The structure of the Pseudomonas aeruginosa T6SS PldB immunity protein PA5086 is reported at 1.9 Å resolution. Comparison of PA5086 with its homologs PA5087 and PA5088 showed great similarities in sequence and structure, but vast divergences in electrostatic potential surfaces.
Keywords: X-ray crystallography, T6SS immunity proteins, stockpiling mechanism, electrostatic potential surface, PA5086, Pseudomonas aeruginosa, type VI secretion system
Abstract
The bacterial type VI secretion system (T6SS) secretes many toxic effectors to gain advantage in interbacterial competition and for eukaryotic host infection. The cognate immunity proteins of these effectors protect bacteria from their own effectors. PldB is a T6SS trans-kingdom effector in Pseudomonas aeruginosa that can infect both prokaryotic and eukaryotic cells. Three proteins, PA5086, PA5087 and PA5088, are employed to suppress the toxicity of PldB-family proteins. The structures of PA5087 and PA5088 have previously been reported, but the identification of further distinctions between these immunity proteins is needed. Here, the crystal structure of PA5086 is reported at 1.90 Å resolution. A structural comparison of the three PldB immunity proteins showed vast divergences in their electrostatic potential surfaces. This interesting phenomenon provides an explanation of the stockpiling mechanism of T6SS immunity proteins.
1. Introduction
As one of the leading opportunistic pathogens of humans, Pseudomonas aeruginosa possesses a powerful type VI secretion system (T6SS) that can secrete a large variety of effector–immunity (E–I) pairs that affect many areas of inter-species competition and host invasion (Russell et al., 2011 ▸; Ho et al., 2014 ▸; Hood et al., 2010 ▸). Two phospholipase D (PLD) family proteins, PldA and PldB, which exhibit dual HKDXXXXD (HKD) motifs, have been identified as trans-kingdom T6SS effectors that can affect both bacterial and human cells (Russell et al., 2013 ▸; Jiang et al., 2014 ▸).
Three consecutive genes immediately adjacent to the pldB gene encode three immunity proteins, PA5086, PA5087 and PA5088, that can suppress the toxicity of PldB. Our group have reported the crystal structures of PA5087 and PA5088, which both consist of several Sel1-like repeats [SLRs, named after the Caenorhabditis elegans sel-1 gene products that form helix–turn–helix (α/α) motifs] that fold into super-ring forms (Grant & Greenwald, 1996 ▸; Das et al., 1998 ▸; Yang et al., 2016 ▸). We observed that PA5087 and PA5088 share great structural similarity but differ in their electrostatic potential surfaces (Yang et al., 2016 ▸).
Here, we present the 1.90 Å resolution crystal structure of PA5086, which possesses the same overall structure as its immunity protein neighbors PA5087 and PA5088 but is quite different from them in electrostatic potential surface. Our work completed the characterization of all three PldB immunity proteins and should aid future functional studies.
2. Materials and methods
2.1. Macromolecule production
The gene encoding PA5086 (without the N-terminal 21-residue signal peptide) was amplified from P. aeruginosa PAO1 genomic DNA. The PCR products were cloned into the pGEX-6T vector, which was a modification of the pGEX-6P-1 vector (GE Healthcare, Marlborough, Massachusetts, USA) by adding an N-terminal His6 tag prior to the GST fusion tag and replacing the PreScission protease site with a Tobacco etch virus (TEV) protease cleavage site. The plasmid was then transformed into Escherichia coli BL21(DE3) cells. Large cultures were induced at an OD600 of 0.6 with a final concentration of 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and were left to express at 290 K for 18 h.
The cells containing PA5086 protein were pelleted, resuspended in buffer consisting of 20 mM Tris pH 8.0, 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF) and disrupted using a high-pressure homogenizer. The lysate was centrifuged at 13 000g for 40 min. The supernatant containing the soluble protein was purified via affinity chromatography with nickel–nitrilotriacetic acid resin (Bio-Rad, Hercules, California, USA) and the His6-GST tag was removed by overnight hydrolysis with TEV protease and reloading with 20 mM imidazole. The PA5086 protein was further purified by gel-filtration chromatography (Superdex 200, GE Healthcare, Marlborough, Massachusetts, USA) with buffer consisting of 20 mM Tris pH 8.0, 150 mM NaCl. The purified protein was collected and ultrafiltered to 19 mg ml−1. Macromolecule-production information is summarized in Table 1 ▸.
