Key Points
Question
Are myopia and intraocular pressure associated with retinal detachment?
Findings
In this 2-sample mendelian randomization analysis including nearly 500 000 participants in the UK Biobank cohort, genetic associations between myopia and intraocular pressure with the risk of retinal detachment were found.
Meaning
These results add weight to existing evidence suggesting that myopia prevention efforts may help prevent retinal detachment.
This mendelian randomization study uses genetic data to assess the association of myopia and intraocular pressure with retinal detachment risk.
Abstract
Importance
Rhegmatogenous retinal detachment is a potentially sight-threatening condition. The role of myopia or intraocular pressure (IOP) in retinal detachment remains unclear.
Objective
To determine if myopia or IOP is associated with retinal detachment risk using genetic data.
Design, Setting, and Participants
Observational analyses and 2-sample mendelian randomization were used to evaluate the associations between myopia, IOP, and retinal detachment risk in European descent participants from the UK Biobank (UKBB) cohort (n = 405 692). For retinal detachment, a genome-wide association study on 4257 cases and 39 181 controls in the UKBB was conducted. Genetic variants associated with mean spherical equivalent (MSE) refractive error (n = 95 827) and IOP (n = 101 939) were derived using independent participants from the retinal detachment genome-wide association study. Recruitment to the UKBB occurred between 2006 and 2010, and data analysis occurred from February 2019 to March 2020.
Main Outcomes and Measures
The odds ratio (OR) of retinal detachment caused by per-unit increases in MSE refractive error (in diopters [D]) and IOP (in mm Hg).
Results
Of the 405 692 participants in the UKBB cohort, the mean (SD) age was 56.87 (7.96) years, the mean (SD) MSE was −0.31 (2.65) D, the mean (SD) corneal-compensated IOP was 16.05 (3.49) mm Hg, and 4253 participants (1.0%) had retinal detachment. Genetic analyses of the 4257 cases and 39 181 controls identified 2 novel retinal detachment genes: COL22A1 (lead single-nucleotide variant rs11992725; P = 4.8 × 10−10) and FAT3 (lead single-nucleotide variant rs10765568; P = 1.2 × 10−15). Genetically assessed MSE refractive error was negatively associated with retinal detachment (per-unit [D] increase in MSE refractive error: OR, 0.72; 95% CI, 0.69-0.76; P = 3.8 × 10−44). For each 6-D decrease in MSE refractive error (representing the move of refractive error from emmetropia to high myopia), retinal detachment risk increased 7.2-fold (95% CI, 5.19-9.27). For per-unit (mm Hg) genetically assessed increase in IOP, the risk of retinal detachment increased by 8% (OR, 1.08; 95% CI, 1.03-1.14; P = .001).
Conclusions and Relevance
This study provides genetic support for the assertion that myopia and IOP are associated with the risk of retinal detachment and that myopia prevention efforts may help prevent retinal detachment.
Introduction
Rhegmatogenous retinal detachment (RRD) is a potentially sight-threatening condition that necessitates timely intervention. It manifests when the neurosensory retina is separated from the underlying retinal pigment epithelium due to a break or tear in the neurosensory retina.1,2 The incidence of RRD is between 6.3 and 17.9 in 100 000 individuals per year and the prevalence is approximately 1%, although these vary across different racial/ethnic groups and geographical regions.3 Epidemiology studies have identified myopia, advancing age, and cataract operations as important risk factors for RRD.3,4,5 For instance, the risk of RRD was increased by 3-fold for individuals with myopia.6
Several observational studies have shown an association of myopia with retinal detachment.5,6,7 However, association is not causation, and there are many examples where the results from observational studies conflict with those from the criterion standard for causal inference, randomized clinical trials.8,9 Previous studies examining the association of myopia with retinal detachment are either case series or case-control studies with a relatively small sample size and could be confounded by other risk factors, such as sex, age, and cataract surgery.3
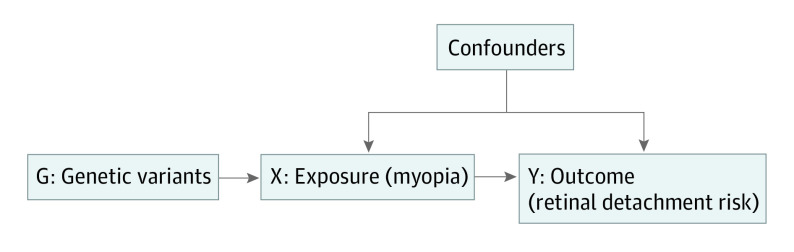
Mendelian randomization (MR) is a statistical method that uses an instrumental variable to investigate the potential causal relationship between risk factors and outcomes.10,11 In MR analysis, genetic variants (G) that are associated with an exposure (X, such as myopia) are randomly allocated at conception. If the genetic variants also affect the risk of an outcome (Y, such as retinal detachment), this helps provide evidence to support a potential causal inference about the association between the exposure and the outcome (Figure 1).11 The MR analysis relies on 3 assumptions: (1) the relevance assumption that genetic variants are associated with the exposure; (2) the independence assumption that genetic variants are not associated with confounders; and (3) the exclusion restriction that genetic variants affect the outcome only through the exposure.10,11
Figure 1. Schematic Representation of Mendelian Randomization Analysis.
