Abstract
The thermostable α-amylase from germinating sword bean (Canavalia gladiata (Jacq.) DC.) seeds (CgAmy) was successfully purified by a combination of ammonium sulphate fractionation and Epoxy-activated Sepharose 6B affinity chromatography. The purified α-amylase showed 507.8 fold with a specific activity of 750.0 U/mg. SDS-PAGE of the purified enzyme revealed a single protein band of 50.0 kDa. Purified enzyme was confirmed as α-amylase type by LC-MS/MS analysis and activity on specific substrate of ethylidene-pNP-G7. The CgAmy revealed extreme activity at a high temperature of 50.0–70.0°C with optimum activity at 70.0°C. The optimal pH of enzyme activity was observed at 6.0. The CgAmy exhibited stability in pH range of 5.0–8.0 and highly thermostable with a temperature of 40.0–60.0°C. The kinetic parameters Km for hydrolysis of starch were found to be 3.12 mg/ml. The α-amylase activity was enhanced in the presence of Co2+ and β-mercaptoethanol. While, Na2+, K2+, Ca2+, Mg2+, Zn2+, Ba2+, Fe2+ and Cd2+ slightly inhibited α-amylase activity. Interestingly, the CgAmy displayed stability towards some organic solvents and detergents. Stability at high temperature and some metal ions, organic solvents and detergents indicated that this enzyme has potential for various applications.
Keywords: α-amylase, characterization, enzymes, purification, Sword bean (Canavalia gladiata (Jacq.) DC.) seeds
Introduction
α-Amylase (1,4-α-D-glucan-4-glucanohydrolase, EC 3.2.1.1) belongs to glycosyl hydrolase (GH-13 family) class which catalyses random the hydrolysis of internal α-1,4 glycosidic bonds in polysaccharides resulting in an α-anomeric configuration of oligosaccharides (van der Maarel et al. 2002). The source of this enzyme is microorganisms, animals, plants, especially in germinating seeds (Muralikrishna and Nirmala 2005; Singh et al. 2017; Sundarram and Murthy 2014). In plants, α-amylase plays a very important role in starch degradation in stored starch during seeds germination releasing sugars for the proper growth of plants (Xie et al. 2007). Enzymatic processes have been used in various industries due to several advantages such as fast in action, specific of the reaction and save energy, raw materials, chemical and provide more environment and consumer safe solution compared to conventional processes (Jegannathan and Nielsen 2013). Cereal α-amylases are a good choice for food and biotechnological application (Muralikrishna and Nirmala 2005). The α-amylases is one of the most critical industrial enzyme which holds the maximum about 25% of the industrial enzyme market with various applications such as food, detergents, textiles, biorefinery, brewing, fermentation, pharmaceuticals, and paper industries etc. (Gupta et al. 2003; Sindhu et al. 2017). The demand for α-amylase has increased in various industries but the production and properties of this enzyme have been limited in terms of specificity, stability and catalytic efficiency (Choi et al. 2015; Sivaramakrishnan et al. 2006). Therefore, there is a need to find novel sources that produce α-amylases with desired characteristics for specific applications. Thermostable enzymes are a desirable characteristic of extreme temperature in various industry processes (Sindhu et al. 2017). In the past, very few researches of α-amylases from plant sources have been reported. However, thermostable α-amylases have been reported from Vicia faba (65°C), red pitaya (Hylocereus polyrhizus) peel (70°C), wheat (Triticum aestivum) seeds (68°C), soybean (Glycine max) seeds (70°C), and mung bean (Vigna radiata) seeds (65°C). Sword bean (Canavalia gladiate (Jacq.) DC.) seeds which widely cultivated in the local areas are interesting for α-amylases source and increasing commercial value. The seed-pods are approximately 20–40 cm long and 3.5–5.0 cm broad including on average 8 to 16 of very large and high stored starch seeds (Vadivel and Janardhanan 2004). The seed germination process is predicted from α-amylases activity.
In this research, we report the purification, biochemical characterization, and discovery of thermostable, surfactant and organic solvent stable α-amylase from newly source germinating sword bean (Canavalia gladiata (Jacq.) DC.) seeds.
Materials and methods
Materials
Seeds of sword bean were collected from Nakhon Ratchasima province, Thailand. They were surface sterilized with 1% (v/v) sodium hypochlorite solution for 10 min followed by thoroughly washing with distilled water. Seeds were imbibed 12 h in distilled water and then placed to germinate in dark over moist filter paper on a moist sand bed for 48 h (Singh et al. 2017).