Table 1. Macromolecule-production information.
| Source organism | P. aeruginosa |
| DNA source | P. aeruginosa PAO1 |
| Forward primer† | CGCGGATCCGCTGACAGCAAGCCTTTTGTC |
| Reverse primer‡ | GGCCTCGAGTTACCGCTCCGGCTTCCAGTC |
| Cloning vector | pGEX-6T |
| Expression vector | pGEX-6T |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | GSADSKPFVCVNEKDHLPPLDPQADAWYREAATLAKPDTLRPWPRIVGLYSKAAERGHWKAMHNLANLYRTGWPGGVEKDTQKALDLYQKMIDLDVPQGFYDMGAMIGNRAGVKNPATDGLTFLDKAASLGNPPALTELGKFYIYVAKKKDLGLAYTHCAASQGYAPASYELGAYYKIVEHNFPKALVYYQVSVSQGGKSAAFFLSRVFGSETPPASAMWYAPDEKLREAYYSIYKKLEADPDLRFPNLIEDYPLPPHPTQGYDADRPDWKPER |
The BamHI site is underlined.
The XhoI site is underlined.
2.2. Crystallization
Crystallization screening of PA5086 was carried out at 293 K using the sitting-drop vapor-diffusion technique. The best crystals of PA5086 were obtained within five weeks using a condition consisting of 0.2 M sodium sulfate decahydrate, 20%(w/v) polyethylene glycol 3350. Crystallization information is summarized in Table 2 ▸.
Table 2. Crystallization.
| Method | Sitting-drop vapor diffusion |
| Plate type | 48-well |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 19 |
| Buffer composition of protein solution | 20 mM Tris pH 8.0, 150 mM NaCl |
| Composition of reservoir solution | 0.2 M sodium sulfate decahydrate, 20%(w/v) polyethylene glycol 3350 |
| Volume and ratio of drop | 1 µl protein solution:1 µl reservoir solution |
| Volume of reservoir (µl) | 80 |
2.3. Data collection and processing
A data set was collected from a PA5086 crystal at 100 K at station 1W2B at Beijing Synchrotron Radiation Facility (BSRF). All data were initially integrated, scaled and merged using the XDS package (Kabsch, 2010 ▸) and were processed using the CCP4 suite (Winn et al., 2011 ▸; Potterton et al., 2018 ▸). Initial phases were acquired by molecular replacement using the structure of PA5087 (PDB entry 5jkp; Yang et al., 2016 ▸) as the search model in phenix.phaser (Liebschner et al., 2019 ▸) and structure determination was performed by phenix.autobuild (Liebschner et al., 2019 ▸).
