Abstract
Despite substantial progress in treatment of T-cell acute lymphoblastic leukemia (T-ALL), mortality remains relatively high, mainly due to primary or acquired resistance to chemotherapy. Further improvements in survival demand better understanding of T-ALL biology and development of new therapeutic strategies. The Notch pathway has been involved in the pathogenesis of this disease and various therapeutic strategies are currently under development, including selective targeting of NOTCH receptors by inhibitory antibodies. We previously demonstrated that the NOTCH1-specific neutralizing antibody OMP52M51 prolongs survival in TALL patient-derived xenografts bearing NOTCH1/FBW7 mutations. However, acquired resistance to OMP52M51 eventually developed and we used patient-derived xenografts models to investigate this phenomenon. Multi-level molecular characterization of T-ALL cells resistant to NOTCH1 blockade and serial transplantation experiments uncovered heterogeneous types of resistance, not previously reported with other Notch inhibitors. In one model, resistance appeared after 156 days of treatment, it was stable and associated with loss of Notch inhibition, reduced mutational load and acquired NOTCH1 mutations potentially affecting the stability of the heterodimerization domain. Conversely, in another model resistance developed after only 43 days of treatment despite persistent down-regulation of Notch signaling and it was accompanied by modulation of lipid metabolism and reduced surface expression of NOTCH1. Our findings shed light on heterogeneous mechanisms adopted by the tumor to evade NOTCH1 blockade and support clinical implementation of antibody-based target therapy for Notch-addicted tumors.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological disease that results from clonal expansion of transformed lymphoid progenitors at different developmental stages.1 Cure rates for pediatric ALL are currently approximately 90%, but prognosis for children who experienced relapse remains poor, and it has only marginally improved over the past two decades. Therefore, more efforts are required for patients with chemotherapy-resistant leukemia to identify effective treatment strategies.2,3
The Notch pathway plays a crucial role in T-cell lineage specification and thymic development and its deregulated activation has been linked to T-ALL development and maintenance.1,4 Notably, about 50-60% of T-ALL samples show activating mutations in the NOTCH1 gene5,6 and 15% of T-ALL cases present mutations or deletions in its ubiquitin ligase FBW7.7 The role of Notch in solid and hematological malignancies suggests targeting of this pathway for therapeutic purposes in Notch-driven malignancies. Consolidated therapeutic approaches include the use of gamma secretase inhibitors (GSI) that block Notch processing, alternative molecules that affect Notch signaling and antibodies or decoy peptides to target specific Notch receptors or their ligands, hypothetically overcoming adverse effects due to the pan-Notch signaling inhibition associated with GSI.8
We have recently shown the therapeutic efficacy of the NOTCH1-specific monoclonal antibody OMP52M51 (Brontictuzumab) in pediatric T-ALL xenografts bearing NOTCH1/FBW7 mutations, including samples derived from relapsed and difficult-to-treat patients.9 OMP52M51 has been in clinical trial in patients with relapsed or refractory lymphoid malignancies (trial ID: NCT01703572) and with relapsed or refractory advanced solid tumors (trial ID: NCT01778439), although it has not been considered for further clinical development. Preliminary experiments in our lab showed that T-ALL PDX acquire resistance to OMP52M51 and this event may likely occur also in patients enrolled in clinical trials with this Notch inhibitor. Moreover, state-of-the-art knowledge about the molecular mechanisms of resistance to Notch blockade stems from various experimental models which utilized almost exclusively GSI to block Notch signaling.10 We were therefore interested in exploiting PDX models to investigate possible novel mechanisms of resistance to antibody-mediated NOTCH1 blockade with the long-term aim to support clinical development of antibodies targeting Notch in cancer.
Methods
Establishment of T-ALL xenografts and development of resistant PDX in vivo
Xenografts (PDTALL) establishment and their genetic characterization are reported elsewhere.9 To develop resistance to Notch blockade, PDTALL cells were intravenously (i.v.) injected in NOD/SCID mice (5×106 cells/mouse; 5-6 mice/group) and animals were intraperitoneally (i.p.) treated with the humanized anti-human NOTCH1 mAb OMP52M51 (Oncomed Pharmaceuticals Inc., Redwood, CA, USA) or control antibody (Rituximab, Roche, Basel, Switzerland) until disease progression. Both antibodies were administrated weekly at 20 mg/Kg, starting two days after i.v. injection of T-ALL cells. In the acute treatment experiment, mice were treated with OMP52M51 four days prior to sacrifice. Procedures involving animals conformed current laws and policies (EEC Council Directive 86/609, OJ L 358, 12/12 1987) and were authorized by the Italian Ministry of Health (894/2016-PR). T-ALL growth was monitored by periodic blood drawings and flow cytometric analysis of CD5 and CD7 (the antibodies used were both from Coulter, Fullerton, CA, USA). Leukemic cells were recovered from the spleen and used for the following analyses.
Reverse transcription-PCR (RT-PCR) and quantitative PCR (qPCR)
Total RNA was isolated using TRIzol Reagent (Life Technologies, Paisley, UK) according to the manufacturer’s instructions. cDNA was synthesized from 1-1.5 μg of total RNA using the High Capacity RNA-to-cDNA kit (Life Technologies). For analysis of the Notch pathway activation, a panel of 21 NOTCH-target genes were evaluated in duplicates by Custom TaqManArray Cards as described before9 using the TaqMan Universal PCR Master Mix (Life Technologies) and ABI Prism 7900 Sequence Detection System. Relative quantification was done using the DDCt method, normalizing to β2-microglobulin mRNA. Primers used for validation of microarray results are reported in the Online Supplementary Material and Methods.
Western blot analysis
Western blot methods used in this study have been previously published.11 Immunoprobing was performed using antibodies reported in the Online Supplementary Material and Methods section. Antigens were identified by luminescent visualization using the Western Lightning Plus ECL (Perkin Elmer) or ECL Select (Amersham, GE Healthcare, Chicago, IL, USA) and signal intensity was measured using a Biorad XRS chemiluminescence detection system. In a set of experiments we used subcellular fractionation, which was performed as previously described in Pinazza et al.12
Preparation of cRNA, GeneChip microarray analysis and data normalization
Samples were prepared with GeneChip® WT PLUS Reagent Kit (Affymetrix, Santa Clara, CA, USA). Total RNA and cRNA quality were evaluated with the Agilent RNA 6000 Nano Kit and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) before microarray hybridization. cDNA was quantified by Implen NanoPhotometer (Implen, München, Germany). Labeled sense-strand cDNA was used for screening of GeneChip® Human Transcriptome Array 2.0 (Affymetrix). Each sample consisted of a single mouse (n=4-5 samples/group). Hybridization and scanning were conducted on the Affymetrix platform. Further experimental details can be found in the Online Supplementary Material and Methods.
