Abstract
Precision cancer medicine aims to classify tumors by site, histology, and molecular testing to determine an individualized profile of cancer alterations. Viruses are a major contributor to oncogenesis, causing 12% to 20% of all human cancers. Several viruses are causal of specific types of cancer, promoting dysregulation of signaling pathways and resulting in carcinogenesis. In addition, integration of viral DNA into the host (human) genome is a hallmark of some viral species. Tests for the presence of viral infection used in the clinical setting most often use quantitative PCR or immunohistochemical staining. Both approaches have limitations and need to be interpreted/scored appropriately. In some cases, results are not binary (virus present/absent), and it is unclear what to do with a weakly or partially positive result. In addition, viral testing of cancers is performed separately from tests to detect human genomic alterations and can thus be time-consuming and use limited valuable specimen. We present a hybrid-capture and massively parallel sequencing approach to detect viral infection that is integrated with targeted genomic analysis to provide a more complete tumor profile from a single sample.
As carcinogens, oncogenic viruses make significant contributions to 12% to 20% of all cancers.1,2 Most infecting oncoviruses carry double-stranded DNA genomes whose introduction to host cells predisposes them to the development of cancer through delivery of viral oncogenes and/or disruption of the host genome by viral genome integration.3 Furthermore, the type of cancer induced is specific to the virus, the presence of which implements a set of cellular characteristics distinct from their noninfected counterparts. For example, infection by a high-risk human papillomavirus (HPV) genotype significantly increases the likelihood of cervical carcinoma through expression of the E6 and E7 oncogenes, which inhibit p53 and pRb, respectively.4 Increased expression of E6 and E7 is associated with viral integration that results in inactivation of the E2 gene, a negative regulator of their expression, frequently through the partial deletion of the genic DNA. Once relieved of E2 regulatory functions, E6/E7 promote cell cycle dysregulation, thereby carving a path toward a malignant state. In addition to cervical carcinoma, HPV is the primary agent responsible for induction of anal, oropharyngeal (a subtype of head and neck cancer), vulvar, vaginal, and penile cancers.5 The need to accurately discriminate between HPV-positive and HPV-negative cancers of the head and neck is highlighted by inferior prognosis for non-HPV cancer patients.6,7 Knowledge of a patient's HPV status is clinically significant, and confirmation of HPV status is most important, as routinely used tests, such as p16 immunohistochemistry (IHC) and HPV in situ hybridization (ISH), can yield false-positive results.
Similarly, hepatitis B virus (HBV) infection is associated with 60% of hepatocellular carcinomas.8 Mechanistically, infection promotes tumorigenesis as a result of chronic inflammation induced by infection. Significantly, risk of developing hepatocellular carcinoma decreases up to 78% when HBV-infected patients are treated, making early detection of the virus a critical factor in cancer prevention.9
Merkel cell polyomavirus (MCPyV) was identified as a causative agent for most (80%) of Merkel cell carcinomas (MCCs) and was the first member of the Polyomaviridae family shown to have oncogenic activity in a human host.10 MCPyV undergoes clonal integration into the tumor genome. Although MCCs that do not harbor MCPyV may have a worse prognosis,11 the ability to discriminate between MCPyV-positive and MCPyV-negative forms of MCC could inform how treatment and monitoring of patients in remission will occur.12
Kaposi sarcoma–associated herpesvirus (KSHV) or human herpesvirus 8 (HHV-8) is the causative agent of Kaposi sarcoma and primary effusion lymphoma.13,14 KSHV is also highly associated with multicentric Castleman disease, an aggressive lymphoproliferative disorder.15 These diseases occur most commonly in immune compromised individuals, including those with AIDS. KSHV latently infects tumor cells to drive cell growth. Although KSHV is a relatively large virus, encoding nearly 100 genes, only a small subset of these are expressed in latent infection.3
Currently, most viral testing is performed using either IHC, PCR restriction fragment length polymorphism (PCR-RFLP), ISH, or real-time PCR. Use of such tests requires manual review, and interpretation of results can be indeterminate for samples of low viral titer. Furthermore, the number of targets that can be simultaneously interrogated is limited. For instance, most clinical HBV testing is done by IHC, which is restricted to a small number of antigens. Similarly, real-time PCR tests, such as the COBAS 4800 HPV test (Roche, Branchburg, NJ), are targeted to discrete regions in 14 HPV subtypes.16 Indeed, many laboratories use only p16 IHC as a surrogate for HPV infection, and there is no standard for confirmation. Such tests offer no insight to a virus's genomic variation, which may contain information that could reveal particular strains or mutations present in specific viral oncogenes, the effects of which may increase carcinogenic potential and/or cancer aggressiveness. Often, the clinical readout of such tests simply notes the presence or absence of a high-risk (ie, HPV16 or HPV18) virus.
Related to the issue of viral genomic variation are co-occurring somatic mutations of the host genome, which cannot be assessed with any of the virus tests mentioned in the previous paragraph and often go unknown because patients may only receive virus testing. Biopsy material is of limited quantity, representing an impediment to the performance of multiple diagnostic assays. Furthermore, most in vitro diagnostic tests are of limited sample throughput and focus on analysis of a small repertoire of analytes. Such testing provides clinicians with an incomplete view of factors that drive a patient's particular form of cancer, driving clinical decisions that may not have the most therapeutic impact.
We report the development of ViroPanel, a hybrid-capture and targeted DNA sequencing panel, which combines our previously published cancer panel (OncoPanel17, 18, 19) covering 447 cancer-related genes, with baits specific for 19 oncoviruses, including HBV, MCPyV, KSHV, and 16 high-risk HPV genotypes.
This allows simultaneous detection and comprehensive, accurate characterization of both variant detection in the cancer genome and the presence of viral sequence with or without integration in the host genome.
We performed a technical feasibility study to determine the sensitivity and specificity of virus detection and concordance of genomic host variants detected with and without viral baits. Our findings demonstrate ViroPanel can reliably detect the presence of infecting oncoviruses without diminishing OncoPanel's ability to characterize the tumor genome. Furthermore, genomic profiling of the viral sequences will allow accurate identification of virus subtypes, delineate the proportion of viral genome present in the tumor sample, permit discovery of viral mutations that may increase oncogenic activity, and allow us to identify sites of viral integration that may facilitate our understanding of their effect(s) on host gene regulation and genomic instability.
Materials and Methods
Panel Design
ViroPanel targets 19 oncogenic viruses (Table 1) reported to be causative cancer agents that disrupt tumor suppressors, facilitate genomic instability, and increase oncogene expression. The full viral genomes of HBV (ayw strain), MCPyV, and the high-risk HPV genotypes, 16, 18, 33, and 45, were targeted because of their being the primary etiologic agents for their respective cancer types. In addition, the coding regions for E6 and E7 oncoproteins were targeted in 12 high-risk HPV genotypes (collectively causally related to >99% of all HPV-related cancers: HPV31; 35H; 39; 51; 52; 56; 58; 59; 66; 68a; 73; and 82), as was LANA1/vCyclin and LANA2 of HHV-8. Targeted viral regions totaled 54 Kb (1276 probes). ViroPanel baits were diluted at a ratio of 1:7, then mixed at equal volume with our clinical tumor profiling bait set, OncoPanel version 3, which targets the coding sequences of 447 genes and 191 intergenic regions from 60 genes for genomic rearrangement detection.19 RNA bait target sites of interest were designed and synthesized through Agilent SureSelect (Agilent Technologies, Santa Clara, CA).
Table 1.
