Abstract
The autosomal dominant disorder Schnyder corneal dystrophy (SCD) is caused by mutations in UbiA prenyltransferase domain-containing protein-1 (UBIAD1), which uses geranylgeranyl pyrophosphate (GGpp) to synthesize the vitamin K2 subtype menaquinone-4 (MK-4). SCD is characterized by opacification of the cornea, owing to aberrant build-up of cholesterol in the tissue. We previously discovered that sterols stimulate association of UBIAD1 with ER-localized HMG-CoA reductase, which catalyzes a rate-limiting step in the synthesis of cholesterol and nonsterol isoprenoids, including GGpp. Binding to UBIAD1 inhibits sterol-accelerated ER-associated degradation (ERAD) of reductase and permits continued synthesis of GGpp in cholesterol-replete cells. GGpp disrupts UBIAD1-reductase binding and thereby allows for maximal ERAD of reductase as well as ER-to-Golgi translocation of UBIAD1. SCD-associated UBIAD1 is refractory to GGpp-mediated dissociation from reductase and remains sequestered in the ER to inhibit ERAD. Here, we report development of a biochemical assay for UBIAD1-mediated synthesis of MK-4 in isolated membranes and intact cells. Using this assay, we compared enzymatic activity of WT UBIAD1 with that of SCD-associated variants. Our studies revealed that SCD-associated UBIAD1 exhibited reduced MK-4 synthetic activity, which may result from its reduced affinity for GGpp. Sequestration in the ER protects SCD-associated UBIAD1 from autophagy and allows intracellular accumulation of the mutant protein, which amplifies the inhibitory effect on reductase ERAD. These findings have important implications not only for the understanding of SCD etiology but also for the efficacy of cholesterol-lowering statin therapy, which becomes limited, in part, because of UBIAD1-mediated inhibition of reductase ERAD.
Keywords: endoplasmic reticulum, Golgi apparatus, vesicular transport, menaquinone-4, cholesterol, UbiA prenyltransferase domain-containing protein-1
Schnyder corneal dystrophy (SCD) is an autosomal dominant disease characterized by progressive opacification of the cornea that results from over-accumulation of cholesterol and other lipids in the tissue (1–4). SCD is caused by mutations in the gene encoding UbiA prenyltransferase domain-containing protein-1 (UBIAD1), an integral membrane UbiA prenyltransferase (5). UBIAD1 transfers the geranylgeranyl group from geranylgeranyl pyrophosphate (GGpp) to vitamin K3 (menadione) released from dietary vitamin K1 (phylloquinone), resulting in production of the vitamin K2 subtype menaquinone-4 (MK-4) (6, 7). Missense mutations altering 21 amino acids in the UBIAD1 protein have been identified in SCD families (6, 8). Structural analyses of archaeal UbiA prenyltransferases reveal that many residues corresponding to SCD-associated mutations in human UBIAD1 localize to the active site of the enzyme, indicating that SCD-associated UBIAD1 variants are enzymatically defective (9, 10). Indeed, several SCD-associated UBIAD1 variants exhibited reduced enzymatic activity (6, 11); however, whether other variants of the enzyme are defective in MK-4 synthesis is unknown.
The initial connection between UBIAD1 and cholesterol synthesis was provided by co-immunoprecipitation studies that revealed an association between the prenyltransferase and HMG-CoA reductase (6), an ER-resident enzyme that catalyzes reduction of HMG-CoA to mevalonate. This reaction is the rate-limiting step in a branched pathway that not only produces cholesterol but also produces the nonsterol isoprenoids, farnesyl pyrophosphate and GGpp. Farnesyl pyrophosphate and GGpp can become covalently attached to many cellular proteins and are intermediates in the production of other nonsterol isoprenoids, such as ubiquinone, heme, dolichol, and MK-4 (12–14). The reductase is subject to stringent control, in part, through a mechanism involving its sterol-induced ubiquitination and ER-associated degradation (ERAD). ERAD of reductase is initiated by its sterol-induced binding to the ER membrane proteins Insig-1 and Insig-2 (15, 16). This binding is mediated entirely by the membrane domain of reductase, which contains eight transmembrane helices and precedes the cytosolic catalytic domain (17, 18). Insig-associated ubiquitin ligases combine to mediate ubiquitination of reductase, which marks the enzyme for extraction from the ER membrane and cytosolic release for proteasome-mediated ERAD (19–23). The ERAD of reductase is enhanced by geranylgeraniol (GGOH), the alcohol derivative of GGpp (16). We postulate that GGpp, derived from phosphorylation of GGOH, augments extraction of ubiquitinated reductase from ER membranes (20, 24).
Sterols also trigger binding of reductase to UBIAD1 (25), which blocks a post-ubiquitination step in reductase ERAD and allows continued production of mevalonate that becomes preferentially incorporated into nonsterol isoprenoids (26). Accumulation of GGpp in ER membranes triggers release of UBIAD1 from reductase, relieving the inhibition of reductase ERAD and allowing vesicular transport of UBIAD1 from the ER to the Golgi. UBIAD1-mediated sensing of membrane embedded GGpp is abrogated in SCD. This is evidenced by the sequestration of SCD-associated UBIAD1 in the ER of GGpp-replete cells, which results in the inhibition of reductase ERAD in a dominant-negative manner (25, 27). This inhibition of ERAD significantly contributes to the dysregulation of cholesterol synthesis that characterizes SCD (26).
In the current studies, we expand on these findings by examining the enzymatic activity of UBIAD1. Our results revealed that SCD-associated variants of UBIAD1 were enzymatically defective and some of them markedly accumulated in cells. Further examination provided evidence that accumulation of SCD-associated UBIAD1 resulted from sequestration in the ER, which protected the mutant proteins from autophagic degradation. Besides providing new insights into the cell biology and biochemistry of UBIAD1 and SCD-associated variants, our studies have important implications for the pathology of SCD and cholesterol-lowering therapies that become limited owing to UBIAD1-mediated inhibition of reductase ERAD.
