Abstract
The long noncoding RNA (lncRNA) Eosinophil Granule Ontogeny Transcript (EGOT) has been reported to inhibit the proliferation and migration of glioma cells, and promote the development and progression of gastric cancer through the Hedgehog (Hh) signaling pathway. This study was conducted to assess the role of EGOT in the progression of breast cancer. We observed that EGOT is significantly down‐regulated in breast cancer tissues and cell lines, and EGOT expression is negatively correlated with the Ki67 expression. Overexpression of EGOT in BT549 cells decreased cell viability and migration. In addition, overexpression of EGOT resulted in decreases in expression of key genes in the Hh pathway, including Gli1, smoothened protein, protein patched homolog 1 and Hedgehog‐interacting protein (HHIP). Breast cancer tissues exhibited an increase in Gli1 expressions. Altered expression of Gli1, smoothened protein, protein patched homolog 1 and HHIP caused by EGOT overexpression were fully restored in cells transfected with plasmid complementory DNA (pcDNA) EGOT and treated with purmorphamine, an agonist of the Hh pathway. Cell viability and migration were also restored by purmorphamine. We conclude that lncRNA EGOT may inhibit breast cancer cell viability and migration via inactivation of the Hh pathway.
Keywords: breast cancer, cell migration, cell proliferation, Hedgehog pathway, lncRNA EGOT
Long noncoding RNA Eosinophil Granule Ontogeny Transcript (EGOT) is greatly down‐regulated in breast cancer tissues and cell lines. Furthermore, the expression of key genes in the Hedgehog (Hh) pathway is reduced at both mRNA and protein levels when EGOT is overexpressed. Purmorphamine restored the expression of Hh pathway genes and reversed the effects on cell viability and migration caused by overexpression of EGOT. EGOT thus may inhibit breast cancer cell viability and migration via inhibition of the Hh pathway.
Abbreviations
- EGOT
Eosinophil Granule Ontogeny Transcript
- ER
estrogen receptor
- HER‐2
human epidermal growth factor receptor 2
- Hh
Hedgehog
- lncRNA
long noncoding RNA
- MTT
3‐(4,5)‐dimethylthiahiazo(‐z‐y1)‐3,5‐di‐phenytetrazoliumromide
- PVDF
poly(vinylidene difluoride)
- qRT‐PCR
real‐time quantitative PCR
- SD
standard deviation
- TNBC
triple‐negative breast cancer
Breast cancer is a malignant tumor caused by a malignant tumor invasion and destruction of normal breast tissue. The global death toll of breast cancer accounts for 14% of all cancer deaths and is the second most common cancer among women. According to cancer statistics, there were more than 2.3 million new cases of invasive breast cancer worldwide in 2015, with 402 000 women having died of breast cancer [1]. In recent years, surgery, chemotherapy and radiotherapy have become the main methods of breast cancer treatment with the continuous development of various treatment methods [2]. However, 4–10% of patients with breast cancer in China are found with distant metastasis every year, with clinical treatments bringing poor results [3]. Early screening and treatment are both essential in preventing and helping to alleviate breast cancer.
Long noncoding RNA (lncRNA) is a type of noncoding RNA of more than 200 nucleotides, which is also a product of the RNA polymerase II transcription and lacks an ORF. The initial study concluded that lncRNA has biological functions. In recent years, studies have found that lncRNA can be involved in the development of tumors as a carcinogenic or tumor suppressor, with its abnormal expression being closely related to tumor cell proliferation, metastasis and apoptosis [4]. Aberrant expressions in lncRNA also play an important role in the development of breast cancer. Current studies have shown that HOTAIR [5], GAS5 [6], PVTl [7], MALATl [8] and other lncRNAs have strong effects on breast cancer cell proliferation, apoptosis and migration. The lncRNA Eosinophil Granule Ontogeny Transcript (EGOT), a human gene, is located in 3p26.1 and is highly conserved at the nucleic acid level. Wagner et al. [9] have found that EGOT is involved in the development of eosinophils and is expressed in mature eosinophils. They also have demonstrated that EGOT is not related to ribosomes and is likely to function as a noncoding RNA through sucrose density gradients. Wagner et al. [9] also additionally reported that EGOT is highly expressed in bone marrow and plays an important role in bone marrow hematopoietic stem cells. For some tumors, such as renal cell carcinoma, EGOT is a tumor suppressor and is likely a potential prognostic biomarker for kidney cancer [10]. Moreover, Wu et al. [11] have found that EGOT inhibits the proliferation and migration of glioma cells and promotes apoptosis in human gliomas.
