Abstract
Rice grain size plays a crucial role in determining grain quality and yield. In this study, two multiparent advanced generation intercross (MAGIC) populations, DC1 and BIM, were evaluated for grain size across three environments and genotyped with 55K array-based SNP detection and genotype-by-sequencing (GBS), respectively, to identify QTLs and SNPs associated with grain length, grain width, grain length–width ratio, grain thickness, and thousand grain weight. A total of 18 QTLs were identified for the five grain size-related traits and explained 6.43–63.35% of the total phenotypic variance. Twelve of these QTLs colocalized with the cloned genes, GS3, GW5/qSW5, GW7/GL7/SLG7, and GW8/OsSPL16, of which the first two genes showed the strongest effect for grain length and grain width, respectively. Four potential new genes were also identified from the QTLs, which exhibited both genetic background independency and environment stability and could be validated in future studies. Moreover, the significant SNP markers identified are valuable for direct utilization in marker-assisted breeding to improve rice grain size.
Keywords: grain size, association mapping, MAGIC population, rice, single nucleotide polymorphism
Introduction
Grain size is one of the key agronomic traits that proceeded from unconscious selective pressure over the course of rice domestication. Improving grain size as an early adaptive response to rice cultivation is a result of unintentional selection for seeds that could survive even with deeper soil cultivation (Purugganan and Fuller, 2009). Interestingly, it was further subject to deliberate selection and breeding as it impacts rice grain quality and yield per se (Tan et al., 2000; Lu et al., 2013). A wide range of grain size characteristics therefore exists in modern-day rice varieties, which mainly defines consumers’ preferences and market value (Fitzgerald et al., 2009).
Rice grain size is characterized by a combination of grain length (GL), grain width (GW), and grain thickness (GT) (Tan et al., 2000; Xing et al., 2002). It is closely associated with grain weight, a major yield component along with the number of panicles per plant and the number of grains per panicle. Many QTL mapping studies using populations derived from various biparental crosses have been conducted. Over 400 QTLs for grain size have been mapped across 12 chromosomes of rice, of which 109, 95, and 107 were associated with GL, GW, and thousand grain weight (TGW), respectively (Huang et al., 2013). A few of these QTLs have been fine-mapped including gw3.1 (Li et al., 2004), qGL3-a (Wan et al., 2006), gw8.1 (Xie et al., 2006), gw5 (Wan et al., 2008), GW1-1 and GW1-2 (Yu et al., 2008), gw9.1 (Xie et al., 2008), GW3 and GW6 (Guo et al., 2009), qGL7 (Bai et al., 2010), qGL4b (Kato et al., 2011), tgw11 (Oh et al., 2011), qSS7 (Qiu et al., 2012), qGS7 (Shao et al., 2012), qGRL1 (Singh et al., 2012), and GS2 (Zhang et al., 2013). Recent advances in rice functional genomics facilitated the cloning and functional characterization of several genes that either positively or negatively regulates grain size. Negative grain size regulators that were previously cloned include GS3 (Fan et al., 2006), GW2 (Song et al., 2007), TGW6 (Ishimaru et al., 2013), GW7/GL7/SLG7 (Wang et al., 2015a, c; Zhou et al., 2015), qGL3/qTGW3 (Ying et al., 2018), and LARGE8 (Xu et al., 2018); whereas positive grain size regulators include GW5/qSW5 (Shomura et al., 2008; Weng et al., 2008), GS5 (Li et al., 2011), GW8/OsSPL16 (Wang et al., 2012b), GS2 (Duan et al., 2015; Hu et al., 2015), BG1 (Liu et al., 2015), WTG1 (Huang et al., 2017; Liu et al., 2018), GS9 (Zhao et al., 2018), and qLGY3/OsLG3b (Yu et al., 2018). It is noteworthy that these cloned genes control grain size by altering cell proliferation and/or cell expansion affecting cell numbers either in latitudinal or longitudinal directions. Furthermore, functional characterization revealed a variety of different proteins involved in a range of signal transduction pathways affecting grain size. However, only a few genes/QTLs are directly useful in breeding. As most of the genetic mapping studies conducted have relied on traditional linkage mapping using populations derived from biparental crosses, the identified QTLs are often not transferable to other genetic backgrounds since the estimated effects are limited to the two parents under study. Given the tiny fraction of the total variation present, different biparental populations yield different QTLs and with varying effects due to epistasis, pleiotropy, and QTL-by-environment interaction.
The genome-wide association study (GWAS) overcomes the limitation of biparental linkage mapping by taking advantage of many years of historical and evolutionary recombination to localize QTL in genetically diverse populations. In rice, the GWAS has been demonstrated to be a powerful complementary strategy to the biparental linkage mapping for grain size (Si et al., 2016; Duan et al., 2017; Yu et al., 2017; Ma et al., 2019). However, the GWAS using natural populations is often associated with complicated population structure and cryptic kinship, which results to spurious marker trait associations (MTAs). The use of multiparent advanced generation intercross (MAGIC) populations offers an alternative approach to the GWAS using natural populations and to linkage mapping using populations derived from biparental crosses. MAGIC has more allelic and phenotypic diversity ensuring that more QTLs segregate within the population (Cavanagh et al., 2008; Huang et al., 2015). Furthermore, it has a better control over population structure and kinship, and has been proven effective in identifying major genes via association mapping (Bandillo et al., 2013; Meng et al., 2016, 2017; Descalsota et al., 2018; Ogawa et al., 2018; Ponce et al., 2018).
In this study, association mapping was conducted in two MAGIC populations tested across three environments for 2 years to identify QTLs associated with the grain size-related traits—GL, GW, grain length–width ratio (GLWR), and TGW. The results in this study could provide valuable information to further elucidate the genetic basis of rice grain size and in marker-assisted breeding.
Materials and Methods
Association Mapping Panel
Two MAGIC populations developed at the International Rice Research Institute (IRRI) were used: (1) a four-parent MAGIC population, DC1, previously characterized by Meng et al. (2016), and (2) an eight-parent MAGIC population reported by Bandillo et al. (2013), which we referred to as Bandillo indica MAGIC (BIM) population in this study. The parents used to develop both populations were presented in Supplementary Table 1. A total of 221 and 378 lines in the DC1 and BIM populations, respectively, were evaluated for five grain size-related traits.
Phenotyping and Phenotypic Analysis
The field trials were conducted in three testing environments, one in the Philippines at the headquarters of the IRRI, Los Baños, Laguna in 2017 and two in China at Henan in 2018 and at Hainan in 2017 and in 2018. Trials in each testing environment were laid out in an incomplete block design. Freshly harvested paddy was dried to moisture content of 12–14% and equilibrated in paper bags at room temperature for 3 months prior to measurement of grain size-related traits. GL (mm) and GW (mm) were evaluated using grain scanner (Seiko Epson, Suwa City, Japan). Grain images (0.042433 mm/pixel) were analyzed using the SmartGrain software program (Tanabata et al., 2012). GLWR was calculated as the ratio of GL and GW. GT (mm) and TGW (g) were measured according to the National Rice Grain Quality Assessment Standard of China (GB/T17891-1999). About 100 grains for each entry were evaluated for GL and GW, and a total of 6 grains were measured for GT.
The population size of DC1 and BIM varied greatly in all the three testing environments (Supplementary Table 2). Phenotypic analysis was conducted using a linear mixed model to properly handle unbalanced data. The best linear unbiased estimates (BLUEs) of each line were obtained using the PBTools (bbi.irri.org). Trait correlations were calculated and plotted using the corrplot package in R.
