Abstract
Virus-induced gene silencing (VIGS) is a useful tool for functional genomics in plants. In this study, we tried to apply cucumber mosaic virus (CMV) to efficient induction of VIGS in spinach. Although VIGS for spinach had been previously developed based on two viruses (beet curly top virus and tobacco rattle virus), they still have some problems with systemic movement and long-term maintenance of VIGS in spinach. Although ordinary CMV strains infect spinach inducing distinct mosaic symptoms, using a CMV pseudorecombinant, we can modify the viral pathogenicity to attenuate viral symptoms that may mask the silencing phenotype. We here successfully demonstrated the viral ability to silence the phytoene desaturase (PDS) and the dihydroflavonol 4-reductase (DFR) genes in spinach. Because CMV could quickly induce VIGS even at 7–10 days postinoculation and the virus did not disappear even at the flowering stage, this CMV-based VIGS system would contribute to functional genomics in spinach and especially to the elucidation of molecular mechanisms for some properties unique to spinach such as plasticity of sex expression; the CMV-induced VIGS can last until the flowering stage after the virus was inoculated onto the seedling.
Keywords: cucumber mosaic virus, spinach, virus-induced gene silencing
Spinach (Spinacia oleracea) is a member of Chenopodioideae, a subfamily of the family Amaranthaceae, and is one of the most nutritious vegetables, rich in vitamins and minerals, being grown in over 50 countries (Fuentes-Bazan et al. 2012). It is commonly considered as dioecious species, although certain cultivars and genotypes can produce individuals with both staminate and pistillate flowers (i.e., monoecious plants) (Janick and Stevenson 1955b; Onodera et al. 2008). The dioecism and monoecism in spinach are utilized for producing commercial hybrid seeds, and hence the elucidation of the mechanisms for controlling sex expression is important for spinach breeding (Janick 1998; Onodera et al. 2011).
Spinach has long served as a model to study plant sex determination and expression as well as flavonoid biosynthesis and chloroplast function (Beerhues and Wiermann 1988; Beerhues et al. 1988; Chailakhyan and Timiriazev 1979; Ellis 1981; Janick and Stevenson 1954, 1955a, 1955b, 1955c; Sherry et al. 1993; Shimada et al. 2004; Yamamoto et al. 2014). The draft genome assemblies and the transcriptome data sets of spinach have been recently released (Okazaki et al. 2019; Xu et al. 2015, 2017), which are useful to identify loci responsible for important agronomic traits (e.g., loci for sex determination, disease resistance, etc.), enhancing the value of spinach as a model plant more and more. As the information on the genomic organization accumulate, functional analysis of spinach genes must be accelerated. However, rapid and efficient tools for studies of functional genomics are still not well established in spinach; although spinach transformation methods have been developed, they are based on callus-mediated regeneration systems, requiring much time and labor to obtain transformants (Chin et al. 2009; Nguyen et al. 2013).
Virus-induced gene silencing (VIGS) is useful for functional genomics especially in plants to which conventional transgenic methods cannot be easily applicable. For spinach, VIGS systems have been developed based on two viruses so far; one is beet curly top virus (BCTV) (Golenberg et al. 2009; Sather et al. 2010) and the other is tobacco rattle virus (TRV) (Lee et al. 2017). Although both BCTV- and TRV-based VIGS systems have been demonstrated to be effective in silencing some spinach genes, there is still room for improvement of VIGS in spinach. In the BCTV-based silencing, the virus vector with an insert was localized only in the inoculated leaves and could not systemically spread in the entire plant when the ribulose bisphosphate carboxylase small subunit, transketolase and homeotic transcription factor genes were targeted (Golenberg et al. 2009); the silenced phenotypes were observed at 6–8 weeks postinoculation (wpi). In the TRV-based silencing, when the phytoene desaturase (PDS) gene was silenced, the silencing phenotype appeared at 3 wpi but eventually disappeared after 4 wpi due to the reduction in viral accumulation in the systemically infected leaves (Lee et al. 2017). It is obvious that viral systemic infection is very important to induce efficient VIGS as demonstrated in some legume species and lily plants (Pflieger et al. 2013; Tasaki et al. 2016). Toward developing a more practical VIGS vector for spinach, we here examined the ability of the cucumber mosaic virus (CMV) vector to systemically spread in infected spinach and to sustain its systemic infection until the reproductive stage of plant. Results presented in this study demonstrate that our CMV vector targeting either the PDS gene or the dihydroflavonol 4-reductase (DFR) gene could induce efficient VIGS in spinach at most 7–10 days postinoculation (dpi) and existed in the infected spinach even after 2 months postinoculation.
