Abstract
1q21 amplification is an important prognostic marker in multiple myeloma. In this study we identified that IL6R (the interleukin-6 membrane receptor) and ADAR1 (an RNA editing enzyme) are critical genes located within the minimally amplified 1q21 region. Loss of individual genes caused suppression to the oncogenic phenotypes, the magnitude of which was enhanced when both genes were concomitantly lost. Mechanistically, IL6R and ADAR1 collaborated to induce a hyper-activation of the oncogenic STAT3 pathway. High IL6R confers hypersensitivity to interleukin-6 binding, whereas, ADAR1 forms a constitutive feed-forward loop with STAT3 in a P150-isoform-predominant manner. In this respect, ADAR1-P150 acts as a direct transcriptional target for STAT3 and this STAT3-induced-P150 in turn directly interacts with and stabilizes the former protein, leading to a larger pool of proteins acting as oncogenic transcription factors for pro-survival genes. The importance of both IL6R and ADAR1-P150 in STAT3 signaling was further validated when concomitant knockdown of both genes impeded IL6-induced-STAT3 pathway activation. Clinical evaluation of various datasets of myeloma patients showed that low expression of either one or both genes was closely associated with a compromised STAT3 signature, confirming the involvement of IL6R and ADAR1 in the STAT3 pathway and underscoring their essential role in disease pathogenesis. In summary, our findings highlight the complexity of the STAT3 pathway in myeloma, in association with 1q21 amplification. This study therefore reveals a novel perspective on 1q21 abnormalities in myeloma and a potential therapeutic target for this cohort of high-risk patients.
Introduction
Multiple myeloma (MM) is a latent type of hematologic malignancy characterized by abnormal accumulation of plasma cells in the bone marrow. It is well established that MM cells are highly dependent on the bone marrow microenvironment enriched with growth factors for support and propagation.1–3 Among these factors, interleukin-6 (IL6), which is secreted in an autocrine and paracrine fashion, is pivotal for the survival and proliferation of MM cells: high expression of IL6 prevents drug-induced-apoptosis.1,4–6 Blood serum from MM patients contains elevated levels of IL6 and this is significantly associated with worse disease outcome.6,7
Mechanistically, IL6 confers oncogenicity through the activation of the Janus kinases (JAK)/signal transducers and activators of transcription 3 (STAT3) pathway, initiated with its binding to the transmembrane receptor IL6R.4,8,9 STAT3 is activated when its tyrosine-705 (Y705) is phosphorylated by JAK upon IL6 stimulation, leading to transcription of various pro-survival and anti-apoptotic genes such as MCL1 and BCL2.10,11 In line with this mechanism of action, primary MM tumors possess constitutive activation of the STAT3 pathway (incidence rate of 40-60%) with close correlation with poor prognosis and chemoresistance,9,12–14 suggesting that IL6/STAT3 signaling is essential for MM therapeutic targeting.
Cytogenetic abnormalities remain one of the main hallmarks of MM. 1q21 amplification [1q21(amp)], the most commonly identified chromosomal aberration, present in 36-48% of newly diagnosed cases of MM, is an important prognostic marker.15–18 Previous studies revealed that CKS1B and PMSD4 are genes that putatively drive disease aggressiveness in 1q21(amp) cases;18–20 nevertheless, biological and functional reports on these genes conferring oncogenic phenotypes are lacking. In reality, the critical genes within the minimally amplified region have yet to be fully characterized.
IL6R, the gene encoding the transmembrane receptor protein for IL6 is located on chromosome 1q21. Although it has been shown that IL6R has a role in predicting patients’ outcome,5,21,22 it remains unknown whether IL6R expression is associated with 1q21(amp) or could cause the hyperactivation of STAT3 signaling that could potentially contribute to adverse disease manifestations.
Importantly, our group and Lazzari et al. have recently reported that overexpression of ADAR1, an RNA editing enzyme, residing in the 1q21 region near the aforementioned IL6R, also has significant prognostic value in MM.23,24 The protein exists in two distinct isoforms, namely the short P110 and the long P150 isoforms. While P110 is constitutively expressed in the nucleus, P150 expression is interferon-inducible and can shuttle between the nucleus and cytoplasm due to the presence of a nuclear export signal on its extended amino terminal domain.25–27 Although interferon and IL6 are both members of the cytokine family and can be upregulated upon infection and inflammation, a possible cross-talk between IL6/STAT3 signaling and ADAR1 expression has not been thoroughly investigated. Aberrant adenosine-to-inosine editing events arising from deregulated ADAR1 been widely reported in cancers,27–31 however, the general knowledge on its upstream regulators and the mechanisms mediating its overexpression have remained elusive, not to mention in MM.
In view of both IL6R and ADAR1 being located in close proximity on 1q21 and having been independently reported to be prognostically important for MM, we sought to delineate their potential collaboration in the pathogenesis of MM and to determine how they are associated with STAT3 signaling. Here, we report that 1q21(amp) leads to elevated expression of IL6R and ADAR1. ADAR1-P150 and STAT3 form a regulatory feedback loop mediating the growth and proliferation of MM cells; the convergence of regulatory signals from both IL6R and ADAR1-P150 confers hyperactivation of STAT3 signaling, potentially driving the malicious evolution of MM. Critically, MM patients with concurrent overexpression of both proteins had a poorer prognosis than those who had no abnormality or a single one.
Methods
Patients’ samples and human multiple myeloma cell lines
Primary samples from the healthy volunteers and MM patients were collected after obtaining informed consent, according to conditions stated by the Institutional Review Board, National University Hospital. All human MM cell lines used have been previously characterized.32 Isolation of patients’ samples and culture conditions for them and human MM cell lines are described in the Online Supplementary Information. The purity of CD138+ cells of the patients’ samples was checked with anti-κ and anti-λ immunofluorescence staining (Online Supplementary Figure S5).
Gene expression profiling, genomic hybridization and biostatistics
Details of the multiple gene expression profiling and array comparative genomic hybridization datasets analyzed are provided in Online Supplementary Table S1. Analyses of IL6R/ADAR1 copy number and expression and STAT3 signature/index are described in the Online Supplementary Information. Statistical analyses between two groups in the patients’ datasets were performed with the Wilcoxon test. P-values between two groups from in vitro studies were computed with an independent t-test, assuming a normal distribution of the means. All experiments were repeated at least twice (biological replicates) and the error bars in the graphs represent the means ± standard deviations.
ADAR1-P150, IL6R and STAT3 knockdown and overexpression
We knocked down ADAR1-P150 and IL6R in human MM cell lines with specific shRNA through lentivirus infection. shRNA sequences are available in the Online Supplementary Information (Online Supplementary Table S3). pCDNA-Flag-tagged vector encoding for P150 protein was transfected into MM cells using an electroporation method (Neon, Invitrogen). siRNA against STAT3 and pIRES-STAT3 plasmids were also introduced into MM cells via Neon transfection. Based on the basal level of the proteins (Figure 1B), we picked H929 and U266 cell lines for P150 and IL6R knockdown and the OCIMY5 and KMS12BM lines for P150 over-expression.
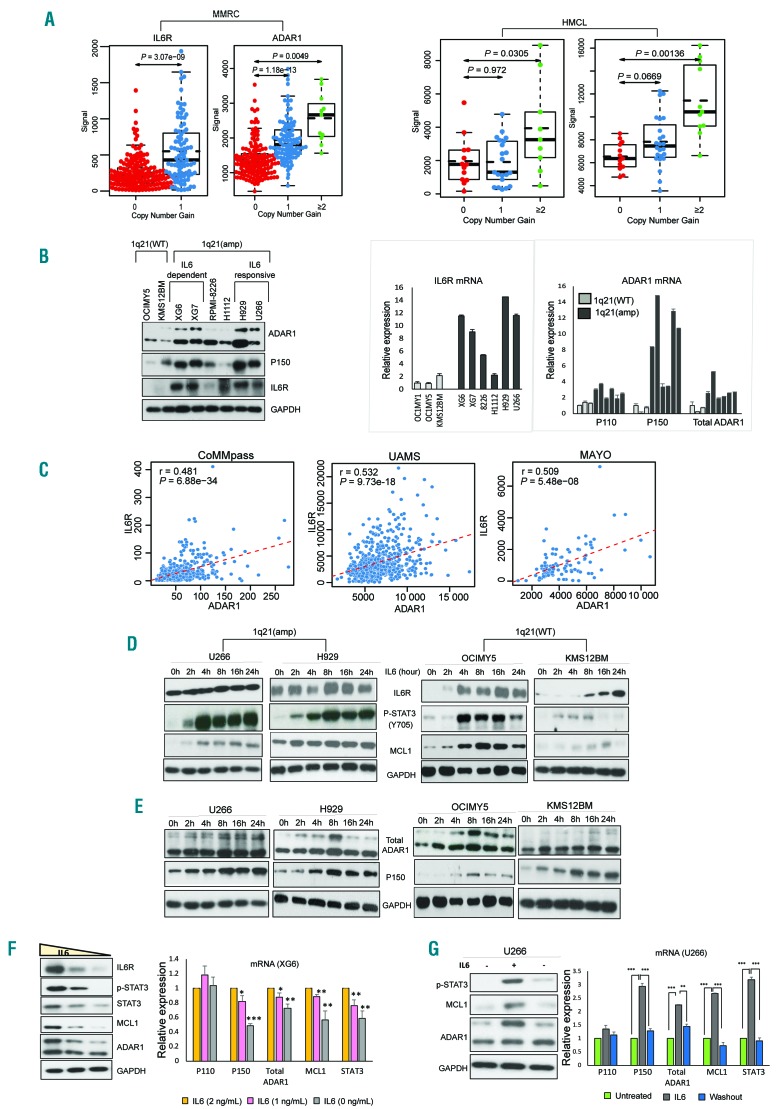
Figure 1.