SNP Genotyping
The DC1 population was sequenced with a high-density array-based SNP platform using the 55K Affymetrix Axiom Rice Genotyping Array at the CapitalBio Technology Beijing, China (Meng et al., 2017), whereas the BIM population was sequenced with a GBS approach using Illumina HiSeq at the Cornell University (Bandillo et al., 2013). A stringent filtering strategy was conducted to choose high-quality SNPs for association. All heterozygous markers were set to missing. Markers with minor allele frequency (MAF) < 0.05 were removed. Highly correlated markers (r2 > 0.95) were also excluded from the SNP data set (Supplementary Datasheet 1).
Population Structure and Linkage Disequilibrium Analysis
Principal component analysis (PCA) was used to infer population structure. The first two principal components (PCs) were plotted using ggplot2 in R to visualize the dispersion of DC1 and BIM lines. The LD analysis was performed by pairwise comparisons in a set of filtered SNP markers (MAF < 0.05) using the LD function in TASSEL v.5.2.19 (Bradbury et al., 2007). Squared allele frequency correlations (r2) between marker pairs were used to estimate LD. The loci with significant LD were identified based on p < 0.0001; the rest were considered not informative. Significant intrachromosomal r2 values were plotted against physical distance, and a smoothering second degree LOESS curve (Cleveland, 1979) was fitted using the ggplot2 package in R (Wickham, 2016). The intersection of the loess curve and the critical r2 value beyond which LD was likely to be caused by genetic linkage was considered as the estimated extent of LD decay (Breseghello and Sorrells, 2006).
Association Analysis
Association analysis was carried out using the GAPIT in R implementing a mixed linear model to account for both population structure and kinship (Yu et al., 2006). The threshold value was set at −log10(P) < 4 as shown in the Manhattan plot to identify the peak association signals. The plots were visualized using the qqman package in R (Turner, 2014). Peaks exhibiting the significance threshold level within a physical distance of ∼2.25 and 1.70 Mb for DC1 and BIM populations, respectively, were considered as a single QTL.
A promising QTL was considered when many SNPs lined up near the peak of the QTL and when it showed both genetic background independency and environment stability. Candidate genes in the promising QTLs were searched in the MSU Rice Genome Annotation database (http://rice.plantbiology.msu.edu/). Potential new genes were then selected based on the following criteria: (1) with significant MTAs accounting for over 10% of the total phenotypic variance; and (2) with significant MTAs corresponding to non-synonymous SNPs in the coding region of the genes.
Results
Phenotypic Variation and Trait Correlation
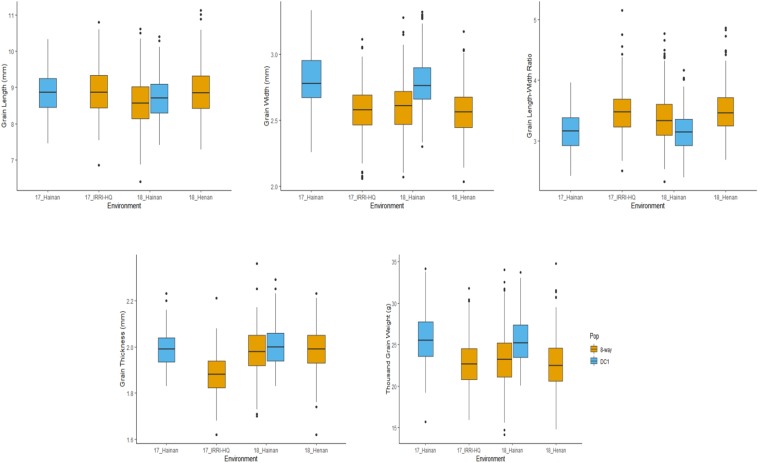
A wide range of values for all the grain size-related traits were observed in both the DC1 and BIM populations including their founder lines across the three testing environments. Wider phenotypic variability was observed in the BIM than in the DC1 population for all the measured traits attributed to the wider phenotypic variation of the BIM founder lines (Supplementary Table 1). Most of the traits appeared to be normally distributed; however, some trait–environment combinations showed skewed distributions as evidently shown by the lopsided boxplot. Prominent data skewness was observed for GW of DC1 and BIM populations tested in Hainan 2017 and Hainan 2018, respectively (Figure 1).
FIGURE 1.
Box plot for the five grain size-related traits of MAGIC population in different environments. 17_Hainan: 2017 in Hainan. 17_IRRI-HQ: 2017 in IRRI. 18_Hainan: 2018 in Hainan. 18_Henan: 2018 in Henan.
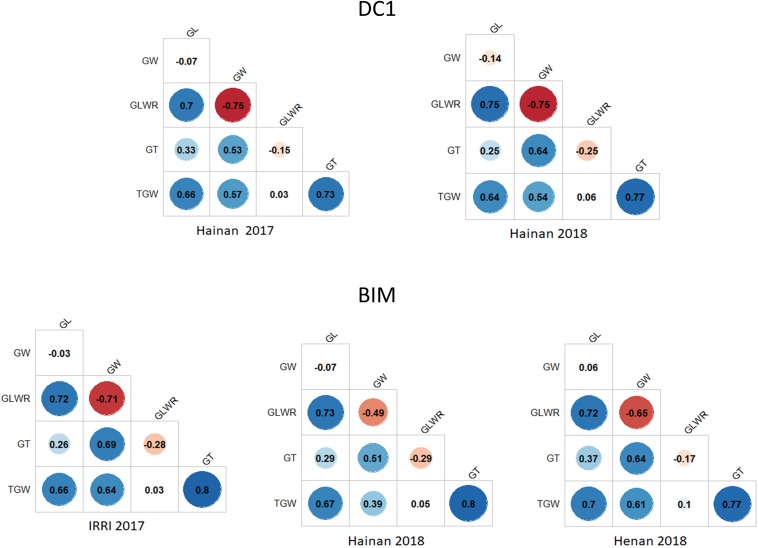
The correlations of TGW with GL, GW, and GT in both populations across different environments were positive ranging from moderate (0.39) to strong (0.80). GT was negatively correlated with GLWR with correlations ranging from −0.15 for the DC1 in Hainan 2017 to −0.29 for the BIM in Hainan 2018. On the contrary, the correlations of GT with GL and GW in both populations and across different environments were positive with low to moderate values (Figure 2). The correlation between GLWR and GL was consistently high in all the trials with values ranging from 0.70 to 0.75. Negative and strong correlation (−0.75) was found between GLWR and GW in the Hainan 2017 and Hainan 2018, whereas for the BIM, it was moderate (−0.49) to strong (−0.71) in Hainan 2018 and IRRI 2017 trials, respectively.
FIGURE 2.
Trait correlations for the five grain size-related traits in DC1 and BIM populations tested across different environments.
Population Structure and Whole Genome Pattern of LD Decay
The first two PCs, which accounted for most of the variation, were plotted to observe any subgroups in both populations under study. Results showed two subgroupings in the DC1 as previously reported by Meng et al. (2016), whereas no subgrouping was observed in the BIM population (Supplementary Figure 1).
