Abstract
To identify genomic alterations contributing to the pathogenesis of high-risk chronic lymphocytic leukemia (CLL) beyond the well-established role of TP53 aberrations, we comprehensively analyzed 75 relapsed/refractory and 71 treatment-naïve high-risk cases from prospective clinical trials by single nucleotide polymorphism arrays and targeted next-generation sequencing. Increased genomic complexity was a hallmark of relapsed/refractory and treatment-naïve high-risk CLL. In relapsed/refractory cases previously exposed to the selective pressure of chemo(immuno)therapy, gain(8)(q24.21) and del(9)(p21.3) were particularly enriched. Both alterations affect key regulators of cell-cycle progression, namely MYC and CDKN2A/B. While homozygous CDKN2A/B loss has been directly associated with Richter transformation, we did not find this association for heterozygous loss of CDKN2A/B. Gains in 8q24.21 were either focal gains in a MYC enhancer region or large gains affecting the MYC locus, but only the latter type was highly enriched in relapsed/refractory CLL (17%). In addition to a high frequency of NOTCH1 mutations (23%), we found recurrent genetic alterations in SPEN (4% mutated), RBPJ (8% deleted) and SNW1 (8% deleted), all affecting a protein complex that represses transcription of NOTCH1 target genes. We investigated the functional impact of these alterations on HES1, DTX1 and MYC gene transcription and found derepression of these NOTCH1 target genes particularly with SPEN mutations. In summary, we provide new insights into the genomic architecture of high-risk CLL, define novel recurrent DNA copy number alterations and refine knowledge on del(9p), gain(8q) and alterations affecting NOTCH1 signaling. This study was registered at ClinicalTrials.gov with number NCT01392079.
Introduction
Advanced understanding of the pathophysiology of chronic lymphocytic leukemia (CLL) has led to targeted therapy approaches such as inhibition of B-cell receptor signaling by BTK inhibitors or PI3K inhibitors and antagonism of BCL-2.1,2 These treatment strategies clearly improved the clinical outcome of high-risk CLL,1,2 although inadequate responses have been observed in as yet insufficiently characterized subgroups of patients.3–5 In the era of chemo(immuno)therapy, high-risk CLL was defined by TP53 deletion/mutation or refractoriness to purine analog-based treatment (no response or progression-free survival <6 months).6 For chemotherapy-free regimens, the prognostic value of TP53 alterations is less clear, but the presence of a complex karyotype, which often occurs together with TP53 deletion/mutation,7 has been identified as an independent risk factor for early progression during venetoclax or ibrutinib treatment.8,9 However, a more recent study has shown that CLL with a complex karyotype is a heterogeneous group with variable clinical behaviors.10
To better understand treatment failure in CLL, a comprehensive characterization of the genomic architecture in high-risk CLL is vital. With 5-10% high-risk cases included in large-scale studies on DNA copy number changes and gene mutations, these cases were underrepresented for systematic analyses restricted to this subgroup.11–14
Available results from single nucleotide polymorphism (SNP)-array profiling of high risk CLL support the notion of increased genomic complexity in the majority of these cases.11–13 However, it should be noted that TP53 dysfunction and defects in other DNA damage response systems such as ATM cause chromosomal instability with random secondary events not necessarily associated with adverse prognosis.15 This constitutes a challenge, to identify those alterations contributing to a high-risk form of disease.
In order to get a more thorough understanding of the pivotal genomic alterations contributing to high-risk CLL biology, we performed high-resolution SNP-array profiling and targeted sequencing on 75 relapsed/refractory CLL cases including 18 cases without TP53 alterations. We extended our cohort by including 71 treatment-naïve, TP53-deficient, primary high-risk cases. All patients’ samples were derived from prospective clinical trials of the French/German CLL study groups (FCLLSG/GCLLSG).
To identify DNA copy number alterations (CNA) occurring more often than would be expected by chance, we applied the Genomic Identification of Significant Targets in Cancer algorithm 2.0 (GISTIC2.0).16 In relapsed/refractory CLL, in which tumor cell clones underwent selective pressure imposed by therapy, CNA with significance assigned by GISTIC2.0 harbored genes with key roles in cell-cycle control. Furthermore, we identified NOTCH1 as a central pathway frequently affected by genomic alterations enhancing its signaling strength.
Methods
Patients and samples
The study included peripheral blood mononuclear cells (PBMC) from 146 high-risk cases (TP53 aberration or refractoriness to purine analogs) enrolled on prospective trials of the GCLLSG/FCLLSG (CLL2O trial, clinicaltrials.gov identifier: NCT01392079; CLL8 trial, NCT00281918; CLL11 trial, NCT01010061). Written informed consent from all patients and ethics committee approval were obtained in accordance with the Declaration of Helsinki.
Selection of cases was guided by sample availability and included 110 of 135 cases from the CLL2O trial,17 27 of 51 cases with 17p deletion from the CLL8 trial18 and nine of 52 cases with 17p deletion from the CLL11 trial.19 All samples were taken at trial enrollment and tumor cells were enriched via CD19 immunomagnetic beads (MACS, Miltenyi Biotec®, Bergisch Gladbach, Germany). CD19 negative PBMC fractions with a tumor cell load <5% were available for paired analysis in 91 cases. Cases lacking matched normal material were analyzed against a pool of ten gender-matched reference samples.
IGHV mutational analysis, fluorescence in situ hybridization (FISH) studies for 11q22.3, 13q14, 12p11.1-q11, 17p13.1, t(11;14)(q13;q23) and TP53 mutational analysis were performed at trial enrollment. Cases positive for t(11;14)(q13;q23) were excluded from the study. Telomere length was determined as described previously.20
Single nucleotide polymorphism array and gene enrichment analysis
Analysis for CNA, including copy neutral losses of heterozygosity, was done using 6.0 SNP arrays (Affymetrix®, Santa Clara, CA, USA). CNA positions and gene locations were determined with the UCSC Genome Browser, assembly March 2006, NCBI36/hg18. CNA frequencies were compared to those observed in treatment-naïve, standard-risk cases (n=304, no TP53 deletion/mutation).13 Microarray raw data were made publicly available at Gene Expression Omnibus (GEO accession number: GSE131114).