Enzyme extraction and purification
All purification steps were carried out at 4°C. Germinated seeds (400 g) were homogenized in 1,200 ml chilled extraction buffer (50 mM sodium acetate buffer, pH 5.5) (Singh et al. 2017) using a laboratory blender. The extract was filleted through two layers of pre-washed cheesecloth. The supernatant was obtained by centrifugation at 21,000×g for 25 min.
Ammonium sulphate fractionation
The crude extract was precipitated in range 35–65% ammonium sulphate saturation (Kumari et al. 2010) and then stirred for 2.5 h at 4°C. The protein precipitate was collected by centrifugation at 21,000×g for 25 min. The pellet was dissolved in minimum volume of extraction buffer (50 mM sodium acetate buffer, pH 5.5) and dialyzed with the same buffer for four buffer changes to remove ammonium sulphate.
Epoxy-activated Sepharose 6B affinity chromatography
The dialyzed supernatant (40 mgProtein) were then loaded onto pre-equilibrated epoxy-activated sepharose 6B ligated with β-cyclodextrin columns with 50 mM acetate buffer (pH 6.0) containing 5 mM CaCl2 (Singh and Kayastha 2014; Vretblad 1974) at flow rate of 0.5 ml/min. The column was washed unbound protein with washing buffer (20 mM acetate buffer (pH 6.0) containing 25 mM CaCl2 and 200 mM NaCl). Elution was carried out with a β-cyclodextrin (10 mg/ml) in the washing buffer. The active fractions (2 ml each) were pooled and then dialysed with 50 mM acetate buffer (pH 6.0) containing 5 mM CaCl2.
Amylase activity assay
The amylase activity was assayed based on the standard method (Miller 1959) with some modification. The reaction mixture contained 0.5 ml of the enzyme and 0.5 ml of 1% w/v of soluble starch in 100 mM sodium acetate buffer pH 5.5. The reaction mixture was incubated for 10 min at 37°C. The reaction was stopped by the addition of 1 ml of DNS reagent and then boiled for 5 min. The released reducing sugar was measured using a spectrophotometer (BioMateTM-3, Thermo Scientific) at 520 nm by using glucose as a standard reducing sugar. One unit of α-amylase activity was defined as the amount of enzyme that produced 1 µmol of glucose per minute under the assay condition.
Determination of protein concentration
Protein concentration was determined according to the Bradford method (Bradford 1976) using bovine serum albumin (BSA) as protein standard. For purification, the protein was determined by measuring the absorbance at 280 nm.
Determination of purity and molecular weight
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the previous method (Laemmli 1970). The gel was stained with Coomassie Brilliant Blue R-250. The molecular weight of the enzyme was estimated using a protein marker with sizes ranging from 17 to 250 kDa (ENZMART Biotech).
Zymographic method
The samples were prepared under non-reducing conditions and separated on 13% polyacrylamide gels containing 0.1% w/v soluble starch (Posoongnoen et al. 2015). The gel was washed with 1% v/v triton X-100 to remove SDS and then incubated with 0.2% w/v starch in 50 mM sodium acetate buffer (pH 5.5) for 6 h at 37°C. The amylase activity band was detected by staining with activity staining solution (1% acetic acid, 10 mM I2 and 14 mM KI).
Optimum pH and pH stability
The optimum pH was determined using amylase activity assay at different pH values using various buffers: 100 mM sodium acetate buffer (pH range 3.7–5.6), 100 mM citrate phosphate buffer (pH range 5.0–6.5), 100 mM potassium phosphate buffer (pH range 6.0–7.6), 100 mM Tris-HCl buffer (pH range 7.0–9.0) at 37°C. The pH stability was evaluated by pre-incubating enzyme with 100 mM buffer at different pH values for 1 h at 4°C. The residual amylase activity was determined by amylase activity assay as mentioned above. The activity of the enzyme before incubation was regarded as 100% relative activity.
Optimum temperature and Heat stability
The optimum temperature was analysed by assaying amylase activity in 100 mM sodium acetate buffer pH 5.5 at temperatures ranging from 30 to 90°C. The heat stability was determined by pre-incubating enzyme at different temperature range in 30 to 90°C in 100 mM sodium acetate buffer pH 5.5 for 1 h. Then, the residual amylase activity was determined by the amylase activity assay. The activity of the enzyme before incubation was regarded as 100% relative activity.
Kinetic studies
The enzyme was determined activity in the presence of varying concentration of starch in a range of 3.0 to 9.0 mg/ml soluble starch in 100 mM potassium phosphate buffer pH 6.0 at 70°C using amylase activity assay. The data was determined of Michaelis constant (Km) and maximum velocity (Vmax) using Lineweaver-Burk plot.