The Hedgehog gene (Hh) was first discovered in Drosophila in 1980 [12] with the family of proteins having crucial functions in embryonic development and cell proliferation [13]. The Hh signaling pathway has been well known as an important signaling pathway and therapeutic target in various kinds of cancers [14]. The protein patched homolog 1 (PTCH) receptor prevents high expressions and activity of the smoothened protein (SMO) without the Hh ligand, whereas the repression of SMO is relieved when Hh is bound, which thus leads to the activation of glioma‐associated oncogene (GLI) transcription factors. These include activators Gli1 and Gli2, along with repressor Gli3. Activated GLI then controls transcriptions of Hh‐targeted genes [15, 16]. GLI can also be activated in a nonclassical way without the Hh ligand and SMO, via receptors in tumor‐associated cytokines. This largely occurs with transforming growth factor β and stromal‐derived factor 1 [17].
Peng et al. [18] have demonstrated that EGOT promotes the development and progression of gastric cancer through the Hh signaling pathway and can be used as a biomarker for the diagnosis and prognosis of gastric cancer. For breast cancer in particular, Xu et al. [19] point out that a down‐regulation of EGOT is associated with malignancy and a poor prognosis of breast cancer through clinical breast cancerous tissues and adjacent noncancerous tissues. They suggest, moreover, that antisense intronic lncRNA EGOT enhancing autophagy has sensitized paclitaxel cytotoxicity via an up‐regulation of ITPR1 expression by RNA–RNA and protein–RNA interactions in breast cancer [20]. Whether EGOT aberrant expressions have an effect on biological activities through Hh signals in breast cancer cells is still unclear.
The aim of this study, therefore, has been to confirm the expression pattern of EGOT in breast cancer cells and adjacent tissues to explore the effects of abnormal expressions in lncRNA EGOT, along with the viability and migration of breast cancer cell lines and the Hh signaling pathway.
Materials and methods
Tissue collection
From October 2016 to July 2018, a total of 50 paired breast tumor and adjacent normal tissues were collected after surgery at The First People’s Hospital of Yunnan Province, China. Tissues were washed in PBS and frozen in liquid nitrogen before being stored at −80 °C. Each patient provided a signed informed consent form. This study was approved by the Ethics Committee of The First People’s Hospital of Yunnan Province. The study conformed to the guidelines set by the Declaration of Helsinki.
Cell lines and culture
Breast cancer cell lines, including BT549, MDA‐MB‐231, MCF7, SKBr3 and HEK293, were purchased from ATCC (Balimore, MD, USA). BT549 cells were cultured with RPMI 1640 (Gibco, Thermo Fisher Scientific, Inc, Grand Island, NY, USA) containing 10% FBS. MDA‐MB‐231 and SKBr3 cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, Inc.) with 10% FBS. MCF‐7 was cultured in minimum Eagle's medium (MEM) including 10% FBS, and HEK293 was also cultured in MEM with 10% FBS. All of the cells were cultured in the constant temperature and humidity chamber at 37 °C with 5% carbon dioxide.
Plasmids, cell transfection and purmorphamine treatment
pcDNA–EGOT plasmids were purchased from GenePharma (Shanghai, China). Plasmids were transfected into BT549 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Purmorphamine (120933; Abcam, Cambridge, UK), the agonist of the Hh pathway, was dissolved in DMSO for this project; the substance was then formulated into a 1 μm solution. After transfection with pcDNA–EGOT or pcDNA (empty vector), cells were treated with 1 μm purmorphamine of DMSO, and treatment with DMSO served as a control.
MTT assay
After cells were counted, cell density was adjusted to 1 × 105 cells·mL−1 with a serum‐free medium. Cells were added to a 96‐well plate and then cultured with 5% CO2 at 37 °C for 24, 48, 72 and 96 h, respectively. 3‐(4,5)‐Dimethylthiahiazo(‐z‐y1)‐3,5‐di‐phenytetrazoliumromide (MTT) was configured as a 5 mg·mL−1 solution in PBS, and 10 μL was added for incubation another 4 h; the absorbance (OD) value was further read at 490 nm by a microplate reader [21].
Migration Transwell
Cells were collected and prepared into a cell suspension of 1 × 105 cells·mL−1. A 600‐μL medium containing 10% FBS was added to the lower chamber, whereas a 200‐μL cell suspension was added to the upper chamber and incubated for 24 h; the liquid from the upper and lower chambers was then discarded. The cells of the lower chamber were fixed by 4% paraformaldehyde for 30 min. After this, the paraformaldehyde was removed, and the cells that did not pass through the membrane were wiped clean with a cotton swab. The lower chamber was stained with 0.1% crystal violet for 10 min, whereas the chamber was washed three times with PBS. Migrated cells were then observed and counted under a microscope [22].
Wound healing assay
Horizontal lines across the well were drawn with a marker pen and ruler on the back of a six‐well plate, while every well passed at least five times. Cell suspensions at the concentration of 1 × 106 were prepared, and 1 × 105 cells were added to each well. Vertical lines to the back lines were drawn with the smaller pipettor and a ruler. Cells were then washed three times with PBS solution, while floating cells were removed completely and serum‐free medium was added. These cells were cultured at 37 °C with 5% CO2 for 24 h; after this, pictures were taken for examination purposes [23].
