Abstract
Autophagy is a genetically regulated process of adaptation to metabolic stress and was recently shown to be involved in the treatment response of chronic myeloid leukemia (CML). However, in vivo data are limited and the molecular mechanism of autophagy regulators in the process of leukemogenesis is not completely understood. Here we show that Beclin-1 knockdown, but not Atg5 deletion in a murine CML model leads to a reduced leukemic burden and results in a significantly prolonged median survival of targeted mice. Further analyses of murine cell lines and primary patient material indicate that active BCR-ABL directly interacts with BECLIN-1 and phosphorylates its tyrosine residues 233 and 352, resulting in autophagy suppression. By using phosphorylation-deficient and phosphorylation-mimic mutants, we identify BCR-ABL induced BECLIN-1 phosphorylation as a crucial mechanism for BECLIN-1 complex formation: interaction analyses exhibit diminished binding of the positive autophagy regulators UVRAG, VPS15, ATG14 and VPS34 and enhanced binding of the negative regulator Rubicon to BCR-ABL-phosphorylated BECLIN-1. Taken together, our findings show interaction of BCR-ABL and BECLIN-1 thereby highlighting the importance of BECLIN-1-mediated autophagy in BCR-ABL+ cells.
Introduction
The BCR-ABL fusion kinase has been identified in more than 95% of chronic myeloid leukemia (CML) and 20% of acute lymphoblastic leukemia (ALL) cases.1,2 Oncogenic BCR-ABL activates several aberrant kinase-dependent pathways including anti-apoptosis, proliferation and differentiation,3,4 leading to the development of several successful tyrosine kinase inhibitors (TKI) in BCR-ABL+ leukemia treatment.5,6 However, there are still unsolved issues in TKI-based therapies for patients with CML: Suppression of the disease relies in most patients on continuous and lifelong TKI therapy7,8 and disease relapse occurs due to emerged TKI resistance.9–11 Thus, identification of additional important mediators could significantly improve CML therapy.
Autophagy is an evolutionarily conserved mechanism for the degradation of cytoplasmic components including organelles and proteins and plays an important role in cellular homeostasis. Because of its potential role in metabolism and cell survival, altered autophagy processes are critical for cancer cell fate. Several reports indicate that autophagy may be a promising target pathway in BCR-ABL+ leukemia treatment.12–18 However, the distinct role of autophagy in the process of leukemogenesis is not completely understood and crucial autophagy mediators have not been evaluated in in vivo leukemogenesis mouse models.
BECLIN-1, a master regulator of autophagy, is essential for the formation of the autophagosome and autolysosome as a part of the Rubicon, VPS15, VPS34, ATG14, UVRAG and BECLIN-1 complex.19–23 An in vitro study has discovered that a treatment strategy combining TKI and inhibitors of BECLIN-1-mediated autophagy may be beneficial for BCR-ABL+ CML therapy,16 but in vivo data are missing and the molecular mechanisms underlying this effect remain unclear.
Methods
GST-pulldown assay, immunoprecipitation and Western blotting
All Beclin-1 fragments were cloned into PGEX-4T2 vector, which were confirmed by Sanger sequencing. Those constructs were transformed into Bl21 competent cells, and a single clone was picked for culture in LB medium at 37°C with vigorous shaking. IPTG was added when the OD600 of the bacterial suspension reached 0.6. After an additional 2 hours (h) of incubation, bacterial cells were harvested, lysed using lysozyme and sonification and incubated for 3 h with glutathione-agarose beads in NETN buffer (0.5% NP40, 20 mM Tris/HCl, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Benzamidin, protease inhibitor cocktail [Roche]) at 4°C. The beads were then incubated with K562 cell lysates over night at 4°C. Immunoprecipitation and Western blotting were performed as described previously.24–26 Briefly, immunoprecipitation was performed by adding IP lysis buffer (40 mM HEPES, 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 50 mM NaF, 0.3% CHAPS, 1 mM sodium orthovanadate, 1 mM glycerolphosphate, protease inhibitor cocktail) to the cells for 1 h on ice. Pre-clearing of the lysates was performed using protein A or G agarose beads, followed by incubation with anti flag beads (Sigma) or antibody plus protein A or G beads (GE healthcare) overnight at 4°C. Protein extraction for Western blotting was performed using protein lysis buffer (10 mM Tris/HCl, 130 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 20 mM Na2HPO4/NaH2PO4, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 20 mM NaF, 1 mM glycerole 2-phosphate, protease inhibitor cocktail).
In vitro kinase assay
In vitro kinase assay was performed as described previously.27 Briefly, recombinant active ABL (ProQinase GmbH) was incubated with 10 μCi [γ-33P]ATP (PerkinElmer) and 1 mg recombinant GST-BECLIN-1 fragment in 50 μL kinase buffer (70 mM HEPES, 25 mM β-glycerophosphate, 3 mM MgCl2, 3 mM MnCl2, 1.2 mM DTT, 50 μg/mL PEG20.000, and 1% DMSO). Reactions were incubated at 30°C for 40 min. Proteins were separated by 10% SDS-PAGE, and phosphorylation was visualized by autoradiography.
Flow cytometry
Flow cytometric staining was performed as previously described.28–30
Mice
Mice were caged in special caging system with autoclaved food and acidified water at the University of Freiburg in accordance with national and institutional guidelines for animal care. All animal studies have been approved by the Ethics committees of the University Medical Center Freiburg and the district government in Freiburg (approval no. 35-9185.81/G-13/05).
Statistics
Statistical comparisons were performed using GraphPad Prism 6 software. Detailed statistical tests and significance cutoffs are indicated in each figure legend. All data represent the mean ± standard error of the mean (SEM).
Study approval
The studies using human samples were conducted according to the Declaration of Helsinki principles. All biological samples were obtained following written informed consent from the patient and upon approval by the Ethics Committee of the University Medical Center Freiburg.
Additional methodology is provided in the Online Supplementary Materials and Methods.