2.4. Structure solution and refinement
The initial model was manually built and adjusted with Coot (Emsley et al., 2010 ▸) and further refined using phenix.refine (Liebschner et al., 2019 ▸). The quality of the final model was checked with MolProbity (Chen et al., 2010 ▸). Data-collection and refinement statistics are given in Tables 3 ▸ and 4 ▸, respectively.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | Beamline 1W2B, BSRF |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 100 |
| Detector | SX165 |
| Crystal-to-detector distance (mm) | 124.3 |
| Rotation range per image (°) | 1 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 10 |
| Space group | C2 |
| a, b, c (Å) | 60.10, 63.20, 66.90 |
| α, β, γ (°) | 90, 105.00, 90 |
| Mosaicity (°) | 0.362 |
| Resolution range (Å) | 32.9–1.9 (1.95–1.90) |
| Total No. of reflections | 138682 (9763) |
| No. of unique reflections | 18481 (1291) |
| Completeness (%) | 96.64 (94.13) |
| Multiplicity | 7.5 (7.56) |
| 〈I/σ(I)〉 | 25.03 (8.91) |
| R r.i.m. | 0.05 |
| Overall B factor from Wilson plot (Å2) | 10.8 |
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 32.9–1.90 (1.95–1.90) |
| Completeness (%) | 96.63 |
| No. of reflections, working set | 17529 |
| No. of reflections, test set | 952 |
| Final R cryst | 0.229 |
| Final R free | 0.266 |
| Cruickshank DPI | 0.222 |
| No. of non-H atoms | |
| Protein | 2085 |
| Ligand | 0 |
| Water | 256 |
| Total | 2341 |
| R.m.s. deviations | |
| Bonds (Å) | 0.005 |
| Angles (°) | 0.745 |
| Average B factors (Å2) | |
| Protein | 13.1 |
| Water | 19.9 |
| Overall | 13.8 |
| Ramachandran plot | |
| Favored regions (%) | 95.8 |
| Additionally allowed (%) | 4.2 |
| Outliers (%) | 0 |
PyMOL (version 1.2; Schrödinger) was used to prepare structural figures. Sequence alignment was performed by ClustalOmega (Sievers et al., 2011 ▸). The figure showing the sequence alignment was generated by ESPript (Robert & Gouet, 2014 ▸). Electrostatic potentials were calculated using the PDB2PQR server with the AMBER force field and were rendered using PyMOL in conjunction with the APBS plugin (Dolinsky et al., 2004 ▸; Baker et al., 2001 ▸).
3. Results and discussion
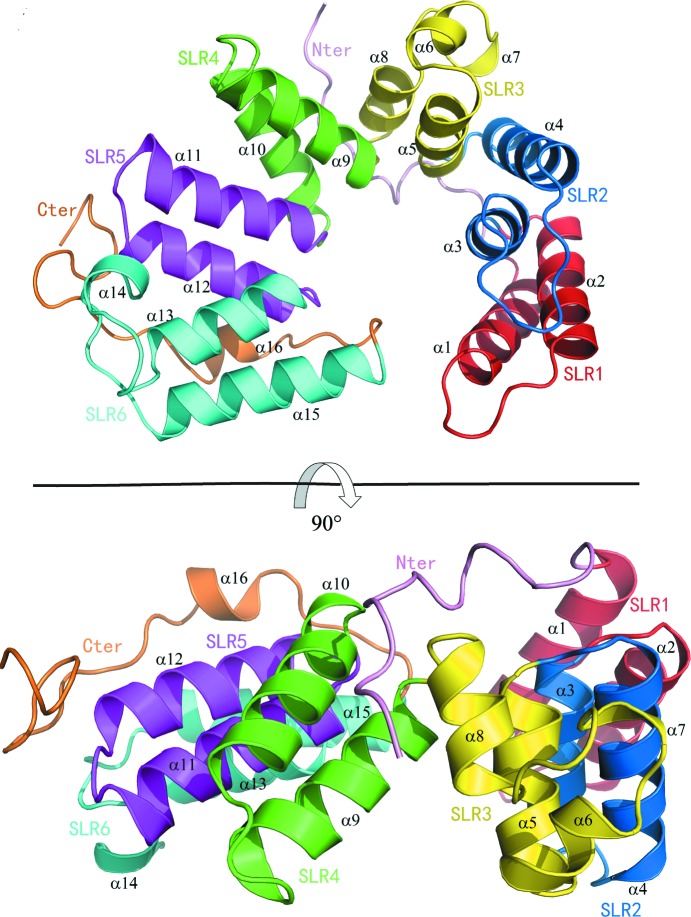
PA5086 is composed of 16 α-helices that can be further categorized into six consecutive SLRs. The overall structure of PA5086 is shown in Fig. 1 ▸. In contrast to the previously reported structures of canonical SLRs, in which the intra-repeat and inter-repeat angles between adjacent helices are always well conserved and the SLRs are always 36 amino-acid residues long (Grant & Greenwald, 1996 ▸; Lüthy et al., 2004 ▸; Urosev et al., 2013 ▸), the six SLRs in PA5086 are distinct in length and conformation, thus folding into a super-ring form rather than the canonical super-helical form (Grant & Greenwald, 1996 ▸; Lüthy et al., 2004 ▸; Urosev et al., 2013 ▸). Similar to the structural features of PA5088 and PA5087, the super-ring fold of PA5086 is stabilized by an ionic bond between Lys55 and Asp262 (Yang et al., 2016 ▸).
