Abstract
Programmed cell death (PCD) and secondary cell wall (SCW) thickening in pear fruit are accompanied by the deposition of cellulose and lignin to form stone cells. Metacaspase is an important protease for development, tissue renewal and PCD. The understanding of the molecular mechanism whereby pear (Pyrus) metacaspase promotes PCD and cell wall lignification is still limited. In this study, the Metacaspases gene family (PbMCs) from P. bretschneideri was identified. PbMC1a/1b was associated with lignin deposition and stone cell formation by physiological data, semiquantitative real-time polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR). Relative to wild-type (WT) Arabidopsis, the overexpression of PbMC1a/1b increased lignin deposition and delayed growth, thickened the cell walls of vessels, xylary fibers and interfascicular fibers, and increased the expression of lignin biosynthetic genes. Yeast two-hybrid (Y2H), bimolecular fluorescence complementation (BiFC) and GST pull-down assays indicated that the PbMC1a/1b protein physically interacted with PbRD21. Simultaneously, the transient expression of PbMC1a/1b and PbRD21 led to significant changes in the expression of genes and lignin contents in pear fruits and flesh calli. These results indicate that PbMC1a/1b plays an important role in cell wall lignification, possibly by interacting with PbRD21 to increase the mRNA levels of some lignin synthesis-associated genes and promote the formation of stone cells in pear fruit.
Subject terms: Plant cell death, Arabidopsis thaliana, Cell wall
Introduction
Pear (Pyrus) is an important fruit commodity around the world, and its quality has great significance for its marketing. Due to their poor quality, the export price of Chinese pears is lower than the market average. Many factors affect the quality of pears, among which stone cells are very important1. Sterling2 and Ranadive and Haard3 described the chemical properties of stone cells and the deposition of lignin in the cell wall. Mature stone cells contain ~18% lignin in pear fruits4. Therefore, it is of great economic significance to study the formation mechanism of lignin and reduce the contents of stone cells to improve the internal quality of pear.
Lignin deposited as lamellar additions to cell walls is the essence of stone cells in pear fruits5. Programmed cell death (PCD) is very important for organism development and tissue renewal6–8. The differentiation of tracheary elements (TEs) and vessel elements involves typical types of PCD, accompanied by the extensive lignification of cell walls9–11. Moreover, the secondary thickening of cell walls and PCD can be repressed with the same inhibitor, which indicates that the two processes are closely related12.
The morphological and biochemical characteristics of the initiation of cell death include cytoplasmic contraction, nuclear morphological changes, cytochrome C release from mitochondria, DNA fragmentation and the involvement of proteases13,14. In animals, the activation of PCD is primarily associated with Cys-dependent Asp-specific peptidase (caspase), and caspase-like enzymes have been found during cell death in plants15. Metacaspases (MCs) are caspase-dependent proteases presented in plants, protozoa, and fungi and show low sequence similarity to caspase16. MCs and caspase cleave substrates at the C-terminus with different amino acid compositions. MCs cut at arginine (Arg) and lysine (Lys), whereas caspase cuts at aspartic acid (Asp)17. MCs are divided into the following two types: type I MCs have an N-terminal prodomain and a zinc finger motif, whereas type II MCs have no prodomain in the N-terminal region but exhibit an inactive short functional pre-region18–20.
In Arabidopsis thaliana, there are three type I MCs (AtMC1–3) and six type II MCs (AtMC4–9)21. AtMC1 positively regulates PCD and has the required caspase catalytic residue, whereas AtMC2 negatively regulates PCD21. AtMC8 is a positive regulator of biotic and abiotic stresses in PCD22. AtMC9 is expressed in differentiated xylem vessels and is involved in PCD in xylem tubular molecules23,24. In pepper (Capsicum annuum L.), Camc9 plays a regulatory role in the process of PCD induced by plant pathogens25. Yeast MC Yca1 regulates PCD induced by oxidative stress, osmotic stress, and various other stimuli26. PttMC13 and PttMC14 are homologs of AtMC9 in hybrid aspen (Populus). Proteomic studies revealed targets of MCs such as cysteine protease RD21, Tudor staphylococcal nuclease, putative aspartic protease3 (PASPA3), heat shock proteins, and 14-3-3 proteins27.
Not all MCs family members in pear (PbMCs) are related to cell wall lignification in fruit. Therefore, it is necessary to identify PbMCs that are associated with cell lignification. The genomic sequencing of P. bretschneideri cv. Dangshan Su pear has been completed28; however, there are few reports of PbMCs functions. The objectives of this study were to identify PbMCs family members in pear, determine the gene(s) involved in the lignification of pear fruit cells, use transgenic technology to verify gene functions, and identify the target protein(s) by using Y2H technology. The results will validate the function of candidate genes and provide a theoretical basis for further understanding the relationship among PCD, lignin synthesis, and cell wall lignification.
Results
Identification and phylogenetic analysis of PbMCs family members
In this study, 15 PbMCs genes were found in the Chinese white pear genome database (Table S1). After considering the corresponding E-values, we selected 11 homologous PbMCs genes to build a phylogenetic tree showing the relationships of pear and Arabidopsis proteins (Fig. S1). The topology of the phylogenetic tree classified the PbMCs proteins into two subgroups, and most clades showed high statistical support (pp > 0.90; bootstrap > 80%). The location on the chromosome, molecular weight and isoelectric point (PI) of the PbMCs were provided in Table S2. Eight genes were mapped onto chromosomes 1, 5, 7, 9, 10, and 12, and other PbMCs were located on scaffold contigs.
Contents of stone cells and lignin in pear fruits during fruit development
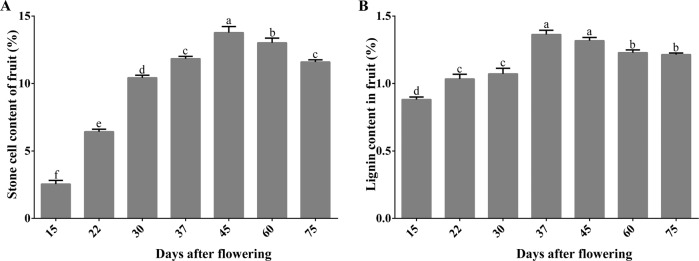
We determined the contents of stone cells and lignin in pear fruits from 15 to 75 days after flowering (DAF) (Fig. 1) because extensive lignin deposition occurred with stone cells in the early developmental stages29. From 15 to 45 DAF, the contents of stone cells increased rapidly, reaching the highest level (13.8%) at 45 DAF, and gradually decreased thereafter (Fig. 1a). The lignin content reached a peak (1.4%) at 37 DAF before the peak of stone cells (Fig. 1b).
Fig. 1.