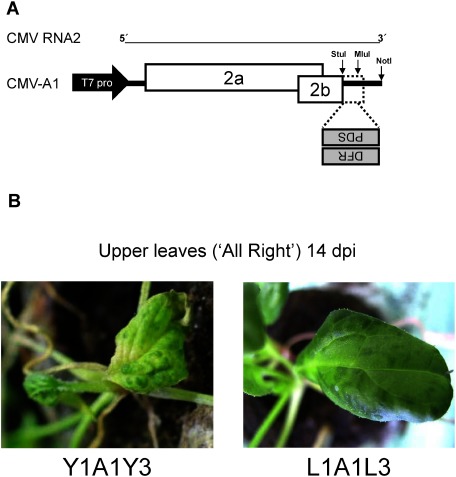
CMV has been developed as a useful VIGS vector for many plants including Nicotiana benthamiana, tomato, pepper, Arabidopsis thaliana, lily, soybean, banana and maize (Hong et al. 2012; Liu et al. 2010; Otagaki et al. 2006; Tzean et al. 2019; Wang et al. 2016). Recently, it has been demonstrated that we could silence even a gene of mycorrhizal fungi that colonized the root of N. benthamiana (Kikuchi et al. 2016). In this study, we used two CMV strains: CMV Y strain (CMV-Y) and CMV legume strain (CMV-L). CMV has tripartite positive-sense RNA genomes (RNA1, RNA2 and RNA3); three genomic RNAs of CMV-Y and CMV-L were named Y1–Y3 and L1–L3, respectively. Full-length cDNAs of all the genomic RNAs of both strains have been cloned into the pUC plasmids (Otagaki et al. 2006; Suzuki et al. 1991). When in vitro transcripts of three genomic RNAs from the recombinant plasmids are mixed, CMV becomes infectious; a pseudorecombinant CMV can be easily created between two CMVs (e.g., CMV-Y and CMV-L) by mixing the viral genomic RNAs. The viral vector construct based on CMV-Y RNA2 is designated A1, which has the cloning site between StuI and MluI for a foreign insert (Figure 1A). Our VIGS system for spinach consists of three sequential steps: (1) cloning of a partial sequence of the spinach gene of target into A1, (2) propagation of the recombinant A1 vector in N. benthamiana by rub-inoculating RNA transcripts, (3) inoculation of leaf sap of infected N. benthamiana onto spinach. In a preliminary experiment, we found that Y1A1Y3 induced severe mosaic symptoms in spinach although the severity varied depending on the tested cultivars. We thus used a pseudorecombinant virus, L1A1L3 that induced relatively mild symptoms on those cultivars (Figure 1B). It is important to use a mild (or symptomless) virus because severe symptoms may mask the silenced phenotype in the infected plants.
Figure 1. Construction of the CMV vector for VIGS. (A) Schematic representation of the CMV vector A1, whose backbone is RNA2 encoding the 2a and 2b genes. The spinach genes (PDS and DFR) were cloned between the StuI and MluI site in the antisense orientation. (B) Symptoms on the upper leaves of spinach (‘All Right’) infected with the original vectors of Y1A1Y3 (left) and L1A1L3 (right).