Figure 1.
Top and side views of PA5086 in cartoon representation. The secondary-structure elements referred to in the text are labeled; SLR1, SLR2, SLR3, SLR4, SLR5 and SLR6 are colored red, blue, yellow, green, magenta and cyan, respectively.
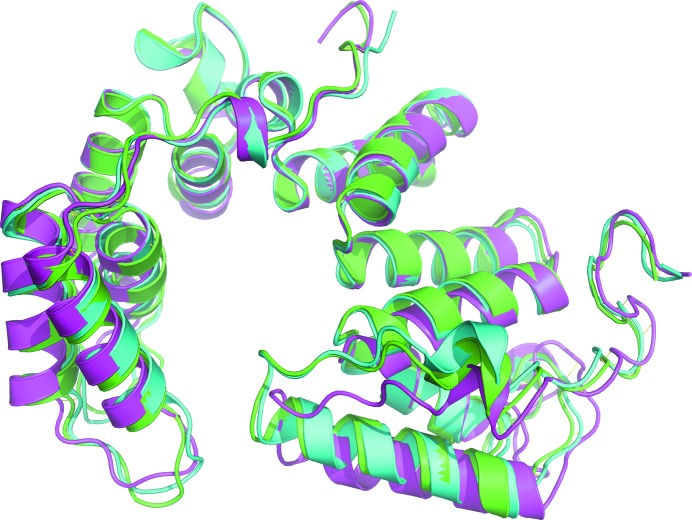
The sequence identity between PA5086 and PA5087 is 85% and that between PA5086 and PA5088 is 73%. With such high sequence identities, structure alignments gave a root-mean-square deviation (r.m.s.d.) of 0.87 Å (260 aligned Cα atoms) between PA5086 and PA5087 (PDB entry 5jkp; Yang et al., 2016 ▸) and an r.m.s.d. of 1.80 Å (252 aligned Cα atoms) between PA5086 and PA5088 (PDB entry 5jjo; Yang et al., 2016 ▸). Structure and sequence alignments among the three PldB immunity proteins are shown in Figs. 2 ▸ and 3 ▸, respectively.
Figure 2.
Structure superposition of PA5086 (green), PA5087 (cyan) and PA5088 (magenta).
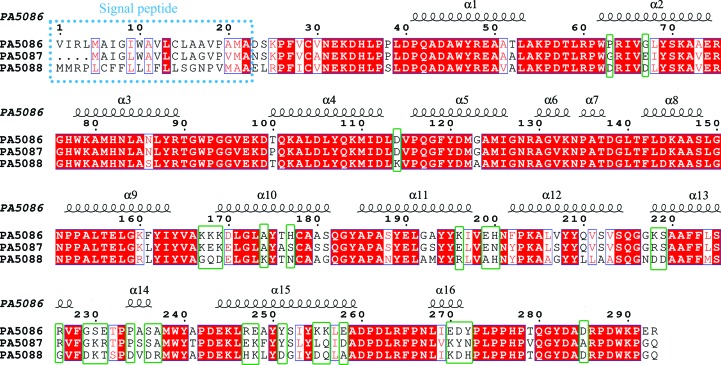
Figure 3.
Sequence alignment of PA5086, PA5087 and PA5088. The α-helices in PA5086 are labeled. The different distributions of basic and acidic residues among the proteins are marked by green rectangles.
The sequence identities among the first nine α-helices of these three proteins, which mainly include the first three SLRs, are quite high, whereas the larger differences among residues are concentrated in their SLR4 and SLR6 units (Fig. 3 ▸). These residue differences do not greatly affect the overall structures, but result in large divergences in the electrostatic potential surfaces among the structures.