IL6R and ADAR1-P150 are closely associated with 1q21 copy number and IL6 stimulation. (A) Gene expression levels of IL6R and ADAR1 in the Multiple Myeloma Research Consortium (MMRC) patients’ dataset (left) and human multiple myeloma cell lines (HMCL) (right) according to 1q21 status. 0- no copy number gain (wildtype, WT), 1- one copy gain, ≥2- two or more copy gains. IL6R and ADAR1 copy number and expression values were determined as described in the Online Supplementary Information. (B) Basal protein (left) and mRNA (right) expression of IL6R and ADAR1 in HMCL as detected by western blot and real time quantitative polymerase chain reaction (RT-qPCR). (C) Correlation of ADAR1 with IL6R in different patients’ datasets computed using the Pearson correlation test. ‘r’ is the correlation coefficient and P-values indicate whether the correlation coefficient is different from 0. (D, E) Western blot analysis of IL6/STAT3 pathway components (D) and ADAR1 expression (E) in cells with 1q21(amp) (U266 and H929) and 1q21(WT) (OCIMY5 and KMS12BM) upon IL6 stimulation (10 ng/mL) at different time points. The top band in total ADAR1 refers to the P150 isoform, while the bottom band is the P110 isoform. The single P150 band in the second blot was probed with a specific antibody that can only detect P150 but not P110 (Ab126745). (F) Western blot and RT-qPCR analysis of protein (left) and mRNA (right) expression of ADAR1 and STAT3 pathway markers in the XG6 cell line which was cultured in conditions with gradual IL6 deprivation (3 ng/mL, 2 ng/mL, 1 ng/mL) for 24 h. (G) U266 was stimulated with IL6 (10 ng/mL) for 8 h before the IL6-containing medium was washed off, and the cells were resuspended in IL6-free medium again for 24 h. ADAR1 and STAT3 pathway components were checked for expression via western blot (left) and qRT-PCR (right). *P<0.05, **P<0.01, ***P<0.0001. MMRC: Multiple Myeloma Research Consortium; HMCL: human multiple myeloma cell lines; WT: wildtype; IL6: interleukin-6.
Assays
Details of the chromatin immunoprecipitation (ChIP), luciferase, co-immunoprecipitation, immunofixation and functional assays (cell growth, colony formation and cell cycle) are available in the Online Supplementary Information. Blots and images presented are representative of multiple repeats (at least two) of the experiments.
RNA-sequencing and analysis of global RNA editing events
Total RNA from IL6-treated MM cells was isolated using the RNeasy kit (Qiagen) according to the manufacturer’s protocol. Whole transcriptome sequencing was done on an Illumina Hi-Seq-4000 platform (Supplier). Detailed information about variant calling and RNA editing analysis are provided in the Online Supplementary Information.
Results
ADAR1-P150 and IL6R are associated with 1q21 copy number and IL6 stimulation
Since IL6R and ADAR1 genes are located in close proximity (Online Supplementary Figure S1A), we postulated that 1q21(amp) could lead to concomitant increases in the expression of both. In silico analyses revealed that 1q21 status was indeed closely associated with IL6R and ADAR1 levels in diverse patients’ datasets and human MM cell lines (Figure 1A and Online Supplementary Figure S1B). Quantitative polymerase chain reaction (qPCR) and western blotting confirmed the upregulation of ADAR1 and IL6R at the mRNA and protein levels, respectively, in cell lines with 1q21(amp) compared to the levels in the wild-type (WT) line (Figure 1B).
With regards to ADAR1, interestingly, we observed that among the 1q21(amp) cells, P150 isoform expression was prominently higher in IL6-responsive and -dependent cells, while the expression of P110 isoform did not vary much (Figure 1B). Importantly, we also observed a consistent positive correlation between ADAR1 and IL6R in patients from several clinical datasets (Figure 1C) suggesting that the ADAR1-P150 isoform, just like IL6R, could be involved in IL6 signaling.
To test this hypothesis, we treated human MM cell lines with IL6 for different periods and investigated the expression profile of various markers involved in IL6-signaling. We observed that IL6 caused an induction of STAT3 signaling in MM cells regardless of the cells’ 1q21 status, albeit at different levels. While there was a gradual and time-dependent accumulation of IL6R in the 1q21(WT) cells, the 1q21(amp) cells showed only a minimal increase, largely due to its already heightened endogenous level, even in the unstimulated state (Figure 1D). As expected, IL6 led to robust upregulation of phospho-STAT3 in both cell types; however, close scrutiny of the time kinetics revealed that 1q21(amp) cells had more rapid and sustainable phospho-STAT3 induction than the 1q21(WT), concomitantly with its downstream target MCL1. This indicated that the STAT3 signaling in 1q21(amp) cells was hypersensitized, potentially in relation to the constitutively expressed IL6R.
With regards to ADAR1 expression, the P150 isoform was evidently upregulated upon IL6 stimulation, whereas, P110 seemed to show minimal changes, at both the protein (Figure 1E) and mRNA levels (Online Supplementary Figure S1C), irrespective of 1q21 status. An antibody specific for the P150 isoform was also used to confirm our observations. We further validated our observations by starving the IL6-dependent-cells lines XG6 and XG7 of IL6, which demonstrated depleted P150, and an attenuation of IL6R, concomitantly with MCL1; P110 was again mildly affected (Figure 1F and Online Supplementary Figure S1D). The specific correlation of IL6 induction with P150 was also recapitulated when the rescue of IL6 stimulation in a IL6-sensitive cell line, U266, returned P150 expression to its basal level (Figure 1G). In corroboration, in vitro IL6 starvation of CD138+ cells harvested from patients’ samples (n=2) compromised the STAT3 pathway, concomitantly with P150 downregulation (Online Supplementary Figure S1E). Collectively, these data suggest that besides potently activating the STAT3 pathway, IL6 has a role in mediating ADAR1-P150 expression in MM cells.
