Abstract
Megakaryoblastic leukemia 1 (MKL1) is a coactivator of serum response factor and together they regulate transcription of actin cytoskeleton genes. MKL1 is associated with hematologic malignancies and immunodeficiency, but its role in B cells is unexplored. Here we examined B cells from monozygotic triplets with an intronic deletion in MKL1, two of whom had been previously treated for Hodgkin lymphoma (HL). To investigate MKL1 and B-cell responses in the pathogenesis of HL, we generated Epstein-Barr virus-transformed lymphoblastoid cell lines from the triplets and two controls. While cells from the patients with treated HL had a phenotype close to that of the healthy controls, cells from the undiagnosed triplet had increased MKL1 mRNA, increased MKL1 protein, and elevated expression of MKL1-dependent genes. This profile was associated with elevated actin content, increased cell spreading, decreased expression of CD11a integrin molecules, and delayed aggregation. Moreover, cells from the undiagnosed triplet proliferated faster, displayed a higher proportion of cells with hyperploidy, and formed large tumors in vivo. This phenotype was reversible by inhibiting MKL1 activity. Interestingly, cells from the triplet treated for HL in 1985 contained two subpopulations: one with high expression of CD11a that behaved like control cells and the other with low expression of CD11a that formed large tumors in vivo similar to cells from the undiagnosed triplet. This implies that pre-malignant cells had re-emerged a long time after treatment. Together, these data suggest that dysregulated MKL1 activity participates in B-cell transformation and the pathogenesis of HL.
Introduction
Hodgkin lymphoma (HL) is a B-cell malignancy of largely unknown etiology. Familial clustering and twin concordance are seen, as are links with viral infections such as Epstein-Barr virus (EBV).1,2 The malignant HL Reed-Sternberg cells have frequently undergone class switch recombination and likely originate from germinal center B cells that fail to undergo apoptosis despite destructive somatic mutations.1,3,4 Various studies have shown the ability of EBV to rescue crippled germinal center B cells from apoptosis, supporting the role of this virus in the pathogenesis of HL.5,6
Megakaryoblastic leukemia 1 (MKL1; also known as MRTF-A, MAL, or BSAC) is a transcriptional coactivator of serum response factor (SRF) and binds to globular (G-)actin via an RPEL motif.7,8 As cytoplasmic G-actin is polymerized into filamentous (F)-actin, the G-actin pool diminishes. This leads to MKL1 translocation into the nucleus where it interacts with SRF to induce transcription of cytoskeleton-related genes, including actin, integrin molecules, and SRF itself.7–10 Indeed, inducible expression of SRF in response to serum stimulation is dependent on SRF and MKL1 activity.9,11 Actin polymerization and MKL1-SRF activity are additionally regulated by extracellular signaling through several integrin molecules which activate the small Rho GTPases, including RhoA.12
MKL1 was initially described as part of a fusion protein in megakaryoblastic leukemia of poor prognosis.13,14 MKL1 expression is detected in malignant cells in breast and liver cancer and is associated with increased cell proliferation, anchorage-independent cell growth, and metastasis.15,16 Small molecule inhibitors of the MKL1-SRF pathway have been identified, facilitating studies on the biological activity of MKL1, and are being tested as potential cancer therapeutic agents.17 One of these compounds is CCG-1423, which was originally identified as a RhoA-MKL1-SRF pathway inhibitor and later discovered to target MKL1 directly.17,18
A loss-of-function mutation in MKL1 was recently identified in a 4-year old girl with severe primary immunodeficiency.19 MKL1 deficiency caused reduced G-actin and F-actin content in the patient’s neutrophils, leading to reduced phagocytosis and migration.19 In 2013, a familial case of two monozygotic triplets who developed HL at the age of 40 and 63 was described.20 Both patients are in remission following HL treatment in 1985 and 2008, respectively, and the third triplet remains undiagnosed. Using microarray comparative genomic hybridization, a 15-31 kb deletion in intron 1 of MKL1 was identified in the triplets.20 The impact of this mutation on MKL1 expression and B-cell function remains unknown.
Here we took the approach of generating EBV-transformed lymphoblastoid cell lines (LCL) from the triplets with the deletion in MKL1 intron 1 (HL0, HL1, and HL2) and from two healthy controls (C1 and C2). We found that the LCL from the undiagnosed triplet had increased MKL1 and SRF expression, and elevated G-actin content. This was associated with hyperproliferation, genomic instability, and tumor formation when the cells were injected into immunocompromised mice. When compared to control LCL with high CD11a expression and capacity to form large aggregates, HL0 LCL expressed low CD11a and had reduced capacity to form aggregates. The HL1 LCL showed a bimodal expression of CD11a and when sorted for CD11a low and CD11a high cells, CD11a high cells mimicked the response of control LCL whereas the H10 CD11a low cells mimicked the response of HL0 cells with increased proliferation and tumor formation. Finally, treatment of HL0 cells with the MKL1 inhibitor CCG-1423 reverted the phenotype and prevented tumor growth in vivo. These data show that unregulated MKL1 alters B-cell cytoskeletal responses leading to B-cell transformation.
Methods
Human blood samples and Epstein-Barr virus transformation
Whole blood samples were obtained from the triplets and age-matched controls after informed consent was given. This study was performed according to the principles expressed in the Helsinki Declaration and with approval from the local ethics committee (Dnr 2015/416-31). For analysis of primary cells, the first experiment included samples from HL0, HL1, and a control (not used for EBV-transformation) collected in February 2015 and the second experiment was conducted on samples from HL2 and C1, collected in May 2015. To establish the EBV-transformed LCL, peripheral blood mononuclear cells from HL0, HL1, and HL2, and two age- and sex-matched controls (C1 and C2), all collected in November 2015, were cultured with supernatant of the virus-producing B95-8 line.21
Mice
NOD/SCID-IL2rγnull (NSG) mice were bred and maintained at the animal facility of the Department of Microbiology, Tumor and Cell Biology at Karolinska Institutet under specific pathogen-free conditions. Female mice were used and all animal experiments were performed after approval from the local ethics committee (the Stockholm District Court, permit N77/13 and N272/14). For inhibitor treatment, 10 μM CCG-1423 or dimethylsulfoxide was injected intratumorally for 6 consecutive days. The volume of the tumor was calculated at the endpoint using a caliper.
Flow cytometry and microscopy
Flow cytometry was performed on peripheral blood mononu-clear cells, LCL, and cultured primary B cells using an LSRFortessa X-20 (BD Biosciences). The results were processed using FlowJo v10 software (TreeStar Inc., St. Ashland, OR, USA). To determine integrin expression at the cell surface, LCL were labeled with an anti-human CD11a antibody (TS2/4; Biolegend) for total CD11a expression, or an anti-human CD11a antibody (Hl111; Biolegend) for inactive/closed conformation-CD11a expression, or an anti-human CD54 antibody (Biolegend), followed by an anti-mouse-Alexa647 antibody (ThermoFisher Scientific). To determine F- and G-actin content in two LCL samples side by side, one sample was incubated with an anti-human CD54 antibody (Biolegend) for 30 min on ice and thereafter labeled with DNaseI-Alexa488 (ThermoFisher Scientific) and phalloidin-Alexa568 (ThermoFisher Scientific).
