Abstract
Bacterial uptake of charged organic pollutants such as the widely used herbicide glyphosate is typically attributed to active transporters, whereas passive membrane permeation as an uptake pathway is usually neglected. For 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes, the pH-dependent apparent membrane permeation coefficients (Papp) of glyphosate, determined by nuclear magnetic resonance (NMR) spectroscopy, varied from Papp (pH 7.0) = 3.7 (±0.3) × 10–7 m·s–1 to Papp (pH 4.1) = 4.2 (±0.1) × 10–6 m·s–1. The magnitude of this surprisingly rapid membrane permeation depended on glyphosate speciation and was, at circumneutral pH, in the range of polar, noncharged molecules. These findings point to passive membrane permeation as a potential uptake pathway during glyphosate biodegradation. To test this hypothesis, a Gram-negative glyphosate degrader, Ochrobactrum sp. FrEM, was isolated from glyphosate-treated soil and glyphosate permeation rates inferred from the liposome model system were compared to bacterial degradation rates. Estimated maximum permeation rates were, indeed, 2 orders of magnitude higher than degradation rates of glyphosate. In addition, biodegradation of millimolar glyphosate concentrations gave rise to pronounced carbon isotope fractionation with an apparent kinetic isotope effect, AKIEcarbon, of 1.014 ± 0.003. This value lies in the range typical of non-masked enzymatic isotope fractionation demonstrating that glyphosate biodegradation was not subject to mass transfer limitations and glyphosate exchange across the cell membrane was rapid relative to enzymatic turnover.
Introduction
Glyphosate (N-(phopshomethyl)glycine) is a nonselective, systemic, postemergent herbicide widely used in agriculture because of its ability to effectively control a broad range of weeds.1−3 One component of its success has been the introduction of transgenic, glyphosate-resistant crops.4,5 The worldwide market share of glyphosate is estimated at USD 5.6 billion, with the USGS estimating glyphosate use at more than 130 000 tons in 2015 alone in the USA.2,6,7 Historically, the acute toxicity of glyphosate has been considered low,3 it appears, however, that the impact of glyphosate on the environment has been underestimated.8−10 Most importantly, the ubiquitous use of glyphosate has been found to affect biodiversity,11 which is aggravated by increased usage due to the planting of glyphosate-resistant crops.12,13 The effect of glyphosate on human health is currently disputed. After the World Health Organization classified glyphosate as “probably carcinogenic” to humans (Group 2A),14 discussion has continued on whether or not glyphosate use poses a cancer risk.15,16 In addition, the detection of glyphosate and its metabolite aminomethylphosphonic acid (AMPA) in surface waters and groundwaters at increasing frequencies lends urgency to the need to more thoroughly explore its environmental fate.17−19 In particular, an improved understanding is warranted on the key drivers that limit its natural microbial degradation, because biodegradation represents the most effective glyphosate remediation pathway.20−23
A series of publications highlights the particular role of pollutant mass transfer into microbial cells as a rate-limiting step for biodegradation, especially at low pollutant concentrations.24,25 The mass transfer of polar and charged species (e.g., zwitterionic glyphosate26) into bacterial cells is currently assumed to occur by active transport.27,28 The passive permeation of charged organic molecules across cell membranes is less well understood than that of neutral nonpolar organic pollutants.29,30 The extent to which the diffusion barrier imposed by the bacterial membrane constitutes an even stronger bioavailability limitation for these charged molecules than for noncharged pollutants is poorly understood.31 Thus, it is important not only to investigate the membrane permeation rate but also to identify whether the rate of glyphosate is determined by the enzymatic reaction or by slow mass transfer of the herbicide across the cell envelope28,32 (where mass transfer can occur by either membrane permeation31 or active transport33).
To investigate membrane permeation processes, a variety of model systems have been used to study the diffusion of drugs and cosmetic ingredients through human epithelia, ranging from n-octanol–water partitioning to more complex systems such as lipid discs and planar lipid membranes.34,−38 However, these model systems are often composed of non-natural lipids, contain organic solvents, or present non-natural lipid–water interfaces. Therefore, membranes resembling biological lipid bilayers (e.g., liposomes with natural lipid composition) are currently the best model to approximate permeation rates valid for natural systems.39,40 Here, we used unilamellar liposomes composed of a single zwitterionic phospholipid, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), as a simplified system to investigate the permeation of glyphosate across phospholipid bilayers.41 The POPC vesicles have a gel-to-liquid crystalline phase transition temperature of −2 °C and, under our experimental condition, are in the liquid crystalline phase, resembling the dominant state of membranes of many bacteria.42,43 This model system lacked additional membrane constituents (e.g., anionic lipids, proteins, lipopolysaccharides) and cell envelope structures (phase-segregated domains in membranes, double membrane in Gram-negative bacteria, peptidoglycan cell wall) that are present in bacterial cells.44 We note that porins in the Gram-negative bacterial outer membrane permit passage of hydrophilic molecules with molecular masses ≲600 Da29 and the large pores of peptidoglycan do not restrict pollutant permeation.45
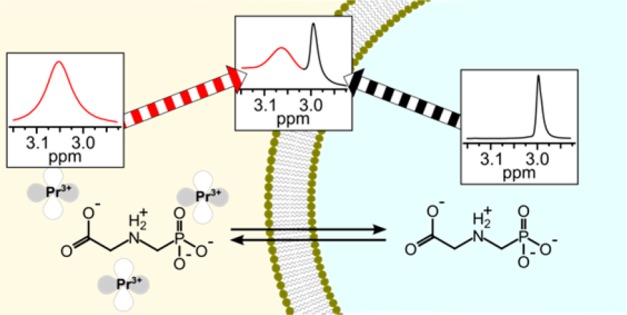
Permeation of the phospholipid bilayer leads to chemical exchange between the outside and the inside of the liposomes. Nuclear magnetic resonance (NMR) spectroscopy offers a direct approach to quantify the permeation process based on the following principle. A nucleus gives rise to an NMR signal at a chemical shift that reflects its chemical environment. Liposomes prepared and suspended in the same solution have roughly equivalent chemical environments inside and outside the liposome. Addition of a non-permeable chemical shift agent (such as a lanthanide ion) to the solution either interior or exterior to the liposome alters the chemical environment inside vis-à-vis outside the liposome (as the shift agent cannot cross the lipid bilayer) and results in distinct peaks in the NMR spectrum for the nucleus inside and outside the lipososmes.37,41,46 (In the present study, praseodymium(III) ions (Pr3+) were added to the solution external to the liposomes as a non-permeating chemical shift agent.) When the apparent exchange rate constant (ktr) between the two chemical environments (here, inside and outside the liposomes) is smaller than the observed frequency difference between the two states (Δυ), dynamic exchange of nuclei between chemical environments at equilibrium leads to line broadening.47 Lineshape analysis subsequently allows quantification of chemical exchange between the two environments based on the evaluation of associated line broadening in the NMR spectrum.47
Complementary to these model systems, we recently advanced compound-specific isotope analysis (CSIA) as an analytical approach to trace limitations of mass transfer across the cell envelope directly in vivo while pollutant biodegradation is ongoing.25,31 The underlying principle is the kinetic isotope effect of the associated enzymatic reaction. As the activation energy during a biochemical reaction is higher for bonds containing a heavy isotope, the turnover of molecules with a heavy isotope in the reactive position is slower. Therefore, as the enzymatic reaction proceeds, molecules containing heavy isotopes become enriched in the residual (nonreacted) substrate relative to those with light isotopes.48 This trend can be evaluated by considering the change in the isotope ratio Rt relative to the initial isotope ratio at the beginning of the degradation R0, and by relating it to the fraction of the remaining pollutant f according to the Rayleigh equation49,50 (1)
| 1 |
where the enrichment factor ε describes how much slower heavy isotopes react compared to light isotopes. Here, the carbon isotope values δ13Ct and δ13C0 at time t and at the beginning of a reaction, respectively, are expressed relative to an international reference material δ13C = (RSample – RReference)/RReference to ensure comparability among laboratories. Thullner et al. delineated a new angle to use the change in isotope ratios as a diagnostic tool to directly observe mass-transfer limitation: strong isotope fractionation is observable, only if the pollutant exchange across the cell envelope is faster than its enzymatic turnover.51 Otherwise, substrate molecules, which experience the isotopic discrimination during the enzymatic reaction in the cytosol, are used up completely so that they do not return to the bulk solution where the isotope ratio is assessed.51−53 As a consequence, the enzymatic isotope fractionation that is observable in solution becomes masked in the presence of mass transfer limitations: i.e., when active transport (or passive membrane permeation) into and out of the cell is the rate-determining step in biodegradation.25,33
For this study, we used a combined approach to gain insight into the role of passive permeation for biodegradation of the zwitterionic pollutant glyphosate, which exists as either a net monoanion (pH < 6) or a net dianion (pH > 6) at circumneutral pH. First, an NMR study was conducted to experimentally determine pH-dependent passive membrane permeation of glyphosate in phosphatidylcholine liposomes as a model system. Second, passive permeation rates were extrapolated and compared to biodegradation rates of different glyphosate degraders to elucidate the role of passive membrane permeation of glyphosate for nutrient uptake. To this end, Ochrobactrum sp. FrEM, a new glyphosate degrader, was isolated from a vineyard soil treated with glyphosate, characterized, and used for degradation experiments. The isotope fractionation measured during glyphosate biodegradation by Ochrobactrum sp. FrEM was explored as a diagnostic tool to directly observe the presence or absence of mass transfer limitations and, thus, to validate the assessment based on the results of the liposome model system and our theoretical considerations.
