Abstract
Rationale: Smoking results in at least a decade lower life expectancy. Mortality among current smokers is two to three times as high as never smokers. DNA methylation is an epigenetic modification of the human genome that has been associated with both cigarette smoking and mortality.
Objectives: We sought to identify DNA methylation marks in blood that are predictive of mortality in a subset of the COPDGene (Genetic Epidemiology of COPD) study, representing 101 deaths among 667 current and former smokers.
Methods: We assayed genome-wide DNA methylation in non-Hispanic white smokers with and without chronic obstructive pulmonary disease (COPD) using blood samples from the COPDGene enrollment visit. We tested whether DNA methylation was associated with mortality in models adjusted for COPD status, age, sex, current smoking status, and pack-years of cigarette smoking. Replication was performed in a subset of 231 individuals from the ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) study.
Measurements and Main Results: We identified seven CpG sites associated with mortality (false discovery rate < 20%) that replicated in the ECLIPSE cohort (P < 0.05). None of these marks were associated with longitudinal lung function decline in survivors, smoking history, or current smoking status. However, differential methylation of two replicated PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta) sites were associated with lung function at enrollment (P < 0.05). We also observed associations between DNA methylation and gene expression for the PIK3CD sites.
Conclusions: This study is the first to identify variable DNA methylation associated with all-cause mortality in smokers with and without COPD. Evaluating predictive epigenomic marks of smokers in peripheral blood may allow for targeted risk stratification and aid in delivery of future tailored therapeutic interventions.
Keywords: DNA methylation, epigenetics, all-cause mortality, survival analysis, chronic obstructive pulmonary disease
At a Glance Commentary
Scientific Knowledge on the Subject
In the United States, smoking results in at least a decade lower life expectancy, and mortality among current smokers is two to three times as high as that of never smokers. Despite significant efforts in genomic analysis, discovery of biomarkers for identification of smokers with the highest risk for death remains elusive. Evaluating predictive epigenomic marks of smokers in peripheral blood may allow for targeted risk stratification and therapeutic interventions to modify smoking-related disease outcomes.
What This Study Adds to the Field
Our genome-wide analysis revealed several methylation sites predictive of all-cause mortality in a cohort of smokers with and without chronic obstructive pulmonary disease, and independent of lung function decline or emphysema progression. We replicated findings, including ones for PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta), in a second cohort and observed differential lung tissue DNA methylation or gene expression signals for a subset of the identified CpG sites. These epigenetic associations with mortality highlight DNA methylation marks that may inform the prediction of smokers at greater risk for smoking-related death, while also highlighting future targets for functional validation.
Smoking has contributed to 25% of deaths among women and men 35 to 69 years of age, resulting in at least a decade lower life expectancy (1). In addition, mortality among current smokers is two to three times as high as never smokers, with the increased mortality linked to a growing list of diseases (2), including chronic obstructive pulmonary disease (COPD). Comorbidity is common among these smoking-linked diseases (3).
Genome-wide association studies of survival in lung cancer (4, 5), heart failure (6), and smoking stages (initiation, persistence, tolerance, and cessation) (7) have identified genetic variants associated with mortality. Epigenome-wide associations between DNA methylation (DNAm) and mortality have been evaluated in peripheral blood leukocyte DNA. In a 9-year longitudinal study of 499 subjects in Italy, which included current and former smokers (56.7% had no history of smoking), 88 methylation marks were found to be associated with mortality in a Cox proportional hazard analysis (8). A study of 9,949 older adults (age 50–75 yr) in Germany produced a mortality risk score based on 10 CpG sites selected from the 58 replicated sites associated with mortality in a Cox regression analysis (9). DNAm levels at 2,806 sites were found to be significantly associated with survival using Cox regression in a cohort of older (age 55–90 yr) Danish twins followed for up to 20 years (10). In a genome-wide study of DNAm in 111 nonagenarians, sites associated with mortality at 4-year follow-up were identified (11). Within a Scottish population of 1,425 individuals, a recent study identified 2,552 CpG sites with DNAm levels associated with all-cause mortality using Cox regression (12). Associations between DNAm and mortality have also been identified in a targeted study of the AHRR gene (13) and in sputum (14). In addition, previous studies have demonstrated the impact of smoking on DNAm in peripheral blood (15, 16), including the effects of smoking intensity and time since quitting (16). Associations between smoking and DNAm have also been observed in buccal mucosa (17). Previous studies have not examined DNAm associations with all-cause mortality in only current and former smokers with and without COPD while controlling for smoking history.
Our study sought to highlight DNAm as a potential biomarker for all-cause mortality in smokers. Although smoking cessation remains a primary clinical intervention in current smokers, our findings in current and former smokers may inform targeted, more intensive interventions for higher risk patients with yet to be recognized impairments (18). We performed genome-wide association tests between DNAm in peripheral blood and all-cause mortality in smokers using a time-to-event analysis. We hypothesized that specific CpG sites in leukocyte DNA would be associated with increased mortality. This analysis was replicated using peripheral blood DNAm in a second cohort predominantly made up of subjects with COPD. Some of the results of this study have been previously reported in the form of an abstract (19).
Methods
COPDGene Study
COPDGene (Genetic Epidemiology of COPD) (20) is a longitudinal cohort study that includes more than 10,000 non-Hispanic white and African American individuals enrolled at 21 centers across the United States. The subjects in this study were current and former smokers with a minimum 10 pack-years smoking history, with and without airflow obstruction. Written informed consent was obtained from each subject, and the study was approved by the institutional review boards of all 21 participating clinical centers.
DNAm
For each of the samples, DNA was extracted from peripheral blood and 1 μg of DNA was bisulfite-converted using the EZ DNA Methylation Gold Kit (Zymo Research). The genome-wide methylation levels of the DNA samples were assayed using the Infinium HumanMethylation450 BeadChip array platform from Illumina. The R statistical environment and the Bioconductor package minfi (21) were used to import and process the methylation data.
Statistical Tests
We performed a time-to-event analysis of mortality with a Cox proportional hazards model (Cox model) using the R package survival (22). The DNAm values for this analysis were z-score transformed. A hazard ratio >1 indicates that increased DNAm (hypermethylation) is associated with a reduced probability of survival. The models included DNAm as the predictor with disease status, age, sex, pack-years of smoking, and categorical variables for current smoking status and processing batch included as covariates. We used the Benjamini–Hochberg procedure, with a false discovery rate (FDR) <5% considered significant. During discovery, we also implemented a less stringent FDR threshold of 20%; these discovery thresholds have been previously applied in studies of DNAm (23–25). Kaplan-Meier curves were produced using the R package survminer. Additional details regarding the methods are provided in an online supplement.