Stone cell content (a) and lignin content (b) of the fruits during the development of ‘Dangshan Su’ pear. Different lowercase letters on a column indicate that the treatment differs significantly at the 1% level
The expression pattern of PbMC1a/1b was consistent with the changes in stone cells and lignin contents
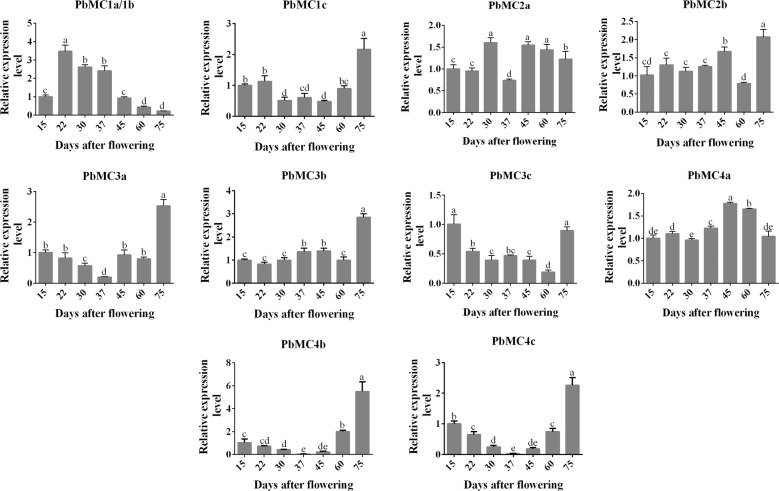
The expressions of PbMCs were measured in different pear tissues (Fig. S2). PbMC1a/1b, PbMC2b, PbMC3c, PbMC4b, and PbMC4c were expressed constitutively in all tissues, whereas the expressions of other PbMCs were tissue specific. PbMC3a was highly expressed in the roots and pollen, and PbMC4c was highly expressed strongly in the roots, leaves, and pollen. PbMC1c, PbMC2a, PbMC3b, and PbMC4a were not significantly expressed in any tissues of pear. The expression profiles of PbMCs during fruit development were also analyzed (Fig. 2). PbMC1a/1b maintained a high expression level from 22 to 37 DAF, was consistent with the trend of stone cell and lignin contents, whereas the other PbMCs exhibited inconsistent expression patterns.
Fig. 2. Relative expression patterns of PbMCs during the development of ‘Dangshan Su’ pear.
Different lowercase letters on a column indicate that the treatment differs significantly at the 1% level
Overexpression of PbMC1a/1b increased lignin deposition in transgenic Arabidopsis plants
PbMC1a/1b may induce PCD by playing a key role in cell wall lignification, and we, therefore, generated transgenic Arabidopsis plants that overexpressed PbMC1a/1b. In the T1 generation, six hygromycin-resistant transgenic plants were identified (Fig. S4a). Lines OE9, OE10, and OE15 exhibited higher levels of PbMC1a/1b expression and were selected for further analyses (Fig. S4b). DNA was extracted again in the T2 generation for identification (Fig. S4c).
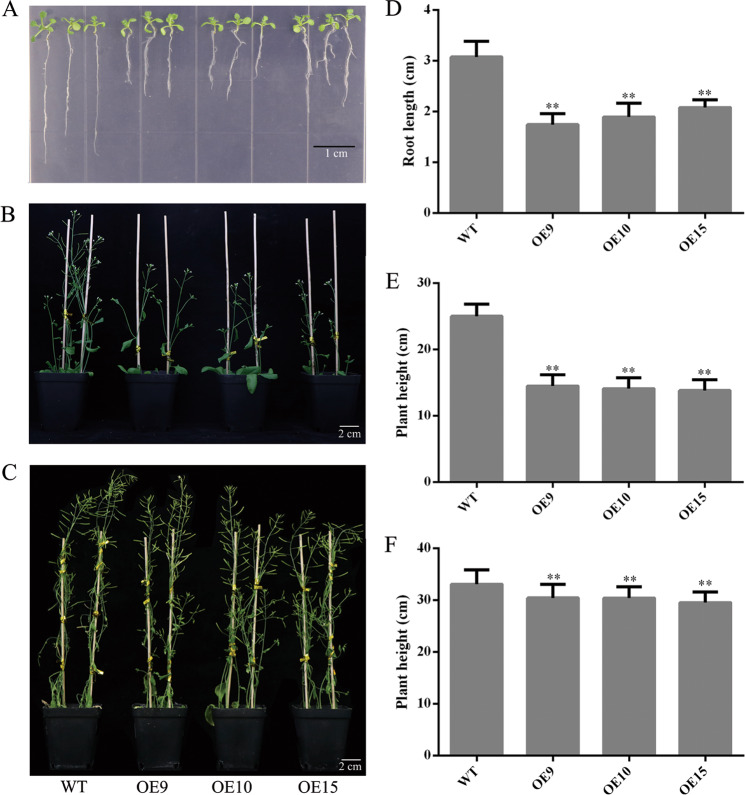
When the T3 generation was homozygous, OE9, OE10, and OE15 were used for plant phenotype examination. Compared with the WT (3.1 cm), the root lengths of transgenic plants were significantly shorter at the seedling stage (1 week old) and were reduced by an average of 1.17 cm (**P < 0.01) (Fig. 3a, d). During the flowering period (5 weeks old), the growth rates of the transgenic plants were slower, and the plant height of OE15 was the shortest compared to the WT (**P < 0.01) (Fig. 3b, e). At the mature stage (8 weeks old), the transgenic plants were an average of 2.94 cm shorter than the WT (Fig. 3c, f). Furthermore, the stems of the WT inflorescences grew faster, and the overexpression of PbMC1a/1b delayed the growth of transgenic Arabidopsis plants.
Fig. 3.