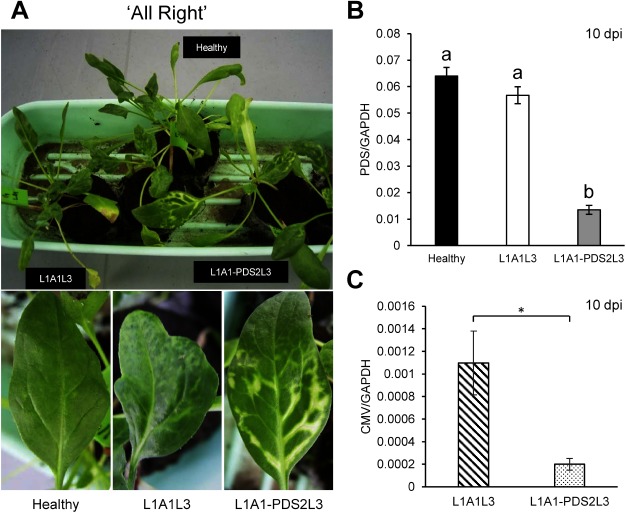
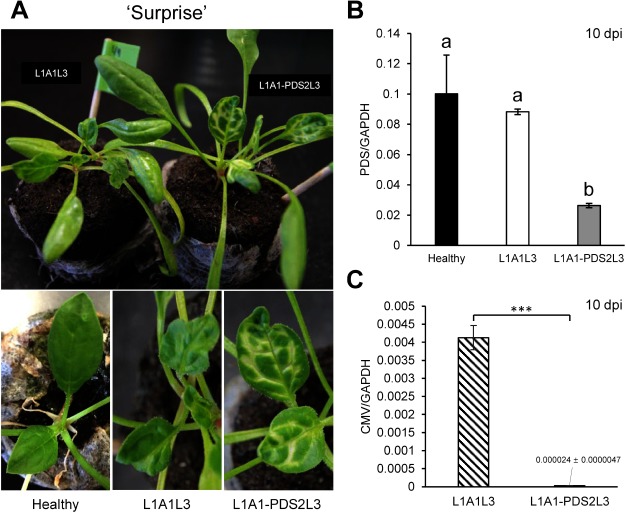
To evaluate the CMV-based VIGS system, the 130-nucleotide (nt) fragment of the PDS gene was first cloned in the A1 vector to create A1-PDS (Figure 1A). Primers used for cloning are shown in the Supplementary Table S1. In vitro transcripts of L1, A1-PDS and L3 were inoculated onto N. benthamiana, and subsequently the leaf sap of infected N. benthamiana was inoculated onto leaves of two spinach cultivars, ‘All Right’ (TAKII & CO., LTD., Kyoto, Japan) and ‘Surprise’ (TOHOKU SEED Co. LTD., Utsunomiya, Japan). In our preliminary experiments, we had found that ‘Surprise’ was a little more susceptible to the pseudorecombinant virus, L1A1L3 than ‘All Right.’ As shown in Figures 2A and 3A, distinct chlorosis was observed at 7–10 dpi along the veins in upper systemically infected leaves. As expected, the PDS mRNA levels were decreased down to less than 1/4 of those of the healthy plants at 10 dpi (Figures 2B, 3B), and photobleaching has lasted over two months (Supplementary Figure S1). There was little difference in photobleaching between the two cultivars when the vector contains the insert fragment, suggesting that the introduction of an insert in the vector may affect the viral pathogenicity. However, the viral accumulation level in ‘Surprise’ was found to be about 1/200 of the control (L1A1L3) while about 1/5 in ‘All Right,’ suggesting that viral accumulation levels are different in different cultivars and that a very low level of viral accumulation can still induce efficient VIGS against PDS (Figures 2C, 3C).