STAT3 transcriptionally regulates ADAR1-P150 expression
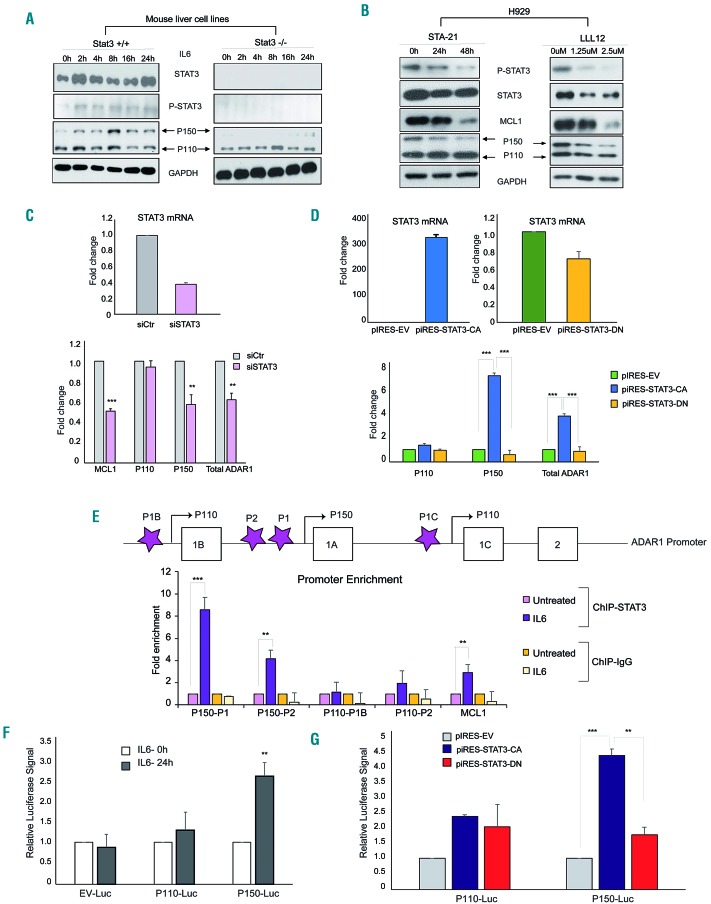
Considering that IL6 robustly activated STAT3 signaling, we next proceeded to investigate the direct role of STAT3 transcription factor on ADAR1-P150. Utilizing mouse liver cells with STAT3-WT+/+ and null−/− genotypes, we observed that the absence of STAT3 (STAT3−/−) grossly compromised the upregulation of IL6-induced-P150 (Figure 2A). Pharmacological inhibition of STAT3 in H929 cells with two specific inhibitors (STA-21 and LLL12) caused a gradual reduction of P150 expression in a time-and dosage-dependent manner (Figure 2B), concomitantly with phospho-STAT3 and MCL1. At the mRNA level, knocking down STAT3 and overexpressing the constitu-tively-activated-STAT3 (CA-Y705), but not the dominant negative mutant derivative, caused a reduction and an increase of P150 mRNA, respectively (Figure 2C, D). These data strongly suggest that STAT3 plays a role in regulating the transcription and expression of P150.
Figure 2.
STAT3 transcriptionally regulates ADAR1-P150 expression. (A) Wildtype (STAT3+/+) and STAT3 knockout (STAT3−/−) mouse liver cell lines were treated with IL6 (10 ng/mL) for different periods and the protein expression profile was checked with western blotting. (B) Western blot analysis of H929 cells treated with STAT3 inhibitors, STA-21 (10 μM for 24 h and 48 h) and LLL12 (for 24 h at 1.25 μM and 2.5 μM). (C) Top panel: real-time quantitative polymerase chain reaction (RT-qPCR) analysis of STAT3 mRNA expression after siRNA-mediated-STAT3 knockdown in H929 cells (24 h). Bottom panel: MCL1 and ADAR1 mRNA expression 24 h after STAT3 knockdown. (D) H929 transfected with pCDNA plasmid transcribing for constitutively-activated STAT3 (STAT3-CA) and its dominant negative mutant (STAT3-DN) (24 h). STAT3-CA is artificially phosphorylated at Y705, while the STAT3-DN is transcriptionally defective and has a gain-of-function of inhibiting endogenous STAT3 expression. Top panel: the transfection efficiency was checked with qRT-PCR. Bottom panel: ADAR1 mRNA expression after STAT3 overexpression. CA: constitutively activated, DN: dominant negative. (E) Top panel: depiction of primers (purple stars) designed to encompass different regions of ADAR1 promoters. Promoters 1B and 1C transcribe for the P110 isoform and promoter 1A transcribes for the P150 isoform. Bottom panel: H929 cells were stimulated with IL6 (10 ng/mL for 6 h) and STAT3 enrichment on the different regions of ADAR1 promoter was investigated with chromatin immunoprecipitation and qPCR with primers depicted in the top panel. (F, G) H929 cells were transfected with a luciferase-reporter vector encoding for either P110 or P150 promoter regions. At 24 h after transfection, the cells were treated with (F) either phosphate-buffered saline (PBS) or IL6 (10 ng/mL) for 8 h or (G) further transfected with either STAT3-CA or STAT3-DN and the cells were left to grow for another 24 h. The luciferase activity was analyzed with a Tecan plate reader. EV: empty vector.
Indeed, our hypothesis was validated when the ChIP-qPCR assay showed an enrichment of STAT3 protein on the promoter regions of P150 and its known target MCL1 (Figure 2E). There was no change in the binding enrichment of STAT3 on the IL6R promoter (Online Supplementary Figure S2B). A luciferase reporter assay provided further evidence of STAT3’s transcriptional regulation of P150 where IL6 stimulation (Figure 2F) and the overexpression of STAT3-CA (but not the dominant negative mutant) (Figure 2G) resulted in enhancement of the luciferase signal as compared to their respective controls.
P150 is important for the growth and proliferation of multiple myeloma cells
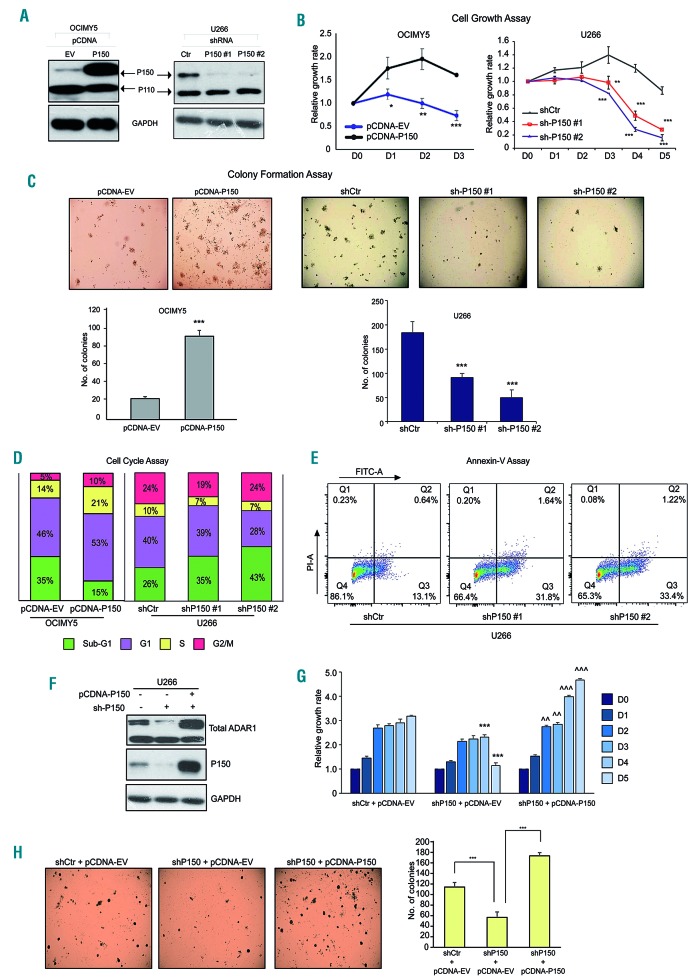
The above results demonstrated that P150 overexpression in MM can arise from the combined effects of 1q21(amp) (Figure 1) and constitutive activation of STAT3 (Figure 2). This double-pronged regulation of P150 suggests that aberrant expression of the isoform may have important biological and functional effects. To elucidate the oncogenic role of IL6/STAT3-induced-P150 in MM, we cloned P150-specific sequences into a pCDNA-overexpressing plasmid and transfected them into OCIMY5 cells, after which we performed various functional assays. Effective overexpression of P150 but not P110 (Figure 3A) allowed us to study the P150-specific oncogenic properties. P150 enrichment in the cells conferred a growth advantage (Figure 3B), increased colony-forming ability (Figure 3C) and a more proliferative cell cycle profile (cellular enrichment in S/G2 phase) (Figure 3D). Conversely, knocking down P150 in U266 cells (Figure 3A) was detrimental to the cells’ survival, with the P150-depleted cells demonstrating a slower growth rate, loss of colony-forming ability and a higher fraction of cells undergoing apoptosis (sub-G1 population enrichment in the cell cycle and increased annexin-V positivity) (Figure 3B-E). These phenotypes were replicated in P150-overexpressing KMS12BM cells (Online Supplementary Figure S3) and shP150-H929 cells (Online Supplementary Figure S4).
Figure 3.