EXPERIMENTAL PROCEDURES
Materials
We obtained GGOH (sc-200858) from Santa Cruz Biotechnology (Dallas, TX); MK-4 (V9378), menadione (M5625), and bafilomycin A1 (SML1661) were obtained from Sigma-Aldrich (St. Louis, MO); GGpp (I-0200) was purchased from Echelon Bioscience (Salt Lake City, UT); and [3H]menadione and [14C]cholesterol were obtained from American Radiolabeled Chemicals (St. Louis, MO). Other reagents including FCS, sodium compactin, and sodium mevalonate were prepared or obtained as previously described (28).
Expression plasmids
All expression plasmids used in this study were previously described (25, 27). Expression plasmid pCMV-Myc-UBIAD1 encodes human UBIAD1 with a Myc epitope at the N-terminus; transcription is driven by the cytomegalovirus (CMV) promoter; pCMV-Myc-UBIAD1 (N102S) encodes Myc-tagged human UBIAD1 with SCD-associated asparagine-102 to serine (N102S) mutation (25); the remaining 19 SCD-associated mutants of Myc-UBIAD1 were described by Schumacher et al. (27).
Cell culture
SV-589 cells, immortalized human fibroblasts expressing the SV40 large T antigen (29), were maintained in medium A (Dulbecco’s modified Eagle’s medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) supplemented with 10% FCS (v/v). UBIAD1-deficient SV-589 (ΔUBIAD1-1) and (ΔUBIAD1-2) cells (26) were grown in medium A supplemented with 10% FCS and 1 mM mevalonate. SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (WT) and SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (N102S), lines of SV-589 (ΔUBIAD1-1) cells that stably express WT or N102S Myc-UBIAD1 (26), were maintained in medium A containing 10% FCS, 1 mM mevalonate, and 700 μg/ml G418. Monolayers of SV-589 and SV-589-derived cell lines were grown at 37°C in a 5% CO2 incubator.
Chinese hamster ovary (CHO)-K1 cells were grown in medium B (1:1 mixture of Ham’s F-12 medium and Dulbecco’s modified Eagle’s medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate) that contained 5% (v/v) FCS. UT-2 cells, reductase-deficient CHO-K1 cells (30), were maintained in medium B supplemented with 5% FCS and 200 μM mevalonate. Monolayers of CHO-K1 and UT-2 cells were grown at 37°C in an 8% CO2 incubator.
Synthesis of [3H]MK-4 in isolated membranes and intact cells
For in vitro studies measuring incorporation of [3H]menadione into MK-4 cells were set up for experiments as described in the figure legends. Following incubations, cells were harvested into medium by scraping and pelleted by centrifugation at 1,000 g for 5 min at 4°C. The cell pellets were washed with cold PBS and resuspended in 1 ml buffer A [100 mM Tris-HCl (pH 8.5) containing 1 mM DTT, and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 0.5 M Pefabloc, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 25 μg/ml ALLN, and 10 μg/ml aprotinin]. Following sonication for 45 s with an intensity of amplitude at 35% (Branson digital sonifier) at 4°C, cell lysates were centrifuged at 3,000 g for 5 min at 4°C to remove unbroken cells and nuclei. The post nuclear supernatants were then centrifuged at 100,000 g for 30 min and the resulting membrane fractions were resuspended in ice-cold buffer A. The protein concentration of resuspended membranes (5 μl) was determined (Pierce™ Coomassie Plus protein assay reagent; Thermo Fisher Scientific, Waltham, MA). Typical reactions were performed in 400 μl buffer A containing 10 μM GGpp, 10 nM [3H]menadione, and 100 μg isolated membrane fractions. Reactions were incubated at 37°C for 3 h, after which they were stopped by the addition of 400 μl methanol containing 16 μg/ml MK-4, 0.025 μCi/ml [14C]cholesterol, and 16 μg/ml cholesterol. Lipids were extracted with 800 μl dichloromethane, dried down, dissolved in dichloromethane, and subjected to TLC in chloroform. Lipids were visualized by iodine vapor staining and incorporation of [3H]menadione into MK-4 was determined by scintillation counting. Values were corrected for recovery using the percent of [14C]cholesterol recovered in each sample.
To measure [3H]MK-4 synthesis in intact cells, we set up cells on day 0 as described in the figure legends. On day 1, the cells were refed medium A with 5% FCS and 2.5 μCi/ml [3H]menadione (unlabeled menadione was added to achieve a final concentration of 50 nM). Some of the cells were incubated in the absence or presence of 10 μM compactin and up to 30 μM GGOH. Following incubation for 16 h at 37°C, cells were washed twice with PBS containing 2% BSA, followed by an additional wash with PBS. The cells were lysed with 0.1 N NaOH; an aliquot of each lysate was removed for protein determination. The remaining lysates were mixed with recovery solution containing 16 μg/ml MK-4, 0.025 μCi/ml [14C]cholesterol, and 16 μg/ml unlabeled cholesterol and extracted with dichloromethane:methanol (2:1). The resulting lipids were subjected to TLC and incorporation of [3H]menadione into MK-4 was determined as described above.