Experimental Section
Chemicals
A list of chemicals used can be found in the Supporting Information.
Liposome Preparation and Characterization
A 25 mg·mL–1 solution of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, transition temperature of −2 °C) in chloroform was prepared, and 50 mg of POPC (2 mL of the POPC solution) was added to a 3 mL screw cap glass vial tested prior to the experiment to withstand exposure to liquid nitrogen (see below). The chloroform was evaporated under a N2 stream, and the lipid film was dried with vacuum for at least 12 h. The dried lipids were hydrated with 1 mL of 20 mM glyphosate in D2O containing a small amount of 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropionic acid (TSP) as internal reference for NMR. The pH of the solution, ranging from pH 4.1 to pH 7.8, was adjusted prior to hydration with 1 M sodium hydroxide (in D2O). (Effective solvent suppression (vide infra) resulted in an insignificant increase in the HOD peak due to NaOH addition.) The vial was vortexed thoroughly; liposomes formed, and the suspension was subjected to three freeze–thaw cycles (freeze in liquid nitrogen for 5 min, thaw in 40 °C water bath for 5 min, and vortex for 30 s) followed by extrusion to yield unilamellar liposomes of uniform size distribution.54,55 The liposomes were extruded in 1000 μL syringes with 11 passages through a 0.2 μm polycarbonate filter with an Avanti Mini-Extruder (Avanti Polar Lipids, Inc., USA). The hydrodynamic size and the zeta potential of the vesicles were determined by dynamic light scattering and laser Doppler electrophoresis with a ZetaSizer Nano ZS (Malvern Instruments Ltd., United Kingdom) in dilutions of 2 μL of liposome solution in 800 μL of D2O. The temperature of the measurement cell was 25 °C. Ten measurements were averaged for each technical replicate (six replicates for dynamic light scattering and five replicates for laser Doppler electrophoresis).
Nuclear Magnetic Resonance Spectroscopy
All measurements were carried out on an Avance III 500 MHz spectrometer equipped with a BBFO+ smartprobe (Bruker, USA) at a sample temperature of 25 °C. NMR spectra were recorded with TopSpin 3.5.6 (Bruker, USA). Apodization, Fourier transformation, phase and baseline corrections, absolute referencing on TSP, spectral analysis, and peak fitting were carried out with MestReNova 11.0.3 (Mestrelab Research, Spain). Standard Bruker pulse sequences were used, and the spectra collection parameters are summarized in Table S1.
Assessing the Line Broadening Due to Chemical Exchange Across the Liposome Membrane
First, a standard 1H spectrum of 550 μL of glyphosate liposome solution was collected to assess the pH-dependent chemical shift of the HOD peak, and the chemical shift of the phosphorus nucleus was determined with proton decoupling (31P{1H}). Then, a proton spectrum with phosphorus decoupling 1H{31P} and solvent suppression was recorded. We added 5.5 μL of a 50 mM PrCl3 solution in D2O to the NMR tube to a final concentration of 0.5 mM PrCl3. Another 1H{31P} spectrum with solvent suppression was recorded, and the glyphosate peaks prior to and after PrCl3 addition were compared by fitting of the peaks. The strong 2JHP coupling of 12.4 Hz between the phosphorus and adjacent protons led to splitting of the peak at 2.99 ppm into a doublet in the 1H NMR spectrum (Figure 1). This doublet, however, complicated peak shape analysis to quantify the rate of glyphosate permeation across liposomes. In the absence of Pr3+, the shape of the separated doublet peaks could be fit. However, addition of Pr3+ led to line broadening due to chemical exchange between the inside and the outside of the liposomes. Thus, the individual peaks of the doublet signal overlapped with each other, rendering peak shape analysis unreliable. We therefore used a 1H NMR pulse sequence that combined solvent suppression (watergate W5) with phosphorus decoupling. As a result, the doublet peak collapsed to a well-resolved singlet that was distinguishable from the POPC liposome signals (Figure 2A).
Figure 1.
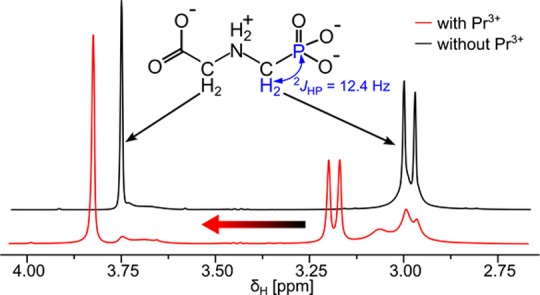
2JHP coupling prevents direct measurement of glyphosate permeation of liposomes with standard 1H-spectra. Glyphosate showed one singlet at 3.74 ppm and one doublet at 2.99 ppm in the 1H NMR spectrum with solvent suppression (black line). Strong 2JHP coupling led to formation of a doublet centered at 2.99 ppm. Upon Pr3+ addition to the liposome suspension, the spectrum changed (red line). The glyphosate peaks outside the liposomes were shifted downfield (doublet at 3.2 ppm and singlet at 3.79 ppm), and the peaks inside the liposomes broadened due to chemical exchange. As a consequence, the individual doublet peaks overlapped, almost coalescing into a singlet and rendering peak shape analysis impossible. Both spectra in this figure were collected at pH 7.5.
Figure 2.
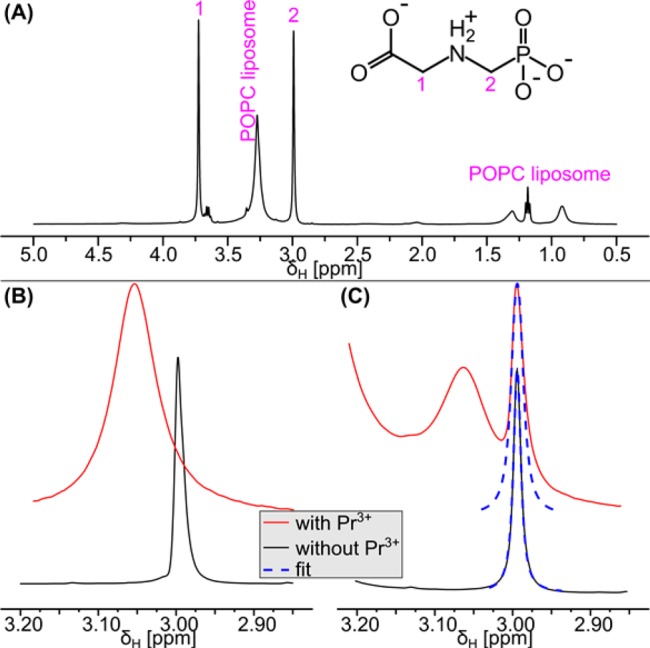
Line broadening due to exchange can be quantified by fitting the peaks in 1H{31P} NMR spectra. (A) Clear separation of the glyphosate signals from the signals of the POPC liposomes in the 1H{31P} NMR spectrum (black line) enabling reliable peak shape analysis. (B) Spectral region showing glyphosate protons attached to carbon 2 with (red line) and without (black line) Pr3+ in the absence of liposomes. The line broadening upon Pr3+ addition was caused by the interaction with the paramagnetic Pr3+. Even though the signal without Pr3+ slightly overlapped with the broad glyphosate signal in the presence of Pr3+, both peaks were well resolved. (C) The glyphosate peaks inside and outside the liposomes remained well resolved when POPC liposomes were present. Fitting the peak shape (blue dashed lines) prior to (black line) and after (red line) the addition of PrCl3 yielded peak widths and, thus, allowed the line broadening to be quantified. The broadening of the glyphosate peak inside the liposomes (2.99 ppm) was caused by chemical exchange of glyphosate between the inside and the outside of the liposomes, because the nonpermeable Pr3+ was not able to interact with glyphosate inside the liposomes. All spectra in this figure were collected at pH 7.5.
