This article reports conformational polymorphisms of the EF-hand protein MCFD2 which is involved in glycoprotein transport..
Keywords: cargo receptor, conformational polymorphism, crystal packing, glycoprotein transport, intracellular lectin, MCFD2, ERGIC-53
Abstract
The transmembrane intracellular lectin ER–Golgi intermediate compartment protein 53 (ERGIC-53) and the soluble EF-hand multiple coagulation factor deficiency protein 2 (MCFD2) form a complex that functions as a cargo receptor, trafficking various glycoproteins between the endoplasmic reticulum (ER) and the Golgi apparatus. It has been demonstrated that the carbohydrate-recognition domain (CRD) of ERGIC-53 (ERGIC-53CRD) interacts with N-linked glycans on cargo glycoproteins, whereas MCFD2 recognizes polypeptide segments of cargo glycoproteins. Crystal structures of ERGIC-53CRD complexed with MCFD2 and mannosyl oligosaccharides have revealed protein–protein and protein–sugar binding modes. In contrast, the polypeptide-recognition mechanism of MCFD2 remains largely unknown. Here, a 1.60 Å resolution crystal structure of the ERGIC-53CRD–MCFD2 complex is reported, along with three other crystal forms. Comparison of these structures with those previously reported reveal that MCFD2, but not ERGIC-53–CRD, exhibits significant conformational plasticity that may be relevant to its accommodation of various polypeptide ligands.
1. Introduction
Intracellular protein trafficking between organelles is ubiquitous in eukaryotic cells. Proteins are transported from the endoplasmic reticulum (ER) to the Golgi complex, undergoing various sugar modifications catalyzed by a variety of glycosidases and glycosyltransferases, and are then delivered to destinations inside and outside the cell (Kornfeld & Kornfeld, 1985 ▸). The delivery of cargo glycoproteins to transport vesicles is mediated by transmembrane cargo receptors with sugar-binding activity (Fiedler et al., 1994 ▸; Hauri et al., 2000 ▸).
In the early secretory pathway, the ER–Golgi intermediate compartment 53 protein (ERGIC-53) and vesicular integral membrane protein of 36 kDa (VIP36) function as cargo receptors between the ER and the Golgi apparatus (Fiedler et al., 1994 ▸; Hauri et al., 2000 ▸). ERGIC-53 and VIP36 share a homologous β-sandwich carbohydrate-recognition domain (CRD) with a structural resemblance (Velloso et al., 2002 ▸; Satoh et al., 2007 ▸) to legume lectins such as concanavalin A (Naismith & Field, 1996 ▸). Our previous sugar-binding profiling data demonstrated that ERGIC-53 and VIP36 interact with high-mannose-type N-linked oligosaccharides containing α-1,2-linked mannobiose or mannotriose structures (Kamiya et al., 2005 ▸, 2008 ▸). ERGIC-53 is known to be involved in the intracellular transport of certain glycoproteins, such as cathepsin C, cathepsin Z, cathepsin-Z-related protein (Appenzeller et al., 1999 ▸), blood coagulation factors V (FV) and VIII (FVIII) (Zhang et al., 2003 ▸), Mac-2-binding protein (Mac-2BP)/galectin-3-binding protein (LGALS3BP) (Chen et al., 2013 ▸; Fukamachi et al., 2018 ▸) and α1-antitrypsin (AAT; Zhu et al., 2018 ▸). ERGIC-53, but not VIP36, forms a stable complex with a 16 kDa soluble EF-hand Ca2+-binding protein called MCFD2 (multi-coagulation factor deficiency 2; Zhang et al., 2003 ▸). Accumulated evidence indicates that MCFD2 substantially contributes to the efficient secretion of several cargo proteins, including FV, FVIII (Zhang et al., 2003 ▸), Mac-2BP/LGALS3BP (Chen et al., 2013 ▸; Fukamachi et al., 2018 ▸) and AAT (Zhu et al., 2018 ▸). Indeed, combined deficiency of FV and FVIII (F5F8D), an autosomal recessive disorder characterized by coordinated reduction in the plasma levels of FV and FVIII, is caused by gene mutations in MCFD2 as well as in LMAN1 encoding ERGIC-53 (Zhang et al., 2006 ▸, 2008 ▸).