Metabolomic analysis
Experimental details can be found in the Online Supplementary Material and Methods.
NGS analysis and Sanger sequencing
Experimental details can be found in the Online Supplementary Material and Methods.
Results
Resistance to OMP52M51 occurs with heterogeneous kinetics and Notch signaling activity in T-ALL PDX
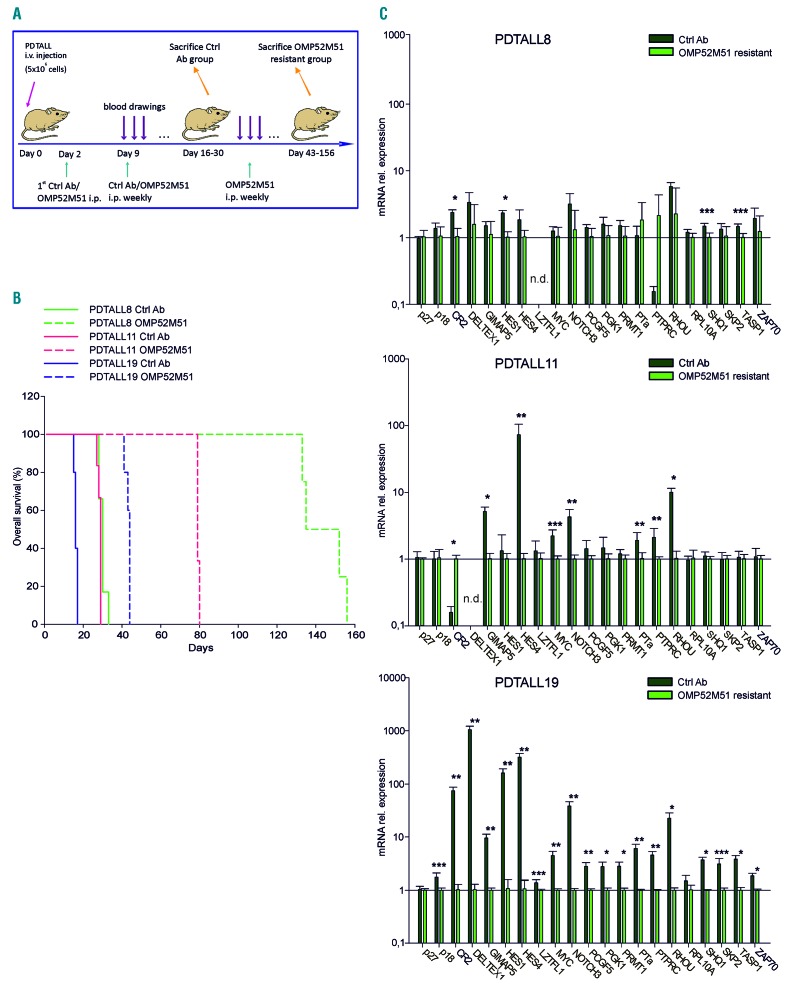
We previously demonstrated that treatment with OMP52M51 delays engraftment of T-ALL cells bearing NOTCH1/FBW7 mutations. Responder PDX disclosed increased T-ALL cell apoptosis, reduction of proliferation and marked inhibition of the Notch transcriptional signature.9 To investigate whether and when resistance to NOTCH1 blockade occurs in a regimen of continuous administration of OMP52M51 mAb, we treated n=3 xenografts bearing activating NOTCH1 mutations9 and initially responsive to OMP52M51 (PDTALL8, PDTALL11, PDTALL19) until the appearance of leukemia. For each PDX, leukemia bearing mice (n=5-6 per group) were treated with OMP52M51 or control antibody once a week. Development of leukemia was evaluated by regular blood drawings and flow cytometric analysis of human CD5 and CD7 T-ALL markers and mice were sacrificed when they presented ~20-25% circulating blasts (Figure 1A). Percentages of T-ALL cells in the spleen were evaluated at sacrifice, confirming an almost complete infiltration (>87%) of this hematopoietic organ by leukemic cells both in control and OMP52M51-resistant mice (Online Supplementary Figure S1A). In PDTALL19 model, leukemia engraftment and development of resistance to OMP52M51 were followed by optical imaging (Online Supplementary Figure S1B). Resistance to OMP52M51 treatment arose in all mice but with different kinetics in the three PDX analyzed, ranging from 43 days of PDTALL19 to 80 days of PDTALL11 and 156 days of PDTALL8 (Figure 1B). Notably, the dynamic tracking of the percentage of T-ALL cells in blood uncovered substantially different profiles in these models. In fact, we observed a steep increase in the case of PDTALL8 model, compared with the mild increase observed in the PDTALL19 model (Online Supplementary Figure S2), suggesting different mechanisms of adaptation to OMP52M51.
Figure 1.
Heterogeneous kinetics and Notch signaling inhibition in PDX resistant to OMP52M51. (A) Outline of the treatment with OMP52M51 or control antibodies (Rituximab). NOD/SCID mice (n=5-6 mice/group) were intraperitoneally (i.p.) treated with OMP52M51 or control antibodies (Ctrl Ab) two days after intravenous (i.v.) injection of T-ALL cells (5×106 cells/mouse). Antibodies were subsequently administrated weekly and leukemia engraftment was tracked by serial blood drawings and flow cytometric analysis. (B) Kaplan-Meier survival curves of leukemic mice after treatment with OMP52M51 (dotted lines) or Ctrl Ab (solid lines) of PDTALL8, PDTALL11 and PDTALL19 xenografts (PDTALL8: P=0.00343; PDTALL11: P=0.00124; PDTALL19: P=0.00264 log-rank test). (C) Notch target genes expression in PDTALL8 (top), PDTALL11 (middle) and PDTALL19 (bottom) resistant compared to control cells (*P≤0.001; **P=0.029; *** P≤0.01. Mean ± standard deviation [SD], 3-5 samples/group).