Genetic variants (G) that are associated with the exposure (X, such as myopia) are used as instrumental variable. The association of genetic variants with the outcome (Y, such as retinal detachment) is only mediated through the exposure.
In this study, we used MR to investigate whether genetic data provides support for a potential causal inference for the association between myopia and retinal detachment risk. Some previous studies have shown that intraocular pressure (IOP) was elevated after retinal detachment surgery.12,13 However, the association of IOP with RRD remains unclear. We also used the MR method to evaluate the association of IOP with retinal detachment. Such assessment of these risk factors could have direct public health and clinical implications for prevention of retinal detachment.
Methods
The study was approved by the National Research Ethics Service Committee North West—Haydock, all participants provided informed written consent, and all study procedures were performed in accordance with the World Medical Association Declaration of Helsinki ethical principles for medical research. Patients were not offered any compensation or incentives to participate in the study.
Study Overview
We first conducted a series of genome-wide association studies (GWASs) for retinal detachment, refractive error, and IOP in the UK Biobank (UKBB) cohort. We then used a 2-sample MR framework to investigate the associations between refractive error, IOP, and retinal detachment. For the refractive error and IOP GWASs, we removed all retinal detachment cases, controls, and their relatives (pi-hat >0.2; identity by descent determined based on autosomal markers) to ensure samples were independent for the 2-sample MR method.11 We also performed observational analyses for the association of myopia or IOP with retinal detachment to compare with the genetically derived (MR) estimates. Recruitment to the UKBB occurred between 2006 and 2010, and data analysis occurred from February 2019 to March 2020.
Retinal Detachment Data
We identified retinal detachment cases using the following criteria: (1) International Classification of Diseases (ICD) diagnosis cases using ICD-10 diagnosis code H33 (retinal detachments and breaks) and ICD-9 diagnosis code 361 (retinal detachments and defects) or (2) self-reported retinal detachment cases from self-reported noncancer illness (UKBB code 1281, retinal detachment; UKBB data field 20002). We excluded participants with nonwhite British ancestry based on principal components.14 Finally, for our retinal detachment GWAS analysis, we included 4257 retinal detachment cases and selected 39 181 controls without any eye problems or disorders from UKBB data field 6148.
For our observational analysis, we identified retinal detachment cases using similar criteria as above and used UKBB participants without retinal detachment as controls. We then removed related samples within and between retinal detachment cases and controls (pi-hat >0.2). Finally, we included 4253 retinal detachment cases and 401 439 controls in our observational analysis.
Mean Spherical Equivalent Data
In UKBB, refractive error was measured by an RC-5000 device (Tomey). We excluded refractometry results that were indicated as unreliable (refractometry result error in UKBB data field 5090 and 5091). We then calculated the mean values of spherical power (UKBB data field 5084 and 5085) and cylindrical power (UKBB data field 5086 and 5087) across both left and right eyes, respectively. Mean spherical equivalent (MSE) refractive error was calculated using the formula Spherical Power + (0.5 × Cylindrical Power). The mean MSE values (across left and right eyes) were used in our analysis. We removed individuals with a history of eye surgery or eye-related complications, including eye surgery (UKBB data field 5181), cataract surgery (UKBB data field 5324), refractive laser eye surgery (UKBB data field 5325), surgery for glaucoma or high eye pressure (UKBB data field 5326), laser treatment for glaucoma or high eye pressure (UKBB data field 5327), and corneal graft surgery (UKBB data field 5328).
For the MSE GWAS analysis, we removed retinal detachment cases, controls, and their relatives (pi-hat >0.2) to avoid sample overlap with the retinal detachment GWAS. This resulted in 95 827 individuals of white British ancestry with MSE refractive error data in the GWAS analysis.
In our sensitivity analysis, we evaluated whether high myopia or myopia was associated with retinal detachment by dichotomizing the MSE measurement. High myopia cases were defined as MSE of −5 diopters (D) or less (n = 5824) and controls as MSE greater than −0.75 D (n = 66 627); myopia cases were defined as MSE of −0.75 D or less (n = 29 200) and controls as MSE greater than −0.75 D (n = 66 627).15
IOP Data
In UKBB, the corneal-compensated IOP was available for 127 455 individuals (UKBB data field 5262 and 5254) measured by the Ocular Response Analyzer (Reichert).14 A detailed description of corneal-compensated IOP phenotype data was presented previously.14 In brief, we calculated the mean corneal-compensated IOP across both the left and right eyes and removed measurements more than 60 mm Hg or less than 5 mm Hg. We also excluded glaucoma cases or participants who had surgery for glaucoma or high eye pressure to prevent reverse causality.