Subcellular fractionation and immunoblot analysis
Cells were set up for experiments on day 0 as described in the figure legends. After incubations, triplicate dishes were harvested and pooled for analysis. Subcellular fractionation of cells by differential centrifugation was performed as described (25), and aliquots of the resulting membrane fractions were subjected to SDS-PAGE and immunoblot analysis. The efficiency of RNAi-mediated knockdown was determined by immunoblot analysis of whole cell lysates. Primary antibodies used for immunoblot analysis included: IgG-A9, a mouse monoclonal antibody against the catalytic domain of hamster reductase (31); rabbit polyclonal anti-calnexin IgG (Novus Biologicals, Littleton, CO); IgG-9E10, a mouse monoclonal antibody against c-Myc purified from the culture medium of hybridoma clone 9E10 (American Type Culture Collection, Manassas, VA); IgG-1H12, a mouse monoclonal antibody against human UBIAD1 (26); rabbit polyclonal anti-LC3-II IgG (Novus Biologicals); rabbit polyclonal anti-ATG-7 IgG (Sigma-Aldrich); rabbit polyclonal anti-ATG-5 IgG, anti-ATG-12 IgG, anti-ATG16L1 IgG, anti-AMBRA1 IgG, and anti-ULK-1 IgG (Cell Signaling Technology, Danvers, MA). Secondary antibodies used for immunoblotting included: peroxidase-conjugated affinity-purified donkey anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) and were visualized using the Super Signal CL-HRP substrate system (Thermo Fisher Scientific). Gels were calibrated with prestained molecular mass markers (Bio-Rad). Filters were exposed to film at room temperature.
Immunofluorescence
SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (WT) and SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (N102S) cells were set up for experiments on day 0 as described in the figure legends. Following incubations described in the figure legends, cells were washed with PBS and fixed for 30 min in methanol at −20°C. After blocking in PBS supplemented with 1% normal goat serum (Life Technologies, Grand Island, NY), coverslips were incubated for 16 h at 37°C with primary antibodies [IgG-9E10, rabbit polyclonal anti-GM130 IgG (32), and rabbit polyclonal anti-LC3-II IgG] diluted in PBS containing 1% normal goat serum. Bound antibodies were visualized with goat anti-mouse IgG conjugated to Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 594 for 1 h. Cells were also stained for 5 min with 300 nM 4′,6-diamidino-2-phenylindole (Life Technologies) to visualize nuclei. Coverslips were mounted in Fluoromount-G solution (Southern Biotech, Birmingham, AL). Fluorescence imaging was performed using a DeltaVision microscopy imaging system (GE Healthcare Life Sciences, Pittsburgh, PA) equipped with a CoolSNAP HQ2 camera (Photometrics, Tucson, AZ) and objective oil lenses 100×/1.40 (Olympus, Waltham, MA). Deconvolved images were generated using DeltaVision SoftWoRx software (GE Healthcare Life Sciences). Brightness levels were adjusted across the entire images using ImageJ software (National Institutes of Health, Bethesda, MD).
RNAi and transient transfection
RNAi was carried out as described previously with minor modifications (25). On-Target plus SMARTpools of siRNAs against ATG-5, ATG-7, ATG-10, ATG-12, ATG16L1, AMBRA1, and ULK-1 were purchased from GE Dharmacon (Lafayette, CO). SV-589 cells were set up for experiments on day 0 as described in the figure legends. On day 1, triplicate dishes of cells were incubated with 120 pmol of siRNA duplexes mixed with Lipofectamine RNAiMAX™ reagent (Thermo Fisher Scientific) diluted in Opti-MEM I reduced serum medium (Life Technologies) according to the manufacturer’s procedure. Following incubation for 5 h at 37°C, the cells received a direct addition of medium A containing 5% FCS (final concentration). On day 2, the cells were transfected with pCMV-Myc-UBIAD1 or pCMV-Myc-UBIAD1 (N102S) as described previously (27). On day 4, the cells were harvested and analyzed as described in the figure legends.
RESULTS
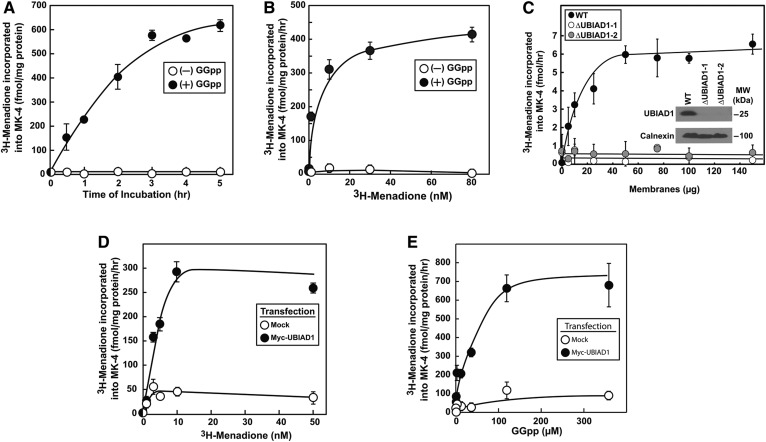
In the experiment shown in Fig. 1A, membranes isolated from SV-589 cells were resuspended in buffer and incubated with [3H]menadione in the absence or presence of GGpp. After the indicated period of time, reactions were terminated; lipids were extracted and fractionated by TLC, followed by scintillation counting. In a time-dependent fashion, a [3H]-labeled product that comigrated with MK-4 on TLC plates was produced when reactions were carried out in the presence (Fig. 1A, closed circles) but not in the absence (open circles) of GGpp. The in vitro synthesis of [3H]MK-4 was also dependent upon the amount of [3H]menadione in the assay (Fig. 1B). Figure 1C shows that while membranes from WT SV-589 cells incorporated [3H]menadione into MK-4 in a dose-dependent fashion (black circles), membranes from two independent lines of UBIAD1-deficient cells [designated SV-589 (ΔUBIAD1-1 and -2)] failed to produce [3H]MK-4 (open and gray circles). Finally, transfection of SV-589 cells with an expression plasmid encoding Myc-tagged human UBIAD1 dramatically enhanced [3H]MK-4 synthesis in membranes that was dependent upon the amount of [3H]menadione (Fig. 1D) and GGpp (Fig. 1E) in reactions.