ViroPanel—Viruses Targeted for Hybrid Capture
| Virus | Accession no. | Target region(s) | Genome coordinates |
|---|---|---|---|
| HBV (strain ayw) | NC_003977.2 | Complete genome | 1–3182 |
| HHV-8 (KSHV) | NC_009333.1 | LANA 1 | 123,010–124,784 |
| HHV-8 (KSHV) | NC_009333.1 | LANA 2 | 126,456–127,446 |
| HPV16 | K02718.1 | Complete genome | 1–7904 |
| HPV18 | X05015.1 | Complete genome | 1–7857 |
| HPV31 | J04353.1 | E6, E7 | 108–856 |
| HPV33 | M12732.1 | Complete genome | 1–7909 |
| HPV35H | X74477.1 | E6, E7 | 110–861 |
| HPV39 | M62849.1 | E6, E7 | 107–921 |
| HPV45 | X74479.1 | Complete genome | 1–7858 |
| HPV51 | M62877.1 | E6, E7 | 96–865 |
| HPV52 | X74481.1 | E6, E7 | 102–852 |
| HPV56 | X74483.1 | E6, E7 | 102–889 |
| HPV58 | D90400.1 | E6, E7 | 110–870 |
| HPV59 | X77858.1 | E6, E7 | 55–865 |
| HPV66 | U31794.1 | E6, E7 | 102–889 |
| HPV68a | DQ080079.1 | E6, E7 | 1–816 |
| HPV73 | X94165.1 | E6, E7 | 102–843 |
| HPV82 | AB027021.1 | E6, E7 | 102–867 |
| MCPyV (R17b isolate) | NC_010277.2 | Complete genome | 1–5387 |
Accessioned reference sequences for each of the 19 targeted viruses are available at https://www.ncbi.nlm.nih.gov/nuccore.
HBV, hepatitis B virus; HHV-8, human herpesvirus 8; HPV, human papillomavirus; KSHV, Kaposi sarcoma–associated herpesvirus; MCPyV, Merkel cell polyomavirus.
Sample Selection
This study was performed in accordance with Dana-Farber Cancer Institute Institutional Review Board guidelines 10-380, 11-104, and 18-240. Samples were derived from formalin-fixed, paraffin-embedded specimens, fresh-frozen material, cultured mammalian cell lines, mouse (tail), and patient-derived xenograft (PDX) samples. Patient samples were chosen on the basis of: i) documented viral status with IHC, ISH, quantitative PCR, or PCR-RFLP testing, ii) cancer type and tissue of origin, and iii) previous clinical massively parallel sequencing profiling with OncoPanel for detection of cancer-associated mutations. A total of 124 samples (Table 2 and Supplemental Table S1) were selected for technical feasibility. Sensitivity and specificity were assessed using 66 samples whose viral status (56 virus-positive and 10 virus-negative samples) was previously determined using clinical or laboratory-developed in vitro diagnostic tests.
Table 2.
ViroPanel Sample List
| Virus | Tumor type | Sample count |
|---|---|---|
| HPV6(+) | Cervical cancer | 1 |
| HPV11(+) | Colorectal cancer | 1 |
| HPV16(+) | Head and neck carcinoma | 9 |
| HPV16(+) | Cervical cancer | 5 |
| HPV18(+) | Cervical cancer | 3 |
| HPV18(+) | Head and neck carcinoma | 1 |
| HPV33(+) | Head and neck carcinoma | 6 |
| HPV35(+) | Head and neck carcinoma | 3 |
| HPV35(+) | None | 1 |
| HPV45(+) | Head and neck carcinoma | 1 |
| HPV53(+) | None | 1 |
| HPV53(+) | Cervical cancer | 1 |
| HPV56(+) | Cervical cancer | 1 |
| HPV61(+) | Cervical cancer | 1 |
| HPV82(+) | Head and neck carcinoma | 1 |
| HHV-8(+); (KSHV) | Kaposi sarcoma | 8 |
| HBV(+) | Hepatocellular carcinoma | 1 |
| HBV(+) | None | 1 |
| HBV(+) | Unknown | 4 |
| MCPyV(+) | Merkel cell carcinoma (cell lines) | 6 |
| Negative, HPV(−) | None (CEPH1408) | 3 |
| Negative, HPV(−) | None (CEPH1347) | 1 |
| Negative, HPV16(−) | Unknown squamous cell carcinoma | 1 |
| Negative, HPV16(−) | Head and neck carcinoma | 1 |
| Negative, MCPyV(−) | Merkel cell carcinoma (cell lines) | 3 |
| Negative | None (mouse tail) | 1 |
| Unknown | Merkel cell carcinoma | 51 |
| Unknown | Merkel cell carcinoma (PDX) | 6 |
| Unknown | Bladder carcinoma | 1 |
Of the 124 samples selected, 66 are of known viral status: 56 virus-positive and 10 virus-negative samples. Also included were 58 of unknown viral status [51 Merkel cell carcinoma (MCC) tumors suspected as MCPyV infected, 6 MCC derived PDX samples, and 1 bladder carcinoma].
HBV, hepatitis B virus; HHV-8, human herpesvirus 8; HPV, human papillomavirus; KSHV, Kaposi sarcoma–associated herpesvirus; MCPyV, Merkel cell polyomavirus; PDX, patient-derived xenograft.
Ten viral negative samples were used: three MCC cell lines negative for MCPyV (MCC13, MCC26, and UISO), one mouse tail (negative control for PDX samples of unknown viral status), one head and neck squamous cell carcinoma (HPV negative; sample 25), one squamous cell carcinoma of unknown type that was negative for HPV 6/11, 16/18, 31/33 (ISH assay; sample 28), and control cell lines GM10831 (CEPH1408; three replicates) and GM10859 (CEPH1347; one sample), both of which were confirmed as HPV negative via PCR-RFLP testing. CEPH1408 and CEPH1347 genomic DNA was purchased from the Coriell Institute for Medical Research (Camden, NJ).
A total of 36 HPV positive samples were included, of which 34 were tumor biopsies. Within the HPV tumor samples are two sets (matched sample sets 7 and 8), each with two samples that are biopsies taken on different dates from their respective patients (Supplemental Tables S1 and S2). Among the 36 HPV positive controls were five samples whose infecting HPV type was not baited for: HPV6, HPV11, HPV53 (two samples), and HPV61. Additional virus positive samples included eight HHV-8 Kaposi sarcoma biopsies, six HBV samples, and six MCC/MCPyV cell lines (BroLi; MKL-1; MKL-2; MS-1; PeTa; and WaGa).
Included for testing were samples of unknown viral status: 51 patients with MCC (suspected MCPyV infection), six PDX (MCC derived), and one bladder cell carcinoma. Among the MCC samples of unknown viral status were three sets of technical replicates to assess reproducibility (matched sample sets 1 to 3) (Supplemental Table S2). Five of the PDX samples had matching primary tumor samples within the MCC samples of unknown viral status (matched sample sets 2 to 6) (Supplemental Table S2).
Library Construction, Hybrid Capture, and Sequencing
Three sets of samples were taken into library construction using either 200 ng (formalin-fixed, paraffin-embedded and fresh-frozen samples) or 100 ng (cell line sample) of extracted genomic DNA. Samples with less than the standard amount underwent library construction using a modified protocol.
Standard library construction: Samples were fragmented to 250 bp using Covaris ultrasonication (LE220 Focused-ultrasonicator; Covaris, Woburn, MA) and size selected with Agencourt AMPure XP beads (Beckman Coulter Inc., Indianapolis, IA) with a bead to sample ratio of 1:1. Samples were prepared for Illumina sequencing using a KAPA HTP or KAPA HyperPrep Library Preparation Kit (Kapa Biosystems, Wilmington, MA) and IDT xGen Dual Index UMI adapters (Integrated DNA Technologies, Coralville, IA).
