Abstract
PURPOSE
To determine the sensitivity and specificity of genetic testing criteria for the detection of germline pathogenic variants in women with breast cancer.
MATERIALS AND METHODS
Women with breast cancer enrolled in a breast cancer registry at a tertiary cancer center between 2000 and 2016 were evaluated for germline pathogenic variants in 9 breast cancer predisposition genes (ATM, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, and TP53). The performance of the National Comprehensive Cancer Network (NCCN) hereditary cancer testing criteria was evaluated relative to testing of all women as recommended by the American Society of Breast Surgeons.
RESULTS
Of 3,907 women, 1,872 (47.9%) meeting NCCN criteria were more likely to carry a pathogenic variant in 9 predisposition genes compared with women not meeting criteria (9.0% v 3.5%; P < .001). Of those not meeting criteria (n = 2,035), 14 (0.7%) had pathogenic variants in BRCA1 or BRCA2. The sensitivity of NCCN criteria was 70% for 9 predisposition genes and 87% for BRCA1 and BRCA2, with a specificity of 53%. Expansion of the NCCN criteria to include all women diagnosed with breast cancer at ≤ 65 years of age achieved > 90% sensitivity for the 9 predisposition genes and > 98% sensitivity for BRCA1 and BRCA2.
CONCLUSION
A substantial proportion of women with breast cancer carrying germline pathogenic variants in predisposition genes do not qualify for testing by NCCN criteria. Expansion of NCCN criteria to include all women diagnosed at ≤ 65 years of age improves the sensitivity of the selection criteria without requiring testing of all women with breast cancer.
INTRODUCTION
The National Comprehensive Cancer Network (NCCN) hereditary cancer testing criteria is one of the current standards for identifying women at increased risk of hereditary breast, ovarian, or pancreatic cancer.1 Although the criteria have expanded over time to be more inclusive, many women with breast cancer currently do not qualify for genetic testing according to NCCN criteria. Two recent studies noted that a significant proportion of carriers of germline pathogenic variants were not identified (up to 50%) if women underwent testing solely based on NCCN criteria (v3.2019).2,3 On this basis, the American Society of Breast Surgeons (ASBrS) recommended that germline genetic testing should be made available to all women with a personal history of breast cancer.4
The implications of the ASBrS recommendations for women with breast cancer not selected for age at diagnosis or family history of cancer have not been adequately studied. Prior studies2,3 were prone to significant ascertainment and selection biases in favor of women with high-risk breast cancer.5 Patients included in prior studies may also have undergone genetic testing because of concern for inherited risk of cancer, despite not meeting NCCN testing criteria. More importantly, the majority of the genes included in the multigene panels in both studies have uncertain clinical relevance for breast cancer,2,3,6,7 which makes it challenging to interpret the significance of the results. Furthermore, these studies and the ASBrS recommendations did not consider the increase in the number of tests needed to detect clinically actionable germline pathogenic variants in all women with breast cancer. In a field with unmet needs for genetic counseling and management, understanding the impact of the ASBrS criteria on genetic testing services is critical for the responsible allocation of resources.6
The conflicting recommendations from NCCN and ASBrS have created a debate on whether all women with breast cancer need to undergo germline genetic testing.8-13 To alleviate some of the confusion associated with these recommendations and to inform genetic testing practice, we evaluated the sensitivity and specificity of NCCN, ASBrS, and other genetic testing criteria for germline pathogenic variants in a large series of women with breast cancer from a breast cancer registry at a tertiary cancer center.
MATERIALS AND METHODS
Sample Selection
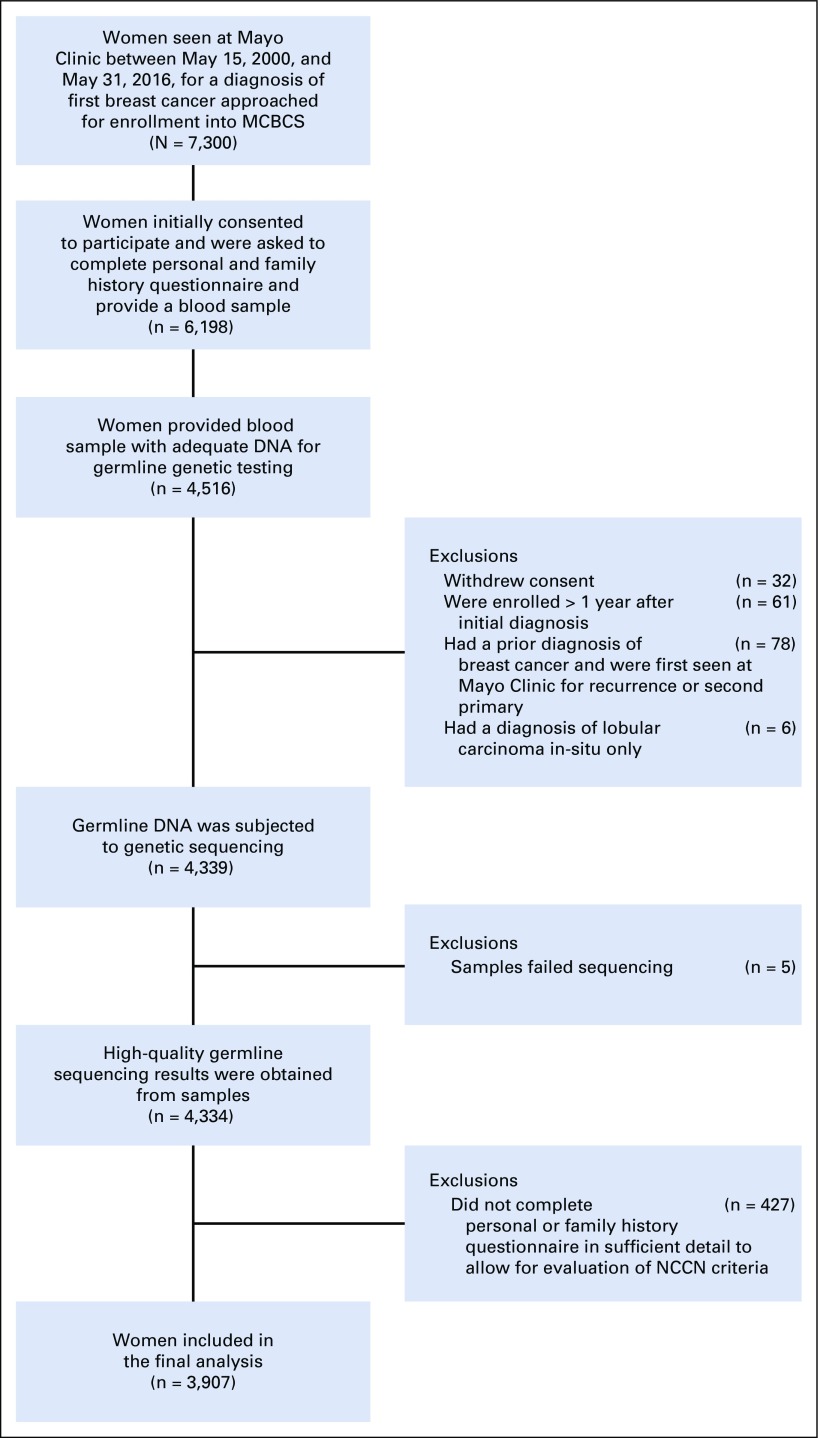
The study sample was derived from the Mayo Clinic Breast Cancer Study (MCBCS), a prospective registry offering participation to all women evaluated at Mayo Clinic Rochester for a diagnosis of first invasive breast cancer or ductal carcinoma in situ between May 15, 2000, and May 31, 2016. Of 7,300 women approached, 6,198 consented to the study and were asked to provide a blood sample and a baseline questionnaire on personal and family history adapted from the Breast Cancer Family Registry.14 Only baseline questionnaire and tumor characteristics from the initial diagnosis were considered in the current study. Patient demographics, tumor characteristics, and family history were also abstracted from electronic medical records to verify existing information. Family history information was available from 4,516 women providing blood samples (Appendix Fig A1, online only). Genetic testing results were offered and/or disclosed to the study participants through pretest and post-test counseling procedures by certified genetic counselors. This study was approved by the Institutional Review Board at Mayo Clinic.