Results
Knockdown of Beclin-1 delays BCR-ABL-mediated leukemogenesis in vivo
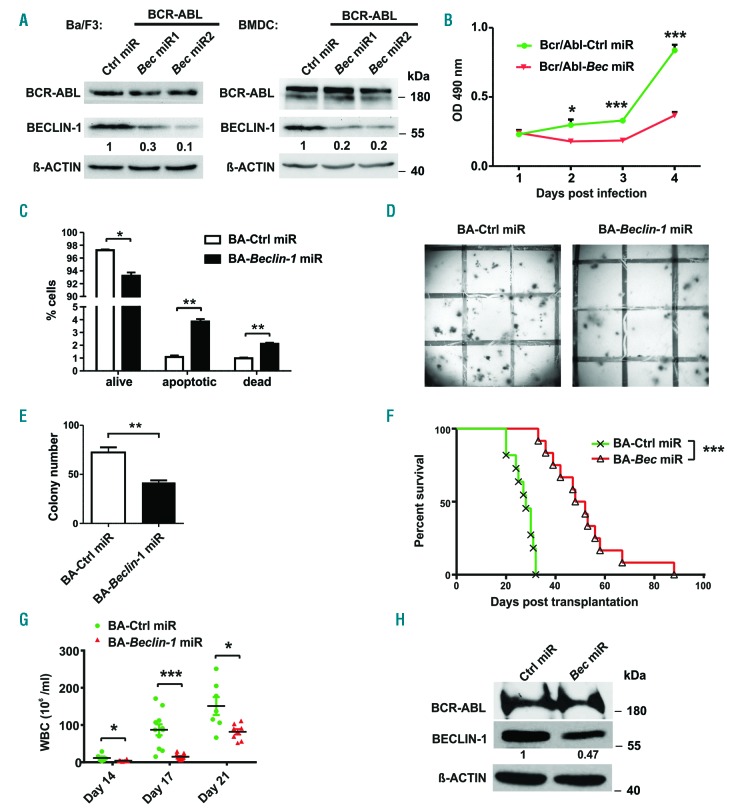
To further investigate the impact of autophagy in CML we examined the role of BECLIN-1, a master autophagy mediator, in BCR-ABL induced transformation and colony forming assays. Beclin-1 was downregulated using a targeted genetic approach with an individualized micro RNA-based knockdown of Beclin-1 in BCR-ABL-overexpressing Ba/F3 cells and bone marrow derived cells (BMDC). Specific knockdown of Beclin-1 with two individually designed siRNA resulted in significantly lower proliferation of BCR-ABL transduced Ba/F3 cells compared to cells infected with a control miR sequence (Figure 1A-B). As the secondly designed Beclin-1 miR resulted in the most efficient Beclin-1 knockdown, we performed all further experiments solely with Beclin-1 miR2. We could detect higher apoptosis levels, but no decrease in cell cycle rate in Beclin-1 miR cells (Figure 1C, Online Supplementary Figure S1A ).
Figure 1.
BECLIN-1 downregulation delays BCR-ABL-mediated proliferation in vitro and in vivo. (A) Immunoblot analyses were used to confirm downregulation of BECLIN-1 using two different Beclin-1 directed miR in Ba/F3 cells and BMDC. (B) Cell proliferation measurement was performed by MTT assay in Ba/F3 cells infected with indicated construct towards IL-3 withdrawal, indicating that BCR-ABL-mediated cell proliferation is impaired by Beclin-1 knockdown. ***P<0.001, *P<0.05, Student’s t-test. (C) Statistical analysis of flow cytometric staining showing Annexin-V−/propidium iodide (PI)- (“alive”), Annexin-V+ / PI− (“apoptotic”) and Annexin-V+ / PI+ (“dead”) Ba/F3 cells with indicated construct. **P<0.01, *P<0.05, Student’s t test. (D) Methylcellulose (MC) colony formation assay of primary 5-FU enriched bone marrow cells showed impaired colony formation upon BCR-ABL expression in Beclin-1 knockdown cells compared to control miR expressing cells. 1,000 EGFP+ BMDC infected with the indicated construct were plated into methylcellulose in the absence of growth factors and colonies were quantified 10 days later. One representative well is shown. Three independent experiments were performed in doublets. Grid size is 5 × 5 mm. (E) Quantitation of the MC shown in (D) (72 vs. 55.7 colony-forming unit [CFU], *P<0.05, and 72 vs. 41.7 CFU, **P<0.01, respectively, student’s t-test). (F) Kaplan-Meier curve demonstrates a significantly prolonged survival of mice transplanted with Beclin-1 knockdown BCR-ABL+ BMDC compared to control mice (Median survival 28 vs. 50 days, ***P<0.001 in two independent transplantations, Log-rank test (n=11, control miR; n=13, Beclin-1 miR)). (G) WBC from peripheral blood (PB) showed a significant reduction of leukemic progression in mice transplanted with Beclin-1 knockdown cells (11.6 vs. 4 million/mL, day 14, *P<0.05; 87.3 vs. 14.8 million/mL, day 17, ***P<0.001; 151.1 vs. 81.9 million/mL, day 21, *P<0.05). (H) Efficient and durable knockdown of Beclin-1 was proven by immunoblot analyses of spleen cells of transplanted mice (day 27).
Furthermore, we could show significantly lower colony formation in BCR-ABL-expressing primary BMDC with Beclin-1 downregulating miR in comparison to control BMDC (Figure 1D-E).