The decay of LD along with physical distances was computed for both the DC1 and BIM populations. A critical value of the determination coefficients r2 > 0.2 was used to be the appropriate threshold for LD (Meng et al., 2016). A scatter r2 against physical distance showed a clean pattern of LD decay in the DC1 and BIM populations. The decline of LD to 50% of its initial value was at 2.25 Mb for the DC1 (Meng et al., 2016) and at 1.70 Mb for the BIM population (Supplementary Figure 2)
QTLs Identified by Association Analysis
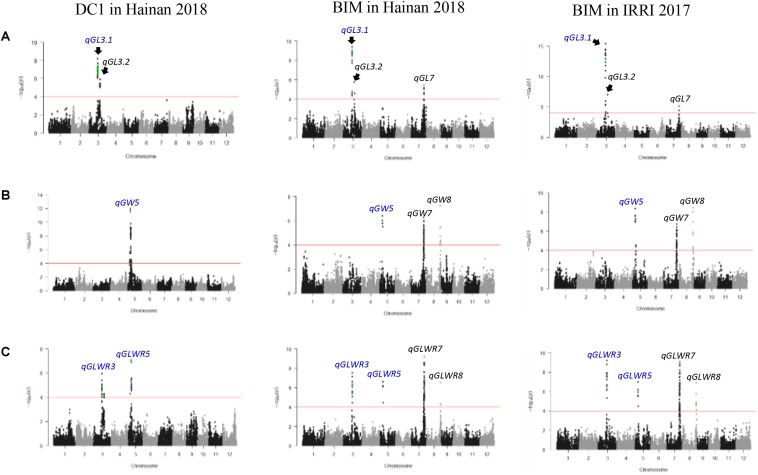
A total of 329 and 334 significant MTAs were identified for the DC1 population in Hainan 2017 and Hainan 2018, respectively. For the BIM population, a total of 480, 413, and 254 significant MTAs were identified in IRRI 2017, Hainan 2017, and Henan 2018, respectively (Supplementary Table 3). These significant MTAs were delineated into a total of 18 QTLs for the five measured traits (Table 1). The genome-wide Manhattan plots for grain size-related traits with QTLs that showed genetic background independency and environment stability were presented in Figure 3. Manhattan plots for all the trait–environment combinations were presented in Supplementary Figure 3.
TABLE 1.
QTLs identified from DC1 and BIM populations for the five grain size-related traits.
| QTL | Panel | Enva | Year | Chrb | Peak SNP | Posc | Allelesd | p-value | %PVEe | Effectf | Known gene |
| qGL3.1 | DC1 | Hainan | 2017 | 3 | AX-154083640 | 16806335 | A/C | 1.93E-09 | 54.73 | 0.3293 | GS3 |
| DC1 | Hainan | 2018 | 3 | AX-154083640 | 16806335 | A/C | 6.58E-09 | 55.28 | 0.0462 | GS3 | |
| BIM | IRRI | 2017 | 3 | rs3_16842769 | 16842769 | C/T | 3.80E-16 | 35.07 | 0.3425 | GS3 | |
| BIM | Hainan | 2018 | 3 | rs3_16842769 | 16842769 | C/T | 4.04E-10 | 21.60 | 0.0127 | GS3 | |
| BIM | Henan | 2018 | 3 | rs3_16729992 | 16729992 | G/C | 5.37E-10 | 21.56 | 0.2809 | GS3 | |
| qGL3.2 | DC1 | Hainan | 2017 | 3 | AX-165099081 | 21547035 | C/T | 8.71E-06 | 54.73 | 0.2209 | – |
| DC1 | Hainan | 2018 | 3 | AX-115865890 | 21602538 | C/T | 1.19E-06 | 55.28 | 0.0330 | – | |
| BIM | IRRI | 2017 | 3 | rs3_20691702 | 20691702 | G/T | 1.01E-07 | 35.07 | −0.1906 | – | |
| BIM | Hainan | 2018 | 3 | rs3_20691702 | 20691702 | G/T | 1.73E-06 | 21.60 | −0.0154 | – | |
| BIM | Henan | 2018 | 3 | rs3_20691702 | 20691702 | G/T | 1.66E-06 | 17.74 | −0.2016 | – | |
| qGL7 | BIM | IRRI | 2017 | 7 | rs7_24645555 | 24645555 | A/G | 6.82E-06 | 35.07 | 0.2911 | GL7/GW7/SLG7 |
| BIM | Hainan | 2018 | 7 | rs7_24669663 | 24669663 | C/G | 6.69E-06 | 21.60 | −0.0117 | GL7/GW7/SLG7 | |
| qGW5 | DC1 | Hainan | 2017 | 5 | AX-165092179 | 5364311 | G/T | 3.08E-15 | 59.78 | −0.1506 | GW5/qSW5 |
| DC1 | Hainan | 2018 | 5 | AX-165092179 | 5364311 | G/T | 1.03E-12 | 24.88 | −0.0021 | GW5/qSW5 | |
| BIM | IRRI | 2017 | 5 | rs5_4753775 | 4753775 | A/G | 4.54E-09 | 22.78 | 0.1164 | – | |
| BIM | Hainan | 2018 | 5 | rs5_4882721 | 4882721 | A/T | 4.08E-07 | 17.96 | −0.0231 | – | |
| BIM | Henan | 2018 | 5 | rs5_4685451 | 4685451 | C/G | 1.37E-05 | 10.42 | −0.0983 | – | |
| qGW7 | BIM | IRRI | 2017 | 7 | rs7_24380529 | 24380529 | A/G | 1.83E-07 | 22.78 | 0.1016 | GL7/GW7/SLG7 |
| BIM | Hainan | 2018 | 7 | rs7_24279896 | 24279896 | C/G | 5.46E-07 | 17.96 | −0.0061 | GL7/GW7/SLG7 | |
| BIM | Henan | 2018 | 7 | rs7_24505268 | 24505268 | A/T | 1.03E-05 | 10.42 | −0.0740 | – | |
| qGW8 | BIM | IRRI | 2017 | 8 | rs8_26573952 | 26573952 | A/C | 3.47E-09 | 22.78 | 0.0933 | GW8/OsSPL16 |
| BIM | Hainan | 2018 | 8 | rs8_26573952 | 26573952 | A/C | 6.04E-08 | 17.96 | 0.0831 | GW8/OsSPL16 | |
| BIM | Henan | 2018 | 8 | rs8_26351058 | 26351058 | C/G | 5.61E-05 | 10.42 | −0.0549 | – | |
| qGLWR3 | DC1 | Hainan | 2017 | 3 | AX-154083640 | 16806335 | A/C | 1.55E-06 | 63.35 | 0.1340 | GS3 |
| DC1 | Hainan | 2018 | 3 | AX-154083640 | 16806335 | A/C | 1.11E-06 | 59.60 | 0.0593 | GS3 | |
| BIM | IRRI | 2017 | 3 | rs3_16842769 | 16842769 | C/T | 5.96E-10 | 29.52 | 0.1453 | GS3 | |
| BIM | Hainan | 2018 | 3 | rs3_16842769 | 16842769 | C/T | 3.04E-08 | 17.95 | 0.0272 | GS3 | |
| BIM | Henan | 2018 | 3 | rs3_16842769 | 16842769 | C/T | 1.27E-05 | 11.98 | 0.1624 | GS3 | |
| qGLWR5 | DC1 | Hainan | 2017 | 5 | AX-165092179 | 5364311 | G/T | 2.31E-09 | 63.35 | 0.1667 | GW5/qSW5 |
| DC1 | Hainan | 2018 | 5 | AX-165085603 | 5368362 | C/T | 8.29E-08 | 59.60 | 0.0046 | GW5/qSW5 | |
| BIM | IRRI | 2017 | 5 | rs5_4685451 | 4685451 | C/G | 1.04E-07 | 29.52 | 0.2049 | – | |
| BIM | Hainan | 2018 | 5 | rs5_4685451 | 4685451 | C/G | 2.40E-07 | 17.95 | 0.0125 | – | |
| qGLWR7 | BIM | IRRI | 2017 | 7 | rs7_24686632 | 24686632 | A/G | 7.88E-10 | 29.52 | 0.2320 | GL7/GW7/SLG7 |
| BIM | Hainan | 2018 | 7 | rs7_24279896 | 24279896 | C/G | 6.46E-10 | 17.95 | −0.0297 | GL7/GW7/SLG7 | |
| BIM | Henan | 2018 | 7 | rs7_24505268 | 24505268 | A/T | 4.29E-07 | 19.23 | 0.1819 | GL7/GW7/SLG7 | |
| qGLWR8 | BIM | IRRI | 2017 | 8 | rs8_26573952 | 26573952 | A/C | 1.22E-07 | 29.52 | −0.1599 | GW8 |
| BIM | Hainan | 2018 | 8 | rs8_26573952 | 26573952 | A/C | 3.09E-07 | 17.95 | 0.0237 | GW8 | |
| qGT1 | BIM | IRRI | 2017 | 1 | rs1_6640911 | 6640911 | C/T | 3.54E-06 | 22.47 | −0.0311 | – |
| qGT3 | BIM | Henan | 2018 | 3 | rs3_17044632 | 17044632 | A/G | 8.04E-05 | 6.43 | −0.0231 | – |
| qGT5 | DC1 | Hainan | 2017 | 5 | AX-155076620 | 5420315 | A/C | 1.01E-05 | 30.87 | −0.0367 | GW5/qSW5 |
| DC1 | Hainan | 2018 | 5 | AX-165092179 | 5364311 | G/T | 2.35E-06 | 33.68 | −0.0009 | GW5/qSW5 | |
| BIM | Hainan | 2018 | 5 | rs5_5391586 | 5391586 | A/C | 8.39E-06 | 21.79 | −0.0004 | – | |