As part of the IOP GWAS analysis, we removed retinal detachment cases, controls, and their relatives (pi-hat >0.2) to avoid sample overlap. Finally, 101 939 individuals with corneal-compensated IOP were included in GWAS analysis.
Statistical Analysis
For the retinal detachment GWAS in UKBB, we used generalized mixed models (R package SAIGE version 0.29.6) to account for sample relatedness and unbalanced case-control ratios.16 In the association analysis, we adjusted for sex, baseline age (age at recruitment), and the first 10 principal components. For the MSE and IOP GWAS analyses, linear mixed models (BOLT-LMM software version 2.3.2)17 were used to account for population stratification and sample relatedness, and sex, baseline age, and the first 10 principal components were adjusted.
Linkage disequilibrium (LD) score regression method was used to estimate the genetic correlation between the traits above using only GWAS summary statistics.18 To calibrate instruments for the MR analyses, we obtained genetic instruments for myopia and IOP from the above GWASs and selected independent genome-wide significant variants (significance set at 2-tailed P < 5 × 10−8 and LD between single-nucleotide variants r2 < 0.05) for each trait of interest. The strength of genetic instruments was evaluated by F statistics and variance explained (R2).19 We then conducted 2-sample MR for myopia and IOP with retinal detachment as implemented in R packages MendelianRandomization and TwoSampleMR.20,21 The inverse-variance weighted (IVW) regression estimates were reported as the main analysis, and we verified the estimates using the MR weighted median and MR-Egger regression methods.22 The intercept from MR-Egger regression was used to assess for horizontal pleiotropy.21 We also used the MR-PRESSO method and funnel plot to evaluate bias from outliers and assess the heterogeneity of genetic instruments.21,23 To assess the association of IOP with retinal detachment risk, we conducted a multivariable MR analysis, a regression-based method, to adjust for the effect of refractive error.24,25,26 For observational analyses, we used a logistic regression model to investigate the association of MSE or IOP with retinal detachment adjusting for sex and baseline age. Analyses were performed with R software version 3.4.1 (The R Foundation).
Results
Observational Analysis
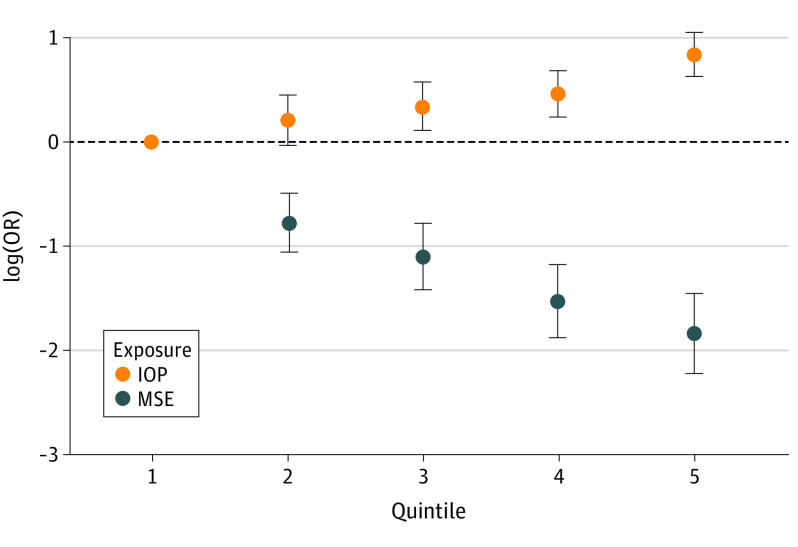
In our observational analysis of 405 692 participants in UKBB, the mean (SD) age was 56.87 (7.96) years, mean (SD) MSE was −0.31 (2.65) D, mean (SD) corneal-compensated IOP was 16.05 (3.49) mm Hg, and 4253 participants (1.0%) had retinal detachment (Table 1). The risk of retinal detachment was linearly increased with decreasing MSE or increasing IOP (Figure 2). On average, for each per-unit (D) increase of MSE, the risk of retinal detachment was decreased by 28% (odds ratio [OR], 0.82; 95% CI, 0.79-0.84; P = 1.1 × 10−47); for each per-unit (mm Hg) increase in IOP, the risk of retinal detachment was increased by 10% (OR, 1.10; 95% CI, 1.08-1.12; P = 6.6 × 10−35). The associations were essentially unchanged after adjusting for sex and age (Table 2). There was a small but significant correlation between IOP and MSE (phenotype Pearson correlation coefficient, −0.076; P = 8.3 × 10−115). In observational logistic regression model, the OR of IOP for retinal detachment risk per 1–mm Hg increase was 1.05 (95% CI, 1.02-1.09; P = 2.5 × 10−4). After adjusting for MSE, the effect size was reduced but still significant (per 1–mm Hg increase: OR, 1.03; 95% CI, 1.00-1.06; P = .04) (Table 2).