a–c WT and transgenic plants grown for 7 d (a), 5 weeks (b) and 8 weeks (c) under a long-day photoperiod. Root lengths at 7 days (d), plant heights at 6 weeks (e) and 8 weeks (f) (**p < 0.01)
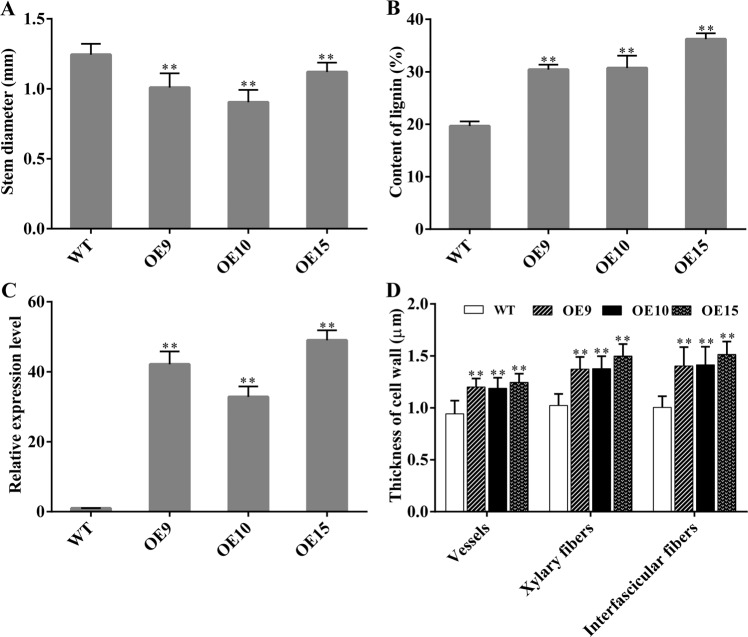
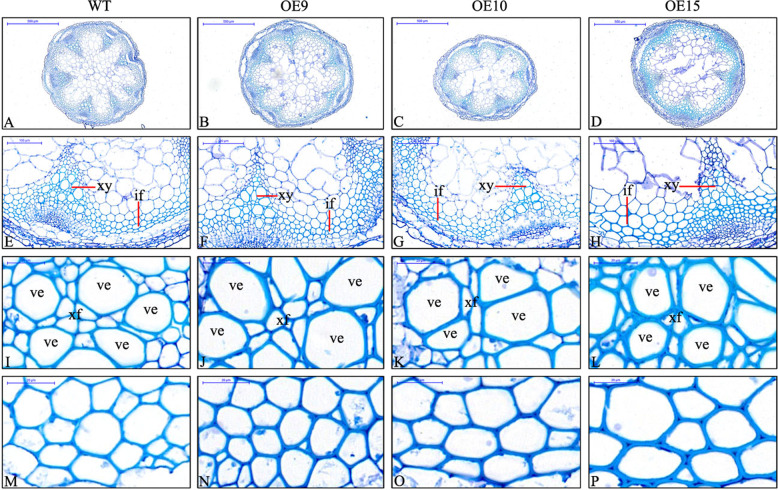
Interfascicular fibers and xylem cells are the main stem tissues supporting the erect growth of inflorescences30, and sections of stems were stained with toluidine blue to observe the changes in cells. The stem diameters of the transgenic plants were significantly smaller than those of the WT (Figs. 4a and 5a–d). The lignin contents of the stems of the transgenic plants were higher than those of the WT, and OE15 was the highest (36.2%), which was 16.57% higher than those of the WT (Fig. 4b). Concurrently, the relative expression of PbMC1a/1b was more than 30 times greater in the transgenic stems than in those of the WT (Fig. 4c). There were no changes in the morphology of vessel cells in the transgenic lines (Fig. 5e–h), but the cell wall thicknesses were increased significantly in vessel elements (28.7%), xylary fibers (44%) and interfascicular fibers (36%) compared to the WT (Figs. 4d and 5i–p). These results indicated that PbMC1a/1b promoted lignin deposition and cell wall lignification.
Fig. 4.
Stem diameters (a), lignin contents of stems (b), relative expression patterns of PbMC1a/1b in stems (c), cell wall thicknesses of vessels, xylary fibers and interfascicular fibers (d) in WT and transgenic plants (**p < 0.01)
Fig. 5. Sections stained with toluidine blue, showing the variation of cell walls in Arabidopsis with PbMC1a/1b overexpression.
a–d: stem cross-sections, e–l: xylem, (M-P): interfascicular fiber. if: interfascicular fiber; xy: xylem; ve: vessel; xf: xylary fiber
The expression levels of genes are related to the synthesis of lignin in SCWs31–33. To elucidate the molecular mechanism of PbMC1a/1b in lignin synthesis, the expression patterns of 14 lignin biosynthetic genes, including 4CL, C3H, C4H, CAD, CCoAOMT, CCR, COMT, F5H, HCT, LAC, and PAL, were also analyzed in stems (Fig. 6). The transcription levels of these genes in transgenic plants were significantly higher than those in WT plants, and LAC4, LAC11, and LAC17 exhibited the highest expression levels. These genes may synergistically promote the synthesis of lignin in the stems, in which LAC4, LAC11, and LAC17 may play key roles.
Fig. 6.
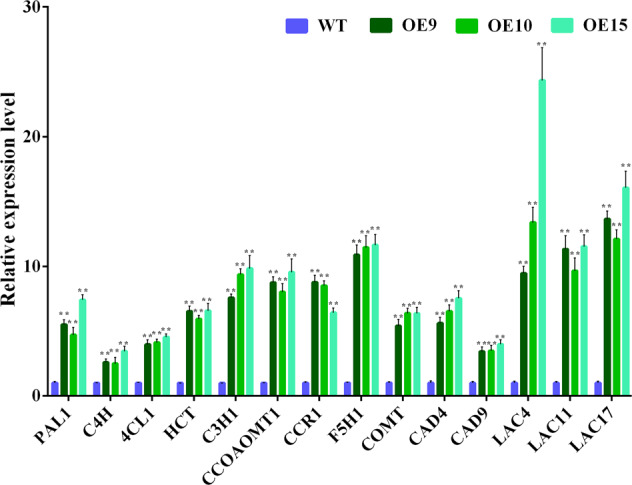
Relative expression patterns of lignin biosynthetic genes in PbMC1a/1b transgenic Arabidopsis plants (**p < 0.01)
PbMC1a/1b physically interacted with PbRD21
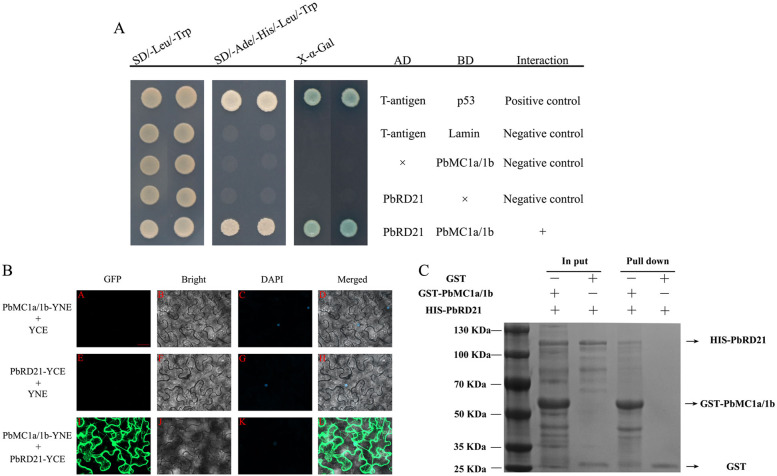
Cysteine protease is a putative target of MCs, and the three peptides of RD21 are highly expressed in the phloem, vascular cambium, and xylem tissues27. Y2H was performed to identify whether PbRD21 may function in a complex with PbMC1a/1b. Only the positive control yeast cells and co-transformants (pGBK-PbMC1a/1b and pGAD-PbRD21) grew normally on SD/-Ade/-Leu/-Trp/-Ura medium, whereas the negative controls did not survive (Fig. 7a). Furthermore, the positive control and co-transformants exhibited blue coloration after X-α-Gal staining.