| qTGW3.1 | DC1 | Hainan | 2017 | 3 | AX-154017324 | 3310128 | A/G | 9.51E-06 | 40.47 | – | – |
| qTGW3.2 | BIM | IRRI | 2017 | 3 | rs3_16996623 | 16996623 | G/T | 6.79E-09 | 31.31 | 1.0800 | GS3 |
| BIM | Hainan | 2018 | 3 | rs3_16736681 | 16736681 | G/T | 4.71E-06 | 21.19 | 0.1362 | GS3 | |
| BIM | Henan | 2018 | 3 | rs3_16729992 | 16729992 | G/C | 4.78E-08 | 19.94 | 1.4490 | GS3 | |
| qTGW5 | DC1 | Hainan | 2017 | 5 | AX-165092179 | 5364311 | A/G | 2.14E-06 | 40.49 | −1.1514 | GW5/qSW5 |
| DC1 | Hainan | 2018 | 5 | AX-165092179 | 5364311 | A/G | 6.23E-06 | 40.80 | −0.0938 | GW5/qSW5 | |
| qTGW8.1 | DC1 | Hainan | 2018 | 8 | AX-115754523 | 861108 | C/T | 9.21E-05 | 40.47 | 0.1782 | – |
| qTGW8.2 | BIM | Henan | 2018 | 8 | rs8_26349349 | 26349349 | G/T | 1.94E-05 | 11.38 | 0.7771 | – |
aEnvironment where the trial was conducted, bChromosome number, cPhysical position (bp), dMajor/minor allele, ePercent phenotypic variance explained by the marker, fAllele effect with respect to minor allele.
FIGURE 3.
Genome-wide Manhattan plots for (A) grain length, (B) grain width, and (C) grain length–width ratio in DC1 and BIM measured in two testing environments. QTLs in blue color exhibited genetic background independency and environment stability. Dots in green color are SNPs corresponding to previously cloned genes (GS3, GW5/qSW5, GL7/GW7/SLG7, and GW8/OsSPL16 on chromosomes 3, 5, 7, and 8, respectively).
Three QTLs, two on chromosome 3 and one on chromosome 7, were identified for GL (Figure 3). qGL3.1 and qGL3.2 were consistently detected in both of the DC1 and BIM populations across the three testing environments. These two closely localized QTLs were in different LD blocks (Supplementary Figure 4) and are therefore independent of each other. Phenotypic variance of qGL3.1 ranged from 21.56 to 55.28%, whereas it ranged from 17.74 to 55.28% for qGL3.2 (Table 1). Four significant SNPs in qGL3.1, one in the coding region and three located at 7.2 to 7.3 kb downstream of the GS3 gene, were consistently identified in the BIM population in the three testing environments. A total of 10 significant SNPs of which 8 were in the intron, 1 in the coding region, and 1 at about 6.2 kb downstream of the GS3 gene were identified significant in the DC1 population. Interestingly, significant non-synonymous SNPs AX-115826214 in the DC1 and rs3_16729992 in the BIM were within the coding region of GS3 gene. qGL7 was detected only in the BIM population, which explained 35.07 and 21.60% of the total phenotypic variance in IRRI 2017 and Hainan 2018, respectively. The peak SNP, rs7_24669663, in this locus was located at 5.2 kb upstream of the GL7/GW7/SLG7 gene (Supplementary Table 3).
A total of three QTLs were detected on chromosomes 5, 7, and 8 for GW (Figure 3). qGW5 was detected in the DC1 and explained 59.78 and 24.88% of the total phenotypic variance in Hainan 2017 and Hainan 2018, respectively. This QTL was also detected in the BIM population in all the three testing environments with the phenotypic variance explained ranging from 10.42 to 22.78%. In this locus, the two significant SNPs, AX-155172546 and AX-165092179, detected in the DC1 were within 1.2 kb upstream of the GW5/qSW5 gene. qGW7 and qGW8 were detected only in the BIM population, both of which explained 22.78, 17.96, and 10.42% of the total phenotypic variation in IRRI 2017, Hainan 2017, and Hainan 2018, respectively. The peak SNP, rs7_24279896, in qGL7 was located at 5.2 kb upstream of the GL7/GW7/SLG7 gene. Moreover, the peak SNP, rs8_26505685, in qGW8 loci was within the coding region of GW8 (Table 1 and Supplementary Table 3).
Four QTLs located on chromosomes 3, 5, 7, and 8 were identified for GLWR (Figure 3). qGLWR3 and qGLWR5 explained very high proportions of the phenotypic variances in the DC1 population in Hainan 2017 (63.35%) and Hainan 2018 (59.60%). On the contrary, relatively low proportions of the phenotypic variations were observed in the BIM population ranging from 11.98 to 29.52% for qGLWR3 and 17.95 to 29.52% for qGLWR5. It is noteworthy that qGLWR3 was stably detected in both populations across all the three testing environments. In the DC1 population, eight, one, and one significant SNP/s were located in the intron, the coding region, and 6.2 kb upstream of the GS3, respectively. For the BIM population, four significant SNPs were within the GS3 gene of which one SNP, rs_16729992, was located in the coding region and three SNPS were within 7.2 to 7.3 kb downstream of the gene. Moreover, two significant SNPs, AX-155172546 and AX-165092179, in qGLWR5 were within 1.2 kb upstream of the GW5/qSW5 gene. The QTLs qGLWR7 and qGLWR8 were both detected in the BIM population. qGLWR7 explained 17.95–29.52% of the total phenotypic variances. The significant SNP, rs7_24669663, in this locus was within 5.2 kb upstream of the GL7/GW7/SLG7 gene. qGLWR8 explained 17.95–29.52% of the total phenotypic variance and it is noteworthy that the significant SNP, rs8_26505685, in this locus was within the coding region of the GW8/OsSPL16 gene (Table 1 and Supplementary Table 3).
Three QTLs located on chromosomes 1, 3, and 5 were detected for GT. qGT1 and qGT3 were detected only in the BIM population in IRRI 2017 and in Henan 2018, respectively. qGT1 explained moderate phenotypic variance (22.47%), whereas qGT3 had relatively low phenotypic variance (6.43%). qGT5 was identified in both populations and explained 21.79–33.68% of the total phenotypic variation. The peak SNP in this QTL, AX-165092179, was located at 1.2 kb upstream of the GW5/qSW5 gene (Table 1 and Supplementary Table 3).