Table 1. Baseline Characteristics of Retinal Detachment Case and Control Sample From UK Biobank.
| Variable | No. (%) | OR (95% CI)a | P valuea | |
|---|---|---|---|---|
| Control (n = 401 439) | Case (n = 4253) | |||
| Sex | ||||
| Male | 184 753 (46.0) | 2483 (58.4) | 1 [Reference] | NA |
| Female | 216 686 (54.0) | 1770 (41.6) | 0.61 (0.57-0.65) | 4.3 × 10−57 |
| Age, mean (SD), y | 56.84 (7.96) | 59.78 (6.65) | 1.05 (1.05-1.06) | 4.1 × 10−124 |
| MSE, mean (SD), D | –0.31 (2.64) | –2.40 (3.75) | 0.82 (0.79-0.84) | 1.1 × 10−47 |
| No. | 90 476 | 340 | NA | NA |
| Myopia | ||||
| No | 62 828 (69.4) | 154 (45.3) | 1 [Reference] | NA |
| Yes | 27 648 (30.6) | 186 (54.7) | 2.74 (2.22-3.40) | 2.3 × 10−20 |
| High myopia | ||||
| No | 62 828 (91.9) | 154 (63.1) | 1 [Reference] | NA |
| Yes | 5531 (8.1) | 90 (36.9) | 6.64 (5.11-8.62) | 1.1 × 10−45 |
| Intraocular pressure, mean (SD), mm Hg | 16.04 (3.48) | 17.39 (3.90) | 1.10 (1.08-1.12) | 6.6 × 10−35 |
| No. | 98 384 | 987 | NA | NA |
Abbreviations: D, diopter; IOP, intraocular pressure; MSE, mean spherical equivalent refractive error; NA, not applicable; OR, odds ratio.
The OR and P value were calculated from simple observational logistic regression analysis without covariates.
Figure 2. Observational Association of Intraocular Pressure (IOP), Mean Spherical Equivalent (MSE) Refractive Error, and Retinal Detachment Risk.
The x-axis is the quintiles for exposures (IOP and MSE), and the y-axis is the natural logarithm of odds ratio (log[OR]) for retinal detachment risk. In the observational logistic regression analysis, we adjusted sex and age in logistic regression models. The figure highlights that the risk of retinal detachment is approximately linearly increased with the decreasing MSE or increasing IOP. The quintiles for MSE are −22.600 to −1.910, greater than −1.910 to −0.217, greater than −0.217 to 0.486, greater than 0.486 to 1.400, and greater than 1.400 to 14.000; the quintiles for IOP are 5.080 to 13.200, greater than 13.200 to 15.000, greater than 15.000 to 16.600, greater than 16.600 to 18.700, and greater than 18.700 to 54.400. Error bars indicate 95% CIs.
Table 2. Mendelian Randomization (MR) and Observational Analysis Estimate of the Associations of Myopia and Intraocular Pressure (IOP) With Retinal Detachment.
| Exposure | MR method | OR (95% CI) | P value | Observational methoda | OR (95% CI) | P value |
|---|---|---|---|---|---|---|
| MSE | IVW | 0.72 (0.69-0.76) | 3.8 × 10−44 | Model 1 | 0.80 (0.78-0.83) | 2.5 × 10−55 |
| IOP | IVW | 1.08 (1.03-1.14) | 1.1 × 10−3 | Model 1 | 1.05 (1.02-1.09) | 2.5 × 10−4 |
| IOP | Multivariable MRb | 1.06 (1.01-1.12) | .03 | Model 2 | 1.03 (1.00-1.06) | .04 |
Abbreviations: IVW, inverse-variance weighted; MSE, mean spherical equivalent refractive error; OR, odds ratio.
In observational model 1, we adjusted for sex and baseline age in logistic regression models. In observational model 2, we adjusted for refractive error, sex, and baseline age.
Multivariable MR analysis, a regression-based MR method, to adjust for the effect of MSE refractive error.