Next, we examined the effects of Beclin-1 knockdown in a CML mouse model in vivo. BMDC from 5-FU pretreated animals were infected with a vector expressing BCR-ABL and the specific Beclin-1 (pMmiRBec–BCR-ABL) or control miR sequence (pMmiRCtrl–BCR-ABL) on one construct and under the LTR promoter. Survival of mice transplanted with BCR-ABL-expressing Beclin-1 knockdown BMDC was sustained and significantly prolonged compared to the control group (median survival 28 vs. 50 days, P<0.0001) (Figure 1F). Furthermore, the white blood cell count (WBC) and the leukemic burden of mice transplanted with Beclin-1 knockdown BCR-ABL BMDC was significantly lower (87.3 vs. 14.8 × 103/μL on day 17, P<0.0001) compared to the control group (Figure 1G). Immunoblotting of splenocyte extracts of transplanted mice confirmed efficient downregulation of Beclin-1 one month after transplantation (Figure 1H). Upon disease induction, fluorescence-activated cell sorting (FACS) analyses of transplanted animals showed no differences in the immune phenotype of the BCR-ABL induced disease by Beclin-1 downregulation (Online Supplementary Figure S1B-D). To test whether the impact of Beclin-1 knockdown on CML cells is due to a general effect of autophagy inhibition or more due to a specific role of BECLIN-1 in BCR-ABL induced diseases, we also deleted another main autophagy regulator, ATG5 in a CML mouse model: Atg5 conditional knockout BMDC31 were infected with a BCR-ABL-Cre fusion vector and transplanted into wild-type (wt) recipient mice. Interestingly, deletion of Atg5 was not able to induce a delay in leukemia induction or progression of BCR-ABL transplanted mice (Online Supplementary Figure S1E). Furthermore, Atg5 deletion had no influence on the WBC of the transplanted animals (Online Supplementary Figure S1F), despite efficient deletion of the floxed Atg5 alleles upon Cre expression in BCR-ABL positive BMDC (Online Supplementary Figure S1G).
In order to exclude toxic effects of Beclin-1 knockdown on normal hematopoiesis, we transplanted solely Beclin-1 miR infected BMDC into mice, which exhibited no differences in survival, WBC or lineage phenotype compared to the control group (Online Supplementary Figure S2A-G).
Our results from the in vivo CML mouse model show a significant and specific impact of Beclin-1 knockdown on CML disease induction.
Active BCR-ABL suppresses autophagy through the BECLIN-1 complex
It has been shown previously that BCR-ABL kinase inhibitors induce autophagy. Accordingly, inhibition of BCR-ABL kinase activity by nilotinib led to an induction of autophagy measured by increased LC3-II expression and punctual LC3 accumulation (Online Supplementary Figure S3A-C). To differentiate, whether the autophagy induction by nilotinib is caused by specific BCR-ABL inhibition or due to an unspecific nilotinib effect, we treated nilotinib-resistant Ba/F3-BCR-ABL-T315I cells with nilotinib and could show that this treatment failed to induce autophagy, suggesting that active BCR-ABL indeed suppresses autophagy. Moreover, addition of interleukin-3 (IL-3), which rescues cells from nilotinib-induced cell death but does not rescue BCR-ABL inhibition, could not block autophagy (Online Supplementary Figure S3D-F).
Based on our mouse model data, we hypothesized that BECLIN-1 may be an essential player in autophagy suppression by BCR-ABL. BECLIN-1 has a crucial role in autophagosome formation as being part of the UVRAG-VPS15-ATG14-VPS34-RUBICON-BECLIN-1 complex. Interestingly, we could show that the formation of the UVRAG-VPS15-ATG14-VPS34-RUBICON-BECLIN-1 complex was altered in a BCR-ABL positive human cell line (K562) after nilotinib treatment (Online Supplementary Figure S3G): recruitment of positive regulators of autophagosome formation (VPS15, VPS34, UVRAG and ATG14) to BECLIN-1 was increased upon BCR-ABL inhibition, whereas the recruitment of the negative regulator RUBICON to BECLIN-1 was impaired after nilotinib treatment. These results indicate that BCR-ABL kinase activity modulates the BECLIN-1 complex composition and thereby leads to autophagy suppression.
BCR-ABL interacts with BECLIN-1
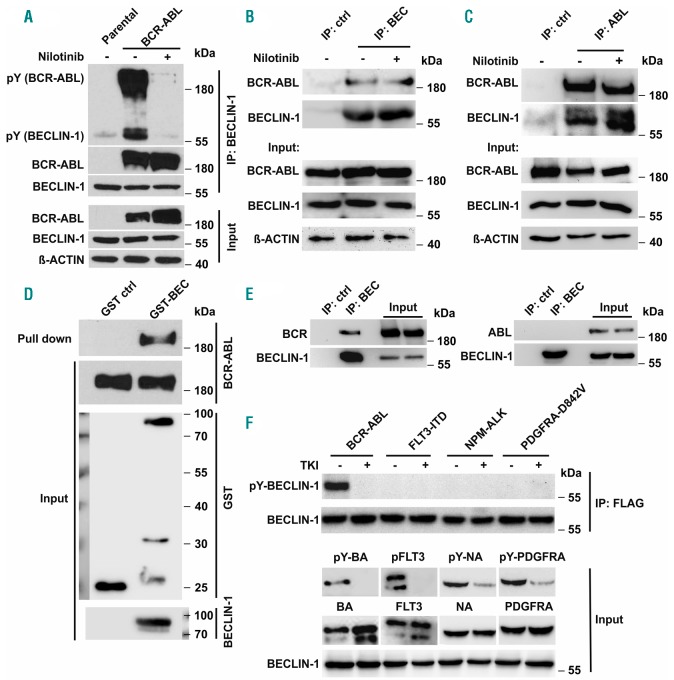
Next, we aimed to investigate how BCR-ABL kinase activity modulates BECLIN-1 complex composition. We found that BCR-ABL strongly binds to BECLIN-1, independent of ABL kinase activity.
Furthermore, BECLIN-1 was tyrosine phosphorylated in a complex with kinase active BCR-ABL indicating that BCR-ABL may directly phosphorylate BECLIN-1 (Figure 2A). Immunoprecipitation of endogenous BCR-ABL in K562 cells confirmed the BCR-ABL/BECLIN-1 interaction (Figure 2B-C) and GST-pulldown-assays using purified BECLIN-1 suggest that BCR-ABL and BECLIN-1 may bind directly to each other (Figure 2D).
Figure 2.