Results
Study Populations
DNAm data were available from peripheral blood leukocyte DNA from COPDGene phase 1 enrollment for 331 COPD cases and 336 control cases. As of January 2018, 566 subjects were alive and 101 were deceased (Table 1 and see Table E1 in the online supplement). There were some notable differences between the subjects at enrollment, including higher smoking intensity (P < 0.0001) and lower lung function (P < 0.0001) among those who died. Although the deceased were slightly younger at enrollment than the survivors, this result was not statistically significant (P = 0.16). Lung-function decline and emphysema progression values were available for those evaluated at follow-up (see Tables 1 and E1). The causes of death for the 101 deceased subjects is summarized in Table E2.
Table 1.
The COPDGene Demographics Stratified by Survival Status
| Demographics | Alive (n = 566) | Deceased (n = 101) | P Value | |
|---|---|---|---|---|
| Age, yr | 63.0 ± 6.7 | 62.0 ± 6.8 | 0.16 | |
| Sex, F/M | 182/384 | 24/77 | 0.10 | |
| Days followed* | 2,409.7 ± 280.8 | 1,361.9 ± 645.5 | <0.0001 | |
| Smoking, current/former | 174/392 | 47/54 | 0.003 | |
| Smoking history, pack-years | 45.2 ± 23.2 | 60.2 ± 29.2 | <0.0001 | |
| Body mass index, kg/m2 | 29.5 ± 5.4 | 26.8 ± 7.2 | 0.0005 | |
| FEV1, % predicted | 71.2 ± 31.2 | 39.7 ± 25.8 | <0.0001 | |
| FEV1/FVC | 0.62 ± 0.19 | 0.42 ± 0.17 | <0.0001 | |
| FEV1 decline, ml/yr† | −37.9 ± 49.0 | −66.6 ± 68.2 | 0.27 | |
| Percent emphysema‡ | 7.4 ± 9.8 | 18.0 ± 15.4 | <0.0001 | |
| Change in percent emphysema§ | 0.9 ± 4.5 | 4.3 ± 6.3 | 0.29 | |
| Airway wall thickness, segmental bronchi‡ | 1.10 ± 0.23 | 1.23 ± 0.26 | <0.0001 | |
| COPD, controls/cases | 321/245 | 15/86 | <0.0001 | |
| School completed | ||||
| 8th grade or less | 12 | 1 | 0.009 | |
| High school, no diploma | 23 | 13 | ||
| High school graduate or General Educational Development | 128 | 26 | ||
| Some college or technical school, no degree | 149 | 25 | ||
| College or technical school (bachelor's or associate’s degree) | 173 | 27 | ||
| Master's or doctoral degree | 81 | 8 | ||
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; COPDGene = Genetic Epidemiology of COPD Study.
Data are shown as mean ± SD or n. Statistical tests: Fisher’s exact test for categorical variables, Welch’s two-sample t test for continuous variables, and the Wilcoxon-Mann Whitney test for school completed.
Number of days to censoring or a mortality event.
n = 417 alive, n = 8 deceased.
n = 546 alive, n = 91 deceased.
n = 384 alive, n = 5 deceased.
For the ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) replication study, DNAm data were available from peripheral blood for 231 subjects. As of January 2019, 185 subjects were alive and 46 were deceased (see Tables E3 and E4). All subjects were former smokers. The COPD cases had higher smoking intensity (P < 0.0001) and lower lung function (P < 0.0001).
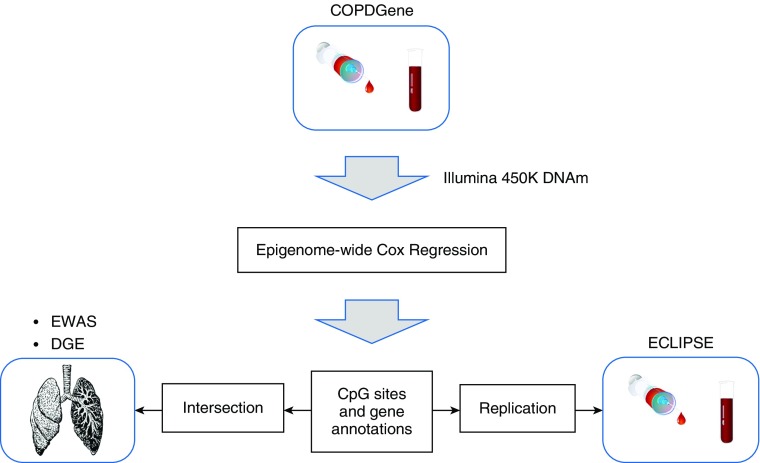
Genome-Wide Cox Regression Analysis
We performed a genome-wide time-to-event analysis of mortality using Cox models with the DNAm data from the 667 COPDGene subjects, followed by replication in ECLIPSE as a second cohort and integration with previous differential methylation and gene expression profiling studies (Figure 1). There were 68 sites associated with mortality at an FDR threshold of 20%; two of these CpG sites annotated to two genes were associated with mortality at an FDR threshold of 5% in the time-to-event analysis in COPDGene (see Tables E5 and E6). For the top site (cg14157855) annotated to ADGRB2, the hazard ratio and 95% confidence interval for 1 SD increase in methylation was 1.42 (1.24–1.61) and these values were 0.68 (0.59–0.79) for the second site (cg13726962) annotated to TSPYL6. A hazard ratio >1 indicates that increased DNAm (hypermethylation) is associated with a reduced probability of survival. Time-to-event analyses of mortality using Cox models were performed in the ECLIPSE population of 231 subjects. For 7 of the 68 (FDR < 20%) sites in the COPDGene population (Table 2), we observed replication among the 6,044 nominal (P < 0.05) results in ECLIPSE (see Table E5).
Figure 1.
Overview of the study illustrating the genome-wide analysis, intersection with previous findings, and replication of the results. COPD = chronic obstructive pulmonary disease; COPDGene = Genetic Epidemiology of COPD; DGE = differential gene expression; DNAm = DNA methylation; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; EWAS = epigenome-wide association study.
Table 2.