Results
The MKL1 intron 1 deletion is associated with increased expression of MKL1 and MKL1-induced genes
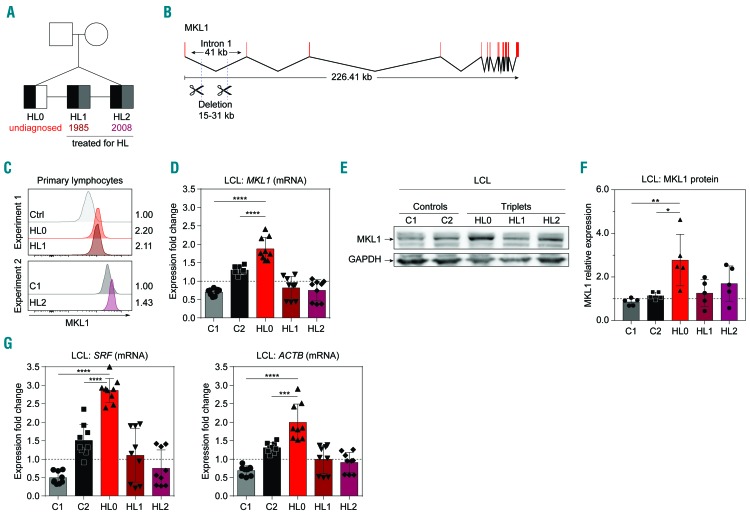
To understand how the deletion in MKL1 intron 1 affected actin cytoskeleton regulation in B cells, we examined freshly isolated cells and LCL from the triplets (HL0, HL1, and HL2) and two healthy controls (C1 and C2) (Figure 1A, B). We reasoned that cells from the undiagnosed HL0 triplet may be in a pre-HL stage, whereas HL1 and HL2 cells may be more similar to control cells because of successful treatment for HL in 1985 and 2008, respectively. MKL1 protein in primary blood lymphocytes was higher in the cells from all triplets than in control cells, as assessed by flow cytometry (Figure 1C and Online Supplementary Figure S1A). Using primer walking and sequencing, we confirmed that the triplets’ cells contained a heterozygous deletion of MKL1 intron 1 (Online Supplementary Figure S2A). The intron 1 is in the 5′ untranslated region of the MKL1 gene. We examined exon boundaries of exons 1-4 in the 5’ untranslated region and found normal expression of adjacent exons, suggesting that the intron 1 deletion did not affect splicing of MKL1 (Online Supplementary Figure S3). MKL1 intron 1 contains many transcription binding sites (Online Supplementary Figure S2B) that may affect MKL1 transcription. We examined MKL1 mRNA expression in LCL by real-time quantitative polymerase chain reaction (RT-qPCR). The LCL from HL0 (the undiagnosed triplet) had higher expression of MKL1 when compared to that of the siblings (HL1 and HL2) and controls (C1 and C2) (Figure 1D). Protein studies indicated that the levels of MKL1 were higher in HL0 than in C1 and C2 (Figure 1E, F and Online Supplementary Figure S1B). To investigate whether increased MKL1 expression was associated with increased MKL1 activity, we examined mRNA expression of specific MKL1-dependent genes including SRF and ACTB.19 HL0 LCL had the highest expression of both these MKL1-dependent genes (Figure 1G). This suggests that the intron 1 deletion in MKL1 directly influences the expression of MKL1 and MKL1 target genes.
Figure 1.
The MKL1 intronic deletion is associated with increased expression of MKL1 and MKL1-induced genes. (A) Pedigree of genetically identical triplets (HL0, HL1, HL2) of whom two have been diagnosed with Hodgkin lymphoma (HL). Presence of the MKL1 intronic deletion is indicated in black, a diagnosis of HL in gray, and undiagnosed in white. Numbers indicate year of treatment. (B) Overview of MKL1 indicating the deletion in intron 1. (C) MKL1 protein expression in primary lymphocytes, determined by flow cytometry, in two separate experiments. Experiment 1: control (Ctrl), HL0, and HL1. Experiment 2: C1 and HL2. Numbers indicate the fold-change in expression normalized to the ctrl and C1 values, respectively. (D) MKL1 mRNA expression. (E) Representative image of MKL1 protein expression. (F) MKl1 expression normalized to GAPDH expression. (G) Expression of MKL1-induced genes by real-time quantitative polymerase chain reaction. (D, F, G) Combined data from three experiments; experiments with primary lymphocytes (C) were performed once. For bar graphs, the dotted line indicates normalization to the mean of C1 and C2. All panels display data from lymphoblastoid cell lines except (C), which displays data from primary lymphocytes. Error bars represent the standard deviation of the measurements. Symbols represent technical replicates from three independent experiments in (D) and (G), and single values from independent experiments in (F). All data were analyzed using analysis of variance with a post-hoc Tukey test. ***P<0.001, ****P<0.0001.
Increased actin content and actin-dependent spreading in HL0 and HL1 cells
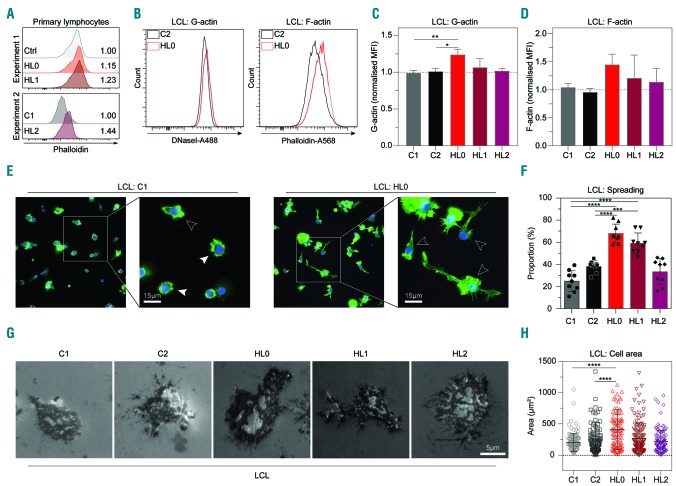
To understand how the deletion in MKL1 intron 1 affected actin content and responses, we examined F-actin content in primary lymphocytes by flow cytometry. No difference was seen when comparing HL0 and HL1 to control lymphocytes while HL2 had a higher F-actin content compared to that of C1 (Figure 2A). In primary monocytes gated based on forward and side scatter profiles by flow cytometry, the triplets’ cells had higher MKL1 expression (Online Supplementary Figure S1C) and higher F-actin content (Online Supplementary Figure S1D). Monocyte-derived dendritic cells from whole blood of the controls and HL patients displayed a similar ability to form podosomes (Online Supplementary Figure S4A, B). To exclude a possible effect of cell size and different hematopoietic cell subsets of primary cells, we compared actin content in cells from the triplets’ LCL with those of controls side by side by alternately labeling one cell population with anti-CD54 antibodies after fixation and before mixing the two populations. Thereafter, we detected G-actin using DNAse1, and F-actin using phalloidin (Figure 2B and Online Supplementary Figure S1E, F). HL0 cells had increased G-actin and a tendency for increased F-actin (Figure 2B-D). To examine whether increased actin content had any impact on cytoskeletal rearrangement, the triplets’ and control LCL were examined microscopically on glass surfaces coated with fibronectin and anti-CD19 antibodies. HL0 and HL1 cells had increased capacity to spread, with formation of long dendritic protrusions (Figure 2E, F and Online Supplementary Figure S5A), and increased adhesive area measured by interference reflection microscopy (Figure 2G, H). This indicates that the deletion in MKL1 intron 1 was associated with increased actin content and actin-dependent B-cell spreading.