Effect of metal ions and chemical reagents on the α-amylase activity
The influence of various metal ions (5 mM CaCl2, MgCl2, MnCl2, BaCl2, CoCl2, CuCl2, ZnCl2, NaCl and KCl) on enzyme activity was analysed. The metal ions were prepared by dissolving the metal ions in deionised water. The reaction mixture of 0.5 ml of purified enzyme with 0.5 ml of metal ion solution was pre-incubated at 4°C and pH 5.5 for 1 h. Then, the residual enzyme activity was determined using amylase activity assay.
For chemical reagents, the enzyme was pre-incubated with chemical reagents of 10 mM β-mercaptoethanol, 10 mM ethylene-diaminetetraacetic acid (EDTA), 5% v/v Triton X-100, 5% v/v Tween 20, 5% w/v SDS and 10% v/v of organic solvent (methanol, ethanol, acetone and hexane) at 4°C and pH 5.5 for 1 h. The residual enzyme activity was measured by the amylase activity assay. The activity of amylase in the absence of any additives was taken as 100% relative activity.
Substrate specificity
The activity of the enzyme against different substrates was determined using amylase activity assay. The enzyme was incubated with 0.1% w/v of substrates (starch, amylopectin, glycogen, β-cyclodextrin, and dextran) in 100 mM sodium acetate buffer (pH 5.5) for 10 min at 37°C. The released reducing sugar was analysed using the DNS method. Soluble starch was used as a reference substrate for enzyme activity comparisons.
Liquid chromatography electrospray ionization tandem mass spectroscopy (LC-ESI-MS/MS) and database searching
The single gel band of purified α-amylase was tryptic digested and analysed with a nano-liquid chromatography system (EASY-nLC II, Bruker) coupled to an ion trap mass spectrometer (Amazon Speed ETD, Bruker) equipped with an ESI nano-sprayer. Tryptic digest of bovine serum albumin (BSA) was used as a standard control of the experiment. Services were provided by Research Instrument Center, Khon Kaen University, Thailand. Proteins identification was performed by searching against the protein database from SwissProt using MASCOT MS/MS Ion Search program (www.matrixscience.com) with the initial searching parameters; Enzyme: Trypsin; carbamidomethylation (C) as fixed modification, and oxidation (HW) and oxidation (M) as variable modification; peptide mass tolerance of 0.5 Da and fragment mass tolerance of 0.5 Da; a peptide charge state of +1, +2, +3; instrument type: ESI-TRAP; and report top: Auto.
Statistical analysis
All samples were analysed three replicates. The results were reported as mean±standard error (SE). Variance was statistical determined by one-way ANOVA. The significant differences between means were defined at p≤0.05.
Results
Purification of α-amylase from sword bean (Canavalia gladiata (Jacq.) DC.) seeds
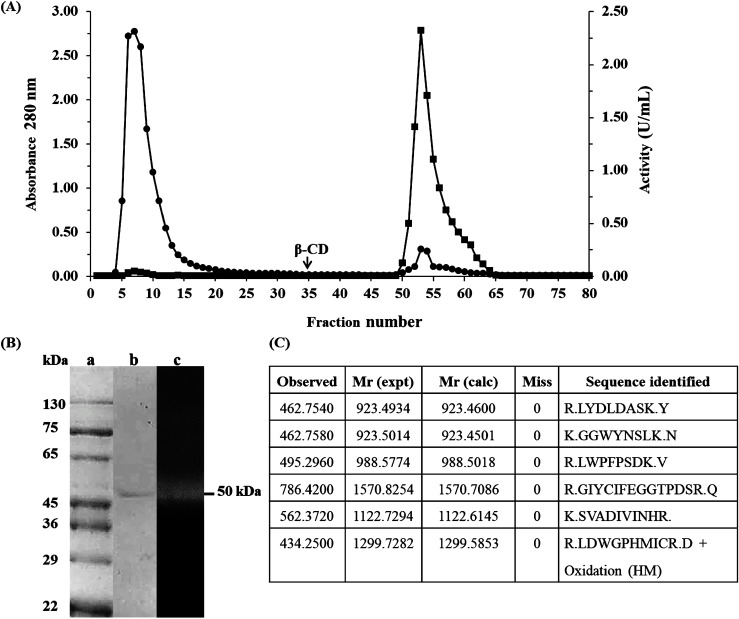
α-Amylase from germinated sword bean (Canavalia gladiata (Jacq.) DC.) seeds (CgAmy) was purified using ammonium sulphate fractionation and affinity chromatography. Ammonium sulphate fractionation revealed a purification fold of 2.92. Then, CgAmy was purified using Epoxy-activated Sepharose 6B affinity chromatography which was eluted as a single peak by β-cyclodextrin (Figure 1(A)). Finally, α-amylase was purified up to 507.78 folds with a specific activity of 750.00 U/mg and a yield of 14.05% (Table 1). The CgAmy was also confirmed α-amylase activity using specific substrate (ethylidene-pNP-G7) which is specifically cleaved by α-amylase. The result revealed that the purified enzyme catalyzed 736.57 nmol/min/ml of ethylidene-pNP-G7 indicating amylase from sword bean (Canavalia gladiata (Jacq.) DC.) seeds is α-type of amylase.