Results from Genome-Wide Cox Proportional Hazard Analysis in the COPDGene* with Nominal† Significance in ECLIPSE
| CpG Site | Gene Symbol | Cox Analysis‡ (P Value) | FDR (Q Value) | Hazard Ratio | Hazard Ratio Confidence |
ECLIPSE Cox Analysis§ (P Value) | FDR (Q Value) | Hazard Ratio | Hazard Ratio Confidence |
Chr | CpG Island Location | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower 0.95 | Upper 0.95 | Lower 0.95 | Upper 0.95 | ||||||||||
| cg08007647 | C1orf146 | 5.40 × 10−7 | 0.055 | 0.69 | 0.60 | 0.80 | 0.043 | 0.440 | 0.74 | 0.56 | 0.99 | 1 | Island |
| cg19828537 | FAM178B | 3.92 × 10−6 | 0.142 | 0.71 | 0.61 | 0.82 | 0.000473 | 0.137 | 0.61 | 0.46 | 0.81 | 2 | N/A |
| cg02618319 | USP2 | 1.41 × 10−5 | 0.145 | 0.60 | 0.48 | 0.76 | 0.000469 | 0.137 | 0.57 | 0.42 | 0.78 | 11 | N/A |
| cg04226365 | CDC5L | 1.82 × 10−5 | 0.152 | 1.32 | 1.16 | 1.49 | 0.000122 | 0.118 | 1.85 | 1.35 | 2.53 | 6 | Island |
| cg24542441 | SEPT10 | 1.88 × 10−5 | 0.153 | 1.43 | 1.21 | 1.68 | 0.0113 | 0.279 | 1.45 | 1.09 | 1.92 | 2 | Island |
| cg03971555 | PIK3CD | 2.80 × 10−5 | 0.182 | 1.58 | 1.27 | 1.95 | 0.00555 | 0.226 | 1.59 | 1.14 | 2.20 | 1 | N_Shore |
| cg12033075 | PIK3CD | 3.62 × 10−5 | 0.196 | 1.56 | 1.26 | 1.93 | 0.0035 | 0.201 | 1.68 | 1.19 | 2.38 | 1 | N_Shore |
Definition of abbreviations: Chr = chromosome; COPD = chronic obstructive pulmonary disease; COPDGene = Genetic Epidemiology of COPD Study; DNAm = DNA methylation; ECLIPSE = Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints; FDR = false discovery rate; N/A = not available; N_Shore = island shore located upstream (north) in chromosome; PIK3CD = phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ.
FDR < 20%.
P < 0.05.
Model 1: ∼ DNAm + COPD + age + sex + pack-years + smoking + batch.
Model 2: ∼ DNAm + COPD + age + sex + pack-years + batch.
The PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta) sites (cg03971555 and cg12033075) separated by two bases had relatively large effect sizes (absolute adjusted mean difference in DNAm between deceased and alive of 5% and 4.7%) with higher DNAm associated with mortality (Table 3 and see Table 2). The hazard ratios and 95% confidence intervals for 1 SD increase in methylation were 1.58 (1.27–1.95) for cg03971555 and 1.56 (1.26–1.93) for cg12033075 (see Table 2). We observed significant (P < 0.05) associations for lung function and emphysema phenotypes in COPDGene for these two PIK3CD sites, where increased DNAm was associated with COPD at enrollment (see Table 3). The absolute adjusted mean difference in DNAm between deceased and alive did not exceed 1% for any of the other replicated sites (see Table 3) nor did it exceed 1% for the two significant sites (0.6% for cg14157855 and 0.5% for cg13726962).
Table 3.
Associations between DNAm at the Replicated Sites from the Genome-Wide Cox Proportional Hazard Analysis* and COPD Status, Baseline Lung Function, Emphysema Phenotypes, and Survival Status
| CpG Site | Gene Symbol | Survival Status† (PValue) | Survival Status (Adjusted Mean Difference in % DNAm‡) | COPD Status§ (P Value) | COPD Status (Adjusted Mean Difference in % DNAm||) | FEV1% Predicted¶ (P Value) | Percent Emphysema** (P Value) |
|---|---|---|---|---|---|---|---|
| cg08007647 | C1orf146 | 0.0066 | −0.2 | 0.76 | −0.02 | 0.7 | 0.97 |
| cg19828537 | FAM178B | 0.00062 | −0.6 | 0.084 | −0.2 | 0.078 | 0.017 |
| cg02618319 | USP2 | 0.012 | −0.2 | 0.36 | −0.05 | 0.42 | 0.59 |
| cg04226365 | CDC5L | 0.00054 | 0.2 | 0.22 | 0.05 | 0.2 | 0.75 |
| cg24542441 | SEPT10 | 0.014 | 0.3 | 0.91 | 0.01 | 0.83 | 0.84 |
| cg03971555 | PIK3CD | 2.31 × 10−5 | 5.0 | 0.036 | 2.0 | 0.023 | 0.036 |
| cg12033075 | PIK3CD | 1.52 × 10−5 | 4.7 | 0.00069 | 2.9 | 0.00029 | 0.00031 |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; DNAm = DNA methylation; PIK3CD = phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ.
Survival status is binary: alive or deceased.
See Table 2.
Model 1: DNAm ∼ survival status + COPD + age + sex + pack-years + smoking + batch.
Negative DNAm difference corresponds to reduced levels (hypomethylation) with mortality; positive DNAm difference corresponds to increased levels (hypermethylation) with mortality.
Model 2: DNAm ∼ COPD + age + sex + pack-years + smoking + batch.
Negative DNAm difference corresponds to hypomethylation with COPD, reduced FEV1 and increased emphysema; positive DNAm difference corresponds to hypermethylation with COPD, reduced FEV1 and increased emphysema.
Model 3: DNAm ∼ FEV1% predicted + age + sex + pack-years + smoking + batch.
Model 4: DNAm ∼ Percent emphysema + age + sex + body mass index + pack-years + smoking + batch.
Box plots illustrate the DNAm distribution for both alive and deceased subjects at the replicated sites (Figure 2 and see Figures E1–E5). The top replicated PIK3CD site (cg03971555) in ECLIPSE had a hazard ratio and 95% confidence interval of 1.59 (1.14–2.20) and the hazard ratio and 95% confidence interval for the second site (cg12033075) was 1.68 (1.19–2.38) (see Table 2), concordant with the COPDGene finding. We further observed evidence of this replication and similarity of effect in the plots of DNAm in the two cohorts, stratified by survival status (see Figure 2).
Figure 2.
Distribution of percent DNAm for the PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ) CpG sites in both COPDGene (Genetic Epidemiology of COPD) and ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints): (A) cg03971555 and (B) cg12033075. The distribution for both alive and deceased subjects is shown. COPD = chronic obstructive pulmonary disease; DNAm = DNA methylation.