Germline Sequencing and Bioinformatics Analysis
Germline DNA extracted from peripheral blood mononuclear cells was analyzed for germline pathogenic variants in the coding regions and consensus splice sites of 37 genes (Appendix Table A1, online only) using a custom amplicon-based QIAseq panel (Qiagen, Hilden, Germany) and sequencing on a HiSeq4000 (Illumina, San Diego, CA) as described previously15 and in the Appendix. Pathogenic and likely pathogenic variants were analyzed together as pathogenic variants. Low penetrance missense variants in CHEK2 were excluded from analyses.
Selection of Genes Based on Clinical Actionability
The primary analysis was restricted to 9 established breast cancer predisposition genes (ATM, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, and TP53)16-18 with clear management recommendations in the NCCN guidelines.1 Analyses were performed separately for pathogenic variants in 6 high-risk genes (BRCA1, BRCA2, CDH1, PALB2, PTEN, and TP53) or pathogenic variants in BRCA1 or BRCA2 only. Separate analysis including BARD1, RAD51C, and RAD51D was performed because pathogenic variants in these genes have been associated with triple-negative breast cancer.1,17,19,20 These genes have recently been included in genetic testing recommendations (NCCN v1.2020),1 although risk management guidelines are not available.
Assessment of NCCN Hereditary Cancer Testing Criteria
Women with a first- or second-degree relative with breast cancer were classified as having a family history of breast cancer. Similar definitions were used for other cancers. Of the NCCN hereditary cancer testing criteria relevant to women with a personal history of breast cancer (v1.2020),1 12 of 15 were fully evaluable (Appendix Table A2, online only). For the other criteria, women provided information on family history of prostate cancer rather than stage or Gleason score of prostate cancer in family members and on number of third-degree relatives with breast, ovarian, pancreatic, and prostate cancer rather than individual-level information. The influence of this information on guideline performance was assessed in a sensitivity analysis. Women meeting any of the evaluated criteria were considered qualified for genetic testing according to NCCN guidelines. Of those with BRCA1 or BRCA2 pathogenic variants who did not meet NCCN criteria, we used the Tyrer-Cuzick risk evaluation tool (v8.0b)21,22 to assess whether these women had > 5% pretest probability of carrying BRCA1 or BRCA2 pathogenic variants.
Statistical Analysis
NCCN, ASBrS, and other age-of-diagnosis– and family-history–based criteria were evaluated for sensitivity and specificity of the testing criteria, the number of women tested, frequency of germline pathogenic variants in the criteria evaluated, and the variant of uncertain significance (VUS)-to-pathogenic-variant ratio (defined in the Appendix, online only). Fisher’s exact test was performed to compare the frequency of pathogenic variants among women meeting and not meeting NCCN criteria, and results were reported as odds ratios with 95% CIs. All tests were 2 sided, and a P value < .05 was considered statistically significant. All analyses were performed using IBM SPSS Statistics (v25; SPSS, Chicago, IL).
RESULTS
Results of Germline Genetic Testing
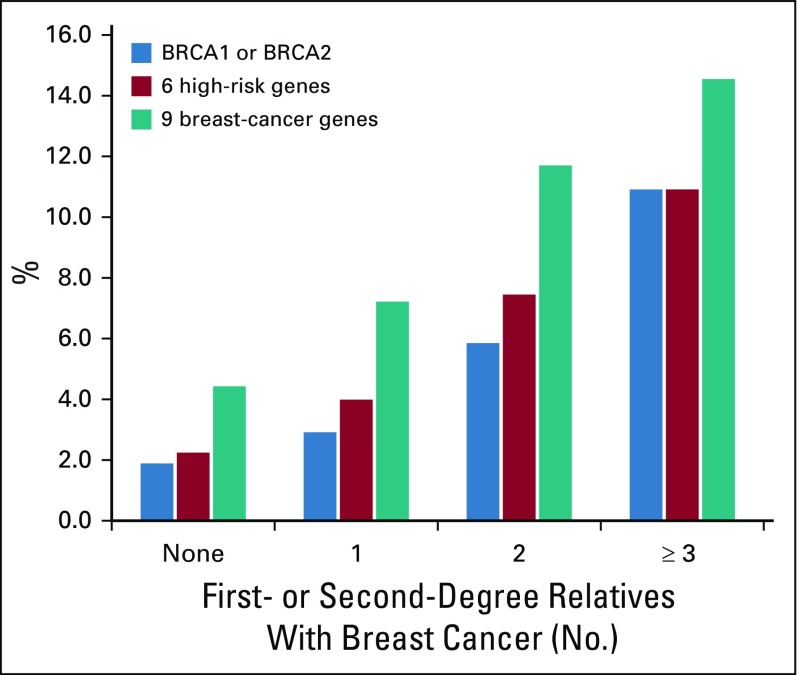
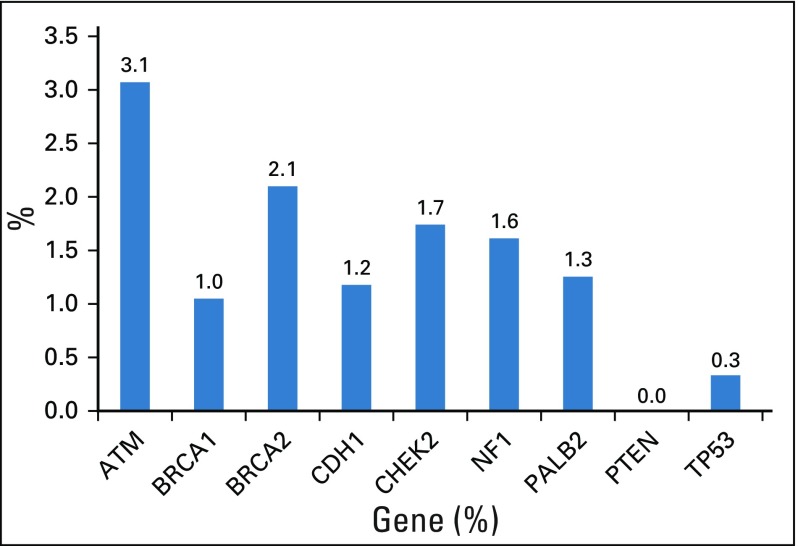
A total of 3,907 women with a diagnosis of invasive breast cancer (84.0%) or ductal carcinoma in situ (16.0%) were included in the final analysis. The median age of breast cancer diagnosis was 57 years (range, 21-94 years). A family history of breast cancer was present in 46.7% of women (Table 1). The majority (40.5%) had only 1 relative with breast cancer. Among the 3,907 women, 241 (6.2%) had germline pathogenic variants in the 9 established predisposition genes. Pathogenic variants in CHEK2 (1.7%), BRCA2 (1.4%), BRCA1 (1.3%), and ATM (1.1%) were the most frequent. The c.1100delC allele accounted for 52 of 67 CHEK2 pathogenic variants (Appendix Table A3, online only). Among women with 2 or more relatives with breast cancer, the frequency of germline pathogenic variants was > 5% for BRCA1 or BRCA2 and > 10% for 9 predisposition genes (Appendix Fig A2, online only). A total of 449 (11.5%) women had a VUS in any of the 9 genes (Appendix Fig A3, online only).