GISTIC2.0 was applied on manually curated DNA copy number data.16 According to default settings, CNA with a q value <0.25 were defined as significant. CNA that reached high confidence levels for being significantly enriched (q value <0.01) were manually curated for minimally affected regions. Genes located within these minimally affected regions were assigned to WikiPathways21,22 and analyzed for pathway enrichments using PathVisio, version 3.2.3.23,24
Next-generation sequencing
Amplicon-based, targeted next-generation sequencing (tNGS) was performed on TP53 exons 2-11, NOTCH1 exon 34, and SF3B1 exons 13-16. In 17 cases TP53, NOTCH1 and SF3B1 mutational status was determined as previously described.25 All coding regions of MGA, SPEN, RBPJ, and SNW1 (in 108 cases each), and CDKN2A and MYC (in 93 cases each) were screened by tNGS.
Quantitative gene expression analysis
Gene expression of CCAT1, MGA, RBPJ, SNW1, HES1, DTX1, MYC, CDKN2A, p14ARF, and p15INK4b was analyzed by quantitative reverse transcription (qRT) polymerase chain reaction (PCR) (TaqMan® Gene Expression Assays; Applied Biosystems®, Foster City, CA, USA). Sample selection was based on highly clonal presence of respective CNA/gene mutations (log2 ratio <−0.8 for deletions and >0.75 for gains; variant allele frequency >0.3 for mutations). Promoter DNA methylation of CDKN2A/B transcripts was assessed by bisulfite PCR followed by Sanger sequencing.
Statistical analysis
Associations between genomic alterations were tested by Fisher exact tests; differences between datasets by Mann Whitney tests. All statistical tests were two-sided and conducted in GraphPad Prism® version 7.0.
Further details are available in the Online Supplementary Methods.
Results
Cohort characteristics
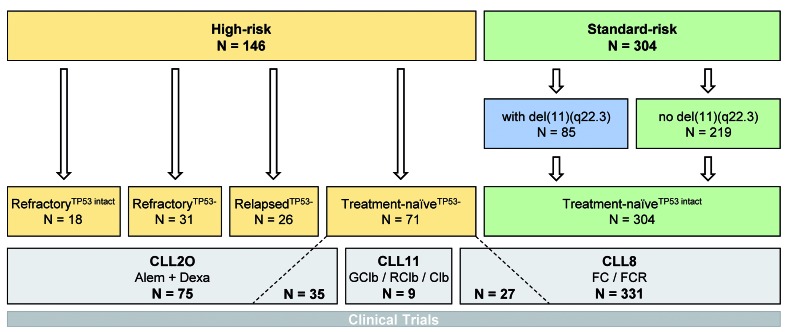
High-resolution genome-wide copy number analysis was performed on CD19-enriched PBMC from 146 CLL patients with high-risk disease. Cases belonged to the following subgroups. (i) Patients refractory to purine analogs from the CLL2O trial (n=49), who had received a median of three previous lines of therapy (range, 1 to 7 lines of therapy). This cohort encompassed patients with TP53 loss and/or mutation (n=31; refractoryTP53−) and patients without TP53 aberration (n=18; refractoryTP53 intact). (ii) Relapsed patients with TP53 loss from the CLL2O trial (n=26; relapsedTP53−), who had received a median of two previous lines of therapy (range, 1 to 4 lines of therapy). (iii) Primary high-risk treatment-naïve patients with TP53 loss (n=71; treatment-naïveTP53−) from the CLL2O (n=35), CLL8 (n=27) and CLL11 trials (n=9). Standard-risk cases used as the reference cohort were derived from the CLL8 trial and included all available cases without a TP53 alteration (n=304; treatment naïveTP53 intact) (Figure 1; characteristics of the patients and samples are provided in Online Supplementary Table S1).
Figure 1.
Description of the sample cohort. High-risk cases of chronic lymphocytic leukemia (CLL) comprised refractory cases (no response or progression-free survival <6 months) with or without TP53 alterations (refractoryTP53−, refractoryTP53 intact, respectively), relapsed cases (response >6 months) with TP53 alterations (relapsedTP53−), and treatment-naïve cases with TP53 alterations (treatment-naïveTP53−). Standard-risk cases were treatment-naïve cases without TP53 alterations (treatment-naïveTP53 intact), comprising cases carrying del(11)(q22.3) and cases not carrying del(11)(q22.3). Patients’ samples were derived from the CLL2O trial, in which patients were treated with alemtuzumab plus dexamethasone, the CLL11 trial, in which treatment with obinutuzumab plus chlorambucil was compared with rituximab plus chlorambucil or chlorambucil monotherapy, and the CLL8 trial, in which treatment with fludarabine/cyclophosphamide (FC) was compared with FC plus rituximab. Alem: alemtuzumab; dexa: dexamethasone; G: obinutuzumab; Clb: chlorambucil; R: rituximab; F: fludarabine; C: cyclophosphamide
Landscape of genomic copy number alterations in high-risk chronic lymphocytic leukemia
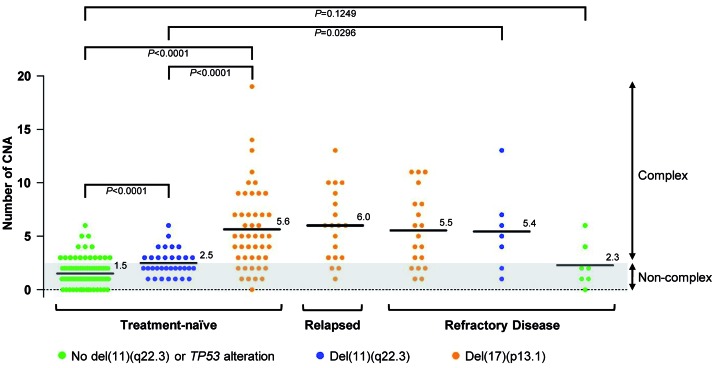
First, we assessed genomic complexity in cases grouped by clinical and genetic characteristics. For this, we used paired cases only to focus on somatically acquired CNA. No difference in median CNA numbers was observed between 39 treatment-naïveTP53− primary high-risk and 38 relapsedTP53−/refractoryTP53− high-risk tumors (5.6 versus 5.8 CNA per case mean) so that genomic complexity with TP53 deficiency was independent of previous therapy. In the absence of TP53 abnormalities, tumors with ATM loss (n=7) had 5.4 CNA per case mean. Refractory high-risk cases lacking TP53 and ATM abnormalities had two or fewer CNA in the majority of cases (5 of 7 cases) (Figure 2).
Figure 2.
Mean number of copy number alterations in clinically and genetically determined subgroups of patients with chronic lymphocytic leukemia. Numbers of copy number alterations (CNA) for all patients who were analyzed against their intra-individual reference DNA (paired). The mean value of CNA numbers is highlighted within each subgroup of patients defined by genetic alterations and disease stage. P values are based on Mann-Whitney tests.