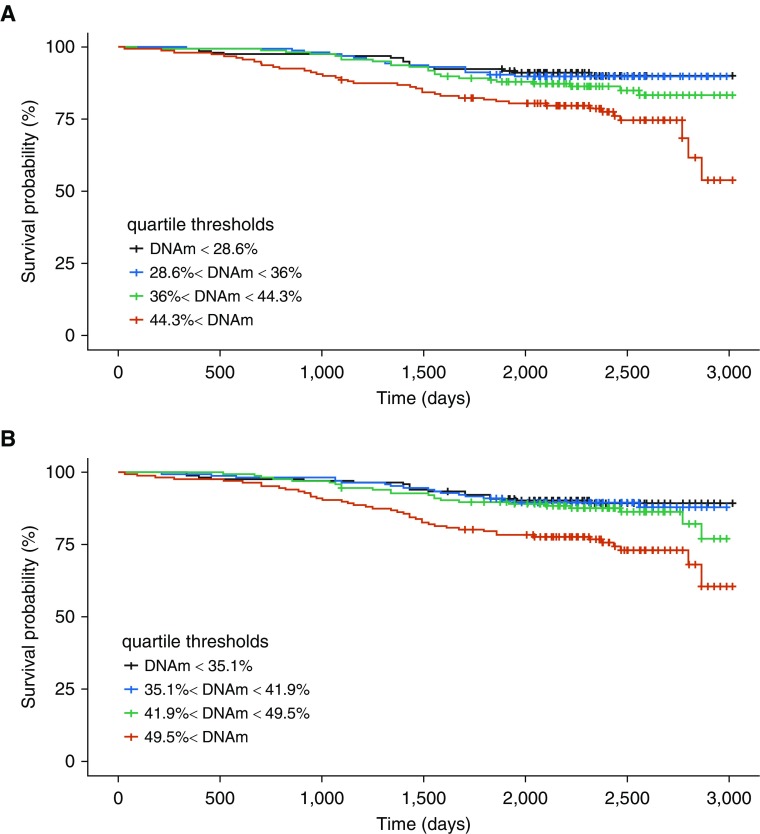
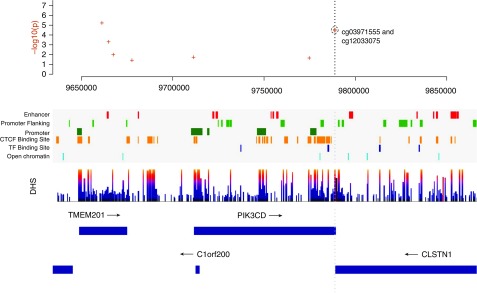
Kaplan-Meier curves illustrate the association between probability of survival and percent DNAm for the PIK3CD (cg12033075 and cg03971555) sites (Figure 3). Similar characteristics are observed in the curves for these sites in ECLIPSE (see Figure E6). These sites are located in genomic regions of DNase hypersensitivity (Figure 4), highlighting the potential regulatory relevance of these DNAm sites as biomarkers of mortality. We also observed evidence that suggests possible TF (transcription factor) binding (ZBTB33, STAT3, POLR2A, and POLR2G) in the region surrounding these two PIK3CD sites (see Figure E7). The scores for these TFs were below the threshold for representation in the composite TF binding signal presented in Figure 4.
Figure 3.
Survival curves for PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ) CpG sites in COPDGene (Genetic Epidemiology of COPD). (A) For the site cg03971555, the subjects alive at enrollment for each bin: n = 160 (red), n = 159 (green), n = 159 (blue), and n = 159 (black). Thirty samples have missing DNAm data (total n = 637). The subjects alive at last time point: n = 121 (red), n = 136 (green), n = 143 (blue), and n = 144 (black). (B) For the site cg12033075, the subjects alive at enrollment for each bin: n = 167 (red), n = 167 (green), n = 166 (blue), and n = 167 (black). The subjects alive at last time point: n = 124 (red), n = 144 (green), n = 148 (blue), and n = 150 (black). Marks show times with data censoring. The time unit is days. COPD = chronic obstructive pulmonary disease; DNAm = DNA methylation.
Figure 4.
Regional genomic plot for the coordinates surrounding the PIK3CD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ) CpG sites (cg12033075 and cg03971555) on chromosome 1. The top plot contains the P values from the Cox proportional hazard analysis. Regulatory information related to promoter and TF (transcription factor) binding for this genomic region is shown, along with boundaries for genes in the region. The vertical gray dotted line passes through the genomic coordinates of the PIK3CD CpG sites to align with the regulatory and gene annotations. The DNase hypersensitivity (DHS) track highlights regions of DHS. The CTCF (CCCTC-binding factor) Binding Site track represents regions of CTCF binding. Additional details regarding the regulatory features are provided in the online supplement.
Sensitivity Analyses
Although the PIK3CD sites and the FAM178B site were associated with lung function or emphysema severity at enrollment, no significant associations with lung function decline or change in quantitative emphysema (phenotypes measured mostly for survivors) were observed for the seven replicated sites (see Table E7). We also did not observe a significant impact of smoking exposure; associations between these sites and pack-years of smoking or current smoking status lacked statistical significance (see Table E7). Six of the seven replicated sites annotated to the genes C1orf146, FAM178B, USP2, SEPT10, and PIK3CD; both significant sites annotated to ADGRB2 and TSPYL6 were nominally significant (P < 0.001) in the analysis of only COPD cases, and the CDC5L site was not significant.
We repeated the genome-wide Cox analysis, including FEV1, percent emphysema, or airway wall thickness, in the Cox model as the disease covariate in place of COPD affection status (see Tables E8–E11). The associations for the replicated sites retained similar significance in these models (see Table E8). The association for the C1orf146 (cg08007647—chromosome 1 open reading frame 146) and USP2 (cg02618319—ubiquitin specific peptidase 2) sites achieved significance (FDR < 5%) when adjusting for percent emphysema; C1orf146 (cg08007647) was also significant when adjusting for airway wall thickness. One site (cg13726962) annotated to TSPYL6 was significant (FDR < 5%) for all four models (see Figure E8). When adjusting for socioeconomic status by including the highest degree or level of education as a covariate, the replicated sites retained similar significance (see Table E8), with the association for C1orf146 (cg08007647) reaching statistical significance (FDR < 5%).
Owing to its association with smoking exposure (26), we replaced the pack-years covariate in our genome-wide Cox analysis with the DNAm methylation values for the AHRR site cg05575921 and observed similar significance in the associations with mortality across the replicated sites (see Table E8). Similar significance was also observed for the replicated sites when adjusting for cellular heterogeneity by including ReFACTor/GLINT (https://github.com/cozygene/glint/releases/) components in the Cox model (see Table E8). With these components in the model, no results met the FDR <5% threshold. When excluding the subjects who died due to lung cancer (11 subjects) or censoring these subjects at the time of death, similar levels of significance were observed (see Table E8), suggesting that the primary driver of these mortality signals is not lung cancer. In these lung cancer sensitivity results, the top replicated finding was statistically significant (C1orf146:cg08007647, q = 0.025 and q = 0.019; see Table E8).