Figure 1. Purification and characterization of α-amylase: (A) Elution profile of purification using epoxy-activated Sepharose 6B linked with β-cyclodextrin (β-CD) for affinity chromatography. The fractions were analyzed enzyme activity (■) and protein concentration at A280 nm (●). (B) Electrophoresis pattern and amylase activity. Molecular weight markers (lane a) and purified enzyme (lane b) using SDS-PAGE, amylase activity by Zymographic method (lane c). (C) Digested peptides which showed match with α-amylase sequence from plant α-amylases.
Table 1. Purification of α-amylase from germinated sword bean (Canavalia gladiata (Jacq.) DC.) seeds.
| Purification steps | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 6,279.00 | 4,251.10 | 1.48 | 1 | 100 |
| 35–65% Ammonium sulphate fractionation | 1,853.18 | 429.14 | 4.32 | 2.92 | 29.51 |
| Epoxy activated sepharose 6B Affinity chromatography | 882.00 | 1.18 | 750.00 | 507.78 | 14.05 |
Electrophoresis and LC-ESI tandem mass spectroscopy
The purified enzyme revealed a single protein band on SDS-PAGE with a mobility equivalent to a molecular mass of 50.0 kDa (Figure 1(B)) indicating its homogeneity. The zymographic method exhibited a single clear band of amylase activity associated with the single protein band on SDS-PAGE (Figure 2 lane B) which confirmed the starch hydrolysis by the α-amylase. Single purified α-amylase band with a molecular mass of 50.0 kDa obtained from SDS-PAGE was excised, tryptic digested and submitted for LC-ESI-MS/MS analysis, resulted in high quality peptide mass fingerprint spectrum. The m/z ratio showed significant matches with mass data of α-amylase from Vigna mungo in the protein database (Figure 1(C)). The results clearly demonstrate that the purified enzyme is α-amylase.
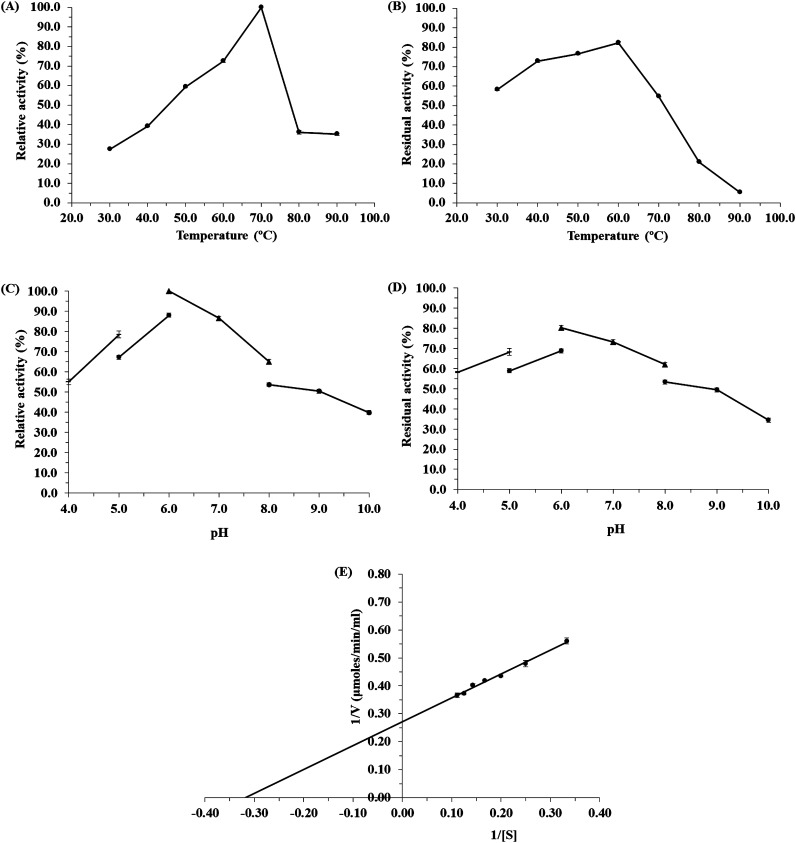
Figure 2. Biochemical properties of α-amylase: (A) The effect of temperature on amylase activity at different temperature ranging from 30.0 to 90.0°C. (B) The effect of temperature on amylase stability. (C) The effect of pH on amylase activity, the pH conditions were adjusted using the following 100 mM sodium acetate buffer pH range 4.0–5.0, 100 mM citrate phosphate buffer pH 5.0–6.0, 100 mM potassium phosphate buffer pH range 6.0–7.0, 100 mM Tris-HCl buffer pH range 7.0–8.0. (D) The effect of pH on amylase stability. (E) Lineweaver-Burk plots in in the presence of different concentration of soluble starch. The data represent the mean±standard deviation from three replicates.