Following treatment of T-ALL PDX with OMP52M51, we previously observed a strong reduction of Notch target gene expression in good responders and we considered this phenomenon as indirect evidence of reduced Notch signaling.9 Therefore, we wondered if this inhibition was still present in mice which developed resistance to OMP52M51 therapy. To investigate this hypothesis, we analyzed expression levels of 21 Notch target genes by qPCR. In the PDTALL8 model, expression levels of Notch target genes were very similar in OMP52M51-resistant and control cells (Figure 1C, top). On the contrary, in PDTALL11 xenograft several genes were significantly downregulated in cells recovered from OMP52M51-resistant compared to control mice (Figure 1C, middle). In the PDTALL19 model, inhibition of Notch signaling was maintained in resistant cells (Figure 1C, bottom) and it was comparable to that previously measured following acute treatment with OMP52M51.9
Serial transplantation experiments disclosed two types of resistance to OMP52M51
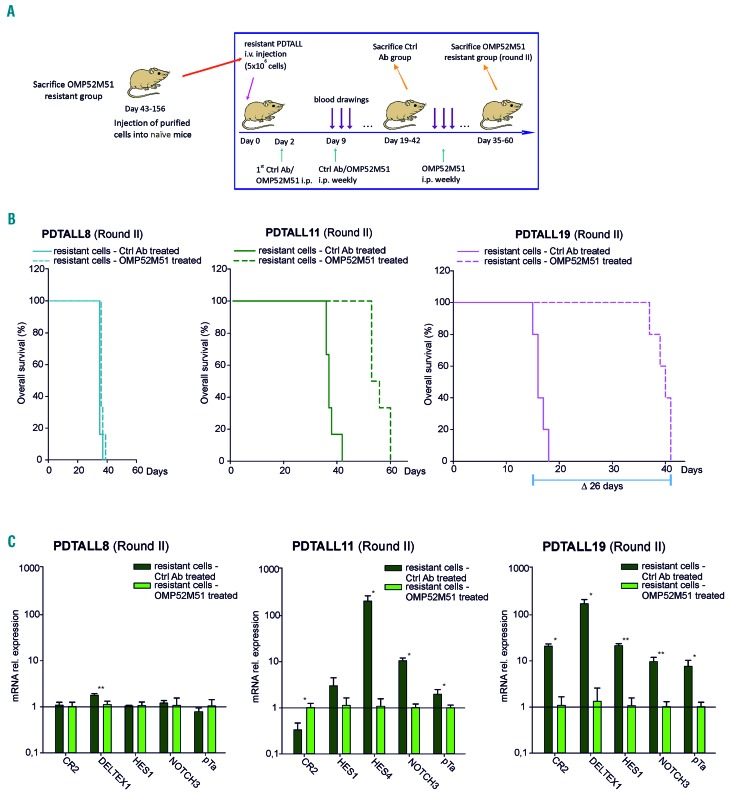
To elucidate whether resistance to OMP52M51 was a stable trait, we performed serial transplantation experiments. T-ALL cells from the spleen of donor mice resistant to OMP52M51 were i.v. injected into naïve recipient mice, which were then weekly treated with OMP52M51 or control antibody according to the standard protocol until the appearance of the signs of disease (Figure 2A). We found that resistance was a stable trait in the PDTALL8 model, the PDX in which resistance developed 156 days after OMP52M51 treatment (late onset). In fact, in the serial transplantation experiment, both OMP52M51-treated and control mice developed leukemia 39 days after T-ALL cell injection (Figure 2B). In contrast, resistance to OMP52M51 was lost upon serial transplantation in PDTALL19 model, the PDX with early onset of resistance. PDTALL19 cells recovered from the spleen of OMP52M51-resistant mice and injected into näїve mice were initially sensitive to OMP52M51 treatment, as no evidence of T-ALL cells was found in the blood of these mice at day 18, when control mice were sacrificed. When administration of OMP52M51 was repeated, however, mice became gradually resistant to therapy and they were eventually sacrificed 41 days after T-ALL cell injection, a time point very similar to that observed in the initial experiment (Figure 2B). It is important to note, however, that following repeated (n=5) cycles of treatment of PDTALL19 cells with OMP52M51, resistance became stable, replacing the initial unstable form of resistance seen in this model (Online Supplementary Figure S3), with possibly different underlying mechanisms. In the PDTALL11 model, the onset of resistance was intermediate between the other two models previously described (Figure 2B). Finally, Notch pathway activation was investigated in secondary treated and non-treated resistant cells. Results disclosed marked differences in the expression of several Notch target genes in the case of PDTALL11 and PDTALL19 and only marginal differences in the case of PDTALL8 (Figure 2C), in line with results of the first round of treatment.
Figure 2.
Serial transplantation experiments disclose different phenotypic traits of resistance. (A) Outline of the experiment. Resistant cells were recovered from the spleen of OMP52M51-treated mice and injected into naїve mice (5-6 mice/group). Recipient mice were then treated weekly with OMP52M51 or control monoclonal antibodies (Ctrl mAb) according to the standard protocol. (B) Kaplan-Meier survival curves of PDTALL8, PDTALL1 and PDTALL19 mice after a second course of treatment with OMP52M51. In these experiments, mice transplanted with PDTALL8, PDTALL11 or PDTALL19 resistant cells from the first round of treatment (PDTALL8: P=0.0441; PDTALL11: P<0,001; PDTALL19: P=0.00181 log-rank test). (C) Expression levels of selected Notch target genes in T-ALL cells obtained from the spleen of mice after the second course of OMP52M51 treatment (*P<0,05; **P<0,001. Mean ± standard deviation [SD], 3-6 samples/group).
Resistance to OMP52M51 is not associated with PTEN or FBW7 alterations
It is well known that the PTEN/PI3K/AKT pathway is frequently altered in T-ALL and that PTEN loss is involved in resistance induced by GSI13 and other therapies.14 Therefore, we analyzed the expression of PTEN in the three PDX models. PTEN was expressed in all models and resistance was not associated with loss of PTEN, since the protein was detectable at comparable levels in treated and control cells (Online Supplementary Figure S4). In line with this finding, comparable levels of AKT activation (determined as pAKTSer473 and pAKTThr308)15 were found in OMP52M51 compared with control cells (Online Supplementary Figure S4). Furthermore, we sequenced the FBW7 gene, since mutations in this gene have also been correlated with GSI resistance.7 Sequencing of FBW7 in PDTALL8, PDTALL11 and PDTALL19 models revealed that neither parental nor resistant cells were harboring a mutated version of FBW7 (Online Supplementary Table S2). We conclude that resistance to OMP52M51 did not involve mechanisms reported in previous studies with GSI.