Real‐time quantitative PCR
RNA extraction in the tissues and cells was performed by TRIzol reagent (Invitrogen) based on the manufacturer’s instructions, with the concentration of RNA being measured by the nanodrop. Genomic DNA was removed, and reverse transcription of 1 μg RNA was performed using a Transcriptor First Strand cDNA Synthesis Kit. After this, the EGOT relative level was tested by the lncRNA qPCR kit (SYBR Green, WH0125‐GUQ), whereas other genes at the mRNA level were measured using the ABI 7500 Fast Real‐Time PCR System (Invitrogen). A method was used to calculate relative levels.
Western blotting
After cells were disrupted, the total protein from cells was extracted. Protein quantification was performed using a BCA kit, and an SDS/PAGE was conducted. The protein from SDS/PAGE gel was transferred onto a poly(vinylidene difluoride) (PVDF) membrane. After this, the PVDF membrane was blocked with 5% nonfat dry milk for 2 h at room temperature. The primary antibodies we used in our research were incubated at a temperature of 4 °C overnight, whereas PVDF membranes were rinsed three times by PBST. The antibodies of Gli1 (ab134906), SMO (ab113438), PTCH1 (ab53715) and HHIP (ab39208) were provided by Abcam. After PVDF membranes were washed once more, they were incubated with the corresponding secondary antibody, whereas the membranes were washed three times with PBST. Finally, an enhanced ECL chemiluminescent kit was used to treat the membrane for color reaction.
Statistical analysis
Data were shown as average ± standard deviation (SD). A t‐test, Spearman’s correlation and one‐tailed ANOVA were implemented using the latest spss, version 25.0 (IBM Corp, Armonk, NY, USA). Relative expression levels were plotted using graphpad prism 6 (San Diego, CA, USA). Quantitative data were expressed as mean ± SD based on three independent experiments. Comparisons between two groups were analyzed using Student’s t‐test. Multiple comparisons among groups were analyzed using Bonferroni posttests followed by two‐way ANOVA. Results were defined at a significance of P < 0.05.
Results
lncRNA EGOT is down‐regulated in breast cancer tissues and cell lines
The relative EGOT level was significantly reduced in breast cancer tissues compared with adjacent tissues (P < 0.0001; Fig. 1A). We examined the correlation between EGOT and a Ki67 expression, and discovered a negative correlation between the two genes. This most likely indicates one glaring insight: the higher the malignancy of the tumor and the larger the Ki67 value, the lower the expression of EGOT (P < 0.0001; Fig. 1B). The quantitative real‐time PCR (qRT‐PCR) results exhibited that the relative level of EGOT expression was significantly down‐regulated in breast cancer cell lines BT549, MDA‐MB‐231, MCF7 and SKBr3, compared with control cell lines in HEK293 (Fig. 1C).
Fig. 1.
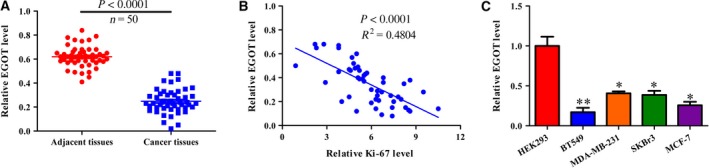
lncRNA EGOT is down‐regulated in breast cancer tissues and cell lines. (A) The relative EGOT level was measured in 50 paired breast cancer and adjacent tissues. (B) A Spearman’s correlation analysis of EGOT and Ki‐67 in 50 breast cancer tissues. (C) Relative EGOT levels were measured in breast cancer cell lines, including BT549, MDA‐MB‐231, MCF7 and SKBr3, and HEK293 was used as a control. Data shown are the mean ± SD for four independent experiments (n = 5) per cell line. Paired t‐test was performed for the comparison between two groups; linear regression analysis was performed to analyze the correlation between EGOT and Ki‐67. *P < 0.05; **P < 0.01 versus HEK293 cells.
Overexpression of EGOT impedes cell proliferation and migration
The expression of EGOT from BT549 was the lowest among these breast cancer cell lines, with BT549 being used for further functional experiments. When the EGOT overexpression system was established, results from the pcDNA–EGOT transfection revealed that EGOT was clearly enhanced in the pcDNA–EGOT group compared with the control group (Fig. 2A). MTT assays were conducted to evaluate cell viability, with results revealing that cell viability is greatly inhibited in the pcDNA–EGOT group compared with pcDNA (Fig. 2B). Cell migration was then detected using a Transwell migration and wound healing assay, with results revealing that the migration ability of cells transfected with pcDNA–EGOT was tremendously impaired compared with the pcDNA group (Fig. 2C,D).
Fig. 2.