A total of five QTLs, two on chromosomes 3 and 8 and one on chromosome 5, were detected for TGW. qTGW3.1 was detected only in the DC1 population in Hainan 2017 and accounted for 40.47% of the total phenotypic variance. qTGW3.2 was detected only in the BIM population and explained 19.94–31.31% of the total phenotypic variance. Four significant SNPs in the qTGW3.2 locus were in the untranslated and coding region of GS3. qTGW5 was detected only in DC1 population with 40.49 and 40.80% of the phenotypic variance in Hainan 2017 and Hainan 2018, respectively. Two significant SNPs, AX-155172546 and AX-165092179, in this QTL were within 1.2 kb upstream of the GW5/qSW5 gene. qTGW8.1 was detected only in the DC1 population in Hainan 2018 and accounted for 40.47% of the total phenotypic variance, whereas qTGW8.2 was detected in the BIM population in Henan 2018 and accounted for 11.38% of the total phenotypic variance (Table 1 and Supplementary Table 3).
Potential Candidate Genes for Promising QTLs
Based on both genetic background independency and stability across different environments, four QTLs on chromosomes 3 and 5 were considered promising (Figure 3). Potential candidate genes in these QTLs regions were narrowed down to a total of four (Table 2) through literature searches and by considering only those with significant non-synonymous SNPs in the coding region. Two potential candidate genes, LOC_Os03g29630 (ulp1) and LOC_Os03g29810 (OsClp6), were identified for qGL3.1 and qGLWR3. Interestingly, OsClp6 was identified in both of the DC1 and BIM populations across all the three testing environments. Two potential genes, LOC_Os05g08850 (putative cytochrome P450) and LOC_Os05g10620 (NAC domain containing protein 75), were identified for qGW5 and qGLWR5.
TABLE 2.
Potential candidate genes from promising QTLs detected for the grain size-related traits.
| QTLs | Gene ID | Gene annotation | SNP marker | Position | SNP |
| qGL3.1, qGLWR3 | LOC_Os03g29630 | ulp1 protease family protein, putative, expressed | AX-115855710 | 16890773 | A/G |
| LOC_Os03g29810 | OsClp6—Putative Clp protease | AX-115799679 | 16996623 | G/T | |
| AX-115815041 | 16997653 | C/T | |||
| AX-165099608 | 16993369 | C/T | |||
| rs3_16996623 | 16996623 | G/T | |||
| qGW5, qGLWR5 | LOC_Os05g08850 | Cytochrome P450, putative, expressed | rs5_4885582 | 4885582 | C/A |
| rs5_4886725 | 4886725 | T/G | |||
| LOC_Os05g10620 | No apical meristem protein, putative, | AX-115819199 | 5785251 | C/A | |
| expressed/NAC domain containing protein 75 | AX-154635971 | 5799150 | C/A |
Discussion
Phenotypic Variation and Trait Correlation
Wide phenotypic variation was observed for all the traits in all the testing environments relative to the founder lines, suggesting the formation of transgressive segregants. Transgressive segregation is of interest for breeders as it provides breeding materials best use for crop improvement. The wide variability for grain size-related traits among the MAGIC lines under study will allow breeders to select superior lines with improved grain size. The high variation observed also suggests that both of the DC1 and BIM populations can be effectively utilized in finding allelic variants responsible for grain size differences.
In the present study, positive and strong correlations were observed between GL and TGW, GT, and GW, which was consistent with the previous studies (Tan et al., 2000; Wang et al., 2012b). This suggests that GL has the largest effect on grain weight compared with other grain size-related traits (Rui and Zhao, 1983; Lin and Wu, 2003; Xing and Zhang, 2010), whereas GW contributes more on GT. Very weak and positive correlation between TGW and GLWR was observed in the two MAGIC populations across the three testing environments. This result was consistent with the findings of Qiu et al. (2015), but different with the result of Xu et al. (2015b).
Population Structure and Whole Genome Pattern of LD Decay
One of the advantages of the MAGIC populations is that they are homogeneous without population structure. Indeed, no substructure was observed for the MAGIC populations in rice (Bandillo et al., 2013; Meng et al., 2016), tomato (Pascual et al., 2015), wheat (Huang et al., 2012a), and barley (Sannemann et al., 2015). In this study, no subpopulation was observed in the BIM population, whereas obvious substructure was observed in the DC1. By carefully inspecting the genotype of the DC1 lines, it was found that substructuring was caused by the presence of a few exceptionally similar lines. Removing these lines resulted to no substructure. It is therefore important in the GWAS that the analysis of population structure must be done prior to association regardless of the type of the populations.
The LD decay of the MAGIC populations under study ranged from 1.70 to 2.25 Mb. This distance was longer compared to indica rice germplasm panels previously reported by Huang et al. (2012b); Mcnally et al. (2009), and Zhao et al. (2011), which ranged from 100 kb to 1 Mb. This is not surprising due to many historical recombination events of indica rice germplasm panels, which contributed to its rapid LD decay. Thus, varying patterns of LD likely reflect the breeding histories and the origins of the germplasm panel used (Flint-Garcia et al., 2003).
Genetic Background and Environment Effects on QTLs Detected
Two of the most important considerations in QTL detection studies are the genetic background effect and QTL-by-environment interaction (QEI), i.e., whether the reported QTLs are robust across different populations and environments. In the present study, the number of QTLs detected significantly varied between two MAGIC populations and across environments, suggesting that GWAS results were indeed highly influenced by both the population’s genetic background and testing environment. Out of the 18 QTLs identified, only 6 QTLs (qGL3.1, qGL3.2, qGW5, qGLWR3, qGLWR5, and qGT5) were consistently detected from the two populations under study. Interestingly, qGL3.1, qGL3.2, qGW5, and qGLWR3 were stably expressed across all the three environments. These QTLs are therefore important in marker-assisted breeding to improve grain size. qGW7, qGW8, qGLWR7, and qTGW3.2 were also stably expressed in all the three testing environments but were detected only in the BIM population. The QTLs for GT (qGT1, qGT3, and qGT5) and TGW (qTGW3.1, qTGW5, qTGW8.1, and qTGW8.2) were detected in only one of the testing environments, suggesting that these two traits were more sensitive to the environment leading to phenotypic plasticity. Phenotypic plasticity is a result of the interaction between QTLs and environment at the molecular level, and therefore, the genotype displays good trait performance only in a specific environment (Sultan, 2000). In this study, inconsistency of the QTLs detected could also be attributed to the threshold level used to declare significant MTAs. For instance, qGL7 and qGLWR8 were not identified in one of the testing environments (Henan 2017) because the p-values of SNPs were slightly higher (−log p = 1.31E-04 to 1.51E-04) than the applied critical threshold (−log p < 1.0E-04). In many QTL detection studies either through linkage or association mapping, inconsistency of the QTLs detected could be due to the type II error arising from the significant threshold level or to the true differential trait expression across environments.
BIM Has Higher Mapping Power and Resolution Than DC1
Nine QTLs (qGT1, qGT3, qTGW3.2, qGL7, qGW7, qGLWR7, qGW8, qGLWR8, and qTGW8.2) were identified only in the BIM population. This is at least partially due to the higher allelic and phenotypic diversity of the BIM population offered by the larger number of parental accessions used compared with the DC1 population. Hence, multiple parents will ensure that several QTLs for the trait of interest segregating within the population could be identified (Kover et al., 2009; Huang et al., 2015). Moreover, high recombination events in the BIM population implies higher genetic mapping resolution, either coarse mapping with low marker densities on early generation lines or fine mapping with high marker densities on late generation lines (Mackay and Powell, 2007). The effect of population size on the GWAS is also well known (Wang et al., 2012a). The larger size of the BIM population (Supplementary Table 2) also contributed to its higher mapping power and resolution. For a QTL to be effectively detected in a small population, it must be in high LD with the tested markers. Furthermore, increasing population size is necessary in detecting loci with low MAF since the power to detect association is a function of allele frequency (Myles et al., 2009). In our study, the size of DC1 population was small and the GL7 and GW8 genes were missed out, since the SNP markers in strong LD with these genes had low MAF and were removed prior to association analysis. Nevertheless, loci with large effects (qGL3.1, qGW5, qGLWR3, and qGLWR5) detected in the BIM population were also detected in the DC1 population. Therefore, if the MAF is high enough, major loci controlling the trait of interest would still be detected regardless of low population size.