Fig. 7.
a Interaction between PbMC1a/1b and PbRD21 verified by Y2H analysis. Selective growth of the Y2H system was performed by using the X-gal activity assay. Yeast cells were cotransformed with pGAD-PbRD21 + pGBKT7 and pGBK-PbMC1a/1b + pGADT7. pGADT7-T + pGBKT7-Lam was used as a negative control, and pGADT7-T + pGBKT7-53 was used as a positive control. b BiFC assay using Nicotiana benthamiana. The negative controls were PbMC1a/1b-YFPN + YFPC and PbRD21-YFPN + YFPC. All images were taken by confocal microscopy. Bar: 40 μm. c Interaction assay via GST pull-down analysis. PbMC1a/1b and PbRD21 fused with GST and HIS tags were mixed and passed through a glutathione column (binding GST tag). After elution with pull-down binding buffer, the samples were separated by SDS-PAGE. GST alone was used as the negative control
To verify the possibility of their interaction, the subcellular localizations of PbMC1a/1b and PbRD21 were identified. Control GFP was distributed evenly throughout tobacco cells, while the PbMC1a/1b-GFP fusion protein was located in the cytoplasm and nucleus, and PbRD21-GFP was only found in the nucleus (Fig. S3). Through subcellular localization analysis, both PbMC1a/1b and PbRD21 were found in the nucleus, which may indicate the possibility of their interaction.
The results of Y2H analysis were verified by BiFC and GST pull-down assays. Green fluorescence was observed in tobacco cells harboring the PbMC1a/1b-pSPYNE and PbRD21-pSPYCE vectors. However, no green fluorescence was observed in cells transformed with the negative control vectors (PbMC1a/1b-pSPYNE and pSPYCE; PbRD21-pSPYCE and pSPYNE) (Fig. 7b). Furthermore, PbMC1a/1b interacted with PbRD21 in vitro according to a GST pull-down assay (Fig. 7c). Y2H, BiFC and GST pull-down assays indicated that PbMC1a/1b interacted with PbRD21 and that their interaction occurred in the cytoplasm and nucleus.
Transient expression of PbMC1a/1b and PbRD21 changed the lignin content in pear fruits and fruit calli
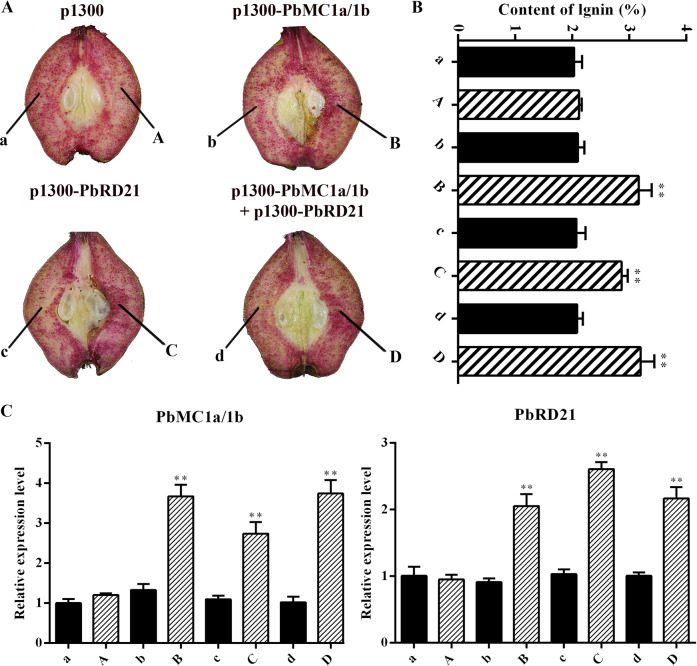
To verify the roles of PbMC1a/1b and PbRD21 in lignin biosynthesis, overexpression vectors were transiently expressed in pear fruit. 7 days after injection, increases in lignin staining were observed at the injection sites used under the overexpression of PbMC1a/1b, PbRD21 and a mixture of the two (Fig. 8a). The lignin contents were increased by 50.6, 38.2, and 52.9% compared with corresponding noninjected points, but no increase was evident in plants injected with the control vector (Fig. 8b). In addition, the injection of the PbMC1a/1b overexpression construct increased the expression of PbRD21 and vice versa (Fig. 8c). At the same time, lignin contents increased significantly after the overexpression of PbMC1a/1b and PbRD21 and decreased after their silencing in fruit calli (Fig. S5). Furthermore, the expression level of PbRD21 was similar to the contents of stone cells, lignin, and PbMC1a/1b (Fig. S6).
Fig. 8. Lignin contents and gene expression levels in fruit tissues infiltrated with PbMC1a/1b and PbRD21 overexpression constructs.
a Transient assay for the overexpression of PbMC1a/1b and PbRD21 in ‘Dangshan Su’ at 37 DAF. For different gene constructs, the injection sites were labeled A, B, C, and D, and the corresponding noninjected points were labeled a, b, c, and d. Images were obtained 7 days after the infiltration of Agrobacterium. b Lignin contents in the fleshy tissues in the fruits around the injection points (A, B, C, and D) and corresponding noninjected points (a, b, c, and d) (**p < 0.01). c The expression levels of PbMC1a/1b and PbRD21 in the fleshy tissues around the injection points and corresponding noninjected points were analyzed using qRT-PCR
Discussion
Stone cells are widely present in various parts of fruit, especially in the flesh, which reduces the quality of fruit34–36. Recent studies have focused on the cloning and regulation of lignin synthesis structural genes based on the pear genome and germplasm resources37,38. Cell wall lignification is associated with PCD, and MCs play a key role in this process27. Only a few MCs have been characterized, mainly from model plants21. The functional roles and potential regulatory mechanisms of MCs in perennial fruit trees remain elusive. Therefore, clarifying the function of MCs in pear will promote our understanding of the multiple roles of MCs. Here, we demonstrated that PbMC1a/1b and its interacting protein (PbRD21) promoted the expression of lignin biosynthetic genes, cell lignification and, thus, likely the formation of stone cells in pear fruit.
MCs play a key role in plant growth, development, and biotic and abiotic stress responses39 and show specific expressions in different tissues and organs in Arabidopsis40, rice41, grape42, and tomato43. In this study, we performed genome-wide analyses and identified 11 PbMCs. PbMC1a/1b, PbMC2b, PbMC3c, PbMC4b and PbMC4c were expressed in pear fruit and may be related to fruit development. During the development of fruit, the peak lignin content appeared before the peak of stone cells34. The development of stone cells is a process associated with the SCW thickening and lignification of parenchyma cells, and the synthesis of lignin provides material for the development of stone cells44. Our results demonstrated that lignin was an important raw material for stone cell formation.
During the formation of stone cells in pear fruits, PCD and SCW thickening are accompanied by cellulose and lignin deposition45. Cell death is regulated by transcription during xylem maturation, including the development of the SCW and lignification of cells46. The sequencing of signaling mutants indicated that the expression of MCs interfered with the biosynthesis of lignin47. RNAi knockdown of PttMC13 and PttMC14 driven by the xylem-specific proPtCOMT promoter inhibits expression more effectively than that driven by the 35S promoter27, and COMT reduces the syringyl lignin content significantly when the expression of COMT is decreased48. In this study, transgenic PbMC1a/1b Arabidopsis grew slowly; the cell walls of the vessels, xylary fibers, and interfascicular fibers were thicker than those of the WT; and the lignin contents of the stems were increased significantly. These results demonstrated that PbMC1a/1b promoted the thickening of the cell walls via lignin deposition.