Genetic Correlation Between Retinal Detachment, Myopia, and IOP
We conducted a retinal detachment GWAS for 4257 cases and 39 181 controls and identified 2 novel retinal detachment genes: COL22A1 (lead single-nucleotide variant rs11992725; P = 4.8 × 10−10) and FAT3 (lead single-nucleotide variant rs10765568; P = 1.2 × 10−15) (eTable 1 and eFigures 1 and 2 in the Supplement). From LD score regression, we found very weak evidence of genomic inflation in our retinal detachment GWAS (genomic control λ, 1.04; LD score regression intercept, 0.98) (eFigure 1 in the Supplement). The genetic correlation between retinal detachment against MSE and IOP were −0.45 (P = 1.3 × 10−15) and 0.28 (P = 1.6 × 10−6), respectively. These results indicated a high proportion of shared genetic associations between retinal detachment and myopia or IOP.
Association of Myopia With Retinal Detachment in MR Analysis
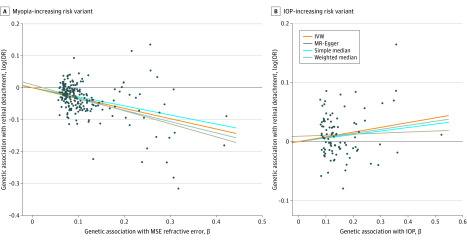
Myopia was measured using MSE refractive error. From the MSE GWAS, we selected 224 independent single-nucleotide variants as genetic instruments (eTable 2 in the Supplement). The F statistics and the proportion of variance-explained R2 by instruments were 54 and 0.12, respectively. In total, 10 myopia variants were associated with retinal detachment risk at a Bonferroni-corrected P value of P < 2.2 × 10−4 (P < .05/224) (eTable 3 in the Supplement). On average, MSE-decreasing alleles were associated with increased risk of retinal detachment (Figure 3) (Table 2). The MR-IVW OR of retinal detachment per 1-D increase was 0.72 (95% CI, 0.69-0.76; P = 3.8 × 10−44) in genetically predicted MSE. The MR estimates from weighted median and MR-Egger methods were consistent with overlapping 95% CIs (eTable 4 in the Supplement). There was no evidence of directional pleiotropy inflating our estimates based on the MR-Egger intercept test (eTable 4 in the Supplement). The MR-PRESSO test also suggested there were no horizontal pleiotropic outliers to distort our result. The funnel plot showed no evidence of asymmetry (eFigure 3 in the Supplement).
Figure 3. Association of Myopia-Increasing and Intraocular Pressure (IOP)–Increasing Risk Variants With Risk of Retinal Detachment.
A, The x-axis shows 224 genetic instruments for mean spherical equivalent (MSE) refractive error and their βs (effect size estimates) with refractive error. The y-axis shows the association of the same variants with retinal detachment; the y-axis unit is the effect size of single-nucleotide variants on retinal detachment in log(odds ratio [OR]). B, The x-axis shows 99 genetic instruments for IOP, and the y-axis shows the association of them with retinal detachment. The mendelian randomization (MR) inverse-weighted (IVW), MR-Egger, simple median, and weighted median regression lines are plotted.
In our sensitivity analysis, we also converted the continuous MSE to high myopia or myopia traits. These results indicate that higher genetic susceptibility to myopia was associated with retinal detachment (eTable 4 and eFigure 4 in the Supplement). For a binary exposure (ie, high myopia), the MR estimate represents a unit increase in the log(OR) of the exposure (exp[1] = 2.72), which means on average the change in the outcome for each 2.72-fold increase in the prevalence of the exposure.27 This makes the interpretation of MR estimate for high myopia incomparable with the OR from observational analysis. However, if we assume there is a linear relationship between MSE and the risk of retinal detachment (Figure 2), we can derive the OR for high myopia based on MSE MR estimation (exp[log(1/0.72) × 6] = 7.2 [95% CI, 5.19-9.27], where 0.72 is the OR for per 1-D increase of genetically predicted MSE and 6 represents the move of refraction error from emmetropia to high myopia).
Association of Higher IOP With Retinal Detachment in MR Analysis
From the IOP GWAS, we selected 99 single-nucleotide variants as genetic instruments (eTable 5 in the Supplement). The F statistics and the proportion of variance-explained R2 by instruments were 51 and 0.05, respectively. Intraocular pressure risk alleles were on average associated with increased risk of retinal detachment (Figure 3) (Table 2). For each unit (mm Hg) of genetically predicted increased IOP, the risk of retinal detachment increased by 8% (OR, 1.08; 95% CI, 1.03-1.14; P = .001). There was no evidence of directional pleiotropy biasing our findings based on the MR-Egger test and MR-PRESSO global test.