ADAR1-P150 is important for the growth and proliferation of multiple myeloma cells. (A) Western blot analysis to check for the efficiency of P150 overexpression in OCIMY5 cells and lentivirus-mediated-shRNA knockdown of P150 in U266 cells. Cells were harvested at 48 h after manipulation for protein isolation. The shRNA sequences are available in the Online Supplementary Information. (B) Daily cell growth assay (CTG assay) depicting the growth profile of multiple myeloma cells with manipulated levels of P150. The reading for day 0 (D0) was taken 48 h after manipulation. (C) Colony-formation assay assessing the ability of the cells to form colonies upon P150 overexpression and knockdown. Top panel: representative images of the soft agar incubated over 7-14 days. Bottom panel: quantification of the colonies formed in triplicate wells of soft agar. (D) Cell cycle analysis (propidium iodide staining) of P150-overexpressed OCIMY5 cells and P150-depleted U266 cells. Cells were harvested 48 h after manipulation for analysis. The percentage on each bar represents the number of the cell population within the cell cycle phase out of the total number of cells captured and analyzed. *P<0.05 **P<0.001 ***P<0.0001 (E) Annexin-V-FITC apoptosis assay of U266 cells 48 h after P150 knockdown. (F) Western blot analysis for the expression of total ADAR1 and P150 24 h after transfection with shP150 or upon co-transfection with shP150 and pCDNA-P150 in U266 cells. (G) Daily cell growth assay of U266 cells with different levels of P150. The reading for day 0 (D0) was taken 48 h after manipulation. **P<0.05 against shCtr+pCDNA-EV, ***P<0.001 against shCtr+pCDNA-EV, ^^P<0.05 against shP150+pCDNA-EV, ^^^P<0.001 against shP150+pCDNA-EV. (H) Left panel: representative photographs of colonies formed in triplicate wells of soft agar over 14 days. Right panel: quantification of the number of colonies in U266 cells with different levels of P150. ***P<0.0001.
To further ensure that our observations thus far were P150-specific, we performed a rescue experiment by introducing the P150 isoform back into the shP150 cells (Figure 3F). The re-introduction of P150 clearly led to a rescue of growth (Figure 3G) and colony-forming ability (Figure 3H), suggesting that the growth disadvantage observed in the shP150 cells in Figure 3B-E was, in part, mediated by the lack of P150.
ADAR1-P150 in turn mediates IL6-induced STAT3 signaling
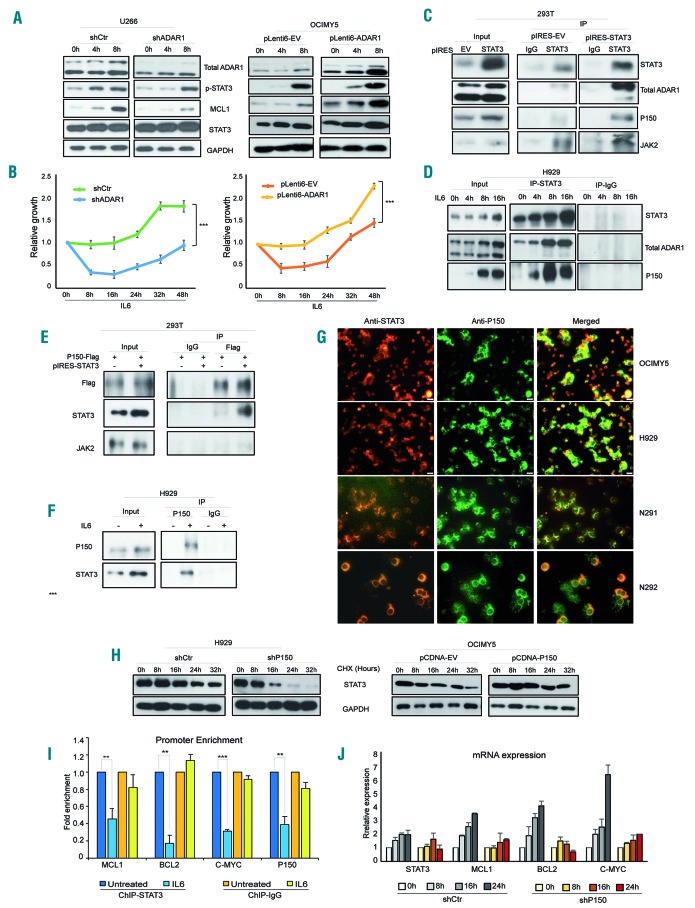
Interestingly, we also observed evidence of STAT3 activity being mediated by ADAR1. In the ADAR1 knockdown cells, IL6 delayed the activation of STAT3 signaling, as shown by slower induction of phospho-STAT3 and its direct target, MCL1. Up to 8 h were needed for the mechanism to start in U266-shADAR1, whereas, U266-shCtr already demonstrated STAT3 activation at 4 h after IL6 stimulation. This was completely reversed in the OCIMY5 cells which overexpressed ADAR1 (Figure 4A). Importantly, this protein expression profile was commensurate with the cellular phenotypes, in which the more rapid STAT3 activation (in U266-shCtr and OCIMY5-pLenti6-ADAR1) was associated with significantly accelerated IL6-induced growth (Figure 4B).
Figure 4.
STAT3-induced-ADAR1-P150 in turn mediates STAT3 signaling. (A) Stable isogenic cell lines (U266 and OCIMY5) with different levels of ADAR1 expression that have been previously established24 were treated with IL6 (10 ng/mL) at different time points before western blotting was done to analyze the protein expression profile. (B) Daily cell growth of IL6-stimulated-U266 and OCIMY5 with differential ADAR1 expression. The reading for day 0 (D0) was taken 48 h after manipulation. ***P<0.0001. (C) 293T cells were transfected with either an empty vector (pIRES-EV) or a STAT3-overexpressing vector (pIRES-STAT3) for 24 h and cells were collected for protein extraction and subsequent co-immunoprecipitation experiments. The total cell lysate was incubated with either IgG control antibody or a specific STAT3 antibody. Its potential interaction with endogenous ADAR1 was checked with western blot analysis. JAK2 served as the positive control. (D) Phospho-STAT3 was induced with IL6 (10 ng/mL) in H929 cells which were harvested to determine their total protein level. Protein was pulled down with either STAT3 or IgG antibodies and western blot was used to check for the potential interaction. (E) 293T cells were transfected with either P150-Flag only or with both P150-Flag and STAT3 and reciprocal immunoprecipitation was done with IgG and Flag-antibody. The interaction between P150-Flag and STAT3 was checked with western blot analysis. (F) H929 cells were stimulated with IL6 (10 ng/mL) for 16 h and endogenous P150 was pulled down with its specific antibody or negative control IgG, for reciprocal immunoprecipitation. Its interaction with STAT3 was assessed with western blot. (G) Cytospin slides consisting of cells from multiple myeloma cell lines (OCIMY5 and H929) and patients’ samples (N291 and N292) were immunos-tained with STAT3- and P150-specific antibodies. Immunofluoresence signals for both proteins were analyzed under a fluorescence microscope. Total magnification used was 400×. Red (Alexa-Fluor-555): STAT3; green (Alexa-Flour-488): P150; yellow: co-localization. (H) A cycloheximide (CHX) chase assay was performed to identify the stability of the STAT3 protein. P150-knockdown U266 cells and P150-overexpressing OCIMY5 cells with their respective control cells were treated with the protein synthesis inhibitor CHX at 50 μg/mL to elucidate the time kinetics for STAT3 protein degradation. The intracellular content of STAT3 protein was analyzed by western blotting. (I) Fold enrichment of STAT3 occupancy on its target gene promoters before and after P150 knockdown in U266 cells, as analyzed with chromatin immunoprecipitation quantitative polymerase chain reaction analysis (qPCR). **P<0.05, ***P<0.001 (J) Reat-time qPCR analysis for the mRNA induction of STAT3 target genes in response to IL6 (10 ng/mL) treatment at different time points in control (shCtr) and P150-knockdown (shP150) U266 cells.
To elucidate the potential mechanism, we first checked for a possible ADAR1-STAT3 interaction. Pulling down STAT3 from STAT3-overexpressing 293T cells showed enrichment of endogenous P150 and its known interacting partner, JAK2, in the immunoprecipitated fraction but not in the IgG control fraction. On the other hand, P110 seemed to form only a loose interaction with STAT3 (Figure 4C). Consistently, in IL6-responsive H929 cells, accumulation of IL6-induced phospho-STAT3 led to a gradual increase of interaction with endogenous ADAR1, particularly P150 (Figure 4D), indicating that it was the main interacting isoform. To confirm this observation, we performed reciprocal immunoprecipitation. Co-transfection of P150-Flag and STAT3 plasmids into 293T cells and pulling down only the P150 isoform also revealed efficient co-precipitation with STAT3 protein (Figure 4E). There was no interaction of P150 with JAK2. Similarly, P150 could be seen co-precipitating with endogenous STAT3 in the IL6-induced fraction of H929 cells (Figure 4F).