Most enzymes involved in lignin monomer biosynthesis, such as C4H, CAD, CSE, and LAC, have been reported49. The expression levels of these genes increased significantly in transgenic PbMC1a/1b plants, and LACs exhibited the highest expression levels. In Arabidopsis, there was no significant change in lignin content in the single LAC11 mutant, and only a slight decrease was observed in the LAC4 LAC11 and LAC4 LAC17 double mutants. However, the lignin content of the LAC4 LAC11 LAC17 triple mutant was reduced significantly, indicating that there is functional redundancy among these genes31,33. Furthermore, LAC11 is the direct target of SND1 and VND7, and the promoter of LAC11 exhibits four secondary wall NAC-binding elements (SNBE)50. Interestingly, the post-translationally activated target genes of VND6 and VND7, such as XCP1, XCP2, and MCs, are related to the formation of secondary wall but also to cell death51,52. The homologous genes of VND7 or other transcription factors regulating MCs and LACs need further verification in pear. Whether PbMCs and PbLACs synergistically promote the formation of lignin in pear fruit also needs further verification.
Arabidopsis RD21 is a pro-death cysteine protease that has an inactive form53. MCs may be responsible for RD21 processing after the rupture of the vacuole membrane in xylem elements. In addition, the activation of MCs has been reported to be responsible for processing RD21 or other proteases during xylem maturation27, but no additional studies have supported this claim. In this study, the interaction between PbMC1a/1b and PbRD21 was verified, and their interaction sites were in both the cytoplasm and nucleus, but their subcellular localizations were different. One explanation for the strong nucleo-cytoplasmic signals of the PbMC1a/1b-PbRD21 interaction could be the differential subcellular localization of PbRD21. Because pre-domain removal occurred during protein extraction from leaves, a change in the subcellular localization of RD21 occurred54. A more likely explanation is that PbMC1a/1b changes the subcellular localization of PbRD21. RD21 shows a change in its subcellular localization when interacting with proteins. Hs4E02 is located in the nucleus, causing RD21A to accumulate in different cell compartments, and interaction is observed in the cytoplasm as well as the nucleus55. AtSerpin1 is a protease that inhibits AtMC1-mediated cell death and colocalizes in the cytoplasm and nucleus56. RD21 is simultaneously targeted by an inhibitor of AtSerpin1. A PCD inducer increases vesicle membrane permeability and leads to the co-localization of RD21 and AtSerpin1 in the cytoplasm57,58. Collectively, our data demonstrated that PbMC1a/1b interacts with PbRD21.
Lignin contents in pear fruits and flesh calli were also changed significantly by the transient expression of PbMC1a/1b and PbRD21, and their expression levels were positively correlated with the lignin content. However, the up- or downregulated contents of lignin biosynthesis genes were similar in PbMC1a/1b and PbRD21 single or co-transformed materials. Although caspase and MCs have different substrate specificities and biochemical properties, they are functionally similar: both proteases can hydrolyze a single conserved substrate53. They may compete for substrates and result in similar upregulation of lignin contents in single or multiple mutants. MCs may be involved in postmortem autolysis, for example, through interacting with RD21 and other proteolytic enzymes. How MCs interact with RD21 or other proteolytic enzymes to promote lignin deposition has not been reported until now; however, the mechanism of their interaction needs further study.
Conclusion
Overall, our results demonstrated that PbMC1a/1b (PCD-associated gene) promoted SCW thickening and increased the expression levels of lignin biosynthetic genes and lignin contents and clarified the involvement of PbMC1a/1b in pear fruit cell lignification. The overexpression of PbMC1a/1b led to high expression of LACs, but further research is needed to identify the synergy between them. The cysteine protease RD21, a putative target protein of MCs, is another protease associated with PCD and was identified as a member of the interaction. PbMC1a/1b and PbRD21 could promote lignin formation, and the mechanism of their interaction requires further exploration. These studies will help reveal the mechanism of SCW thickening and lignin deposition to form stone cells in pear fruits from the perspective of PCD. In the future, a single-cell approach may be needed to decipher the regulatory mechanisms of MCs, RD21, and PCD at the early stage of lignification, to identify the different activities between parenchyma cells and lignifying cells and to prevent the formation of stone cells in pear fruit.
Materials and methods
Plant materials
The study was conducted in a 50-year-old ‘Dangshan Su’ Pear (P. bretschneideri) orchard in Xiaji Town, Gaoyou City, Jiangsu Province, and healthy fruit trees were selected. One hundred fruit samples were collected on ice at 15, 22, 30, 37, 45, 60, and 75 DAF. Fruits were chopped after peeling, some flesh was used for the determination of physiological indicators, and the remaining flesh samples were frozen in liquid nitrogen and stored in a refrigerator at −80 °C for subsequent tests. Fresh root, stem, leaf, pollen and pulp tissues, and organs were sampled from the same plant, quickly frozen in liquid nitrogen and stored at −80 °C. The wild-type of A. thaliana was the Columbia ecotype.
Stone cell content in pear flesh
Stone cell content analysis was performed based on the method of Syros et al.59. The operation steps were as follows: twenty fruits at 15, 22, 30, 37, and 45 DAF and three fruits at 60 and 75 DAF of uniform sizes were selected and peeled, and the edible parts of the fruits were reserved for testing. One hundred grams of flesh was frozen at −20 °C for 24 h, then thawed at room temperature, and homogenized with distilled water in a blender for 10 min, after which the sample was stirred for 1 min, allowed to stand for 3 min, and the upper layer suspension was discarded. The remainder of the suspension was precipitated with 0.5 mM hydrochloric acid (HCl) for 30 min, decanted and washed with distilled water. The procedures were repeated several times until there were only stone cells.
Lignin determination
Twenty fruits at 15, 22, 30, 37, and 45 DAF, three fruits at 60 and 75 DAF, twenty Arabidopsis stems and twenty fruit calli were oven dried. After being ground to powder, 0.01 g of the samples with 3 replicates per sample was used to determine lignin content according to the method described by Tao et al.5. Samples were ground with 95% ethanol, washed 3 times with 95% ethanol and ethanol:hexane (1:2, v/v), and dried. The dried precipitates were digested in 2 ml of 25% (v/v) acetyl bromoacetic acid solution and crystallized at 70 °C for 30 min. The reaction was terminated by the addition of 0.9 ml of 2 N NaOH, after which 5 ml of acetic acid and 0.1 ml of 7.5 M hydroxylamine hydrochloride were added. The volume was adjusted to 10 ml with acetic acid, and the absorbance at A280 was determined. Lignin contents were calculated from a lignin standard sample (Sigma-Aldrich, USA) curve.