Low-input library construction with KAPA HTP kit (<30 ng; n = 5): Briefly, end-repair reaction volumes were reduced by 65% while both A-tailing and ligation volumes were at 40% of manufacturer recommended volumes. Similarly, enzyme amounts used were reduced (end repair, 40% reduction; A-tailing, 33% reduction; ligation, 40% reduction) as was the amount of adapter/ligation (60% reduction; adapter stock concentration was 4 μmol/L).
Hybrid capture and sequencing: Library aliquots were pooled by equal volume and sequenced on an Illumina MiSeq Nano version 2 (Illumina Inc., San Diego, CA) flow cell to estimate each library's concentration on the basis of the number of index reads per sample. For hybrid capture, libraries were pooled (approximately 12 libraries/pool) by equal mass (total of 750 ng of input) and captured using either SureSelectXT HS or SureSelectXT Fast Target Enrichment System (Agilent Technologies) using ViroPanel baits that were diluted 1:7 and then mixed with OncoPanel baits at equal volume. Paired-end sequencing (2 × 101 nucleotides; 14-nucleotide index read for i7 (8-nucleotide index plus a 6-nucleotide unique molecular index sequence) and 8-nucleotide read for i5 were performed on an Illumina HiSeq 2500 in Rapid Run Mode.
Bioinformatics and Human Read Analysis
Pooled samples were demultiplexed and sorted using Illumina's bcl2fastq software version 2.17. Reads were aligned to the b37 reference sequence from the Human Genome Reference Consortium (GRCh37) as well as viral genomes targeted by the Virus Capture Bait set using bwa mem.20 Targeted viral and human genomes were combined into one alignment reference to map reads to the best matching reference sequence (Table 1). Reference sequences were included for HPV6 (X00203.1), HPV11 (M14119.1), HPV53 (X74482.1), and HPV61_E6 (U31793.1), viruses that were not baited but for which infected samples were included.
Duplicate reads were identified using unique molecular indexes using fgbio (http://fulcrumgenomics.github.io/fgbio, last accessed November 19, 2019) and marked using the Picard tools (http://broadinstitute.github.io/picard, last accessed November 19, 2019). The alignments were further refined using the Genome Analysis Toolkit for localized realignment around insertion/deletion sites and base quality score recalibration.21,22
Mutation analysis for single nucleotide variants (SNVs) was performed using MuTect version 1.1.4 in paired mode using CEPH as normal and annotated by Variant Effect Predictor version 79.23,24 SomaticIndelDetector tool (part of Genome Analysis Toolkit version 4.1.4.0, https://software.broadinstitute.org/gatk, last accessed November 19, 2019) was used for insertion/deletion calling. Additional filters were applied using Exome Sequencing Project and gnomAD data sets25 to flag common single-nucleotide polymorphisms (SNPs). Variants with frequency >10% in any gnomAD or Exome Sequencing Project population were considered to be a common SNP irrespective of presence in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. Copy number variants were identified using an in-house algorithm-RobustCNV.18 Large structural variations were detected using BreaKmer.26
Viral Analysis, Detection, and Integration
Viral presence was determined by counting the number of properly aligned read pairs that mapped to the viral genome with mapping qualities greater than Q40 using pysam (http://pysam.readthedocs.io/en/latest/api.html, last accessed November 19, 2019). To adjust for differences in sequencing depth, viral read counts were normalized per million mapped reads in the sample. For overall detection of viral genome presence, the normalized aligned read count to the entire viral genomes interrogated was used. Significant reductions in false-positive virus detection were accomplished by identifying reads whose dual-matched sample barcodes were not identical.27 Evidence of viral integration was detected using structural variation and insertion/deletion analysis by assembly (SvABA, https://github.com/walaj/svaba, last accessed November 19, 2019).28 This tool performs de novo sequence assemblies on soft clipped and discordantly aligned reads that could provide evidence for viral integrations. Assembled contigs are then aligned back to the reference genome to validate the event and obtain break point coordinates. We performed alignment on two separate references where the artificial break point of the viral genome was located in unique regions (linearizing the circular genome leads to false positive for structural variant detection). By aligning twice and taking overlapping events, those events caused by the artificial break point are removed. Because SvABA's filtering rules were not optimized for viral detection, the unfiltered output was parsed for our analysis.
OncoPanel Concordance Analysis and Sensitivity and Specificity Calculations
BAM files were reprocessed using a nonviral analysis workflow. SNV calling was performed with MuTect version 1.1.4. Sequential MCC cases had been collected and processed by the clinical OncoPanel assay over several years, resulting in testing with three different versions of the OncoPanel gene panel.17, 18, 19 To account for those differences, SNVs were selected from regions common to all iterations of OncoPanel (>672 Kb of bait territory).17, 18, 19 Furthermore, SNV filtering done with a panel of normals was specific to each version of OncoPanel used. Therefore, the union of all panels of normal SNVs that occurred in all OncoPanel versions was used. Sensitivity, positive predictive value, false discovery rate, and false negative rate were calculated by use of the true-positive (TP), false-negative (FN), and false-positive (FP) counts in the following formula's: sensitivity = TP/(TP + FN), positive predictive value = TP/(TP + FP), false discovery rate = FP/(FP + TP), and false negative rate = FN/(FN + TP).
Global Variant Concordance (Host Genome)
Sensitivity and specificity were determined using reproducible variants, with an allele fraction of ≥10% and a coverage of ≥50×, as previously described.17 Variants recovered with ViroPanel were deemed to be true variants if they were previously detected with OncoPanel alone (without viral baits) and were classified as false positive if no prior detection was noted.
Clinical Variant Analysis
Manually reviewed variants detected with OncoPanel were used in a concordance analysis with variants detected by ViroPanel.18 Only variants that occurred in baited regions common to all OncoPanel versions were considered.
Evaluate Potential of Improper HPV Alignment in Absence of Correct Reference Sequence and SNV Identification
To investigate whether viral alignments were accurate or the result of being forced by the aligner, a pairwise sequence alignment was performed on the viral reference genomes for the E6 and E7 genes using ClustalW (Clustal 2.1 Multiple Sequence Alignments with the default parameters). The alignment quality of samples was further characterized with an identified primary infection by running them through MuTect 1.1.4 to identify somatic variations against the reference in the viral genomes.
For samples 18 and 20, the reads aligned to the primary infections viral genome were extracted and realigned to references where the primary infection genome was removed using bwa. The resulting alignments were subsequently deduplicated using Picard tools and then run through MuTect and CollectHsMetrics, as the previous samples were, to characterize the aligned reads.
Bioinformatic Calculation of Viral Genome Percentage Present
Viral genome coverage was determined by binning viral genomes into 40-bp intervals and using Picard CollectHsMetrics to detect the coverage per interval. Interval coverages are reported as a percentage of the viral genome targeted that has ≥10× coverage on the basis of the CollectHsMetrics output.
Results
Sequencing Metrics
Unlike OncoPanel genes, targets for the viral bait set are not present in all samples. Therefore, sequencing metrics were calculated using only the OncoPanel bait set. Sample failures for the OncoPanel assay have previously been defined as those samples that have a mean target coverage (MTC) of ≤50× or <80% on-target bases covered at or equal to 30× MTC.17 Using these criteria, a single, expected failure was observed, corresponding to mouse tail (sample 92), which was included as a filtering control for PDX samples. For sample sets 1 (samples 1 to 32), 2 (samples 33 to 101), and 3 (samples 102 to 124), MTC of 253× (range, 56.32× to 361.80×), 179× (range, 91.63× to 222.25×), and 233.51× (range, 82.96× to 357.02×) were observed, respectively (Table 3 and Supplemental Table S3). Mean duplication rates for the three sample sets were similar (30.8%, 39%, and 56.14%) as were the mean number of pass filtering reads per sample: 20.8, 16.33, and 23.98 million/sample for sets 1 to 3, respectively (Table 3 and Supplemental Table S3). Percentage target bases for each set was >80% (mean value). Data generated and analyzed for the current study are derived from patient samples containing identifiable sequencing data and are not publicly available. However, the corresponding author will make every effort to share data based on reasonable request.