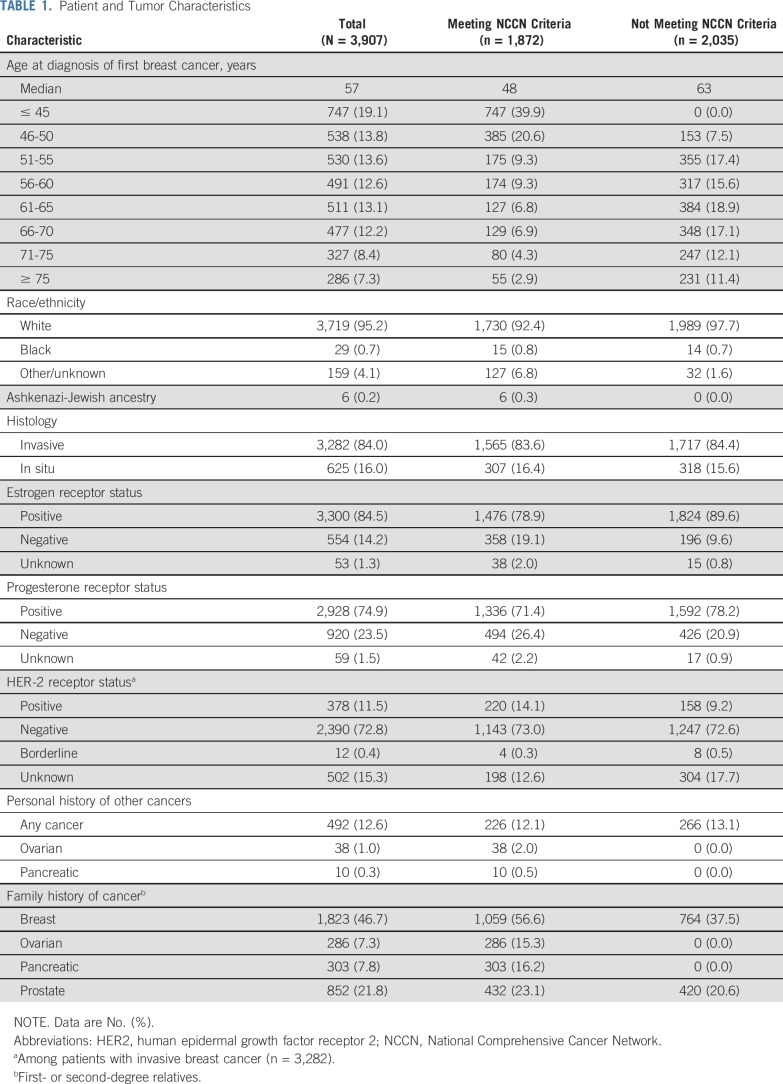
TABLE 1.
Patient and Tumor Characteristics
Comparison of NCCN and ASBrS Criteria
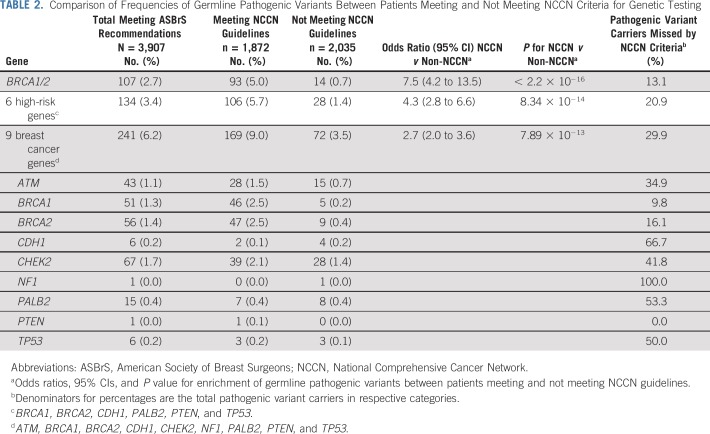
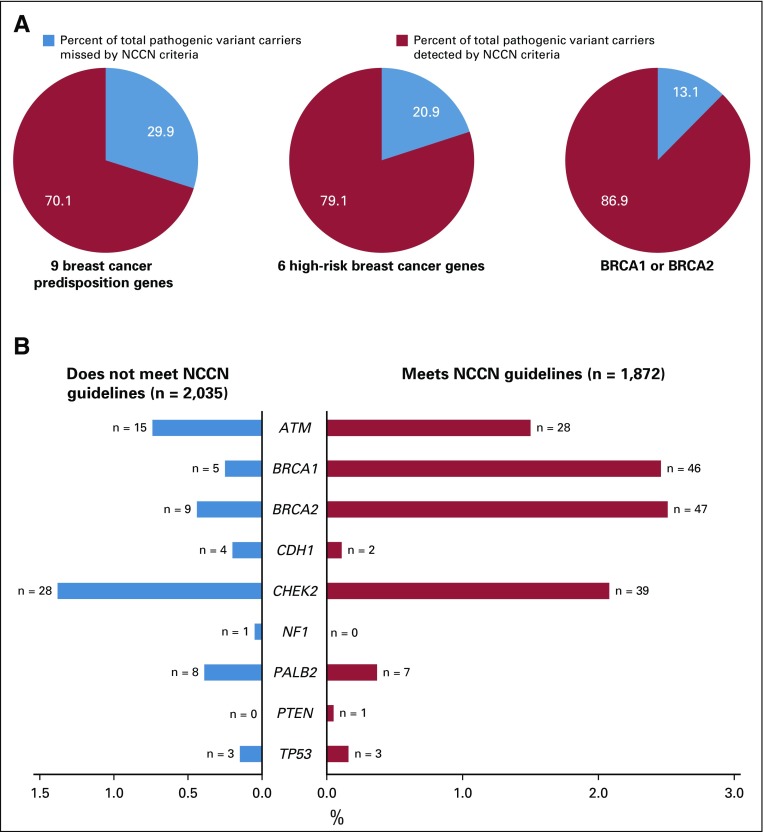
Of the 3,907 women, 1,872 (47.9%) met NCCN testing criteria, whereas 2,035 (52.1%) did not. The characteristics of women in these categories are shown in Table 1. Testing of all women as recommended by ASBrS identified pathogenic variants in 9 actionable predisposition genes in 6.2% of women, in 6 high-risk genes in 3.4% of women, and in BRCA1 or BRCA2 in 2.7% of women (Table 2). Those meeting NCCN criteria were more likely to carry a pathogenic variant in the 9 genes than women not meeting criteria (9.0% v 3.5%; P < .001). Similar results were observed for the 6 high-risk genes (5.7% v 1.4%; P < .001) and for BRCA1 or BRCA2 (5.0% v 0.7%; P < .001). However, 72 (29.9%) of the 241 women with pathogenic variants in the 9 genes, 28 (20.9%) of 134 with pathogenic variants in the 6 high-risk genes, and 14 (13.1%) of 107 with pathogenic variants in BRCA1 or BRCA2 did not qualify for genetic testing based on NCCN criteria (Table 2; Fig 1). The NCCN v1.2020 testing guidelines differ from the v3.2019 guidelines by recommending testing of those with > 5% pretest probability of a BRCA1 or BRCA2 pathogenic variant. None of the 14 women with BRCA1 or BRCA2 pathogenic variants who did not qualify for testing under previous NCCN v3.2019 criteria had > 5% pretest probability (all14 women had < 2.5% probability) of a BRCA1/2 pathogenic variant, as measured using the Tyrer-Cuzick model. Thus, this additional criterion did not improve the sensitivity of the testing guidelines in this study. Additional analyses considering all women with a family history of prostate cancer (Appendix Table A4, online only) or all women with third-degree relatives with breast, ovarian, pancreatic or prostate cancer (Appendix Table A5, online only) did not substantially alter the 70.1% sensitivity from the primary analysis. The specificity of NCCN criteria was approximately 53% for pathogenic variants in 9 predisposition genes, 6 high-risk genes, and BRCA1 or BRCA2 (Table 3). The VUS rate for the 9 genes was only marginally higher in women meeting NCCN criteria compared with women not meeting criteria (12.7% v 10.4%; P = .02). However, this resulted in a higher VUS-to-pathogenic variant ratio for women not meeting NCCN criteria (2.9 vs. 1.4). Interestingly, only 2 of 6 carriers of TP53 pathogenic variants and 1 of 6 CDH1 carriers met testing criteria for Li-Fraumeni syndrome23 or Hereditary Diffuse Gastric Cancer criteria,24 respectively.
TABLE 2.
Comparison of Frequencies of Germline Pathogenic Variants Between Patients Meeting and Not Meeting NCCN Criteria for Genetic Testing
FIG 1.