Subsequently, we checked for the localization of ADAR1 and STAT3 in human MM cell lines (OCIMY5 and H929) and patients’ samples (N291 and N292), and found that STAT3 was localized in both the nucleus and cytoplasm, whereas, P150 was predominant in the cytoplasm. Merged images of individual proteins clearly showed co-localization of STAT3 and P150 in the cytoplasm (Figure 4G). It is therefore plausible that this cytoplasmic co-localization could have rendered the environment conducive for their physical interaction.
Next, we investigated how this interaction could enhance STAT3 activity. A cycloheximide chase assay revealed that P150 could promote the stability of STAT3 protein. Knocking down P150 enhanced STAT3 degradation (Figure 4H). At 8 h, STAT3 already showed signs of degradation and was almost completely degraded at 24 h after cycloheximide treatment in the shADAR1 cells, in contrast to the shCtr cells which required up to 16 h for a mild cycloheximide effect to be visible. Similarly, the P150-overexpressed cells also took longer (up to 24 h) to show signs of STAT3 protein degradation.
These data suggest that P150 could mediate the activity of STAT3 signaling via a direct physical interaction, which leads to stabilization of the latter, thus, more is shuttled into the nucleus to transcribe various pro-oncogenic genes. This hypothesis was validated when the loss of P150 led to compromised STAT3 binding to its target gene promoters (Figure 4I) and sub-optimal induction of its target genes (Figure 4J).
Combined IL6R and P150 suppression leads to reduced IL6-induced oncogenicity
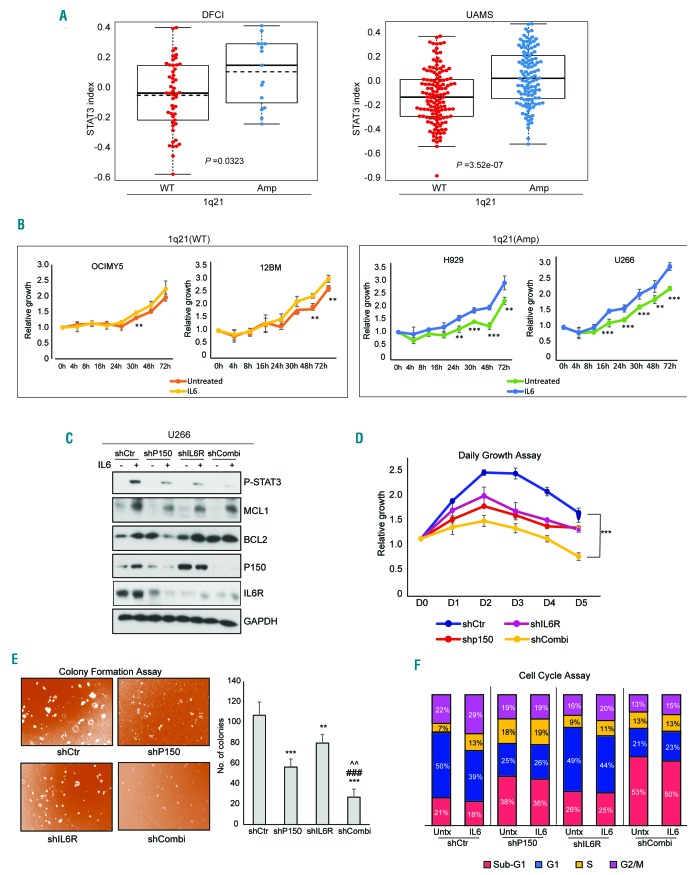
The more rapid and sustainable STAT3 signaling in the 1q21(amp) cells (which express IL6R constitutively) (Figure 1D), coupled with the STAT3- P150 feedback regulatory loop (Figure 4) led us to hypothesize that the concomitant gain of IL6R and ADAR1 may lead to hyperactivation of STAT3 signaling, conferring a more malignant state to 1q21(amp) cells. Indeed, the observation of 1q21(amp) patients having significantly higher STAT3 indices (STAT3 pathway activation) further supports the basis of our hypothesis (Figure 5A).
Figure 5.
Combined IL6R and P150 suppression led to reduced IL6-induced oncogenicity. (A) The association between 1q21 status and STAT3 signature in Dana Farber Cancer Institute (DFCI) and University for Arkansas Medical School (UAMS) patient’s datasets. WT- wildtype, Amp- two or more copies. (B) CTG assay assessing the growth rate of multiple myeloma cell lines with different 1q21 status over a 72 h time course. **P<0.05, ***P<0.001 (C) At 48 h after lentivirus infection, U266 cells with shRNA-mediated P150 and/or IL6R knockdown were treated with IL6 (10 ng/mL) for 8 h and then the STAT3 pathway protein expression profile was analyzed by western blotting. (D-F) Daily growth assays (D), colony formation assays (E) and cell cycle assays (F) were done 48 h after lentivirus infection of shRNA to elucidate the phenotypes associated with loss of either one or both P150 and IL6R. For the cell cycle assays, the cells were stimulated with IL6 (10 ng/mL) for 24 h (to induce STAT3 signaling) before being fixed for fluorescence activated cell sorting analysis. This was to study the direct effects of loss of expression of these genes on the STAT3-mediated cell cycle profile. **P<0.05 against shCtr, ***P<0.001 against shCtr, ###P<0.001 against shP150, ^^P<0.05 against shIL6R.
We further investigated this by first comparing the growth profile of 1q21(WT) and 1q21(amp) upon IL6 stimulation. Not surprisingly, exogenous IL6 conferred more active growth to all tested cell lines compared to untreated ones (Figure 5B). Importantly, however, 1q21(amp) cells demonstrated more pronounced growth than the WT cells. The growth rate of the IL6-stimulated 1q21(amp) cells already surpassed that of the unstimulated ones as soon as 4 h after IL6 induction. In contrast, the 1q21(WT) cells required up to 24 h after IL6 stimulation to display a differential growth phenotype. This implies that the 1q21(amp) cells had a sensitized IL6/STAT3 pathway and that there was a possibility that the pathway-related genes within 1q21 could be driving this phenotype.
To elucidate the potential importance of IL6R and P150, we performed single and co-depletion of IL6R and P150 and assessed the functional consequences for STAT3 activity and MM cell growth. Suppression of IL6R or P150 alone obliterated the IL6-induced STAT3 pathway, manifested by the weaker induction of phospho-STAT3 and its downstream factors (Figure 5C). Inhibition of this pathway was detrimental to MM cells, which experienced growth retardation (Figure 5D), impeded colony formation (Figure 5E) and halted IL6-induced-cell cycle progression (as compared to IL6-treated shCtr cells which progressed through the S/G2 phase) (Figure 5F), suggesting that both factors are individually important for IL6-induced STAT3 activation. Notably, the combined loss of IL6R and P150 resulted in more deleterious cellular phenotypes as compared to the loss of just one respective protein, consistent with the more prominent attenuation of IL6-induced STAT3 signaling (Figure 5C). A similar trend was also observed in H929 cells (Online Supplementary Figure S6).