Effect of temperature on α-amylase activity
The CgAmy was highly activity in a wide range of temperatures from 50 to 70°C with optimum activity at 70°C (Figure 2(A)). The enzyme has activity more than 35% relative activity in the range of 80–90°C. The CgAmy also exhibited high stability at a wide temperature range of 30–70°C which retained more than 50% of the initial activity (Figure 2(B)). Furthermore, the residual activity of α-amylase at a high temperature of 70°C and 80°C was 54.58% and 20.91% of residual activity, respectively. The result demonstrated that the CgAmy is a thermostable amylase.
Effect of pH on α-amylase activity
According to the optimum pH profile (Figure 2(C)), the CgAmy revealed appropriate activity in a broad range of pH 5.0–7.0 with an optimum activity at pH 6.0. This enzyme has more than 50% activity at pH 4.0–8.0. The CgAmy displayed highly stable activity in the pH range of 5.0–8.0 retaining over 50% residual activity (Figure 2(D)). The enzyme retained about 80.31% of its maximal activity at pH 6.0.
Enzyme kinetic constants
Kinetic studies were carried out under standard conditions using soluble starch as substrate. Km and Vmax of the CgAmy which was determined for starch as the substrate from Lineweaver-Burk plots were 3.70 μmoles/min/ml and 3.12 mg/ml, respectively (Figure 2(E)). The lower Km value is related to the higher affinity to the substrate.
Substrate specificity
The substrate specificity of the α-amylase using different polysaccharides as substrates revealed that the CgAmy has the highest activity with starch as a substrate. The enzyme also was active on amylopectin and glycogen at 96.59% and 59.58% activity, respectively (Table 2). The enzyme showed no hydrolysis activity of dextran and β-cyclodextrin. Substrates containing the α-1,4-linkage were better substrates for α-amylase.
Table 2. Substrate specificity of α-amylase.
| Substrates | Activity (%) |
|---|---|
| Starch | 100.00±0.01e |
| Amylopectin | 96.59±0.13d |
| Glycogen | 59.58±0.20c |
| β-cyclodextrin | 0.00±0.00a |
| Dextran | 0.51±0.10b |
Mean value followed by different letters differs significantly (p≤0.05).
Effect of metal ions and chemical reagents on α-amylase activity
The metal ions and chemical reagents had different effects on the enzyme activity. The activity of the CgAmy was slightly activated in the presence of Co2+ ions (107.84%) (Table 3). The metal ions of 5 mM Na+, K+, Ca2+, Mg2+, Zn2+, Ba2+, Fe2+, and Cd2+ were slightly affected to α-amylase activity retaining over 70% residual activity. However, the CgAmy activity was inhibited in the presence of 5 mM Cu2+ and Ni2+ with 35.77% and 53.27% of its activity, respectively. β-Mercaptoethanol had no significant inhibit on the enzyme activity. However, the chelating agent EDTA displays significant (p≤0.05) inhibited the enzyme. The stability of the CgAmy was also investigated in the presence of surfactant (SDS, Triton X-100 and Tween-20). The enzyme was highly tolerance in the presence of the non-ionic surfactants of Triton X-100 (95.17%) and Tween-20 (114.10%) but in the presence of SDS, the enzyme was inactivated. Furthermore, the CgAmy was highly stable in the presence of methanol, ethanol, acetone, and hexane, retaining 84.06%, 82.87%, 117.64% and 92.80% of initial activity, respectively.