Bacterial Isolation and Characterization
For bacterial isolation from soil, mineral salt medium at pH 7.0 containing 60 mM sodium glutamate as carbon source and 38 mM ammonium chloride as nitrogen source was used. Glyphosate (3 mM) was the sole phosphorus source. A detailed description of the bacterial isolation from vineyard soil can be found in the Supporting Information.
Biodegradation of Glyphosate by Ochrobactrum sp. FrEM
The biodegradation of glyphosate by Ochrobactrum sp. FrEM was carried out in two biological replicates. We inoculated 50 mL of medium (see the Supporting Information) with O. sp. FrEM and incubated the culture at 30 °C at 160 rpm overnight. Cells were harvested by centrifugation (2100g, Heraeus Megafuge 1.0R, Germany), washed twice with medium, and transferred to 50 mL of fresh medium lacking phosphorus for phosphorus depletion. After incubation at 30 °C for 48 h, cells were harvested by centrifugation (2100g, Heraeus Megafuge 1.0R, Germany) and used to inoculate 150 mL of medium containing 120 μM glyphosate as the only phosphorus source. Bacterial growth was monitored at OD600 with a Cary 50 Bio UV–vis spectrometer (Varian Medical Systems, Inc., USA). During glyphosate biodegradation, samples for isotope analysis (10 mL) and the reaction was stopped by adding 1 mL of 2 M NaOH. The samples were lyophilized and reconstituted in water. The water volume for reconstitution was decreased from 5 to 2 mL as glyphosate was consumed for glyphosate preconcentration to be within the working range of the isotope measurements (see below). The isotope ratio (expressed using the delta notation δ13C in per mille (‰) relative to Vienna PeeDee Belemnite (V-PDB)) and the concentration of glyphosate were determined by liquid chromatography Isolink-isotope ratio mass spectrometry (LC-IRMS) (Thermo Fisher, Germany). The method used for carbon isotope analysis of glyphosate was modified from Kujawinski et al.56 as follows: A mixed-phase Primesep 100 column 100 × 5.6 mm, 3 μm particle size (SIELC Technologies, USA) was used as stationary phase, and 2.5 mM phosphate buffer at pH 3.1 was used as mobile phase. Separation was achieved with 300 μL·min–1 isocratic flow. The injection volume was 25 μL. The reagents for chemical conversion to CO2 at 99.9 °C were 1.5 M phosphoric acid and 0.84 M peroxodisulfate at a flow rate of 50 μL·min–1 each. The helium (grade 5.0) flow rate in the separation unit was set to 2.3 mL·min–1. The glyphosate concentration was determined with the area of the glyphosate CO2 peak in the LC Isolink-IRMS chromatogram via external calibration with glyphosate standards in water (0.03, 0.06, 0.12, and 0.30 mM).
Results and Discussion
Praseodymium(III) Ions Interact with Glyphosate As Well As the Liposome Surface
The liposome were of a uniform size with a hydrodynamic diameter of 204 ± 5 nm (median ± standard deviation, range: 194 nm to 239 nm). The median polydispersity index was 0.093 indicating a uniform and narrow size distribution of the individual liposome preparations. The near zero zeta potential of the liposomes composed of lipids bearing zwitterionic phosphatidylcholine headgroups57 changed to +29 ± 6 mV upon PrCl3 addition, because the strongly positively charged Pr3+ associated with the negatively charged phosphate group of POPC. Praseodymium(III) was added to produce a chemical environment outside the liposomes differing from that inside to allow glyphosate exchange across the membrane to be quantified. Indeed, addition of Pr3+ resulted in an interaction of glyphosate with the chemical shift agent which led to a position-specific downfield shift (ΔδΗ) of the glyphosate 1H NMR signals. The chemical shift change produced by a 1 mM PrCl3 solution was ΔδΗ = 0.06 ppm for the PO32––CH2–NH2+–CH2–COO– protons and ΔδΗ = 0.16 ppm for the PO32––CH2–NH2+–CH2–COO– protons in the 1H NMR spectrum of glyphosate (Figure S1). The phosphorus peak was shifted downfield by ΔδP = 1.29 ppm in the 31P{1H} spectrum (Figure S1). The chemical shift change was strongest for the phosphorus peak and weakest for the protons adjacent to the carboxyl group which indicated that Pr3+ directly interacted with the negatively charged phosphate group and not with the negatively charged carboxyl group of the zwitterionic glyphosate.
Addition of Praseodymium(III) and Subsequent Peak Shape Analysis Quantified Chemical Exchange of Glyphosate
Subsequent addition of 0.5 mM Pr3+ to a glyphosate solution without liposomes moved the chemical shift of the collapsed singlet of the PO32––CH2–NH2+–CH2–COO– protons downfield from 2.99 to 3.06 ppm. Interaction with the paramagnetic Pr3+ changes the local magnetic field, leading to the shift in frequency; the association/dissociation of Pr3+ and glyphosate produced line broadening (Figure 2B). We observed a similarly strong chemical shift change when adding Pr3+ to a glyphosate solution containing liposomes (Figure 2C). While the interaction of nonpermeable Pr3+ with glyphosate outside the liposomes influenced the chemical shift, the shift agent could not enter the liposomes as demonstrated by the unchanged chemical shift for glyphosate inside the liposomes. As a consequence, two distinct peaks appeared in the spectrum, and the glyphosate peak outside the liposomes was well-resolved from the peak inside. This indicated that the exchange was slow on the NMR time scale; that is, the ratio ktr/ΔδΗ is smaller than one (ktr/ΔδΗ < 1), where ktr is the apparent rate constant of exchange and ΔδΗ is the chemical shift difference in 1H NMR.47 The glyphosate exchange across the liposome bilayer was fast enough, however, to lead to considerable line broadening, Δυ, of the peak corresponding to glyphosate inside the liposomes. The line broadening (Δυ) depends on the apparent rate constant of exchange according to eq 2(37) and ranged from Δυ = 2.6 Hz at neutral pH to Δυ = 40.8 Hz at pH 4.
| 2 |
The glyphosate peaks inside the liposomes were fitted to determine the peak width prior to (υ0) and after addition of PrCl3 (υex). The resultant line broadening Δυ = υex – υ0 (Figure 2C) was used to calculate ktr for each liposome preparation.