Table 3.
ViroPanel Sequencing Metrics
| Sample set | Samples | Total reads | PF reads | % Selected bases | MTC | % Target bases at 30× | % Target bases at 50× | % Duplication |
|---|---|---|---|---|---|---|---|---|
| 1 | 1–32 | 20,852,400.19 | 20,852,400.19 | 82.31 | 253.88 | 90.58 | 87.12 | 30.78 |
| 2 | 33–101 | 16,524,325.79 | 16,524,325.79 | 85.06 | 179.76 | 88.86 | 84.75 | 39.25 |
| 3 | 102–124 | 24,764,409.30 | 23,919,764.70 | 87.85 | 233.50 | 90.77 | 87.36 | 56.14 |
Data are expressed as means. All sequencing metrics were calculated using human OncoPanel targets, which are expected to be present in all samples irrespective of viral status. Therefore, depending on the level of viral sequences captured, percentage selected bases will be slightly lower than their actual value for OncoPanel. Percentage target bases, specific to OncoPanel baits, is a more accurate reflection of overall capture performance. Similarly, duplication rates will be slightly lower because of the greater capture area introduced by the viral baits. Mouse tail was used as a filtering control to remove nonhuman/nonviral reads from patient-derived xenograft samples. Therefore, because of the extremely low number of on-target reads (Supplemental Table S3), mouse tail was not included for calculating the mean sequencing metrics.
MTC, mean target coverage; PF, pass filtering.
OncoPanel Variant Concordance for Capture with and without Viral Baits
Addition of viral baits to OncoPanel may affect its ability to detect host mutations. To assess this issue, all tier 1 to 418 (ie, all nonsynonymous, presumed somatic) host variants detected by ViroPanel in 45 samples (sets 1 and 2) were compared with prior OncoPanel results to determine the level of concordance. These samples were previously profiled with various versions of OncoPanel (versions 1 to 3).17, 18, 19 To account for differences between versions, the concordance analysis was restricted to baited regions common to all versions of OncoPanel. Mean sensitivity and specificity were 99.00% (95% CI, 97.97%–99.54%) and 99.98% (95% CI, 99.98%–99.99%), respectively, demonstrating ViroPanel's ability to detect clinically relevant variants is unaffected by addition of baits targeting oncoviruses. The true positive rate of the test is 84.4% (95% CI, 81.9%–86.6%), the true negative rate is 0.000008% (95% CI, 0.000004%–0.000016%), and the F1 score (the harmonic mean of precision and sensitivity) is 0.9113.
Virus Subtype Detection
Samples were classified as positive for virus infection if ≥10 viral reads mapped to 50% of the baited target region at a depth ≥10×. However, manual review of sequencing coverage using Integrated Genome Viewer (IGV) with SvABA integration data allowed further assessment of samples whose viral read counts and depth of coverage were below thresholds for a positive designation. Such samples may still be classified as positive for viral infection if manual review shows coverage at <10× depth is uniform over approximately 50% of baited target territory, indicating low-level infection or perhaps a heterogeneous sample with a limited number of infected cells. In addition, the presence of viral and host integration break points can be used to further support a positive designation for such samples.
ViroPanel found 100 of 124 samples to be positively infected (Table 4 and Supplemental Table S1). Six of the positive samples (33, 38, 42, 93, 103, and 121) were initially assigned as negative for virus infection because of viral read counts that were <10 reads/million. However, inspection of the sequencing reads in IGV demonstrated low coverage throughout most of their respective bait territories, supporting the presence of the detected virus. Of the samples whose viral status had been previously determined, ViroPanel results were concordant for 61 of the 66 samples. Manual interrogation of discrepant samples (see Discrepant Sample Review and Sensitivity and Specificity) shows actual concordance to be 65 of 66 samples (Table 4). The number of viral reads observed showed a large range from sample to sample, irrespective of the virus type (Supplemental Table S1).
Table 4.
ViroPanel Concordance
| Previous virus test result | ViroPanel—primary virus detected | Concordant samples, n | Total virus-positive sample count |
|---|---|---|---|
| HHV-8 (+) | HHV-8 | 8/8 | 8 |
| HBV (+) | HBV | 6/6 | 6 |
| HPV6 (+) | HPV6 | 1/1 | 1 |
| HPV11 (+) | HPV11 | 1/1 | 1 |
| HPV45 (+) | HPV16∗ | 1/1∗ | 1 |
| HPV16 (+) | HPV16 | 13/13 | 13 |
| HPV16 (+) | HPV18† | 1/1† | 1 |
| HPV18 (+) | HPV18 | 4/4 | 4 |
| HPV33 (+) | HPV33 | 6/6 | 6 |
| HPV35 (+) | HPV35 | 4/4 | 4 |
| HPV53 (+) | HPV53 | 2/2 | 2 |
| HPV56 (+) | HPV56 | 1/1 | 1 |
| HPV61 (+) | HPV61 | 1/1 | 1 |
| HPV82 (+) | HPV82 | 1/1 | 1 |
| MCPyV (+) | MCPyV | 6/6 | 6 |
| HPV (−) | HPV16 (1 sample) | 6/7 | 1 |
| MCPyV (−) | None | 3/3 | 0 |
| None | MCPyV | NA | 43 |
| Total | 65/66 | 100 |
Number of concordant samples is listed by virus type along with the total number of controls concordant and total number of samples determined to be positive for virus infection. ViroPanel found evidence to support positive virus infection for 100 samples. Of 58 samples with no previous virus testing, ViroPanel showed 43 to be positively infected for MCPyV. Sample 28 previously tested negative for HPV was found by ViroPanel to be positive for HPV16 (listed as discordant). Of 66 virus controls, 65 were concordant.
HBV, hepatitis B virus; HHV-8, human herpesvirus 8; HPV, human papillomavirus; MCPyV, Merkel cell polyomavirus.
Sample 16 was previously reported to be HPV45 positive but found by ViroPanel to be HPV16 positive (see Discrepant Sample Review and Sensitivity and Specificity).
Sample 123 was previously assigned as positive for HPV16 on the basis of p16 expression. ViroPanel found this sample to be co-infected with HPV16, HPV18, and HPV33 (see Co-Infection Status), with HPV18 representing the primary infecting virus.
HBV Detection
ViroPanel confirmed the presence of HBV in all six samples (21 to 24, 116, and 117) previously found to be HBV positive using IHC. For these samples, the number of sequence reads aligning to the HBV genome ranged from 502 to 146,677 viral reads/million (Table 4 and Supplemental Table S1); viral reads per million are calculated [viral reads/(total reads/1 × 106)] using viral and total reads having a proper pair of >Q40 that are not duplicates. Of the remaining 118 samples, 72 and 77 contained a limited number of reads to HBV occurring as putative co-infections and primary infections, respectively. Review of sequencing coverage for sample 72 showed insufficient evidence to support HBV co-infection (Table 5 and Supplemental Table S1). MCC sample 77 was not previously tested for virus infection, had five HBV reads/million. Manual review of the sequencing coverage in IGV shows low coverage at two discrete sites (<50% of genome), resulting in classification as negative. Significantly, all negative controls (10 samples) were found to be negative for HBV, demonstrating no off-target capture.
Table 5.