Figure 2.
Increased actin content and actin-dependent spreading in HL0 and HL1 cells. (A) Phalloidin expression in primary lymphocytes determined by flow cytometry. Experiment 1: Control (Ctrl), HL0, and HL1. Experiment 2: C1 and HL2. Numbers indicate fold-change in expression normalized to ctrl and C1, respectively. (B) Representative flow cytometry plot of G-actin and F-actin content in C2 and HL0 and (C, D) quantification of G- and F-actin content in control and the triplets’ lymphoblastoid cell lines (LCL). (E) Immunocytochemistry of LCL adhering to glass slides coated with fibronectin and anti-CD19 antibody. Spread cells are defined as those with lamellipodia-like structures and/or long protrusions. White arrowheads indicate examples of spread cells; black arrowheads indicate non-spread cells. Cells stained with phalloidin-Alexa488 (green color) and mounted in Vectashield Antifade Mounting Medium with 4′,6-diamidino-2- phenylindole (DAPI, blue color). Original magnification ×400. (F) Proportion of spread LCL. The experiment was repeated three times and cell spreading was determined each time in three separate fields of view. Total cells counted: C1: 800; C2: 1356; HL0: 1081; HL1: 1062; HL2: 983; and 200 cells or more were counted per sample per experiment. (G) Interference reflection microscopy of LCL adhering to cover slips coated with fibronectin and anti-CD19 antibody. Original magnification ×630. (H) Area of LCL spread on coverslips. Total cells counted: C1: 119; C2: 109; HL0: 109; HL1: 131; HL2: 123. (C, D, F) Combined data from three experiments. (H) Combined data from two experiments. (A) The experiments with primary lymphocytes were performed once. (B) Representative histograms. (E, G) Representative images. For bar graphs, the dotted line indicates normalization to the mean of C1 and C2. All panels display data from LCL except (A), which displays data from primary lymphocytes. Error bars represent the standard deviation of the measurements. Symbols represent technical replicates from three independent experiments in (F), and pooled single values from three independent experiments in (H). All data were analyzed using analysis of variance with the post-hoc Tukey test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
HL0 cells display decreased aggregation and reduced CD11a integrin expression
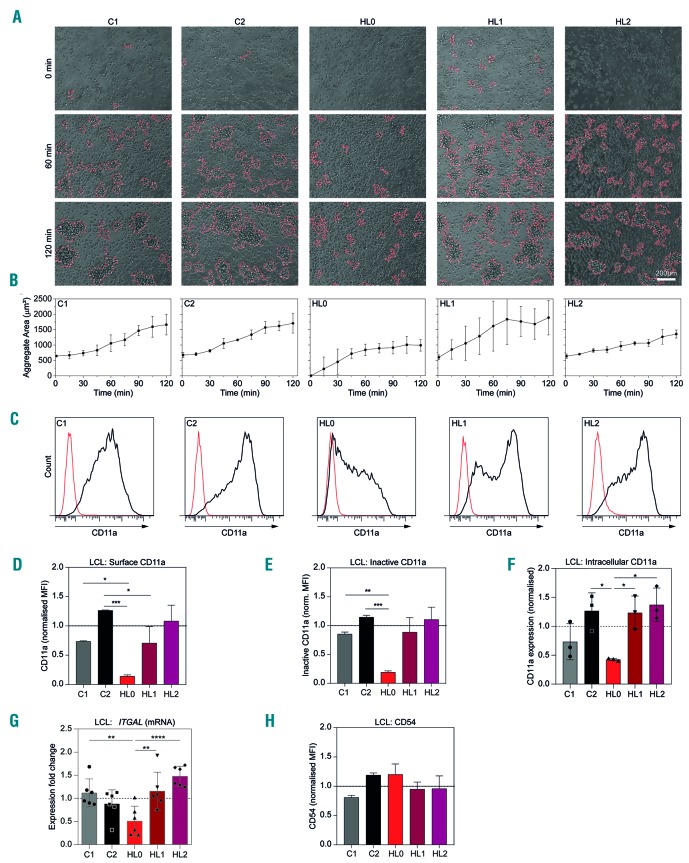
LCL grow in clusters by homotypic aggregation dependent on leukocyte function antigen-1 (LFA-1, consisting of subunits CD11a/integrin αL and CD18/integrin β2) and intercellular adhesion molecule-1 (ICAM-1/CD54) (Online Supplementary Figure S6).22–24 When culturing LCL, we observed decreased clustering of the triplets’ cells, an effect that was especially pronounced for HL0 LCL (Online Supplementary Figure S5B). To quantify this, we measured aggregation from single cell suspensions by live cell imaging over 2 h. LCL from C1, C2, HL1, and HL2 formed aggregates instantly, whereas those from HL0 displayed impaired aggregation with delayed formation of small aggregates over 2 h (Figure 3A, B). We investigated whether decreased aggregation resulted from altered surface expression of adhesion receptors. C1, C2, HL1, and HL2 cells displayed similar expression of total surface and intracellular CD11a, and inactive CD11a (Figure 3C-G and Online Supplementary Figure S5C). HL0 cells displayed lower expression of total surface and intracellular CD11a, and inactive CD11a as well as lower CD11a mRNA (ITGAL) (Figure 3C-G and Online Supplementary Figure S5C). Interestingly, cells lacking MKL1 expression19 showed higher surface CD11a expression when compared to control cells (Online Supplementary Figure S4C). LCL from the triplet and controls had similar expression of CD54 (Figure 3H and Online Supplementary Figure S5C). This shows that HL0 cells had reduced capacity to form aggregates and this was associated with low expression of CD11a.
Figure 3.
HL0 cells display decreased aggregation and reduced CD11a integrin expression. (A) Aggregation of lymphoblastoid cell lines (LCL), representative images. (B) Average area of aggregates observed in (A). (C) Representative flow cytometry plots of total CD11a surface expression in LCL. (D) Quantification of CD11a surface expression as seen in (C). (E, F) Quantification of inactive CD11a surface expression and intracellular CD11a expression as seen in Online Supplementary Figure S3. (G) Expression of ITGAL (CD11a) by real-time quantitative polymerase chain reaction. (H) Quantification of CD54 surface expressio. (B, D-F) Combined data from three experiments. (G-H) Combined data from two experiments. For bar graphs, the dotted line indicates normalization to the mean of C1 and C2. All panels display data from LCL. Error bars represent the standard deviation of the measurements. Symbols represent technical replicates from three independent experiments in (G), and single values from three independent experiments in (F). All data were analyzed using analysis of variance with the post-hoc Tukey test. *P<0.05, **P<0.01, ***P<0.001.