The two replicated PIK3CD sites associated with survival status in our Cox model also demonstrated an association (P < 0.05) with COPD status (see Table 3). In addition, we observed that COPD status is significantly (P = 4.39 × 10−8; hazard ratio, 5.30) associated with survival status in a Cox regression analysis, controlling for age, sex, pack-years of smoking, current smoking status, and processing batch (see Table E12). We sought insight into the relationship between DNAm of the replicated sites and survival in the context of COPD through adjustments to our models. We tested whether the association between COPD status and survival is maintained when controlling for DNAm (see Table E12) and observed significance (P < 1. 0 × 10−5) for all seven replicated sites. In addition, we sought to highlight the effects of COPD status in our associations between DNAm and survival (see Table 2) by evaluating models without adjustment for COPD status. For this model, we observed significance (P < 1.0 × 10−3) for all seven sites. Taken together, these findings suggest DNAm of the PIK3CD sites and COPD status may have shared effects on survival.
Functional Context across Tissues
To provide additional functional insight for the findings, we sought to reveal the relationship between DNAm and gene expression in blood (ECLIPSE) (27), and lung tissue (28, 29) for the replicated (see Table 2) and significant sites (ADGRB2:cg14157855 and TSPYL6:cg13726962). We performed linear regression analysis using the DNAm data from this ECLIPSE cohort and the peripheral blood gene expression data for the same subjects from a previous ECLIPSE network-based clustering study (27) (GEO Accession GSE76705). For lung tissue, the analysis was performed for the 107 COPD cases and 40 control cases with both DNAm and gene expression data from two previous studies (28, 29). Plots of gene expression and DNAm were created for the significant (P < 0.05) associations (see Figures E9–E15). DNAm of both PIK3CD sites (cg03971555 and cg12033075) were associated with gene expression in both peripheral blood and lung tissue. In blood, DNAm and gene expression were correlated. In lung tissue, they were anticorrelated.
To provide lung disease context, we queried the results from a previous lung tissue epigenome-wide association study of 114 COPD cases and 46 control smokers, and a differential gene expression analysis of 111 COPD cases and 40 control smokers (28, 29) for the seven replicated sites (see Table E13). DNAm levels at the mortality-associated PIK3CD (cg12033075) and FAM178B (cg19828537—family with sequence similarity 178 member B) CpG sites were associated (P < 0.05) with COPD status in lung tissue, with a direction of effect opposite what we observed in blood (decrease in DNAm of PIK3CD [cg12033075] corresponds to increased disease severity in lung tissue). Additionally, differential gene expression by COPD status was observed (P < 0.05) for a probe mapped to SEPT10 (septin 10). This SEPT10 gene expression probe was different than the one demonstrating a significant association with DNAm (see Figure E13).
Discussion
Our genome-wide time-to-event analysis of mortality in 667 carefully phenotyped current and former smokers from the COPDGene study has revealed epigenetic markers of mortality in peripheral blood. Two differentially methylated CpG sites were found to be significantly associated with mortality, using models controlling for COPD affection status and smoking history. Seven of the nominally significant findings were replicated in an independent cohort. The hazard ratios and confidence intervals were reproduced in ECLIPSE for each of the seven sites, recapitulating the epigenetic mortality signal from the COPDGene discovery cohort. We did not specifically observe associations between these mortality-associated sites and lung function decline or changes in percent emphysema. These decline-related phenotypes were evaluated predominantly in survivors; fewer than 10 subjects had phase 2 follow-up visits before death. We did observe associations for two sites with baseline lung function and emphysema. Taken together, this suggests relevance for these mortality markers in a population of smokers at risk for lung disease.
The two replicated sites (cg03971555 and cg12033075) annotated to PIK3CD with larger effect sizes and perhaps are more applicable because biomarkers are located in the northern shores of CpG islands. The CpG shore region has been identified as more regulatory in nature (30–32). Regulatory relevance is also suggested by the presence of these sites in DNase hypersensitive regions of the genome. The significant correlation between percent DNAm and gene expression for PIK3CD implies functional relevance of these sites in peripheral blood. Concordant with our prior observations regarding opposite directions of effect in COPD associations with DNAm in blood and lung tissue, we observed anticorrelation between DNAm and gene expression in lung tissue for the PIK3CD sites. Similar links in peripheral blood and normal lung tissue have been observed for DNAm in a study of patients with cancer and smokers (33). These PIK3CD sites are in the 3′ untranslated region of the gene. This location may contribute to the specific DNAm and gene expression relationship observed, and to the opposite effect we observe across tissue, because the complex regulatory effects of DNAm are yet to be fully understood (34–36). More concretely, we observed evidence hinting at possible binding by the TF ZBTB33 at these sites and it has been suggested that the direction of the effect of ZBTB33 on expression can be cell-specific (37). We identified the PIK3CD gene in our previous study of the genetic control of DNAm (31). However, an alternate CpG site, also located at the north shore of a CpG island (cg26573321), was the PIK3CD site highlighted among the significant methylation quantitative trail loci (association between DNAm and genetic variants) in that study of colocalization with COPD genome-wide association studies.
In this study, increased DNAm of PIK3CD (cg03971555 and cg12033075) in blood was associated with COPD. Decreased DNAm of PIK3CD (cg12033075) was associated with COPD in lung tissue (28), providing disease context for the peripheral blood mortality signals. Increased PI3K-delta (phosphoinositide-3-kinase-delta) contributes to corticosteroid resistance in COPD (38). We observed positive correlation between DNAm (cg03971555 and cg12033075) and gene expression of PIK3CD in ECLIPSE. We also observed increased DNAm at these sites in COPD. Taken together, this supports the previous PI3K-delta findings because increased PIK3CD expression is expected in the blood of the ECLIPSE COPD cases. Although we observed anticorrelation between DNAm at cg12033075 and PIK3CD gene expression in lung tissue, significant differential expression of PIK3CD with COPD status was not previously observed (29). However, the significant anticorrelation of PIK3CD DNAm (cg03971555 and cg12033075) and gene expression does hint at regulatory relevance in the PI3K-delta pathways for these mortality-associated sites. Additional data, including protein levels and survival outcomes, will be required in a lung tissue cohort to enable more extensive observations.