Viral Co-Infection
| Sample | ViroPanel primary virus | Second virus detected (viral reads/million) | Virus co-infection status |
|---|---|---|---|
| 5 | HPV16 | HPV33 = 6 | Y |
| 7 | HPV18 | HPV16 = 10 | S |
| 9 | HPV18 | HPV33 = 1 | S |
| 11 | HPV33 | HPV16 = 97 | Y |
| 12 | HPV33 | HPV56 = 1 | S |
| 14 | HPV35 | MCPyV = 21 | Y |
| 14 | HPV35 | HPV33 = 1 | N |
| 17 | HPV53 | HPV18 = 8 | S |
| 17 | HPV53 | HPV33 = 28; HPV58 = 4 | N |
| 18 | HPV56 | HPV66 = 157 | Y |
| 21 | HBV | MCPyV = 1 | Y |
| 21 | HBV | HHV-8 = 1 | N |
| 28 | HPV16 | HPV18 = 2 | N |
| 29 | HHV-8 (KSHV) | MCPyV = 9 | Y |
| 50 | MCPyV | HPV18 = 2 | N |
| 55 | MCPyV | HPV33 = 10 | S |
| 65 | MCPyV | HPV16 = 56 | Y |
| 65 | MCPyV | HPV56 = 4 | S |
| 72 | MCPyV | HBV = 1 | N |
| 102 | HPV16 | MCPyV = 5 | S |
| 107 | HPV18 | HPV16 = 10; HPV33 = 69 | Y |
| 108 | HPV33 | HPV16 = 2 | S |
| 113 | HHV-8 (KSHV) | HPV16 = 1 | S |
| 115 | HHV-8 (KSHV) | HPV16 = 8 | Y |
| 115 | HHV-8 (KSHV) | HPV33 = 1 | N |
| 116 | HBV | HPV16 = 111 | Y |
| 116 | HBV | MCPyV = 2 | N |
| 117 | HBV | HPV16 = 3 | Y |
| 123 | HPV18 | HPV16 = 2 | Y |
| 124 | HHV-8 (KSHV) | HPV16 = 1 | S |
Samples with secondary viruses detected are listed along with the primary virus present. Positive co-infection status was assigned by manual review of the secondary virus sequencing coverage in Integrated Genome Viewer. Co-infections were designated as positive on the basis of there being approximately 50% of coverage over the bait territory.
HBV, hepatitis B virus; HHV-8, human herpesvirus 8; HPV, human papillomavirus; KSHV, Kaposi sarcoma–associated herpesvirus; MCPyV, Merkel cell polyomavirus; N, negative; S, suspected infection; Y, positive infection.
HHV-8 (KSHV) Detection
ViroPanel included baits directed to the LANA1/vCyclin and LANA2 regions of HHV-8 and confirmed the presence of HHV-8 in all eight samples (29 to 32, 113 to 115, and 124) previously shown to be HHV-8 (IHC test) positive (Table 4 and Supplemental Table S1). HHV-8 viral read counts ranged in these samples from 238 to 9124 reads/million (Supplemental Table S1). Similar to HBV, none of the negative controls (10 samples) detected HHV-8. Of the remaining 116 samples, sample 21 (primary infection with HBV) was reported as having one viral read/million (Table 5 and Supplemental Table S1). Inspection of sequencing coverage in IGV did not support co-infection with HHV-8.
HPV Detection
HPV detection using ViroPanel was performed on 36 samples that had previously been found positive for high-risk HPV genotypes using clinical IHC, ISH, or a PCR-RFLP assay (Supplemental Table S1). Among the 36 HPV positive samples, five that were not targeted by ViroPanel [HPV6, HPV11, HPV53 (2 samples), and HPV61] were included to assess if conserved regions within E6 and E7 would be sufficient for hybrid capture. ViroPanel detected the presence of an HPV genotype in all 36 of the HPV positive controls (Table 4 and Supplemental Table S1). Samples showed a wide distribution of viral reads/million, with the highest observed in an HPV33 infected sample (sample 12) having 483,197 reads/million. Despite the high viral read count, OncoPanel MTC was 250×, well above the minimum required threshold of 50× (Supplemental Table S3). Furthermore, all samples with detected viral reads showed OncoPanel MTC ≥50× regardless of viral read count number (Supplemental Table S3).
Sample 16 was found to be positive for an oncogenic HPV type. However, ViroPanel detected 93,400 viral reads/million that supported HPV16 infection rather than the previously reported presence of HPV45 (Figure 1 and Supplemental Table S1), further detailed in Discrepant Sample Review and Sensitivity and Specificity.
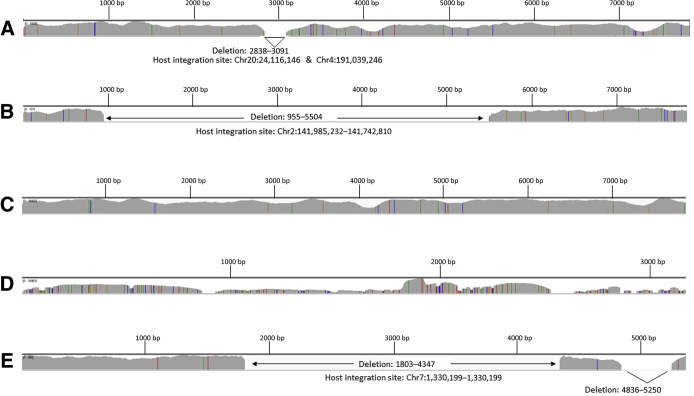
Figure 1.
Virus target coverage obtained with ViroPanel. Integrated Genome Viewer (IGV) coverage plots of selected ViroPanel samples. A: ViroPanel detected 5581 viral reads/million to support presence of human papillomavirus (HPV) 16 in sample 28 (previously tested HPV negative via in situ hybridization) occurring with a 253-bp deletion (HPV16: 2838 to 3091 bp) and evidence of viral integration. B: Sample 7 shows evidence of HPV18 integration and a 4549-bp deletion (HPV18: 955 to 5504 bp), identical integration features observed in a second biopsy taken from the same patient on a later date (sample 9). C: ViroPanel detected 94,340 viral reads/million supporting HPV16 infection in sample 16, which was previously tested and designated as HPV45 because of a G > A mutation at position HPV16: 7008 that eliminated a Pst1 restriction enzyme site used for PCR–restriction fragment length polymorphism analysis (see Discrepant Sample Review and Sensitivity and Specificity). D: Hepatitis B virus coverage observed in sample 21. E: Merkel cell polyomavirus coverage observed in Merkel cell carcinoma sample 80, with evidence of viral integration; viral and host break points observed in matching patient-derived xenograft, sample 95. Genomic start positions in IGV plots correspond to first nucleotide in each of the virus genomic references (Table 1). Chr, chromosome.
Clinically, p16 expression is routinely used to infer the presence of HPV16 (Supplemental Table S1). Ten HPV16 positive controls were previously assigned their viral status on the basis of IHC testing for p16. Of these samples, three (samples 103, 121, and 123) had viral read counts that were at or below our criteria for designation as being positively infected, leading to initial designation as discordant samples. However, review of the sequencing data in IGV showed low coverage throughout their HPV16 genomes, resulting in redesignation as positive for HPV16 infection. Interestingly, sample 123 was co-infected with HPV18, whose viral read count of 47,549 viral reads/million classifies it as the true primary infecting virus.
The five samples (1, 2, 17, 19, and 112) infected by HPV types for which there were no ViroPanel baits present were all reported as having >10 viral reads/million (Supplemental Table S1). Review of their sequencing coverage in IGV shows the samples to have low coverage that supports a positive designation, which is discussed further below.
Of the 10 viral negatives, nine contained no viral reads to any of the 16 HPV types baited or the four unbaited HPV types. Sample 28, which had previously tested negative by ISH (negative for HPV6/11, HPV16/18, and HPV31/33), was found to have 5581 reads/million aligning throughout the HPV16 genome, supporting the presence of HPV16 (Figure 1 and Supplemental Table S1); discrepant sample is discussed below.