Resistance to OMP52M51 is associated with a distinct transcriptional signature in PDTALL19 model
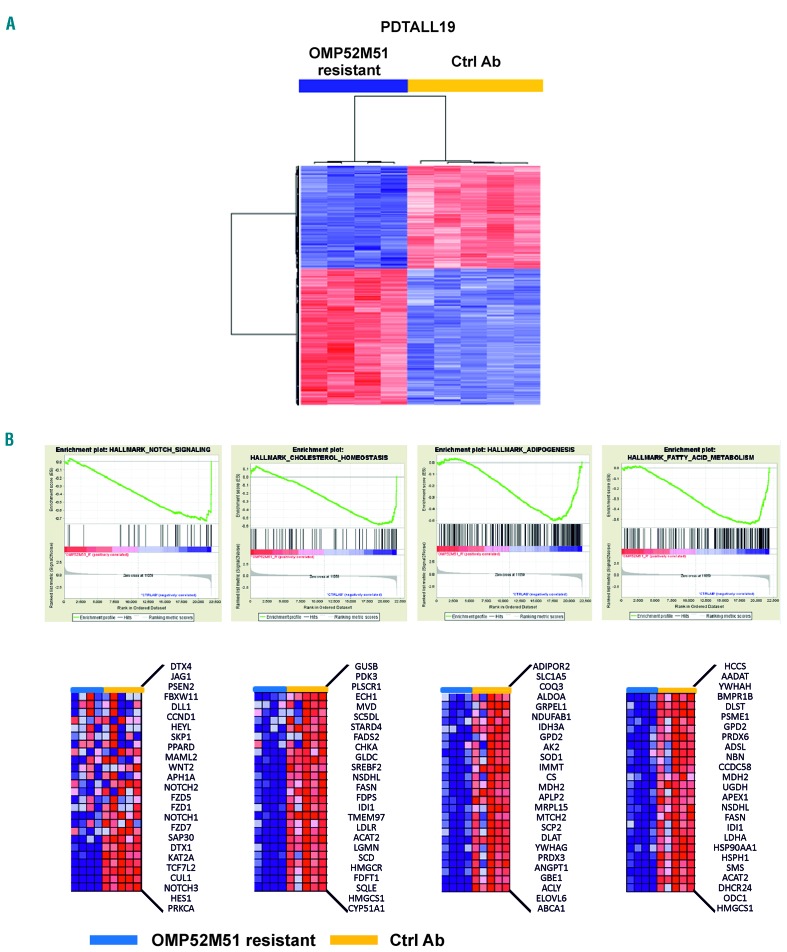
We exploited transcriptome analysis to investigate the mechanisms of resistance to OMP52M51 in the PDTALL8 and PDTALL19 models, which presented completely different phenotypes of resistance. In the PDTALL8 model – the PDX characterized by late onset and stable resistance to OMP52M51 – only a few ProbeSet ID without an associated gene annotation were modulated, so we concluded that the gene expression profiles of resistant cells were superimposable to control ones (Online Supplementary Figure S5). In contrast, in the PDTALL19 model – the PDX characterized by early onset and transient resistance to OMP52M51 – we identified 327 up- and 257 down-regulated genes in the comparison between OMP52M51-resistant and control cells (Figure 3A, Online Supplementary Table S3-4). Gene set enrichment analysis (GSEA) identified 20 pathways significantly down-regulated in OMP52M51-resistant TALL samples (Online Supplementary Figure S6). Among them, we found the Notch pathway (Figure 3B), confirming the inhibition measured by TaqMan arrays, oxidative phosphorylation, unfolded protein response, pathways related to proliferative processes - such as the G2/M checkpoint and mitotic spindle – and MYC targets, a well-known mediator of Notch activity. Interestingly, GSEA disclosed also metabolic pathways significantly down-regulated in resistant compare to control cells, including cholesterol homeostasis, adipogenesis and fatty acid metabolism (Figure 3B). Selective transcriptome findings were validated by quantitative RT-PCR (Online Supplementary Figure S7). In order to unravel if these modulations were due to inhibition of Notch signaling or were hallmarks of resistance to OMP52M51, we performed gene expression profile analysis in mice engrafted with PDTALL19 cells and acutely treated with OMP52M51. The majority of pathways identified by GSEA were present also in these samples (Online Supplementary Figure S8-9), but with a higher level of down-regulation compared to OMP52M51 resistant cells (Online Supplementary Figure S10), suggesting that the inhibitory effect of the antibody on signaling is partially lost when resistance develops.
Figure 3.
Transcriptome analysis of OMP52M51-resistant PDTALL19 cells. (A) Heatmap of genes modulated comparing OMP52M51 resistant and control cells (4-5 samples/group). Red and blue colors indicate higher and lower expression levels, respectively. The columns represent individual samples (B) top: Gene set enrichment analysis (GSEA) plots of four enrichment sets down-regulated in resistant cells. Bottom: Heatmaps and lists of the top 25 differentially expressed genes of the corresponding plots reported above.
Resistance to OMP52M51 is associated with rewiring of lipid metabolism in PDTALL19 cells
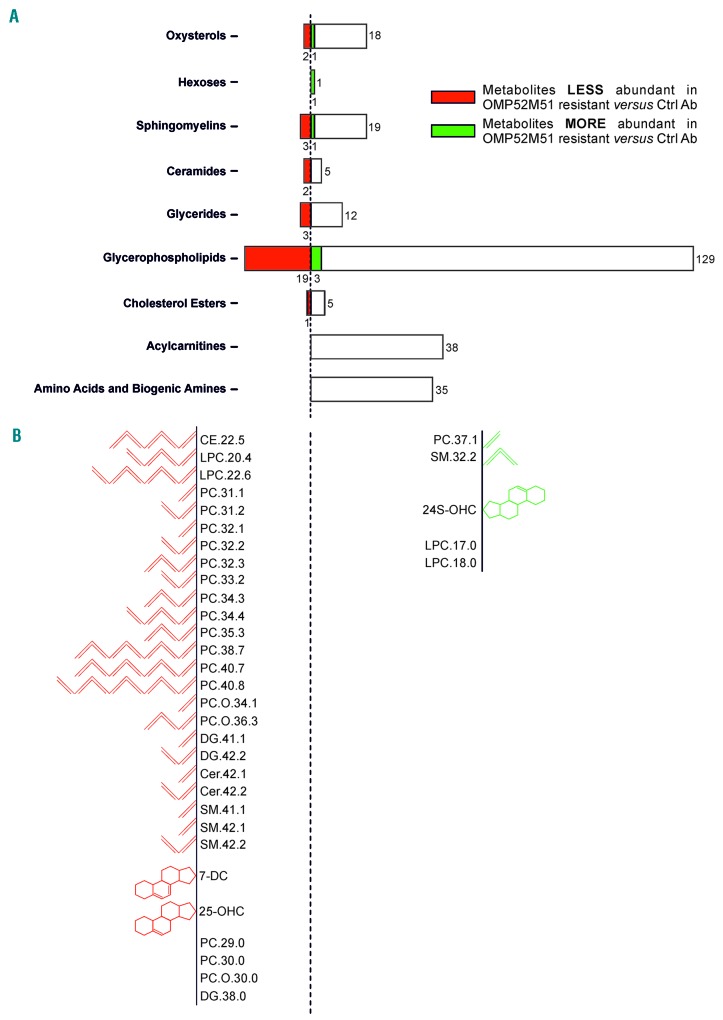
These results prompted investigation of the effects of Notch blockade on lipid metabolism. To this end, we performed tandem mass spectrometry (MS/MS) for different classes of metabolites. This analysis revealed that numerous constituents of cell membranes were less abundant in OMP52M51-resistant versus control PDTALL19 samples, including members of glycerophospholipids, sphingomyelins and ceramides families, as well as oxysterols and one cholesterol ester. Only few metabolites in these classes were enriched in OMP52M51-resistant cells (Figure 4A). Quantification of the cellular contents of lipid droplets by flow cytometry confirmed a significant reduction of lipid storage in OMP52M51-resistant cells compared to controls (Online Supplementary Figure S11). Moreover, MS/MS analysis discovered an imbalance between saturated and (poly-) unsaturated lipids and identified a trend towards reduction of the unsaturation degree in OMP52M51-resistant samples (Figure 4B). Notably, acute treatment of PDTALL19 mice with OMP52M51 did not cause significant alterations in lipids (Online Supplementary Figure S12), indicating that dis-regulated lipid metabolism is a feature of resistance.