More than half of the high-risk cases showed complex CNA with two or more switches between two copy number states on at least one chromosome (87/146 cases, 60%). Thirteen CNA in 12/146 cases (8%) fulfilled the formal criteria of chromothripsis, which is defined by ten or more switches between two or more copy number states on an individual chromosome.26 Reducing the required number of copy number switches to eight and six increased numbers to 23 CNA in 16 cases (11%) and 50 CNA in 32 cases (22%), respectively.
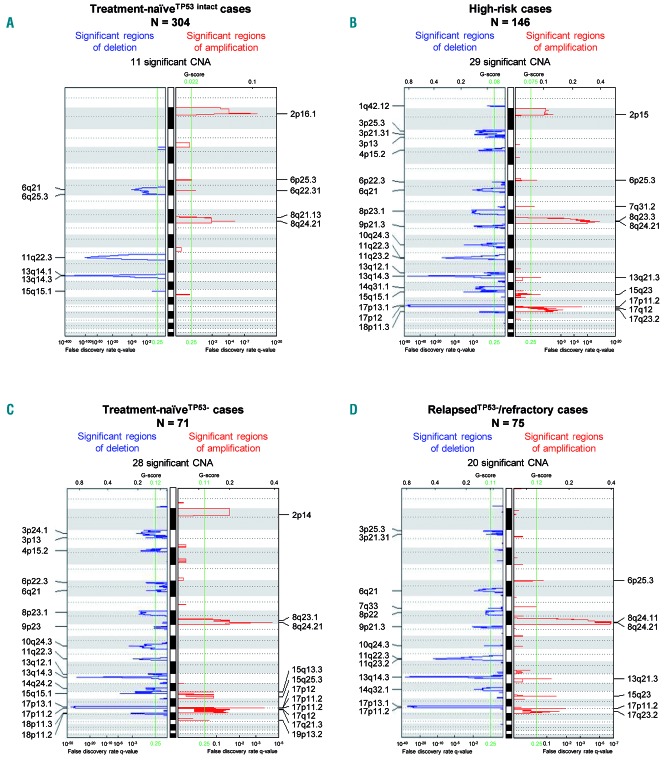
Overall, the cohort of high-risk CLL cases comprised more than 1,500 individual copy number changes (Online Supplementary Table S2). To identify CNA that were significantly enriched, we conducted GISTIC2.0 analysis. While therapy-naïveTP53 intact patients only had 11 significantly enriched CNA, GISTIC2.0 assigned significance to 28 CNA within the treatment-naïveTP53− cohort and to 20 CNA within the relapsedTP53−/refractory cohort when using the default q value of 0.25 as the cut-off for significance (Figure 3).
Figure 3.
Identification of significant copy number gains and losses by GISTIC2.0. (A-D) GISTIC2.0 results of four separate analyses conducted on the following cohorts: (A) treatment-naïveTP53 intact standard-risk cases (n=304); (B) all high-risk cases (n=146); (C) treatment-naïveTP53− primary high-risk cases (n=71); and (D) relapsedTP53−/refractory high-risk cases (n=75). False discovery rate q values are plotted along the x axis. Chromosomal positions are plotted along the y axis. Altered regions with significance levels exceeding 0.25 (marked by vertical green lines) were deemed to be significant.
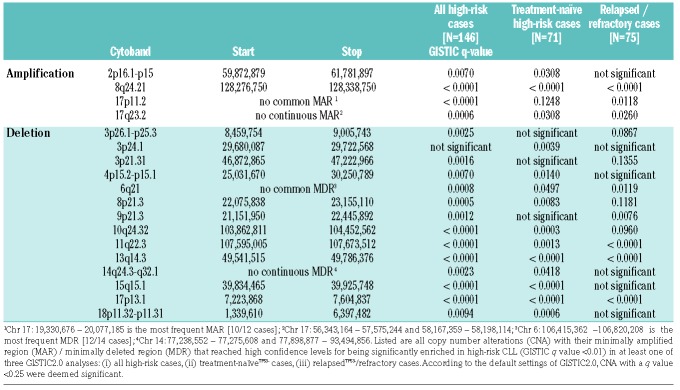
To further reduce complexity, we focused our analysis on CNA that reached high confidence levels for being significantly enriched. High confidence was defined by a GISTIC q value <0.01 in at least one of three GISTIC2.0 analyses: (i) the entire high-risk cohort, (ii) the treatment naïveTP53− cohort, and (iii) the relapsedTP53−/refractory cohort (for minimally affected regions and confidence levels see Table 1). We compared frequencies observed in high-risk cases with those observed in treatment-naïve standard-risk cases. Apart from +(2)(p16.1 p15) and del(6)(q21), all CNA listed in Table 1 occurred at least two times more often in the cohort of high-risk cases: +(8)(q24.21) in 16.4% versus 3.6%, del(8)(p23.1) in 15.8% versus 0.3%, del(18)(p11.31) in 13.7% versus 2.0%, del(10)(q24.32) in 11.0% versus 1.3%, del(15)(q15.1) in 10.3% versus 3.6%, del(3)(p21.31) in 10.3% versus 0.3%, and del(14q) in 7.5% versus 0.7%. Losses in 14q were heterogeneous and a continuous minimally deleted region could not be identified. The two minimally deleted regions in 14q24.3 and 14q31.3 were defined by one case with a discontinuous deletion.
Table 1.
DNA copy number alterations with high confidence levels for being significantly enriched in high-risk chronic lymphocytic leukemia.
GISTIC2.0 also assigned q values <0.01 to novel CNA: +(17)(p11.2) found in 8.2%, +(17)(q23.2) in 7.5%, del(3)(p25.3) in 9.6%, del(3)(p24.1) in 8.9%, del(4)(p15.2) in 8.2%, and del(9)(p21.3) in 8.9% of high-risk cases. Boundaries of minimally affected chromosomal regions in 3p25.3, 3p24.1, and 17q23.2 derived from complex discontinuous CNA.
GISTIC2.0 results for the treatment naïveTP53− and relapsedTP53−/refractory cohort shared similarities, but the latter group had fewer significant CNA. This suggested a selection of clones with CNA contributing to CLL high-risk biology and failure of previous chemo(immuno)therapy. In the relapsedTP53−/refractory cohort, only gain(8)(q24.21) and del(9)(p21.3) retained GISTIC q values <0.01 beyond CNA routinely assessed by FISH (Table 1). Of note, the minimally affected regions in 8q24.21 and 9p21.3 both contained key regulators of cell cycle progression, namely MYC and CDKN2A/B.
In contrast, del(18p), del(15)(q15.1), del(4)(p15), del(14q) and gain(2p) failed to reach significant GISTIC q values in the relapsedTP53−/refractory cohort, so that these CNA less likely conferred refractoriness.