To obtain structural insights into the transport functions of the ERGIC-53CRD–MCFD2 complex, crystal structures of ERGIC-53CRD bound to MCFD2 in the presence and absence of mannosyl oligosaccharide ligands have been determined (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸). These structures, when compared with an uncomplexed MCFD2 structure in solution (Guy et al., 2008 ▸), suggest that MCFD2, but not ERGIC-53CRD, undergoes a conformational transition upon formation of the ERGIC-53CRD–MCFD2 complex. The complexed structure provides a working model of cooperative interplay between ERGIC-53 and MCFD2. ERGIC-53 binds to N-linked glycans, whereas the allosterically activated MCFD2 binds polypeptide segments of cargo glycoproteins (Nishio et al., 2010 ▸; Kamiya et al., 2012 ▸). Crystal structures of sugar-bound ERGIC-53CRD–MCFD2 complexes provide a structural basis for the recognition of N-linked glycoproteins by ERGIC-53 (Satoh et al., 2014 ▸; Zheng et al., 2013 ▸). A recent NMR study showed that MCFD2 interacts with peptide segment 807–816 of the B domain of FVIII through its canonical ligand-binding site (Yagi et al., 2020 ▸). Although MCFD2 recognizes a similar sequence in the B domain of FV and presumably in other cargo glycoproteins, how MCFD2 recognizes various polypeptide ligands remains largely unsolved. Conformational variations of MCFD2, but not ERGIC-53CRD, have been identified in ERGIC-53CRD–MCFD2 complexes (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸), suggesting conformational plasticity of MCFD2. However, these structures should be interpreted with great care because they could have been affected by the extent of crystal packing observed in their crystal structures.
We determined an improved 1.60 Å resolution crystal structure of the ERGIC-53CRD–MCFD2 complex, in addition to three other crystal forms that contain MCFD2 structures with smaller crystal packing compared with previous crystal structures (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸). Although the global structures of the complexes were similar to previously characterized structures, in our study we observed significant conformational polymorphism in MCFD2, but not in ERGIC-53CRD, in their complexed states.
2. Materials and methods
2.1. Macromolecule production
Protein expression and purification of complexes between ERGIC-53CRD (residues 31–269) and MCFD2 (full-length, residues 27–146; N-terminally truncated, residues 67–146) were performed as described previously (Nishio et al., 2010 ▸; Satoh et al., 2014 ▸; Table 1 ▸). The purified ERGIC-53CRD–MCFD2 complexes were dissolved in 10 mM Tris–HCl pH 8.0 containing 10 mM CaCl2.
Table 1. Macromolecule-production information.
| Forms 1 and 4 | Forms 2 and 3 | |
|---|---|---|
| Expression construct | ||
| ERGIC-53CRD | Residues 31–269 | Residues 31–269 |
| MCFD2 | Residues 27–146 (full-length) | Residues 67–146 (N-terminally truncated) |
| Complete amino-acid sequence† | ||
| ERGIC-53CRD | MNHKVHMDGVGGDPAVALPHRRFEYKYSFKGPHLVQSDGTVPFWAHAGNAIPSSDQIRVAPSLKSQRGSVWTKTKAAFENWEVEVTFRVTGRGRIGADGLAIWYAENQGLEGPVFGSADLWNGVGIFFDSFDNDGKKNNPAIVIIGNNGQIHYDHQNDGASQALASCQRDFRNKPYPVRAKITYYQNTLTVMINNGFTPDKNDYEFCAKVENMIIPAQGHFGISAATGGLADDHDVLSFLTFQLTE | |
| MCFD2 | MGHHHHHHHHHHSSGHIEGRHMLEEPAASFSQPGSMGLDKNTVHDQEHIMEHLEGVINKPEAEMSPQELQLHYFKMHDYDGNNLLDGLELSTAITHVHKEEGSEQAPLMSEDELINIIDGVLRDDDKNNDGYIDYAEFAKSLQ | MGHHHHHHHHHHSSGHIEGRHMLEMSPQELQLHYFKMHDYDGNNLLDGLELSTAITHVHKEEGSEQAPLMSEDELINIIDGVLRDDDKNNDGYIDYAEFAKSLQ |
The expression-tag sequence is underlined.