In our sensitivity analysis, we removed MSE loci from IOP genetic instruments (single-nucleotide variants with P < .001 in MSE GWAS), but our results were essentially unchanged (eTable 4 in the Supplement). We also conducted a multivariable MR analysis to assess the association of IOP with retinal detachment after adjusting for the effects from MSE. The association of IOP with retinal detachment did not essentially change after the adjustment of MSE (OR, 1.06; 95% CI, 1.01-1.12; P = .03) (Table 2). The results from multivariable MR analysis were also similar to an observational logistic regression model adjusting for the effects of MSE exhibiting overlapping 95% CIs (Table 2).
Discussion
We have conducted an observational and an MR analysis to investigate the associations between myopia and IOP with retinal detachment risk. Our MR analysis provided support for a potential causal inference for the associations of myopia and IOP with retinal detachment; the risk of retinal detachment was linearly increased with a higher level of myopia or IOP. Our sensitivity analyses support our main results, providing support for a potential causal association between myopia and retinal detachment.
Myopia is a leading cause of vision impairment worldwide, and its prevalence is increasing rapidly.28 Myopia is often considered benign because it could be easily corrected with contact lenses, glasses, or surgery.29 Clinically, pathological myopia with pathological signs of myopic maculopathy, myopic crescent, posterior staphyloma, or myopic optic neuropathy is often observed in eyes of high myopia rather than in eyes with a mild to moderate degree of myopia.28,30 Previous epidemiological studies have shown an association of myopia with retinal detachment.5,6,7 However, these studies are either case series reports or case-control studies with a relatively small sample size. In our study, the large sample size and well-measured MSE refractive error data exhibited a linear association between the degree of myopia and the risk of retinal detachment. In traditional observational studies, the association of myopia with retinal detachment could be confounded by other risk factors, such as sex, age, eye trauma, and cataract surgery. For instance, in our UKBB observational analysis, participants with high myopia had 2.5-fold increased risk of cataract compared with participants without myopia. Since cataract operation is also a risk factor for retinal detachment, the association of high myopia with retinal detachment could be partly confounded by cataract in observational analysis. The advantage of this study is that we used the MR method to investigate the association of myopia with retinal detachment. The robustly associated genetic instruments from several myopia proxy traits yielded similar results. These results reinforce that myopia is not a benign condition that can be trivially fixed with spectacles, and even a mild to moderate degree of myopia could cause serious eye complications.
To our knowledge, there are no previous studies investigating the association of IOP with retinal detachment. However, some studies showed that IOP was elevated after retinal detachment surgery.12,13 In this study, we used genetic data to evaluate the association of IOP with retinal detachment among adults of European descent. Our observational and MR results indicated higher levels of IOP were associated with retinal detachment. In our sensitivity analysis where we adjusted for MSE or removed MSE-related single-nucleotide variants, the effect size of IOP on retinal detachment risk attenuated very slightly (multivariable MR per 1–mm Hg increase: OR, 1.06; 95% CI, 1.01-1.12; P = .03). A previous study indicated that elevated IOP may increase the stretching stress on the sclera, choroid, and retina.31 However, further functional studies are warranted to investigate the underlying biological mechanism(s) between IOP and retinal detachment.
A concern in MR analysis is that a valid causal inference relies on the 3 MR assumptions. In this study, we investigated these assumptions using both biological knowledge and various statistical methods. First, we conducted GWASs for refractive error and IOP with large sample size, and the genetic variants are strong genetic instruments (relevance assumption). To our knowledge, there are no alternative biological pathways that can influence the associations of genetic instruments and retinal detachment risk (biological knowledge). We also used various MR methods (MR-Egger, weighted median, MR-PRESSO, and multivariable MR methods) that yield consistent estimates in the presence of invalid instruments in our sensitivity analysis to triangulate our causal inference. Notably, we found very weak evidence of pleiotropic effects (independence assumption and exclusion restriction). These analyses indicated that our findings were unlikely to be biased by violation of MR assumptions.
Limitations
Our results should be interpreted in light of their limitations. In our retinal detachment GWAS, we identified retinal detachment cases using both hospital health records and self-reported retinal detachment cases, and retinal detachment cases could include different subtype cases. However, a 2020 study32 showed that self-reported retinal detachment cases displayed similar epidemiologic features as ICD-defined cases. When we restricted retinal detachment cases to ICD-defined cases, the MR results were essentially unchanged (eFigure 5 in the Supplement). We further only used cases identified as ICD-10 code H330 (retinal detachment with retinal break). The MR-IVW OR of retinal detachment (only including cases using ICD-10 code H330) was 0.69 (95% CI, 0.64-0.74; P = 5.6 × 10−24) per unit (D) increase in genetically predicted MSE. For each unit (mm Hg) of genetically predicted increased IOP, the risk of RRD increased by 10% (OR, 1.10; 95% CI, 1.03-1.19; P = .007). The MR estimates were essentially unchanged, although as expected with lower power (and consequentially, the confidence intervals were wider). Second, we only conducted myopia, IOP, and retinal detachment GWASs in participants of European ancestry; thus, the generalizability of our findings in other racial/ethnic groups still needs further investigation. Third, the presence of horizontal pleiotropy (genetic instruments affecting the outcome through another pathway rather than the exposure) may violate one of the MR assumptions.11 However, in our MR sensitivity analysis, we conducted the MR-Egger intercept test and the MR-PRESSO global test and found no evidence for directional horizontal pleiotropy inflating our estimates. Given the large effect size for the association between myopia and RRD and the weak correlation between myopia and IOP, it remains possible that the effect size for the association IOP and RRD is confounded by the association of myopia with RRD. Our multivariable MR analysis suggests the IOP association was independent of the myopia association, although we cannot rule out the IOP association being driven by myopia (or another unknown confounder). A further specific limitation of our multivariable MR method is that the association between myopia and IOP was assumed to be linear; since this is difficult to assess, we cannot be certain that the associations for myopia and IOP with retinal detachment are truly independent.