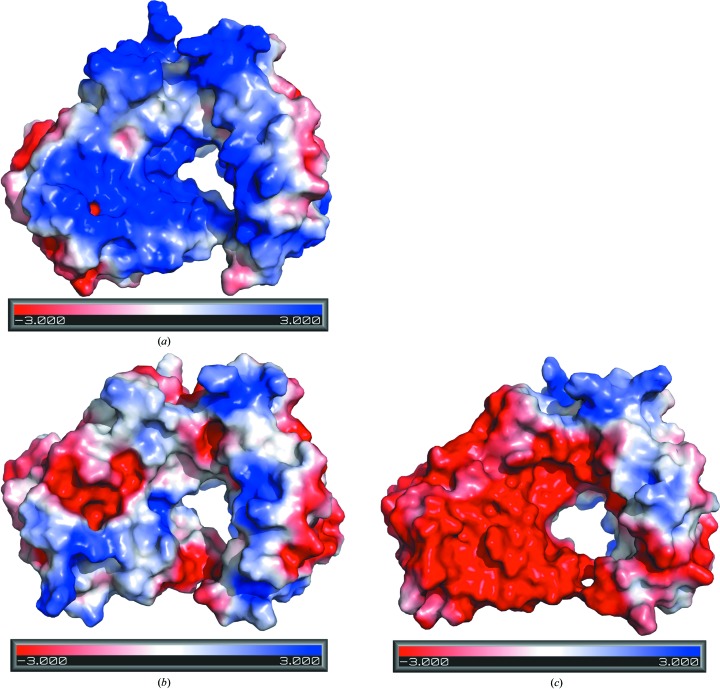
The theoretical pI values of PA5086, PA5087 and PA5088 are 6.67, 7.11 and 5.49, respectively. The electrostatic potential surface of PA5086 from the viewpoint of Fig. 1 ▸ (top) is largely electropositive (Fig. 4 ▸ a), whereas the electrostatic potential for the same surface area of PA5088 is predominantly negative (Fig. 4 ▸ c). In the case of PA5087, the same electrostatic potential surface area possesses both electropositive and electronegative patches (Fig. 4 ▸ b). The divergences in the electrostatic surfaces are mainly caused by differences in the distributions of basic and acidic residues (Fig. 3 ▸), especially for residues in SLR4, SLR5 and SLR6.
Figure 4.
Electrostatic potential surfaces of PA5086 (a), PA5087 (b) and PA5088 (c).
The solvent-accessible surface areas of the PA5086, PA5087 and PA5088 structures are 13 491, 13 671 and 13 627 Å2, respectively. The structure formed by canonical SLRs usually creates an amphipathic groove that functions as a scaffold for protein–protein interactions, and this groove might be suitable for the recognition of target proteins (Das et al., 1998 ▸). In the case of PA5086, PA5087 and PA5088 a hole is formed in the center of each super-ring fold, with different electrostatic potentials for passage through each hole (Fig. 4 ▸), which might play an important role in the interactions with toxins.
PldA and PldB are two trans-kingdom effectors that can affect both prokaryotic and eukaryotic cells (Jiang et al., 2014 ▸). Although they are both categorized into the same Tle5 protein family, their cognate immunity proteins show great structural divergence. PA3488, the cognate immunity protein of PldA, adopts a crab claw-shaped structure (Yang et al., 2017 ▸), whereas PA5086, PA5087 and PA5088 have super-ring folds formed by six SLR units. This suggests that the structures of PldA and PldB may also have major differences.
PA5086, PA5087 and PA5088 function equally well in suppressing PldB toxicity and they combine to contribute to inhibition of PldB in a dose-dependent manner (Jiang et al., 2014 ▸). In our previous study, we performed molecular-docking experiments using the structures of PA5088 and PA5087 against the structure of Streptomyces PLD (PDB entry 2ze4; A. Suzuki, K. Kakuno, R. Saito, Y. Iwasaki, T. Yamane & T. Yamane, unpublished work). We proposed two toxin–antitoxin interaction models via aromatic stacking by the conserved aromatic residues Tyr189, Tyr209, Phe222, Phe223 and Tyr254 on SLR5 and SLR6, or Tyr282 and Trp289 on the C-terminal tails of PA5088 and PA5087 (Yang et al., 2016 ▸). We found that these aromatic residues were also conserved in PA5086 in both the sequence and structure, and therefore they might provide similar PldB-binding affinities.