BCR-ABL interacts with and phosphorylates BECLIN-1. (A) Co-immunoprecipitation analyses in BCR-ABL-tranfected HEK293T cells demonstrate, that BCR-ABL strongly phosphorylates and binds BECLIN-1. A phosphotyrosine antibody was used for phospho-BECLIN-1 and phospho-BCR-ABL detection. (B) Immunoprecipitation of BCR-ABL with BECLIN-1 and (C) BECLIN-1 with ABL in K562 cells confirms the interaction reciprocally and at endogenous level. (D) GST-pull-down of BCR-ABL with recombinant GST-Beclin-1 in K562 cells corroborated the interaction between BCR-ABL and BECLIN-1. (E) BECLIN-1 immunoprecipitation in Beclin-1 transfected HEK293T cells demonstrates that BECLIN-1 is interacting with BCR but not with ABL. (F) BECLIN-1 is exclusively phosphorylated by BCR-ABL among several oncogenic tyrosine kinases in HEK293T cells. For TKI treatment, specific inhibitors (nilotinib for BCR-ABL, sorafenib for FLT3-ITD and PDGFRA-D842V, and TAE684 for NPM-ALK) were added into medium four hours before cell harvest.
We could also detect BCR-ABL/BECLIN-1 co-localization using immunofluorescence staining (Online Supplementary Figure S4A). To investigate which region of BCR-ABL binds to BECLIN-1, we performed binding assays by overexpressing either BCR or ABL together with Beclin-1 in 293T cells. Immunoprecipitations revealed that BCR interacts with BECLIN-1 but not ABL (Figure 2E).
BCR-ABL directly phosphorylates BECLIN-1 at specific tyrosine residues Y233 and Y352
We next investigated, whether BECLIN-1 is exclusively phosphorylated by BCR-ABL. Interestingly, all other tested oncogenic kinases (FLT3-ITD, NPM-ALK and PDGFRA-D842V) failed to induce BECLIN-1 phosphorylation, implying that BECLIN-1 is a specific substrate of BCR-ABL and not a general target of oncogenic tyrosine kinase signaling (Figure 2F). Moreover, we were able to confirm BECLIN-1 tyrosine phosphorylation in all tested samples of primary CML patient material, whereas BECLIN-1 phosphorylation was absent in healthy control samples (Online Supplementary Figure S4B).
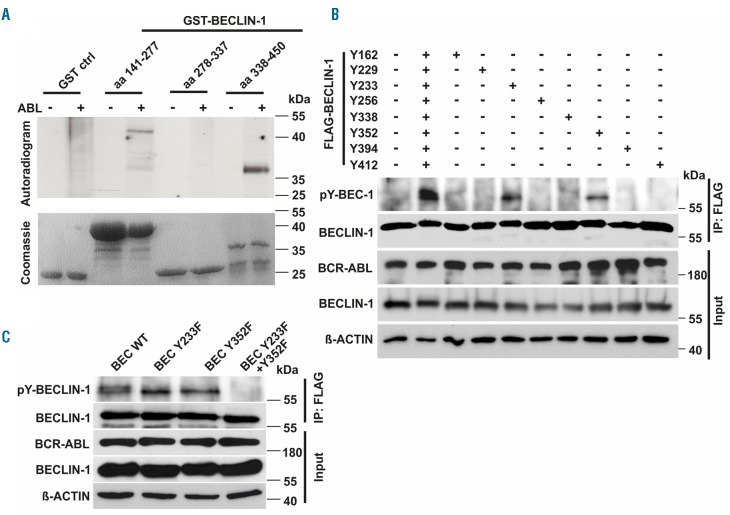
To test whether BECLIN-1 is a direct target of BCR-ABL we performed an in vitro kinase assay, and identified specific phosphorylation in two distinct regions of BECLIN-1: One spanning amino acid (aa) region 141 - 277 and another aa region 338 - 450 (Figure 3A). Furthermore, we generated a series of tyrosine residue mutants to determine specific BECLIN-1 tyrosine residues phosphorylated by BCR-ABL. Strong phosphorylation by BCR-ABL could be detected on BECLIN-1 tyrosine residues Y233 and Y352, whereas Y162 and Y338 show minor phosphorylation (Figure 3B). Western blot analyses of single and double phosphorylation-deficient mutants of those distinct BECLIN-1 tyrosine residues validated our results (Figure 3C) and demonstrated that BCR-ABL phosphorylates BECLIN-1 specifically at tyrosine residues Y233 and Y352. Interestingly, tyrosine Y352 (AA352-355 YCSG) is part of a STAT5 Src Homology 2 (SH2) domain binding motif (Y[VLTFIC]xx).37
Figure 3.
BCR-ABL phosphorylates BECLIN-1 at tyrosine residues Y233 and Y352. (A) In vitro kinase assay with active ABL and indicated recombinant GST-Beclin-1-fragments reveals specific BECLIN-1 phosphorylation at amino acids (aa) 141-277 and aa 338-450 by ABL. (B) Site directed mutagenesis identifies Beclin-1 Y233 and Y253 as crucial for BCR-ABL phosphorylation. HEK293T cells were co-transfected with FLAG-tagged-Beclin-1 mutants and BCR-ABL, thereafter FLAG-immunoprecipitated and phosphotyrosine-immunoblotted. (C) Expression of a phosphorylation-deficient BECLIN-1 Y233F/Y352F mutant in HEK293T cells confirms tyrosine residues Y233/Y352 as essential sites for BCR-ABL phosphorylation.