2.2. Crystallization
Crystals were obtained under several crystallization conditions in the presence and absence of FV-derived and FVIII-derived peptides (Table 2 ▸). Initial crystallization screening was conducted by sitting-drop vapor diffusion using 1.0 µl volumes of protein solution and precipitant solution equilibrated against 100 µl of the latter. The crystallization conditions were optimized by hanging-drop vapor diffusion in the same manner as the sitting-drop experiments using a mother-liquor volume of 500 µl.
Table 2. Crystallization.
| Form 1 | Form 2 | Form 3 | Form 4 | |
|---|---|---|---|---|
| Protein concentration (mg ml−1) | 9.0 | 11.0 | 11.0 | 14.0 |
| Buffer composition of protein solution | 10 mM Tris–HCl pH 8.0, 10 mM CaCl2 | 10 mM Tris–HCl pH 8.0, 10 mM CaCl2, 2 mM FV peptide (929–937, SDLLLLKQS) | 10 mM Tris–HCl pH 8.0, 10 mM CaCl2, 2 mM FV peptide (929–937, SDLLLLKQS) | 10 mM Tris–HCl pH 8.0, 10 mM CaCl2,, 2.5 mM FVIII peptide (776–816) |
| Composition of reservoir solution | 1.2 M sodium malonate, 0.5%(v/v) Jeffamine ED-2001, 50 mM Tris–HCl pH 7.0 | 17% PEG 8000, 8% ethylene glycol, 0.1 M Tris–HCl pH 8.5 | 19% PEG 3350, 8% ethylene glycol, 0.1 M bis-Tris pH 6.0 | 1.1 M sodium malonate, 0.5%(v/v) Jeffamine ED-2001, 0.1 M HEPES pH 7.0 |
2.3. Data collection and processing
Crystals were cryoprotected with the crystallization solution containing 20% glycerol. Diffraction data were collected on the BL5A beamline equipped with an ADSC Quantum 315r detector at the Photon Factory, Japan and on the Osaka University BL44XU beamline equipped with a Rayonix MX225HE CCD detector at SPring-8, Japan. Intensity integration and data scaling were performed using the HKL-2000 suite (Otwinowski & Minor, 1997 ▸). Data-collection statistics are given in Table 3 ▸.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Form 1 | Form 2 | Form 3 | Form 4 | |
|---|---|---|---|---|
| Diffraction source | BL5A, Photon Factory | BL44XU, SPring-8 | BL44XU, SPring-8 | BL44XU, SPring-8 |
| Wavelength (Å) | 1.0000 | 0.9000 | 0.9000 | 0.9000 |
| Temperature (K) | 95 | 95 | 95 | 95 |
| Detector | ADSC Quantum 315r CCD | Rayonix MX225HE CCD | Rayonix MX225HE CCD | Rayonix MX225HE CCD |
| Space group | P21 | P21 | P21 | P3121 |
| a, b, c (Å) | 57.92, 116.93, 58.07 | 73.62, 168.89, 73.96 | 103.06, 58.77, 119.39 | 113.13, 113.13, 157.60 |
| α, β, γ (°) | 90, 120.13, 90 | 90, 119.86, 90 | 90, 109.01, 90 | 90, 90, 120 |
| Resolution range (Å) | 50–1.60 (1.63–1.60) | 50–2.40 (2.44–2.40) | 50–2.50 (2.54–2.50) | 50–3.05 (3.10–3.05) |
| Total No. of reflections | 318800 | 223803 | 167632 | 162674 |
| No. of unique reflections | 88266 | 61522 | 46552 | 22770 |
| Completeness (%) | 98.9 (100) | 99.7 (100) | 99.4 (97.8) | 100 (100) |
| Multiplicity | 3.7 (3.6) | 3.7 (3.7) | 3.6 (3.3) | 7.1 (7.2) |
| 〈I/σ(I)〉 | 39.2 (3.4) | 38.6 (9.8) | 34.9 (8.8) | 27.3 (7.6) |
| R r.i.m. † | 0.069 (0.548) | 0.066 (0.220) | 0.059 (0.213) | 0.126 (0.364) |
| Overall B factor from Wilson plot (Å2) | 19.58 | 32.98 | 28.84 | 25.54 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
2.4. Structure solution and refinement
Initial phase determinations were performed by the molecular-replacement method using MOLREP (Vagin & Teplyakov, 2010 ▸) and Phaser (McCoy et al., 2007 ▸) with the reported trigonal structure (PDB entry 3a4u; Nishio et al., 2010 ▸) as the search model. Further model building was performed manually using Coot (Emsley et al., 2010 ▸). Subsequent refinements were performed using REFMAC5 (Murshudov et al., 2011 ▸). The stereochemical quality of the final models was assessed using MolProbity (Chen et al., 2010 ▸). The refinement statistics are summarized in Table 4 ▸. Structural superposition was performed with SUPERPOSE (Krissinel & Henrick, 2004 ▸). Molecular graphics were prepared using PyMOL (Schrödinger). Crystal contact areas were analyzed using PISA (Krissinel & Henrick, 2007 ▸).
Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| Form 1 | Form 2 | Form 3 | Form 4 | |
|---|---|---|---|---|
| Resolution range (Å) | 20.00–1.60 (1.64–1.60) | 20.00–2.40 (2.46–2.40) | 20.00–2.51 (2.57–2.51) | 20.00–3.05 (3.13–3.05) |
| Completeness (%) | 98.8 | 99.6 | 98.6 | 99.9 |
| σ Cutoff | None | None | None | None |
| No. of reflections | ||||
| Working set | 82482 (6092) | 57782 (4293) | 43802 (2948) | 21558 (1536) |
| Test set | 4370 (328) | 3081 (203) | 2318 (157) | 1162 (80) |
| Final R cryst | 0.180 (0.233) | 0.205 (0.261) | 0.193 (0.242) | 0.173 (0.236) |
| Final R free | 0.199 (0.264) | 0.246 (0.317) | 0.232 (0.302) | 0.173 (0.236) |
| No. of non-H atoms | ||||
| Protein | 4062 | 9087 | 9252 | 6889 |
| Ca2+ | ||||
| ERGIC-53CRD | 0 [chains A/C] | 8 [chains A/C/E/G] | 8 [chains A/C/E/G] | 3 [chains A/C/E] |
| MCFD2 | 4 [chains B/D] | 8 [chains B/D/F/H] | 8 [chains B/D/F/H] | 6 [chains B/D/F] |
| Ligand | 12 | 0 | 0 | 0 |
| Water | 239 | 238 | 293 | 10 |
| Total | 4317 | 9345 | 9564 | 6911 |
| R.m.s. deviations | ||||
| Bonds (Å) | 0.011 | 0.013 | 0.013 | 0.013 |
| Angles (°) | 1.437 | 1.509 | 1.533 | 1.642 |
| Average B factors (Å2) | ||||
| Protein | 30.2 | 45.3 | 38.0 | 38.3 |
| Ca2+ | 23.6 | 43.0 | 35.2 | 40.5 |
| Ligand | 27.7 | 0.0 | 0.0 | 0.0 |
| Water | 39.6 | 40.1 | 32.8 | 17.2 |
| Ramachandran plot | ||||
| Favored regions (%) | 97.7 | 98.1 | 97.1 | 95.8 |
| Additionally allowed (%) | 2.1 | 1.8 | 2.8 | 4.1 |
3. Results and discussion
3.1. Structures of the ERGIC-53CRD–MCFD2 binary complex
In this study, we determined four crystal structures of the ERGIC-53CRD–MCFD2 complex, including an improved 1.60 Å resolution structure (Fig. 1 ▸), and found significant structural variations in MCFD2. Crystals of the ERGIC-53CRD–MCFD2 complex were obtained under various crystallization conditions (termed ‘crystal forms 1–4’). The ERGIC-53CRD–MCFD2 crystals in crystal forms 1 and 4 contained two and three binary-complex molecules (termed ‘complexes 1A–1B’ and ‘complexes 4A–4C’) per asymmetric unit, respectively, whereas those in crystal forms 2 and 3 contained four binary-complex molecules (termed ‘complexes 2A–2D’ and ‘complexes 3A–3D’). The ERGIC-53CRD–MCFD2 complex contains four Ca2+ ions. Two Ca2+ ions bound to the sugar-binding site of ERGIC-53CRD and two Ca2+ ions bound to MCFD2 (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸). In the crystal form 1 structure (complexes 1A and 1B), the Ca2+-binding sites of ERGIC-53CRD were disordered despite having sufficient space for Ca2+ ions in crystallo. In crystal form 4 the Ca2+-binding sites in ERGIC-53CRD were partially disordered: complexes 4A and 4B contained two and one Ca2+ ions, respectively, whereas complex 4C had no bound Ca2+ ions. In contrast, in the crystal form 2 and 3 structures (complexes 2A–2D and 3A–3D) all four Ca2+ ions were observed in the ERGIC-53CRD–MCFD2 complexes (Table 4 ▸). These data suggest that ERGIC-53CRD tends to lose Ca2+ ions upon crystallization in comparison with MCFD2, consistent with the previous crystallographic study (Satoh et al., 2014 ▸).