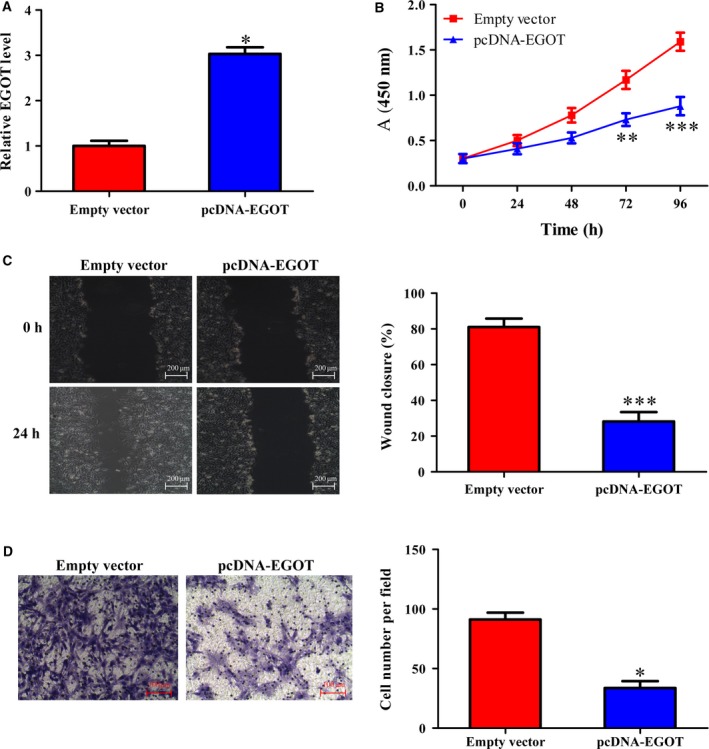
Overexpression of EGOT affects cell viability and migration. (A) The relative level of EGOT in cells transfected with pcDNA–EGOT or pcDNA was detected by qRT‐PCR. (B) An MTT assay was conducted to assess cell viability. (C, D) Wound healing assays and Transwell migration assays were used to detect cell migration after an overexpression of EGOT. Scale bars: 200 μm (C); 100 μm (D). The results were the average from at least three independent experiments, mean ± SD; paired t‐test was performed for the comparison between two groups; Bonferroni posttests followed by two‐way ANOVA were used to assess the cell viability. *P < 0.05; **P < 0.01; ***P < 0.001 versus pcDNA.
Gli1 is up‐regulated in the breast cancer tissues, and Gli1, SMO, PTCH1 and HHIP were all down‐regulated after an EGOT overexpression
Interestingly, we found that Gli1 was significantly elevated at the mRNA level in breast cancer tissues compared with adjacent tissues (Fig. 3A). Not only this, but the expression of Gli1, SMO, PTCH1 and HHIP at mRNA and protein levels was detected in either the pcDNA–EGOT or pcDNA group (Fig. 3B,C). These results demonstrated that Gli1, SMO, PTCH1 and HHIP were all reduced at transcriptional and translational levels in cells transfected with pcDNA–EGOT. We therefore speculate that lower EGOT expression levels may be related to the Hh pathway.
Fig. 3.
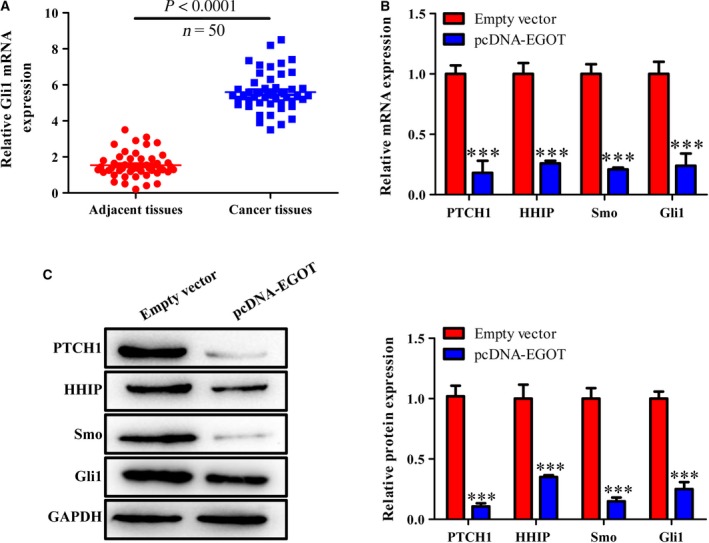
Gli1 is up‐regulated in breast cancer tissues, and Gli1, SMO, PTCH1 and HHIP were down‐regulated after EGOT overexpression. (A) The relative Gli1 mRNA level was measured by qRT‐PCR in 50 breast cancer and adjacent tissues. (B) The expression of Gli1, SMO, PTCH1 and HHIP at mRNA levels was detected in cells transfected with pcDNA–EGOT or pcDNA. (C) The expression of Gli1, SMO, PTCH1 and HHIP at protein level was detected in cells transfected with pcDNA–EGOT or pcDNA. The results were the average from at least three independent experiments, mean ± SD; paired t‐test was performed for the comparison between two groups. ***P < 0.001 versus pcDNA.