HPV co-infections were observed in 21 samples, most of them with low viral read counts (Table 5 and Supplemental Table S1); this is discussed in Co-Infection Status.
Merkel Cell Polyomavirus Detection
ViroPanel confirmed the presence of MCPyV in all six samples (cell line samples 86 to 91) known to be MCPyV positive (Supplemental Table S1). Furthermore, no MCPyV sequence reads were detected in the MCC cell lines (samples 83 to 85) known to be MCPyV negative or any of the other negative virus controls [CEPH controls (n = 4), mouse tail control (n = 1), and patient samples (n = 2)].
In addition to the samples of known viral status, 57 Merkel cell carcinoma samples of unknown viral status were tested and MCPyV detected in 43. Four of the samples (33, 38, 42, and 93) had <10 viral/reads million and were redesignated as MCPyV positive after review of the sequencing data in IGV demonstrated low coverage throughout most of the virus genome. MCPyV read counts showed a wide distribution, with the highest being 503,370 reads/million observed in sample 40 (Supplemental Table S1), OncoPanel MTC = 151×. ViroPanel found evidence of secondary MCPyV co-infections in five samples (14, 21, 29, 102, and 116), all with low viral read counts (Table 5 and Supplemental Table S1); this is discussed in Co-Infection Status.
Discrepant Sample Review and Sensitivity and Specificity
For discrepant samples 16 and 28, their prior documentation status and viral test histories were reviewed. Sample 16 (previously documented as being positive for HPV45) had no sequencing reads mapping to HPV45. However, it showed 93,400 reads distributed throughout the genome of HPV16 (Supplemental Table S1 and Figure 1). This sample was previously tested using an HPV PCR-RFLP assay. Review of the gel image (Supplemental Figure S1) shows a digestion pattern different from both HPV45 and HPV16. Looking at the sequencing data in IGV, we find that the aberrant restriction pattern was due to the presence of a G > A mutation in 71% of the covering reads at HPV16 genomic coordinate 7008. The G > A mutation occurred within a Pst1 restriction site used to cleave the amplicon, leading to a misinterpretation of the digestion pattern. For sensitivity and specificity calculations, this sample has been classified as concordant because of agreement between the genomic data and the predicted and observed effect in the PCR-RFLP assay.
The second discordant sample (sample 28) was documented as being viral negative on the basis of an ISH assay targeting HPV 6/11, 16/18, and 31/33 that was conducted at an outside facility. ViroPanel detected 5581 reads covering all genes of HPV16 (Figure 1 and Supplemental Table S1). An aliquot of the original source material was submitted for a third HPV test, which assesses HPV status with a PCR-RFLP assay. Results supported the original ISH test result (negative). Detection of viral reads throughout the HPV16 genome (Figure 1) tends to support the notion that the sample tested is weakly positive for HPV16, with next-generation sequencing being a more sensitive test than PCR; alternatively, this is a false-positive result.
Furthermore, virus integration analysis identified viral and host break points in sample 28, indicating the presence of an integrated virus genome (Figure 1 and Supplemental Table S4). Finally, SNP analysis did not detect any unexpected matches with any of the other samples tested (data not shown). However, to assess concordance, we have designated sample 28 as discordant from previous test results. Therefore, with the exception of sample 28, ViroPanel was 98.5% concordant with previous test results for 65 of the 66 control samples; as discussed in HPV Detection, samples 103, 121, and 123 were reclassified as HPV16 positive after review of their sequencing coverage in IGV showed support for virus presence.
Detection of Viral Integration in the Host Genome
Samples found to be positive for viral infection were further analyzed using structural variation analysis by assembly (SvABA) to search for evidence of viral integration events in soft clipped and discordant reads.28 Our definition for viral integration required detection of two or more viral/host break points. Integration events were observed in MCPyV (42 samples), HPV16 (nine samples), HPV18 (five samples), HPV33 (four samples), and HBV (one sample) samples (Supplemental Table S4). HPV and MCPyV genomes displayed a wide range in the size of their genomic (viral) deletions, some of which had lost >50% of their genetic material (Figure 1). Interestingly, within several samples, ViroPanel detected the presence of multiple viral integration variants defined by different break points and alternative viral deletions (Supplemental Table S4). In addition, several examples where sequencing coverage was present throughout the viral genome while co-occurring with clear evidence supporting viral break points whose genomic locations co-incided with dramatic alterations in coverage depth were noted, suggesting a deletion of the viral genome had occurred with possible co-occurrence of viral species of varying genomic lengths (data not shown).
Among the cohort were two sets of biopsy time points occurring in HPV16 (set 7; samples 7 and 9) and HPV18 (set 8; samples 109 and 110). Significantly, the samples within each set shared the same integration break points, demonstrating their common origin (Figure 1 and Supplemental Tables S2 and S4). Similarly, among the samples whose viral status was unknown were five sets of PDX samples with matching primary tumors and one set of technical replicates, all of which shared identical integration loci among the samples of their respective set (Supplemental Table S2).
Detection of Nontargeted HPV Subtypes through Capture of Conserved Regions
The high level of sequence identity shared among clinically important HPV subtypes, especially for the E6 and E7 coding regions, could result in some level of capture for nontargeted subtypes. To test this hypothesis, five samples positive for HPV 6, 11, 53 (2 samples), and 61, HPV genotypes that were not targeted by the bait set, were included to ascertain if they could be captured and correctly detected (Supplemental Table S1). After inclusion of the untargeted HPV genomes to our viral and host references, reads were detected for all five of the nontargeted HPV subtypes. However, the number of reads was significantly lower than their targeted counterparts, probably reflecting capture of only the most conserved viral sequences (Supplemental Table S1). Manual review of sequencing coverage in IGV demonstrates evidence supporting positive infection status for all five samples (samples 1, 2, 17, 19, and 112).
Given the off-target capture of these unbaited HPV samples, we investigated the likelihood of incorrect assignment of an HPV type in the event its correct reference genome (bait territory) file is absent. The E6 and E7 genes were targeted in all HPV types, baited for. A ClustalW analysis was performed to assess the nucleotide similarity of these genes between HPV types, and the mean percentage nucleotide identity was found to be between E6 and E7 for different HPV types to be 46.82% and 44.94%, respectively (Supplemental Table S5). To further assess the likelihood of an improper alignment, samples 18 and 20 were realigned using reference files that lacked their genomes HPV56 and HPV82, respectively. These samples were chosen because they contained the greatest number of viral SNVs (Supplemental Table S6). No alignment reads were reported for sample 18 to any HPV type. Sample 20 showed alignment to a single discrete region of the baited territory of HPV51, with 1.9% of the baited region covered at a depth of 10×, which would be insufficient for a positive infection designation (Supplemental Table S7).
Given i) the strict alignment criteria used, ii) the moderate nucleotide identity of E6 and E7 between HPV types, and iii) the fact that HPV types are generally defined as containing >10% genome variation from one another, the probability of an incorrect alignment is low.
Percentage of Viral Genome Present
Using sample sets 1 and 2 (samples 1 to 101), interval bait coverage was used to calculate the percentage of viral genome present at sequencing depths of 10×, 30×, 50×, and 100× for each sample’s primary infecting virus. Therefore, the percentage of virus genome reported is a calculation of the bait territory captured at a given depth of coverage. Analysis was restricted to the virus territory baited for. HBV-infected samples (full genome baited) 21 to 24 retained most of their viral genomes, ranging from 95.31% to 100% present at a depth of 10× (Supplemental Table S6). In contrast, samples infected with MCPyV, HPV16, HPV18, and HPV33 (full genomes targeted) showed a wide distribution in the level of viral genomic material retained (Supplemental Table S6), as has previously been reported.29, 30
SvABA integration analysis identified integration events for 42 MCPyV infected samples. Within the integrated cohort, the percentage of MCPyV present at 10× coverage ranged from 25% to 100%, with 22 samples having lost ≥50% of their total genome.