One of our findings meeting the strict FDR threshold was a site annotated to ADGRB2 (cg14157855-adhesion G protein–coupled receptor B2). DNAm of a different site (cg12211691) annotated to ADGRB2 in peripheral blood mononuclear cells was found to be associated with age in African American individuals (39). The other site meeting this FDR threshold was annotated to TSPYL6 (cg13726962-TSPY–like 6). In a previous study in leukocytes, DNAm of another TSPYL6 site (cg25249300) was found to be associated with the intrapair difference in lung function of middle-aged monozygotic twins (40). These previous DNAm associations with age and lung function suggest a link between both sites and factors relevant to mortality.
Five previous genome-wide studies have investigated the association between blood DNAm and mortality using the Illumina 450K platform (8–11). We observed additional replication of our findings in a recent DNAm study of late-life survival in the peripheral blood mononuclear cells of 111 nonagenarians (11). In that study, the PIK3CD sites (cg12033075; P = 0.0056 and cg03971555; P = 0.0066) were among their nominal 4-year follow-up results. Although the study of DNAm and mortality by Zhang and colleagues was in a population of >50% former or current smokers, we did not replicate their top findings (9). In that study, they controlled for baseline disease and smoking status in the validation phase. However, during discovery they did not include disease status, smoking status, or smoking history in their models. This impacts our ability to replicate because our discovery models include smoking history in addition to disease and smoking status. The presence of approximately 30% never smokers in their study also impacts our ability to replicate their findings. Our replicated sites were not found among the 2,552 significant (FDR < 5%) results in a recent study of all-cause mortality and aging (12). The prevalence of smoking in the population from that study was not presented and disease status was not included in their models. Additional efforts are required to assess the specific influences of smoking, COPD status, and comorbidities on the identification of DNAm biomarkers of mortality.
Aging Pathways
PIK3CD and other PI3K family members play an important role in a broad range of regulatory processes, including cell growth and metabolism. Disruption of PI3K signaling has been implicated in cancer, immunological disorders, diabetes, and cardiovascular disease, motivating development of therapies targeting PI3K pathways, particularly in cancer (41). In lungs of patients with COPD, it has been suggested that accelerated aging is enhanced through PI3K signaling, whereby oxidative stress signals lead to senescence, increased inflammation, and further oxidative stress (42, 43). In leukocytes, the PI3Ks typically phosphorylate inositol lipids as part of their involvement in immune responses. These kinases are believed to contribute to disease progression in the airway through disruption of normal signaling (44). This disruption of PIK3CD activity may lead to immunodeficiency and subsequent airway damage (45), with possible involvement in lung cancer development (46). Associated pathways are important in multiple cell types, impacting airway epithelial function and B-lymphocyte and T-lymphocyte development, with implications in oxidative stress, inflammation, and corticosteroid effectiveness. The use of mTOR inhibitors, targeting a PI3K-related pathway in vitro, has demonstrated restoration of corticosteroid sensitivity (47) and reduced cell senescence (48) in COPD. Together, these results suggest existing therapeutic approaches targeting methylation could address aging and longevity within particular subpopulations (49), and that PIK3CD has an important role in this context for smokers with or without lung disease.
There are several limitations to our analysis that should be acknowledged. The ascertainment of COPDGene study participants based on at least a 10 pack-year smoking history may limit the generalizability to populations with less smoke exposure. The smoking status in our discovery and replication cohorts are different. The subset of ECLIPSE subjects with methylation data is all former smokers. The number of subjects in the ECLIPSE study subset is smaller (231 for ECLIPSE and 667 for COPDGene), limiting our power to detect statistically significant genome-wide findings. However, the proportion of deaths (46 of 231; 20%) after 8 years of follow-up is similar (chi-squared test, P = 0.11) to the COPDGene discovery cohort (101 of 667; 15%). Although less stringent discovery thresholds have been previously applied in studies of DNAm (23–25), our use of an FDR threshold of 20% during discovery could influence replication in later studies. Our analysis of decline phenotypes was performed primarily in survivors, given the limited number of subjects with longitudinal evaluation before death. Samples from African American subjects were not available, limiting our ability to generalize across populations. The lack of associations of our replicated sites with smoking status or history suggests that we may be capturing a signal relevant to nonsmokers. However, future studies in a population of nonsmokers will be required to assess applicability. Comorbidities are expected in cohorts of COPD cases and smoking control cases (3), with a broad range of causes of death. However, we are only adjusting for lung-related measures of disease. In genome-wide DNAm studies, polymorphic regions with rare single nucleotide genetic variants can bias methylation levels. We did not observe compelling evidence of these effects in our study. Although we have corrected for cellular heterogeneity using principal component analysis methods in our sensitivity analysis, the DNAm values may remain partially dependent on the cellular distributions. Having this variability present in our analyses is a limitation. However, the variation in cell type composition captured by the DNAm measurements could also be considered part of a relevant biomarker signal. Last, we are not able to determine the temporal nature of the DNAm sites in this cross-sectional study. Hypermethylation of these sites may have preceded the onset of COPD and subsequent death, or this variation may be concomitant with COPD onset. Our sensitivity analysis suggested DNAm of PIK3CD and COPD status have shared effects on survival. However, understanding the molecular details of this relationship will require functional validation. Future studies of longitudinal DNAm data will also address this limitation and assess the prognostic value of these putative biomarkers, as well as their value as risk biomarkers in a population of healthy smokers.
Conclusions
We identified DNAm sites predictive of all-cause mortality in a cohort of current and former smokers, with and without COPD, and independent of longitudinal lung function decline or emphysema progression based on associations primarily in survivors. These sites were associated with COPD status at enrollment. However, they were not associated with smoking history or status. Among our replicated findings was the aging-pathway gene PIK3CD. Although smoking cessation and abstinence are important for decreasing mortality related to smoking, former smokers still demonstrate elevated mortality, suggesting additional therapeutic interventions are necessary to prevent smoking-related diseases. This study hints at predictive relevance of methylation marks for the identified sites in a population of smokers, irrespective of lung disease presentation, helping to inform the selection of smokers at greater risk of death, while highlighting targets for functional validation and therapeutic intervention.
Supplementary Material
Acknowledgments
Acknowledgment
ECLIPSE Steering Committee: Harvey Coxson (Canada), Lisa Edwards (GlaxoSmithKline, USA), Katharine Knobil (Co-chair, GlaxoSmithKline, United Kingdom), David Lomas (United Kingdom), William MacNee (United Kingdom), Edwin Silverman (United States), Ruth Tal-Singer (GlaxoSmithKline, United States), Jørgen Vestbo (Co-chair, Denmark), and Julie Yates (GlaxoSmithKline, United States).