Conclusions
Our study provides evidence supportive of a potential causal association between both myopia and IOP on retinal detachment risk. Such assessment of these risk factors could have direct public health and clinical implications for prevention of retinal detachment. Further research is required to determine whether reducing progression of myopia of an individual through interventions such as topical atropine33 or time spent outdoors34 will modify the risk of retinal detachment.
eFigure 1. Quantile-quantile plot for UK Biobank retinal detachment genome-wide association.
eFigure 2. Manhattan plot of retinal detachment genome-wide association study in UK Biobank.
eFigure 3. Funnel plots of myopia and intraocular pressure single genetic variant mendelian randomization analyses on retinal detachment risk.
eFigure 4. Myopia-increasing and high myopia–increasing risk variants are associated with increased risk of retinal detachment.
eFigure 5. Myopia-increasing and intraocular pressure (IOP)–increasing risk variants are associated with increased risk of International Classification of Diseases (ICD)–defined retinal detachment.
eTable 1. Two retinal detachment loci from UK Biobank.
eTable 2. Genetic instruments for mean spherical equivalent refractive error.
eTable 3. Myopia genome-wide significant variants that associated with retinal detachment risk at a Bonferroni-corrected level.
eTable 4. Mendelian randomization estimated associations between myopia and intraocular pressure (IOP) risk variants with retinal detachment.
eTable 5. Genetic instruments for intraocular pressure.
References
- 1.Kuhn F, Aylward B. Rhegmatogenous retinal detachment: a reappraisal of its pathophysiology and treatment. Ophthalmic Res. 2014;51(1):15-31. doi: 10.1159/000355077 [DOI] [PubMed] [Google Scholar]
- 2.Feltgen N, Walter P. Rhegmatogenous retinal detachment—an ophthalmologic emergency. Dtsch Arztebl Int. 2014;111(1-2):12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitry D, Charteris DG, Fleck BW, Campbell H, Singh J. The epidemiology of rhegmatogenous retinal detachment: geographical variation and clinical associations. Br J Ophthalmol. 2010;94(6):678-684. doi: 10.1136/bjo.2009.157727 [DOI] [PubMed] [Google Scholar]
- 4.Javitt JC, Street DA, Tielsch JM, et al. ; Cataract Patient Outcomes Research Team . National outcomes of cataract extraction. retinal detachment and endophthalmitis after outpatient cataract surgery. Ophthalmology. 1994;101(1):100-105. doi: 10.1016/S0161-6420(13)31251-2 [DOI] [PubMed] [Google Scholar]
- 5.Cambiaggi A. Myopia and retinal detachment: statistical study of some of their relationships. Am J Ophthalmol. 1964;58:642-650. doi: 10.1016/0002-9394(64)91383-2 [DOI] [PubMed] [Google Scholar]
- 6.Austin KL, Palmer JR, Seddon JM, et al. . Case-control study of idiopathic retinal detachment. Int J Epidemiol. 1990;19(4):1045-1050. doi: 10.1093/ije/19.4.1045 [DOI] [PubMed] [Google Scholar]
- 7.The Eye Disease Case-Control Study Group Risk factors for idiopathic rhegmatogenous retinal detachment. Am J Epidemiol. 1993;137(7):749-757. doi: 10.1093/oxfordjournals.aje.a116735 [DOI] [PubMed] [Google Scholar]
- 8.Ziff OJ, Lane DA, Samra M, et al. . Safety and efficacy of digoxin: systematic review and meta-analysis of observational and controlled trial data. BMJ. 2015;351:h4451. doi: 10.1136/bmj.h4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young SS, Karr A. Deming, data and observational studies: a process out of control and needing fixing. Significance. 2011;8(3):116-120. doi: 10.1111/j.1740-9713.2011.00506.x [DOI] [Google Scholar]
- 10.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925-1926. doi: 10.1001/jama.2017.17219 [DOI] [PubMed] [Google Scholar]
- 11.Davies NM, Holmes MV, Davey Smith G. Reading mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. doi: 10.1136/bmj.k601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto K, Iwase T, Terasaki H. Long-term changes in intraocular pressure after vitrectomy for rhegmatogenous retinal detachment, epi-retinal membrane, or macular hole. PLoS One. 2016;11(11):e0167303. doi: 10.1371/journal.pone.0167303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koreen L, Yoshida N, Escariao P, et al. . Incidence of, risk factors for, and combined mechanism of late-onset open-angle glaucoma after vitrectomy. Retina. 2012;32(1):160-167. doi: 10.1097/IAE.0b013e318217fffb [DOI] [PubMed] [Google Scholar]