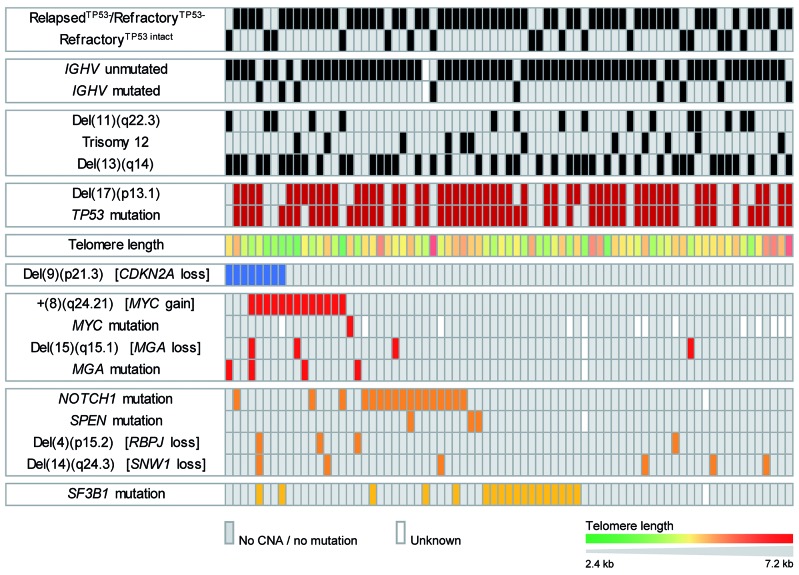
We next analyzed the 146 high-risk cases for associations between genomic lesions. The distribution of genomic lesions across samples is depicted in Figure 4 for relapsedTP53−/refractory cases and in Online Supplementary Figure S1 for treatment naïveTP53− cases. Comparing cases with and without TP53 alteration, only del(11)(q22.3) significantly associated with refractoryTP53 intact cases (P<0.01). Testing for dependence of a genomic lesion on the presence of a TP53 alteration was hampered by the low number of refractoryTP53 intact cases included in the study. Loss of CDKN2A/B was associated with gain of the MYC gene locus, with MGA mutation, and with loss of MGA by del(15)(q15.1) (P=0.028, P=0.044, and P=0.03, respectively). Interestingly, cases with co-occurring CDKN2A/B loss and MYC gain were among those with the shortest telomeres as a potential sign of higher proliferation rates in these tumors. Co-occurrence of coding NOTCH1 and SF3B1 mutations was lower than would have been expected to occur by chance (3/145 cases, P=0.034), although both mutations were frequent. NOTCH1 mutations occurred in 24% relapsedTP53−/refractory and 21% treatment-naïveTP53− cases and SF3B1 mutations in 24% relapsedTP53−/refractory and 23% treatment naïveTP53− cases.
Figure 4.
Distribution of genetic characteristics across relapsedTP53−/refractory cases. CNA: copy number alteration.
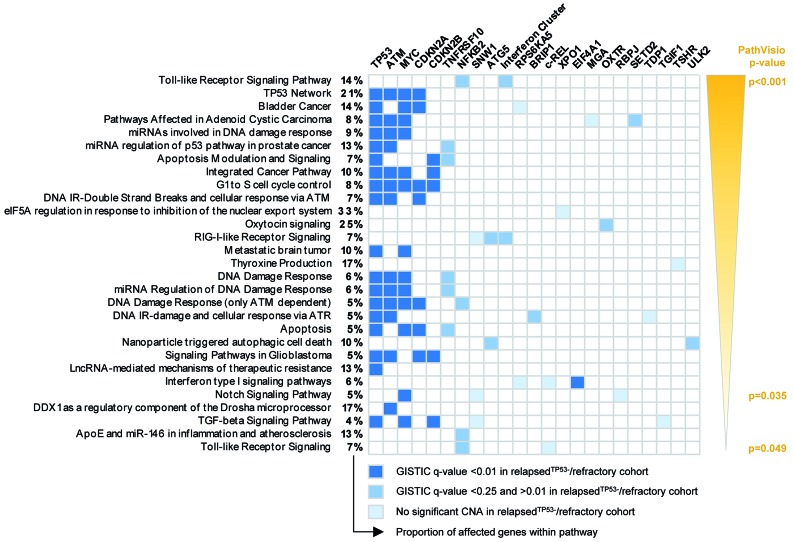
Lastly, to identify distinct molecular pathways affected by CNA with significant GISTIC q values, we analyzed genes located in minimally involved regions as shown in Table 1 by PathVisio. PathVisio assigned significance to 29 pathways. Due to TP53, ATM, and CDKN2A/B loss and MYC gain, 12 of 29 pathways were related to DNA damage response, apoptosis or cell cycle control. Of note, “NOTCH1 signaling” was identified as one significant pathway based on loss of two genes associated with NOTCH1 target gene repression (RPBJ and SNW1) and gain of the NOTCH1 target gene MYC. Toll like receptor signaling was the pathway with the most significant P value, but this was due to loss of an interferon cluster located in 9p21.3 (Figure 5).
Figure 5.
Biological pathways significantly affected by copy number alterations. Significant biological pathways as identified by PathVisio. All genes located within the minimally affected regions of copy number alterations (CNA) that are listed in Table 1 were included in the analysis and genes were assigned to WikiPathways. Pathways are listed by P values as determined via PathVisio. The percentages provided refer to the proportion of affected genes within the respective pathway. The color coding for affected genes refers to the GISTIC q value, which was assigned to the respective CNA.
Since our results drew attention to CDKN2A/B, MYC and NOTCH1 signaling, we next shed more light on genetic lesions relating to them.