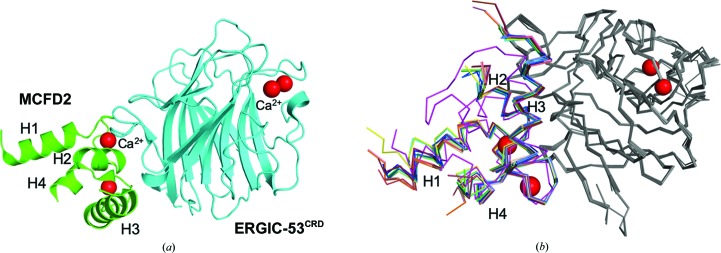
Figure 1.
Structures of MCFD2 complexed with ERGIC-53CRD. (a) A ribbon model of the ERGIC-53CRD–MCFD2 complex is shown. ERGIC-53CRD and MCMD2 are colored cyan and green, respectively. Bound Ca2+ ions are shown as red spheres. The positions of the helices (H1–H4) of MCFD2 are indicated. (b) Backbone-atom superimposition of ERGIC-53CRD–MCFD2 crystal structures together with the apo MCFD2 NMR structure (PDD entry 2vrg, lowest-penalty model 1; Guy et al., 2008 ▸). ERGIC-53CRD structures are colored gray. MCFD2 structures are colored as follows: complex 1A, wheat; complex 1B, olive; complex 2A, blue; complex 2B, marine blue; complex 2C, slate; complex 2D, sky blue; complex 3A, raspberry; complex 3B, salmon; complex 3C, deep salmon; complex 3D, chocolate; complex 4A, green; complex 4B, lime; complex 4C, forest; PDB entry 3a4u (Nishio et al., 2010 ▸), yellow; PDB entry 3lcp (Wigren et al., 2010 ▸), chain C, orange; PDB entry 3lcp, chain D, pink; PDB entry 3wht (Satoh et al., 2014 ▸), cyan. The solution structure of apo MCFD2 is colored magenta.
3.2. Crystal packing of MCFD2 complexed with ERGIC-53CRD
To validate the structural variability of the ERGIC-53CRD–MCFD2 complexes, the crystal contacts observed in molecules in the asymmetric and symmetry-related units were analyzed together with previously reported structures (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸). The crystal contact areas are summarized in Supplementary Table S1. Notable differences existed in contact areas among all MCFD2 structures (445–992 Å2). In complex 4A the total contact area was 444.8 Å2 for MCFD2, suggesting that the MCFD2 structure in this crystal form is least affected by crystal packing in crystallo.