Phospho-mimic mutant Beclin-1 Y233E/Y352E suppresses autophagy through BECLIN-1 complex alterations whereas the phospho-deficient Beclin-1 Y233F/Y352F mutant induces autophagy
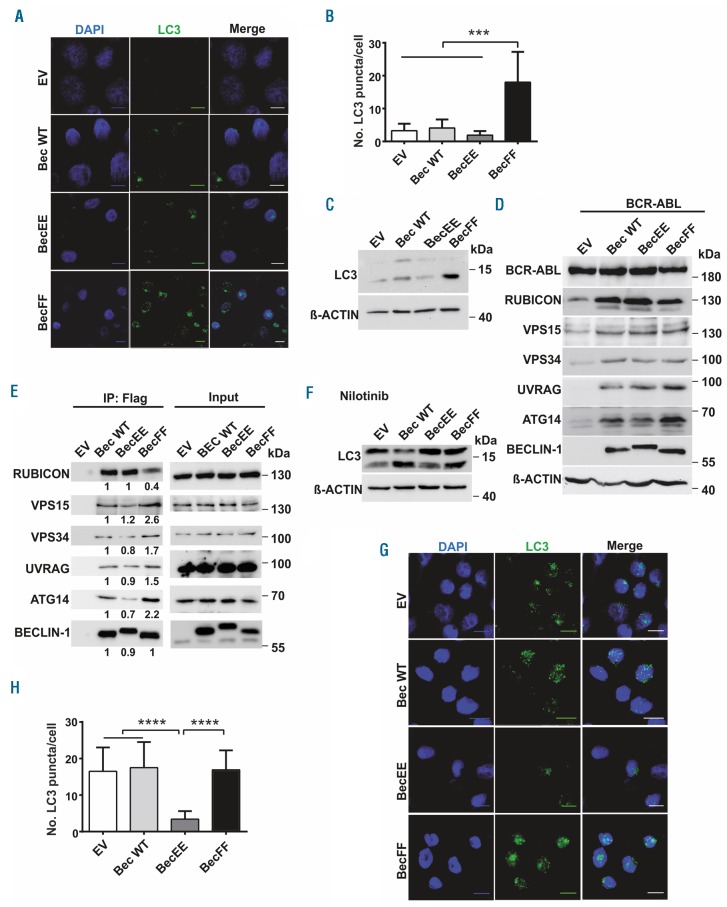
To evaluate whether phosphorylation of BECLIN-1 regulates autophagy, we generated a BECLIN-1 phosphorylation-mimic (Y233E/Y352E) and a phosphorylation-deficient mutant (Y233F/Y352F). In an LC3 puncta assay in K562 cells, we found that the phosphorylation-mimic BECLIN-1 mutant suppresses autophagy, whereas the phosphorylation-deficient BECLIN-1 mutant induces increased autophagy (Figure 4A-B). By immunoblotting, we could confirm that the phosphorylation-mimic mutant Y233E/Y352E decreases autophagy, whereas expression of the phosphorylation-deficient mutant Y233F/Y352F induces autophagy (Figure 4C). Our findings therefore suggest that phosphorylation of BECLIN-1 by BCR-ABL suppresses autophagy induction.
Figure 4.
BCR-ABL mediated BECLIN-1 phosphorylation leads to suppression of autophagy and altered BECLIN-1 complex formation. (A) Confocal microscopy of K562 cells reveals significant induction of autophagy in BECLIN-1 Y233F/Y352F-expressing cells indicated by increased number of LC3 puncta. (B) Statistical analysis of LC3 puncta count in Beclin-1-mutant-transduced K562 cells (3.3, 4.1, 1.9, and 18 units respectively; ****P<0.0001 Student’s t-test; Scale bar 10 μm). (C) Upregulation of LC3-II in K562 cells transduced with the phosphorylation-deficient Beclin-1 Y233F/Y352F mutant. (D) Immunoblot analyses were used for the detection of ATG14, UVRAG, VPS34 and RUBICON expression levels in Beclin-1 knockout MEF transduced with BCR-ABL and the indicated Beclin-1 construct. (E) Immunoprecipitation of FLAG-tagged phosphorylation-deficient BECLIN-1 Y233F/Y352F mutant (BecFF) in K562 cells indicates altered autophagy complex formation compared to the phosphorylation-mimic BECLIN-1 mutant (BecEE). (F) Beclin-1 tyrosine phosphorylation-mimic mutation blocks nilotinib-induced autophagy seen by reduced LC3-II levels compared to empty vector, Beclin-1 wt, and Beclin-1 FF. LC3 levels were evaluated by immunoblotting in the indicated Beclin-1 mutant-trans-duced K562 cells. (G) LC3 puncta formation was measured using confocal microscopy in K562 cells transduced with mCherry-EGFP-LC3 and the indicated Beclin-1 construct upon nilotinib treatment. Cells were stained with anti-LC3 antibody and nuclei were counterstained with DAPI. Scale bar, 10 μm. (H) Quantitation of LC3 puncta (16.5, 17.5, 3.4, and 16.9 dots/cell, respectively; P<0.0001 for comparison of Beclin-1 EE to empty vector [EV], Beclin-1 wt and Beclin-1 FF). K562 cells trans-duced with mCherry-EGFP-LC3 and the indicated Beclin-1 constructs were used for LC3 puncta measurement via Olympus ScanR screening station. Bars represent the mean±SD. ****P<0.0001 by Student’s t-test. EV: empty vector; wt: wild-type FLAG-Beclin-1; EE: FLAG-Beclin-1 Y233E/Y352E; FF: FLAG-Beclin-1 Y233F/Y352F.
Next we sought to know, whether the impaired autophagy induction of the phospho-mimic mutant Y233E/Y352E may be due to an altered recruitment of complex components to BECLIN-1. It has been shown recently, that lack of BECLIN-1 leads to downregulation of the BECLIN-1 complex binding partners.38 Indeed, Beclin-1 deficient, BCR-ABL expressing MEF showed downregulation of BECLIN-1 binding partners, which could be rescued by re-expression of either wt Beclin-1 or both phospho-mutants ((BEC FF and EE). Furthermore, expression of the Beclin-1 Y233E/Y352E mutant leads to decreased UVRAG and ATG14 levels, whereas Rubicon levels were increased compared to phosphorylation-deficient BECLIN-1 cells (Figure 4D).