Fig. 1.
Development of in vitro assay for UBIAD1-mediated synthesis of MK-4. A–C: SV-589, SV-589 (ΔUBIAD1-1), and SV-589 (ΔUBIAD1-2) cells were set up on day 0 at 7 × 105 cells per 100 mm dish in medium A containing 5% FCS. On day 2, cells were harvested for subcellular fractionation as described in the Experimental Procedures. Aliquots of the resulting membrane fractions (100 μg protein per reaction in A and B; 0–150 μg per reaction in C) were resuspended in reaction buffer containing 10 nM (A, C) (0.5 μCi per reaction) or 0–80 nM (B) (0–4 μCi per reaction) [3H]menadione in the absence or presence of 50 μM GGpp. Following incubation at 37°C for the indicated period of time (A) or 4 h (B, C), lipids were extracted and analyzed by TLC as described in the Experimental Procedures. Incorporation of [3H]menadione into MK-4 was determined by scintillation counting. Each value is the mean of triplicate incubations (± standard error). In C, aliquots of membrane fractions (micrograms of protein per lane) from SV-589, SV-589 (ΔUBIAD1-1), and SV-589 (ΔUBIAD1-2) cells were subjected to SDS-PAGE and immunoblot analysis was carried out with IgG-1H12 (against human UBIAD1) and polyclonal anti-calnexin IgG. D, E: SV-589 cells were set up on day 0 as described in A–C. On day 1, the cells were transfected with 3 μg per dish pCMV-Myc-UBIAD1 or empty pCDNA3.1 vector (Mock) in 5% FCS as described in the Experimental Procedures. On day 3, cells were harvested; membrane fractions were prepared, resuspended in reaction buffer (10 μg protein/reaction), and incubated for 4 h at 37°C with 0–50 nM [3H]menadione (0–2.5 μCi per reaction) and 100 μM GGpp (D) or 0–300 μM GGpp and 10 nM [3H]menadione (0.5 μCi per reaction) (E). Mean values of triplicate reactions are presented. Results shown in this and subsequent figures are representative of at least two independent experiments.
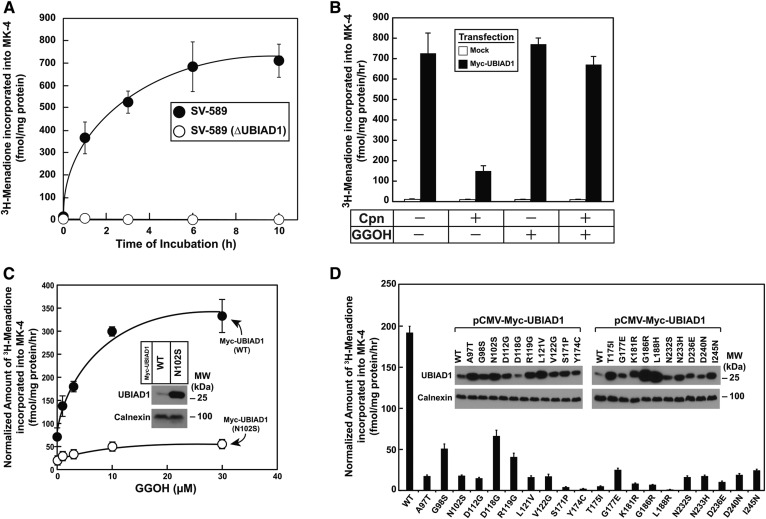
Figure 2A shows an experiment in which intact SV-589 and SV-589 (ΔUBIAD1-1) cells were labeled with [3H]menadione for various periods of time, after which cells were harvested and incorporation of the radiolabel into MK-4 was measured. The results show that when intact WT SV-589 cells were incubated in the presence of [3H]menadione, [3H]MK-4 was produced in a time-dependent fashion (Fig. 2A, closed circles). However, [3H]MK-4 synthesis was completely absent in SV-589 (ΔUBIAD1-1) cells (open circles). Synthesis of [3H]MK-4 was restored in SV-589 (ΔUBIAD1-1) cells upon overexpression of Myc-UBIAD1 (Fig. 2B). The reaction was inhibited by compactin, which depletes cells of isoprenoids by competitively inhibiting reductase. [3H]MK-4 synthesis was restored by the addition to cells of GGOH. Figure 2C compares [3H]MK-4 synthetic activity in SV-589 (ΔUBIAD1-1) cells transfected with WT UBIAD1 and the SCD-associated N102S variant. Although cells were transfected with equivalent amounts of plasmid encoding WT and N102S UBIAD1, the SCD-associated protein markedly accumulated (Fig. 2C, see inset). When normalized to the amount of UBIAD1 expression, GGOH-dependent synthesis of [3H]MK-4 in UBIAD1 (N102S)-transfected cells was reduced relative to that observed in cells transfected with WT UBIAD1 (Fig. 2C). Similar results were observed with the remaining 19 SCD-associated mutants of UBIAD1 (Fig. 2D). Most of the SCD-associated mutants of UBIAD1 were markedly stabilized compared with the WT protein (Fig. 2D, see inset) and exhibited reduced [3H]MK-4 synthetic activity, although stabilization and reduced enzymatic activity varied among the SCD-associated UBIAD1 mutants. Similar results were obtained in in vitro experiments that measured GGpp-dependent MK-4 synthetic activity in membranes of cells transfected with WT UBIAD1 or various SCD-associated mutants of the enzyme (supplemental Fig. S1).