It is interesting to consider why PldB requires three immunity proteins instead of one as found in most other T6SS effectors. One theory suggests that the ‘stockpiling’ of similar immunity proteins may protect bacteria against similar effectors from different species (Jiang et al., 2014 ▸). Our findings have greatly supported this theory by showing the vast divergences in the electrostatic potential surfaces of similar immunity proteins. PA5086, PA5087 and PA5088 may have different affinities for different PLD effectors from different species owing to their distinct surface potentials, and thus they should provide P. aeruginosa with better protection in competition with bacterial rivals.
Supplementary Material
PDB reference: PA5086, 6lch
Acknowledgments
We are grateful to the staff members at BSRF for sample testing and data collection.
Funding Statement
This work was funded by National Natural Science Foundation of China grant 31700651. National Basic Research Program of China (973 Program) grant 2017YFA0504900. Beijing Municipal Science and Technology Commission grant Z191100007219007.
References
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001). Proc. Natl Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Das, A. K., Cohen, P. W. & Barford, D. (1998). EMBO J. 17, 1192–1199. [DOI] [PMC free article] [PubMed]
- Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. (2004). Nucleic Acids Res. 32, W665–W667. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Grant, B. & Greenwald, I. (1996). Genetics, 143, 237–247. [DOI] [PMC free article] [PubMed]
- Ho, B. T., Dong, T. G. & Mekalanos, J. J. (2014). Cell Host Microbe, 15, 9–21. [DOI] [PMC free article] [PubMed]
- Hood, R. D., Singh, P., Hsu, F., Güvener, T., Carl, M. A., Trinidad, R. R., Silverman, J. M., Ohlson, B. B., Hicks, K. G., Plemel, R. L., Li, M., Schwarz, S., Wang, W. Y., Merz, A. J., Goodlett, D. R. & Mougous, J. D. (2010). Cell Host Microbe, 7, 25–37. [DOI] [PMC free article] [PubMed]
- Jiang, F., Waterfield, N. R., Yang, J., Yang, G. & Jin, Q. (2014). Cell Host Microbe, 15, 600–610. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Lüthy, L., Grütter, M. G. & Mittl, P. R. (2004). J. Mol. Biol. 340, 829–841. [DOI] [PubMed]
- Potterton, L., Agirre, J., Ballard, C., Cowtan, K., Dodson, E., Evans, P. R., Jenkins, H. T., Keegan, R., Krissinel, E., Stevenson, K., Lebedev, A., McNicholas, S. J., Nicholls, R. A., Noble, M., Pannu, N. S., Roth, C., Sheldrick, G., Skubak, P., Turkenburg, J., Uski, V., von Delft, F., Waterman, D., Wilson, K., Winn, M. & Wojdyr, M. (2018). Acta Cryst. D74, 68–84. [DOI] [PMC free article] [PubMed]
- Robert, X. & Gouet, P. (2014). Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed]
- Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W. & Mougous, J. D. (2011). Nature, 475, 343–347. [DOI] [PMC free article] [PubMed]
- Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., Wai, S. N. & Mougous, J. D. (2013). Nature, 496, 508–512. [DOI] [PMC free article] [PubMed]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D. & Higgins, D. G. (2011). Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed]
- Urosev, D., Ferrer-Navarro, M., Pastorello, I., Cartocci, E., Costenaro, L., Zhulenkovs, D., Maréchal, J. D., Leonchiks, A., Reverter, D., Serino, L., Soriani, M. & Daura, X. (2013). BMC Struct. Biol. 13, 19. [DOI] [PMC free article] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Yang, X.-Y., Li, Z.-Q., Gao, Z.-Q., Wang, W.-J., Geng, Z., Xu, J.-H., She, Z. & Dong, Y.-H. (2017). Protein Sci. 26, 2083–2091. [DOI] [PMC free article] [PubMed]
- Yang, X.-Y., Li, Z.-Q., She, Z., Geng, Z., Xu, J.-H., Gao, Z.-Q. & Dong, Y.-H. (2016). FEBS Lett. 590, 2787–2796. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PA5086, 6lch