Characterization of del(9)(p21.3) as a novel recurrent copy number alteration in high-risk chronic lymphocytic leukemia
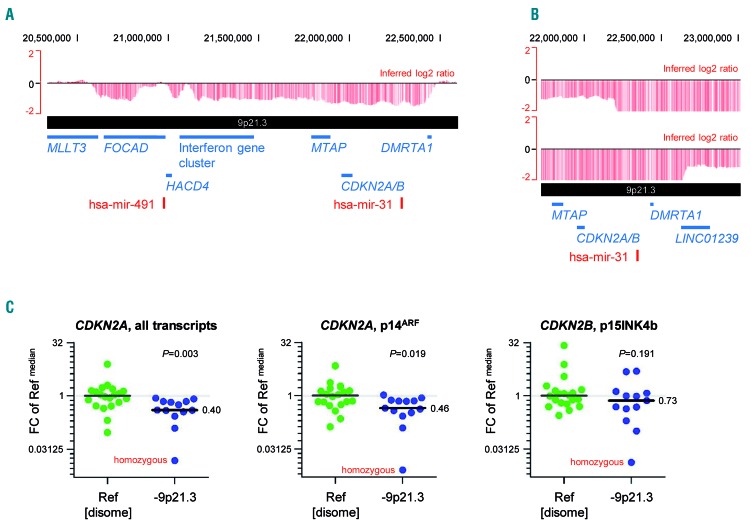
Deletions in 9p21.3 were found in 13 patients (7% treatment-naïveTP53−; 11% relapsedTP53−/refractory cases). This high frequency was surprising, since del(9)(p21.3) has not been observed in standard-risk cases at treatment initiation.13 The minimally deleted region encompassed CDKN2A, CDKN2B, MTAP, DMRTA1 and an interferon gene cluster (Figure 6A). Two cases had focal homozygous deletions within larger monoallelic deletions, but the CDKN2A/B loci were covered by the homozygous deletion in only one case. The minimally deleted homozygous region contained only DMRTA1 coding for a transcription factor with unknown target genes (Figure 6B). Somatic CDKN2A mutations were not found in the high-risk cases screened by tNGS.
Figure 6.
Del(9)(p21.3) leads to loss of CDKN2A/B. (A) Minimal consensus region of monoallelic del(9)(p21.3). Raw log2 ratio, chromosome 9, case #2O_CLL047 displayed with the UCSC genome browser (hg18). Red bars represent determined log2 ratios of single probe sets sorted by their physical position along the chromosome. The minimal consensus region of monoallelic del(9)(p21.3) harboring the CDKN2A/B gene loci is shown. (B) Minimal consensus region of biallelic del(9)(p21.3). Raw log2 ratio, chromosome 9, cases #2O_CLL011 and #2O_CLL050 displayed with the UCSC genome browser (hg18). The minimal consensus region of biallelic del(9)(p21.3) harboring the DMRTA1 gene locus only is shwon. (C) Expression levels of CDKN2A/B transcripts in cases with del(9)(p21.3). Gene expression levels of CDKN2A transcripts (left: all transcripts, middle: p14ARF transcript alone) as well as the CDKN2B transcript p15INK4B (right) were calculated relative to ACTB expression levels. Fold changes (FC) were calculated towards the median ΔCt value of all reference samples (Ref). Median expression levels within each group of samples are highlighted and differences between groups were analyzed by the Mann-Whitney test. All available 9p-deleted cases were included in the analysis.
Consistent with a role of CDKN2A/B in proliferation control, loss of their gene loci has been directly associated with Richter transformation (RT), since del(9)(p21) had so far been only observed in lymph node biopsies with histologically confirmed RT and not in corresponding peripheral blood samples acquired during the CLL phase.27,28 We, therefore, next investigated associations between del(9)(p21.3) and the development of RT, the presence of which had been an exclusion criterion at enrollment on each clinical trial. During the observation period, only one patient with del(9)(p21.3) developed histologically confirmed RT. This patient was under alemtuzumab maintenance therapy and was diagnosed with RT 557 days after trial enrollment. The observation periods for the other 11 patients ranged from 15 to 1387 days. Short periods of less than 4 months, observed in five of the 11 patients, were related to progressive disease (n=2; 38 and 75 days after trial enrollment), fatal infections (n=3), or early patient dropout (n=1). The first patient with progressive disease had marked lymphadenopathy (abdominal lymph nodes >10 cm) but a normal lactate dehydrogenase level of 227 U/L. The second patient had a homozygous CDKN2A deletion with a concurrent MYC gain, rapidly progressive lymphadenopathy, and a vastly increased lactate dehydrogenase level of 1026 U/L. Lymph node biopsies were not taken in either case.
We also searched for patients who developed RT during their observation period and identified 14 patients with histologically confirmed RT all of whom belonged to the relapsedTP53−/refractory cohort. The diagnosis of RT was made at a median of 407 days after trial enrollment (range, 21-568 days). In general, genomic profiles obtained from tumor cell-enriched PBMC taken at trial enrollment did not differ between patients who developed RT during follow up and the other relapsedTP53−/refractory cases (Online Supplementary Figure S2). Three tumors transformed during the first 6 weeks after trial enrollment (#2O_CLLL033 at day 21; #2O_CLL056 at day 36; and #2O_CLL037 at day 41). Interestingly, these three tumors had alterations affecting NOTCH1 as well as MYC signaling alongside TP53 dysfunction (Online Supplementary Tables S1 and S2).
Overall, del(9)(p21.3) was less frequently associated with RT than previous publications had suggested. Our next aim was, therefore, to understand whether CDKN2A/B expression was compensated by the second allele in cases with heterozygous loss. In 9p disome cases, gene expression levels of CDKN2A and CDKN2B were variable. Although the median expression levels of CDKN2A transcripts were significantly lower in cases with del(9)(p21.3), the respective values in cases with heterozygous loss did not fall below the range of values observed in 9p disome cases (Figure 6C, details in Online Supplementary Table S3). Promoter methylation, serving as an explanation of abnormally reduced CDKN2A/B expression in 9p-disome cases, was not found for p14ARF, p16INK4A and p15INK4B transcripts (based on 8 selected 9p disome cases including 2 cases with noticeably low expression levels of CDKN2A transcripts and p14ARF; data not shown).
Characterization of genomic lesions relating to MYC
Gains on 8q were found in 16% of high-risk cases, with a comparable distribution between treatment-naïveTP53− and relapsedTP53−/refractory cases (15% and 17%, respectively). Two types of gains were identified: (i) broad gains covering the MYC locus; (ii) focal gains with a size <500 kb in 8q24.21 affecting a super enhancer region proximal of the MYC locus. The broad type of gains was far more frequent in high-risk cases than in standard-risk ones (14.4% versus 2.2%). The frequency of focal gains was comparable in both risk groups (1.4% and 1.3%) and no relapsedTP53−/refractory case was affected. Only one COSMIC-listed MYC mutation was found in 93 high-risk cases screened.
Despite their low frequency, the focal gains in 8q24.21 raised our interest, since they were the only recurrent gains in non-coding DNA regions found throughout the entire CLL genome. Their minimally gained region encompassed three long non-coding RNA, namely CASC19, CCAT1, and CASC21 (Online Supplementary Figure S3). Of these, Colon Cancer Associated 1 (CCAT1) has been associated with adverse risk in several solid cancers,29,30 since its transcript has been described to stabilize a chromosome loop between MYC and the enhancer region.31 However, CCAT1 expression in CLL cases with 8q gain did not exceed normal expression levels (Online Supplementary Figure S4).