From our results we hypothesized that the phosphorylation status of BECLIN-1 is important for the stabilization and recruitment of the different binding partners to the BECLIN-1 complex. Interestingly, co-immunoprecipitation of BECLIN-1 complex components revealed that autophagy activating proteins like VPS15, VPS34 and ATG14 were recruited less to the phospho-mimic BECLIN-1 (BEC EE) complex compared to the phospho-deficient BECLIN-1 (BEC FF) complex (Figure 4E, Online Supplementary Figure S4C). These results indicate that the altered autophagy by the two mutants is due to altered binding capacities of positive regulators to the BECLIN-1 core complex and thereby alters BECLIN-1 complex activity.
Next we asked, whether expression of the phospho-mimic Beclin-1 mutant could overcome the TKI-induced BCR-ABL inhibition-mediated autophagy induction and indeed, expression of Beclin-1 EE Y233E/Y352E impaired nilotinib-induced autophagy measured by LC3-II expression (Figure 4F) and LC3 puncta accumulation (Figure 4G-H). BCR-ABL inhibition by nilotinib was not able to enhance the autophagy-stimulatory effect of the phosphorylation-deficient BECLIN-1 Y233F/Y352F mutant compared to wt BECLIN-1, pointing to a BECLIN-1-specific autophagy regulation by BCR-ABL (Figure 4F-H).
In further BECLIN-1 complex analyses, we could demonstrate that the resistance of the phospho-mimic Beclin-1 mutant to nilotinib-induced autophagy is caused by an altered composition of the BECLIN-1 core complex with an impaired recruitment of the activation components ATG14, UVRAG, VPS15 and a gain of the negative regulator RUBICON to the BECLIN-1 core complex (Online Supplementary Figure S5A). BECLIN-1 phosphorylation with subsequent resistance to TKI-induced autophagy may thereby provide a novel explanation of how leukemic cells can escape autophagy-induced cell death and develop TKI resistance.
Recently, it has been shown that BECLIN-1 S90 phosphorylation is involved in starvation-mediated autophagy.39 To test whether BECLIN-1 phosphorylation at Y233/Y352 can influence starvation-mediated or rapamycin-mediated autophagy, we starved K562 cells or treated them with rapamycin and found that cellular autophagy is induced in Beclin-1 wt cells (Online Supplementary Figure S5B-D) and no differences could be demonstrated in K562 cells expressing either BECLIN-1 Y233E/Y352E or BECLIN-1 Y233F/Y352F (Online Supplementary Figure S5E-H). These results indicate that tyrosine phosphorylation of BECLIN-1 at Y233 and Y352 is not involved in starvation- or rapamycin-mediated autophagy but rather seems to be specific for tyrosine kinase-mediated autophagy processes.
Discussion
Recently, several studies have suggested that autophagy, a mechanism maintaining cellular homeostasis, plays an essential role in CML. However, the precise machinery of autophagy in CML development is not completely understood and crucial autophagocytotic mediators have not been investigated in vivo for their role in leukemogenesis in relevant CML mouse models.
Here, we define a molecular mechanism of autophagy suppression by BCR-ABL-specific BECLIN-1 phosphorylation. Silencing of Beclin-1 by siRNA technology led to a significantly prolonged survival of BCR-ABL transplanted mice, whereas no profound differences could be found for Atg5 deletion. Binding of BECLIN-1 to BCR-ABL led to phosphorylation at tyrosine residues Y233 and Y352, alteration of the BECLIN-1 interactome, and suppression of autophagy function.
Several active oncogenic kinases were demonstrated to serve as negative regulators of autophagy processes, whereas inhibition of oncogenic tyrosine kinases can reverse this effect. Until now, some links of BCR-ABL to autophagy processes have been described: BCR-ABL activates the PI3K/AKT signaling pathway, which is considered as a pathway inhibiting autophagy. Furthermore, TKI treatment itself triggers autophagy in BCR-ABL+ cells and TKI-induced cell death can potentially be increased by targeting autophagy proteins in addition. Recently, it was demonstrated that Ponatinib-resistant CML cells can acquire BCL-ABL-independent resistance through autophagy inhibition by activation of mTOR.46 Therefore, we hypothesize that BECLIN-1 may play similar roles in the resistance-acquired signaling cascade. BECLIN-1 is a central autophagy mediating protein in mammalian cells, and EGFR and AKT kinase have been shown to phosphorylate BECLIN-1 leading to inactivation of the protein and suppression of autophagy. Interestingly, active EGFR has been shown to phosphorylate the same tyrosine residues Y233/Y352 as BCR-ABL, further highlighting the importance of these tyrosine residues for autophagy suppression.42 In line with these previous findings, we have demonstrated in the present study that BCR-ABL phosphorylates BECLIN-1 and thereby suppresses autophagy. Interestingly, this effect is mediated by the alteration of BECLIN-1 affinity to BECLIN-1 binding partners (ATG14, VPS34 and VPS15), known as the BECLIN-1 core complex. Our results therefore provide a novel explanation for the suppression of autophagy in CML and expand our knowledge regarding BECLIN-1-associated pathogenic mechanisms in BCR-ABL+ leukemia.
A role for BECLIN-1 has been proposed in various malignancies, such as breast or lung carcinomas. However, the precise role of BECLIN-1 in tumorigenesis remains unclear: On one hand, Beclin-1 is considered as a tumor suppressor and its overexpression is favorable for treatment of various solid tumors.51–54 Furthermore, low expression of Beclin-1 is a marker of poor prognosis and enhanced aggressiveness in breast cancer55 and loss of one Beclin-1 allele leads to enhanced tumor development in mice. On the other hand, our present study demonstrates that knockdown of Beclin-1 prolongs the survival of BCR-ABL+ leukemic mice, which is consistent with a previous in vitro study in CML cell lines.16 Gene array analysis of CML patient samples revealed upregulated Beclin-1 levels in CML patients compared to healthy controls (fold change: 1.22; q-value: <0.1%).56 Importantly, we could show that CML patients exhibit a significant increase of phosphorylated BECLIN-1 levels. Targeting BECLIN-1 in specific approaches might thereby represent an elegant and alternative treatment option for TKI-resistant or intolerant CML patients by rendering CML cells sensitive to targeted therapies.