Identification of PbMCs genes in the white pear genome and bioinformatics analyses
The general feature formats (GFFs), proteome sequences and complete genome of pear (P. bretschneideri) were downloaded from http://peargenome.njau.edu.cn28. The protein sequences of Arabidopsis were obtained from http://www.arabidopsis.org/60. If two or more protein sequences at the same locus were identical in two proteome datasets, the longest sequence was selected where they overlapped. The HMM configuration file for the NAM domain (PF00656) was downloaded from http://pfam.xfam.org/family/PF00656/. HMMER was used to search a custom database containing proteomes with a threshold set to the Pfam GA aggregation cutoff value61. Proteins selected by HMMER were used for BLASTp queries in the original protein database. The NAM domains of the BLASTp hits were scanned using InterPro (http://www.ebi.ac.uk/interpro/)62, and those showing an E-value less than 1e−10 and the protein domains were considered. PbMCs genes were named based on homologous genes in Arabidopsis. Finally, a phylogenetic tree was constructed by the maximum likelihood (ML) method using MEG 6.0.
RNA extraction, semiquantitative RT-PCR and qRT-PCR analyses
Total RNA was extracted with the Polysaccharide Polyphenol Plant RNA Extraction Kit (Chengdu Fuji Biotechnology, China). According to the instruction manual, ~1 µg of total RNA was transcribed into cDNA using HiScript Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). The primer sequences for the semiquantitative RT-PCR and qRT-PCR assays were designed using Primer 5 (in the program EMBOSS Explorer) and were listed in Table S3. The method and program of Shi et al.63 were employed for semiquantitative RT-PCR. qRT-PCR was performed using a SYBR® Green Premix kit (TaKaRa Biotechnology, China). The program of Jin et al.64 was used for qRT-PCR. Each cDNA was analyzed in triplicate, after which the average threshold cycle (Ct) was calculated for each sample. Relative expression levels were calculated with the 2−ΔΔCt method65. Tubulin was analyzed in parallel as a reference control for P. bretschneideri, and AtActin was used for Arabidopsis.
Gene cloning and vector construction
The full-length PbMC1a/1b without the stop codon was cloned by RT-PCR with primer pairs containing Xba I and BamH I restriction sites (Table S3) using the Phanta® Max High-Fidelity PCR Enzyme (Vazyme, China). The product was introduced to the pCAMBIA1300 vector to generate a fusion construct (p35S-PbMC1a/1b-GFP) using a One-step Rapid Cloning Kit (Vazyme, China). The same methods were used for cloning and introduction into the pTRV2 vector (containing Xba I and Sac I restriction sites) to generate the pTRV2-PbMC1a/1b fusion construct. The fusion constructs (p35S-PbMC1a/1b-GFP and pTRV2-PbMC1a/1b) were transferred to E. coli strain DH5α by a freeze-thaw method. After sequence confirmation, the fusion constructs and control vector (pCAMBIA1300) were transferred to Agrobacterium tumefaciens strain GV3101 via the same freeze-thaw method. Finally, the same methods were used to construct the p35S-PbRD21-GFP and pTRV2-PbRD21 fusion expression vectors.
Subcellular localization of PbMC1a/1b and PbRD21
Transient transformation of tobacco leaves was performed using the method described by Yang et al.66. Agrobacterium was cultured at a ratio of 1:100 in LB liquid medium (containing 50 μg/mL kanamycin and 50 μg/mL rifampicin)64. The bacterial suspension was incubated at 28 °C on a 200 r/min shaker and grown to an OD600 of 0.6–0.8. The cells were collected by centrifugation at 4500 × g for 10 min and resuspended in a solution containing 10 mmol/L MgCl2, 10 mmol/L MES and 0.15 mmol/L AS. The bacterial suspension liquid was adjusted to an OD600 of 0.8–1.2 and cultured for 3 h in the dark. The inoculum was injected into the lower surface of 5-week-old N. benthamiana leaves. The transformed leaves were grown for 48 h and then subjected to live cell imaging using an inverted confocal microscope (Zeiss LSM 780, Germany).
Transient expression of pear fruit and fruit calli
Agrobacterium cells were collected and activated using the same method as that used for subcellular localization. Cells were injected into the flesh of ‘Dangshan Su’ pears at 37 DAF using needleless syringes. Six fruits were injected with each construct. The transformed fruits were placed in a dark environment at 25 °C overnight and then transferred to a growth chamber (25 °C, 16 h light/8 h dark photoperiod) for 7 days under low light conditions. Thereafter, the fruits were stained with phloroglucinol-HCl (Wiesner reagent)29, and lignin contents were determined. The expression levels of genes were determined by qRT-PCR, and the primers were listed in Table S3. Twenty fruit calli were also soaked in the Agrobacterium suspensions, cultured for 1 h in the dark, and placed on MS medium without growth regulators for 7 d in the dark, after which they were dried, and the lignin content was determined as previously described.
Transformation and characterization of transgenic plants
Arabidopsis plants (ecotype Columbia) were transformed by the vacuum infiltration of whole plants in an Agrobacterium suspension according to Bechtold et al.67. The plants were self-pollinated, and the seeds were harvested after the stems were completely dry. The T1 generations were selected on Arabidopsis medium containing 20 mg/L hygromycin36. After transgenic Arabidopsis seedlings were obtained, genomic DNA was extracted from in vitro-grown T1 and WT plants using cetyltrimethyl ammonium bromide (CTAB)36. To verify the transgenic Arabidopsis plants, PCR assays were performed, in which each 20 μl reaction contained 10 μl of buffer (Vazyme, China), 1 μl of DNA, 1 μl of each primer and 7 μl of sterile distilled water. The conditions described for PCR and electrophoresis in agarose gels by Jin et al.63 were followed. The relative expression levels of PbMC1a/1b in the leaves of T1 plants were determined by qRT-PCR with three technical replicates for each sample. The line showing the highest expression among the three lines was selected for subsequent verification, and the T3 generation of homozygous Arabidopsis was used for functional verification.
Transgenic T3 generation and WT plants were transferred to MS medium without growth regulators. The root lengths of 50 seedlings from each line were measured after 1 week of germination. Seedlings were transplanted into plastic pots with a mixture of vermiculite and soil (1:2) under 16 h of light and 40% relative humidity in the same chamber. The plant heights of 30 plants for each line were determined at 5 and 8 weeks. Stems were harvested at 8 weeks68, and toluidine blue staining, which was repeated three times for each line, was performed using the method of Berthet et al.31. ImageJ software was used to measure stem diameters as well as the cell wall thicknesses of vessels, xylary fibers, and interfascicular fibers in 100 cells. Lignin contents in the stems of 20 plants were determined at the mature stage. RNAs were extracted from the stems of the WT and transgenic T3 generations, and the expression levels of genes involved in phenylpropanoid biosynthesis (pxb00940) were quantitatively determined by qRT-PCR (primers listed in Table S3).