HL0 cells display increased proliferation and genomic instability
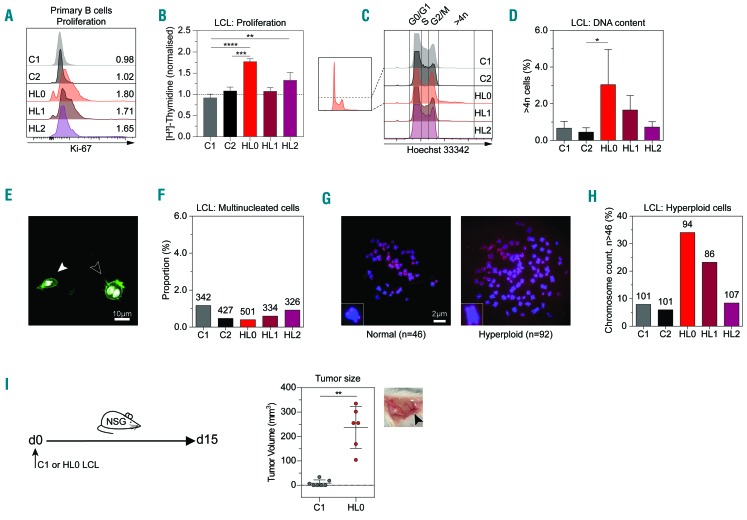
HL has previously been associated with immunodeficiency and altered proliferation.25,26 To examine for immunodeficiency, we used flow-cytometric assay of specific cell-mediated immune response in activated whole blood (FASCIA) analysis to detect B- and T-cell responses to various antigens.27 We found that the triplets’ B cells displayed a decreased response to poke-weed mitogen but had normal T-cell responses (Online Supplementary Table S1). To address how increased MKL1 expression correlated to proliferation, primary B cells were cultured with anti-CD40 antibodies and interleukin-4 for 72 h and labeled with the proliferation marker Ki-67 (Figure 4A and Online Supplementary Figure S5D). The triplets’ samples had more Ki-67+ proliferating B cells than those of the controls (Figure 4A). To exclude that any variation was due to sample collection time when using Ki-67, we measured proliferation in LCL by DNA synthesis over 20 h using 3H-thymidine incorporation. Compared to controls, HL0 cells had greater DNA synthesis (Figure 4B). To examine whether proliferation rate was associated with cell cycle alterations, we used DNA-specific Hoechst labeling to evaluate cells in the G0/G1 (2n DNA content), S, and G2/M (4n DNA content) phases. The proportion of cells in S or G2/M phase was higher in HL0 LCL (Figure 4C). Three percent of HL0 cells had more than 4n DNA content, compared to 0–1% of control cells (Figure 4D). Since EBV has been shown to induce multiple nuclei in transformed cells,28 we counted nuclei in the LCL from the triplets and controls to evaluate whether multinuclearity could explain the proportion of cells with more than 4n DNA content. No correlation was seen between multinuclearity and DNA content (Figure 4E, F), indicating that increased DNA content could have resulted from genomic instability. When arresting LCL from the triplets and controls in metaphase, HL0 and HL1 had higher proportions of cells (approximately 25–35%) with more than 46 chromosomes (Figure 4G, H). To investigate the growth of HL0 and C1 cells in vivo, the cells were injected subcutaneously into NSG mice that lack a functional immune response and cell growth was examined as tumor mass on day 9. C1 cells failed to form a tumor mass whereas HL0 cells formed tumors with distinct angiogenesis (Figure 4I). Together these observations suggest that increased MKL1 expression and activity is directly associated with hyperproliferation and genomic instability leading to formation of a tumor mass in vivo.
Figure 4.
HL0 cells display increased proliferation and genomic instability. (A) Expression of the proliferation marker Ki-67 by flow cytometry on primary B cells cultured 48 h with interleukinIL-4 and anti-CD40 antibodies. Numbers indicate fold-change of expression normalized to the mean of C1 and C2. (B) DNA synthesis rate as measured by radiation from incorporated 3H-thymidine after 48 h. (C) Proportions of cell cycle phases (G0/G1, S and G2/M) determined by Hoechst 33342 staining, measured by flow cytometry (representative histograms). Left graph shows how the Y-axis was cut to allow for emphasized visualization of hyperploid cells with >4n of DNA content, quantified in (D). (E) Comparison of mononuclear (indicated by white arrowhead) and multinuclear (black arrowhead) cells of lymphoblastoid cell lines (LCL). White color: 4′,6-diamidino-2- phenylindole (DAPI); green color: phalloidin-Alexa488. (F) Proportion of multinucleated LCL cells, as assessed by manual microscopical counting of nuclei stained with DAPI. Numbers indicate counted cells for each sample. (G) Telomere-fluorescence in situ hybridization on chromosomes in metaphase. Representative images of a normal cell with 46 chromosomes and a hyperploid cell with 92 chromosomes; insets show magnified representative chromosomes. Original magnification ×1000. Chromosomes hybridized with TelG-Cy3 PNA probe (red color) and mounted in Vectashield Antifade Mounting Medium with DAPI (blue color). (H) Proportion of metaphases with a hyperploid amount of chromosomes (>46). Numbers indicate counted metaphases for each sample. (I) Subcutaneous injection of C1 and HL0 LCL in NSG mice and measurement of tumor mass at day 9. Each circle represents one mouse. Right subpanel: representative image of tumor mass assessed at the endpoint. Black arrowhead indicates angiogenesis. (A) Data from one experiment. (B, D, F, H) Combined data from three experiments. (I) Combined data from two experiments. For bar graphs, the dotted line indicates normalization to the mean of C1 and C2. All panels display data from LCL except (A), which displays data from primary B cells. Error bars represent the standard deviation of the measurements. All data were analyzed using analysis of variance with the post-hoc Tukey test, except (I) that was analyzed using a t-test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
HL1 cells contain two distinct populations of which one shows an HL0 phenotype
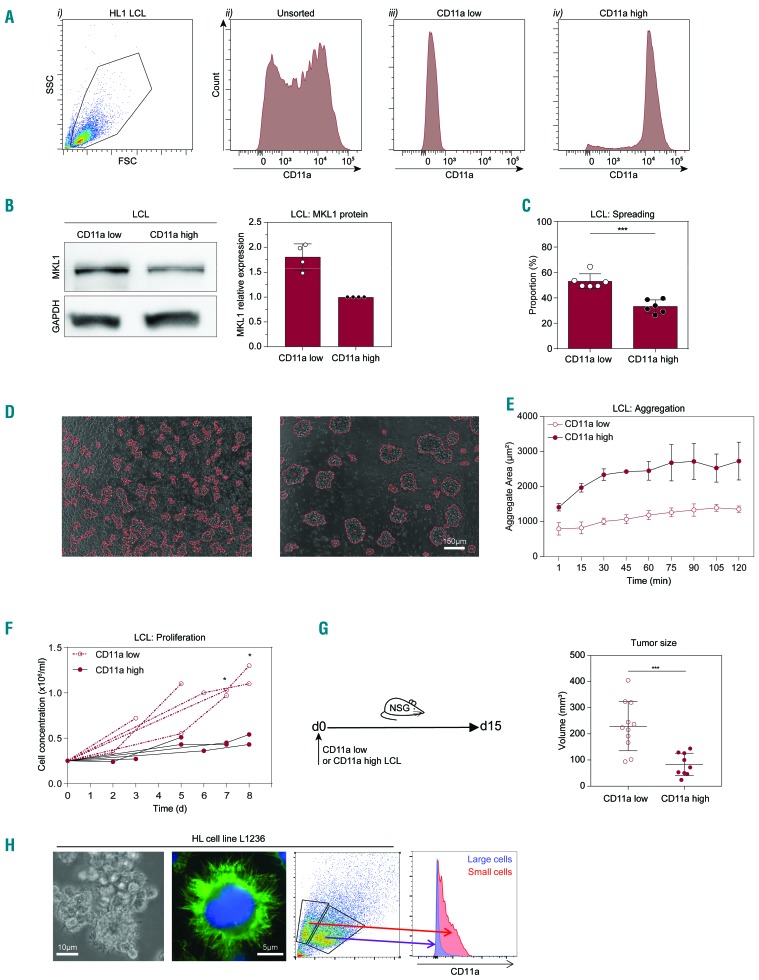