As an orthogonal check, the percentage of virus genome present was determined using the SvABA integration data relative to the IGV sequencing coverage, and excellent agreement was found with the interval coverage method. Nine HPV samples were found to be integrated and were observed to have a wide range in the genome percentage present at 10× depth, ranging from 43% to 100% (Supplemental Table S6).
Co-Infection Status
ViroPanel detected viral reads for co-infection in 24 samples (Table 5 and Supplemental Table S1). Review of sequencing coverage in IGV confirmed 12 of the samples to possess low coverage throughout most of their baited territory, providing support for the presence of a second, and in one case a third, infecting virus. Among these samples, six (samples 11, 14, 18, 65, 107, and 116) had particularly strong evidence of co-infection.
The primary infections and co-infections (primary/co-infection) for samples 5, 11, 14, 18, and 123 were HPV16/HPV33, HPV33/HPV16, HPV35/MCPyV, HPV56/HPV66, and HPV18/HPV16, respectively. Interestingly, sample 107 was co-infected by three HPV types, HPV18 (primary infection; 2293 viral reads/million), HPV16, and HPV33. Furthermore, HPV16 occurred as co-infections in sample 116 and 117, whose primary infection was HBV. A third HBV infected sample, sample 21, was found to be co-infected with MCPyV. Similarly, HHV-8 infected samples 29 and 115 were co-infected with MCPyV and HPV16, respectively. Finally, sample 65 was infected by MCPyV (primary virus; 1740 viral reads/million) and HPV16.
Of the remaining samples, nine had sequencing coverage that warrants their designation as suspected for positive infection. Significantly, none of these co-infected samples showed detectable SNP profiles (on the basis of host SNP analysis; data not shown) that matched any other samples, providing further confidence that these are real viral co-infections native to the biopsied specimen.
Virus SNV Detection
Virus-positive samples from sets 1 and 2 (samples 1 to 101) were assessed for SNVs within their viral reads using MuTect version 1.0.47986. Table 6 summarizes the range of SNV variants detected along with the mean number of SNVs per virus type and the mean nucleotide variation relative to the reference sequence (calculated using the captured bait territory size; SNVs with an allele fraction >30%/captured bait territory). Supplemental Table S6 lists the number of SNVs occurring with an allele fraction ≥30% for each sample and the percentage nucleotide variation based on the actual bait territory captured. The significance of these viral SNVs is uncertain. The percentage of nucleotide variation for all viruses was <10%, with the highest observed in sample 1 (6.6% nucleotide variation), which was infected for HPV6, a virus that was not baited for. Designation as a unique HPV type generally requires >10% nucleotide variation at the genomic level. The low variation rate in combination with our concordance data further supports the conclusion that the correct identity of the infecting virus has been determined by ViroPanel.
Table 6.
SNV Summary
| Virus—ViroPanel (samples, n) | Target region | Target size, bp | Mean captured territory, bp | SNV range | Mean SNVs/virus∗ | Mean % nucleotide variation† |
|---|---|---|---|---|---|---|
| HBV (4) | Full genome | 3182 | 3182 | 6–30 | 14 | 0.44 |
| HHV-8 (KSHV) (4) | LANA1/2 | 2764 | 2764 | 0–32 | 9.5 | 0.34 |
| HPV16 (6) | Full genome | 7904 | 7732 | 4–35 | 18.33 | 0.24 |
| HPV18 (3) | Full genome | 7857 | 4844 | 14–19 | 17.33 | 0.44 |
| HPV33 (3) | Full genome | 7909 | 6987 | 1–10 | 5 | 0.07 |
| HPV35 (3) | E6, E7 | 751 | 751 | 9–17 | 13.33 | 1.78 |
| HPV56 (1) | E6, E7 | 787 | 787 | NA | 45 | 5.72 |
| HPV82 (1) | E6, E7 | 765 | 765 | NA | 33 | 4.31 |
| HPV6 (1) | Unbaited | NA | 528 | NA | 32 | 6.06 |
| HPV11 (1) | Unbaited | NA | 724 | NA | 25 | 3.45 |
| HPV53 (1) | Unbaited | NA | 784 | NA | 34 | 4.34 |
| HPV61 (1) | Unbaited | NA | 703 | NA | 6 | 0.85 |
| MCPyV (49) | Full genome | 5387 | 3644 | 0–20 | 5.16 | 0.15 |
SNV range and mean SNVs per virus type listed for samples 1 to 101. Mean percentage nucleotide variation calculated using SNVs with an allele fraction >30% and the mean captured territory. Individual sample values listed in Supplemental Table S6.
HBV, hepatitis B virus; HHV-8, human herpesvirus 8; HPV, human papillomavirus; KSHV, Kaposi sarcoma–associated herpesvirus; MCPyV, Merkel cell polyomavirus; NA, not applicable; SNV, single nucleotide variant.
On the basis of SNVs with an allele fraction >30%.
SNVs/captured territory.
Discussion
In this study, we demonstrate that ViroPanel can be used to genomically profile somatic host mutations while simultaneously detecting HHV-8, HBV, MCPyV, and multiple HPV types. In addition, our analysis workflow allows us to identify virus integration events and define virus/host break points from which we can determine the length of associated deletions of the host and/or virus genomes. With respect to oncoviruses, accurate identification of the integration status is clinically relevant because it can affect virus oncogene and host tumor suppressor expression. For instance, in the context of an HPV infection, integration is often accompanied by partial deletion of the E2 gene, which negatively regulates E6 and E7 viral oncogenes. Once relieved of the E2 transcriptional repression, E6 and E7 increase the carcinogenic potential of the virus by inactivating p53 and pRb, respectively. Clinically, the ability to discern an integration event could truly indicate causality, beyond tests like p16+ IHC and viral PCR/ISH. This is relevant as some nonoropharynx tumors, like paranasal sinus cancers, nasal cavity tumors, some oral tongue cancers, and even Epstein-Barr virus nasopharyngeal tumors can be HPV+ (by other methods), but it is not known if the presence of virus impacts outcomes or relates to causality. It also may be important to understand HPV co-infection and whether one or more HPV strains contribute to carcinogenesis in a given tumor, or if one is a bystander infection.
Furthermore, analysis of virus SNV data will allow us to identify subtypes that are more carcinogenic, which can be used to better inform clinical treatment.25 For instance, ViroPanel found significant evidence of HPV16 presence in a sample that was previously documented as being infected by HPV45, a genotype with lower risk of promoting cancer, a diagnosis that could influence treatment strategy. Moreover, some recent protocol treatments use HPV-16 E7 directed vaccine therapies, so this knowledge is relevant. Expansion of our analysis will allow us to use virus SNV data to phylogenetically classify infecting viruses and identify genomic alterations of viral oncogenes documented as having increased carcinogenic activity. Inclusion of viral status in association with genomic profiling of the patient's tumor will present a more unified and comprehensive view of the genetic alterations driving the cancer, allowing for more personalized patient treatment and care.
Acknowledgments
We thank Profile (Brigham and Women's Hospital and Dana-Farber Cancer Institute) and the Center for Cancer Genomics (Dana-Farber Cancer Institute) for providing assistance with sequence analysis and sample procurement; Amanda Paskavitz, Haley Coleman, Audrey Dalgarno, and Meredith Bemus (Center for Cancer Genomics) for assistance with sample identification and sequencing; and our patients for participating in this research.
Footnotes
Supported by the Dana-Farber Cancer Institute and the Department of Pathology, Brigham and Women’s Hospital.
Disclosures: Salary support for G.J.S. comes from a National Cancer Institute Cancer Research Training Award.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2019.12.010.