Purmorphamine restores the expression of key genes in the Hh signaling pathway in breast cancer cell line BT549 overexpressing EGOT
We further discovered that validating the role of the Hh pathway in EGOT overexpressions triggered inhibitive effects on cell viability and migration. Purmorphamine, the agonist of the Hh pathway, was used to treat cells with an overexpression of EGOT. The results showed that there was no significant difference at mRNA and protein levels in PTCH1 and HHIP in the pcDNA group with or without purmorphamine molecules. The mRNA and protein levels of Gli1 and SMO were up‐regulated with purmorphamine in the pcDNA empty vector group. However, there was an obvious increase in mRNA and protein levels in PTCH1, Gli1, SMO and HHIP in pcDNA–EGOT cells with purmorphamine compared with those who had only pcDNA–EGOT cells (Fig. 4A,B).
Fig. 4.
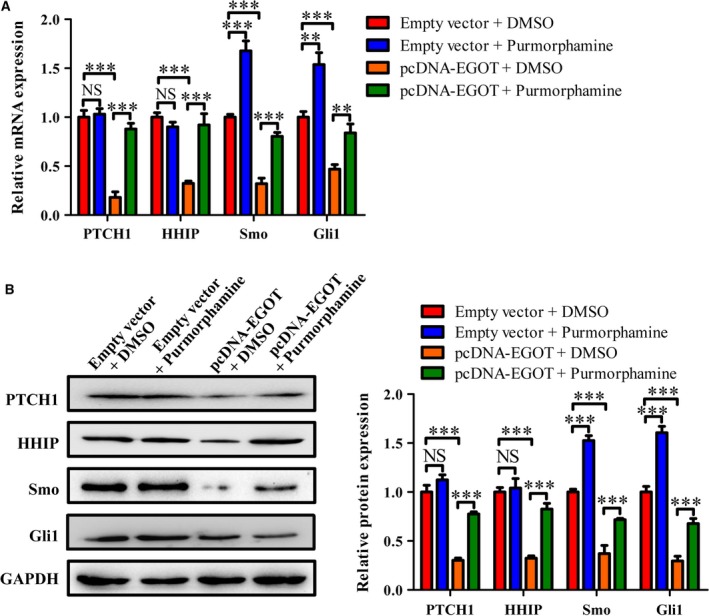
Effects of EGOT overexpression on the Hh signaling pathway‐related genes in breast cancer cell line BT549. BT549 cells were transfected with pcDNA–EGOT or pcDNA, and then treated with purmorphamine or DMSO. (A) The relative EGOT level was measured by qRT‐PCR in various treatment groups. (B) The expression of Gli1, SMO, PTCH1 and HHIP at protein levels was detected in these groups. The results were the average from at least three independent experiments, mean ± SD; Bonferroni posttests followed by two‐way ANOVA were used to compare the four groups. **P < 0.01; ***P < 0.001. NS, not significant.
Purmorphamine restores viability and migration in breast cancer cell line BT549, with an overexpression of EGOT
As shown in Fig. 5A, the viability of cells transfected with pcDNA–EGOT and incubated with DMSO pcDNA was lower than that in pcDNA + DMSO groups, although the viability of these cells enhanced after cells with EGOT overexpression were treated with purmorphamine. Moreover, the results of cell viability and migration showed a similar tendency to the cell viability during two other methods of treatment: the wound healing test (Fig. 5B) and the Transwell assay (Fig. 5C).
Fig. 5.