Glyphosate Permeation of Lipid Bilayers Depends Strongly on pH
Because ktr strongly depends on the surface area and on the size of the liposomes, ktr is not suitable to compare the chemical exchange of different liposome preparations and at different pH values. Therefore, Males and Herring derived the apparent permeation coefficient, Papp [m·s–1], by including the inner liposome volume and the volume-to-surface ratio according to eq 3,37 where dlip is the diameter of the liposome and δ is the membrane thickness (4 nm); note that this membrane thickness δ should not be confused with the chemical shift δΗ (or δP) in the NMR spectrum or the δ13C value. Our NMR approach is not subject to unstirred water layer effects (i.e., the permeation coefficient so determined reflects primarily diffusion across the lipid bilayer alone). The NMR experiments measured a dynamic exchange process under equilibrium conditions. The ktr does, however, include a contribution from glyphosate diffusing a short distance through water to arrive at the lipid bilayer surface. We therefore refer to our rate constant of exchange as an apparent value and the permeation coefficient derived from it as an apparent permeation coefficient, Papp.
| 3 |
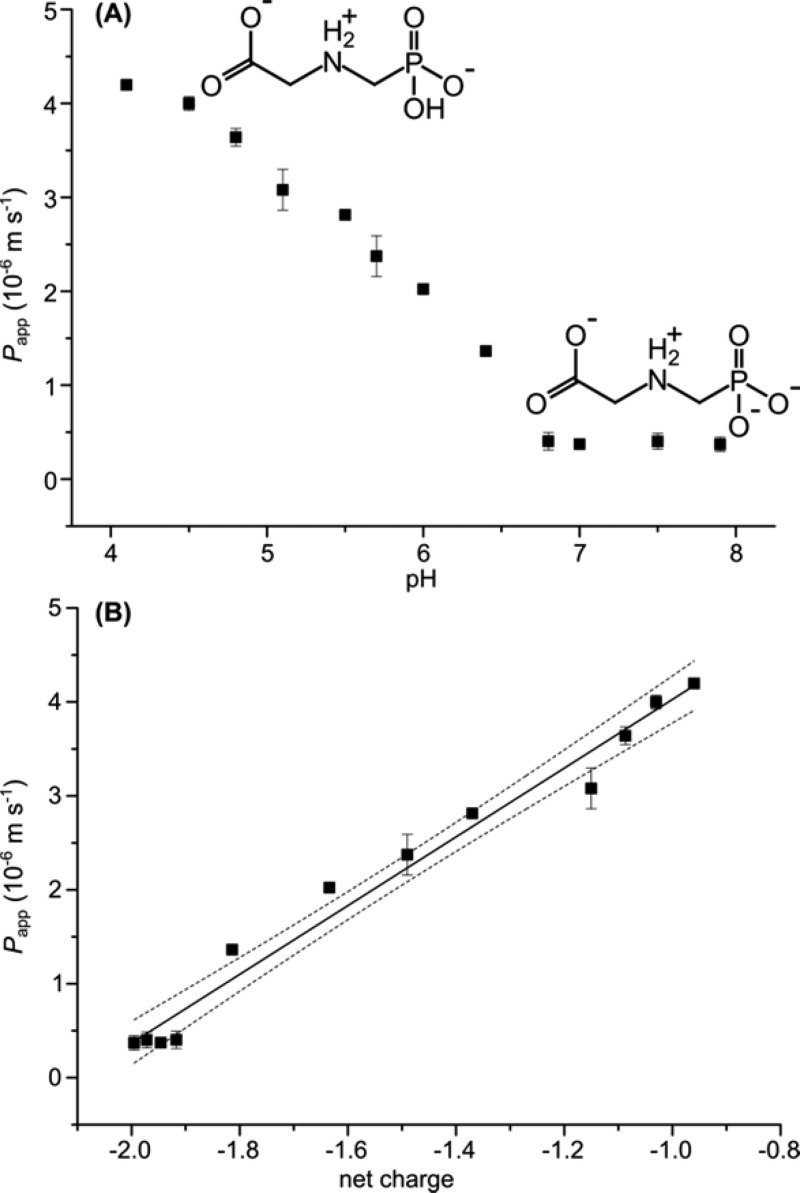
The permeation coefficient describes how rapidly glyphosate permeates a hypothetical two-dimensional POPC membrane sheet and was much higher than expected (Figure 3A). At circumneutral pH, the apparent permeation coefficient of glyphosate (double negatively charged, molecular weight (MW) = 167 g·mol–1) Papp (pH 7.0) = 3.7 (±0.3) × 10–7 m·s–1 was considerably higher than that of maleate46 (net dianion, MW = 114 g·mol–1) and in the same range as the permeation coefficient of the polar, neutral serotonin species (MW = 176 g·mol–1).58 With decreasing pH, the permeation rate increased, with an apparent permeation coefficient of Papp(pH 4.1) = 4.2 (±0.1) × 10–6 m·s–1 at pH 4.1. The pH dependence correlated linearly with the average degree of ionization and thus the average charge of glyphosate (Figure 3B). The net charge of −2 (one positive and three negative charges) of glyphosate at neutral pH slowed passive membrane permeation. Protonation of the phosphate group at pH 4.1 reduced the net charge of glyphosate to −1 and, consequently, accelerated membrane permeation. This also indicates that the effect of Pr3+ on the measured permeation coefficient is negligible. If the change toward positive liposome surface potential due to Pr3+ addition (vide supra) facilitated permeation due to attraction of the strongly negatively charged glyphosate, the doubly negatively charged glyphosate species would permeate faster. However, the opposite is the case. Furthermore, two distinct glyphosate peaks appear in the NMR spectrum (vide supra), demonstrating that Pr3+ does not enter the liposomes together with glyphosate, which could lead to changed permeation characteristics. Therefore, we hypothesize that the zwitterionic structure of glyphosate facilitates glyphosate permeation, whereas the increased negative charge at neutral pH slowed passive permeation of glyphosate.
Figure 3.
The pH dependence of the permeation coefficient Papp (black squares) correlated with the net charge of glyphosate. (A) Papp depended strongly on the pH of the liposome solution. The permeation at neutral pH (Papp (pH 7.0) = 3.7 (±0.3) × 10–7 m·s–1) was 1 order of magnitude lower than at slightly acidic pH (Papp (pH 4.1) = 4.2 (±0.1) × 10–6 m·s–1). (B) Permeation correlated with the ionization of glyphosate which can be explained by species-specific permeation coefficients for the net monoanion and net dianion. Both panels show the mean values with error bars denoting one standard deviation.
Membrane Permeation Can Lead to Considerable Glyphosate Uptake into Bacterial Cells
The entry of non-polar pollutants into bacterial cells by passive permeation of the cell envelope is well recognized,30,59 and charged, polar molecules like glyphosate are commonly assumed to be taken up almost exclusively by active transport or porin-assisted permeation.60,61 Contrary to this expectation, in a liposome model system that lacked transporters or porins we observed membrane permeation coefficients for glyphosate in the same range as those for noncharged molecules (see above).58 This observation suggests that passive membrane permeation of glyphosate mono- and dianions may provide sufficient influx into bacterial cells for the herbicide to serve as a phosphorus source. Glyphosate diffusion through water is fast compared to that through the lipid membrane.62,63 The membrane therefore represents a significant diffusive barrier influences the rate the amount of substrate outside the bacteria is reduced via passive membrane permeation at the rate (dnout/dt)permeation, where nout denotes the number of moles of glyphosate outside the bacterial cell. This process is driven by the concentration gradient across the membrane and is defined by the linear exchange term in eq 4 as proposed by Males et al.64
| 4 |
Here, Klip-w is the membrane lipid–water partition coefficient, and [Sout] and [Sin] are the glyphosate concentrations outside and inside the bacterial cell. The products Klip-w[Sout] and Klip-w[Sin] are the concentrations within the lipid membrane (outside and inside), respectively. With the definition of the diffusion coefficient across the membrane (lipid bilayer) Dlip (eq 5), the apparent rate constant of exchange (ktr) can be calculated for a single bacterial cell by eq 6
| 5 |
| 6 |
where Acell ≈ 3 μm2 is the estimated surface area of one bacterial cell and δ is the membrane thickness (one 4 nm membrane in Gram-positive and two 4 nm thick membranes in Gram-negative bacteria). Together with eq 7, a term is obtained for the concentration gradient-dependent glyphosate influx into a single bacterial cell.