Figure 4.
Lipidomic analysis of OMP52M51-resistant PDTALL19 cells. (A) The length of the columns is proportional to the abundance of analytes in the corresponding class. Numbers near columns represent the number of metabolites included in the analysis (right) and the significant ones (middle) for each class. Metabolites significantly less abundant in resistant cells are represented in red, instead the more abundant are in green (adjusted P-value<0.05, five samples/group). (B) An overview of significant less abundant (left) and more abundant (right) lipid metabolites in resistant cells. Lipids are schematically represented depending on their saturation degree.
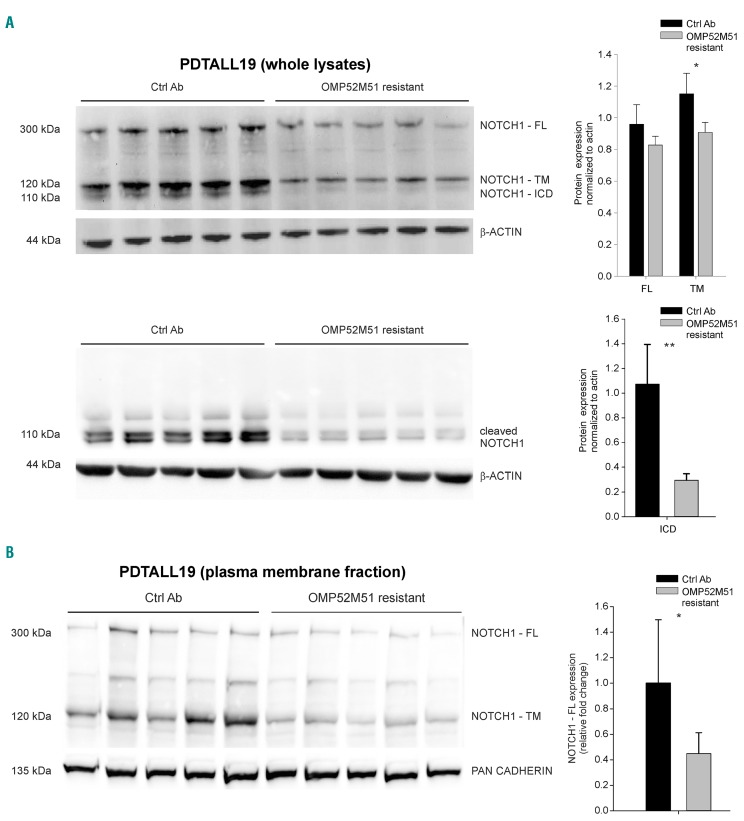
Since NOTCH1 is a membrane-bound protein, we hypothesized that a different composition of lipid constituents could affect the fluidity of the membrane and the surface levels of the receptor. To this end, we analyzed NOTCH1 protein levels by Western blot (WB) analysis both in whole cell lysates (Figure 5A) and in plasma membrane fractions (Figure 5B). OMP52M51-resistant samples revealed a clear reduction in trans-membrane (TM) and intracellular domain (ICD) forms, a result that was expected since binding of the antibody should prevent activation of the pathway. These results were confirmed by WB analysis of whole cell lysates using the NOTCH1 ICD-specific Val1744 monoclonal antibody (mAb) (Figure 5A). Interestingly, we also observed a decrease in the full-length NOTCH1 form, which was not anticipated and suggested lower levels of NOTCH1 on the cell surface (Figure 5A-B). This modulation in NOTCH1 protein levels was not observed in OMP52M51-resistant PDTALL8 and PDTALL11 cells, consistent with marginal variations in genes involved in lipid metabolism in these models (Online Supplementary Figure S13). However, acute treatment of PDTALL19 mice with OMP52M51 was also associated with reduced NOTCH1 FL and ICD levels (Online Supplementary Figure S12), indicating that these changes were related to NOTCH1 blockade rather than to resistance. Finally, given the marked changes in lipid metabolism seen in the PDTALL19 model, we investigated other possible phenotypic changes associated with resistance. By flow cytometry analysis, we found that OMP52M51-resistant cells exhibited a smaller size compared with control cells and there were also significant changes in surface expression of the T-cell markers CD3 and CD7 and the pan-leucocyte marker CD11a (Online Supplementary Figure S14).
Figure 5.
OMP52M51-resistant PDTALL19 cells show reduction of surface NOTCH1 protein. (A) Left: NOTCH1 full-length (FL), transmembrane (TM) and intra-cellular domain (ICD) expression levels were measured by Western blot analysis on whole cell lysates. Cleaved NOTCH1 levels (mid panel) were determined by probing the filters with the Val1744 antibody. Right: quantification of NOTCH1 FL, TM and ICD levels normalized to β-actin expression (mean ± standard deviation [SD]) of control and resistant cells (five samples/group. *P=0.005; **P<0.001). (B) Left: Analysis of NOTCH1 expression on plasma membrane lysates obtained by subcellular fractionation. Right: Quantification of NOTCH1-FL expression in control antibodies (Ctrl Ab) and resistant cells, normalized to the corresponding housekeeping protein pan-cadherin and normalized to controls (mean ± SD, 10 samples/group.*P=0.001).