Potential Candidate Genes Controlling Grain Size Were Identified
Two potential candidate genes, the ubiquitin-like protein 1 (ulp1) and OsCLP6, were identified for QTLs qGL3.1 and qGLWR3. The upl1 (LOC_Os03g29630) is likely to be involved in the ubiquitin-proteasome proteolytic pathway controlling grain size. Several ubiquitin-proteasome pathway-related genes were previously characterized and involved in ubiquitin-mediated control of seed size. The ubiquitin-proteasome pathway has been recently shown to play a crucial role in regulating seed size in different crops (Li and Li, 2016). In rice, the protein of unknown function encoded by GW5 has been suggested to be involved in the pathway through its interaction with polyubiquitin (Weng et al., 2008). The RING-type E3 ubiquitin ligase encoded by GW2 positively regulates GW and grain weight by restricting cell division (Song et al., 2007). HGW, which encodes a novel plant specific ubiquitin-associated (UBA) domain protein, functions as a key upstream regulator of GW (Li et al., 2012). The rice deubiquitinase gene OsOTUB1/WTG1 encodes an otubain-like protease and affects grain size (Huang et al., 2017; Liu et al., 2018). Recently, LG1 encoding for ubiquitin specific protease 15 (OsUBP15) was reported to function as a positive regulator of GW in rice (Shi et al., 2019). The other potential candidate gene, OsCLP6 (LOC_Os03g29810), encoding a putative chitinase-like protein (Clp) protease homolog was identified in both populations across all the three testing environments. Chitinases are classic pathogenesis-related proteins involved in plant growth and development regulation with the first genetic evidence observed in the Arabidopsis AtCTL1 gene (Graham and Sticklen, 1994; Zhong et al., 2002). In rice, secreted chitinase-like proteins (OsCLP) have been shown to play a pivotal role in root and shoot growth by regulating the intracellular calcium concentrations. Interestingly, Wu et al. (2017) reported that OsCLP negatively regulates rice GL with the seeds of CLP overexpression lines being shorter and rounder than those of the wild type and the osclp mutant.
Two genes, putative cytochrome P450 (Cyp/LOC_Os05g08850) and putative no apical meristem (NAC) gene (LOC_Os05g10620), were identified as potential candidate genes for the QTLs qGW5 and qGLWR5. Cytochrome proteins (CYPs) have been reported to play a crucial role in a variety of biosynthetic pathways including brassinosteroid (BR) signaling. In rice, CYP724B1 encoded by the D11 gene showed homology to enzymes involved in BR biosynthesis. Loss of function of D11 impaired BR biosynthesis and ultimately resulted to reduction in GL (Tanabe et al., 2005). BR-mediated grain size control in rice has been reported for several QTLs/genes including GS5 (Li et al., 2011), qGL3/GL3.1 (Qi et al., 2012; Zhang et al., 2012), GW5/qSW5 (Wan et al., 2008; Weng et al., 2008; Liu et al., 2017), GS2 (Che et al., 2015), and GS9 (Zhao et al., 2018). Defects in BR biosynthesis produce smaller seeds, but it is still uncertain how BRs promote seed growth (Hong et al., 2002, 2003). It is noteworthy that several CYP family members are involved in seed size control, including the Arabidopsis CYP78A5, CYP78A6, and CYP78A9 (Adamski et al., 2009; Fang et al., 2012), and soybean CYP78A72 and CYP78A10 (Wang et al., 2015b; Zhao et al., 2016), which all act as positive regulators of grain size, whereas the rice CYP78A13 and CYP704A3 (Xu et al., 2015a; Tang et al., 2016) act as negative regulators. NAC genes have been implicated in the control of seed size via the control of various stages of seed development (Agarwal et al., 2011). Three NAC genes, ONAC020 (LOC_Os01g01470), ONAC026 (LOC_Os01g29840), and ONAC023 (LOC_Os02g12310), were reported to be highly upregulated during seed development and were strongly associated with rice grain size from the analysis of sequence variations in the upstream regulatory region (Mathew et al., 2016).
Conclusion
Many grain size-related genes have been identified and cloned in the last decades; however, their molecular roles and their interaction between different signaling pathways are still fragmented. Therefore, identification and functional characterization of new genes are of significance. Using two MAGIC populations tested in three environments, the present study provided additional insight on the genetic architecture of rice grain size. Identification of QTLs with genetic background independency and environment stability is essential because grain size-related traits show phenotypic plasticity. The QTLs and the significant SNPs identified in this study, particularly those that exhibited genetic background independency and environment stability, will be useful for breeding indica rice to improve grain size. Moreover, the potential candidate genes reported are also important targets for future functional characterization studies to fill up the gaps and/or build up genetic framework of signaling pathways regulating grain size in rice. Methodologies such as gene editing and transferred DNA insertion mutant screens could be used to validate the effect of these genes and their functional variants. It is also noteworthy that populations derived from multiparent crosses provide more phenotypic and allelic diversity than the conventional biparental populations and have lesser confounding effects in terms of population structure and genetic relatedness than natural populations. The present study proved that the BIM population was generally better than the DC1 population.
Data Availability Statement
All datasets generated and analyzed for this study are included in the article/Supplementary Material.
Author Contributions
YL, GY, and LG designed the experiment. KP, YZ, and YL performed all the phenotypic evaluations. KP performed the data analysis and drafted the manuscript. YL and GY edited the manuscript. All authors approved the content of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Hei Leung (International Rice Research Institute) for providing the seeds and genotype data of the BIM lines; Pengfei Lu, Yaxin Huang (Jining Normal University), Junhui Zan, and Xiaofei Shi (Henan Agricultural University) for the assistance on phenotyping; and Jindong Liu and Jiaolong Li for helping with data analysis.