We noticed that HL1 cells recurrently showed an intermediate phenotype when compared to HL0 and control cells. HL1 cells showed a bimodal expression of CD11a and we examined whether this represented two populations of cells with different phenotypes. We used fluorescence activated cell sorting (FACS) to sort out CD11a low and CD11a high cells from the HL1 cells (Figure 5A). PCR of genomic DNA showed that sorted HL1 CD11a low and HL1 CD11a high cells contained the heterozygous deletion of MKL1 intron 1 (Online Supplementary Figure S2A). CD11a low cells had increased MKL1 protein and increased cell spreading when compared to CD11a high cells (Figure 5B, C). We quantified aggregation over 2 h and found that CD11a low cells had a reduced capacity to aggregate compared to that of CD11a high cells (Figure 5D, E). To examine proliferation, we sorted 250,000 CD11a low and CD11a high cells and counted them between days 3 and 8. The CD11a high cells showed only modest proliferation whereas the CD11a low cells expanded 3- to 4-fold in the period between day 3 and day 8 (Figure 5F). To examine proliferation of cells in vivo, we injected CD11a low and CD11a high cells into NSG mice and determined tumor growth on day 15. CD11a low cells formed a large tumor mass with visible angiogenesis, whereas CD11a high cells formed a smaller tumor mass (Figure 5G). To address whether the control C1 and C2 cells also contained a stable CD11a low population, we used FACS and cultured the C1 and C2 CD11a low and CD11a high cells for 17 days. The C1 and C2 CD11a low cells gained expression of CD11a during the culture, suggesting that the CD11a phenotype in control cells was unstable (Online Supplementary Figures S7 and S8). We next examined whether alterations in CD11a expression were present in the L1236 HL cell line derived from a primary isolate of a patient with advanced HL.29 The L1236 cells contained large Reed-Sternberg-like cells and smaller cells (Figure 5H). Using flow cytometry, small and large cells were identified based on forward and side scatter. Small cells had low expression of CD11a and the large cells were devoid of CD11a expression (Figure 5H). Together, these data suggest that low expression of CD11a may be a characteristic feature of pre-malignant B cells in HL.
Figure 5.
HL1 cells comprise two distinct populations of which one shows an HL0 phenotype. (A) (i) Representative flow cytometry plots from eight experiments showing gating on lymphoblastoid cell lines (LCL) using side scatter (SSC) vs. forward scatter (FSC). (ii) CD11a expression in HL1 cells. HL1 cells were sorted according to CD11a surface expression resulting in the two populations, CD11a low and CD11a high, cells shown in (iii) and (iv), respectively. (B) MKL1 expression in CD11a low and CD11a high LCL was evaluated by western blot. Left subpanel: representative western blot showing MKL1 expression in CD11a low and CD11a high cells. Right subpanel: quantification by densitometry of MKL1 expression relative to GAPDH expression in four independent experiments. (C) Proportion of spread LCL. CD11a low and high cells were allowed to spread on glass slides coated with fibronectin and anti-CD19 antibody. The experiment was repeated two times and cell spreading was determined each time in three separate fields of view. Total cells counted: CD11a low: 689; CD11a high: 579; and 200 cells or more were counted per sample per experiment. (D) Aggregation of CD11a low and high LCL, representative images. (E) Average area of aggregates. Results from three independent experiments. (F) Concentration of CD11a low and CD11a high LCL cells over 8 days. Results from five independent experiments. (G) Left subpanel: CD11a low or CD11a high LCL cells were injected subcutaneously into NSG mice and the tumor size was measured 15 days later. Right subpanel: quantification of the tumor size in NSG mice injected with either CD11a low or high LCL cells. Results from three independent experiments. (H) Microscopy of the L1236 HL cell line. Green: F-actin; blue: nuclear stain. CD11a expression determined by flow cytometry. Symbols represent single values from four independent experiments in (B) and (F), and technical replicates from two and three independent experiments in (C) and (G), respectively. Error bars represent the standard deviation of the measurements. All data were analyzed using t-tests. *P<0.05, ***P<0.001.
Inhibition of MKL1 activity in HL0 cells induces a phenotype similar to that of control cells
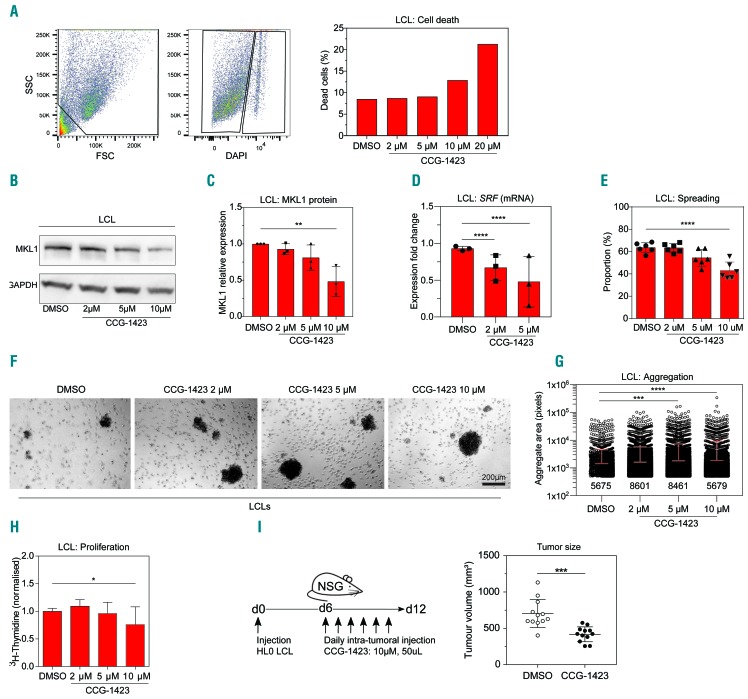
We next investigated whether we could revert the phenotype of HL0 cells to that of control cells using the MKL1 small molecule inhibitor CCG-1423.17,18 To define the dose range, HL0 cells were treated with different doses of CCG-1423 and cell death measured by flow cytometry. At doses of 2-10 μM CCG-1423, 90% of cells were viable (Figure 6A). HL0 cells treated with CCG-1423 displayed dose-dependent lowering of MKL1 protein and decreased SRF expression (Figure 6B-D). The spreading capacity of HL0 cells was reduced upon CCG-1423 treatment (Figure 6E). Lowering MKL1 protein and activity led to increased aggregate formation and reduced proliferation (Figure 6F-H). To examine whether the MKL1 inhibitor could suppress tumor growth in vivo, HL0 cells were injected into NSG mice by subcutaneous injections. From day 6 to day 12, a daily dose of 10 μM CCG-1423 was injected intratumorally. Compared to treatment with vehicle (dimethylsulfoxide), treatment with the MKL1 inhibitor led to reduced HL0 tumor mass in mice (Figure 6I).