Assessment of the genetic background of OMP52M51 resistance in the PDTALL8 model
In the case of PDTALL8 model, transcriptional data suggested that mechanisms underlying the stable resistance to OMP52M51 treatment could involve an “on target” mutation of the Notch pathway selected during treatment and serial transplantation experiments, leading to the loss of sensitivity to OMP52M51.
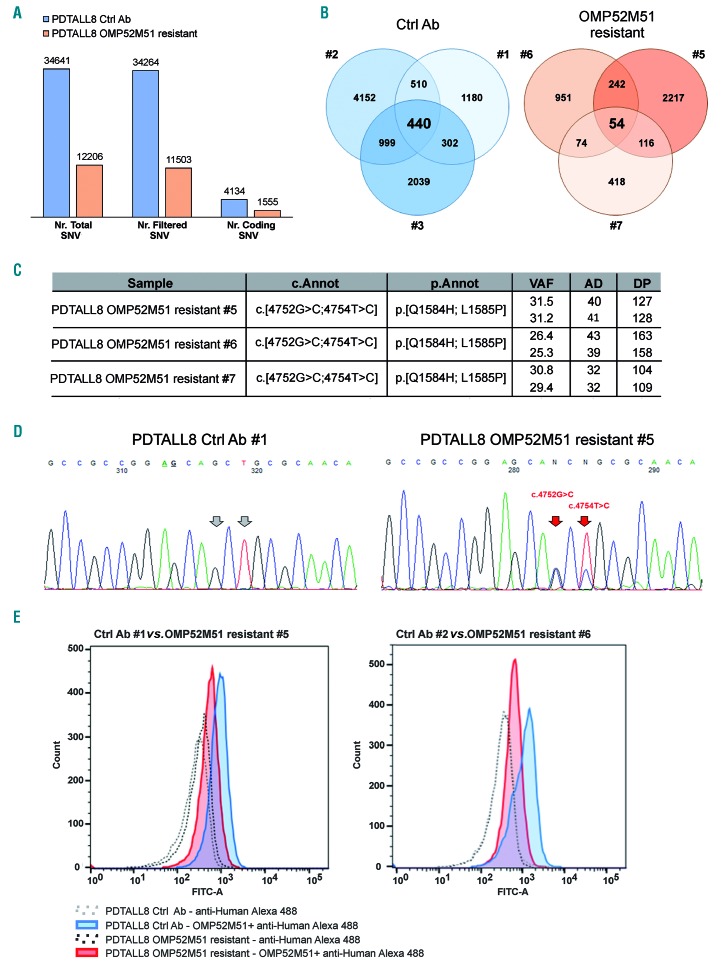
To investigate this hypothesis, we performed single nucleotide polymorphism (SNP) arrays and whole exome sequencing (WES) of paired control-resistant mice (see Online Supplementary Table S5 for WES metrics details), allowing the identification of variants that could be not detected by Sanger sequencing due to a relatively low variant frequency. Cytoscan arrays failed to identify copy number variations associated with resistance to OMP52M51 in PDTALL8 cells (Online Supplementary Figure S15). However, bioinformatics analysis of WES revealed that control mice displayed a higher tumor mutational load than OMP52M51-resistant samples, both considering the total number of variants (34,641 variants in controls compared to 12,206 in OMP52M51-resistant samples) and shared confident non-synonymous calls (440 vs. 54; Figure 6A-B and Online Supplementary Table S6). This difference could be explained by the “tumor clonal selection” model. Speculatively, the OMP52M51 antibody could act as a selective agent, favouring outgrowth of a subpopulation of cells from the initial tumour. Interestingly, WES analysis highlighted the presence of two NOTCH1 activating variants mapping to the heterodimerization domain (HD),16 i.e. p.Q1584H and p.L1585P, present only in the OMP52M51-resistant mice (Figure 6C). We validated these mutations by Sanger sequencing and extended analysis to additional samples from the same experiment (four to five mice/group; Online Supplementary Figure S16). Sanger sequencing confirmed that both mutations were present in OMP52M51 resistant mice and were lacking in controls (Figure 6D).
Figure 6.
Molecular profiling of PDTALL8 control and OMP52M51 resistant T-cell acute lymphoblastic leukemia cells. (A) Comparison between the average mutational burden in PDTALL8 untreated and resistant cells based on whole exome sequencing (WES) results. Total: total number of single-nucleotide variant (SNV); filtered: variants with a variant allele fraction (VAF) >10% and coverage depth (DP) >30 excluding common polymorphisms; coding: filtered coding variants excluding synonymous genetic alterations. (B) Venn diagrams of coding variants for each mouse. 440 SNV and 54 SNV are shared respectively between control (#1-3) and resistant mice (#5-7) inside each group. (C) NOTCH1 variants found only in OMP52M51 resistant samples. cDNA coordinates, amino acidic changes, VAF, alternative allele depth (AD) and DP are reported. (D) Direct sequencing of exon 26 in a representative control (Ctrl Ab #1) and resistant (OMP52M51 resistant #5) pair. Resistance-related mutations are indicated with the red arrows. (E) Flow cytometric analysis of surface expression of NOTCH1 in T-cell acute lymphoblastic leukemia (T-ALL) cells from the spleen of PDTALL8 mice treated with either OMP52M51 or control antibody. Two representative samples per group are shown.
To investigate whether these mutations occurred at low level in parental cells, we performed targeted sequencing analysis. All treated xenograft (three replicates/group) presented p.L1585P and p.Q1584H variants in cis (Online Supplementary Figure S17). On the other hand, at a depth of 300X the p.L1585P and p.Q1584H variants were not identified in control samples and therefore, if present prior to treatment, must have been present at a frequency of less than 5%, which was the estimated sensitivity of the next-generation sequencing (NGS) assay.
Finally, according to the literature, these mutations destabilize the structure of the HD domain17 and therefore could affect binding of OMP52M51. This was indeed shown by flow cytometry experiments showing that the binding of OMP52M51 is lower in PDTALL8 resistant compared with control cells (Figure 6E).