Fig. 2.
SCD-associated variants of UBIAD1 are defective in MK-4 synthetic activity. A: SV-589 and SV-589 (ΔUBIAD1) cells were set up on day 0 at 2.5 ± 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, the cells were switched to medium A supplemented with 5% FCS and 10 μM compactin. Following incubation for 16 h at 37°C, cells were refed identical medium containing 50 nM [3H]menadione (2.5 μCi per reaction) and 100 μM GGOH for the indicated period of time. Following incubations, the cells were washed, lysed in 0.1 N NaOH, and lipids were extracted as described in the Experimental Procedures. The amount of [3H]menadione incorporated into MK-4 was determined by TLC, followed by scintillation counting. The values represent the mean of triplicate reactions (standard error). B: SV-589 (ΔUBIAD1) cells were set up on day 0 at 4 × 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, the cells were transfected with 1.5 μg per dish pCMV-Myc-UBIAD1 or empty pcDNA 3.1 vector (Mock) in medium A supplemented with 5% FCS. On day 2, cells were switched to medium A containing 5% FCS in the absence or presence of 10 μM compactin. After incubation for 16 h at 37°C, cells were treated for 5 h with 50 nM [3H]menadione (2.5 μCi per reaction) in the absence or presence of 10 μM compactin and/or 30 μM GGOH as indicated. Cells were subsequently harvested and the incorporation of [3H]menadione into MK-4 was determined as described in A. Values represent the mean of triplicate reactions (standard error). C: SV-589 (ΔUBIAD1) cells were set up on day 0 at 2.5 × 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, the cells were transfected with 1.5 μg per dish pCMV-Myc-UBIAD1 (WT) or pCMV-Myc-UBIAD1 (N102S) in medium A containing 5% FCS. On day 2, cells were switched to medium A supplemented with 5% FCS and 10 μM compactin. Following incubation for 16 h at 37°C, the cells were refed identical medium containing 50 nM [3H]menadione (2.5 μCi per reaction) and the indicated concentrations of GGOH. Following incubation for 5 h, cells were harvested and incorporation of [3H]menadione into MK-4 was determined as described in A. Aliquots of membranes (30 μg protein per lane) isolated from transfected cells were subjected to SDS-PAGE, and immunoblot analysis was carried out with IgG-9E10 (against Myc-UBIAD1) and anti-calnexin IgG (see inset). The band corresponding to Myc-UBIAD1 was quantified using ImageJ software and used to normalize values obtained for [3H]MK-4 synthetic activity. Values represent the mean of triplicate reactions (standard error). D: SV-589 (ΔUBIAD1) cells were set up on day 0 as described in A and transfected on day 2 with 1.5 μg per dish pCMV-Myc-UBIAD1 (WT) or the indicated SCD-associated variant of Myc-UBIAD1 in medium A supplemented with 5% FCS and 10 μM compactin. On day 3, cells received identical medium containing 50 nM [3H]menadione (2.5 μCi per reaction) and 30 μM GGOH. Following incubation for 6 h at 37°C, cells were harvested and incorporation of radioactivity into MK-4 was determined as described in A. Aliquots of membranes (30 μg protein per lane) isolated from transfected cells were subjected to SDS-PAGE, and immunoblot analysis was carried out with IgG-9E10 (against Myc-UBIAD1) and anti-calnexin IgG (see inset). The normalized amount of [3H]MK-4 synthetic activity was determined as described in C. Values represent the mean of triplicate incubations (standard error).
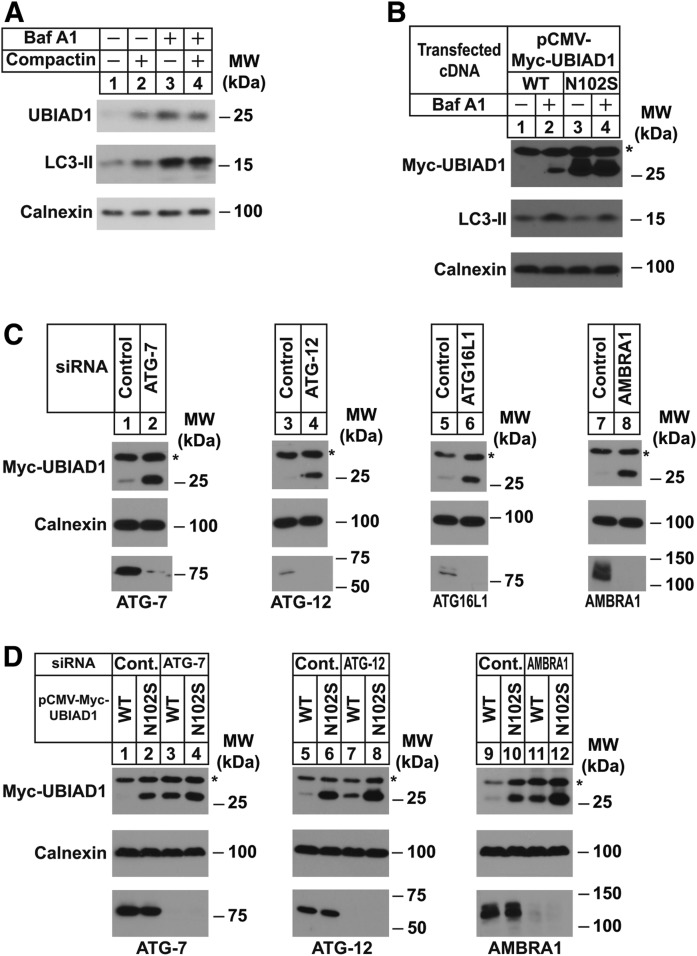
Next, we sought to examine the mechanism for stabilization of SCD-associated variants of UBIAD1. Considering the differential subcellular localization of WT and SCD-associated UBIAD1 in isoprenoid-replete cells (27), we reasoned that ER-sequestration protects the mutant enzymes from basal degradation that requires transport to or through the Golgi. This notion is supported by the results of Fig. 3A, which show that chronic treatment (3 days) of SV-589 cells with a low concentration of compactin-stabilized (1 μM) endogenous UBIAD1 (compare lanes 1 and 2). Stabilization of UBIAD1 was also observed when cells were treated with bafilomycin A1 (lane 3), an inhibitor of vacuolar type H+-ATPases (33, 34). We also observed marked stabilization of Myc-UBIAD1 in transfected SV-589 cells (Fig. 3B, lanes 1 and 2); Myc-UBIAD1 (N102S) accumulated in cells as expected (compare lanes 1 and 3); however, this accumulation was not enhanced by bafilomycin A1 (lane 4).