Figure 6.
Inhibition of MKL1 activity in HL0 cells induces a phenotype similar to that of control cells. (A) Left and middle subpanels: representative flow cytometry plots showing gating for size, granularity, and 4′,6-diamidino-2- phenylindole (DAPI) fluorescence intensity as a measure of the proportion of dead cells of HL0 lymphoblastoid cell lines (LCL) treated with 10 μM of CCG-1423 for 24 h. Right subpanel: quantification of death of HL0 cells treated with dimethylsulfoxide (DMSO), as the control, or CCG-1423 for 24 h. Representative graph of three independent experiments. (B) Representative image of MKL1 protein expression in HL0 LCL treated with DMSO or CCG-1423 for 24 h. (C) Quantification of MKL1 expression levels normalized to GAPDH in HL0 LCL treated with DMSO or CCG-1423 for 24 h. Results from three independent experiments. (D) Expression of SRF mRNA, determined by real-time quantification polymerase chain reaction, in HL0 LCL treated with DMSO or CCG-1423 for 24 h. Results from three independent experiments. (E) Proportion of spread HL0 LCL after treatment with DMSO or CCG-1423 for 48 h. The experiment was repeated twice and cell spreading was determined each time in three separate fields of view. Total cells counted: DMSO: 541; 2 μM: 517; 5 μM: 454; 10 μM: 271; and 100 cells or more were counted per sample per experiment. (F) Aggregation in culture of HL0 LCL treated with DMSO or CCG-1423 for 24 h. More than 15 fields were randomly chosen for quantification. Original magnification ×40. (G) Quantification of aggregation observed in (F) using ImageJ. Data representative of two independent experiments. Numbers indicate counted aggregates for each sample. (H) DNA synthesis rate in HL0 LCL as measured by radiation from incorporated 3H-thymidine after 48 h of treatment with DMSO or CCG-1423. (I) HL0 LCL cells were injected into NSG mice. From day 6, 50 μL of DMSO or 10 μM of CCG-1423 were injected intratumorally daily for 6 days. Tumor size was measured on day 12. Error bars represent the standard deviation of the measurements. Symbols represent single values from three independent experiments in (C), mean values in (D), and technical replicates from two independent experiments in (C) and (G). All data were analyzed using analysis of variance with the post-hoc Tukey test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
Dysregulation of MKL1 expression and the actin cytoskeleton has been implicated in hematologic malignancies, although the exact mechanism has not been determined (Online Supplementary Figure S9).30,31 Mutated MKL1 was originally described in patients with acute megakaryoblastic leukemia in whom MKL1 was fused with RBM15 by a chromosomal translocation.13,14 The role of MKL1 in megakaryocyte differentiation, migration, and in the formation of proplatelets was subsequently described in MKL1-deficient mice.32,33 In humans, MKL1 deficiency has a profound affect on the hematopoietic cell actin cytoskeleton resulting in severely impaired cell migration and phagocytosis.19 Genetically identical triplets with a large deletion in MKL1 intron 1, of whom two affected by HL and one undiagnosed, provided a unique opportunity to investigate MKL1 and B-cell responses in the pathogenesis of HL. We found that increased activity of MKL1 in B cells led to the classical hallmarks of cancer cells: hyperproliferation, genomic instability, and formation of tumors with induction of angiogenesis.34
In this study we mainly used EBV-transformed B cells (LCL) from two controls and the triplets. To minimize possible variation due to EBV transformation, all samples were EBV-transformed under the same conditions with regards to day of transduction, virus batch and concentration. The EBV cells enabled an extensive investigation of the impact of the MKL1 intron 1 deletion on B cells and importantly they allowed us to perform controlled experiments in which cells from the controls and the triplets could be compared side by side. A limitation with EBV transformation is that highly proliferative B cells are favored during EBV transformation.35 Moreover, the donor distribution of naïve, non-switched memory, and switched memory B cells is reflected in the EBV-transformed B cells.35,36 We cannot exclude that the EBV transformation induced expansion of a dominant subclone; however, CD11a staining revealed variable expression of CD11a in each sample although with different distributions. In fact, the variable CD11a expression allowed us to define different subpopulations in the HL0 and HL1 samples and argue against a dominant subclone upon EBV transformation.
Since the deletion in MKL1 is intronic, we did not anticipate any changes in the amino acid sequence and found that the MKL1 expressed was of normal size. The first intron is often essential for the regulation of gene expression,37,38 which is why we investigated possible alterations of MKL1 expression in the triplets’ cells and found that those from the undiagnosed triplet (HL0) displayed the highest expression of MKL1 while cells from HL1 and HL2, the triplets who were successfully treated for HL, showed an intermediate phenotype. This suggests a phenotypic difference between the triplets based on the onset and treatment of the disease. MKL1, via its interaction with SRF, regulates expression of hundreds of genes involved in cell migration, adhesion, and differentiation.15,39 We show that the high mRNA and protein expression of MKL1 in HL0 cells correlated with high expression of the MKL1 target genes SRF and ACTB. MKL1 activity promotes migration and proliferation of cancer cells,16,40,41 suggesting that HL0 LCL could be pre-malignant.
Because the absence of MKL1 results in a severely impaired actin cytoskeleton,19 we reasoned that overexpression of MKL1 could result in a more active actin cytoskeleton. Actin cytoskeleton proteins are upregulated in invasive cancer,42 suggesting that the increased activity of the actin cytoskeleton in the HL0 cells could enhance cell migration and cell division, promoting invasion of cells and growth of tumors. In support of this, HL0 cells with the highest MKL1 expression had higher G- and F-actin content and displayed increased spreading, a process that depends on actin cytoskeleton dynamics. The migration and hyperproliferation of cancer cells are also regulated by alteration of integrin expression at the cell surface and modification of the subsequent intracellular integrin signaling.43 EBV-transformed B cells aggregate in cell culture via the interaction between the integrin CD11a (a subunit of LFA-1) and the adhesion molecule ICAM-1 (this study).22–24 We examined LCL aggregation in vitro and observed that HL0 cells aggregated poorly, while HL1 cell aggregation varied and HL2 cell aggregation was similar to that of control cells. We found that the levels of surface and total protein as well as mRNA expression of CD11a were low in HL0 cells, but similar to control levels in HL2 cells. Interestingly, CD11a expression in HL1 cells showed a bimodal distribution with CD11a low and CD11a high cells, which might be the cause of the high variation of HL1 cell aggregation results. It is possible that the overactive actin cytoskeleton in HL0 cells prevented correct translocation of adhesion receptors to the cell surface or affected integrin inside-out and outside-in signaling.