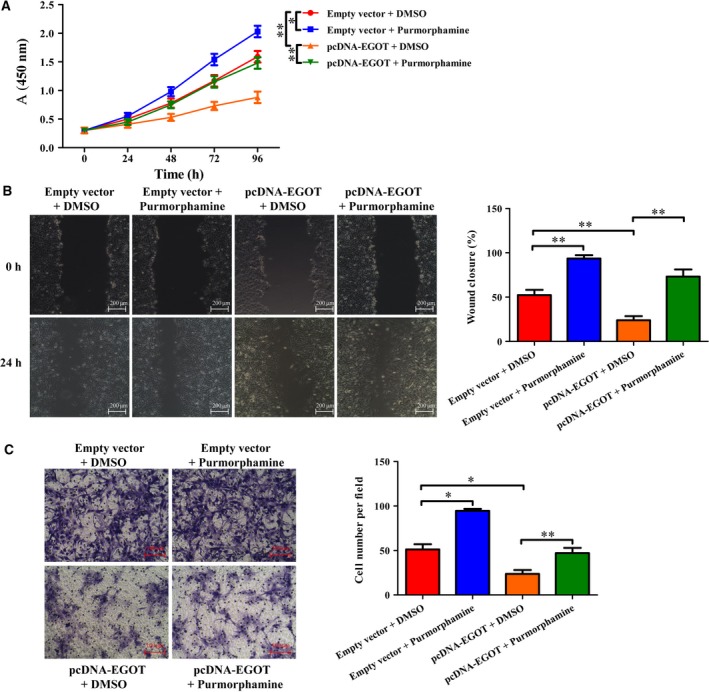
Effect of Hh signaling pathway on cell viability and migration in breast cancer cell line BT549. BT549 cells were transfected with pcDNA–EGOT or pcDNA, then treated with purmorphamine or DMSO. (A) An MTT assay was used to measure cell viability in various treatment groups. (B, C) Wound healing (B) and Transwell assays (C) were used to detect cell migration in these groups. Scale bars: 200 μm (B); 100 μm (C). The results were the average from at least three independent experiments, mean ± SD; paired t‐test was performed for the comparison between two groups; Bonferroni posttests followed by two‐way ANOVA were used to assess cell viability. *P < 0.05; **P < 0.01.
Discussion
With advances in RNA sequencing and transcriptome analysis, researchers have surprisingly found that up to 76% of the human genome can be transcribed into RNA molecules, including lncRNAs [24, 25]. According to the latest version of lncRNA database LNCipedia 5.2, there are 127 802 transcripts and 56 946 human annotated lncRNAs [26]. In the past two decades, lncRNAs have been discovered to be aberrantly expressed, regulating the development and progression of tumors [27, 28]. A number of lncRNAs have been deregulated in cancers, controlled by epigenetic, genetic and transcriptional factors [29]. Up until to now, moreover, numerous abnormal expression lncRNAs have been found in breast cancer that are specifically involved in the cancer’s initiation and development [30, 31, 32, 33, 34].
In our study, lncRNA EGOT was detected in breast cancer tissues and cell lines (Fig. 1), which was further consistent with Xu et al.’s results [19], whereby the expression of EGOT was negatively correlated with Ki67. As expected, overexpression in EGOT negatively affected cell proliferation and migration (Fig. 2). Therefore, the role of EGOT in breast cancer progression has been identified as a tumor suppressor.
In recent publications, aberrant expressed lncRNAs were reported to strengthen cancer progression via activation of the Hh signaling pathway, including gastric cancer, prostate cancer, breast cancer and pancreatic cancer [16, 18, 35, 36]. To further uncover the role of EGOT in breast cancer, we studied the relationship between EGOT and the Hh signaling pathway. As the terminal effector of the typical Hh signaling pathway, the Gli1 family functions as a transcription factor, and abnormal regulation of Gli1 protein leads to tumorigenesis [17, 37]. Interestingly, we found that Gli1 was significantly up‐regulated in breast cancer tissues, whereas PTCH1, SMO, Gli1 and HHIP at mRNA and protein levels were all down‐regulated after controlling for EGOT overexpression (Fig. 3). We hypothesize that an overexpression of EGOT decreased the expression of PTCH1, thus suppressing its binding to the Hh ligand, which has proved to lead to low expressions of SMO and Gli1. As a result, the expression of the Hh interacting protein HHIP in nucleus was down‐regulated. According to these results, we deduce that the Hh signaling pathway may be involved in breast cancer progression.
Onishi and Katano [38] and Rubin and de Sauvage [14] have concluded that abnormal activation in the Hh signaling pathway is closely correlated with the initiation and development of various cancers. Purmorphamine is an Hh agonist that directly targets SMO transmembrane proteins [39]. Activation of Hh signaling by purmorphamine promotes the transcription of various genes, including Gli1, PTCH1 and alkaline phosphatase [34]. To confirm whether purmorphamine can reverse the expression of genes in the Hh signaling pathway caused by an overexpression in EGOT in breast cancer, we transfected BT549 cells with pcDNA–EGOT or pcDNA. After this, cells were treated with purmorphamine. The results demonstrate that purmorphamine reversed the expression of PTCH1, SMO, Gli1 and HHIP in the overexpression system of EGOT (Fig. 4), which was consistent with the finding by Lin et al. [34]. Purmorphamine inhibited osteoblast differentiation in human multipotent adipose‐derived stem cells and mesenchymal stem cells (MSCs) from bone marrow by activating the Hh signaling pathway [40]. Purmorphamine was then used to measure cell proliferation and migration ability in the overexpression system of EGOT. Results indicated that purmorphamine reversed cell proliferation and migration abilities in breast cancer (Fig. 5).