| 7 |
The glyphosate influx is at its maximum (dnout/dt)cell-permeation-max when the concentration gradient is large ([Sin] = 0). We compared this maximum permeation rate with the glyphosate degradation rate of Achromobacter sp. MPS 12A described by Sviridov et al.22 The glyphosate degradation rate of a single Achromobacter sp. MPS 12A cell (dn/dt)deg-cell = −1.4 × 10–21 mol·s–1·cell–1 at a concentration of 3 mM22 was estimated by correlating the number of cells with the optical density OD600 and the bulk glyphosate degradation rate. While this correlation strongly depends on the strain and the growth conditions, the previously reported value of 8 × 108 cells·mL–1·OD600–1 provides a good first estimate.65,66 The comparison showed that the calculated maximum membrane permeation rate at pH 7 (dnout/dt)cell-perm-max = −1.9 × 10–18 mol·s–1·cell–1 was 2 orders of magnitude higher than the degradation rate. As a consequence, even though glyphosate has a net charge of −2 at pH 7, its passive membrane permeation is predicted to be fast enough to sustain bacterial transformation at the maximum degradation rate. This hypothesis clearly warrants further testing. If true, confirmation should be possible by (a) the observation of similarly rapid biodegradation per cell in a different strain and (b) applying compound-specific isotope fractionation as a diagnostic tool to observe the absence of mass transfer limitations directly. If permeation is indeed faster than enzymatic conversion, glyphosate molecules inside and outside the cell are expected to be in rapid equilibrium. Thus, glyphosate molecules enriched in heavy isotopes due to the enzymatic reaction in the cytosol will pass out of the cell into the bulk solution. This would lead to the isotope effect of the enzyme reaction being observable outside the cell, resulting in strong isotope fractionation during biodegradation. A new bacterium was, therefore, isolated from soil, and isotope fractionation was measured during glyphosate degradation.
Isolation and Glyphosate Degradation Activity of Orchrobactrum sp. FrEM
Repeated subculturing of an inoculum from soil samples in a medium containing 3 mM glyphosate as the sole phosphorus source resulted in the isolation of a bacterial strain with glyphosate-degrading activity. Glyphosate was used only as phosphorous source, consistent with Shushkova et al. how faced difficulties when using glyphosate as the sole carbon and phosphorus source for isolation.67 The bacteria were rod-shaped as observed by light microscopy (Figure S2). Sequence alignment (BLAST) of the 16S rRNA showed a 99% homology with Ochrobactrum anthropic, O. rhizosphaerae, O. pituitosum, and O. intermedium, which all belong to the family of Brucellaceae in Alphaproteobacteria, and 70% homology with Ochrobactrum hematophilum. The strain was termed Ochrobactrum sp. FrEM (Figure S3). The fastest glyphosate degradation by O. sp. FrEM occurred after 4.5 days when the cell density was high (OD600 ≈ 0.8). Within 12 h, the glyphosate concentration decreased from 104 to 55 mM equaling a maximum glyphosate degradation rate (dn/dt)deg-cell = −1.7 × 10–21 mol·s–1·cell–1 (Figure 4A) which was as high as that of Achromobacter sp. MPS 12A (see above).22 Furthermore, just as for Achromobacter sp. MPS 12A, the calculated maximum membrane permeation rate at pH 7 (dnout/dt)cell-perm-max = −7.5 × 10–20 mol·s–1·cell–1 at a concentration of 0.12 mM was larger than the degradation rate indicating that passive permeation of the cell envelope is likely not rate limiting for glyphosate biodegradation. We subsequently aimed to verify this hypothesis by compound-specific isotope fractionation analysis.
Figure 4.
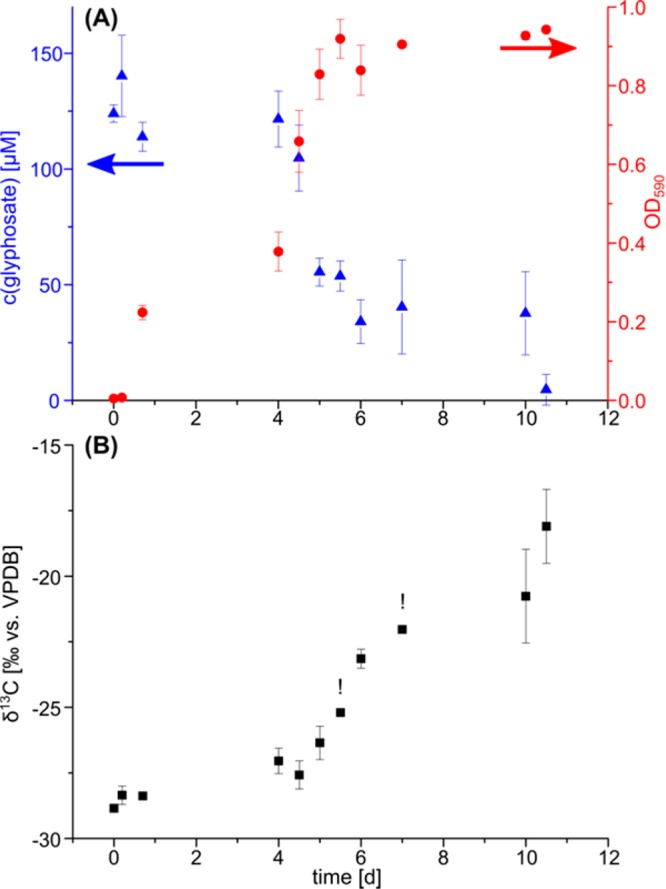
Glyphosate biodegradation was accompanied by growth and strong isotope fractionation. (A) Glyphosate degradation by Ochrobactrum sp. FrEM. Consumption of glyphosate (blue triangles) as source of phosphorus led to bacterial growth (red circles). (B) During this biodegradation, 13C/12C ratios of glyphosate increased, as indicated by less negative δ13C values. All graphs show the mean and the error bars indicating the range of two biological replicates. The exclamation marks (!) above two isotope data points indicate that a reliable isotope value could be measured for only one biological replicate at the respective time points. Isotope values were measured in technical triplicates for each sample and are associated with an analytical uncertainty of ±0.3‰.
Carbon Isotope Fractionation Revealed Rapid Glyphosate Mass Transfer Across the Cell Wall
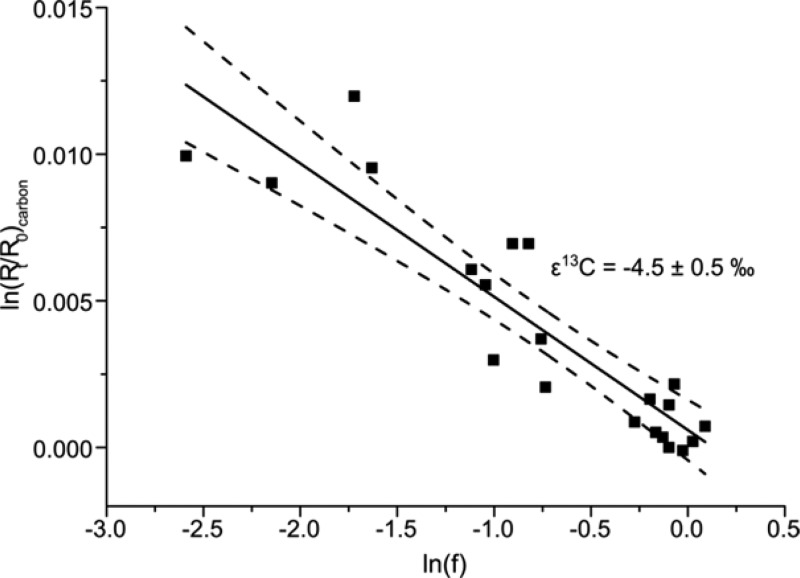
Glyphosate biodegradation by O. sp. FrEM (Figure 4A) was accompanied by significant carbon isotope fractionation. The δ13C values of glyphosate increased from −28 (±0.5)‰ in the beginning to −19 (±0.5)‰ after 90% glyphosate conversion reflecting an enrichment of 13C over 12C. The corresponding enrichment factor ε13C = −4.5 (±0.5)‰ was determined according to the Rayleigh equation (Figure 5 and eq 1). The primary apparent kinetic isotope effect, AKIE, a measure for the isotope effect at the reactive position, allows the direct comparison of isotope effects of different reactions and reactants and was calculated according to eq 8(68)
| 8 |
where n denotes the total number of carbon atoms and x is the number of carbon atoms at the reactive position. With n = 3 and x = 1, the primary apparent kinetic isotope effect for glyphosate degradation (via breaking a single bond between carbon and phosphorus) was AKIEcarbon = 1.014 ± 0.003, which is in the range of chemical reactions that involve breaking a single bond to a carbon atom.68,69
Figure 5.