Because of the possible pre-malignant stage of HL0 cells, we investigated cell proliferation in vitro and in vivo through formation of tumor mass. HL0 cells were hyper-proliferative in vitro and also in vivo where they rapidly formed tumors in immunocompromised NSG mice. The HL0 population showed increased DNA content and a higher percentage of hyperploid cells. Because polyploidy is a result of incomplete cytokinesis,44 the increased hyperploidy of HL0 cells could be due to an elevated rate of cytokinesis failure, possibly connected to overactivity of the actin cytoskeleton as shown previously when the actin regulator WASp is overactive.45,46 Reed-Sternberg cells originate from failed cytokinesis and re-fusion of HL daughter cells.47,48 This suggests that the failure of HL0 cells to complete cytokinesis could lead to the formation of giant multinucleated cells similar to Reed-Sternberg cells.
The variable phenotype of the HL1 and HL2 triplets treated for HL in 1985 and 2008, respectively, was at first puzzling. Since all triplets contained the heterozygous deletion of MKL1 intron 1, we reasoned that whether a cell expresses the healthy MKL1 allele or the intron 1-deleted allele of MKL1 must be a stochastic event. Transcription of the MKL1 gene is quite complex, with several sets of transcripts. The transcriptional start site is in exon 4, placing intron 1 in the 5’ untranslated region of MKL1. To understand the MKL1-associated phenotype, we took advantage of the bimodal expression of CD11a in HL1 cells to sort out CD11a low and CD11a high cells. CD11a high cells behaved similarly to the control cells, displaying low expression of MKL1, low spreading and proliferation, together with a strong aggregation response. In contrast, CD11a low cells behaved similarly to HL0 cells with high expression of MKL1, decreased aggregation, as well as increased proliferation in vitro and tumor formation in vivo. We also sorted out the small population of CD11a low cells from control (C1 and C2) cells; however, control CD11a low cells were not stable in culture and started to express CD11a during 17 days of culture. This suggests that the stable CD11a low phenotype in HL0 cells and sorted HL1 cells is associated with increased MKL1 expression. Interestingly, HL Reed-Sternberg cells are characterized by low mRNA expression of CD11a.49,50 Thus, since the HL0 triplet has not received HL treatment we reason that HL0 cells could represent a pre-HL stage. The presence of CD11a low cells in HL1, who was treated for HL with mustargen, oncovin, procarbazine, prednisone/adriamycin, bleomycin, vinblastine, and dacarbazine in 1985,20 may indicate de novo HL with the CD11a low cells possibly outgrowing the healthy CD11a high cells. With this reasoning, the HL2 patient who received adriamycin, bleomycin, vinblastine and dacarbazine to treat HL in 200820 should have predominantly CD11a high cells, assuming that the highly proliferative CD11a low cells would have been mostly eradicated by the HL treatment.
Finally, we investigated whether the HL0 cell phenotype could be reverted by inhibition of MKL1 using the small molecule CCG-1423.17,18 HL0 cells treated with this MKL1 inhibitor showed decreased expression of MKL1 protein and the MKL1 target gene SRF, as well as reduced cell spreading. Importantly, using the MKL1 inhibitor we could revert the hyperproliferation of HL0 cells in vitro and also, by intratumoral injections, in vivo.
We present here the first example of an intron mutation in MKL1, resulting in increased MKL1 expression, increased actin content, decreased aggregation, hyperproliferation, and genomic instability in B cells. Of the triplets’ samples, those from HL0 showed the most pronounced difference in both MKL1 expression and activity, as well as in cellular responses when compared to controls. Cells from HL1 and HL2, successfully treated for HL, had a phenotype closer to that of healthy controls. This finding, together with the known role of MKL1 in metastasis15 and with high mRNA expression of MKL1 in many types of lymphomas, suggests that increased MKL1 expression actively participates in B-cell transformation and the pathogenesis of HL.
Acknowledgments
We are grateful to the family and healthy controls for providing blood samples. We thank Dr Kaisa Lehti, Dr Peter Bergman, and Dr Andreas Lundqvist for valuable input. This work was supported by a postdoctoral fellowship from the Swedish Childhood Cancer Fund and the Swedish Society of Medical Research to JR, an MD-PhD (CSTP) fellowship and a clinical internship (research AT) from Karolinska Institutet to AS, a clinical postdoctoral fellowship from the Swedish Society of Medical Research to HB, a postdoctoral fellowship from Olle Engqvist Byggmästare to MH, a PhD fellowship from Fundação para a Ciência e a Tecnologia to MMSO, funds from the Swedish Medical Society to HB and LSW, Groschinsky Foundation and Åke Wiberg Foundation to HB and LSW, the Swedish Cancer Society to MB and LSW, as well as the Swedish Research Council, the Swedish Childhood Cancer Fund, Karolinska Institutet, the European Commission 7th Framework Program, Åke Olsson Foundation, Jeansson Foundation, and Bergvall Foundation to LSW. LSW is a Ragnar Söderberg Fellow in Medicine.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/5/1339
References
- 1.Borchmann S, Engert A. The genetics of Hodgkin lymphoma: an overview and clinical implications. Curr Opin Oncol. 2017;29(5):307–314. [DOI] [PubMed] [Google Scholar]
- 2.Goldin LR, Bjorkholm M, Kristinsson SY, et al. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009;146(1):91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanzler H, Kuppers R, Hansmann ML, et al. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184(4):1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin-Subero JI, Klapper W, Sotnikova A, et al. Chromosomal breakpoints affecting immunoglobulin loci are recurrent in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Cancer Res. 2006;66(21):10332–10338. [DOI] [PubMed] [Google Scholar]
- 5.Bechtel D, Kurth J, Unkel C, et al. Transformation of BCR-deficient germinal-center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and posttransplantation lymphomas. Blood. 2005;106(13):4345–4350. [DOI] [PubMed] [Google Scholar]
- 6.Mancao C, Altmann M, Jungnickel B, et al. Rescue of “crippled” germinal center B cells from apoptosis by Epstein-Barr virus. Blood. 2005;106(13):4339–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang DZ, Li S, Hockemeyer D, et al. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci U S A. 2002;99(23):14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miralles F, Posern G, Zaromytidou AI, et al. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113(3):329–342. [DOI] [PubMed] [Google Scholar]
- 9.Cen B, Selvaraj A, Burgess RC, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23(18):6597–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer JA, Misra RP. Expression of the serum response factor gene is regulated by serum response factor binding sites. J Biol Chem. 1996;271(28):16535–16543. [DOI] [PubMed] [Google Scholar]
- 12.Hermann MR, Jakobson M, Colo GP, et al. Integrins synergise to induce expression of the MRTF-A-SRF target gene ISG15 for promoting cancer cell invasion. J Cell Sci. 2016;129(7):1391–1403. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Morris SW, Valentine V, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28(3):220–221. [DOI] [PubMed] [Google Scholar]