Table 3. Effect of metal ions and chemical reagents on the α-amylase activity.
| Different additives | Concentration | Relative activity (%) |
|---|---|---|
| Control (Non-component) | — | 100±0.02i |
| Metal ions | ||
| NaCl | 5 mM | 95.40±1.00gh |
| KCl | 5 mM | 90.71±0.81f |
| CaCl2 | 5 mM | 89.47±0.97j |
| MgCl2 | 5 mM | 82.04±1.21e |
| ZnCl2 | 5 mM | 81.57±0.64e |
| BaCl2 | 5 mM | 90.48±0.33f |
| FeCl2 | 5 mM | 78.88±0.48e |
| CdCl2 | 5 mM | 74.56±1.74d |
| CoCl2 | 5 mM | 107.84±1.14m |
| CuCl2 | 5 mM | 35.77±0.36b |
| NiCl2 | 5 mM | 53.27±0.23c |
| Chemical reagents | ||
| EDTA | 10 mM | 0.00±0.00a |
| β-mercaptoethanol | 10 mM | 117.65±2.58l |
| SDS | 5% | 0.00±0.00a |
| Triton X-100 | 5% | 95.17±0.54gh |
| Tween-20 | 5% | 114.10±1.04k |
| Methanol | 10% | 84.06±1.56fg |
| Ethanol | 10% | 82.87±0.69h |
| Acetone | 10% | 117.64±2.36l |
| Hexane | 10% | 92.80±0.35fg |
Mean value followed by different letters differs significantly (p≤0.05).
Discussion
The CgAmy was successfully purified using two steps of ammonium sulphate fractionation and affinity chromatography. α-Amylase inhibitor in crude extract was possibly removed by ammonium sulphate fractionation step (Wisessing et al. 2010). The less number of enzyme purification steps showed high fold and yield of enzyme purification. The affinity chromatography used in this study has also been reported successfully for other plant amylases (Kumari et al. 2010; Singh et al. 2017; Tripathi et al. 2007). The purified α-amylase from Vigna radiata was purified 599 fold to a final specific activity of 437 U/mg and an overall recovery of 9% (Tripathi et al. 2007). Kumari et al. (2010) purified α-amylase from glycine max having specific activity of 384.61 Units/mg, 6.6% yield and purification of 400.63 fold. The purification procedure in this report is a simple and effective method for α-amylase purification. Recently, Singh et al. (2017) reported α-amylase from Vicia faba with a specific activity of 369 U/mg, purification fold of 2050 and 24% yield. The purified enzyme revealed a molecular mass of 50.0 kDa which is very close to α-amylase from Vigna radiata (46 kDa) (Tripathi et al. 2007). In plants (Table 4), α-amylases have been reported molecular mass range from 32 kDa to 84 kDa. The CgAmy was in accordance with this range.
Table 4. Biochemical properties of some plant α-amylases.
| Plant sources | Molecular weight (kDa) | Optimum pH (% relative activity) | Optimum temperature (% relative activity) | Km | Reference |
|---|---|---|---|---|---|
| Safflower (Carthamus tinctorius L.) seeds | 35.0 | 6.0 | 55 | — | Ben Elarbi et al. 2009 |
| Apple (Malus pumila) | 51.2 | 6.8 | 37 | 2.0×10−3 g/ml | Kanwal et al. 2004 |
| Pachyrhizus erosus L. tuber | 40.0 | 7.3 | 37 | 0.29% | Noman et al. 2006 |
| Mung bean (Vigna radiata) seeds | 46.0 | 5.6 | 65 | 1.60 mg/ml | Tripathi et al. 2007 |
| Soybean (Glycine max) seeds | 84.0 | 5.5 | 70 | 0.71 mg/ml | Kumari et al. 2010 |
| Wheat (Triticum aestivum) seeds | 32.0 | 5.0 | 68 | 1.56 mg/ml | Singh and Kayastha 2014 |
| Red pitaya (Hylocereus polyrhizus) peel | 42.1 | 5.0 | 70 | 2.70 mg/ml | Amid and Manap 2014 |
| Vicia faba | 45.0 | 6.0 | 65 | 4.60 mg/ml | Singh et al. 2017 |
| Sword bean (Canavalia gladiata (Jacq.) DC.) seeds | 50.0 | 6.0 | 70 | 3.12 mg/ml | This study |
The optimum temperature of the CgAmy was at a high temperature of 70°C that is consistent with purified α-amylase from Glycine max (70°C) and is higher compared to Vigna radiata (65°C) and safflower (55°C) (Table 4). The CgAmy also revealed high stability at high temperature range of 30–70°C. The results demonstrated that the α-amylase is a thermostable amylase which can be used to degrade gelatinized starch at high temperature of the starch industry. After pre-incubation at higher temperatures of 90°C, the CgAmy activity dropped rapidly which was probably due to denaturation of the enzyme. The optimum pH (pH 6.0) of the CgAmy is in accordance with the data reported for other plant α-amylases (Table 4) and values reported for α-amylase from Glycine max (pH 5.5), Vigna radiata (pH 5.6) and Vicia faba (pH 6.0) (Table 4). In the starch hydrolysis industry, most of the α-amylase is inactive at acidic pH of 3.2–6.0 of native starch slurry for starch processing (Sivaramakrishnan et al. 2006). Amylase with low pH value can be used as a substitute for currently liquefied amylase (optimum pH of 6.8) and reduced expenses and time of pH adjusting from its native pH to match with the optimum pH of the enzyme (Wu et al. 2018). The Km value of α-amylase from other plants has been reported ranging from 0.71 to 4.6 mg/ml (Table 4). However, the kinetic values of the amylases are difficult to compare with other amylases reports because of variation in substrates and assay conditions. The CgAmy also was active on amylopectin and glycogen. Amid and Manap (2014) reported that α-amylase from pitaya peel revealed the highest activity toward starch followed by amylopectin (90%) and glycogen (82%). The CgAmy showed no hydrolysis of dextran indicating that the enzyme has the capability of cleaving α-1,4 bonds but not α-1,6 bonds. The enzyme also did not hydrolysed β-cyclodextrin with was reported as a competitive inhibitor of α-amylase (Hamilton et al. 2000). The no hydrolytic activity with β-cyclodextrin and dextran supported that CgAmy prefers α-1,4 glycosidic linkage for cleaving and is α-amylase type. The α-amylase from pitaya peel (Amid and Manap 2014) and mung beans (Tripathi et al. 2007) also reported no significant hydrolysis of β-cyclodextrin.