Y2H and BiFC assays
To explore the molecular mechanism whereby the overexpression of PbMC1a/1b to increases lignin content, the full-length cDNA of PbRD21 was amplified using a primer pair (Table S3) and cloned into the pGADT7 vector to obtain AD-PbRD21. PbMC1a/1b was amplified with primers (Table S3) and inserted into pGBDT7 to generate BD-PbMC1a/1b. The two constructs were transformed together into AH109 yeast cells, followed by culturing in SD/-Ade/-His/-Leu/-Trp plates to identify DNA-protein interactions.
For BiFC analysis, PbRD21 without a stop codon was cloned with a primer pair (Table S3) and introduced into pUC-pSPYCE-35S(nYFP) to obtain PbRD21-nYFP. Simultaneously, PbMC1a/1b without a stop codon was amplified using a primer pair (Table S3) and inserted into pUC-pSPYNE-35S(cYFP) to produce PbMC1a/1b-cYFP. Transient expression analyses of PbMC1a/1b-cYFP and PbRD21-nYFP in tobacco leaves were carried out according to Zhao et al.33. YFP fluorescence was observed using a confocal laser scanning microscope (LSM510 Meta, Zeiss, Germany).
In vitro GST pull-down assay
The GST-PbMC1a/1b and His-PbRD21 constructs were introduced into the E. coli BL21 (DE3) strain. One mM isopropyl sulfo-beta-galactoside (IPTG) was added at 37 °C to induce the expression of the fusion proteins. Glutathione-agarose 4B (GE Healthcare) beads and Ni-agarose were used according to the manufacturer’s instructions (GE Healthcare) to purify GST-PbMC1a/1b and His-PbRD21. For the protein pull-down assay, the GST fusion proteins were combined in a glutathione Sepharose 4B column. The loaded matrix was incubated with the purified His fusion protein in binding buffer for 4 h at 4 °C. The beads were centrifuged at 2000 × g for 1 min at 4 °C and washed three times with protein pull-down wash buffer, and the bound proteins were eluted and fractionated by 10% SDS-PAGE.
Statistical analysis
Each treatment was repeated at least three times with consistent results. Data are presented as the means ± SE of at least three independent replicates from one representative experiment. The data were analyzed with the ANOVA program of SPSS (IBM SPSS 22), where **P < 0.01 was considered to indicate a significant difference.
Supplementary information
Acknowledgements
We thank Xuebing Zhang and Yiming Shen (technicians of Xiaji Orchard) for selecting the pear samples. This work was financially supported by the Major Science and Technology Project of Corps (2017DB006), the National Natural Science Foundation of China (31972361, 31672105, and 31372044), and the National High-Technology Research, Development Program (2011AA10020602), for the design of the study and the collection, analyses, and interpretation of data.
Author contributions
S.T.T. contributed to the experimental design and management, data analysis, and paper review. X.G. contributed to the performance of laboratory work and manuscript preparation. K.J.Q., Z.H.X., L.Y.Z., and Y.Z.Y. prepared the plant materials and performed tissue collection. X.G., L.Y.Z. and J.H.X. contributed to the data analysis. W.K.R. prepared laboratory materials during the experiments. K.S., S.K., and S.L.Z. provided suggestions and reviewed and modified this paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-0280-x).
References
- 1.Cai Y, et al. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic.-Amst. 2010;125:374–379. doi: 10.1016/j.scienta.2010.04.029. [DOI] [Google Scholar]
- 2.Sterling C. Sclereid development and the texture of Bartlett pears. J. Food Sci. 1954;19:433–443. doi: 10.1111/j.1365-2621.1954.tb17474.x. [DOI] [Google Scholar]
- 3.Ranadive AS, Haard NF. Chemical nature of stone cells from pear fruit. J. Food Sci. 1973;38:331–333. doi: 10.1111/j.1365-2621.1973.tb01418.x. [DOI] [Google Scholar]
- 4.Lee SH, Park YS. Effects of calcium chloride spray on peroxidase activity and stone cell development in pear fruit (Pyrus pyrifolia ‘Niitaka’) 園芸学会雑誌. 2007;76:191–196. [Google Scholar]
- 5.Tao S, Khanizadeh S, Zhang H, Zhang S. Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci. 2009;176:413–419. doi: 10.1016/j.plantsci.2008.12.011. [DOI] [Google Scholar]
- 6.Conway, T. J. & McCabe, P. F. Plant Programmed Cell Death (PCD): Using Cell Morphology as a Tool to Investigate Plant PCD (Springer, Cham, 2018).
- 7.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locato, V. & De Gara, L. Programmed cell death in plants: An overview. Plant Programmed Cell Death (Humana Press, New York, NY, 2018). [DOI] [PubMed]
- 9.Escamez S, Tuominen H. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. J. Exp. Bot. 2014;65:1313–1321. doi: 10.1093/jxb/eru057. [DOI] [PubMed] [Google Scholar]
- 10.Kwon SI, et al. Overexpression of constitutively active Arabidopsis RabG3b promotes xylem development in transgenic poplars. Plant Cell Environ. 2011;34:2212–2224. doi: 10.1111/j.1365-3040.2011.02416.x. [DOI] [PubMed] [Google Scholar]
- 11.Pyo H, Demura T, Fukuda H. TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 2007;51:955–965. doi: 10.1111/j.1365-313X.2007.03180.x. [DOI] [PubMed] [Google Scholar]
- 12.Groover A, Jones AM. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 1999;119:375–384. doi: 10.1104/pp.119.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam E. Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Bio. 2004;5:305. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- 14.Wang CL, et al. S-RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro. Plant J. 2009;57:220–229. doi: 10.1111/j.1365-313X.2008.03681.x. [DOI] [PubMed] [Google Scholar]
- 15.Bursch W, Ellinger A, Gerner CH, Fröhwein U, Schulte-Hermann R. Programmed cell death (PCD): apoptosis, autophagic PCD, or others? Ann. Ny. Acad. Sci. 2000;926:1–12. doi: 10.1111/j.1749-6632.2000.tb05594.x. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 17.Tsiatsiani L, et al. Metacaspases. Cell Death Differ. 2011;18:1279. doi: 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi CJ, Berges JA. New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death Dis. 2013;4:e490. doi: 10.1038/cddis.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uren AG, et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 20.Vercammen D, Declercq W, Vandenabeele P, Van Breusegem F. Are metacaspases caspases? J. Cell Biol. 2007;179:375–380. doi: 10.1083/jcb.200705193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coll NS, et al. Arabidopsis type I metacaspases control cell death. Science. 2010;330:1393–1397. doi: 10.1126/science.1194980. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe N, Lam E. Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 2011;66:969–982. doi: 10.1111/j.1365-313X.2011.04554.x. [DOI] [PubMed] [Google Scholar]
- 23.Bollhöner B, et al. Post mortem function of AtMC9 in xylem vessel elements. N. Phytol. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- 24.Turner S, Gallois P, Brown D. Tracheary element differentiation. Annu. Rev. Plant Biol. 2007;58:407–433. doi: 10.1146/annurev.arplant.57.032905.105236. [DOI] [PubMed] [Google Scholar]