Figure 2. VIGS against the PDS gene in the spinach cultivar ‘All Right.’ (A) Photobleaching phenotype observed on the upper systemic leaves of spinach inoculated with L1A1-PDS2L3 at 10 dpi. The pictures of leaves of healthy plants and empty vector-infected plants are shown for comparison. (B) PDS expression levels in the upper non-inoculated leaves at 10 dpi. Real-time RT-PCRs were conducted using primer pairs as in Supplementary Table S1. The glyceraldehyde 3-phosphate (GAPDH) gene was used as a reference gene. Means (±SE) among the infected plants were analyzed for significant differences using Tukey’s multiple comparison test (* p<0.05); different letters above the bars indicate a significant difference among isolates. (C) CMV accumulation levels in upper non-inoculated leaves at 10 dpi. Real-time RT-PCR for the virus accumulation was conducted using primer pair of CMV-DET-5-340 and CMV-DET-3-340 (Supplementary Table S1). The CMV-DET-5-340 primer hybridizes to both CMV-L RNA3 (positions 1864–1884) and the subgenomic RNA from RNA3 designated RNA4, which is the mRNA for the coat protein. The CMV-DET-3-340 primer hybridizes to the 3′ end of RNA3 and RNA4. The size of the PCR product is 353 bp. The GAPDH gene was used as a reference (Supplementary Table S1). Statistical analysis was conducted by Student’s t-test (* p<0.05).
Figure 3. VIGS against the PDS gene in the spinach cultivar ‘Surprise.’ (A) Photobleaching observed on the upper leaves of spinach inoculated with L1A1-PDS2L3 at 10 dpi. The pictures of leaves of healthy plants and empty vector-infected plants are shown for comparison. (B) PDS expression levels in the upper non-inoculated leaves at 10 dpi. Real-time RT-PCRs were conducted using primer pairs as in Supplementary Table S1. Statistical analysis for real-time RT-PCR of PDS was conducted using Tukey’s multiple comparison test (* p<0.05); different letters above the bars indicate a significant difference among isolates. (C) CMV accumulation levels in upper non-inoculated leaves at 10 dpi. Real-time RT-PCR was conducted as explained in Figure 2. Statistical analysis was performed using Student’s t-test (*** p<0.001).
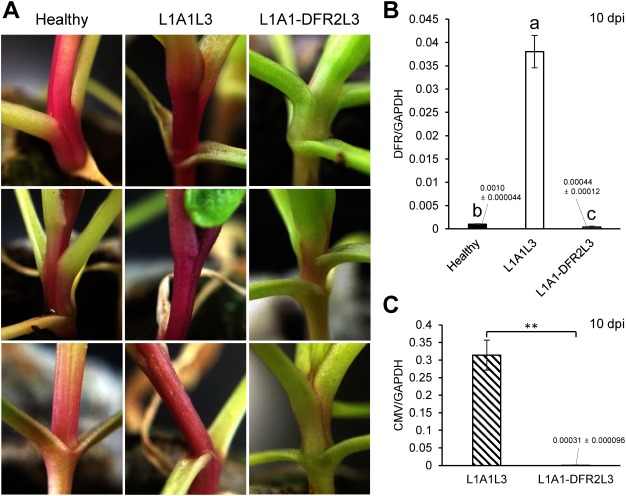
To further test whether this vector can be used for functional genomics of spinach genes, we chose the dihydroflavonol 4-reductase (DFR) gene, which is one of the key genes in the anthocyanin pathway (Shimada et al. 2004). In green spinach, anthocyanins are not normally synthesized but instead, betacyanins accumulate. However, it has been reported that some red-purple cultivars produce anthocyanins in leaves and stem although all red-purple cultivars cannot necessarily synthesize anthocyanins (Cai et al. 2018). We here cloned the 130-nt spinach DFR fragment into the A1 vector to produce A1-DFR using primer pair shown in the Supplementary Table S1 and induced VIGS by inoculating A1-DFR onto several red-purple spinach cultivars, whose seeds were purchased on the Japanese market: ‘Kuroshio,’ Kobayashi-Shubyo, Hyogo; ‘Akajiku-Salad,’ Musashino-Shubyo, Tokyo; ‘Wase-Salad-Akari,’ TAKII, Kyoto; ‘Akakuki-Minster,’ Nakahara-Saishujyo, Fukuoka; ‘Miyabi,’ TOHOKU, Utsunomiya. Plants were grown in the plant incubator at 24°C under 12 h day. These inoculated plants did not show any severe symptoms but did very mild symptoms in upper leaves (Supplementary Figure S2). Although the red pigmentation was not decreased in most of the tested cultivars (five cultivars, 4 individual plants tested for each cultivar) by A1-DFR VIGS, we observed clear decoloration of the red pigment in the stem of one of the used cultivars (cultivar ‘Kuroshio,’ Figure 4A), suggesting that this cultivar may produce anthocyanins. We also confirmed that the DFR mRNA levels in the A1-DFR-infected plants were greatly reduced at 10 