The activity of CgAmy was slightly activated in the presence of Co2+ ions. The α-amylase from Aspergillus oryzae IFO-30103 also found Co2+ as an activator (Bhanja Dey and Banerjee 2015). The inactivation of the enzyme by Cu2+ and Ni2+ may be due to their binding to the catalytic residues in the active site of the enzyme. The chelating agent EDTA showed the enzyme inhibition, suggesting that CgAmy is a metalloenzyme. This result is in agreement with α-amylase from Pachyrhizus erosus L. tuber, soybean, mung bean, wheat which was inhibited in the presence of EDTA (Kumari et al. 2010; Noman et al. 2006; Singh and Kayastha 2014; Tripathi et al. 2007). The CgAmy was inactivated in the presence SDS which suggested that SDS disrupts the protein’s higher ordered structure resulting in inactivation of the enzyme (Singh and Kayastha 2014). The CgAmy also was highly stable in some organic solvents and detergents. The thermostable enzymes have been reported that is inconsistent with more resistant to organic solvents and detergents (Klibanov 2001). The resistant in the reagents suggested that the enzyme has a well-packed structure and rigid native conformation (Chakraborty et al. 2011). Due to the good enzymatic properties of the enzyme in organic solvents, it may have a wide range of industrial applications such as the treatment of carbohydrate-polluted industrial wastewater contaminated with organic solvent (Wu et al. 2018), alcohol production from starch material, brewing of alcoholic beverages.
In the present investigation, thermostable α-amylase from germinating Sword bean (Canavalia gladiata (Jacq.) DC.) seeds (CgAmy) have discovered. Furthermore, it exhibited good stability in the presence of some detergents and organic solvents. Therefore, sword bean (Canavalia gladiata (Jacq.) DC.) seeds should be considered as novel sources and an important candidate for producing thermostable α-amylase that are desirable characteristics in various industries.
Acknowledgments
Authors gratefully appreciate the financial support of this work by Nakhon Ratchasima Rajabhat University. We are also thankful to the division of Chemistry, division of Biology and science center, Faculty of science and technology, Nakhon Ratchasima Rajabhat University for providing the infrastructure and facility.