Besides chromosomal gains on 8q, we and others have previously identified MGA deletion/mutation as a potential alternative mechanism for increased MYC activity in CLL.13,32 MGA is a protein that forms a heterodimer with MAX and this heterodimer antagonizes MYC induced transcriptional changes.33 Against the background of frequent MYC gain in relapsedTP53−/refractory cases, loss of a significant GISTIC q value for del(15)(q15.1) in the relapsed/refractory cohort was surprising, since its minimally deleted region encompasses the MGA gene locus. We performed qRT PCR to determine MGA gene expression in cases with heterozygous 15q deletion relative to 15q disome cases and found that MGA transcription in cases with heterozygous MGA loss was fully compensated by the remaining allele (Online Supplementary Figure S5). Nonetheless, we found an increased frequency of truncating MGA mutations with five mutations in 4/108 high-risk cases (3.7%). As all MGA mutated cases belonged to the relapsedTP53−/refractory cohort, they were clearly enriched in this cohort (4/74 cases; 5.4%).
Characterization of genomic lesions related to NOTCH1 signaling
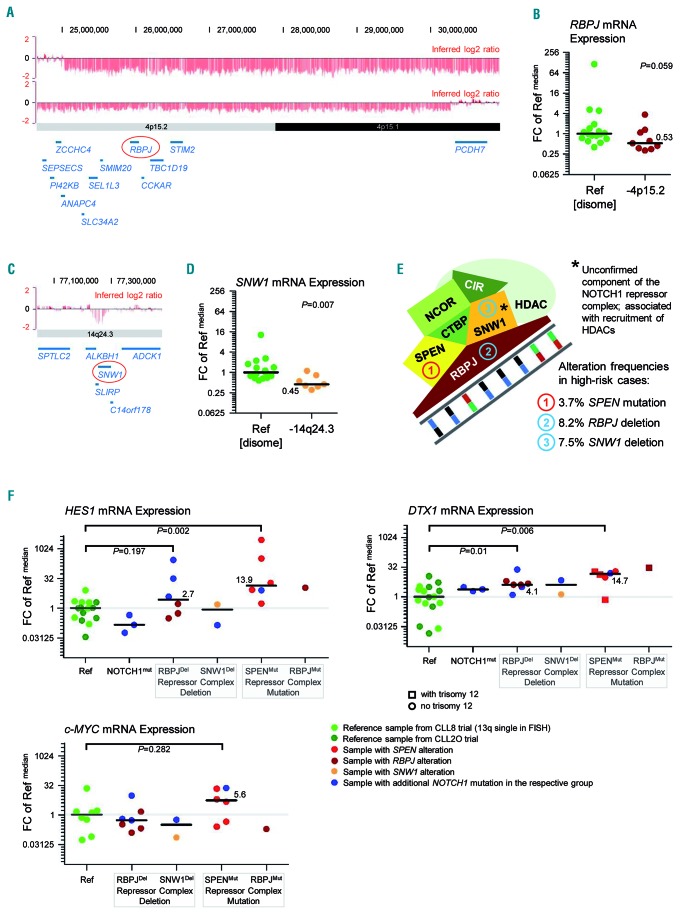
Based on our PathVisio results, we also investigated genomic alterations associated with NOTCH1 signaling. As a novel finding, three components of a protein complex repressing NOTCH1 target genes were recurrently affected: RBPJ in 4p15.2 was deleted in 8.2%; SPEN was mutated in 3.7% (4/108 cases); and SNW1 in 14q24.3 coding for an unconfirmed component of the repressor complex was deleted in 7.5% of cases (Figure 7A, C, E).
Figure 7.
Disruption of the NOTCH1 transcription repressor complex adds to the overall frequency of altered NOTCH1 signaling in high-risk chronic lymphocytic leukemia. (A) Minimal consensus region of del(4)(p15) covering the RBPJ gene locus. Raw log2 ratio, chromosome 4, cases #2O_CLL036 and #2O_CLL068 displayed with the UCSC genome browser (hg18). Red bars represent determined log2 ratios of single probe sets sorted by their physical position along the chromosome. The minimal consensus region of del(4)(p15) harboring the RBPJ gene locus is shown. (B) RBPJ expression levels in cases with del(4)(p15). RBPJ gene expression levels were calculated relative to ACTB expression levels and fold changes (FC) were calculated towards the median ΔCt value of all reference samples (Ref). Median expression levels within each group of samples are highlighted and the difference between the two groups was analyzed by the Mann-Whitney test. Only 4p-deleted cases with a log2 ratio lower than 0.89 were included. (C) Minimal consensus region of del(14)(q24.3) covering the SNW1 gene locus. Raw log2 ratio, chromosome 14, case #2O_CLL027. The minimal consensus region of del(14)(q24.3) harboring the SNW1 gene locus is shown. (D) SNW1 expression levels in cases with del(14)(q24.3). SNW1 gene expression levels were calculated relative to ACTB expression levels and FC were calculated towards the median ΔCt value of all reference samples (Ref). Median expression levels within each group of samples are highlighted and the difference between the two groups was analyzed by the Mann-Whitney test. Only 14q-deleted cases with a log2 ratio lower than 0.87 were included. (E) Composition of the NOTCH1 transcription repressor complex. Simplified illustration of the NOTCH1 transcription repressor complex. The mutation frequency of SPEN is based on targeted next-generation sequencing results on 108 cases from the high-risk cohort. The deletion frequencies of RBPJ and SNW1 are based on the entire high-risk cohort (n=146). SNW1/SKIP is an unconfirmed component of the NOTCH1 repressor complex, which has been associated with the recruitment of histone deacetylases. (F) HES1, DTX1 and MYC expression levels in cases with genomic alterations affecting the NOTCH1 transcription repressor complex. HES1, DTX1 and MYC gene expression levels were calculated relative to 18S expression levels and FC were calculated towards the median ΔCt value of all reference samples (Ref). Median expression levels within each group of samples are highlighted and differences between groups were analyzed by the Mann-Whitney tests. Non-purified peripheral blood mononuclear cells with a tumor cell load >70% were used for the experiment. Reference samples without evidence of a genetic alteration affecting NOTCH1 signaling were taken from the CLL8 trial (favorable risk cases with 13q deletion as sole abnormality in routine fluorescence in situ hybridization analysis; light green) and from the CLL2O trial (high risk cases; dark green). Additionally, three cases with a highly clonal NOTCH1 mutation were included (blue). Cases with RBPJ alteration are shown in dark red, samples with SNW1 alteration in orange, and samples with SPEN alteration in pale red. Samples with additional NOTCH1 mutation are indicated in blue within their respective sample group. The DTX1 gene locus is located on chromosome 12. In the figure illustrating DTX1 expression levels, cases without trisomy 12 are indicated by a round symbol and cases with trisomy 12 are indicated by a square symbol. Trisomy 12 appeared to be enriched in samples with mutations affecting the repressor complex. A gene dosage effect on DTX1 gene expression was not apparent.