3.3. Comparison of MCFD2 structures derived from different crystal forms
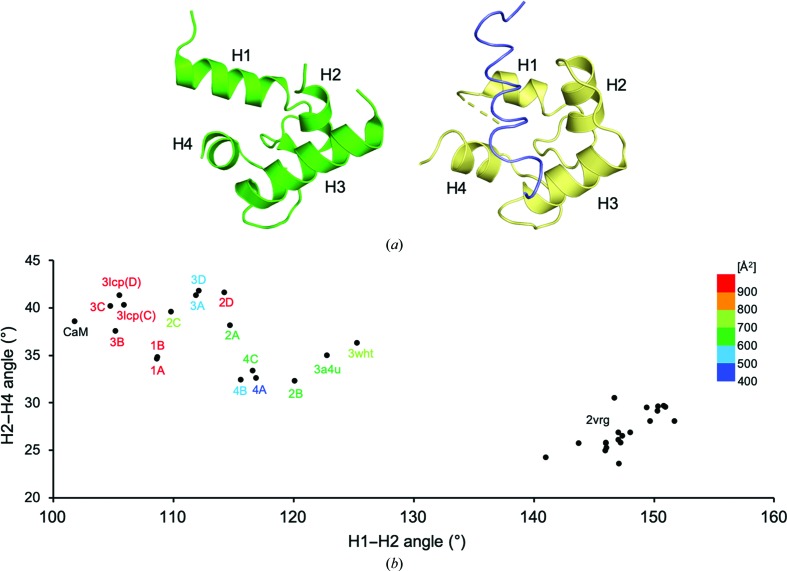
The crystal structures of ERGIC-53CRD in crystal forms 1–4 were quite similar to one another, with root-mean-square deviation (r.m.s.d.) values of 0.02–0.30 Å for the Cα atoms of the commonly observed residues (comprising amino-acid residues 44–129, 144–154, 162–170 and 186–268; Fig. 1 ▸ b). In the improved 1.60 Å resolution structure, an alternative conformation of the Arg111 side chain of ERGIC-53CRD was newly observed (Supplementary Fig. S1). In contrast, the crystallographic data from crystal forms 1–4 showed significant conformational variations of MCFD2, with r.m.s.d. values of 0.02–0.60 Å for the Cα atoms of the commonly observed residues (comprising amino-acid residues 76–97 and 112–142). In particular, the MCFD2 residues constituting the H1 and H4 helices and containing the Ca2+-binding sites involved in interactions with ERGIC-53CRD exhibited marked structural variations (Fig. 1 ▸ b). Intriguingly, the potentially mobile segments are located in the ligand-binding site of MCFD2 (Yagi et al., 2020 ▸), which corresponds to that of EF-hand proteins, as exemplified by calmodulin (CaM; Elshorst et al., 1999 ▸; Fig. 2 ▸ a). In the ligand-bound CaM complex, the H1, H2 and H4 helices are mainly involved in peptide-ligand binding (Elshorst et al., 1999 ▸). These observations prompted an examination of the correlative helix angles (H1–H2 and H2–H4 helices) of MCFD2 derived from the 13 currently determined binary-complex structures together with previously reported structures, including apo MCFD2 solution structures (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸). The comparative data indicated that the H1–H2 and H2–H4 helix angles are highly distributed around 32–42° (H1–H2) and 105–125° (H2–H4) (Fig. 2 ▸ b). The correlative helix angles differed significantly between ERGIC-53CRD-bound and unbound MCFD2 (Fig. 2 ▸ b), as elucidated previously (Nishio et al., 2010 ▸). The deviation of the helix angles of ERGIC-53CRD-bound MCFD2 is higher than that of the unbound form, suggesting conformational plasticity of MCFD2 in the binary complex. The distribution pattern was not well correlated with the crystallization conditions, as exemplified by complexes 2A–2D and 3A–3D. However, there seems to be a relationship between the correlative helix angle and the crystal contact area, especially in the structures that possess larger contact areas (colored in red in Fig. 2 ▸ b). Notably, the correlative helix angle derived from a ligand-bound CaM complex (Elshorst et al., 1999 ▸) is similar to those from MCFD2 complexes with greater crystal-packing areas. Therefore, it is possible that the neighboring molecule in the crystal contacts the peptide-binding site of MCFD2, facilitating its conformational change towards the ligand-bound state in crystallo. Taken together with previous crystallographic observations (Satoh et al., 2014 ▸; Nishio et al., 2010 ▸; Wigren et al., 2010 ▸), together with solution NMR data (Guy et al., 2008 ▸), our findings indicate that the ligand-binding site of MCFD2 is allosterically activated by ERGIC-53CRD upon complex formation, as suggested in our previous study (Nishio et al., 2010 ▸). In this study, we revealed conformational variability of MCFD2, especially at its ligand-binding site, when in complex with its binding partner ERGIC-53. The conformational polymorphism of MCFD2 in the cargo-receptor complex may be relevant to its conformational adjustability in the recognition of various glycoprotein ligands.