Evaluation of sensitivity of National Comprehensive Cancer Network (NCCN) criteria for pathogenic variant carriers in 3,907 women with breast cancer. (A) Percent of total pathogenic variant carriers missed by NCCN criteria in 9 breast cancer predisposition genes, 6 high-risk predisposition genes, and BRCA1 or BRCA2 (9 genes: ATM, BRCA1, BRCA2, CDH1, CHEK2, NF1, PALB2, PTEN, and TP53; 6 genes: BRCA1, BRCA2, CDH1, PALB2, PTEN, and TP53). (B) Comparison of germline pathogenic variant frequencies between women meeting and not meeting NCCN guidelines.
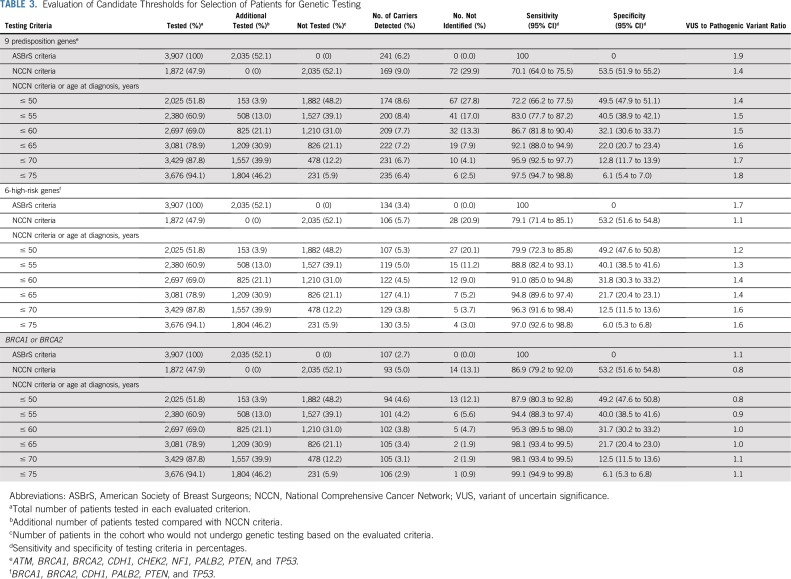
TABLE 3.
Evaluation of Candidate Thresholds for Selection of Patients for Genetic Testing
Alternative Selection Criteria for Genetic Testing
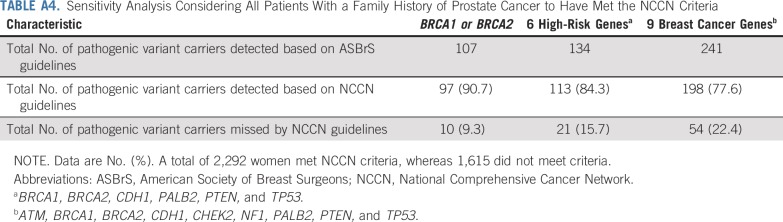
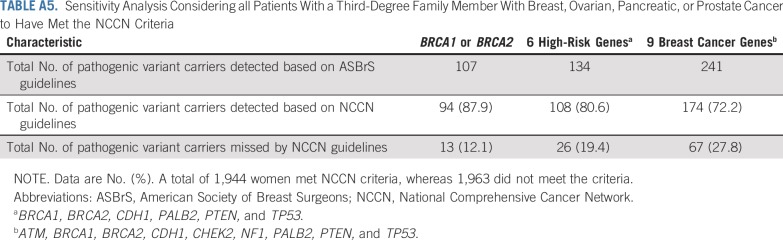
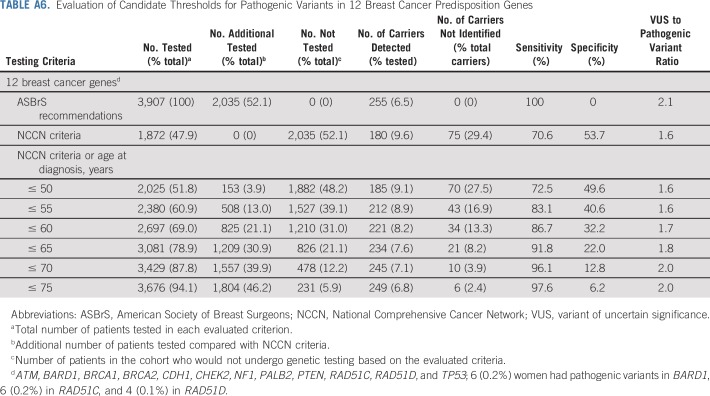
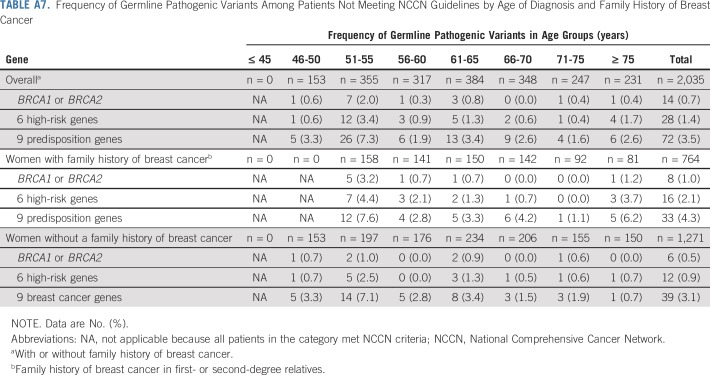
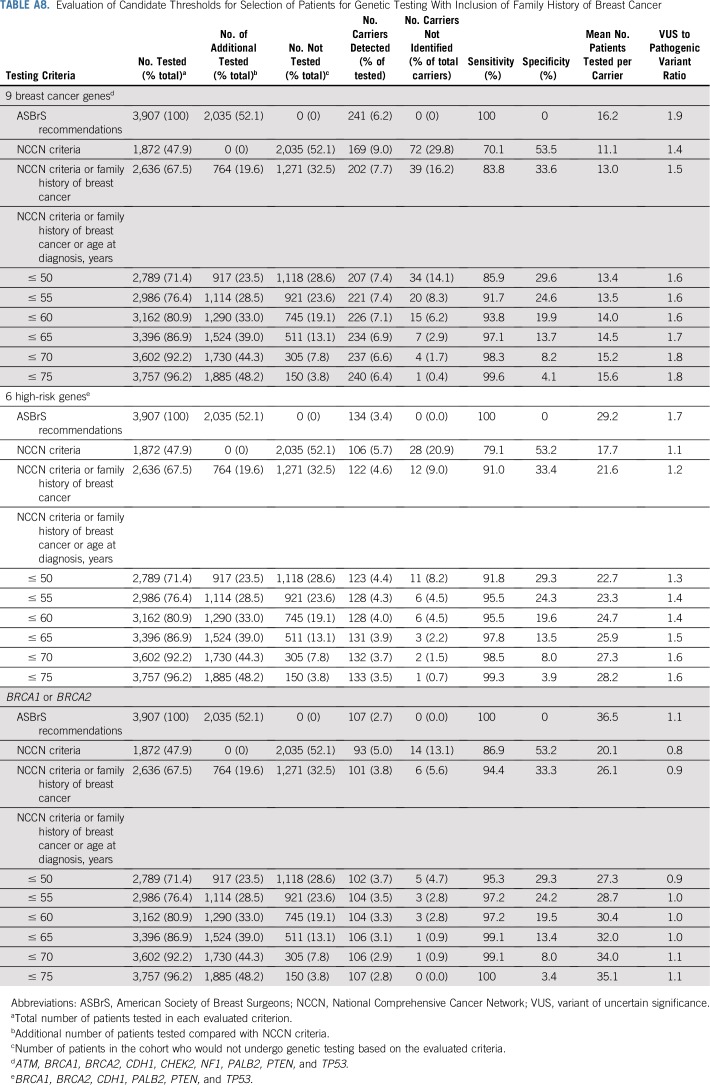
Next, we explored the potential impact of combining additional age at diagnosis of breast cancer and family history of breast cancer criteria with the NCCN criteria on the sensitivity of the selection guidelines (Table 3). Expansion of NCCN criteria to include all women diagnosed with breast cancer at ≤ 65 years of age increased the sensitivity for pathogenic variants in BRCA1 or BRCA2 to > 98%. These criteria required testing of an additional 31% of women, leaving 21% untested, and yielding a specificity of 22%. Similar results were seen when evaluating 12 predisposition genes (Appendix Table A6, online only). Importantly, v1.2020 guidelines recommend excluding women with a breast cancer diagnosis at > 65 years of age and no family history of cancer from testing. Among 511 women in this category in this study, germline pathogenic variants were detected in only 0.2% in BRCA1 or BRCA2, 0.6% in high-risk genes, and 1.4% in the 9 predisposition genes (Appendix Table A7, online only). Separately, expansion of NCCN criteria to include all women with a family history of breast cancer increased the sensitivity to 84% for pathogenic variants in the 9 predisposition genes, 91% for the 6 high-risk genes, and 94% for BRCA1 or BRCA2 (Appendix Table A8, online only). When combining both breast cancer family history and age at diagnosis of breast cancer with NCCN criteria, > 90% sensitivity for the 9 predisposition genes was observed for women diagnosed at ≤ 55 years of age. However, several carriers of pathogenic variants < 65 years of age did not qualify for testing (Appendix Table A8, online only). Appendix Table A9 (online only) shows a comparison of patient and tumor characteristics between women in the MCBCS and SEER Iowa Registry.