Recently, a study uncovered a kinase-independent role of EGFR in autophagy, showing that inactive oncogenic EGFR reversely triggers autophagy.57 These findings support the hypothesis that cells can develop TKI-resistance through autophagy induction, which might be caused or even triggered through the inactive oncogenic kinase itself. This sheds some light on the role of autophagy on cellular survival rather than cell death and gives a rationale to explore the combinatory effect of kinase inhibitors with autophagy inhibitors. A number of drugs (Chloroquine, Bafilomycin A1, MAPK inhibitors and PI3K inhibitors) were described to have inhibitory effects on autophagy, however, most of them are poorly selective, limiting their therapeutic application. Therefore, the development of highly specific and selective autophagy inhibitors remains a mandatory necessity for the successful evaluation of the therapeutic combination therapy with TKI in CML. Moreover, accurate target identification among major autophagy players is fundamental for successful therapeutic application: BECLIN-1 seems an attractive target, as its role in kinase-driven cancer is not only shown for solid cancers but also for hematopoietic malignancies in our study. For ATG5, we were not able to prove essential function in CML development in mice, whereas Liu et al. were able to demonstrate a crucial role of the protein in MLL-AF9 mediated AML induction in mice.61 ATG3 expression was shown to be indispensable for effective CML progression.14 These results indicate that autophagy induction is highly specific and stringently regulated, not only depending on the particular malignancy, but also on the involvement of the master regulators.
Taken together, in this study we uncover a crucial role of BECLIN-1 in BCR-ABL mediated transformation in vivo and were able to identify a molecular mechanism by which BCR-ABL kinase activity regulates autophagy. We show that BCR-ABL binds and phosphorylates BECLIN-1 on tyrosine residues 233 and 352, thereby leading to alterations of the UVRAG-VPS15-ATG14-VPS34-RUBICON-BECLIN-1 complex. Moreover, the BCR-ABL/BECLIN-1 interaction suppresses autophagy and thereby bypasses the negative effect of autophagy on cancer cell survival and proliferation. Importantly, these data may be of clinical relevance, as CML patients exhibit upregulated BECLIN-1 phosphorylation levels. Our findings provide a novel link between BCR-ABL and BECLIN-1 and shed some light on how specific oncogenes influence autophagy.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/5/1285
Funding
The authors would like to thank their funding agencies. This work was supported by research grants from the Deutsche José Carreras Leukämie-Stiftung (DJCLS R14/22 to JD and ALI and DJCLS 02 FN/2017 to TAM). ALI was supported by a research grant from the University Medical Center Freiburg and from the government Baden-Württemberg (BSL). JD and TAM were supported by a DFG grant (FOR 2033). TBH was supported by the DFG (CRC1140, CRC 992, HU 1016/8-1), by the BMBF (01GM1518C), by the Else-Kröner Fresenius Stiftung (NAKSYS), by the European Research Council-ERC grant 616891 and by the H2020-IMI2 consortium BEAt-DKD.
References
- 1.Marcucci G, Perrotti D, Caligiuri MA. Understanding the molecular basis of imatinib mesylate therapy in chronic myeloge-nous leukemia and the related mechanisms of resistance. Commentary re: A. N. Mohamed et al., The effect of imatinib mesylate on patients with Philadelphia chromosome-positive chronic myeloid leukemia with secondary chromosomal aberrations. Clin. Cancer Res., 9: 1333-1337, 2003. Clin Cancer Res. 2003; 9(4):1248–1252. [PubMed] [Google Scholar]
- 2.Talpaz M, Shah NP, Kantarjian H, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354(24):2531–2541. [DOI] [PubMed] [Google Scholar]
- 3.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007; 7(6):441–453. [DOI] [PubMed] [Google Scholar]
- 4.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–183. [DOI] [PubMed] [Google Scholar]
- 5.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat Rev Cancer. 2007;7(5):345–356. [DOI] [PubMed] [Google Scholar]
- 6.An X, Tiwari AK, Sun Y, Ding PR, Ashby CR, Jr, Chen ZS. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: a review. Leuk Res. 2010; 34(10):1255–1268. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia R, Holtz M, Niu N, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003; 101(12):4701–4707. [DOI] [PubMed] [Google Scholar]
- 8.Mahon FX, Rea D, Guilhot J, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029–1035. [DOI] [PubMed] [Google Scholar]
- 9.de Lavallade H, Apperley JF, Khorashad JS, et al. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008; 26(20):3358–3363. [DOI] [PubMed] [Google Scholar]
- 10.Holyoake TL, Helgason GV. Do we need more drugs for chronic myeloid leukemia? Immunol Rev. 2015;263(1):106–123. [DOI] [PubMed] [Google Scholar]
- 11.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. [DOI] [PubMed] [Google Scholar]
- 12.Helgason GV, Karvela M, Holyoake TL. Kill one bird with two stones: potential efficacy of BCR-ABL and autophagy inhibition in CML. Blood. 2011;118(8):2035–2043. [DOI] [PubMed] [Google Scholar]
- 13.Sheng Z, Ma L, Sun JE, Zhu LJ, Green MR. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118(10):2840–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altman BJ, Jacobs SR, Mason EF, et al. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene. 2011; 30(16):1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellodi C, Lidonnici MR, Hamilton A, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119(5):1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y, Yang L, Zhao M, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26(8):1752–1760. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell R, Hopcroft LEM, Baquero P, et al. Targeting BCR-ABL-independent TKI resistance in chronic myeloid leukemia by mTOR and autophagy inhibition. J Natl Cancer Inst. 2018;110(5):467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karvela M, Baquero P, Kuntz EM, et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy. 2016; 12(6):936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22(2):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009; 5(5):713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phos-phatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Y, Wang QJ, Yue Z. Atg14L and Rubicon: yin and yang of Beclin 1-mediated autophagy control. Autophagy. 2009; 5(6):890–891. [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009; 11(4):385–396. [DOI] [PubMed] [Google Scholar]