According to the expression of estrogen receptor (ER), progesterone receptor and human epidermal growth factor receptor 2 (HER‐2), breast cancer is identified as four molecular subtypes: basal‐like, HER‐2 positive, luminal A and luminal B, with the first two subtypes from negative ER tumors and the last two from positive ER tumors [41]. The basal‐like tumors are mainly composed of triple‐negative breast cancer (TNBC), tumors that lack expression of ER, progesterone receptor and HER‐2 [42]. We collected a total of 50 breast cancer samples, 18 of which were TNBC tumors, 9 HER‐2 tumors, 13 luminal A tumors and 10 luminal B tumors. TNBC takes up 15–20% in all invasive breast cancers and occurs more frequently in young women; meanwhile, the survival rate of TNBC is the worst [43]. In our study, BT549 and MDA‐MB‐231 cells belong to the TNBC subtype, whereas MCF7 is a luminal A subtype and SKBr3 is an HER‐2‐positive subtype. In experiments involved in the Hh signaling pathway, the cell line BT549 was used to further real molecular mechanisms in this aggressive subtype of breast cancer.
Conclusions
EGOT was greatly down‐regulated in breast cancer tissues and cell lines, and the relative level of EGOT was negatively correlated with the expression of Ki67. As we inferred, the overexpression of EGOT impaired cell viability and migration in BT549 cell lines. Furthermore, the expression of Gli1 was significantly increased in breast cancer tissues. The relative expressions of PTCH1, SMO, Gli1 and HHIP at both mRNA and protein levels were reduced in the overexpression of EGOT. Purmorphamine molecules were used to treat cells with an overexpression of EGOT to ensure two things: first, that the Hh pathway was activated; and second, that the roles of Hh signaling in EGOT‐induced inhibitive effects on breast cancer cells were found. The results found that the relative expression of PTCH1, SMO, Gli1 and HHIP, and cell viability and migration caused by an overexpression of EGOT were reversed by purmorphamine. In total, lncRNA EGOT is proved to inhibit TNBC cell viability and migration via modulation of the Hh pathway. Our present research supports helpful evidence to the anti‐oncogene of EGOT in breast cancer and may extend novel knowledge of therapeutic methods for TNBC.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
SQ and GBC conducted experiments and were responsible for data acquisition and manuscript writing. JP, JL and JMC were responsible for data interpretation and data analysis. JJW and LL helped in statistical analysis. KXY conceived and designed the study, and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by Joint Program of Applied Basic Research of Yunnan Provincial Department of Science and Technology–Kunming Medical University [Grant No. 2017FE467(‐153)].
References
- 1. Siegel RL, Miller KD and Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65, 5–29. [DOI] [PubMed] [Google Scholar]
- 2. Swain SM, Baselga J, Kim S‐B, Ro J, Semiglazov V, Campone M, Ciruelos E, Ferrero JM, Schneeweiss A, Heeson S et al (2015) Pertuzumab, trastuzumab, and docetaxel in HER2‐positive metastatic breast cancer. N Engl J Med 372, 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fredholm H, Magnusson K, Lindström LS, Garmo H, Fält SE, Lindman H, Bergh J, Holmberg L, Pontén F, Frisell J et al (2016) Long‐term outcome in young women with breast cancer: a population‐based study. Breast Cancer Res Treat 160, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen YM, Liu Y, Wei HY, Lv KZ and Fu PF (2016) Large intergenic non‐coding RNA‐ROR reverses gemcitabine‐induced autophagy and apoptosis in breast cancer cells. Oncotarget 7, 59604–59617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC and Yu J (2016) LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 35, 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mourtada‐Maarabouni M, Pickard MR, Hedge VL, Farzaneh F and Williams GT (2009) GAS5, a non‐protein‐coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28, 195–208. [DOI] [PubMed] [Google Scholar]
- 7. Guan Y, Kuo W‐L, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao J‐H, Yu M, Miller MA, Santos JL et al (2007) Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res 13, 5745–5755. [DOI] [PubMed] [Google Scholar]
- 8. Cho SF, Chang YC, Chang CS, Lin S‐F, Liu YC, Hsiao HH, Chang JG and Liu TC (2014) MALAT1 long non‐coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer 14, 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, Esplin MS, Weiss RB and Gleich GJ (2007) EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 109, 5191–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin L, Quan J, Pan X, He T, Hu J, Li Y, Gui Y, Yang S, Mao X, Chen Y et al (2017) Identification of lncRNA EGOT as a tumor suppressor in renal cell carcinoma. Mol Med Rep 16, 7072–7079. [DOI] [PubMed] [Google Scholar]
- 11. Wu Y, Liang S, Xu B, Zhang R, Zhu M, Zhou W, Zhang S, Guo J, Xu L and Zhu H (2017) Long noncoding RNA eosinophil granule ontogeny transcript inhibits cell proliferation and migration and promotes cell apoptosis in human glioma. Exp Ther Med 14, 3817–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nüsslein‐Volhard C and Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. [DOI] [PubMed] [Google Scholar]