DISCUSSION
In a series of women with breast cancer recruited in a tertiary care center, the performance of NCCN hereditary cancer testing criteria was compared with ASBrS recommendations. The detection of pathogenic variants in 9 established predisposition genes in 9.0% of women meeting NCCN criteria was consistent with results from clinically tested cohorts.17,18,25 However, in contrast to prior studies,2,3,26 women meeting NCCN criteria had a higher frequency of germline pathogenic variants than women not meeting criteria (9.0% v 3.5%). Despite this, approximately 30% of women with pathogenic variants in the 9 predisposition genes and 13% with pathogenic variants in BRCA1 or BRCA2 did not qualify for testing by NCCN criteria. Thus, this study confirms that NCCN criteria are not optimal for selection of women with breast cancer for breast cancer predisposition gene testing.
Differences in results between studies may be explained by the study participants and the genes evaluated. Prior studies included women who may have undergone genetic testing despite not meeting NCCN criteria and also included several genes, such as MUTYH, that are not associated with increased breast cancer risk. In addition, some studies included women referred for testing over several years without taking updates in the NCCN criteria into account. The evaluation of the most recent version of the NCCN criteria (v1.2020) for pathogenic variants in established breast cancer genes using a uniform set of variables is a significant strength of this study. In addition, to fully evaluate the clinical utility of testing criteria, the sensitivity for germline pathogenic variants in BRCA1 or BRCA2 only, 6 high-risk genes, and 9 actionable predisposition genes were considered. Identification of a pathogenic variant in BRCA1 or BRCA2 can lead to significant changes in patient management through additional mammographic and magnetic resonance imaging screening, risk-reducing prophylactic surgeries,27-29 and systemic treatment,30-32 which may lead to improved survival. However, changes in management are limited to additional cancer screening for the moderate-risk predisposition genes.1
Although ASBrS criteria detect a substantially larger number of germline pathogenic variants than the NCCN criteria, there are challenges associated with testing all women with breast cancer.6 First, the substantially higher number of women tested, estimated at another 52% in this study, will lead to increased costs. Second, the added volume may exacerbate current unmet needs for genetic services and counseling.33,34 Third, more VUS will be detected, which may lead to anxiety35 and unwarranted interventions.36 Fourth, the clinical utility of testing women diagnosed with breast cancer > 65 years of age is not fully understood. Similar concerns about testing everyone with a breast cancer diagnosis have been raised in several commentaries,6,12,37 including a recent position statement by the American College of Medical Genetics and Genomics.5
As an alternative to adopting testing of all women, this study demonstrates that expanding the current NCCN criteria to include all women diagnosed with breast cancer at age ≤ 65 years has the potential to achieve > 90% sensitivity for 9 predisposition genes and 6 high-risk genes, and > 98% sensitivity for BRCA1 or BRCA2. These criteria reduced the proportion of women tested by 21% and decreased the VUS-to-pathogenic variant ratio compared with the ASBrS recommendations, which may translate into cost savings and lesser burden on genetic services. Importantly, this approach captured all young pathogenic variant carriers, and only older women with a low likelihood of pathogenic variants did not qualify for testing.38 The recently updated NCCN criteria (v1.2020) recommend against genetic testing in women > 65 years of age with no family history of cancer.1 This study found frequencies of germline pathogenic variants in the BRCA1 or BRCA2 and 6- or 9-gene categories of < 1.5% for women > 65 years of age without a family history and supports these recommendations.
Recently, the US Preventive Services Task Force (USPSTF) also recommended that asymptomatic women with a personal history of breast cancer should be screened by primary care clinicians for a referral to genetic counseling services.39 Although we acknowledge the significance of the USPSTF recommendations in cancer-free women in the primary care setting, the sensitivity of the risk assessment tools recommended by USPSTF in women with breast cancer is not clearly defined. In addition, several of these tools do not take a personal history of breast cancer into account. This may result in undertesting and failure to detect those with pathogenic variants among women with breast cancer.40-42 Probability models have also been added to the v1.2020 NCCN guidelines. Women who do not meet NCCN criteria but have a probability of > 5% of a BRCA1 or BRCA2 pathogenic variant based on prior-probability models now qualify for genetic testing, and those with 2.5% probability can be considered for testing. However, in this study, neither the 5% or 2.5% probability thresholds based on the Tyrer-Cuzick model changed the sensitivity of testing criteria for BRCA1 or BRCA2 in women with breast cancer. Thus, the utility of the probability models for selecting more women with a family history of cancer for testing is unclear. Additional studies will be needed to address this question.
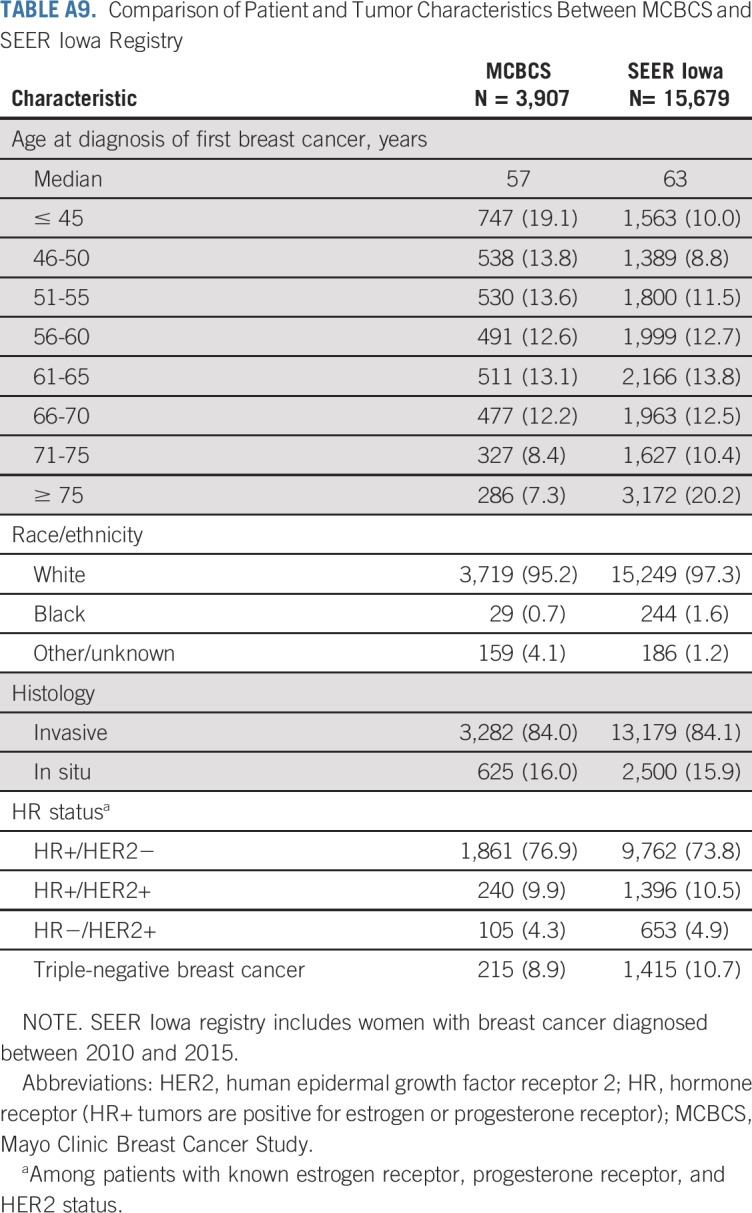
The study sample was representative of women with breast cancer evaluated at a tertiary cancer center but was enriched for women diagnosed at < 46 years of age compared with patients with breast cancer reported in the SEER Iowa Registry43 (Appendix Table A9, online only). However, the proportion of women diagnosed between the ages of 50 and 75 years, the racial composition, and the distribution of clinical tumor subtype defined by hormone receptor status were similar to patients with breast cancer in the SEER Iowa Registry (Appendix Table A9). In addition, the proportion of women with a family history of breast cancer was similar to other studies of unselected women with breast cancer from tertiary medical centers44,45 and a population-based study of women with breast cancer.46 Additional studies will be needed to determine whether these findings can be applied to patients with breast cancer in the general population. Meanwhile, this study of unselected patients with breast cancer from a tertiary cancer center provides much-needed information on sensitivity and specificity of testing criteria, which will help guide personalized decision making on genetic testing.