dpi in spite of the fact that the empty vector (L1A1L3) infection itself increased the DFR expression (Figure 4B). Our real-time RT-PCRs showed that the viral accumulation levels in L1A1-DFR2L3-infected plants with the silenced phenotype were about 1/1000 of the control (L1A1L3), suggesting that the high level of viral accumulation of L1A1L3 may have caused the huge increase in DFR expression (Figure 4B, C). There are two possible explanations for the observation that the viral accumulation level of L1A1-DFR2L3 was very low at 10 dpi in the systemic tissues. The CMV vector itself is degraded by VIGS in infected tissues but normally tolerant to VIGS to some extent because of its secondary structure. If the insertion of a target sequence in the vector alters such a secondary structure, then the viral genomic RNA would become much more susceptible to VIGS, resulting in a great decrease in viral titer. As VIGS is efficiently induced against the target gene, the virus containing the target sequence will be also efficiently degraded. Alternatively, the insertion of a foreign sequence itself sometimes directly interferes with the viral replication, depending on both the size and sequence. We assume that either or both of the two cases may have occurred on L1A1-DFR2L3. In addition, to examine whether L1A1-DFR2L3 loses the insert sequence in systemically infected upper leaves, we analyzed the leaves just beneath bolted flowers of the plants at 60 dpi. The results showed that the virus still retained the complete insert, and the accumulation level was about 1/15 of the control (L1A1L3), suggesting that the virus can maintain its systemic infection even after two months postinoculation (Supplementary Figure S3). These results altogether demonstrate that the CMV-based VIGS vector can be used for functional genomics in spinach without inducing severe symptoms.
Figure 4. Expected silencing phenotype (decoloration) observed in the stem of spinach inoculated with L1A1-DFR2L3 at 10 dpi. The set of pictures is VIGS against the DFR gene in the red-purple spinach cultivar ‘Kuroshio.’ The pictures of the stems from three individual plants (healthy plants and L1A1L3-infected plants) are shown for comparison. The GAPDH gene was used as a reference gene. (B) Real-time RT-PCR was conducted using primer pairs as in Supplementary Table S1. To maintain homogeneity of variance among the sample groups, the raw data was converted to log data for the statistical analysis. Means (±SE) among the infected plants were analyzed for significant differences using Tukey’s multiple comparison test (* p<0.05); different letters above the bars indicate a significant difference among isolates. (C) Comparison of viral accumulation by real-time RT-PCR. Total RNA was extracted from stem. Real-time RT-PCR was conducted as explained in Figure 2. Statistical analysis was performed using Student’s t-test (** p<0.01).
In conclusion, we here tried to develop another VIGS system for spinach, applying the CMV vector to VIGS against spinach PDS and DFR, which would be more practical than the other two BCTV- and TRV-based VIGS systems. This CMV construct can systemically infect many spinach cultivars and efficiently accumulate in infected plants. Because we keep some CMV strains isolated directly from spinach, we can use those viral RNA genomes to adjust the viral pathogenicity and symptoms. For example, even though the initial construct does not efficiently infect a cultivar of interest due to a resistant gene, we can test other pseudorecombinant viruses using CMV spinach strains that can overcome the resistance.
Acknowledgments
The authors thank Tohoku Seed Co. Ltd. (Utsunomiya, Tochigi, Japan) for providing the spinach cultivar used in this study. We also thank Masahide Kawano for technical assistance.