Figure 2.
Conformational variations of the ligand-binding site of MCFD2. (a) Relative positions of helices H1–H4 in the EF-hand proteins MCFD2 and calmodulin. Structures of MCFD2 in complex with ERGIC-53CRD (PDB entry 3a4u; Nishio et al., 2010 ▸) and the ligand-bound calmodulin complex (PDB entry 1cff; Elshorst et al., 1999 ▸) are shown on the left and right, respectively. The calmodulin and Ca2+-pump peptide ligand structures are colored yellow and slate, respectively. (b) Correlative helix-angle distributions in the ligand-binding site of MCFD2. Vertical and horizontal axes indicate H2–H4 and H1–H2 helix angles, respectively. The data represent helix-angle conformations derived from currently determined MCFD2 structures together with previously reported MCFD2 and calmodulin structures, which are labeled with ‘complex name’, ‘PDB code’ or ‘CaM’, respectively, with a color gradient according to their crystal contact areas. For the apo MCMD2 NMR structure (PDB entry 2vrg; Guy et al., 2008 ▸), the angles derived from the 20 lowest target-function structures are shown.
Supplementary Material
PDB reference: ERGIC-53–MCFD2, form 1, 4ygb
PDB reference: form 2, 4ygc
PDB reference: form 3, 4ygd
PDB reference: form 4, 4yge
Supplementary Figure S1 and Table S1. DOI: 10.1107/S2053230X20005452/nw5099sup1.pdf
Acknowledgments
We thank Kiyomi Senda and Kumiko Hattori for their help in the preparation of the recombinant proteins. We thank the beamline staff at the Photon Factory and SPring-8 for providing data-collection facilities and support.
Funding Statement
This work was funded by Japan Society for the Promotion of Science grant JP19H03361 to Tadashi Satoh. National Institutes of Natural Sciences grant 19-315 to Tadashi Satoh.
References
- Appenzeller, C., Andersson, H., Kappeler, F. & Hauri, H.-P. (1999). Nat. Cell Biol. 1, 330–334. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Chen, Y., Hojo, S., Matsumoto, N. & Yamamoto, K. (2013). Glycobiology, 23, 904–916. [DOI] [PubMed]
- Elshorst, B., Hennig, M., Försterling, H., Diener, A., Maurer, M., Schulte, P., Schwalbe, H., Griesinger, C., Krebs, J., Schmid, H., Vorherr, T. & Carafoli, E. (1999). Biochemistry, 38, 12320–12332. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fiedler, K., Parton, R. G., Kellner, R., Etzold, T. & Simons, K. (1994). EMBO J. 13, 1729–1740. [DOI] [PMC free article] [PubMed]
- Fukamachi, M., Kasamatsu, A., Endo-Sakamoto, Y., Fushimi, K., Kasama, H., Iyoda, M., Minakawa, Y., Shiiba, M., Tanzawa, H. & Uzawa, K. (2018). Exp. Cell Res. 368, 119–125. [DOI] [PubMed]
- Guy, J. E., Wigren, E., Svärd, M., Härd, T. & Lindqvist, Y. (2008). J. Mol. Biol. 381, 941–955. [DOI] [PubMed]
- Hauri, H.-P., Kappeler, F., Andersson, H. & Appenzeller, C. (2000). J. Cell Sci. 113, 587–596. [DOI] [PubMed]
- Kamiya, Y., Kamiya, D., Yamamoto, K., Nyfeler, B., Hauri, H.-P. & Kato, K. (2008). J. Biol. Chem. 283, 1857–1861. [DOI] [PubMed]