Fig. 3.
Inhibition of autophagy stabilizes WT but not SCD-associated UBIAD1. A: SV-589 cells were set up on day 0 at 2.5 × 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, the cells were switched to medium A supplemented with 5% FCS in the absence or presence of 1 μM compactin or 10 nM bafilomycin A1. Following incubation for 36 h at 37°C, cells were harvested for subcellular fractionation. Aliquots of the resulting membrane fractions (10 μg protein per lane) were subjected to SDS-PAGE, followed by immunoblot analysis with IgG-1H12 (against UBIAD1), anti-LC3 IgG, and anti-calnexin IgG. B: SV-589 cells were set up on day 0 at 2.5 × 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, the cells were transfected with 1.5 μg per dish pCMV-Myc-UBIAD1 (WT) or (N102S) in medium A supplemented with 5% FCS. On day 2, the cells were switched to medium A supplemented with 5% FCS in the absence or presence of 100 nM bafilomycin A1. After incubation for 16 h at 37°C, cells were harvested for subcellular fractionation and aliquots of the resulting membrane fractions (30 μg protein per lane) were analyzed by immunoblot with IgG-9E10 (against Myc-UBIAD1), anti-LC3 IgG, and anti-calnexin IgG. C: SV-589 cells were set up on day 0 at 2.5 × 105 cells per 60 mm dish in medium A containing 5% FCS. On day 1, cells were transfected in medium A containing 5% FCS with siRNAs targeting mRNAs encoding GFP (Control), ATG-7, ATG-12, ATG-16L1, and AMBRA1 as indicated and described in the Experimental Procedures. On day 2, cells were transfected in identical medium with 1.5 μg per dish pCMV-Myc-UBIAD1. On day 4, the cells were harvested for subcellular fractionation; aliquots of membrane fractions (30 μg protein per lane) were analyzed by immunoblot with IgG-9E10 (against Myc-UBIAD1), anti-ATG-7 IgG, anti-ATG12 IgG, anti-ATG16L1, anti- AMBRA1 IgG, and anti-calnexin IgG. D: SV-589 cells were set up on day 0, transfected on day 1 with siRNAs against GFP (Control), ATG-7, ATG-12, and AMBRA1, and transfected on day 2 with 1.5 μg per dish pCMV-Myc-UBIAD1 (WT) or (N102S) as described in C. On day 4, cells were harvested for subcellular fractionation and aliquots of the membrane fractions (30 μg protein per lane) were subjected to SDS-PAGE, followed by immunoblot analysis with IgG-9E10 (against Myc-UBIAD1), anti-ATG-7 IgG, anti-ATG12 IgG, anti-AMBRA1 IgG, and anti-calnexin IgG. The * denotes a nonspecific cross-reacting band.
The autophagy-lysosomal pathway, which plays a key role in cellular homeostasis, involves engulfment of proteins or organelles by autophagosomes that subsequently deliver their contents to lysosomes for degradation (35). Bafilomycin A1 inhibits autophagic degradation by preventing the acidification of endosomes and lysosomes (36). Bafilomycin A1-induced accumulation of the autophagosome marker LC3-II (see Fig. 3A, lanes 3 and 4; Fig. 3B, lanes 2 and 4) led us to examine a role for the autophagy-lysosome pathway in degradation of WT UBIAD1. Figure 3C shows an RNAi experiment in which SV-589 cells were transfected with siRNAs against mRNAs encoding GFP (which is not expressed in the cells) or several proteins of the autophagy-lysosomal pathway (37, 38). Immunoblot analysis of the siRNA-transfected cells revealed that Myc-UBIAD1 was significantly stabilized by the knockdown of Atg-7 (Fig. 3C, lane 2), Atg-12 (lane 4), Atg-16L1 (lane 6), and AMBRA1 (lane 8). Importantly, RNAi-mediated knockdown of Atg-7, Atg-12, and AMBRA1 stabilized WT Myc-UBIAD1 (Fig. 3D, lanes 3, 7, and 11); however, the treatments failed to stabilize Myc-UBIAD1 (N102S) to a similar extent (lanes 4, 8, and 12).