Footnotes
Funding. This research was supported by the National Natural Science Foundation of China (Grant No. 31801337), the China Postdoctoral Science Foundation (Grant No. 2018M631640), The Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (Grant No. CAAS-XTCX2016001), the CAAS Innovative Team Award to GY’s team, Shenzhen Science and Technology Projects (Grant No. JSGG20160608160725473), and Dapeng District Industry Development Special Funds (No. KY20180217).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00395/full#supplementary-material
References
- Adamski N. M., Anastasiou E., Eriksson S., O’Neill C. M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106 20115–20120. 10.1073/pnas.090702410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P., Kapoor S., Tyagi A. K. (2011). Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33 189–202. 10.1002/bies.201000107 [DOI] [PubMed] [Google Scholar]
- Bai X., Luo L., Yan W., Kovi M. R., Zhan W., Xing Y. (2010). Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11:16. 10.1186/1471-2156-11-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A. L., Lobina I. T., Dilla-Ermita C. J., et al. (2013). Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6:E11. 10.1186/1939-8433-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., Buckler E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M. E. (2006). Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetic 172 1165–1177. 10.1534/genetics.105.044586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell M. (2008). From mutations to MAGIC: resources for gene discovery validation and delivery in crop plants. Curr. Opin. Biol. 11 215–221. 10.1016/j.pbi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. 10.1038/nplants.2015.195 [DOI] [PubMed] [Google Scholar]
- Cleveland W. S. (1979). Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 74 829–836. 10.2307/2286407 [DOI] [Google Scholar]
- Descalsota G. I., Swamy M. B. P., Zaw H., Inabangan-Asilo M. A., Amparado A., Mauleon R., et al. (2018). Genome-wide association mapping in rice MAGIC plus population detects QTLs and Genes useful for biofortification. Front. Plant. Sci. 9:1347. 10.3389/fpls.2018.01347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. 10.1038/nplants.2015.203 [DOI] [PubMed] [Google Scholar]
- Duan P., Xu J., Zeng D., Zhang B., Geng M., Zhang G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10 685–694. 10.1016/j.molp.2017.03.009 [DOI] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112 1164–1171. 10.1007/s00122-006-0218-1 [DOI] [PubMed] [Google Scholar]
- Fang W., Wang Z., Cui R., Li J., Li Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70 929–939. 10.1111/j.1365-313X.2012.04907.x [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. A., McCouch S. R., Hall R. D. (2009). Not just a grain of rice: the quest for quality. Trends Plant Sci. 14 133–139. 10.1016/j.tplants.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Flint-Garcia S. A., Thornsberry J. M., Buckler E. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54 357–374. 10.1146/annurev.arplant.54.031902.134907 [DOI] [PubMed] [Google Scholar]
- Graham L. S., Sticklen M. B. (1994). Plant chitinases. Can. J. Bot. 72 1057–1083. 10.1139/b94-132 [DOI] [Google Scholar]
- Guo L., Ma H., Jiang H., Zheng D., Hu J., Wu L., et al. (2009). Genetic analysis and fine mapping of two genes for grain shape and weight in rice. J. Integr. Plant Biol. 51 45–51. 10.1111/j.1744-7909.2008.00793.x [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., Inukai Y., Fujioka S., Yukihisa S. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32 495–508. 10.1046/j.1365-313X.2002.01438.x [DOI] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 15 2900–2910. 10.1105/tpc.014712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant. 8 1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Huang B. E., George A. W., Forrest K. L., Kilian A., Hayden M. J., Morell M. K., et al. (2012a). A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10 826–839. 10.1111/j.1467-7652.2012.00702.x [DOI] [PubMed] [Google Scholar]
- Huang B. E., Verbyla K. L., Verbyla A. P., Raghavan C., Singh V. K., Gaur P., et al. (2015). MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet. 128 999–1017. 10.1007/s00122-015-2506-0 [DOI] [PubMed] [Google Scholar]
- Huang K., Wang D., Duan P., Zhang B., Xu R., Li N., et al. (2017). WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size in rice. Plant J. 91 849–860. 10.1111/tpj.13613 [DOI] [PubMed] [Google Scholar]
- Huang R., Jiang L., Zheng J., Wang T., Wang H., Huang Y., et al. (2013). Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18 218–226. 10.1016/j.tplants.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., et al. (2012b). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44 32–39. 10.1038/ng.1018 [DOI] [PubMed] [Google Scholar]
- Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., Onodera H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45 707–711. 10.1038/ng.2612 [DOI] [PubMed] [Google Scholar]
- Kato T., Segami S., Toriyama M., Kono I., Ando T., Yano M., et al. (2011). Detection of QTLs for grain length from large grain rice (Oryza sativa L.). Breed. Sci. 61 269–274. 10.1270/jsbbs.61.269 [DOI] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., Purugganan M. D., et al. (2009). A multiparent advanced generation inter-cross to fine-map quantitative traits in arabidopsis thaliana. PLoS Genet. 5:1–15. 10.1371/journal.pgen.1000551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chu H., Zhang Y., Mou T., Wu C., Zhang Q., et al. (2012). The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight. PLoS One 7:e34231. 10.1371/journal.pone.0034231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Thomsom M., McCouch S. R. (2004). Fine mapping of a grain-weight quantitative trait locus in the pericentromeric region of rice chromosome 3. Genetics 168 2187–2195. 10.1534/genetics.104.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Li Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33 23–32. 10.1016/j.pbi.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Lin L. H., Wu W. R. (2003). Mapping of QTLs underlying grain shape and grain weight in rice. Mol. Plant Breed. 1 337–342. [Google Scholar]
- Liu J., Chen J., Zheng X., Wu F., Lin Q., Heng Y., et al. (2017). GW5 acts in the brassinosteroid signaling pathway to regulate grain width and weight in rice. Nat. Plants 3:7043. 10.1038/nplants.2017.43 [DOI] [PubMed] [Google Scholar]
- Liu L., Tong H., Xiao Y., Che R., Xu F., Hu B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112 11102–11107. 10.1073/pnas.1512748112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Han R., Wu K., Zhang J., Ye Y., Wang S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Comm. 9:852. 10.1038/s41467-018-03047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Shao D., Qiu X., Sun L., Yan W., Zhou X., et al. (2013). Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 200 1269–1280. 10.1111/nph.12430 [DOI] [PubMed] [Google Scholar]
- Ma X., Feng F., Zhang Y., Elesawi I. E., Xu K., Li T., et al. (2019). A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 15:e1008191. 10.1371/journal.pgen.1008191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I., Powell W. (2007). Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 12 57–63. 10.1016/j.tplants.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Mathew I. E., Das S., Mahto A., Agarwal P. (2016). Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci. 7:1638. 10.3389/fpls.2016.01638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnally K. L., Childs K. L., Bohnert R., Davidson R. M., Zhao K., Ulat V. J., et al. (2009). Genome wide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. U.S.A. 106 12273–12278. 10.1073/pnas.0900992106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Wang B., Zhao X., Ponce K., Qian Q., Ye G. (2017). Association mapping of ferrous, zinc, and aluminum tolerance at the seedling stage in indica rice using MAGIC populations. Front. Plant Sci. 8:1822. 10.3389/fpls.2017.01822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Zhao X., Ponce K., Ye G., Leung H. (2016). QTL mapping for agronomic traits using multi-parent advanced generation inter-cross (MAGIC) populations derived from diverse elite indica rice lines. Field Crops Res. 189 19–42. 10.1016/j.fcr.2016.02.004 [DOI] [Google Scholar]
- Myles S., Peiffer J., Brown P. J., Ersoz E. S., Zhang Z., Costich D. E., et al. (2009). Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21 20194–22202. 10.1105/tpc.109.068437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Nonoue Y., Tsunematsu H., Kanno N., Yamamoto T., Yonemaru J. (2018). Discovery of QTL alleles for grain shape in the Japan-MAGIC rice population using haplotype information. G3-Genes Genom Genet 8 3559–3565. 10.1534/g3.118.200558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Balkunde S., Yang P., Yoon D. B., Ahn S. N. (2011). Fine mapping of grain weight QTL, tgw11 using near isogenic lines from a cross between Oryza sativa and O. grandiglumis. Genes Genom. 33 259–265. 10.1007/s13258-011-0038-9 [DOI] [Google Scholar]