Pronounced isotope fractionation indicated rapid glyphosate exchange across the bacterial cell envelope. The carbon isotope fractionation factor (ε13C = −4.5 (±0.5)‰) was determined according to the Rayleigh equation (eq 1). The corresponding AKIEcarbon = 1.014 ± 0.003 (see eq 8) was in the range of typical carbon isotope effects. This indicated that the enzymatic isotope fractionation was not masked by mass transfer limitations and that exchange of glyphosate across the cell envelope was comparatively rapid during bacterial degradation by Ochrobactrum sp. FrEM.
This suggests that if any additional rate-determining steps such as active transport33 or slow passive membrane permeation31 masked the enzymatic isotope fractionation, such an effect was small. As a consequence, we conclude that, indeed, glyphosate exchanged rapidly across the cell envelope consistent with our hypothesis that passive permeation across the cell envelope may be an important, and until now underestimated, driver to facilitate biodegradation of glyphosate or other charged pollutants (C, N, and P sources). Future research should not only address the role of pH in the permeation of whole cells during biodegradation but also elucidate the possible role of transporters or porins (e.g., by studying isotope fractionation during glyphosate degradation in cell-free extracts of O. sp. FrEM or with liposomes containing the degrading enzyme).
Possibility of Mass Transfer Limitations at Low Concentrations in the Environment
While passive membrane permeation has previously been associated with nonpolar molecules, our results suggest that in addition to protein-assisted uptake (e.g., by active transporters70,71 or assisted diffusion via porins29,72), charged species like glyphosate can enter bacterial cells by passive permeation of the cell membrane and that this can occur more rapidly than commonly thought. This can facilitate glyphosate biodegradation and lead to rapid turnover at high concentrations in water and soil.10 A different situation must be considered, however, if the concentration gradient across the cell envelope is shallower, that is, than in our experiments. While the degradation-associated isotope fractionation was determined at high concentrations (>25 μM), the concentration in soil, groundwaters, and surface waters is much lower (<2 μM, <15 nM, and <0.5 μM, respectively).18,73 We recently demonstrated that mass transfer across the cell membrane becomes rate-limiting for atrazine biodegradation at trace concentrations.25 Similarly, at a glyphosate concentration of 1 μM, the calculated maximum membrane permeation rate is only (dnout/dt)cell-perm-max = −6.5 × 10–22 mol·s–1·cell–1, which is lower than the respective degradation rate per cell. At these concentrations, acceleration of cell wall transfer of glyphosate with high affinity active transporters may become necessary to boost biodegradation. Interestingly, Pipke et al. described such an active glyphosate transporter with an uptake rate of (dnout/dt)cell-transport = −1.8 × 10–21 mol·s–1·cell–1 which is just in the range of observed glyphosate degradation rates.74 However, its affinity constant KM = 0.125 mM for glyphosate uptake is rather high, resulting in low transporter activity at trace concentrations. This increased mass transfer limitation at trace concentrations may cause biodegradation to stall and might explain the frequent detection of glyphosate in the environment.17
Acknowledgments
This work was funded by an ERC consolidator grant (“MicroDegrade”, grant no. 616861) awarded by the European Research Council to M.E. The NMR instrumentation was supported by a generous gift from Paul J. and Margaret M. Bender to the Magnetic Resonance Facility in the Chemistry Department of the University of Wisconsin-Madison. J.A.P. gratefully acknowledges the support of the William A. Rothermel Bascom Professorship.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.8b01004.
A list of chemicals, media composition, strain isolation procedure, figures of the glyphosate spectra, micrograph of Ochrobactrum sp. FrEM, phylogenetic tree, LC-IRMS chromatogram, and a table summarizing spectra collection parameters (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Franz J. E.; Mao M. K.; Sikorski J. A.. Glyphosate: a unique global herbicide; American Chemical Society: Washington, DC, 1997. [Google Scholar]
- Dill G. M.; Sammons R. D.; Feng P. C. C.; Kohn F.; Kretzmer K.; Mehrsheikh A.; Bleeke M.; Honegger J. L.; Farmer D.; Wright D.; Haupfear E. A.. Glyphosate: discovery, development, applications, and properties. In Glyphosate Resistance in Crops and Weeds; John Wiley & Sons, Inc.: New York, 2010; pp 1–33. [Google Scholar]
- Duke S. O.; Powles S. B. Glyphosate: a once-in-a-century herbicide. Pest Manage. Sci. 2008, 64 (4), 319–325. 10.1002/ps.1518. [DOI] [PubMed] [Google Scholar]
- Dill G. M. Glyphosate-resistant crops: history, status and future. Pest Manage. Sci. 2005, 61 (3), 219–224. 10.1002/ps.1008. [DOI] [PubMed] [Google Scholar]
- Bøhn T.; Cuhra M.; Traavik T.; Sanden M.; Fagan J.; Primicerio R. Compositional differences in soybeans on the market: Glyphosate accumulates in roundup ready GM soybeans. Food Chem. 2014, 153, 207–215. 10.1016/j.foodchem.2013.12.054. [DOI] [PubMed] [Google Scholar]
- Woodburn A. T. Glyphosate: Production, pricing and use worldwide. Pest Manage. Sci. 2000, 56 (4), 309–312. . [DOI] [Google Scholar]
- USGS. Estimated Annual Agricultural Pesticide Use; https://water.usgs.gov/nawqa/pnsp/usage/maps/show_map.php?year=2015&map=GLYPHOSATE&hilo=H&disp=Glyphosate (accessed 05.04.2018).