Supplemental Data
Analysis of sample 16 human papillomavirus (HPV) PCR–restriction fragment length polymorphism (RFLP) data used to assign HPV45 status. Sample 16 discrepancy is caused by a variant in one of the RFLP digest sites required for genotyping the HPV subtypes. A: Degenerate primers amplify a consensus region of the HPV L1 gene. A and B: The HPV16 amplicon contains two PstI restriction digest sites (yellow; A), whereas the HPV 45 amplicon contains only one PstI site (B); the reverse primer sequence (PrimerR) is underlined. C: Gel image and expected/observed Pst1 fragment lengths of the HPV RFLP assay for HPV16-positive, HPV-negative (NEG), and sample 16 amplicons. HPV16 predominant Pst1 bands at 216 and 210 nucleotides (nt) are indicated by blue and orange arrows, respectively. Sample 16 resembled and was interpreted as HPV45 because of the 242-nt band (green arrow), but a faint band at 216 nt is visible in the digest (blue arrow). D: A G/A mutation in the second PstI site (boxed area) at position 7008 (asterisk) is present in 71% of the reads, resulting in an aberrant restriction digest pattern that was misinterpreted as HPV45. The faint band at 216 nt is due to the 30% wild-type sequence in this sample. ViroPanel finds supporting evidence only for HPV16 infection.
References
- 1.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.NTP (National Toxicology Program) U.S. Department of Health and Human Services; Research Triangle Park, NC: 2016. Report on Carcinogens, Fourteenth Edition. [Google Scholar]
- 3.Mesri E.A., Feitelson M.A., Munger K. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe. 2014;15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J.J. Genomic instability induced by human papillomavirus oncogenes. N Am J Med Sci (Boston) 2010;3:43–47. doi: 10.7156/v3i2p043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saraiya M., Unger E.R., Thompson T.D., Lynch C.F., Hernandez B.Y., Lyu C.W., Steinau M., Watson M., Wilkinson E.J., Hopenhayn C., Copeland G., Cozen W., Peters E.S., Huang Y., Saber M.S., Altekruse S., Goodman M.T., Workgroup, HPVToC US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst. 2015;107:djv086. doi: 10.1093/jnci/djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C., Westra W.H., Li S., Cmelak A., Ridge J.A., Pinto H., Forastiere A., Gillison M.L. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 7.Ang K.K., Harris J., Wheeler R., Weber R., Rosenthal D.I., Nguyen-Tan P.F., Westra W.H., Chung C.H., Jordan R.C., Lu C., Kim H., Axelrod R., Silverman C.C., Redmond K.P., Gillison M.L. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 9.Sung J.J., Tsoi K.K., Wong V.W., Li K.C., Chan H.L. Meta-analysis: treatment of hepatitis B infection reduces risk of hepatocellular carcinoma. Aliment Pharmacol Ther. 2008;28:1067–1077. doi: 10.1111/j.1365-2036.2008.03816.x. [DOI] [PubMed] [Google Scholar]
- 10.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moshiri A.S., Doumani R., Yelistratova L., Blom A., Lachance K., Shinohara M.M., Delaney M., Chang O., McArdle S., Thomas H., Asgari M.M., Huang M.L., Schwartz S.M., Nghiem P. Polyomavirus-negative merkel cell carcinoma: a more aggressive subtype based on analysis of 282 cases using multimodal tumor virus detection. J Invest Dermatol. 2017;137:819–827. doi: 10.1016/j.jid.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker J.C., Stang A., DeCaprio J.A., Cerroni L., Lebbe C., Veness M., Nghiem P. Merkel cell carcinoma. Nat Rev Dis Primers. 2017;3:17077. doi: 10.1038/nrdp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 14.Cesarman E., Chang Y., Moore P.S., Said J.W., Knowles D.M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 15.Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d'Agay M.F., Clauvel J.P., Raphael M., Degos L., Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 16.Heideman D.A., Hesselink A.T., Berkhof J., van Kemenade F., Melchers W.J., Daalmeijer N.F., Verkuijten M., Meijer C.J., Snijders P.J. Clinical validation of the cobas 4800 HPV test for cervical screening purposes. J Clin Microbiol. 2011;49:3983–3985. doi: 10.1128/JCM.05552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia E.P., Minkovsky A., Jia Y., Ducar M.D., Shivdasani P., Gong X., Ligon A.H., Sholl L.M., Kuo F.C., MacConaill L.E., Lindeman N.I., Dong F. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–758. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 18.Sholl L.M., Do K., Shivdasani P., Cerami E., Dubuc A.M., Kuo F.C. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna G.J., Lizotte P., Cavanaugh M., Kuo F.C., Shivdasani P., Frieden A., Chau N.G., Schoenfeld J.D., Lorch J.H., Uppaluri R., MacConaill L.E., Haddad R.I. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight. 2018;3:e98811. doi: 10.1172/jci.insight.98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abo R.P., Ducar M., Garcia E.P., Thorner A.R., Rojas-Rudilla V., Lin L., Sholl L.M., Hahn W.C., Meyerson M., Lindeman N.I., Van Hummelen P., MacConaill L.E. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using Kmers. Nucleic Acids Res. 2015;43:e19. doi: 10.1093/nar/gku1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacConaill L.E., Burns R.T., Nag A., Coleman H.A., Slevin M.K., Giorda K., Light M., Lai K., Jarosz M., McNeill M.S., Ducar M.D., Meyerson M., Thorner A.R. Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics. 2018;19:30. doi: 10.1186/s12864-017-4428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wala J.A., Bandopadhayay P., Greenwald N.F., O'Rourke R., Sharpe T., Stewart C., Schumacher S., Li Y., Weischenfeldt J., Yao X., Nusbaum C., Campbell P., Getz G., Meyerson M., Zhang C.Z., Imielinski M., Beroukhim R. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akagi K., Li J., Broutian T.R., Padilla-Nash H., Xiao W., Jiang B., Rocco J.W., Teknos T.N., Kumar B., Wangsa D., He D., Ried T., Symer D.E., Gillison M.L. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starrett G.J., Marcelus C., Cantalupo P.G., Katz J.P., Cheng J., Akagi K., Thakuria M., Rabinowits G., Wang L.C., Symer D.E., Pipas J.M., Harris R.S., DeCaprio J.A. Merkel cell polyomavirus exhibits dominant control of the tumor genome and transcriptome in virus-associated merkel cell carcinoma. mBio. 2017;8:e02079-16. doi: 10.1128/mBio.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of sample 16 human papillomavirus (HPV) PCR–restriction fragment length polymorphism (RFLP) data used to assign HPV45 status. Sample 16 discrepancy is caused by a variant in one of the RFLP digest sites required for genotyping the HPV subtypes. A: Degenerate primers amplify a consensus region of the HPV L1 gene. A and B: The HPV16 amplicon contains two PstI restriction digest sites (yellow; A), whereas the HPV 45 amplicon contains only one PstI site (B); the reverse primer sequence (PrimerR) is underlined. C: Gel image and expected/observed Pst1 fragment lengths of the HPV RFLP assay for HPV16-positive, HPV-negative (NEG), and sample 16 amplicons. HPV16 predominant Pst1 bands at 216 and 210 nucleotides (nt) are indicated by blue and orange arrows, respectively. Sample 16 resembled and was interpreted as HPV45 because of the 242-nt band (green arrow), but a faint band at 216 nt is visible in the digest (blue arrow). D: A G/A mutation in the second PstI site (boxed area) at position 7008 (asterisk) is present in 71% of the reads, resulting in an aberrant restriction digest pattern that was misinterpreted as HPV45. The faint band at 216 nt is due to the 30% wild-type sequence in this sample. ViroPanel finds supporting evidence only for HPV16 infection.