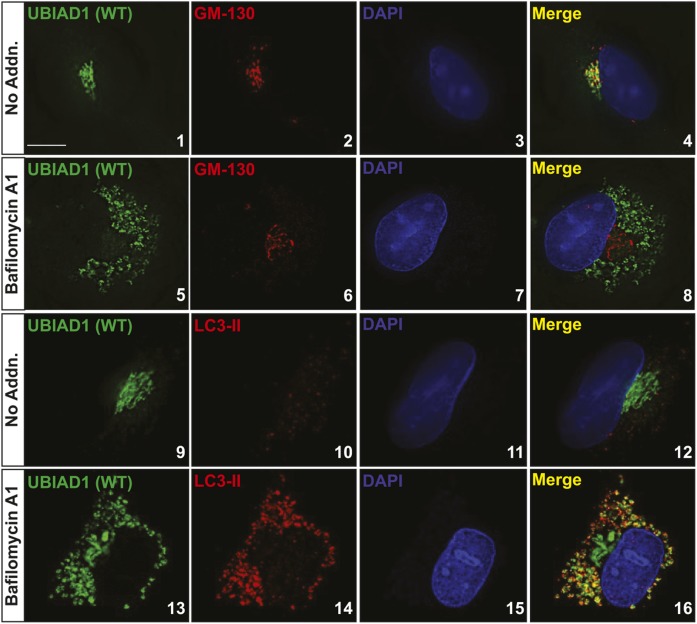
Figure 4 examines the effects of bafilomycin A1 on the subcellular localization of Myc-UBIAD1 in SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (WT) cells, which stably overexpress the protein. Consistent with our previous results (25, 27), Myc-UBIAD1 colocalized with the Golgi protein GM-130 (Pearson correlation coefficient = 0.668) when cells were cultured in isoprenoid-replete medium containing FCS (Fig. 4, panels 1–4; supplemental Fig. S2). In the presence of bafilomycin A1, Myc-UBIAD1 was redistributed to punctate structures scattered throughout the cytosol (Fig. 4, panels 5 and 13), whereas the subcellular localization of GM-130 remained unchanged (Fig. 4, panel 6; supplemental Fig. S2). Expression of the autophagosome marker LC3-II was markedly enhanced by bafilomycin A1 (Fig. 4, compare panels 10 and 14) and the protein colocalized with Myc-UBIAD1 (Pearson correlation coefficient = 0.594) in the punctate cytosolic structures (Fig. 4, panels 13–16; supplemental Fig. S3).
Fig. 4.
Accumulation of UBIAD1 in autophagosomes upon treatment of cells with bafilomycin A1. SV589/pMyc-UBIAD1(WT) cells were set up for experiments on day 0 at 4 × 104 cells per well of 12-well plates with glass coverslips in medium A containing 5% FCS. On day 2, cells received identical medium in the absence or presence of 100 nM bafilomycin A1. Following incubation for 16 h at 37°C, cells were fixed and analyzed by immunofluorescence microscopy using IgG-9E10 (against Myc-UBIAD1), anti-GM130 IgG, and anti-LC3 IgG as described in the Experimental Procedures. For all imaging experiments, results are representative of at least 100 cells. Pearson correlation coefficients were determined using ImageJ colocalization without thresholding for the entire image. Scale bar = 5 microns.
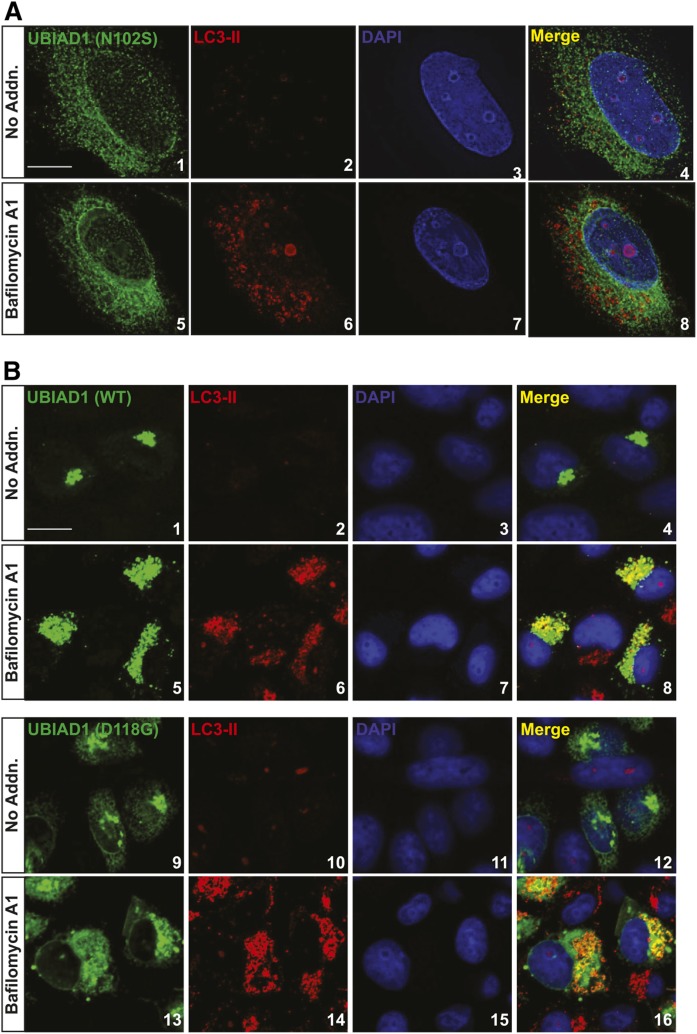
The subcellular localization of Myc-UBIAD1 (N102S) in SV-589 (ΔUBIAD1)/pMyc-UBIAD1 (N102S) cells exhibited a reticular pattern, indicative of localization to ER membranes (Fig. 5A, panel 1; supplemental Fig. S4). Importantly, the SCD-associated mutant remained in the ER when the cells were treated with bafilomycin A1 (lanes 5–8; supplemental Fig. S4). We showed previously that several SCD-associated variants of UBIAD1, including UBIAD1 (D118G), partially localized to the Golgi of isoprenoid-replete cells (27). Figure 2D (see inset) shows that UBIAD1 (D118G) was not markedly stabilized compared with other SCD-associated variants. Consistent with this, supplemental Fig. S5 shows that UBIAD1 (D118G) became stabilized in the presence of bafilomycin A1. In the experiment of Figure 5B, SV-589 cells were transfected with expression plasmids encoding WT or D118G versions of Myc-UBIAD1. As expected, Myc-UBIAD1 (WT) localized to the Golgi in the absence of bafilomycin (Fig. 5B, panels 1 and 4) and colocalized with LC3-II in the presence of the drug (panel 8). Myc-UBIAD1 (D118G) was distributed to both the ER and Golgi in untreated cells (Fig. 5B, panels 9 and 12); significant colocalization between the mutant protein and LC3-II was observed upon bafilomycin treatment (panel 16).