Abbreviations
- BCTV
beet curly top virus
- CMV
cucumber mosaic virus
- CMV-L
CMV legume strain
- CMV-Y
CMV Y strain
- DFR
dihydroflavonol 4-reductase
- dpi
days postinoculation
- GAPDH
glyceraldehyde 3-phophate dehydrogenase
- PDS
phytoene desaturase
- RT-PCR
reverse transcription polymerase chain reaction
- SE
standard error
- TRV
tobacco rattle virus
- VIGS
virus-induced gene silencing
Supplementary Data
References
- Beerhues L, Robenek H, Wiermann R (1988) Chalcone synthases from spinach (Spinacia oleracea L.): II. Immunofluorescence and immunogold localization. Planta 173: 544–553 [DOI] [PubMed] [Google Scholar]
- Beerhues L, Wiermann R (1988) Chalcone synthases from spinach (Spinacia oleracea L.): I. Purification, peptide patterns, and immunological properties of different forms. Planta 173: 532–543 [DOI] [PubMed] [Google Scholar]
- Cai X, Lin L, Wang X, Xu C, Wang Q (2018) Higher anthocyanin accumulation associated with higher transcription levels of anthocyanin biosynthesis genes in spinach. Genome 61: 487–496 [DOI] [PubMed] [Google Scholar]
- Chailakhyan MK, Timiriazev KA (1979) Genetic and hormonal regulation of growth, flowering, and sex expression in plants. Am J Bot 66: 717–736 [Google Scholar]
- Chin DP, Bao JH, Mii M (2009) Transgenic spinach plants produced by Agrobacterium-mediated method based on the low temperature-dependent high plant regeneration ability of leaf explants. Plant Biotechnol 26: 243–248 [Google Scholar]
- Ellis RJ (1981) Chloroplast proteins: synthesis, transport, and assembly. Annu Rev Plant Physiol 32: 111–137 [Google Scholar]
- Fuentes-Bazan S, Uotila P, Borsch T (2012) A novel phylogeny-based generic classification for Chenopodium sensu lato, and a tribal rearrangement of Chenopodioideae (Chenopodiaceae). Willdenowia 42: 5–24 [Google Scholar]
- Golenberg EM, Sather DN, Hancock LC, Buckley KJ, Villafranco NM, Bisaro DM (2009) Development of a gene silencing DNA vector derived from a broad host range geminivirus. Plant Methods 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-S, Rhee S-J, Kim E-J, Kim T-S, Ryu K-H, Masuta C, Lee G-P (2012) Application of a reassortant cucumber mosaic virus vector for gene silencing in tomato and chili pepper plants. Plant Pathol J 28: 81–86 [Google Scholar]
- Janick J (1998) Hybrids in horticultural crops. In: Lamkey KR, Staub JE (eds) Concepts and Breeding of Heterosis in Crop Plants. Crop Scinece Society of America, pp 45–56
- Janick J, Stevenson EC (1954) A genetic study of the heterogametic nature of the staminate plant in spinach. Proc Am Soc Hortic Sci 63: 444–446 [Google Scholar]
- Janick J, Stevenson EC (1955a) Environmental influences on sex expression in monoecious lines of spinach. Proc Am Soc Hortic Sci 65: 416–422 [Google Scholar]
- Janick J, Stevenson EC (1955b) Genetics of the monoecious character in spinach. Genetics 40: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janick J, Stevenson EC (1955c) The effects of polyploidy on sex expression in spinach. J Hered 46: 151–156 [Google Scholar]
- Kikuchi Y, Hijikata N, Ohtomo R, Handa Y, Kawaguchi M, Saito K, Masuta C, Ezawa T (2016) Aquaporin-mediated long-distance polyphosphate translocation directed towards the host in arbuscular mycorrhizal symbiosis: application of virus-induced gene silencing. New Phytol 211: 1202–1208 [DOI] [PubMed] [Google Scholar]