ECLIPSE Scientific Committee: Alvar Agusti (Spain), Peter Calverley (United Kingdom), Bartolome Celli (United States), Courtney Crim (GlaxoSmithKline, United States), Bruce Miller (GlaxoSmithKline, United Kingdom), William MacNee (Chair, United Kingdom), Stephen Rennard (United States), Ruth Tal-Singer (GlaxoSmithKline, United States), Emiel Wouters (the Netherlands), and Julie Yates (GlaxoSmithKline, United States).
ECLIPSE Investigators: Y. Ivanov (Pleven, Bulgaria), K. Kostov (Sofia, Bulgaria), J. Bourbeau (Montreal, Canada), M. Fitzgerald (Vancouver, British Columbia, Canada), P. Hernandez (Halifax, Nova Scotia, Canada), K. Killian (Hamilton, Ontario, Canada), R. Levy (Vancouver, British Columbia, Canada), F. Maltais (Montreal, Canada), D. O′Donnell (Kingston, Ontario, Canada), J. Krepelka (Prague, Czech Republic), J. Vestbo (Hvidovre, Denmark), E. Wouters (Horn-Maastricht, the Netherlands), D. Quinn (Wellington, New Zealand), P. Bakke (Bergen, Norway), M. Kosnik (Golnik, Slovenia), A. Agusti (Spain), J. Sauleda (Spain), P. de Mallorca (Spain), Y. Feschenko (Kiev, Ukraine), V. Gavrisyuk (Kiev, Ukraine), L. Yashina (Kiev, Ukraine), N. Monogarova (Donetsk, Ukraine), P. Calverley (Liverpool, United Kingdom), D. Lomas (Cambridge, United Kingdom), W. MacNee (Edinburgh, United Kingdom), D. Singh (Manchester, United Kingdom), J. Wedzicha (London, United Kingdom), A. Anzueto (San Antonio, Texas), S. Braman (Providence, Rhode Island), R. Casaburi (Torrance, California), B. Celli (Boston, Massachusetts), G. Giessel (Richmond, Virginia), M. Gotfried (Phoenix, Arizona), G. Greenwald (Rancho Mirage, California), N. Hanania (Houston, Texas), D. Mahler (Lebanon, New Hampshire), B. Make (Denver, Colorado), S. Rennard (Omaha, Nebraska), C. Rochester (New Haven, Connecticut), P. Scanlon (Rochester, Minnesota), D. Schuller (Omaha, Nebraska), F. Sciurba (Pittsburgh, Pennsylvania), A. Sharafkhaneh (Houston, Texas), T. Siler (St. Charles, Missouri), E. Silverman (Boston, Massachusetts), A. Wanner (Miami, Florida), R. Wise (Baltimore, Maryland), and R. ZuWallack (Hartford, Connecticut).
Footnotes
Supported by NIH grants K25 HL136846 (J.D.M.), P01 HL105339 (E.K.S.), P01 HL114501 (A.M.K.C.), R01 HL111759 (J.Q.), U01 HL089856 (E.K.S.), U01 HL089897, and P01 HL132825. The COPDGene study (U01 HL089856 and U01 HL089897) (NCT00608764) was funded by the NIH and also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, GSK, and Sunovion. The National Emphysema Treatment Trial was supported by NHLBI N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119, the Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The ECLIPSE study (NCT00292552; GSK code SCO104960) was funded by GlaxoSmithKline.
Author Contributions: J.D.M.: analysis and interpretation of data, manuscript preparation, and approval of the final version. B.M.: acquisition of data, review of the manuscript, and approval of the final version of the manuscript. E.R.: acquisition of data, review of the manuscript, and approval of the final version of the manuscript. M.H.: acquisition of data, review of the manuscript, and approval of the final version of the manuscript. C.P.H.: analysis and interpretation of data, manuscript preparation, and approval of the final version. R.T.-S.: acquisition of data, review of the manuscript, and approval of the final version of the manuscript. J.Q.: analysis and interpretation of data, review of the manuscript, and approval of the final version of the manuscript. A.M.K.C.: acquisition of data, review of the manuscript, and approval of the final version of the manuscript. E.K.S.: analysis and interpretation of data, manuscript preparation, and approval of the final version. D.L.D.: acquisition of data, analysis and interpretation of data, manuscript preparation, and approval of the final version.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201902-0439OC on January 29, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Jha P, Ramasundarahettige C, Landsman V, Rostron B, Thun M, Anderson RN, et al. 21st-century hazards of smoking and benefits of cessation in the United States. N Engl J Med. 2013;368:341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 2.Carter BD, Abnet CC, Feskanich D, Freedman ND, Hartge P, Lewis CE, et al. Smoking and mortality: beyond established causes. N Engl J Med. 2015;372:631–640. doi: 10.1056/NEJMsa1407211. [DOI] [PubMed] [Google Scholar]
- 3.Sin DD, Anthonisen NR, Soriano JB, Agusti AG. Mortality in COPD: role of comorbidities. Eur Respir J. 2006;28:1245–1257. doi: 10.1183/09031936.00133805. [DOI] [PubMed] [Google Scholar]
- 4.Bossé Y, Amos CI. A decade of GWAS results in lung cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:363–379. doi: 10.1158/1055-9965.EPI-16-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CC, Bush WS, Crawford DC, Wenzlaff AS, Schwartz AG, Wiencke JK, et al. Germline genetic variants and lung cancer survival in African Americans. Cancer Epidemiol Biomarkers Prev. 2017;26:1288–1295. doi: 10.1158/1055-9965.EPI-16-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JG, Felix JF, Morrison AC, Kalogeropoulos A, Trompet S, Wilk JB, et al. CHARGE-SCD Consortium; EchoGen Consortium; QT-IGC Consortium; CHARGE-QRS Consortium. Discovery of genetic variation on chromosome 5q22 associated with mortality in heart failure. PLOS Genet. 2016;12:e1006034. doi: 10.1371/journal.pgen.1006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Pitkäniemi J, Heikkilä K, Chou Y-L, Madden PAF, Korhonen T, et al. Genome-wide time-to-event analysis on smoking progression stages in a family-based study. Brain Behav. 2016;6:e00462. doi: 10.1002/brb3.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore AZ, Hernandez DG, Tanaka T, Pilling LC, Nalls MA, Bandinelli S, et al. Change in epigenome-wide DNA methylation over 9 years and subsequent mortality: results from the InCHIANTI study. J Gerontol A Biol Sci Med Sci. 2016;71:1029–1035. doi: 10.1093/gerona/glv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wilson R, Heiss J, Breitling LP, Saum K-U, Schöttker B, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svane AM, Soerensen M, Lund J, Tan Q, Jylhävä J, Wang Y, et al. DNA methylation and all-cause mortality in middle-aged and elderly Danish twins. Genes (Basel) 2018;9:78. doi: 10.3390/genes9020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jylhävä J, Kananen L, Raitanen J, Marttila S, Nevalainen T, Hervonen A, et al. Methylomic predictors demonstrate the role of NF-κB in old-age mortality and are unrelated to the aging-associated epigenetic drift. Oncotarget. 2016;7:19228–19241. doi: 10.18632/oncotarget.8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund JB, Li S, Baumbach J, Svane AM, Hjelmborg J, Christiansen L, et al. DNA methylome profiling of all-cause mortality in comparison with age-associated methylation patterns. Clin Epigenetics. 2019;11:23. doi: 10.1186/s13148-019-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bojesen SE, Timpson N, Relton C, Davey Smith G, Nordestgaard BG. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax. 2017;72:646–653. doi: 10.1136/thoraxjnl-2016-208789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leng S, Diergaarde B, Picchi MA, Wilson DO, Gilliland FD, Yuan J-M, et al. Gene promoter hypermethylation detected in sputum predicts FEV1 decline and all-cause mortality in smokers. Am J Respir Crit Care Med. 2018;198:187–196. doi: 10.1164/rccm.201708-1659OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol Genet. 2014;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]