This study has several limitations. The cost effectiveness of testing criteria was not evaluated. Although few models have suggested cost effectiveness of population-based testing,47-49 these models have not been validated in clinical practice.50 Another limitation of the study is that specific criteria within the NCCN guidelines were not evaluated, allowing for the potential erroneous classification of some of the women. However, sensitivity analyses evaluating the impact of the missing information yielded results similar to the primary analyses. Finally, the study sample was predominantly white, which limits the application of the findings to a racially diverse population.
Among women with breast cancer, those meeting NCCN criteria were more likely to carry a germline pathogenic variant. However, a substantial proportion of carriers do not qualify for testing by NCCN criteria. Although ASBrS recommendations are more sensitive than NCCN criteria, a substantially larger proportion of women with breast cancer must undergo testing. In a large unselected series of women with breast cancer, we demonstrate that expanding the NCCN testing criteria to include all women diagnosed with breast cancer at or before the age of 65 years has the potential to improve the sensitivity of germline genetic testing without the need for evaluation of all women with breast cancer.
APPENDIX
Information
QIAsEquation 37 gene custom amplicon panel.
Germline DNA samples were analyzed for pathogenic variants in cancer predisposition genes using a custom QIAseq 37-gene amplicon panel Table A1. Genomic DNA samples were subjected to multiplex amplicon-based analysis of 746 target regions covering all coding regions and consensus splice sites from 37 cancer predisposition genes, including the 12 established breast cancer predisposition genes, including BRCA1 and BRCA2 (BRCA1, BRCA2, ATM, BARD1, CDH1, CHEK2, NF1, PALB2, PTEN, RAD51C, RAD51D, TP53). The QIAseq protocol has been optimized for high-throughput robotic processing of DNA samples (Hu C, et al: JAMA 319:2401-2409, 2018). Libraries were individually bar coded by dual indexing and sequenced in pools of 768 on a HiSeq4000. The median sequence read depth per nucleotide was 200×, with 99.7% of target regions yielding > 20× reads in all samples.
Three validation studies were conducted to assess the accuracy of the QIAseq custom panel. The first blinded study of 48 samples containing known pathogenic variants in the predisposition genes identified all 48 pathogenic variants, including 2 BRCA1 large genomic rearrangements for the sensitivity of 100%. No false-positive variants were identified for a specificity of 100% (Hu C, et al: JAMA 319:2401-2409, 2018). The second study evaluated 48 samples from patients with pancreatic cancer selected based on results from prior genetic testing. All 32 pathogenic variants, including 2 large genomic rearrangements, were identified in a blinded analysis. The sensitivity and specificity for this study were 100% (Hu C, et al: JAMA 319:2401-2409, 2018). The third study involved QIAseq analysis of 96 samples from patients with breast cancer in a Mayo Clinic breast cancer registry that had received clinical genetic testing. All 100 pathogenic variants, including 8 large genomic rearrangements, were identified in a blinded analysis for 100% sensitivity and specificity. Overall, the QIAseq assay has greater than 99% analytical sensitivity and specificity for single nucleotide variants, insertions and deletions < 15 bp in length, and exon-level deletions and duplications.
Methods
Germline sequencing and bioinformatics analysis.
Pooled sample libraries from 768 samples were subjected to paired-end 150 bp sequencing in each lane of a HiSeq4000. The median nucleotide coverage was 200×. Reads were trimmed with Cutadapt v1.10 (Martin M, https://doi.org/10.14806/ej.17.1.200) and aligned with bwa-mem (Li H, https://arxiv.org/abs/1303.3997). Sequence realignment, recalibration, haplotype calling, and depth of coverage were conducted using Genome Analysis Toolkit v3.4-46 (DePristo MA, et al: Nat Genet 43:491-498, 2011). Copy number variation (CNV) was detected with Pattern CNV v1.1.3 (Wang C, et al: Bioinformatics 30:2678-2680, 2014). Annotation of variants was provided through the BioR toolkit (Kocher JP, et al: Bioinformatics 30:1920-1922, 2014) leveraging dbNSFP v3.0 (Liu X, et al: Hum Mutat 37:235-241, 2016), ClinVar (Landrum MJ, et al: Res 44:D862-D868, 2016), and CAVA (Münz M, et al: Genome Med 7:76, 2015). Population frequencies of pathogenic variants were derived from Genome Aggregation Database (gnomAD: http://gnomad.broadinstitute.org) and Exome Aggregation Consortium non-TCGA controls. Pathogenic variants were viewed with VCF-Miner (Hart SN, et al: Brief Bioinform 17:346-351, 2016). A 5-tier system was used to classify pathogenic variants based on the American College of Medical Genetics and the Association for Molecular Pathology guidelines (Richards S, et al: Genet Med 17:405-424, 2015).
Definition of terms.
Sensitivity was defined as the ability of the testing criteria to correctly designate women with germline pathogenic variants as qualifying for genetic testing. Sensitivity of different testing criteria was separately evaluated for pathogenic variants in BRCA1 or BRCA2, 6 high-risk genes, and 9 breast cancer predisposition genes. It was estimated as follows:
Specificity was defined as the ability of the testing criteria to correctly reject women without a germline pathogenic variant as not qualifying for genetic testing. Specificity of different testing criteria was separately evaluated for pathogenic variants in BRCA1 or BRCA2, 6 high-risk genes, and 9 breast cancer predisposition genes. It was estimated as follows:
The variant of uncertain significance (VUS)-to-pathogenic variant ratio was defined as the ratio of the number of women with VUS results to the number of women with germline pathogenic variants identified by the testing criteria. This was estimated separately for BRCA1 or BRCA2, 6 high-risk genes, and 12 breast cancer predisposition genes.
FIG A1.
Sample selection. MCBCS, Mayo Clinic Breast Cancer Study; NCCN, National Comprehensive Cancer Network.
FIG A2.
Frequency of germline pathogenic variants by family history of breast cancer according to the number of first- or second-degree relatives with breast cancer.
FIG A3.
Frequencies of variants of uncertain significance by gene.
TABLE A1.
List of Genes Included in the QIAseq Panel
TABLE A2.
Subcategories of BRCA1/2 Testing Criteria Evaluated in This Study Along With the Number of Patients in the Study Who Met the Criteria
TABLE A3.