- Lee J, Cao D-V, Kim J, Pamplona RS, Ahn J, Cho S-K, Yang S-W, Riu K-Z, Boo K-H (2017) Development of a virus-induced gene silencing (VIGS) system for Spinacia oleracea L. In Vitro Cell Dev Biol Plant 53: 97–103 [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, et al. (2010) Soybean stem growth habit gene Dt1 is an orthologue of Arabidopsis TERMINAL FLOWER1. Plant Physiol 153: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QV, Boo KH, Sun HJ, Cao DV, Lee D, Ko SH, Kang S, Yoon S, Kim SC, Park SP, et al. (2013) Evaluation of factors influencing Agrobacterium-mediated spinach transformation and transformant selection by EGFP fluorescence under low-selective pressure. In Vitro Cell Dev Biol Plant 49: 498–509 [Google Scholar]
- Okazaki Y, Takahata S, Hirakawa H, Suzuki Y, Onodera Y (2019) Molecular evidence for recent divergence of X- and Y-linked gene pairs in Spinacia oleracea L. PLoS One 14: e0214949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Yonaha I, Masumo H, Tanaka A, Niikura S, Yamazaki S, Mikami T (2011) Mapping of the genes for dioecism and monoecism in Spinacia oleracea L.: evidence that both genes are closely linked. Plant Cell Rep 30: 965–971 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Yonaha I, Niikura S, Yamazaki S, Mikami T (2008) Monoecy and gynomonoecy in Spinacia oleracea L.: Morphological and genetic analyses. Sci Hortic (Amsterdam) 118: 266–269 [Google Scholar]
- Otagaki S, Arai M, Takahashi A, Goto K, Hong J-S, Masuta C, Kanazawa A (2006) Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant Biotechnol 23: 259–265 [Google Scholar]
- Pflieger S, Richard MMS, Blanchet S, Meziadi C, Geffroy V (2013) VIGS technology: An attractive tool for functional genomics studies in legumes. Funct Plant Biol 40: 1234–1248 [DOI] [PubMed] [Google Scholar]
- Sather DN, Jovanovic M, Golenberg EM (2010) Functional analysis of B and C class floral organ genes in spinach demonstrates their role in sexual dimorphism. BMC Plant Biol 10: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry RA, Eckard KJ, Lord EM (1993) Flower development in dioecious Spinacia oleracea (Chenopodiaceae). Am J Bot 80: 283–291 [Google Scholar]
- Shimada S, Takahashi K, Sato Y, Sakuta M (2004) Dihydroflavonol 4-reductase cDNA from non-anthocyanin-producing species in the Caryophyllales. Plant Cell Physiol 45: 1290–1298 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kuwata S, Kataoka J, Masuta C, Nitta N, Takanami Y (1991) Functional analysis of deletion mutants of cucumber mosaic virus RNA 3 using an in vitro transcription system. Virology 183: 106–113 [DOI] [PubMed] [Google Scholar]
- Tasaki K, Terada H, Masuta C, Yamagishi M (2016) Virus induced gene silencing (VIGS) in Lilium leichtlinii using the Cucumber mosaic virus vector. Plant Biotechnol 33: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzean Y, Lee M-C, Jan H-H, Chiu Y-S, Tu T-C, Hou B-H, Chen H-M, Chou C-N, Yeh H-H (2019) Cucumber mosaic virus-induced gene silencing in banana. Sci Rep 9: 11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Yang X, Wang N, Liu X, Nelson RS, Li W, Fan Z, Zhou T (2016) An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J 86: 102–115 [DOI] [PubMed] [Google Scholar]
- Xu C, Jiao C, Sun H, Cai X, Wang X, Ge C, Zheng Y, Liu W, Sun X, Xu Y, et al. (2017) Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8: 15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Jiao C, Zheng Y, Sun H, Liu W, Cai X, Wang X, Liu S, Xu Y, Mou B, et al. (2015) De novo and comparative transcriptome analysis of cultivated and wild spinach. Sci Rep 5: 17706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Oda Y, Haseda A, Fujito S, Mikami T, Onodera Y (2014) Molecular evidence that the genes for dioecism and monoecism in Spinacia oleracea L. are located at different loci in a chromosomal region. Heredity 112: 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.