IL6R, ADAR1 and STAT3 demonstrate good clinical correlation
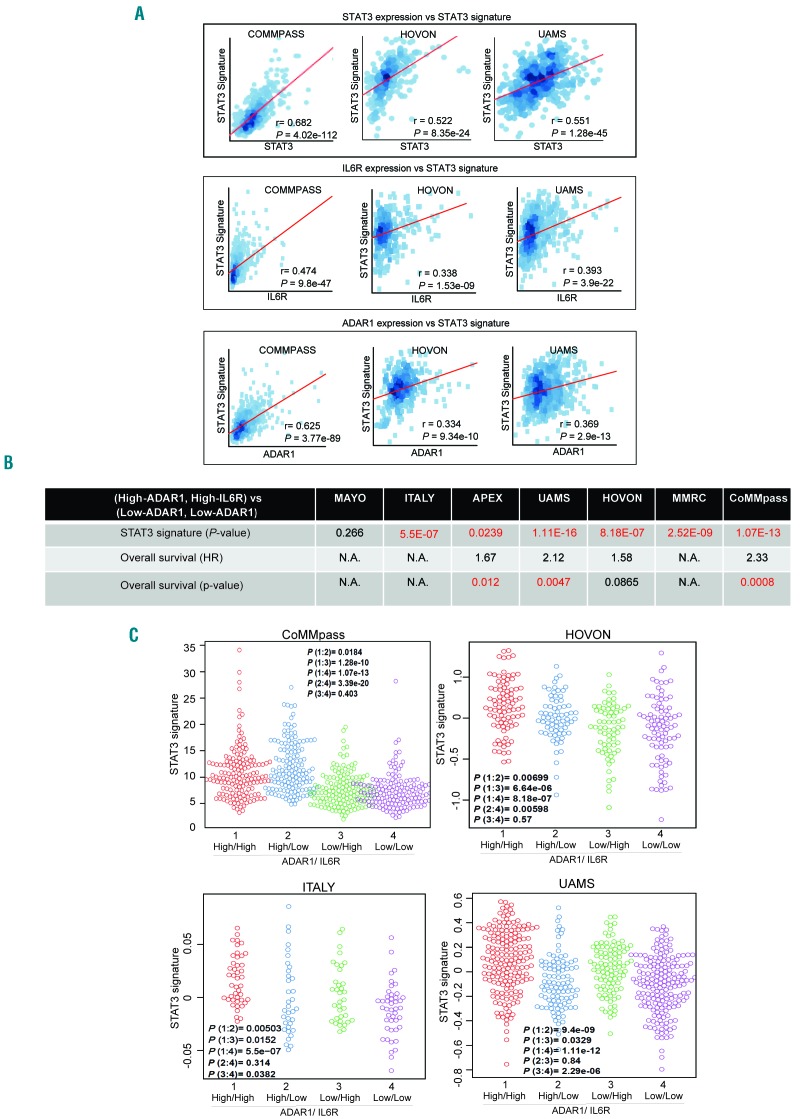
Having shown the biological association of both IL6R and ADAR1-P150 with STAT3, we then investigated the clinical correlations by analyzing publicly available patients’ datasets. Positive correlations were consistently observed between the STAT3 signatures and the expression of STAT3, IL6R and ADAR1 in various patients’ datasets (Figure 6A).
Figure 6.
IL6R, ADAR1 and STAT3 demonstrated good clinical correlation. (A) Pearson correlation between the STAT3 signature and STAT3 (top panel), IL6R (central panel) and ADAR1 (bottom panel) expression in different patients’ datasets. r is the correlation coefficient and P-values indicate whether the correlation coefficient is different from 0. (B) Comparison of the STAT3 signature and overall survival between patients with high expression of both ADAR1 and IL6R and patients with low expression of both genes. P-values highlighted in red indicate statistical significance (P<0.05). (High-ADAR1, High-IL6R indicates high expression of both genes; Low-ADAR1 and Low-IL6R indicates low expression of both genes. HR: hazard ratio. N.A.: information not available in that particular dataset, therefore analysis cannot be done. (C) Comparison of the STAT3 signature in patients with different levels of ADAR1 and IL6R expression. High/high: the expression of both genes is high. High/low: ADAR1 expression is high, IL6R expression is low. Low/high: ADAR1 expression is low, IL6R expression is high, Low/low: the expression of both genes is low. P(1:2) represents the P-value between group 1 (high/high) and group 2 (high/low). Similarly, the rest of the comparisons are done according to the group numbers. The STAT3 signature reflects the activity of the signaling pathway.
To demonstrate the importance of the combined gain of both genes within 1q21, we computed the correlation between STAT3 signature enrichment and overall survival in patients with differential expression of these genes. Six out of seven datasets revealed a significantly enriched STAT3 signature in the high-ADAR1+high-IL6R as compared to the low-ADAR1+low-IL6R groups of patients (Figure 6B). We also found that lack of expression of either gene resulted in a lower STAT3 signature expression and, more importantly, loss of both genes caused an even more significant suppression of the signature (Figure 6C), signifying that both factors are influential players in the STAT3 pathway. Importantly, this differential STAT3 activity in different groups was also closely associated with the patients’ prognosis. Patients with high expression of both genes had a worse overall survival than those with low expression of both genes (Figure 6B and Online Supplementary Figure S7) further supporting our hypothesis that the concomitant gain of IL6R and ADAR1 is critical in driving poor disease outcome in the 1q21(amp) cases.
Potential therapeutic implications of the IL6R-P150-STAT3 interaction
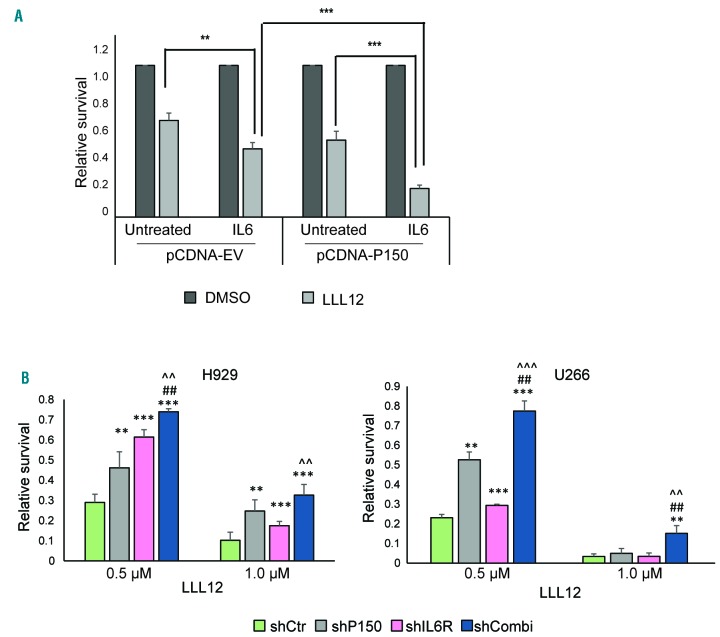
The STAT3 inhibitor, LLL12, was reported to be effective in MM cells in vitro,33 but the study did not highlight its association with a differential degree of STAT3 activity. We postulated that the prolonged and amplified STAT3 signals from P150-induced STAT3 stabilization could have an impact on cellular responsiveness to LLL12. Here, we demonstrated that cells with ectopically expressed P150 were indeed more sensitive to LLL12 treatment and that the cellular inhibition effect was further enhanced under IL6 stimulation (Figure 7A). This suggests that the lengthened STAT3 signaling event from IL6 induction and stabilization of the protein by the downstream P150 provides an ample target for the drug, rendering the cells hypersensitive to LLL12. To determine whether this principle holds true in 1q21(amp) cells, which have another hit of STAT3 activation through IL6R overexpression, we knocked down P150, IL6R or both and compared the cells’ responsiveness to LLL12. Depletion of either gene did indeed compromise the sensitivity of the cells (under persistent IL6 stimulation) to the drug and loss of both genes caused the cells of both lines tested to be even more resistant (Figure 7B).
Figure 7.
Potential therapeutic implications of the IL6R-P150-STAT3 interaction. (A) P150 was overexpressed in OCIMY5 cells and 48 h later the cells were stimulated with IL6 (10 ng/mL) for 8 h before treatment with LLL12 (1.0 μM) for 48 h. Relative survival was quantified with the CTG assay and was normalized to that of the dimethylsulfoxide (DMSO)-control. **P<0.05, ***P<0.001. (B) H929 and U266 cells were infected with lentivirus shRNA against either P150 or IL6R or both and these cells with different levels of P150 and IL6R expression at 48 h after infection were treated with 0.5 μM and 1.0 μM LLL12 for another 48 h. Cell survival was assessed with the CTG assay. Relative survival was normalized to the survival of the DMSO control. **P<0.05 against shCtr, ***P<0.001 against shCtr, ##P<0.05 against shP150, ^^P<0.05 aga inst shIL6R, ^^^P<0.001 against shIL6R.
Discussion
The growth privilege of MM cells is widely attributed to their concerted interactions with the bone marrow microenvironment and the growth factors enriched within the bone marrow are, therefore, deemed indispensable for MM survival.3,5 Studies over the years have provided solid evidence that among these growth factors, IL6 is one of the key cytokines driving the growth and proliferation of MM cells, and its oversecretion could lead to drug resistance.1,4,13 IL6 induces STAT3 activation and this signaling pathway has been a long-standing oncogenic player.1,33 Various therapies targeting this pathway have been developed and although in vitro and in vivo laboratory testing in MM had shown some potential,34–36 the outcome of clinical trials on anti-IL6 antibodies was less meaningful.37,38 The stumbling block was probably the lack of complete biological understanding of the IL6/STAT3 pathway itself. Our current work focuses on further dissecting this pathway and here we report novel IL6-induced oncogenicity in myeloma. Our data unveil a close interplay between IL6R, ADAR1 and STAT3 proteins, which could contribute to the hyper-activation of STAT3 signaling, consequently causing a more proliferative cellular profile in MM.