References
- Amid M, Manap MYA (2014) Purification and characterization of a novel amylase enzyme from red pitaya (Hylocereus polyrhizus) peel. Food Chem 165: 412–418 [DOI] [PubMed] [Google Scholar]
- Ben Elarbi M, Khemiri H, Jridi T, Ben Hamida J (2009) Purification and characterization of alpha amylase from safflower (Carthamus tinctorius L.) germinating seeds. C R Biol 332: 426–432 [DOI] [PubMed] [Google Scholar]
- Bhanja Dey T, Banerjee R (2015) Purification, biochemical characterization and application of α-amylase produced by Aspergillus oryzae IFO-30103. Biocatal Agric Biotechnol 4: 83–90 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Khopade A, Biao R, Jian W, Liu XY, Mahadik K, Chopade B, Zhang L, Kokare C (2011) Characterization and stability studies on surfactant, dertergent and oxidant stable α-amylase from marine haloalkaliphilic Saccharopolyspora sp. A9. J Mol Catal B Enzym 68: 52–58 [Google Scholar]
- Choi JM, Han SS, Kim HS (2015) Industrial applications of enzyme biocatalytic: Current status and future aspect. Biotechnol Adv 33: 1443–1454 [DOI] [PubMed] [Google Scholar]
- Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B (2003) Microbial α-amylase: A biotechnological perspective. Process Biochem 38: 1599–1616 [Google Scholar]
- Hamilton LM, Kelly CT, Fogarty WM (2000) Review: Cyclodextrins and their interaction with amylolytic enzymes. Enzyme Microb Technol 26: 561–567 [DOI] [PubMed] [Google Scholar]
- Jegannathan KR, Nielsen PH (2013) Environmental assessment of enzyme use in industrial production: A literature review. J Clean Prod 42: 228–240 [Google Scholar]
- Kanwal B, Zia MA, Yasin M, Rahman K, Sheikh MA (2004) Purification and characterization of α-amylase from apple (Malus pumila). Int J Agric Biol 6: 233–236 [Google Scholar]
- Klibanov AM (2001) Improving enzymes by using them in organic solvent. Nature 409: 241–246 [DOI] [PubMed] [Google Scholar]
- Kumari A, Singh VK, Fitter J, Polen T, Kayastha AM (2010) α-Amylase from germinating soybean (Glycine max) seeds-purification, characterization and sequential similarity of conserved and catalytic amino acid residues. Phytochemistry 71: 1657–1666 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural protein during the assembly of head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31: 426–428 [Google Scholar]
- Muralikrishna G, Nirmala M (2005) Cereal α-amylases-an overview. Carbohydr Polym 60: 163–173 [Google Scholar]
- Noman ASM, Hoque MA, Sen PK, Karim MR (2006) Purification and some properties of α-amylase from post-harvest Pachyrhizus erosus L. tuber. Food Chem 99: 444–449 [Google Scholar]
- Posoongnoen S, Ubonbal R, Thammasirirak S, Daduang J, Minami H, Yamamoto K, Daduang S (2015) α-Amylase from Mon Thong durian (Durio zibethinus Murr. cv. Mon Thong)-nucleotide sequence analysis, cloning and expression. Plant Biotechnol 32: 1–10 [Google Scholar]
- Sindhu R, Binod P, Madhavan A, Beevi US, Mathew AK, Abraham A, Pandey A, Kumar V (2017) Molecular improvements in microbial α-amylases for enhanced stability and catalytic efficiency. Bioresour Technol 245(Pt B): 1740–1748 [DOI] [PubMed] [Google Scholar]
- Singh K, Ahmad F, Singh VK, Kayastha K, Kayastha AM (2017) Purification, biochemical characterization and Insilico modelling of α-amylase from Vicia faba. J Mol Liq 234: 133–141 [Google Scholar]
- Singh K, Kayastha AM (2014) α-amylase from wheat (Triticum aestivum) seeds: Its purification, biochemical attributes and active site studies. Food Chem 162: 1–9 [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A (2006) α-amylases from microbial sources: An overview on recent developments. Food Technol Biotechnol 44: 173–184 [Google Scholar]
- Sundarram A, Murthy TPK (2014) α-Amylase production and applications: A review. J Appl Environ Microbiol 2: 166–175 [Google Scholar]
- Tripathi P, Lo Leggio L, Mansfeld J, Ulbrich-Hofmann R, Kayastha AM (2007) α-Amylase from mung beans (Vigna radiata): Correlation of biochemical properties and tertiary structure by homology modelling. Phytochemistry 68: 1623–1631 [DOI] [PubMed] [Google Scholar]
- Vadivel V, Janardhanan K (2004) The nutritional and antinutritional attributes of sword bean (Canavalia gladiata (Jacq.) DC.): An under-utilized tribal pulse from south India. Int J Food Sci Technol 39: 917–926 [Google Scholar]
- van der Maarel MJEC, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94: 137–155 [DOI] [PubMed] [Google Scholar]
- Vretblad P (1974) Immobilization of ligands for biospecific affinity chromatographic via their hydroxyl group, the cyclohexa amylose-β-amylase system. FEBS Lett 47: 86–89 [DOI] [PubMed] [Google Scholar]
- Wisessing A, Engkagul A, Wongpiyasatid A, Choowongkomon K (2010) Biochemical characterization of the α-amylase inhibitor in mung beans and its application in inhibiting the growth of Callosobruchus maculatus. J Agric Food Chem 58: 2131–2137 [DOI] [PubMed] [Google Scholar]
- Wu X, Wang Y, Tong B, Chen X, Chen J (2018) Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4–423. Int J Biol Macromol 109: 329–337 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Hanzlik S, Cook E, Shen QJ (2007) Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol Biol 64: 293–303 [DOI] [PubMed] [Google Scholar]