Discussion
In the last 10 years, personalized treatment of cancer has improved substantially thanks to the identification of specific genetic alterations and consequent development of target therapies against oncogenic drivers. In this landscape, the resistance of cancer cells to pharmacological treatment remains the major challenge to face in order to increase the efficacy of new drugs.18 Although we are aware of some intrinsic limitations of xenografts, such as the lack of the immune system, the systemic T-ALL models used in this study are suitable to study effects of drugs directly targeting tumors cells, and we used them to investigate the molecular mechanisms of resistance to Notch targeted therapy. Our results demonstrate that the resistance appeared following long-term administration of OMP52M51 antibody in each of the three PDX models tested, though with different timing. In fact, PDTALL8, the PDX with a late onset of resistance and loss of Notch signaling inhibition, was characterized by a stable resistance. On the contrary, in the PDTALL19 model, resistance appeared earlier, Notch signaling was inhibited and, importantly, it was an unstable trait. Analysis of the slope of the percentage of T-ALL cells in blood of these mice during serial drawings suggested that adaptation to OMP52M51 in this model consisted mainly of a delayed but constant growth of the PDX without the development of true resistance during the first round of treatment. However, upon repeated rounds of treatment with OMP52M51 stable resistance eventually occurred, suggesting a two-stage form of resistance in this model. PDTALL11 disclosed an intermediate behavior both regarding the time of development of resistance, the stability of resistance and Notch signaling inhibition, likely due to a mixture of different mechanisms. Speculatively, an additional round of treatment might lead to the selection of a completely resistant clone also in the PDTALL11 model, although this was not investigated here.
Intriguingly, previous studies elegantly addressed the issue of clonality of T-ALL xenografts and correlated the genetic complexity of T-ALL cells with the speed of leukemia development in xenograft models.19,20 Specifically, in the case of delayed T-ALL growth, leukemia cells were in part genetically diverse, the resulting xenograft leukemia arising from different but branched subclones present in the original sample. Although not investigated in our study, it could be speculated that the clonal architecture of the PDX might have an influence on the time of development and the type of resistance to anti-NOTCH1 therapy.
In the PDX tested, the resistance was never associated with the loss of PTEN, AKT activation or mutations in FBW7 gene, which represent some of the previously described mechanisms of resistance to Notch inhibition by GSI.7,13 The strikingly different phenotypes of the PDTALL8 and PDTALL19 models underscored two novel mechanisms of resistance. In the case of PDTALL19, we found that tumor adaptation to anti-NOTCH1 therapy was accompanied by a distinct transcriptomic and metabolic signature. Alterations converged on lipid metabolism, hinting at significant alterations in constituents of cell membranes such as phosphatidylcholines, lysophosphatidylcholines, sphingomyelins and ceramides. Moreover, there was a trend towards the reduction of the unsaturation degree of lipids in OMP52M51-resistant samples, hypothetically suggesting a modulation of membrane fluidity, since saturated fatty acids are more densily packed.21 Some years ago, a well-designed study correlated a different composition of phospholipids with an increase in endocytosis and therefore a reduction in EGFR and Notch signaling, due to the fact that these receptors were less abundant on the cell membrane.22 Albeit performed in Drosophila, we speculated that the different composition and/or unsaturation degree of membrane components could affect the amount of NOTCH1 receptor on cell surface and, therefore, the amount of target available for binding the therapeutic antibody. For a linear pathway such as Notch, the exposure of the receptor on cell surface is fundamental and, therefore, the number of NOTCH1 receptors are strictly regulated.23–25 However, we found reduced NOTCH1 levels also after short-term treatment with OMP52M51, a condition where lipid alterations were not detected, indicating that the alterations in lipid metabolism are in fact a feature associated with the resistance in the PDTALL19 model, but this does not likely cause reduced NOTCH1 FL levels in these cells. In any case, recent findings gave prominence to new functions of membrane components, beyond the structural one. Phosphatidylcholines, lysophosphatidylcholines, sphingomyelins and ceramides could, in fact, be involved in cell signaling and behave as messengers frequently altered in pathologic conditions such as cancer.26–29 Therefore, it could be possible that the variations in membrane components observed in PDTALL19 model could influence the microenvironment and/or T-ALL cell behaviour. Along this line, our finding that T-ALL cells resistant to OMP52M51 had reduced size and showed lower surface levels of CD3, CD4 and CD11a compared with control cells supports this hypothesis.
In a recent paper, Herranz and colleagues identified glucose metabolism and glutaminolysis as key determinants in cell resistance to Notch blockade with GSI.30 In our model, we also found an increase in the amount of hexoses in resistant cells, but the innovative finding was represented by the association between the resistance to OMP52M51 antibody and the modulations of the sterol biosynthetic pathway. Moreover, in our model we did not observe the activation of autophagy (data not shown), which other groups previously detected in cells after Notch inhibition by GSI.30 Regarding the PDTALL19 model, finally, the comparison between resistant cells and acute treatment with OMP52M51 disclosed that the antibody inhibited the same GSEA pathways but with a different strength.
Another possible hypothesis behind the reduction of surface NOTCH1 receptor regards DELTEX1. Many studies performed in Drosophila have shown that the activation, degradation and recycling of Notch receptors could depend on their ubiquitination pattern.31,32 DELTEX1 is an ubiquitin-ligase that could act as a positive or a negative regulator of Notch signaling depending on the cellular context and could influence endocytic trafficking of the receptor.33–35 Notably, in our models DELTEX1 was one of the most down-regulated genes in PDTALL19 cells, whereas it was not detectable in PDTALL11 cells and was not modulated in PDTALL8 cells (Figure 1C), PDX in which we did not observe any reduction of surface NOTCH1 protein.
In the PDTALL8 model, given the stability of the resistance and the loss of Notch signaling inhibition, the most likely explanation for the observed resistance was the selection of a mutated clone no longer responsive to therapy. To address this possibility, NGS analysis performed in control and resistant cells disclosed a selection of clones of the initial tumour, since treated samples presented a lower mutational burden compared to control. Moreover, NGS analysis revealed the presence of two activating mutations in NOTCH1 gene - p.Q1584H and p.L1585P - in the OMP52M51 resistant mice. These variants, reported as T-ALL associated in CIViC database (36), were in cis and cause the introduction of a positive charge (p.Q1584H) and a break (p.L1585P) to the α2-helix in the HD domain,16 adding steric bulk in the HD core. These modifications potentially affect the stability of HD domain and could influence binding of the OMP52M51 antibody, causing the loss of responsiveness to the therapy. Notably, we confirmed the p.Q1584H and the p.L1585P mutations by targeted sequencing but could not find them in the parental cells prior to treatment. However, as all of the independently derived resistant PDTALL8 tumor samples disclosed the same p.Q1584H and p.L1585P variants, it is extremely likely that these variants must have been present in a minor sub-clone prior to selective pressure with OMP52M51.