- Pascual L., Desplat N., Huang B. E., Desgroux A., Bruguier L., Bouchet J. P., et al. (2015). Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13 565–577. 10.1111/pbi.12282 [DOI] [PubMed] [Google Scholar]
- Ponce K., Ye G., Zhao X. (2018). QTL identification for cooking and eating quality in indica rice using multi-parent advanced generation intercross (MAGIC) population. Front. Plant Sci. 9:868. 10.3389/fpls.2018.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan M. D., Fuller D. Q. (2009). The nature of selection during plant domestication. Nature 457 843–848. 10.1038/nature07895 [DOI] [PubMed] [Google Scholar]
- Qi P., Lin Y. S., Song X. J., Shen J. B., Huang W., Shan J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22 1666–1680. 10.1038/cr.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Pang Y., Yuan Z., Xing D., Xu J., Dingkuhn M., et al. (2015). Genome-wide association study of grain appearance and milling quality in a worldwide collection of indica rice germplasm. PLoS One 10:e0145577. 10.1371/journal.pone.0145577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Rong G., Tan Y., Yu S. (2012). Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 125 1717–1726. 10.1007/s00122-012-1948-x [DOI] [PubMed] [Google Scholar]
- Rui C. Q., Zhao A. C. (1983). Genetic analysis of weight and shape F1’s grain by diallel crossing methods in indica rice. Sci. Agric. Sin. 5 14–20. [Google Scholar]
- Sannemann W., Huang B. E., Mathew B., Leon J. (2015). Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 35 1–16. 10.1007/s11032-015-0284-7 [DOI] [Google Scholar]
- Shao G., Wei X., Chen M., Tang S., Luo J., Jiao G., et al. (2012). Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor. Appl. Genet. 125 1303–1312. 10.1007/s00122-012-1914-7 [DOI] [PubMed] [Google Scholar]
- Shi C., Ren Y., Liu L., Wang F., Zhang H., Tian P., et al. (2019). Ubiquitin specific protease 15 has an important role in regulating width and size in rice. Plant Physiol. 180 381–391. 10.1104/pp.19.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40 1023–1028. 10.1038/ng.169 [DOI] [PubMed] [Google Scholar]
- Si L., Chen J., Huang X., Gong H., Luo J., Huo Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48 447–457. 10.1038/ng.3518 [DOI] [PubMed] [Google Scholar]
- Singh R., Singh A. K., Sharma T. R., Singh A., Singh N. K. (2012). Fine mapping of grain length QTLs on chromosomes 1 and 7 in Basmati rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 21 157–166. 10.1007/s13562-011-0080-3 [DOI] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Sultan S. (2000). Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 5 537–542. 10.1016/j.tplants.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Tan Y. F., Xing Y. Z., Li J. X., Yu S. B., Xu C. G., Zhang Q. F. (2000). Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 101 823–829. 10.1007/s001220051549 [DOI] [PubMed] [Google Scholar]
- Tanabata T., Shibaya T., Hori K., Ebana K., Yano M. (2012). SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160 1871–1880. 10.1104/pp.112.205120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17 776–790. 10.1105/tpc.104.024950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wu T., Ye J., Sun J., Jiang Y., Yu J., et al. (2016). SNP-based analysis of genetic diversity reveals important alleles associated with seed size in rice. BMC Plant Biol. 16:93. 10.1186/s12870-016-0779-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. D. (2014). qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv [Preprint]. 10.1101/005165 [DOI] [Google Scholar]
- Wan X., Weng J., Zhai H., Wang J., Lei C., Liu X., et al. (2008). Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetic 179 2239–2252. 10.1534/genetics.108.089862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X. Y., Wan J. M., Jiang L., Wang K., Zhai H. Q., Weng F., et al. (2006). QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor. Appl. Genet. 112 1258–1270. 10.1007/s00122-006-0227-0 [DOI] [PubMed] [Google Scholar]
- Wang H., Smith K. P., Combs E., Blake T., Horsely R. D., Muehlbauer G. J. (2012a). Effect of population size and unbalanced data sets on QTL detection using genome-wide association mapping in barley breeding germplasm. Theor. Appl. Genet. 124 111–124. 10.1007/s00122-011-1691-8 [DOI] [PubMed] [Google Scholar]
- Wang S., Li S., Liu Q., Wu K., Zhang J., Wang Y., et al. (2015a). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47 949–954. 10.1038/ng.3352 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., et al. (2012b). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44 950–954. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- Wang X., Li Y., Zhang H., Sun G., Zhang W., Qiu L. (2015b). Evolution and association analysis of GmCYP78A10 gene with seed size/weight and pod number in soybean. Mol. Biol. Rep. 42 489–496. 10.1007/s11033-014-3792-3 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., et al. (2015c). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47 944–948. 10.1038/ng.3346 [DOI] [PubMed] [Google Scholar]
- Weng J., Gu S., Wan X., Gao H., Guo T., Su N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18 1199–1209. 10.1038/cr.2008.307 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wu J., Wang Y., Kim S. G., Jung K. H., Gupta R., Park Y., et al. (2017). A secreted chitinase-like protein (OsCLP) supports root growth through calcium signaling in Oryza sativa. Physiol. Plantarum. 161 273–284. 10.1111/ppl.12579 [DOI] [PubMed] [Google Scholar]
- Xie X. B., Jin F., Song M. H., Suh J. P., Hwang H. G., Kim Y. G., et al. (2008). Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa x O. rufipogon cross. Theor. Appl. Genet. 116 613–622. 10.1007/s00122-007-0695-x [DOI] [PubMed] [Google Scholar]
- Xie X. B., Song M. H., Jin F., Ahn S. N., Suh J. P., Hwang H. G., et al. (2006). Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor. Appl. Genet. 113 885–894. 10.1007/s00122-006-0348-5 [DOI] [PubMed] [Google Scholar]
- Xing Y., Tan Y., Hua J., Sun X., Xu C., Zhang Q. (2002). Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor. Appl. Genet. 105 248–257. 10.1007/s00122-002-0952-y [DOI] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular basis of rice yield. Annu. Rev. Plant Biol. 61 1–22. 10.1146/annurev-arplant-042809-112209 [DOI] [PubMed] [Google Scholar]
- Xu F., Fang J., Ou S., Gao S., Zhang F., Du L., et al. (2015a). Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 38 800–811. 10.1111/pce.12452 [DOI] [PubMed] [Google Scholar]
- Xu F., Sun X., Chen Y., Huang Y., Tong C., Bao J. (2015b). Rapid identification of major QTLs associated with rice grain weight and their utilization. PLoS One 10:e0122206. 10.1371/journal.pone.0122206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Yu H., Wang J., Duan P., Zhang B., Li J., et al. (2018). A mitogen-activated protein kinase phosphatase influences grain size and weight in rice. Plant J. 95 937–946. 10.1111/tpj.13971 [DOI] [PubMed] [Google Scholar]
- Ying J. Z., Ma M., Bai C., Huang X. H., Liu J. L., Fan Y. Y., et al. (2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11 750–753. 10.1016/j.molp.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Yu J., Miao J., Zhang Z., Xiong H., Zhu X., Sun X., et al. (2018). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotech. J. 16 1667–1678. 10.1111/pbi.12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W. H., Bi I. V., Yamasaki M., Doebley J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- Yu J., Xiong H., Zhu X., Zhang H., Li H., Miao J., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 15:28. 10.1186/s12915-017-0365-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Yang C., Fan Y., Zhuang J., Li X. (2008). Genetic dissection of a thousand-grain weight quantitative trait locus on rice chromosome 1. Chin. Sci. Bull. 53 2326–2332. 10.1007/s11434-008-0281-x [DOI] [Google Scholar]
- Zhang W., Sun P., He Q., Shu F., Wang J., Deng H. (2013). Fine mapping of GS2, a dominant gene for big grain rice. Crop J. 1 160–165. 10.1016/j.cj.2013.10.003 [DOI] [Google Scholar]
- Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109 21534–21539. 10.1073/pnas.1219776110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Dai A., Wei H., Yang S., Wang B., Jiang N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90 33–47. 10.1007/s11103-015-0392-0 [DOI] [PubMed] [Google Scholar]
- Zhao D. S., Li Q. F., Zhang C. Q., Zhang C., Yang Q. Q., Pan L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. 10.1038/s41467-018-03616-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C. W., Eizenga G. C., Wright M. H., Ali M. L., Price A. H., et al. (2011). Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2:E467. 10.1038/ncomms1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Kays S. J., Schroeder B. P., Ye Z. H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14 165–179. 10.1105/tpc.010278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Miao J., Gu H., Peng X., Leburu M., Yuan F., et al. (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201 1591–1599. 10.1534/genetics.11581115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated and analyzed for this study are included in the article/Supplementary Material.