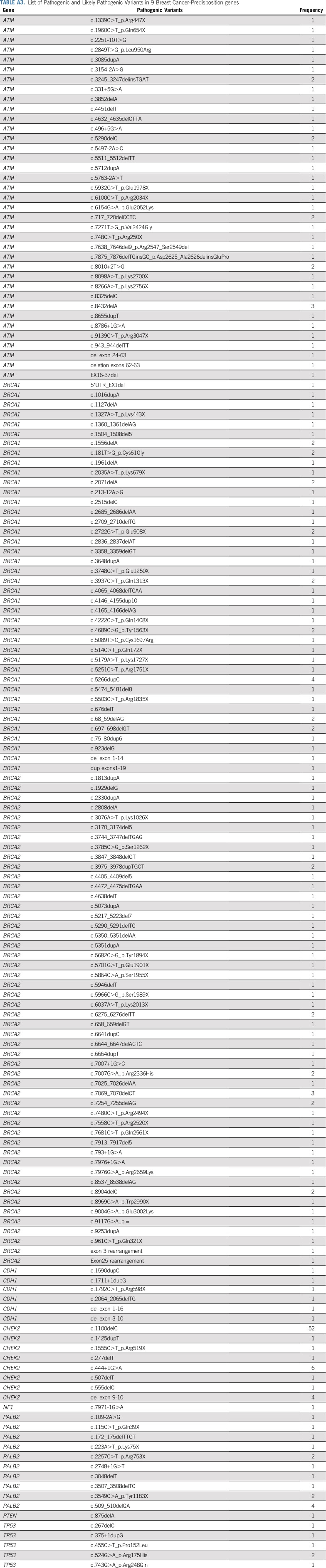
List of Pathogenic and Likely Pathogenic Variants in 9 Breast Cancer-Predisposition genes
TABLE A4.
Sensitivity Analysis Considering All Patients With a Family History of Prostate Cancer to Have Met the NCCN Criteria
TABLE A5.
Sensitivity Analysis Considering all Patients With a Third-Degree Family Member With Breast, Ovarian, Pancreatic, or Prostate Cancer to Have Met the NCCN Criteria
TABLE A6.
Evaluation of Candidate Thresholds for Pathogenic Variants in 12 Breast Cancer Predisposition Genes
TABLE A7.
Frequency of Germline Pathogenic Variants Among Patients Not Meeting NCCN Guidelines by Age of Diagnosis and Family History of Breast Cancer
TABLE A8.
Evaluation of Candidate Thresholds for Selection of Patients for Genetic Testing With Inclusion of Family History of Breast Cancer
TABLE A9.
Comparison of Patient and Tumor Characteristics Between MCBCS and SEER Iowa Registry
SUPPORT
Supported in part by National Institutes of Health (NIH) Grants No. CA116167, CA176785, CA192393, and CA225662, an NIH Specialized Program of Research Excellence in Breast Cancer (CA116201), and the Breast Cancer Research Foundation.
Listen to the podcast by Dr Cobain at http://ascopubs.org/journal/jco/podcasts
AUTHOR CONTRIBUTIONS
Conception and design: Siddhartha Yadav, Steven N. Hart, Eric C. Polley, Nicole Sandhu, Matthew P. Goetz, Judy C. Boughey, Fergus J. Couch
Financial support: Matthew P. Goetz, Fergus J. Couch
Administrative support: Patrick Fitz-Gibbon, Fergus J. Couch
Provision of study materials or patients: Daniela L. Stan, Lonzetta Neal, Janet E. Olson, Matthew P. Goetz, Fergus J. Couch
Collection and assembly of data: Siddhartha Yadav, Chunling Hu, Kun Y. Lee, Tricia Lindstrom, Sebastian Armasu, Patrick Fitz-Gibbon, Karthik Ghosh, Sandhya Pruthi, Lonzetta Neal, Christine Klassen, Janet E. Olson, Tanya L. Hoskin, Matthew P. Goetz, Kathryn J. Ruddy, Fergus J. Couch
Data analysis and interpretation: Siddhartha Yadav, Chunling Hu, Steven N. Hart, Nicholas Boddicker, Eric C. Polley, Jie Na, Rohan Gnanaolivu, Kun Y. Lee, Karthik Ghosh, Daniela L. Stan, Sandhya Pruthi, Nicole Sandhu, Deborah J. Rhodes, Prema P. Peethambaram, Tufia C. Haddad, Janet E. Olson, Tanya L. Hoskin, Matthew P. Goetz, Susan M. Domchek, Judy C. Boughey, Fergus J. Couch
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Evaluation of Germline Genetic Testing Criteria in a Hospital-Based Series of Women With Breast Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/journal/jco/site/ifc.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Eric C. Polley
Research Funding: Grail
Sandhya Pruthi
Patents, Royalties, Other Intellectual Property: Mytonomy (Inst)
Tufia C. Haddad
Consulting or Advisory Role: Tersera
Research Funding: Takeda (Inst)
Matthew P. Goetz
Consulting or Advisory Role: Lilly, bioTheranostics, Genomic Health, Novartis, Eisai, Sermonix, Context Therapeutics, Pfizer
Research Funding: Lilly, Pfizer
Patents, Royalties, Other Intellectual Property: Methods and materials for assessing chemotherapy responsiveness and treating cancer, methods and materials for using butyrylcholinesterases to treat cancer, development of human tumor xenografts from women with breast cancer treated with neoadjuvant chemotherapy (Inst).
Travel, Accommodations, Expenses: Lilly
Susan M. Domchek
Honoraria: AstraZeneca, Clovis Oncology, Bristol-Myers Squibb
Research Funding: AstraZeneca (Inst), Clovis Oncology (Inst), Pharmamar (Inst)
Judy C. Boughey
Research Funding: Myriad Genetics (Inst)
Patents, Royalties, Other Intellectual Property: Patent pending: Methods and materials for assessing chemotherapy responsiveness and treating cancer (Inst).
Kathryn J. Ruddy
Stock and Other Ownership Interests: Merck, Pfizer
Patents, Royalties, Other Intellectual Property: My husband is a co-inventor of technology licensed by Mayo Clinic to AliveCor (Mountain View, CA), which makes a smartphone-enabled remote ECG monitoring system (I).
Fergus J. Couch
Consulting or Advisory Role: AstraZeneca
Speakers' Bureau: Ambry Genetics, Qiagen
Research Funding: Grail
Travel, Accommodations, Expenses: Grail, Qiagen
Other Relationship: Ambry Genetics
No other potential conflicts of interest were reported.