- Helander M.; Saloniemi I.; Saikkonen K. Glyphosate in northern ecosystems. Trends Plant Sci. 2012, 17 (10), 569–574. 10.1016/j.tplants.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Relyea R. A. The lethal impact of roundup on aquatic and terrestrial amphibians. Ecological Applications 2005, 15 (4), 1118–1124. 10.1890/04-1291. [DOI] [Google Scholar]
- Giesy J. P.; Dobson S.; Solomon K. R.. Ecotoxicological risk assessment for roundup herbicide. In Reviews of Environmental Contamination and Toxicology: Continuation of Residue Reviews; Ware G. W., Ed.; Springer New York: New York, NY, 2000; pp 35–120. [Google Scholar]
- Sullivan T. P.; Sullivan D. S. Vegetation management and ecosystem disturbance: Impact of glyphosate herbicide on plant and animal diversity in terrestrial systems. Environ. Rev. 2003, 11 (1), 37–59. 10.1139/a03-005. [DOI] [Google Scholar]
- Mamy L.; Gabrielle B.; Barriuso E. Comparative environmental impacts of glyphosate and conventional herbicides when used with glyphosate-tolerant and non-tolerant crops. Environ. Pollut. 2010, 158 (10), 3172–3178. 10.1016/j.envpol.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Firbank L. G.; Forcella F. Genetically modified crops and farmland biodiversity. Science 2000, 289 (5484), 1481–1482. 10.1126/science.289.5484.1481. [DOI] [PubMed] [Google Scholar]
- WHO . IARC Monographs: Evaluation of Five Organophosphate Insecticides and Herbicides; WHO: Geneva, 2015; Vol. 112. [Google Scholar]
- Chang E. T.; Delzell E. Systematic review and meta-analysis of glyphosate exposure and risk of lymphohematopoietic cancers. J. Environ. Sci. Health, Part B 2016, 51 (6), 402–434. 10.1080/03601234.2016.1142748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti G.; Koutros S.; Hofmann J. N.; Sandler D. P.; Lubin J. H.; Lynch C. F.; Lerro C. C.; De Roos A. J.; Parks C. G.; Alavanja M. C.; Silverman D. T.; Beane Freeman L. E. Glyphosate use and cancer incidence in the agricultural health study. JNCI: Journal of the National Cancer Institute 2018, 110, 509. 10.1093/jnci/djx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamy L.; Barriuso E.; Gabrielle B. Environmental fate of herbicides trifluralin, metazachlor, metamitron and sulcotrione compared with that of glyphosate, a substitute broad spectrum herbicide for different glyphosate-resistant crops. Pest Manage. Sci. 2005, 61 (9), 905–916. 10.1002/ps.1108. [DOI] [PubMed] [Google Scholar]
- Battaglin W. A.; Meyer M. T.; Kuivila K. M.; Dietze J. E. Glyphosate and its degradation product ampa occur frequently and widely in u.s. Soils, surface water, groundwater, and precipitation. J. Am. Water Resour. Assoc. 2014, 50 (2), 275–290. 10.1111/jawr.12159. [DOI] [Google Scholar]
- Horth H.; Blackmore K.. Survey of Glyphosate and Ampa in Groundwaters and Surface Waters in Europe; No.: UC8073; WRc plc: Swindon, Wiltshire, United Kingdom, 2009; Vol. 2. [Google Scholar]
- Ermakova I.; Kiseleva N.; Shushkova T.; Zharikov M.; Zharikov G.; Leontievsky A. Bioremediation of glyphosate-contaminated soils. Appl. Microbiol. Biotechnol. 2010, 88 (2), 585–594. 10.1007/s00253-010-2775-0. [DOI] [PubMed] [Google Scholar]
- Pipke R.; Amrhein N. Degradation of the phosphonate herbicide glyphosate by Arthrobacter atrocyaneus ATCC 13752. Appl. Environ. Microbiol. 1988, 54 (5), 1293–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviridov A. V.; Shushkova T. V.; Zelenkova N. F.; Vinokurova N. G.; Morgunov I. G.; Ermakova I. T.; Leontievsky A. A. Distribution of glyphosate and methylphosphonate catabolism systems in soil bacteria Ochrobactrum anthropi and Achromobacter sp. Appl. Microbiol. Biotechnol. 2012, 93 (2), 787–96. 10.1007/s00253-011-3485-y. [DOI] [PubMed] [Google Scholar]
- Borggaard O. K.; Gimsing A. L. Fate of glyphosate in soil and the possibility of leaching to ground and surface waters: A review. Pest Manage. Sci. 2008, 64 (4), 441–456. 10.1002/ps.1512. [DOI] [PubMed] [Google Scholar]
- Bosma T. N. P.; Middeldorp P. J. M.; Schraa G.; Zehnder A. J. B. Mass transfer limitation of biotransformation: Quantifying bioavailability. Environ. Sci. Technol. 1997, 31 (1), 248–252. 10.1021/es960383u. [DOI] [Google Scholar]
- Ehrl B. N.; Kundu K.; Gharasoo M.; Marozava S.; Elsner M.. Rate limiting mass transfer in micropollutant degradation revealed by isotope fractionation. Environ. Sci. Technol., submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprankle P.; Meggitt W.; Penner D. Adsorption, mobility, and microbial degradation of glyphosate in the soil. Weed Science 1975, 23, 229–234. [Google Scholar]
- Button D. K. Kinetics of nutrient-limited transport and microbial growth. Microbiol. Rev. 1985, 49, 270–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T.; Robert K. P.. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. In Advances in Microbial Physiology; Academic Press: New York, 2008; Vol. 53, pp 169–315. [DOI] [PubMed] [Google Scholar]
- Delcour A. H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta, Proteins Proteomics 2009, 1794 (5), 808–816. 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales R. E.; Ditty J. L.. Substrate transport. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis K. N., Ed.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2010; pp 1545–1553. [Google Scholar]
- Ehrl B. N.; Gharasoo M.; Elsner M. Isotope fractionation pinpoints membrane permeability as a barrier to atrazine biodegradation in gram-negative Polaromonas sp. Nea-C. Environ. Sci. Technol. 2018, 52 (7), 4137–4144. 10.1021/acs.est.7b06599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick L. M.; Quadroni M.; Egli T. Short- and long-term changes in proteome composition and kinetic properties in a culture of Escherichia coli during transition from glucose-excess to glucose-limited growth conditions in continuous culture and vice versa. Environ. Microbiol. 2001, 3 (9), 588–599. 10.1046/j.1462-2920.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- Qiu S.; Gözdereliler E.; Weyrauch P.; Lopez E. C. M.; Kohler H.-P. E.; Sørensen S. R.; Meckenstock R. U.; Elsner M. Small 13C/12C fractionation contrasts with large enantiomer fractionation in aerobic biodegradation of phenoxy acids. Environ. Sci. Technol. 2014, 48 (10), 5501–5511. 10.1021/es405103g. [DOI] [PubMed] [Google Scholar]
- Beate B. I.; Sigg L.. Chemical speciation of organics and of metals at biological interphases. In Physicochemical Kinetics and Transport at Biointerfaces; Hermann P.; Van Leeuwen W. K., Ed.; Wiley: New York, 2004; Vol. 9, pp 205–269. [Google Scholar]
- Wohnsland F.; Faller B. High-throughput permeability pH profile and high-throughput alkane/water log P with artificial membranes. J. Med. Chem. 2001, 44 (6), 923–930. 10.1021/jm001020e. [DOI] [PubMed] [Google Scholar]
- Passeleu-Le Bourdonnec C.; Carrupt P.-A.; Scherrmann J. M.; Martel S. Methodologies to assess drug permeation through the blood–brain barrier for pharmaceutical research. Pharm. Res. 2013, 30 (11), 2729–2756. 10.1007/s11095-013-1119-z. [DOI] [PubMed] [Google Scholar]
- Males R. G.; Herring F. G. A 1H-NMR study of the permeation of glycolic acid through phospholipid membranes. Biochim. Biophys. Acta, Biomembr. 1999, 1416 (1–2), 333–338. 10.1016/S0005-2736(98)00236-3. [DOI] [PubMed] [Google Scholar]
- Naderkhani E.; Vasskog T.; Flaten G. E. Biomimetic PVPA in vitro model for estimation of the intestinal drug permeability using fasted and fed state simulated intestinal fluids. Eur. J. Pharm. Sci. 2015, 73, 64–71. 10.1016/j.ejps.2015.03.017. [DOI] [PubMed] [Google Scholar]
- Lande M. B.; Donovan J. M.; Zeidel M. L. The relationship between membrane fluidity and permeabilities to water, solutes, ammonia, and protons. J. Gen. Physiol. 1995, 106 (1), 67–84. 10.1085/jgp.106.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C.; Bagatolli L. A.; Volovyk Z. N.; Thompson N. L.; Levi M.; Jacobson K.; Gratton E. Lipid Rafts Reconstituted in Model Membranes. Biophys. J. 2001, 80 (3), 1417–1428. 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xlang T.-X.; Anderson B. D. Development of a combined NMR paramagnetic ion-induced line-broadening/dynamic light scattering method for permeability measurements across lipid bilayer membranes. J. Pharm. Sci. 1995, 84 (11), 1308–1315. 10.1002/jps.2600841110. [DOI] [PubMed] [Google Scholar]