Finally, the possible role of the microenvironment must be taken into consideration, since different tumours could differ, not only in intrinsic lesions but also in their dependence on the niche – therefore, different T-ALL cases might respond distinctly to the same treatment. A better characterization of both cell-autonomous lesions and cues from the microenvironment, which was apart from the purpose of this paper, would extend the complete understanding of how the malignancy progresses and resistance develops.1
In conclusion, our study highlights heterogeneity in the phenotypic and molecular features of resistance to OMP52M51. Notably, traits associated with resistance differ from those previously described following treatment with GSI, suggesting that leukemia cells can adopt several strategies to evade Notch inhibition according to the therapeutic drug used.
Acknowledgments
We thank Dr. Erich Piovan (University of Padova, Padova, Italy) for providing the PTEN antibody and Dr. Tim Hoey (Oncomed Pharmaceuticals Inc., Redwood, CA, USA) for providing the OMP52M51 antibody.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/5/1317
Funding
This work was supported by AIRC (Grant n. IG18803 to SI).
References
- 1.Oliveira ML, Akkapeddi P, Alcobia I, et al. From the outside, from within: Biological and therapeutic relevance of signal transduction in T-cell acute lymphoblastic leukemia. Cell Signal. 2017;38:10–25. [DOI] [PubMed] [Google Scholar]
- 2.Pierro J, Hogan LE, Bhatla T, Carroll WL. New targeted therapies for relapsed pediatric acute lymphoblastic leukemia. Expert Rev Anticancer Ther. 2017;17(8):725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pui CH, Yang JJ, Hunger SP, et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2015;33(27):2938–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8(5):380–390. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. [DOI] [PubMed] [Google Scholar]
- 6.Sulis ML, Williams O, Palomero T, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008; 112(3):733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204(8):1813–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Platonova N, Lesma E, Basile A, et al. Targeting Notch as a therapeutic approach for human malignancies. Curr Pharm Des. 2017;23(1):108–134. [DOI] [PubMed] [Google Scholar]
- 9.Agnusdei V, Minuzzo S, Frasson C, et al. Therapeutic antibody targeting of Notch1 in T-acute lymphoblastic leukemia xenografts. Leukemia. 2014;28(2):278–288. [DOI] [PubMed] [Google Scholar]
- 10.Hales EC, Taub JW, Matherly LH. New insights into Notch1 regulation of the PI3K-AKT-mTOR1 signaling axis: targeted therapy of gamma-secretase inhibitor resistant T-cell acute lymphoblastic leukemia. Cell Signal. 2014;26(1):149–161. [DOI] [PubMed] [Google Scholar]
- 11.Pinazza M, Borga C, Agnusdei V, et al. An immediate transcriptional signature associated with response to the histone deacetylase inhibitor Givinostat in T acute lymphoblastic leukemia xenografts. Cell Death Dis. 2016;6:e2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinazza M, Ghisi M, Minuzzo S, et al. Histone deacetylase 6 controls Notch3 trafficking and degradation in T-cell acute lymphoblastic leukemia cells. Oncogene. 2018;37(28):3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown KK, Toker A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep. 2015;7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Z, Shi YX, Tsao T, et al. Targeting of mTORC1/2 by the mTOR kinase inhibitor PP242 induces apoptosis in AML cells under conditions mimicking the bone marrow microenvironment. Blood. 2012; 120(13):2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon WR, Roy M, Vardar-Ulu D, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113(18):4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breit S, Stanulla M, Flohr T, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4):1151–1157. [DOI] [PubMed] [Google Scholar]
- 18.Pazarentzos E, Bivona TG. Adaptive stress signaling in targeted cancer therapy resistance. Oncogene. 2015;34(45):5599–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clappier E, Gerby B, Sigaux F, et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J Exp Med. 2011;208(4):653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poglio S, Lewandowski D, Calvo J, et al. Speed of leukemia development and genetic diversity in xenograft models of T cell acute lymphoblastic leukemia. Oncotarget. 2016;7(27):41599–41611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rysman E, Brusselmans K, Scheys K, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70(20):8117–8126. [DOI] [PubMed] [Google Scholar]
- 22.Weber U, Eroglu C, Mlodzik M. Phospholipid membrane composition affects EGF receptor and Notch signaling through effects on endocytosis during Drosophila development. Dev Cell. 2003; 5(4):559–570. [DOI] [PubMed] [Google Scholar]
- 23.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16(5):633–647. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto S, Charng WL, Bellen HJ. Endocytosis and intracellular trafficking of Notch and its ligands. Curr Top Dev Biol. 2010;92:165–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortini ME, Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19(4):323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drzazga A, Sowińska A, Koziołkiewicz M. Lysophosphatidylcholine and lysophos-phatidylinosiol–novel promissing signaling molecules and their possible therapeutic activity. Acta Pol Pharm. 2014;71(6):887–899. [PubMed] [Google Scholar]
- 27.Furse S, de Kroon AI. Phosphatidylcholine’s functions beyond that of a membrane brick. Mol Membr Biol. 2015;32(4):117–119. [DOI] [PubMed] [Google Scholar]
- 28.Ridgway ND. The role of phosphatidyl-choline and choline metabolites to cell proliferation and survival. Crit Rev Biochem Mol Biol. 2013;48(1):20–38. [DOI] [PubMed] [Google Scholar]
- 29.Ryland LK, Fox TE, Liu X, Loughran TP, Kester M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol Ther. 2011;11(2):138–149. [DOI] [PubMed] [Google Scholar]
- 30.Herranz D, Ambesi-Impiombato A, Sudderth J, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21(10):1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Regulation of ligand-independent Notch signal through intracellular trafficking. Commun Integr Biol. 2012;5(4):374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanwar R, Fortini ME. Notch signaling: a different sort makes the cut. Curr Biol. 2004; 14(24):R1043–1045. [DOI] [PubMed] [Google Scholar]
- 33.Hori K, Fostier M, Ito M, et al. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004; 131(22):5527–5537. [DOI] [PubMed] [Google Scholar]
- 34.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol. 2011; 195(6):1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkin M, Tongngok P, Gensch N, et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev Cell. 2008;15(5):762–772. [DOI] [PubMed] [Google Scholar]
- 36.Griffith M, Spies NC, Krysiak K, et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat Genet. 2017;49(2): 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]