- 24.Grundler R, Thiede C, Miething C, Steudel C, Peschel C, Duyster J. Sensitivity toward tyrosine kinase inhibitors varies between different activating mutations of the FLT3 receptor. Blood. 2003;102(2):646–651. [DOI] [PubMed] [Google Scholar]
- 25.Illert AL, Kawaguchi H, Antinozzi C, et al. Targeted inactivation of nuclear interaction partner of ALK disrupts meiotic prophase. Development. 2012;139(14):2523–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illert AL, Zech M, Moll C, et al. Extracellular signal-regulated kinase 2 (ERK2) mediates phosphorylation and inactivation of nuclear interaction partner of anaplastic lymphoma kinase (NIPA) at G2/M. J Biol Chem. 2012;287(45):37997–38005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundler R, Brault L, Gasser C, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med. 2009;206(9):1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller TA, Grundler R, Istvanffy R, et al. Lineage-specific STAT5 target gene activation in hematopoietic progenitor cells predicts the FLT3(+)-mediated leukemic phenotype. Leukemia. 2016;30(8):1725–1733. [DOI] [PubMed] [Google Scholar]
- 29.Illert AL, Albers C, Kreutmair S, et al. Grb10 is involved in BCR-ABL-positive leukemia in mice. Leukemia. 2015; 29(4):858–868. [DOI] [PubMed] [Google Scholar]
- 30.Albers C, Illert AL, Miething C, et al. An RNAi-based system for loss-of-function analysis identifies Raf1 as a crucial mediator of BCR-ABL-driven leukemogenesis. Blood. 2011;118(8):2200–2210. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, Hartleben B, Kretz O, et al. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy. 2012; 8(5):826–837. [DOI] [PubMed] [Google Scholar]
- 32.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ultimo S, Simioni C, Martelli AM, et al. PI3K isoform inhibition associated with anti Bcr-Abl drugs shows in vitro increased anti-leukemic activity in Philadelphia chromosome-positive B-acute lymphoblastic leukemia cell lines. Oncotarget. 2017;8(14):23213–23227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita M, Nishinaka Y, Kato I, et al. Dasatinib induces autophagy in mice with Bcr-Abl-positive leukemia. Int J Hematol. 2017;105(3):335–340. [DOI] [PubMed] [Google Scholar]
- 35.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011; 18(4):571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKnight NC, Zhenyu Y. Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr Pathobiol Rep. 2013;1(4):231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uyar B, Weatheritt RJ, Dinkel H, Davey NE, Gibson TJ. Proteome-wide analysis of human disease mutations in short linear motifs: neglected players in cancer? Mol Biosyst. 2014;10(10):2626–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKnight NC, Zhong Y, Wold MS, et al. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 2014;10(10):e1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Y, An Z, Zou Z, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goussetis DJ, Gounaris E, Wu EJ, et al. Autophagic degradation of the BCR-ABL oncoprotein and generation of antileukemic responses by arsenic trioxide. Blood. 2012;120(17):3555–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Rothe K, Yen R, et al. A novel AHI-1-BCR-ABL-DNM2 complex regulates leukemic properties of primitive CML cells through enhanced cellular endocytosis and ROS-mediated autophagy. Leukemia. 2017; 31(11):2376–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Y, Zou Z, Becker N, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang RC, Wei Y, An Z, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338(6109):956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kharas MG, Janes MR, Scarfone VM, et al. Ablation of PI3K blocks BCR-ABL leukemogenesis in mice, and a dual PI3K/mTOR inhibitor prevents expansion of human BCR-ABL+ leukemia cells. J Clin Invest. 2008;118(9):3038–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klejman A, Rushen L, Morrione A, Slupianek A, Skorski T. Phosphatidylinositol-3 kinase inhibitors enhance the anti-leukemia effect of STI571. Oncogene. 2002;21(38):5868–5876. [DOI] [PubMed] [Google Scholar]
- 46.Mitchell R, Hopcroft LEM, Baquero P, et al. Targeting BCR-ABL-Independent TKI Resistance in Chronic Myeloid Leukemia by mTOR and Autophagy Inhibition. J Natl Cancer Inst. 2018;110(5):467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploin-sufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999; 402(6762):672–676. [DOI] [PubMed] [Google Scholar]
- 49.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26(9):877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiqui-tination activity of USP10 and USP13. Cell. 2011;147(1):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn CH, Jeong EG, Lee JW, et al. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115(12):1344–1349. [DOI] [PubMed] [Google Scholar]
- 53.Shen Y, Li DD, Wang LL, Deng R, Zhu XF. Decreased expression of autophagy-related proteins in malignant epithelial ovarian cancer. Autophagy. 2008;4(8):1067–1068. [DOI] [PubMed] [Google Scholar]
- 54.Won KY, Kim GY, Kim YW, Song JY, Lim SJ. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum Pathol. 2010;41(1):107–112. [DOI] [PubMed] [Google Scholar]
- 55.Tang H, Sebti S, Titone R, et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine. 2015;2(3):255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diaz-Blanco E, Bruns I, Neumann F, et al. Molecular signature of CD34(+) hematopoietic stem and progenitor cells of patients with CML in chronic phase. Leukemia. 2007;21(3):494–504. [DOI] [PubMed] [Google Scholar]
- 57.Tan X, Thapa N, Sun Y, Anderson RA. A kinase-independent role for EGF receptor in autophagy initiation. Cell. 2015;160(1-2):145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and pro-survival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6(3):322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Hu Q, Shen HM. Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol Res. 2016; 105:164–175. [DOI] [PubMed] [Google Scholar]
- 60.Pasquier B. Autophagy inhibitors. Cell Mol Life Sci. 2016;73(5):985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Q, Chen L, Atkinson JM, Claxton DF, Wang HG. Atg5-dependent autophagy contributes to the development of acute myeloid leukemia in an MLL-AF9-driven mouse model. Cell Death Dis. 2016; 7(9):e2361. [DOI] [PMC free article] [PubMed] [Google Scholar]