- 14.Mercher T, Coniat MB, Monni R, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci U S A. 2001;98(10):5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medjkane S, Perez-Sanchez C, Gaggioli C, et al. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11(3):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muehlich S, Hampl V, Khalid S, et al. The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene. 2012;31(35):3913–3923. [DOI] [PubMed] [Google Scholar]
- 17.Evelyn CR, Wade SM, Wang Q, et al. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6(8):2249–2260. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K, Watanabe B, Nakagawa Y, et al. RPEL proteins are the molecular targets for CCG-1423, an inhibitor of Rho signaling. PLoS One. 2014;9(2):e89016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Record J, Malinova D, Zenner HL, et al. Immunodeficiency and severe susceptibility to bacterial infection associated with a loss-of-function homozygous mutation of MKL1. Blood. 2015;126(13):1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjorkholm M, Sjoberg J, Nygell UA, et al. Development of Hodgkin lymphoma in homozygotic triplets with constitutional deletion in MKL1. Blood. 2013;121(23):4807. [DOI] [PubMed] [Google Scholar]
- 21.Nagy N, Adori M, Rasul A, et al. Soluble factors produced by activated CD4+ T cells modulate EBV latency. Proc Natl Acad Sci U S A. 2012;109(5):1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patarroyo M, Beatty PG, Nilsson K, Gahmberg CG. Identification of a cell-surface glycoprotein mediating cell adhesion in EBV-immortalized normal B cells. Int J Cancer. 1986;38(4):539–547. [DOI] [PubMed] [Google Scholar]
- 23.Gronberg A, Halapi E, Ferm M, Petersson M, Patarroyo M. Regulation of lymphocyte aggregation and proliferation through adhesion molecule CD54 (ICAM-1). Cell Immunol. 1993;147(1):12–24. [DOI] [PubMed] [Google Scholar]
- 24.Rothlein R, Springer TA. The requirement for lymphocyte function-associated antigen 1 in homotypic leukocyte adhesion stimulated by phorbol ester. J Exp Med. 1986;163(5):1132–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merk K, Bjorkholm M, Tullgren O, Mellstedt H, Holm G. Immune deficiency in family members of patients with Hodgkin’s disease. Cancer. 1990;66(9):1938–1943. [DOI] [PubMed] [Google Scholar]
- 26.Bjorkholm M, Holm G, Mellstedt H. Persisting lymphocyte deficiences during remission in Hodgkin’s disease. Clin Exp Immunol. 1977;28(3):389–393. [PMC free article] [PubMed] [Google Scholar]
- 27.Marits P, Wikstrom AC, Popadic D, Winqvist O, Thunberg S. Evaluation of T and B lymphocyte function in clinical practice using a flow cytometry based proliferation assay. Clin Immunol. 2014;153(2):332–342. [DOI] [PubMed] [Google Scholar]
- 28.Shumilov A, Tsai MH, Schlosser YT, et al. Epstein-Barr virus particles induce centrosome amplification and chromosomal instability. Nat Commun. 2017;8:14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf J, Kapp U, Bohlen H, et al. Peripheral blood mononuclear cells of a patient with advanced Hodgkin’s lymphoma give rise to permanently growing Hodgkin-Reed Sternberg cells. Blood. 1996;87(8):3418–3428. [PubMed] [Google Scholar]
- 30.Verrills NM, Liem NL, Liaw TY, Hood BD, Lock RB, Kavallaris M. Proteomic analysis reveals a novel role for the actin cytoskeleton in vincristine resistant childhood leukemia–an in vivo study. Proteomics. 2006;6(5):1681–1694. [DOI] [PubMed] [Google Scholar]
- 31.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13(10):900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilles L, Bluteau D, Boukour S, et al. MAL/SRF complex is involved in platelet formation and megakaryocyte migration by regulating MYL9 (MLC2) and MMP9. Blood. 2009;114(19):4221–4232. [DOI] [PubMed] [Google Scholar]
- 33.Smith EC, Thon JN, Devine MT, et al. MKL1 and MKL2 play redundant and crucial roles in megakaryocyte maturation and platelet formation. Blood. 2012;120(11):2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 35.Dorner M, Zucol F, Berger C, et al. Distinct ex vivo susceptibility of B-cell subsets to epstein-barr virus infection according to differentiation status and tissue origin. J Virol. 2008;82(9):4400–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heath E, Begue-Pastor N, Chaganti S, et al. Epstein-Barr virus infection of naive B cells in vitro frequently selects clones with mutated immunoglobulin genotypes: implications for virus biology. PLoS Pathog. 2012;8(5):e1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang GL, Li BK, Zhang MY, et al. Allele loss and down-regulation of heparanase gene are associated with the progression and poor prognosis of hepatocellular carcinoma. PLoS One. 2012;7(8):e44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol. 2017;91(Pt B):145–155. [DOI] [PubMed] [Google Scholar]
- 39.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340(6134):864–867. [DOI] [PubMed] [Google Scholar]
- 40.Cheng X, Yang Y, Fan Z, et al. MKL1 potentiates lung cancer cell migration and invasion by epigenetically activating MMP9 transcription. Oncogene. 2015;34(44):5570–5581. [DOI] [PubMed] [Google Scholar]
- 41.Hu Q, Guo C, Li Y, Aronow BJ, Zhang J. LMO7 mediates cell-specific activation of the Rho-myocardin-related transcription factor-serum response factor pathway and plays an important role in breast cancer cell migration. Mol Cell Biol. 2011;31(16):3223–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773(5):642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamidi H, Pietila M, Ivaska J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br J Cancer. 2016;115(9):1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen HG, Ravid K. Polyploidy: mechanisms and cancer promotion in hematopoietic and other cells. Adv Exp Med Biol. 2010;676:105–122. [DOI] [PubMed] [Google Scholar]
- 45.Moulding DA, Moeendarbary E, Valon L, Record J, Charras GT, Thrasher AJ. Excess F-actin mechanically impedes mitosis leading to cytokinesis failure in X-linked neutropenia by exceeding Aurora B kinase error correction capacity. Blood. 2012;120(18):3803–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westerberg LS, Meelu P, Baptista M, et al. Activating WASP mutations associated with X-linked neutropenia result in enhanced actin polymerization, altered cytoskeletal responses, and genomic instability in lymphocytes. J Exp Med. 2010;207(6):1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rengstl B, Newrzela S, Heinrich T, et al. Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells. Proc Natl Acad Sci U S A. 2013;110(51): 20729–20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuceu C, Hempel WM, Sabatier L, Bosq J, Carde P, M’Kacher R. Chromosomal instability in Hodgkin lymphoma: an in-depth review and perspectives. Cancers. 2018;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwering I, Brauninger A, Klein U, et al. Loss of the B-lineage-specific gene expression program in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;101(4):1505–1512. [DOI] [PubMed] [Google Scholar]
- 50.Tiacci E, Doring C, Brune V, et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood. 2012;120(23):4609–4620. [DOI] [PubMed] [Google Scholar]