresh S, Stamataki Z, et al. Monocyte subsets in human liver disease show distinct phenotypic and functional characteristics. Hepatology 2013;57:385‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strauss O, Phillips A, Ruggiero K, Bartlett A, Dunbar PR. Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Sci Rep 2017;7:44356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uhrig A, Banafsche R, Kremer M, Hegenbarth S, Hamann A, Neurath M, et al. Development and functional consequences of LPS tolerance in sinusoidal endothelial cells of the liver. J Leukoc Biol 2005;77:626‐633. [DOI] [PubMed] [Google Scholar]
- 20. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol 1995;22:696‐699. [DOI] [PubMed] [Google Scholar]
- 21. Ju C, Tacke F. Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell Mol Immunol 2016;13:316‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017;17:306‐321. [DOI] [PubMed] [Google Scholar]
- 23. Friedman SL, Ratziu V, Harrison SA, Abdelmalek MF, Aithal GP, Caballeria J, et al. A randomized, placebo‐controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology 2018;67:1754‐1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reid DT, Reyes JL, McDonald BA, Vo T, Reimer RA, Eksteen B. Kupffer cells undergo fundamental changes during the development of experimental NASH and are critical in initiating liver damage and inflammation. PLoS One 2016;11:e0159524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang R, Wu H, Liu Y, Yang C, Pan Z, Xia J, et al. Increase of infiltrating monocytes in the livers of patients with chronic liver diseases. Discov Med 2016;21:25‐33. [PubMed] [Google Scholar]
- 26. Bility MT, Nio K, Li F, McGivern DR, Lemon SM, Feeney ER, et al. Chronic hepatitis C infection‐induced liver fibrogenesis is associated with M2 macrophage activation. Sci Rep 2016;6:39520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weston CJ, Zimmermann HW, Adams DH. The role of myeloid‐derived cells in the progression of liver disease. Front Immunol 2019;10:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heymann F, Tacke F. Immunology in the liver—from homeostasis to disease. Nat Rev Gastroenterol Hepatol 2016;13:88‐110. [DOI] [PubMed] [Google Scholar]
- 29. Saha B, Kodys K, Szabo G. Hepatitis C virus‐induced monocyte differentiation into polarized M2 macrophages promotes stellate cell activation via TGF‐beta. Cell Mol Gastroenterol Hepatol 2016;2:302‐316.e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gadd VL, Melino M, Roy S, Horsfall L, O'Rourke P, Williams MR, et al. Portal, but not lobular, macrophages express matrix metalloproteinase‐9: association with the ductular reaction and fibrosis in chronic hepatitis C. Liver Int 2013;33:569‐579. [DOI] [PubMed] [Google Scholar]
- 31. Zimmermann HW, Seidler S, Nattermann J, Gassler N, Hellerbrand C, Zernecke A, et al. Functional contribution of elevated circulating and hepatic non‐classical CD14CD16 monocytes to inflammation and human liver fibrosis. PLoS One 2010;5:e11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tippett E, Cheng WJ, Westhorpe C, Cameron PU, Brew BJ, Lewin SR, et al. Differential expression of CD163 on monocyte subsets in healthy and HIV‐1 infected individuals. PLoS One 2011;6:e19968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akil A, Endsley M, Shanmugam S, Saldarriaga O, Somasunderam A, Spratt H, et al. Fibrogenic gene expression in hepatic stellate cells induced by HCV and HIV replication in a three cell co‐culture model system. Sci Rep 2019;9:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan‐Garcia A, Wai LE, Zheng D, Ceccarello E, Jo J, Banu N, et al. Intrahepatic CD206. J Hepatol 2017;67:490‐500. [DOI] [PubMed] [Google Scholar]
- 35. Triantafyllou E, Woollard KJ, McPhail MJW, Antoniades CG, Possamai LA. The role of monocytes and macrophages in acute and acute‐on‐chronic liver failure. Front Immunol 2018;9:2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol 2017;66:1300‐1312. [DOI] [PubMed] [Google Scholar]
- 37. Krenkel O, Puengel T, Govaere O, Abdallah AT, Mossanen JC, Kohlhepp M, et al. Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology 2018;67:1270‐1283. [DOI] [PubMed] [Google Scholar]
- 38. Zaretsky JM, Garcia‐Diaz A, Shin DS, Escuin‐Ordinas H, Hugo W, Hu‐Lieskovan S, et al. Mutations associated with acquired resistance to PD‐1 blockade in melanoma. N Engl J Med 2016;375:819‐829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu XD, Hoang A, Zhou L, Kalra S, Yetil A, Sun M, et al. Resistance to antiangiogenic therapy is associated with an immunosuppressive tumor microenvironment in metastatic renal cell carcinoma. Cancer Immunol Res 2015;3:1017‐1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mlecnik B, Bindea G, Kirilovsky A, Angell HK, Obenauf AC, Tosolini M, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med 2016;8:327ra326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