RBPJ is essential for DNA binding of the NOTCH1 intracellular domain (NICD1), which is released as a transcription factor upon activation of the NOTCH1 cell surface receptor. In the absence of NICD1 in the nucleus, RBPJ forms a protein complex with SHARP encoded by SPEN and other proteins that recruit histone deacetylases to condense the chromatin around NOTCH1 target genes.34–36 Disruption of this repressor complex has been associated with tumorigenesis.37 SKIP encoded by SNW1 is an unconfirmed complex component that has been associated with recruitment of histone deacetylases to NOTCH1-regulated genes.34
Median gene expression levels of RBPJ and SNW1 were lower in cases with deletion, supporting a potential functional relevance of RBPJ and SNW1 loss (Figure 7B, D). This led us to hypothesize that disruption of the NOTCH1 repressor complex might lead to de-repression of NOTCH1 target genes. We therefore measured expression levels of HES1, DTX1 and MYC as genes known to be NOTCH1-regulated in primary CLL samples.38,39 Non-CD19-enriched PBMC samples were used for this analysis of NOTCH1 target gene expression, since EDTA-containing sorting buffer used for cell enrichment can activate NOTCH1 signaling ex vivo (Online Supplementary Figure S6).40 Sample suitability was determined by the following selection criteria: (i) tumor cell count ≥70%; (ii) variant allele frequency >0.3 for SPEN and RBPJ mutations; and (iii) log2 ratio <−0.8 for del(4)(p15.2) or del(14)(q24.3)
Six of nine available SPEN mutant samples and one available RBPJ mutant sample fulfilled these requirements. Target gene expression did not correlate with the number of non-B cells per sample (Online Supplementary Figure S7). In line with our hypothesis, SPEN mutant tumors had significantly higher median expression levels of HES1 and DTX1 and four SPEN mutant cases had remarkably high expression levels of MYC.
Of 14 RBPJ-deleted samples with available RNA, seven fulfilled our requirements. In this highly selected group of cases, RBPJ deletion occurred together with an activating NOTCH1 mutation in three out of the seven cases. HES1 expression was very variable, but in cases with combined RBPJ and NOTCH1 disruption it was highly increased. DTX1 expression was more independent of the NOTCH1 mutation status and its median was significantly higher in cases with RBPJ loss. However, expression of MYC was not altered in 4p deleted cases.
Of 12 SNW1-deleted cases with available RNA, only two cases fulfilled our requirements (Figure 7F; details in Online Supplementary Table S4).
Discussion
Genome-wide screening for CNA in high-risk CLL revealed complex genomic aberrations in the majority of cases, which was in contrast to the high genomic stability observed in treatment-naïve, standard-risk CLL without TP53 or ATM alterations.13 Our cohort consisted mainly of TP53-deficient cases and we found genomic instability to the same degree in patients who had or had not been previously treated.
Our focus was to identify CNA within complex genomic rearrangements relevant to the development of high-risk CLL. Using GISTIC2.0 as a systematic approach to distinguish meaningful chromosomal aberrations from random background alterations,16 we identified CDKN2A/B loss, MYC gain and abnormally strong NOTCH1 signaling as significantly enriched alterations. The genomic profiles observed in peripheral blood CLL cells taken from high-risk patients shared extensive similarities with the profiles found in RT, in which CDKN2A/B loss, MYC gain and activating NOTCH1 mutations have also been identified as hallmark genomic lesions alongside TP53 alterations.27,28 However, none of the respective alterations could be directly associated with transformation so that extrinsic stimuli from the microenvironment and/or additional intrinsic stimuli are required to induce transformation.
Loss of CDKN2A/B is a sparsely described CNA in CLL,41 although homozygous deletion has recently been associated with resistance to venetoclax treatment.5 The role in CLL progression of the signaling network around MYC, which comprises transcriptional inhibitors such as MGA,42 is poorly understood. The CDKN2A/B gene loci encode cell-cycle regulators decelerating cell proliferation at multiple levels.43 The CDKN2A gene product p14ARF tightly controls pro-proliferative activities of MYC so that reduced CDKN2A expression in combination with increased MYC expression can result in accelerated proliferation.44 Enrichment of CDKN2A/B loss and MYC gain in the relapsedTP53−/refractory cohort and frequent co-occurrence of both aberrations therefore hints at a key role for deficient cell-cycle control in the development of high-risk CLL.
The small focal gains within the 8q24.21 super-enhancer region are relevant with regard to them harboring binding sites for the NICD1 transcription factor.38 MYC is a well established target gene of NOTCH1,39 and tight bonds between aberrantly strong NOTCH1 signaling and increased MYC activity have been observed in T-cell acute lymphoblastic leukemia and were also shown to exist in CLL.38,39 Hence, genomic aberrations with an activating effect on NOTCH1 signaling strength can indirectly increase the number of cases with enhanced MYC activity. Besides coding activating NOTCH1 mutations, which prolong NICD1 transcription factor activity,45 various other genomic alterations were identified to interfere with NOTCH1 signaling. These alterations include non-coding NOTCH1 mutations in the 3’ untranslated region,46 FBXW7 mutations,47 MED12 mutations,48 as well as SPEN mutations and probably also RBPJ alterations.
The fact that NOTCH1 and SF3B1 mutations are often mutually exclusive raises the question as to what extent SF3B1 mutation can disturb the physiological balance between NOTCH1 signaling and MYC transcription. This question is based on the observation that the strength of NOTCH1 signaling depends on DVL2, which inhibits transcriptional activation by NOTCH1.49 Mutations in SF3B1 lead to alternative splicing of DVL2 and the resulting splice variant has been shown to lack its ability to modulate NOTCH1 signaling.50 SF3B1 mutations may, therefore, constitute another frequent mechanism to strengthen the NOTCH1-MYC signaling axis.