- 16.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan ES, Qiu W, Carey VJ, Morrow J, Bacherman H, Foreman MG, et al. Smoking-associated site-specific differential methylation in buccal mucosa in the COPDGene study. Am J Respir Cell Mol Biol. 2015;53:246–254. doi: 10.1165/rcmb.2014-0103OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regan EA, Lynch DA, Curran-Everett D, Curtis JL, Austin JHM, Grenier PA, et al. Genetic Epidemiology of COPD (COPDGene) Investigators. Clinical and radiologic disease in smokers with normal spirometry. JAMA Intern Med. 2015;175:1539–1549. doi: 10.1001/jamainternmed.2015.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrow J, Qiu W, Make BJ, Regan EA, Han MK, Hersh CP, et al. Blood DNA methylation prospectively predicts mortality in former and current smokers in the COPDGene study [abstract] Am J Respir Crit Care Med. 2018;197:A4159. [Google Scholar]
- 20.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. New York, NY: Springer Science & Business Media; 2013. [Google Scholar]
- 23.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 2015;16:8. doi: 10.1186/s13059-014-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong Doo N, Makalic E, Joo JE, Vajdic CM, Schmidt DF, Wong EM, et al. Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature B-cell neoplasms. Epigenomics. 2016;8:55–66. doi: 10.2217/epi.15.97. [DOI] [PubMed] [Google Scholar]
- 26.Philibert RA, Beach SRH, Lei M-K, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenetics. 2013;5:19. doi: 10.1186/1868-7083-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y, Glass K, Liu Y-Y, Silverman EK, Crapo JD, Tal-Singer R, et al. COPD subtypes identified by network-based clustering of blood gene expression. Genomics. 2016;107:51–58. doi: 10.1016/j.ygeno.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrow JD, Cho MH, Hersh CP, Pinto-Plata V, Celli B, Marchetti N, et al. DNA methylation profiling in human lung tissue identifies genes associated with COPD. Epigenetics. 2016;11:730–739. doi: 10.1080/15592294.2016.1226451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrow JD, Zhou X, Lao T, Jiang Z, DeMeo DL, Cho MH, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232. doi: 10.1038/srep44232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. Genome-wide methylation analysis of human colon cancer reveals similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrow JD, Glass K, Cho MH, Hersh CP, Pinto-Plata V, Celli B, et al. Human lung DNA methylation quantitative trait loci colocalize with chronic obstructive pulmonary disease genome-wide association loci. Am J Respir Crit Care Med. 2018;197:1275–1284. doi: 10.1164/rccm.201707-1434OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stueve TR, Li W-Q, Shi J, Marconett CN, Zhang T, Yang C, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet. 2017;26:3014–3027. doi: 10.1093/hmg/ddx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 36.van Eijk KR, de Jong S, Boks MPM, Langeveld T, Colas F, Veldink JH, et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics. 2012;13:636. doi: 10.1186/1471-2164-13-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozner A, Terooatea TW, Buck-Koehntop BA. Cell-specific kaiso (ZBTB33) regulation of cell cycle through cyclin D1 and cyclin E1. J Biol Chem. 2016;291:24538–24550. doi: 10.1074/jbc.M116.746370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.To Y, Ito K, Kizawa Y, Failla M, Ito M, Kusama T, et al. Targeting phosphoinositide-3-kinase-δ with theophylline reverses corticosteroid insensitivity in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:897–904. doi: 10.1164/rccm.200906-0937OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tajuddin SM, Hernandez DG, Chen BH, Noren Hooten N, Mode NA, Nalls MA, et al. Novel age-associated DNA methylation changes and epigenetic age acceleration in middle-aged African Americans and whites. Clin Epigenetics. 2019;11:119. doi: 10.1186/s13148-019-0722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolund ACS, Starnawska A, Miller MR, Schlünssen V, Backer V, Børglum AD, et al. Lung function discordance in monozygotic twins and associated differences in blood DNA methylation. Clin Epigenetics. 2017;9:132. doi: 10.1186/s13148-017-0427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes PJ. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2016;138:16–27. doi: 10.1016/j.jaci.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Mercado N, Ito K, Barnes PJ. Accelerated ageing of the lung in COPD: new concepts. Thorax. 2015;70:482–489. doi: 10.1136/thoraxjnl-2014-206084. [DOI] [PubMed] [Google Scholar]
- 44.Stokes CA, Condliffe AM. Phosphoinositide 3-kinase δ (PI3Kδ) in respiratory disease. Biochem Soc Trans. 2018;46:361–369. doi: 10.1042/BST20170467. [DOI] [PubMed] [Google Scholar]
- 45.Angulo I, Vadas O, Garçon F, Banham-Hall E, Plagnol V, Leahy TR, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitani A, Ito K, Vuppusetty C, Barnes PJ, Mercado N. Restoration of corticosteroid sensitivity in chronic obstructive pulmonary disease by inhibition of mammalian target of rapamycin. Am J Respir Crit Care Med. 2016;193:143–153. doi: 10.1164/rccm.201503-0593OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Houssaini A, Breau M, Kebe K, Abid S, Marcos E, Lipskaia L, et al. mTOR pathway activation drives lung cell senescence and emphysema. JCI Insight. 2018;3:93203. doi: 10.1172/jci.insight.93203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann JW, Zhao X, De Cecco M, Peterson AL, Pagliaroli L, Manivannan J, et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–488. doi: 10.1016/j.cell.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.