- 25.Kim SM, Bae C, Oh SK, Choi D. A pepper (C apsicum annuum L.) metacaspase 9 (Camc9) plays a role in pathogen-induced cell death in plants. Mol. Plant Pathol. 2013;14:557–566. doi: 10.1111/mpp.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee RE, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl Acad. Sci. USA. 2010;107:13348–13353. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bollhöner B, et al. The function of two type II metacaspases in woody tissues of Populus trees. New Phytol. 2018;217:1551–1565. doi: 10.1111/nph.14945. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue C, et al. PbrmiR397a regulates lignification during stone cell development in pear fruit. Plant Biotechnol. J. 2019;17:103. doi: 10.1111/pbi.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, et al. Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-directed transcriptional network in Arabidopsis. Plant Cell. 2018;30:2512–2528. doi: 10.1105/tpc.18.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berthet S, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao X, et al. Gene regulatory networks for lignin biosynthesis in switchgrass (Panicum virgatum) Plant Biotechnol. J. 2019;17:580–593. doi: 10.1111/pbi.13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Q, et al. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng X, et al. Characterization and analysis of CCR and CAD gene families at the whole-genome level for lignin synthesis of stone cells in pear (Pyrus bretschneideri) fruit. Biol. Open. 2017;6:1602–1613. doi: 10.1242/bio.026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng X, et al. Bioinformatic and expression analysis of the OMT gene family in Pyrus bretschneideri cv. Dangshan Su. Genet Mol. Res. 2016;15:gmr.15038664. doi: 10.4238/gmr.15038664. [DOI] [PubMed] [Google Scholar]
- 36.Xue C, et al. PbrMYB169 positively regulates lignification in fruit stone cells of pear (Pyrus bretschneideri) J. Exp. Bot. 2019;70:1801–1814. doi: 10.1093/jxb/erz039. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, Han Y, Li D, Lin Y, Cai Y. Systematic analysis of the 4-Coumarate: coenzyme a ligase (4CL) related genes and expression profiling during fruit development in the Chinese pear. Genes-Basel. 2016;7:89. doi: 10.3390/genes7100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y, et al. Structural, evolutionary, and functional analysis of the class III peroxidase gene family in Chinese Pear (Pyrus bretschneideri) Front. Plant Sci. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubey N, et al. Genome-wide characterization, molecular evolution and expression profiling of the metacaspases in potato (Solanum tuberosum L.) Heliyon. 2019;5:e01162. doi: 10.1016/j.heliyon.2019.e01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon SI, Hwang DJ. Expression analysis of the metacaspase gene family in Arabidopsis. J. Plant Biol. 2013;56:391–398. doi: 10.1007/s12374-013-0290-4. [DOI] [Google Scholar]
- 41.Wang L, Zhang H. Genomewide survey and characterization of metacaspase gene family in rice (Oryza sativa) J. Genet. 2014;93:93–102. doi: 10.1007/s12041-014-0343-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhang C, et al. The metacaspase gene family of Vitis vinifera L.: characterization and differential expression during ovule abortion in stenospermocarpic seedless grapes. Gene. 2013;528:267–276. doi: 10.1016/j.gene.2013.06.062. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, Liu J, Wei Y. Identification and analysis of the metacaspase gene family in tomato. Biochem. Bioph. Res. Co. 2016;479:523–529. doi: 10.1016/j.bbrc.2016.09.103. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, et al. Stone cell distribution and lignin structure in various pear varieties. Sci. Hortic.-Amst. 2014;174:142–150. doi: 10.1016/j.scienta.2014.05.018. [DOI] [Google Scholar]
- 45.Smith WW. The course of stone cell formation in pear fruits. Plant Physiol. 1935;10:587. doi: 10.1104/pp.10.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bollhöner B, Prestele J, Tuominen H. Xylem cell death: emerging understanding of regulation and function. J. Exp. Bot. 2012;63:1081–1094. doi: 10.1093/jxb/err438. [DOI] [PubMed] [Google Scholar]
- 47.Lundström, M. (ed.) Identification and characterization of upstream regulators of Arabidopsis Metacaspase 9 (Linköping Univ. Press, 2011).
- 48.Humphreys JM, Chapple C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002;5:224–229. doi: 10.1016/S1369-5266(02)00257-1. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Q. Lignification: flexibility, biosynthesis and regulation. Trends Plant Sci. 2016;21:713–721. doi: 10.1016/j.tplants.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Zhong R, Lee C, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant. 2010;3:1087–1103. doi: 10.1093/mp/ssq062. [DOI] [PubMed] [Google Scholar]
- 51.Ohashi-Ito K, Oda Y, Fukuda H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell. 2010;22:3461–3473. doi: 10.1105/tpc.110.075036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi M, et al. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011;66:579–590. doi: 10.1111/j.1365-313X.2011.04514.x. [DOI] [PubMed] [Google Scholar]
- 53.Balakireva A, Zamyatnin A. Indispensable role of proteases in plant innate immunity. Int. J. Mol. Sci. 2018;19:629. doi: 10.3390/ijms19020629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu C, et al. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS ONE. 2012;7:e32422. doi: 10.1371/journal.pone.0032422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pogorelko GV, et al. Re-targeting of a plant defense protease by a cyst nematode,effector. Plant J. 2019;98:1000–1014. doi: 10.1111/tpj.14295. [DOI] [PubMed] [Google Scholar]
- 56.Lema Asqui S, et al. AtSERPIN1 is an inhibitor of the metacaspase AtMC1-mediated cell death and autocatalytic processing in planta. N. Phytol. 2018;218:1156–1166. doi: 10.1111/nph.14446. [DOI] [PubMed] [Google Scholar]
- 57.Lampl N, Alkan N, Davydov O, Fluhr R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013;74:498–510. doi: 10.1111/tpj.12141. [DOI] [PubMed] [Google Scholar]
- 58.Rustgi S, Boex-Fontvieille E, Reinbothe C, von Wettstein D, Reinbothe S. Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana. P. Nat. Acad. Sci. USA. 2017;114:2212–2217. doi: 10.1073/pnas.1621496114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Syros T, Yupsanis T, Zafiriadis H, Economou A. Activity and isoforms of peroxidases, lignin and anatomy, during adventitious rooting in cuttings of Ebenus cretica L. J. Plant Physiol. 2004;161:69–77. doi: 10.1078/0176-1617-00938. [DOI] [PubMed] [Google Scholar]
- 60.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2007;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 62.Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2011;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi SL, et al. Identification of S-genotypes in 18 pear accessions and exploration of the breakdown of self-incompatibility in the pear cultivar Xinxue. Scientic-Amst. 2018;238:350–355. [Google Scholar]
- 64.Jin C, et al. A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 2017;8:1049. doi: 10.3389/fpls.2017.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- 67.Bechtold N. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris. Life Sci. 1993;316:1194–1199. [Google Scholar]
- 68.Boyes DC, et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.