Taken together, our results raise the hypothesis that multiple genetic lesions in high risk CLL converge in upregulated MYC activity. Testing this hypothesis requires an integrative analysis of the genome, transcriptome and proteome in samples strictly processed at 4°C and in the absence of calcium chelators. For translation of our results into clinical practice, a systematic record of genomic alterations identified as meaningful in our study needs to be obtained in more recent, prospective clinical trials including treatment arms based on BTK or Pi3K inhibition and/or antagonism of BCL-2. Novel potential markers must be tested for relevance in each treatment arm and markers proving to be relevant must be utilized for the assembly of a genomic clinical database. In the long-term, such an approach will allow an estimation of the likelihood of benefit or disadvantage from a given treatment regimen, hence paving the way towards more personalized treatment choices in the future management of CLL patients.
Acknowledgments
The authors would like to thank the patients who participated in the CLL2O, CLL8, and CLL11 trials; the investigators who treated patients and submitted samples; and Christina Galler, Sabrina Rau, and Jacqueline Fiegel for excellent assistance and support.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/5/1379
Funding
This study was supported by the German Research Foundation (DFG: ED 256/1 1; SFB 1074/B2 and Z1), by grants from the German Federal Ministry of Education and Research (BMBF, PRECiSe) and by the Barts Charity Fund. Central genetic diagnostics were partly funded by F. Hoffmann-La Roche.
References
- 1.Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet. 2018;391(10129):1524–1537. [DOI] [PubMed] [Google Scholar]
- 2.Parikh SA. Chronic lymphocytic leukemia treatment algorithm 2018. Blood Cancer J. 2018;8(10):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woyach JA, Johnson AJ. Targeted therapies in CLL: mechanisms of resistance and strategies for management. Blood. 2015;126(4):471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger JA, Landau DA, Taylor-Weiner A, et al. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herling CD, Abedpour N, Weiss J, et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat Commun. 2018;9(1):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stilgenbauer S, Zenz T. Understanding and managing ultra high-risk chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2010;2010:481–488. [DOI] [PubMed] [Google Scholar]
- 7.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442–2451. [DOI] [PubMed] [Google Scholar]
- 8.Anderson MA, Tam C, Lew TE, et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood. 2017;129(25): 3362–3370. [DOI] [PubMed] [Google Scholar]
- 9.Thompson PA, O’Brien SM, Wierda WG, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121(20):3612–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133(11):1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunnarsson R, Isaksson A, Mansouri M, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24(1):211–215. [DOI] [PubMed] [Google Scholar]
- 12.Ouillette P, Collins R, Shakhan S, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118(11):3051–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelmann J, Holzmann K, Miller F, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120(24): 4783–4794. [DOI] [PubMed] [Google Scholar]
- 14.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. [DOI] [PubMed] [Google Scholar]
- 16.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GIS-TIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilgenbauer SCF, Leblond V, Delmer A, et al. Alemtuzumab combined with dexamethasone, followed by alemtuzumab maintenance or allo-SCT in “ultra high-risk” CLL: final results from the CLL2O phase II study. Blood. 2014;124(21):1991. [Google Scholar]
- 18.Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376(9747):1164–1174. [DOI] [PubMed] [Google Scholar]
- 19.Goede V, Fischer K, Busch R, et al. Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med. 2014;370(12):1101–1110. [DOI] [PubMed] [Google Scholar]
- 20.Steinbrecher D, Jebaraj BMC, Schneider C, et al. Telomere length in poor-risk chronic lymphocytic leukemia: associations with disease characteristics and outcome. Leuk Lymphoma. 2018;59(7):1614–1623. [DOI] [PubMed] [Google Scholar]
- 21.Kelder T, van Iersel MP, Hanspers K, et al. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40(Database issue):D1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutmon M, Riutta A, Nunes N, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44(D1):D488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Iersel MP, Kelder T, Pico AR, et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutmon M, van Iersel MP, Bohler A, et al. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015;11(2): e1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. [DOI] [PubMed] [Google Scholar]
- 26.Rausch T, Jones DT, Zapatka M, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1-2):59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chigrinova E, Rinaldi A, Kwee I, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122(15): 2673–2682. [DOI] [PubMed] [Google Scholar]
- 28.Fabbri G, Khiabanian H, Holmes AB, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013;210(11):2273–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49(3):255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Xie H, Gao Q, Zhan H, Xiao H, Zou Y, et al. Colon cancer associated transcripts in human cancers. Biomed Pharmacother. 2017;94:531–540. [DOI] [PubMed] [Google Scholar]
- 31.Kim T, Cui R, Jeon YJ, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111(11):4173–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Paoli L, Cerri M, Monti S, et al. MGA, a suppressor of MYC, is recurrently inactivated in high risk chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54(5): 1087–1090. [DOI] [PubMed] [Google Scholar]
- 33.Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18(24):7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao HY, Ordentlich P, Koyano-Nakagawa N, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12(15):2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oswald F, Kostezka U, Astrahantseff K, et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21(20):5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66(10):1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulic I, Robertson G, Chang L, et al. Loss of the Notch effector RBPJ promotes tumorigenesis. J Exp Med. 2015;212(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbri G, Holmes AB, Viganotti M, et al. Common nonmutational NOTCH1 activation in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2017;114(14):E2911–E2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48): 18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rand MD, Grimm LM, Artavanis-Tsakonas S, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20(5):1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martel V, Guerci A, Humbert JC, et al. De novo methylation of tumour suppressor genes CDKN2A and CDKN2B is a rare finding in B-cell chronic lymphocytic leukaemia. Br J Haematol. 1997;99(2):320–324. [DOI] [PubMed] [Google Scholar]
- 42.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Ann Rev Cell Dev Biol. 2000;16:653–699. [DOI] [PubMed] [Google Scholar]
- 43.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371(6494):257–261. [DOI] [PubMed] [Google Scholar]
- 44.Cleveland JL, Sherr CJ. Antagonism of Myc functions by Arf. Cancer Cell. 2004;6(4):309–311. [DOI] [PubMed] [Google Scholar]
- 45.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. [DOI] [PubMed] [Google Scholar]
- 46.Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519–524. [DOI] [PubMed] [Google Scholar]
- 47.Close V, Close W, Kugler SJ, et al. FBXW7 mutations reduce binding of NOTCH1, leading to cleaved NOTCH1 accumulation and target gene activation in CLL. Blood. 2019;133(8):830–839. [DOI] [PubMed] [Google Scholar]
- 48.Wu B, Slabicki M, Sellner L, et al. MED12 mutations and NOTCH signalling in chronic lymphocytic leukaemia. Br J Haematol. 2017;179(3):421–429. [DOI] [PubMed] [Google Scholar]
- 49.Collu GM, Hidalgo-Sastre A, Acar A, et al. Dishevelled limits Notch signalling through inhibition of CSL. Development. 2012;139(23):4405–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Brooks AN, Fan J, et al. Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell. 2016;30(5):750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]