To date, 1q21(amp) has remained a high-risk prognostic factor in MM. This poses a serious hurdle in myeloma management since a high percentage of newly diagnosed patients present with this abnormality and they are reported to be resistant to standard and novel therapies.16,17,39,40 Although the clinical significance of CKS1B, the most commonly reported candidate driver gene,18,39 has already been established, we believe that other genes within the amplified region may also be of pathogenic importance. Here, we impart novel information about the pathogenesis of 1q21(amp) and provide compelling evidence that IL6R and ADAR1 on chromosome 1q21 are critical genes for myeloma pathogenesis. Concomitant gain of their genomic loci conferred growth and proliferation privilege to MM cells, working through hyperactivation of the STAT3 pathway. With regards to ADAR1, we saw an isoform-predominant phenomenon in which the P150 isoform was found to be intertwined in the STAT3 pathway by being its downstream transcriptional target and its upstream mediator. This feed-forward regulatory loop causes an augmented oncogenic effect, with both STAT3-induced P150 expression and P150-induced STAT3 protein stabilization acting in concert to promote manifestation of an aggressive disease. Coupled with the overexpression of IL6R that drives the hypersensitivity of the STAT3 pathway, it is plausible that these multi-oncogenic hits could contribute to the poor prognosis in the 1q21(amp) patients.
Although IL6R has been documented to be a predictor of poor outcome in myeloma patients,5,21,22,41 there have not been proper biological studies associating it to myeloma-genicity. Our notion of high IL6R acting as an ever-ready receptor for the IL6 ligand was proven when 1q21(amp) cells which constitutively express endogenous IL6R showed hastened and sustainable STAT3 pathway activation and enhanced IL6-induced-growth. This suggests that high levels of IL6R hypersensitize the cells to IL6 binding. Consequently, activated STAT3 leads to the transcription of not only its classical target gene, thereby mediating growth and proliferation, but also ADAR1-P150, which also has oncogenic roles, thereby augmenting proliferative phenotypes.
Since ADAR1 is a well-known RNA editing enzyme, we also investigated whether our observations were related to the canonical function of this enzyme. IL6 treatment of MM cells for up to 24 h did not lead to much change in the total number of global editing events at the whole transcriptome level (Online Supplementary Figure S8A). A more complex picture emerged at the gene-specific level, whereby, genes of hyper-editing, hypo-editing and no-change of editing frequency appeared without distinct functional segregation among them (Online Supplementary Figure S8B). Analysis on the CoMMpass dataset revealed that the STAT3 signature, an indicator of collective STAT3 activity, was poorly correlated with the number of global editing events (r=0.115) (Online Supplementary Figure S8C). While we could not definitively conclude that the RNA editing’s role of the IL6-induced P150 was not involved in conferring growth benefits to the cells, the fact that the localization of P150 was predominantly cytoplasmic even after IL6 stimulation (Online Supplementary Figure S8D) suggests that RNA editing is not likely to be the prevailing mechanism, as this process takes place in the nucleus where double-stranded mRNA are abundant.
Instead, we opine that the oncogenic effects are likely driven by P150-induced hyperactivation of the STAT3 pathway. The close association between ADAR1 and STAT3 signature strongly indicates that high ADAR1 expression contributes to STAT3 pathway activation, culminating in a conducive growth-promoting environment within the bone marrow. ADAR1 has been reported before to have an editing-independent role,42–45 albeit not an extensive one, thus, our finding adds to the important pool of knowledge on the novel non-canonical function of ADAR1, specifically of its P150 isoform.
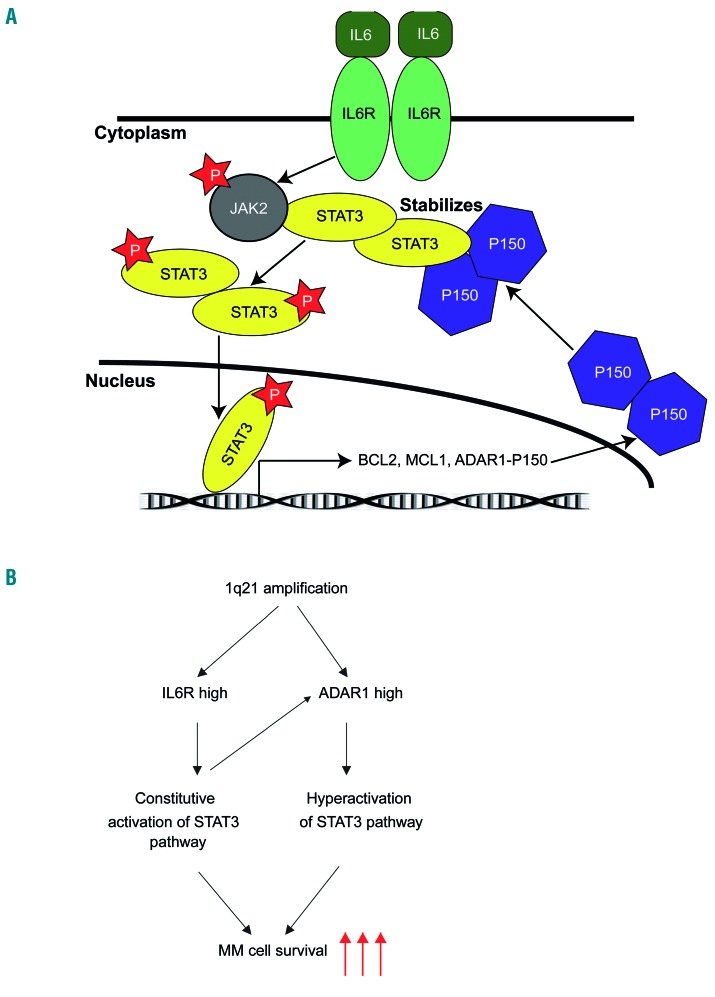
The fact that we also found a close correlation between IL6R and ADAR1, and the STAT3 signature, as well as between these and patients’ survival, gives the strong impression that there is a close interplay between these factors which drives disease prognosis. We summarize our findings in a schematic diagram illustrating how 1q21(amp) can result in the overexpression of IL6R and ADAR1, eventually leading to more intensified oncogenic effects from the aberrantly amplified STAT3 activity (Figure 8A, B).
Figure 8.
Signaling model of the IL6R-ADAR1-STAT3 interplay in multiple myeloma. (A) High levels of IL6 in multiple myeloma (MM) cells within the bone marrow microenvironment leads to persistent activation of the STAT3 pathway. Activated STAT3 (phosphorylated at Y705 by JAK) will drive the transcription of its target genes, including ADAR1-P150. P150 shuttles in and out of nucleus. Cytoplasmic P150 in turn regulates STAT3 activity in a positive feedback loop by forming a physical interaction with STAT3, resulting in protein stabilization of the latter. More STAT3 can therefore be translocated into the nucleus for its transcriptional function, leading to a cascade of oncogenicity deriving from the actions of its downstream targets. (B) 1q21 amplification leads to high expression of both IL6R and ADAR1. Cells with high expression of IL6R are highly sensitized to IL6 stimulation, leading to a constitutive activation of the STAT3 pathway. High expression of ADAR1-P150 mediates STAT3 activity in a feedback loop, as described in (A), causing hyperactivation of the STAT3 pathway. Convergence of mechanisms from concomitant high expression of both IL6R and ADAR1 culminates in enhanced survival of MM cell survival, potentially contributing to the poor disease prognosis in patients with 1q21 amplification.
In conclusion, our current work highlights the complexity of the STAT3 pathway in MM and for the very first time we report its important association with the 1q21(amp) phenotype. It should, however, be noted that our data do not demonstrate a direct causal effect of 1q21 abnormalities, but rather a close relationship, which we showed through our detailed mechanistic study on human cell lines, supported by robust clinical data. Our data suggesting that 1q21(amp) cells are more responsive to the STAT3 inhibitor (LLL12) indicates that these cells have a close association and dependency on STAT3 pathway activity. This study further highlights the potential complex biological impact imparted by 1q21(amp), of which oncogenic phenotypes are conferred by potentially more than just CSK1B. It is plausible that a cascade of survival signals, produced by the gain of other critical oncogenes within this region, and in our case, IL6R and ADAR1-P150, led to the alteration of cytokine-mediated signaling and the intrinsic cellular phenotypes. Our results could explain why targeting IL6 may not be enough BUT targeting STAT3 activity may ACTUALLY be more beneficial. This study provides a new perspective on the STAT3 pathway in MM and the potential therapeutic means for high-risk 1q21(amp) patients.