REFERENCES
- 1.NCCN Clinical Practice Guidelines in Oncology . Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic Version 1. National Comprehensive Cancer Network: ; https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf [DOI] [PubMed] [Google Scholar]
- 2.Beitsch PD, Whitworth PW, Hughes K, et al. Underdiagnosis of hereditary breast cancer: Are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37:453–460. doi: 10.1200/JCO.18.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang S, Axilbund JE, O’Leary E, et al. Underdiagnosis of hereditary breast and ovarian cancer in Medicare patients: Genetic testing criteria miss the mark. Ann Surg Oncol. 2018;25:2925–2931. doi: 10.1245/s10434-018-6621-4. [DOI] [PubMed] [Google Scholar]
- 4.Manahan ER, Kuerer HM, Sebastian M, et al. Consensus guidelines on genetic’ testing for hereditary breast cancer from the American Society of Breast Surgeons. Ann Surg Oncol. 2019;26:3025–3031. doi: 10.1245/s10434-019-07549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. doi: 10.1038/s41436-019-0712-x. Pal T, Agnese D, Daly M, et al: Points to consider: Is there evidence to support BRCA1/2 and other inherited breast cancer genetic testing for all breast cancer patients? A statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med . [Epub ahead of print on December 13, 2019] [DOI] [PubMed] [Google Scholar]
- 6.Milliron KJ, Griggs JJ. Advances in genetic testing in patients with breast cancer, high-quality decision making, and responsible resource allocation. J Clin Oncol. 2019;37:445–447. doi: 10.1200/JCO.18.01952. [DOI] [PubMed] [Google Scholar]
- 7.Tung N, Domchek SM, Stadler Z, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13:581–588. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copur MS, Jonglertham P, Zusag T. Should all patients with a diagnosis of breast cancer undergo expanded panel testing? J Clin Oncol. 2019;37:2175–2176. doi: 10.1200/JCO.19.00064. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopal PS, Catenacci DVT, Olopade OI. The time for mainstreaming germline testing for patients with breast cancer is now. J Clin Oncol. 2019;37:2177–2178. doi: 10.1200/JCO.19.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beitsch P, Hughes K, Whitworth P. Reply to M.S. Copur et al, A. Taylor et al, and P.S. Rajagopal et al. J Clin Oncol. 2019;37:2178–2180. doi: 10.1200/JCO.19.00798. [DOI] [PubMed] [Google Scholar]
- 11.Sorscher S. Universal multigene panel testing in all breast cancer patients. Am J Med. 2019;132:e765–e766. doi: 10.1016/j.amjmed.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Robson M, Domchek S. Broad application of multigene panel testing for breast cancer susceptibility—Pandora’s box is opening wider. JAMA Oncol. 2019;5:1687. doi: 10.1001/jamaoncol.2019.4004. [DOI] [PubMed] [Google Scholar]
- 13.Wood ME, Bedrosian I. Hot topic: Should all women with breast cancer undergo genetic testing? Curr Breast Cancer Rep. 2019;11:381–384. [Google Scholar]
- 14.John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: An infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319:2401–2409. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadav S, Couch FJ. Germline genetic testing for breast cancer risk: The past, present, and future. Am Soc Clin Oncol Educ Book. 2019;39:61–74. doi: 10.1200/EDBK_238987. [DOI] [PubMed] [Google Scholar]
- 17.Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buys SS, Sandbach JF, Gammon A, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 19.Shimelis H, LaDuca H, Hu C, et al. Triple-negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J Natl Cancer Inst. 2018;110:855–862. doi: 10.1093/jnci/djy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. doi: 10.1200/PO.16.00066. Kurian AW, Hughes E, Handorf EA, et al: Breast and ovarian cancer penetrance estimates derived from germline multiple-gene sequencing results in women. JCO Precis Oncol 10.1200/PO.16.00066. [DOI] [PubMed] [Google Scholar]
- 21.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23:1111–1130. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]
- 22.IBIS Breast Cancer Risk Evaluation Tool http://www.ems-trials.org/riskevaluator/
- 23.Chompret A, Abel A, Stoppa-Lyonnet D, et al. Sensitivity and predictive value of criteria for p53 germline mutation screening. J Med Genet. 2001;38:43–47. doi: 10.1136/jmg.38.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.1136/jmg.2009.074237. Fitzgerald RC, Hardwick R, Huntsman D, et al: Hereditary diffuse gastric cancer: Updated consensus guidelines for clinical management and directions for future research. J Med Genet 47:436-444, 2010 [Erratum: J Med Genet 48:216, 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurian AW, Ward KC, Hamilton AS, et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA Oncol. 2018;4:1066–1072. doi: 10.1001/jamaoncol.2018.0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neben CL, Zimmer AD, Stedden W, et al. Multi-gene panel testing of 23,179 individuals for hereditary cancer risk identifies pathogenic variant carriers missed by current genetic testing guidelines. J Mol Diagn. 2019;21:646–657. doi: 10.1016/j.jmoldx.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Kaas R, Verhoef S, Wesseling J, et al. Prophylactic mastectomy in BRCA1 and BRCA2 mutation carriers: Very low risk for subsequent breast cancer. Ann Surg. 2010;251:488–492. doi: 10.1097/SLA.0b013e3181c3c36d. [DOI] [PubMed] [Google Scholar]
- 28.van Sprundel TC, Schmidt MK, Rookus MA, et al. Risk reduction of contralateral breast cancer and survival after contralateral prophylactic mastectomy in BRCA1 or BRCA2 mutation carriers. Br J Cancer. 2005;93:287–292. doi: 10.1038/sj.bjc.6602703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalfe K, Gershman S, Ghadirian P, et al. Contralateral mastectomy and survival after breast cancer in carriers of BRCA1 and BRCA2 mutations: Retrospective analysis. BMJ. 2014;348:g226. doi: 10.1136/bmj.g226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. doi: 10.1056/NEJMoa1706450. Robson M, Im SA, Senkus E, et al: Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 377:523-533, 2017 [Erratum: N Eng J Med 377:1700, 2017] [DOI] [PubMed] [Google Scholar]
- 31.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoll K, Kubendran S, Cohen SA. The past, present and future of service delivery in genetic counseling: Keeping up in the era of precision medicine. Am J Med Genet C Semin Med Genet. 2018;178:24–37. doi: 10.1002/ajmg.c.31602. [DOI] [PubMed] [Google Scholar]
- 34.American Society of Clinical Oncology The state of cancer care in America, 2016: A report by the American Society of Clinical Oncology. J Oncol Pract. 2016;12:339–383. doi: 10.1200/JOP.2015.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culver JO, Brinkerhoff CD, Clague J, et al. Variants of uncertain significance in BRCA testing: Evaluation of surgical decisions, risk perception, and cancer distress. Clin Genet. 2013;84:464–472. doi: 10.1111/cge.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman-Andrews L. The known unknown: The challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci. 2018;4:648–657. doi: 10.1093/jlb/lsx038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. doi: 10.1038/s41436-019-0721-9. Domchek SM: Germline genetic testing for breast cancer: Which patients? What genes? Genet Med 10.1038/s41436-019-0721-9 [Epub ahead of print on December 13, 2019] [DOI] [PubMed] [Google Scholar]
- 38.Chavarri-Guerra Y, Hendricks CB, Brown S, et al. The burden of breast cancer predisposition variants across the age spectrum among 10 000 patients. J Am Geriatr Soc. 2019;67:884–888. doi: 10.1111/jgs.15937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. doi: 10.1001/jama.2019.10987. Owens DK, Davidson KW, Krist AH, et al: Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer: US Preventive Services Task Force recommendation statement. JAMA 322:652-665, 2019 [Erratum: JAMA 322:1830, 2019] [DOI] [PubMed] [Google Scholar]
- 40.Domchek S, Robson M. Broadening criteria for BRCA1/2 evaluation: Placing the USPSTF Recommendation in context. JAMA. 2019;322:619–621. doi: 10.1001/jama.2019.9688. [DOI] [PubMed] [Google Scholar]
- 41.Rajagopal PS, Nielsen S, Olopade OI. USPSTF recommendations for BRCA1 and BRCA2 testing in the context of a transformative national cancer control plan. JAMA Netw Open. 2019;2:e1910142. doi: 10.1001/jamanetworkopen.2019.10142. [DOI] [PubMed] [Google Scholar]
- 42.Newman L. US Preventive Services Task Force breast cancer recommendation statement on risk assessment, genetic counseling, and genetic testing for BRCA-related cancer. JAMA Surg. 2019;154:895. doi: 10.1001/jamasurg.2019.3184. [DOI] [PubMed] [Google Scholar]
- 43. National Cancer Institute: Iowa Registry. https://seer.cancer.gov/registries/iowa.html.
- 44.Kim S-W, Lee CS, Fey JV, et al. Prevalence of BRCA2 mutations in a hospital based series of unselected breast cancer cases. J Med Genet. 2005;42:e5. doi: 10.1136/jmg.2004.025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziogas A, Gildea M, Cohen P, et al. Cancer risk estimates for family members of a population-based family registry for breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:103–111. [PubMed] [Google Scholar]
- 47.Manchanda R, Patel S, Gordeev VS, et al. Cost-effectiveness of population-based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 mutation testing in unselected general population women. J Natl Cancer Inst. 2018;110:714–725. doi: 10.1093/jnci/djx265. [DOI] [PubMed] [Google Scholar]
- 48.Manchanda R, Gaba F. Population based testing for primary prevention: A systematic review. Cancers (Basel) 2018;10:E424. doi: 10.3390/cancers10110424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L, Brentnall A, Patel S, et al. A cost-effectiveness analysis of multigene testing for all patients with breast cancer. JAMA Oncol. 2019;5:1718. doi: 10.1001/jamaoncol.2019.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wentzensen N, Berg CD. Population testing for high penetrance genes: Are we there yet? J Natl Cancer Inst. 2018;110:687–689. doi: 10.1093/jnci/djx282. [DOI] [PMC free article] [PubMed] [Google Scholar]