- Russell N. J.; Fukunaga N. A comparison of thermal adaptation of membrane lipids in psychrophilic and thermophilic bacteria. FEMS Microbiol. Lett. 1990, 75 (2–3), 171–182. 10.1111/j.1574-6968.1990.tb04093.x. [DOI] [Google Scholar]
- Vanounou S.; Pines D.; Pines E.; Parola A. H.; Fishov I. Coexistence of domains with distinct order and polarity in fluid bacterial membranes. Photochem. Photobiol. 2002, 76 (1), 1–11. . [DOI] [PubMed] [Google Scholar]
- Epand R. M.; Epand R. F. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta, Biomembr. 2009, 1788 (1), 289–294. 10.1016/j.bbamem.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Vollmer W.; Blanot D.; De Pedro M. A. Peptidoglycan structure and architecture. FEMS Microbiology Reviews 2008, 32 (2), 149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Prestegard J. H.; Cramer J. A.; Viscio D. B. Nuclear magnetic resonance determinations of permeation coefficients for maleic acid in phospholipid vesicles. Biophys. J. 1979, 26 (3), 575–584. 10.1016/S0006-3495(79)85272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet O.; Loria J. P.; Kroenke C. D.; Pons M.; Palmer A. G. The static magnetic field dependence of chemical exchange linebroadening defines the nmr chemical shift time scale. J. Am. Chem. Soc. 2000, 122 (12), 2867–2877. 10.1021/ja993511y. [DOI] [Google Scholar]
- Melander L.; Saunders W. H.. Reaction Rates of Isotopic Molecules; John Wiley: New York, 1980; p 331. [Google Scholar]
- Hoefs J.Theoretical and Experimental Principles. In Stable Isotope Geochemistry, 3rd ed.; Wyllie P. J., Ed.; Springer-Verlag: Chicago, 1987; pp 1–25. [Google Scholar]
- Schmidt H. L. Fundamentals and systematics of the non-statistical distributions of isotopes in natural compounds. Naturwissenschaften 2003, 90 (12), 537–552. 10.1007/s00114-003-0485-5. [DOI] [PubMed] [Google Scholar]
- Thullner M.; Kampara M.; Richnow H. H.; Harms H.; Wick L. Y. Impact of bioavailability restrictions on microbially induced stable isotope fractionation. 1. Theoretical calculation. Environ. Sci. Technol. 2008, 42 (17), 6544–6551. 10.1021/es702782c. [DOI] [PubMed] [Google Scholar]
- Kampara M.; Thullner M.; Richnow H. H.; Harms H.; Wick L. Y. Impact of bioavailability restrictions on microbially induced stable isotope fractionation. 2. Experimental evidence. Environ. Sci. Technol. 2008, 42 (17), 6552–6558. 10.1021/es702781x. [DOI] [PubMed] [Google Scholar]
- Thullner M.; Fischer A.; Richnow H. H.; Wick L. Y. Influence of mass transfer on stable isotope fractionation. Appl. Microbiol. Biotechnol. 2013, 97 (2), 441–452. 10.1007/s00253-012-4537-7. [DOI] [PubMed] [Google Scholar]
- MacDonald R. C.; MacDonald R. I.; Menco B. P. M.; Takeshita K.; Subbarao N. K.; Hu L.-R. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim. Biophys. Acta, Biomembr. 1991, 1061 (2), 297–303. 10.1016/0005-2736(91)90295-J. [DOI] [PubMed] [Google Scholar]
- Hope M. J.; Bally M. B.; Webb G.; Cullis P. R. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta, Biomembr. 1985, 812 (1), 55–65. 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- Kujawinski D. M.; Wolbert J. B.; Zhang L.; Jochmann M. A.; Widory D.; Baran N.; Schmidt T. C. Carbon isotope ratio measurements of glyphosate and AMPA by liquid chromatography coupled to isotope ratio mass spectrometry. Anal. Bioanal. Chem. 2013, 405 (9), 2869–2878. 10.1007/s00216-012-6669-0. [DOI] [PubMed] [Google Scholar]
- Moncelli M. R.; Becucci L.; Guidelli R. The intrinsic pKa values for phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine in monolayers deposited on mercury electrodes. Biophys. J. 1994, 66 (6), 1969–1980. 10.1016/S0006-3495(94)80990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscio D. B.; Prestegard J. H. NMR studies of 5-hydroxytryptamine transport through large unilamellar vesicle membranes. Proc. Natl. Acad. Sci. U. S. A. 1981, 78 (3), 1638–1642. 10.1073/pnas.78.3.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S.; Escher B. I.; Goss K.-U. Capacities of membrane lipids to accumulate neutral organic chemicals. Environ. Sci. Technol. 2011, 45 (14), 5912–5921. 10.1021/es200855w. [DOI] [PubMed] [Google Scholar]
- Kallimanis A.; Frillingos S.; Drainas C.; Koukkou A. I. Taxonomic identification, phenanthrene uptake activity, and membrane lipid alterations of the PAH degrading Arthrobacter sp. strain Sphe3. Appl. Microbiol. Biotechnol. 2007, 76 (3), 709–717. 10.1007/s00253-007-1036-3. [DOI] [PubMed] [Google Scholar]
- Kell D. B.; Dobson P. D.; Oliver S. G. Pharmaceutical drug transport: the issues and the implications that it is essentially carrier-mediated only. Drug Discovery Today 2011, 16 (15–16), 704–714. 10.1016/j.drudis.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Culbertson C. T.; Jacobson S. C.; Michael Ramsey J. Diffusion coefficient measurements in microfluidic devices. Talanta 2002, 56 (2), 365–373. 10.1016/S0039-9140(01)00602-6. [DOI] [PubMed] [Google Scholar]
- Worch E. A new equation for the calculation of diffusion coefficients for dissolved substances. Vom Wasser 1993, 81, 289–297. [Google Scholar]
- Males R. G.; Phillips P. S.; Herring F. G. Equations describing passive transport through vesicular membranes. Biophys. Chem. 1998, 70 (1), 65–74. 10.1016/S0301-4622(97)00107-5. [DOI] [PubMed] [Google Scholar]
- Volkmer B.; Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS ONE 2011, 6 (7), e23126. 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke F.-D.; Georgi A.; Roland U. Isotope fractionation in phase-transfer processes under thermodynamic and kinetic control – Implications for diffusive fractionation in aqueous solution. Sci. Total Environ. 2018, 610–611 (Supplement C), 495–502. 10.1016/j.scitotenv.2017.08.063. [DOI] [PubMed] [Google Scholar]
- Shushkova T. V.; Ermakova I. T.; Sviridov A. V.; Leontievsky A. A. Biodegradation of glyphosate by soil bacteria: Optimization of cultivation and the method for active biomass storage. Microbiology 2012, 81 (1), 44–50. 10.1134/S0026261712010134. [DOI] [PubMed] [Google Scholar]
- Elsner M.; Zwank L.; Hunkeler D.; Schwarzenbach R. P. A new concept linking observable stable isotope fractionation to transformation pathways of organic pollutants. Environ. Sci. Technol. 2005, 39 (18), 6896–6916. 10.1021/es0504587. [DOI] [PubMed] [Google Scholar]
- Mancini S. A.; Hirschorn S. K.; Elsner M.; Lacrampe-Couloume G.; Sleep B. E.; Edwards E. A.; SherwoodLollar B. Effects of trace element concentration on enzyme controlled stable isotope fractionation during aerobic biodegradation of toluene. Environ. Sci. Technol. 2006, 40 (24), 7675–7681. 10.1021/es061363n. [DOI] [PubMed] [Google Scholar]
- Muller R. H.; Hoffmann D. Uptake kinetics of 2,4-dichlorophenoxyacetate by Delftia acidovorans MC1 and derivative strains: Complex characteristics in response to pH and growth substrate. Biosci., Biotechnol., Biochem. 2006, 70 (7), 1642–1654. 10.1271/bbb.60011. [DOI] [PubMed] [Google Scholar]
- Nichols N. N.; Harwood C. S. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 1997, 179 (16), 5056–61. 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button D. K.; Robertson B.; Gustafson E.; Zhao X. M. Experimental and theoretical bases of specific affinity, a cytoarchitecture-based formulation of nutrient collection proposed to supercede the Michaels-Menten paradigm of microbial kinetics. Appl. Environ. Microbiol. 2004, 70 (9), 5511–5521. 10.1128/AEM.70.9.5511-5521.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis J.; Kantiani L.; Llorca M.; Rubio F.; Ginebreda A.; Fraile J.; Garrido T.; Farre M. Determination of glyphosate in groundwater samples using an ultrasensitive immunoassay and confirmation by on-line solid-phase extraction followed by liquid chromatography coupled to tandem mass spectrometry. Anal. Bioanal. Chem. 2012, 402 (7), 2335–2345. 10.1007/s00216-011-5541-y. [DOI] [PubMed] [Google Scholar]
- Pipke R.; Schulz A.; Amrhein N. Uptake of Glyphosate by an Arthrobacter sp. Appl. Environ. Microbiol. 1987, 53 (5), 974–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.