Acknowledgment
WJC is supported by NMRC Singapore Translational Research (STaR) Investigatorship. This research is partly supported by the National Research Foundation Singapore and the Singapore Ministry of Education under the Research Centers of Excellence initiative as well as the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s Tier 3 grants, grant number MOE2014-T3-1-006. The computational work for this article was partially performed with resources of the National Supercomputing Center, Singapore (https://www.nscc.sg). We thank Dr Chen Leilei for providing experimental advice and vital reagents. We would also like to thank Dr Zhou Jianbiao for kindly contributing the STAT3 plasmids for our experiments.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/5/1391
References
- 1.Matthes T, Manfroi B, Huard B. Revisiting IL-6 antagonism in multiple myeloma. Crit Rev Oncol hematol. 2016;105:1–4. [DOI] [PubMed] [Google Scholar]
- 2.Lee C, Oh J-I, Park J, et al. TNFα mediated IL-6 secretion is regulated by JAK/STAT pathway but not by MEK phosphorylation and AKT phosphorylation in U266 multiple myeloma cells. Biomed Res Int. 2013; 2013:580135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mondello P, Cuzzocrea S, Navarra M, Mian M. Bone marrow micro-environment is a crucial player for myelomagenesis and disease progression. Oncotarget. 2017;8(12): 20394–20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gado K, Domjan G, Hegyesi H, Falus A. Role of interleukin-6 in the pathogenesis of multiple myeloma. Cell Biol Int. 2000;24(4):195–209. [DOI] [PubMed] [Google Scholar]
- 5.Barille S, Bataille R, Amiot M. The role of interleukin-6 and interleukin-6/interleukin-6 receptor-alpha complex in the pathogenesis of multiple myeloma. Eur Cytokine Netw. 2000;11(4):546–551. [PubMed] [Google Scholar]
- 6.Bataille R, Jourdan M, Zhang XG, Klein B. Serum levels of interleukin 6, a potent myeloma cell growth factor, as a reflect of disease severity in plasma cell dyscrasias. J Clin Invest. 1989;84(6):2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelliniemi TT, Irjala K, Mattila K, et al. Immunoreactive interleukin-6 and acute phase proteins as prognostic factors in multiple myeloma. Finnish Leukemia Group. Blood. 1995;85(3):765–771. [PubMed] [Google Scholar]
- 8.Song Z, Ren D, Xu X, Wang Y. Molecular cross-talk of IL-6 in tumors and new progress in combined therapy. Thorac Cancer. 2018;9(6):669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sikka S, Shanmugam MK, Kannaiyan R, et al. Suppression of essential pro-inflammatory signaling pathways by natural agents for the therapy of multiple myeloma. Phytochem Rev. 2014;13(1):79–106. [Google Scholar]
- 10.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel). 2014;6(2):926–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. [DOI] [PubMed] [Google Scholar]
- 12.Jung YY, Lee JH, Nam D, et al. Anti-myeloma effects of Icariin are mediated through the tttenuation of JAK/STAT3-dependent signaling cascade. Front Pharmacol. 2018; 9:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlett-Falcone R, Landowski TH, Oshiro MM, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10(1):105–115. [DOI] [PubMed] [Google Scholar]
- 14.Quintanilla-Martinez L, Kremer M, Specht K, et al. Analysis of signal transducer and activator of transcription 3 (Stat 3) pathway in multiple myeloma: Stat 3 activation and cyclin D1 dysregulation are mutually exclusive events. Am J Pathol. 2003;162(5):1449–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An G, Acharya C, Deng S, et al. Cytogenetic and clinical marks for defining high-risk myeloma in the context of bortezomib treatment. Exp Hematol. 2015;43(3):168–176. [DOI] [PubMed] [Google Scholar]
- 16.An G, Xu Y, Shi L, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014;99(2):353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W, Guo R, Qu X, et al. The amplification of 1q21 is an adverse prognostic factor in patients with multiple myeloma in a Chinese population. Onco Targets Ther. 2016;9:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonseca R, Van Wier SA, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20(11):2034–2040. [DOI] [PubMed] [Google Scholar]
- 19.Chang H, Qi X, Jiang A, Xu W, Young T, Reece D. 1p21 deletions are strongly associated with 1q21 gains and are an independent adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Bone Marrow Transplant. 2010;45(1):117–121. [DOI] [PubMed] [Google Scholar]
- 20.Shaughnessy JD, Jr, Qu P, Usmani S, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pulkki K, Pelliniemi TT, Rajamaki A, Tienhaara A, Laakso M, Lahtinen R. Soluble interleukin-6 receptor as a prognostic factor in multiple myeloma. Finnish Leukaemia Group. Br J Haematol. 1996;92(2):370–374. [DOI] [PubMed] [Google Scholar]
- 22.Ohtani K, Ninomiya H, Hasegawa Y, et al. Clinical significance of elevated soluble interleukin-6 receptor levels in the sera of patients with plasma cell dyscrasias. Br J Haematol. 1995;91(1):116–120. [DOI] [PubMed] [Google Scholar]
- 23.Lazzari E, Mondala PK, Santos ND, et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat Commun. 2017;8(1):1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teoh PJ, An O, Chung TH, et al. Aberrant hyperediting of myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood. 2018;132(12):1304–1317. [DOI] [PubMed] [Google Scholar]
- 25.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Ann Rev Biochem. 2010;79(1):321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song C, Sakurai M, Shiromoto Y, Nishikura K. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases. Genes (Basel). 2016;7(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2013;110(3):1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Li Y, Lin CH, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19(2):209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan TH, Qamra A, Tan KT, et al. ADAR-Mediated RNA editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151(4):637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz N, Levanon EY, Amariglio N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17(11):1586–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fumagalli D, Gacquer D, Rothé F, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13(2):277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teoh PJ, Chung TH, Sebastian S, et al. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia. 2014;14(10):102. [DOI] [PubMed] [Google Scholar]
- 33.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Benson DM, DeAngelis S, et al. A small molecule, LLL12 inhibits constitutive STAT3 and IL-6-induced STAT3 signaling and exhibits potent growth suppressive activity in human multiple myeloma cells. Int J Cancer. 2012;130(6):1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scuto A, Krejci P, Popplewell L, et al. The novel JAK inhibitor AZD1480 blocks STAT3 and FGFR3 signaling, resulting in suppression of human myeloma cell growth and survival. Leukemia. 2011;25(3):538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bataille R, Barlogie B, Lu ZY, et al. Biologic effects of anti-interleukin-6 murine monoclonal antibody in advanced multiple myeloma. Blood. 1995;86(2):685–691. [PubMed] [Google Scholar]
- 37.Voorhees PM, Manges RF, Sonneveld P, et al. A phase 2 multicentre study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with relapsed or refractory multiple myeloma. Br J Haematol. 2013;161(3):357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.San-Miguel J, Bladé J, Shpilberg O, et al. Phase 2 randomized study of bortezomib-melphalan-prednisone with or without siltuximab (anti–IL-6) in multiple myeloma. Blood. 2014;123(26):4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemec P, Zemanova Z, Greslikova H, et al. Gain of 1q21 is an unfavorable genetic prognostic factor for multiple myeloma patients treated with high-dose chemotherapy. Biology Blood Marrow Transplant. 2010;16(4):548–554. [DOI] [PubMed] [Google Scholar]
- 40.Smetana J, Berankova K, Zaoralova R, et al. Gain(1)(q21) is an unfavorable genetic prognostic factor for patients with relapsed multiple myeloma treated with thalidomide but not for those treated with bortezomib. Clin Lymphoma Myeloma Leuk. 2013;13(2):123–130. [DOI] [PubMed] [Google Scholar]
- 41.Kim SY, Min HJ, Park HK, et al. Increased copy number of the interleukin-6 receptor gene is associated with adverse survival in multiple myeloma patients treated with autologous stem cell transplantation. Biol Blood Marrow Transplant. 2011;17(6):810–820. [DOI] [PubMed] [Google Scholar]
- 42.Nemlich Y, Baruch EN, Besser MJ, et al. ADAR1-mediated regulation of melanoma invasion. Nat Commun. 2018;9(1):2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi L, Song Y, Chan Tim Hon M, et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017;45(18):10436–10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie Y, Ding L, Kao PN, Braun R, Yang J-H. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25(16): 6956–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013;5(3):849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]