- Kamiya, Y., Satoh, T. & Kato, K. (2012). Biochim. Biophys. Acta, 1820, 1327–1337. [DOI] [PubMed]
- Kamiya, Y., Yamaguchi, Y., Takahashi, N., Arata, Y., Kasai, K., Ihara, Y., Matsuo, I., Ito, Y., Yamamoto, K. & Kato, K. (2005). J. Biol. Chem. 280, 37178–37182. [DOI] [PubMed]
- Kornfeld, R. & Kornfeld, S. (1985). Annu. Rev. Biochem. 54, 631–664. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Naismith, J. H. & Field, R. A. (1996). J. Biol. Chem. 271, 972–976. [DOI] [PubMed]
- Nishio, M., Kamiya, Y., Mizushima, T., Wakatsuki, S., Sasakawa, H., Yamamoto, K., Uchiyama, S., Noda, M., McKay, A. R., Fukui, K., Hauri, H.-P. & Kato, K. (2010). Proc. Natl Acad. Sci. USA, 107, 4034–4039. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Satoh, T., Cowieson, N. P., Hakamata, W., Ideo, H., Fukushima, K., Kurihara, M., Kato, R., Yamashita, K. & Wakatsuki, S. (2007). J. Biol. Chem. 282, 28246–28255. [DOI] [PubMed]
- Satoh, T., Suzuki, K., Yamaguchi, T. & Kato, K. (2014). PLoS One, 9, e87963. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Velloso, L. M., Svensson, K., Schneider, G., Pettersson, R. F. & Lindqvist, Y. (2002). J. Biol. Chem. 277, 15979–15984. [DOI] [PubMed]
- Wigren, E., Bourhis, J.-M., Kursula, I., Guy, J. E. & Lindqvist, Y. (2010). FEBS Lett. 584, 878–882. [DOI] [PubMed]
- Yagi, H., Yagi-Utsumi, M., Honda, R., Ohta, Y., Saito, T., Nishio, M., Ninagawa, S., Suzuki, K., Anzai, T., Kamiya, Y., Aoki, K., Nakanishi, M., Satoh, T. & Kato, K. (2020). Nat. Commun. 11, 1368. [DOI] [PMC free article] [PubMed]
- Zhang, B., Cunningham, M. A., Nichols, W. C., Bernat, J. A., Seligsohn, U., Pipe, S. W., McVey, J. H., Schulte-Overberg, U., de Bosch, N. B., Ruiz-Saez, A., White, G. C., Tuddenham, E. G., Kaufman, R. J. & Ginsburg, D. (2003). Nat. Genet. 34, 220–225. [DOI] [PubMed]
- Zhang, B., McGee, B., Yamaoka, J. S., Guglielmone, H., Downes, K. A., Minoldo, S., Jarchum, G., Peyvandi, F., de Bosch, N. B., Ruiz-Saez, A., Chatelain, B., Olpinski, M., Bockenstedt, P., Sperl, W., Kaufman, R. J., Nichols, W. C., Tuddenham, E. G. & Ginsburg, D. (2006). Blood, 107, 1903–1907. [DOI] [PMC free article] [PubMed]
- Zhang, B., Spreafico, M., Zheng, C., Yang, A., Platzer, P., Callaghan, M. U., Avci, Z., Ozbek, N., Mahlangu, J., Haw, T., Kaufman, R. J., Marchant, K., Tuddenham, E. G., Seligsohn, U., Peyvandi, F. & Ginsburg, D. (2008). Blood, 111, 5592–5600. [DOI] [PMC free article] [PubMed]
- Zheng, C., Page, R. C., Das, V., Nix, J. C., Wigren, E., Misra, S. & Zhang, B. (2013). J. Biol. Chem. 288, 20499–20509. [DOI] [PMC free article] [PubMed]
- Zhu, M., Zheng, C., Wei, W., Everett, L., Ginsburg, D. & Zhang, B. (2018). Blood Adv. 2, 1014–1021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ERGIC-53–MCFD2, form 1, 4ygb
PDB reference: form 2, 4ygc
PDB reference: form 3, 4ygd
PDB reference: form 4, 4yge
Supplementary Figure S1 and Table S1. DOI: 10.1107/S2053230X20005452/nw5099sup1.pdf