- 13. McMahon AP (2000) More surprises in the Hedgehog signaling pathway. Cell 100, 185–188. [DOI] [PubMed] [Google Scholar]
- 14. Rubin LL and de Sauvage FJ (2006) Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 5, 1026–1033. [DOI] [PubMed] [Google Scholar]
- 15. Jiang J and Hui CC (2008) Hedgehog signaling in development and cancer. Dev Cell 15, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stanton BZ and Peng LF (2010) Small‐molecule modulators of the Sonic Hedgehog signaling pathway. Mol Biosyst 6, 44–54. [DOI] [PubMed] [Google Scholar]
- 17. Skoda AM, Simovic D, Karin V, Kardum V, Vranic S and Serman L (2018) The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci 18, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng W, Wu J, Fan H, Lu J and Feng J (2019) LncRNA EGOT promotes tumorigenesis via Hedgehog pathway in gastric cancer. Pathol Oncol Res 25, 883–887. [DOI] [PubMed] [Google Scholar]
- 19. Xu SP, Zhang JF, Sui SY, Bai NX, Gao S, Zhang GW, Shi QY, You ZL, Zhan C and Pang D (2015) Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol 36, 9807–9812. [DOI] [PubMed] [Google Scholar]
- 20. Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J, Wang Q, Qiao K, Yang W, Xu H et al (2019) Ai‐lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA‐RNA and RNA‐protein interactions in human cancer. Mol Cancer 18, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Meerloo J, Kaspers GJ and Cloos J (2011) Cell sensitivity assays: the MTT assay. Methods Mol Biol 731, 237–245. [DOI] [PubMed] [Google Scholar]
- 22. Justus CR, Leffler N, Ruiz‐Echevarria M and Yang LV (2014) In vitro cell migration and invasion assays. J Vis Exp 88, e51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriguez LG, Wu X and Guan JL (2005) Wound‐healing assay. Methods Mol Biol 294, 23–29. [DOI] [PubMed] [Google Scholar]
- 24. Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A and Meissner A (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R et al (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P and Vandesompele J (2019) LNCipedia 5: towards a reference set of human long non‐coding RNAs. Nucleic Acids Res 47, D135–D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D et al (2015) A cytoplasmic NF‐κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381. [DOI] [PubMed] [Google Scholar]
- 28. Yang F, Zhang L, Xs Huo, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ et al (2011) Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 54, 1679–1689. [DOI] [PubMed] [Google Scholar]
- 29. Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21, 1253–1261. [DOI] [PubMed] [Google Scholar]
- 30. Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, Weedon J, Diallo R, Böcker W and Tiedge H (2004) BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis 25, 2125–2133. [DOI] [PubMed] [Google Scholar]
- 31. Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ (2002) Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23, 1885–1895. [DOI] [PubMed] [Google Scholar]
- 32. Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M and Mo YY (2014) Long non‐coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 5, e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hansji H, Leung EY, Baguley BC, Finlay GJ, Cameron‐Smith D, Figueiredo VC and Askarian‐Amiri ME. ZFAS1: a long noncoding RNA associated with ribosomes in breast cancer cells. Biology Direct 11, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y et al (2016) The LINK‐A lncRNA activates normoxic HIF1α signalling in triple‐negative breast cancer. Nat Cell Biol 18, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du Y‐e, Wen S, Xu L, Tang X, Tang S et al (2016) LncRNA‐Hh strengthen cancer stem cells generation in twist‐positive breast cancer via activation of Hedgehog signaling pathway. Stem Cells 34, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katoh Y and Katoh M (2005) Hedgehog signaling pathway and gastric cancer. Cancer Biol Ther 4, 1050–1054. [DOI] [PubMed] [Google Scholar]
- 37. Gupta S, Takebe N and Lorusso P (2010) Targeting the Hedgehog pathway in cancer. Ther Adv Med Oncol 2, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Onishi H and Katano M (2011) Hedgehog signaling pathway as a therapeutic target in various types of cancer. Cancer Sci 102, 1756–1760. [DOI] [PubMed] [Google Scholar]
- 39. Sinha S and Chen JK (2006) Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem Biol 2, 29–30. [DOI] [PubMed] [Google Scholar]
- 40. Plaisant M, Fontaine C, Cousin W, Rochet N, Dani C and Peraldi P (2009) Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells 27, 703–713. [DOI] [PubMed] [Google Scholar]
- 41. Yersal O and Barutca S (2014) Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J Clin Oncol 5, 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. William D, Foulkes IES and Reis‐Filho Jorge S (2010) Triple negative breast cancer. N Engl J Med 363, 1938–1948. [DOI] [PubMed] [Google Scholar]
- 43. Howlader N, Cronin KA, Kurian AW and Andridge R (2018) Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomarkers Prev 27, 619–626. [DOI] [PubMed] [Google Scholar]