- 14.MacGregor S, Ong J-S, An J, et al. . Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50(8):1067-1071. doi: 10.1038/s41588-018-0176-y [DOI] [PubMed] [Google Scholar]
- 15.Guggenheim JA, Williams C; UK Biobank Eye and Vision Consortium . Role of educational exposure in the association between myopia and birth order. JAMA Ophthalmol. 2015;133(12):1408-1414. doi: 10.1001/jamaophthalmol.2015.3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Nielsen JB, Fritsche LG, et al. . Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335-1341. doi: 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50(7):906-908. doi: 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40(3):740-752. doi: 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734-1739. doi: 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S, Thompson SG. Interpreting findings from mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377-389. doi: 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. doi: 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess S, Thompson SG. Multivariable mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181(4):251-260. doi: 10.1093/aje/kwu283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemani G, Zheng J, Elsworth B, et al. . The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48(3):713-727. doi: 10.1093/ije/dyy262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol. 2018;33(10):947-952. doi: 10.1007/s10654-018-0424-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan IG, Ohno-Matsui K, Saw S-M. Myopia. Lancet. 2012;379(9827):1739-1748. doi: 10.1016/S0140-6736(12)60272-4 [DOI] [PubMed] [Google Scholar]
- 29.Bourke CM, Loughman J, Fltcroft DI, Loskutova E, O’Brien C. We can’t afford to turn a blind eye to myopia. QJM. Published online March 26, 2019. doi: 10.1093/qjmed/hcz076 [DOI] [PubMed] [Google Scholar]
- 30.Wong TY, Ferreira A, Hughes R, Carter G, Mitchell P. Epidemiology and disease burden of pathologic myopia and myopic choroidal neovascularization: an evidence-based systematic review. Am J Ophthalmol. 2014;157(1):9-25.e12. doi: 10.1016/j.ajo.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 31.McMonnies CW. An examination of the relation between intraocular pressure, fundal stretching and myopic pathology. Clin Exp Optom. 2016;99(2):113-119. doi: 10.1111/cxo.12302 [DOI] [PubMed] [Google Scholar]
- 32.Boutin TS, Charteris DG, Chandra A, et al. ; UK Biobank Eye & Vision Consortium; 23andMe Research Team . Insights into the genetic basis of retinal detachment. Hum Mol Genet. 2020;29(4):689-702. doi: 10.1093/hmg/ddz294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chia A, Lu Q-S, Tan D. Five-year clinical trial on atropine for the treatment of myopia 2: myopia control with atropine 0.01% eyedrops. Ophthalmology. 2016;123(2):391-399. doi: 10.1016/j.ophtha.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 34.He M, Xiang F, Zeng Y, et al. . Effect of time spent outdoors at school on the development of myopia among children in China: a randomized clinical trial. JAMA. 2015;314(11):1142-1148. doi: 10.1001/jama.2015.10803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Quantile-quantile plot for UK Biobank retinal detachment genome-wide association.
eFigure 2. Manhattan plot of retinal detachment genome-wide association study in UK Biobank.
eFigure 3. Funnel plots of myopia and intraocular pressure single genetic variant mendelian randomization analyses on retinal detachment risk.
eFigure 4. Myopia-increasing and high myopia–increasing risk variants are associated with increased risk of retinal detachment.
eFigure 5. Myopia-increasing and intraocular pressure (IOP)–increasing risk variants are associated with increased risk of International Classification of Diseases (ICD)–defined retinal detachment.
eTable 1. Two retinal detachment loci from UK Biobank.
eTable 2. Genetic instruments for mean spherical equivalent refractive error.
eTable 3. Myopia genome-wide significant variants that associated with retinal detachment risk at a Bonferroni-corrected level.
eTable 4. Mendelian randomization estimated associations between myopia and intraocular pressure (IOP) risk variants with retinal detachment.
eTable 5. Genetic instruments for intraocular pressure.