Fig. 5.
Accumulation of UBIAD1 in autophagosomes upon treatment of cells with bafilomycin A1. A: SV589/pMyc-UBIAD1 (N102S) cells were set up for experiments on day 0 at 4 × 104 cells per well of 12-well plates with glass coverslips in medium A containing 5% FCS. On day 2, cells received identical medium in the absence or presence of 100 nM bafilomycin A1. Following incubation for 16 h at 37°C, cells were fixed and analyzed by immunofluorescence microscopy using IgG-9E10 [against Myc-UBIAD1 (N102S)] and anti-LC3 IgG as described in the Experimental Procedures. B: SV589 cells were set up for experiments on day 0 at 4 × 104 cells per well of 12-well plates with glass coverslips in medium A containing 5% FCS. On day 2, cells were transfected with plasmids encoding Myc-UBIAD1 (WT) or (D118G) as described in the Experimental Procedures. On day 3, cells received identical medium in the absence or presence of 100 nM bafilomycin A1. Following incubation for 16 h at 37°C, cells were fixed and analyzed by immunofluorescence microscopy using IgG-9E10 [against Myc-UBIAD1 (N102S)] and anti-LC3 IgG as described in A. The nature of the nuclear inclusions observed in A and supplemental Fig. S4 is unclear. Scale bars = 5 microns.
DISCUSSION
The current studies were designed to compare the MK-4 synthetic activity of WT UBIAD1 with that of SCD-associated variants of prenyltransferase. Our studies began with the establishment of an assay that measures incorporation of [3H]menadione into MK-4 in isolated cell membranes. In vitro synthesis of [3H]MK-4 was reconstituted in membranes from SV-589 cells in a manner that was dependent upon time of incubation (Fig. 1A) and the amount of [3H]menadione in the assay (Fig. 1B). Moreover, the reaction exhibited an absolute requirement for the presence of UBIAD1 (Fig. 1C); activity was markedly enhanced in membranes of cells overexpressing Myc-UBIAD1 (Fig. 1D, E). We also measured [3H]MK-4 synthesis in intact cells that were metabolically labeled with [3H]menadione. Incorporation of [3H]menadione into MK-4 occurred in intact WT cells, but not in UBIAD1-deficient SV-589 cells (Fig. 2A). Overexpression of Myc-UBIAD1 restored [3H]MK-4 synthesis in UBIAD1-deficient cells (Fig. 2B); the reaction was inhibited by the addition to cells of compactin (which depletes intracellular levels of GGpp) and fully rescued by GGOH (which restores GGpp).
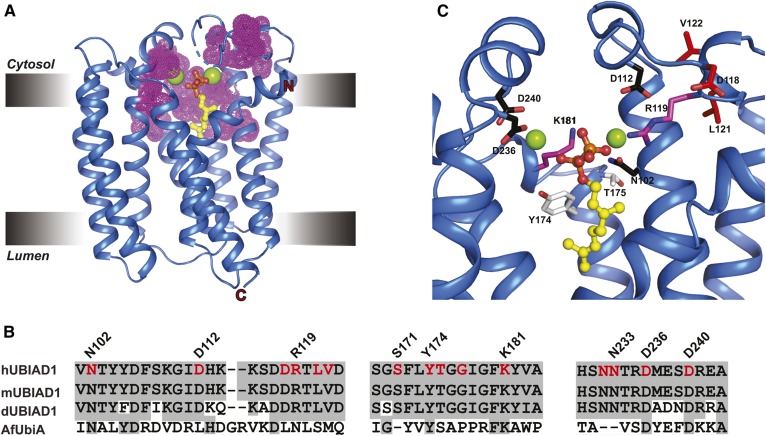
Our previous studies revealed that all 20 SCD-associated variants of UBIAD1 were refractory to GGpp-induced transport to the Golgi and remained sequestered in the ER where they bound to and inhibited ERAD of reductase (27). Our current studies show that SCD-associated UBIAD1 was also defective in GGpp-mediated synthesis of MK-4 (Fig. 2C, D; supplemental Fig. S1). We reason that for some SCD-associated UBIAD1 variants, ER sequestration and reduced enzymatic activity result from reduced affinity of the enzymes for GGpp. This notion is supported by the structural analysis of archaeal UbiA prenyltransferases (9, 10); a model of the Archaeoglobus fulgidus UbiA prenyltransferase (AfUbiA) bound to Mg2+ and geranyl pyrophosphate (Gpp) is shown in Fig. 6A. SCD-associated mutations in human UBIAD1 occur in regions that are conserved in UbiA prenyltransferases (Fig. 6B). Analysis of a model for the human UBIAD1 active site built using AfUbiA as a template (Fig. 6C) reveals that SCD-associated mutations map to a region surrounding the active site of the enzyme and to a catalytic lid that, in AfUbiA, undergoes substrate-induced conformational changes (9, 10). Some of the SCD-associated residues surrounding the active site mediate coordination of either Mg2+ ions (N102, D112, D236, and D240) or the isoprenyl phosphate groups (R119 and K181), whereas others (Y174 and T175) are positioned within the active site. D118, L121, and V122 localize to the mobile lid and likely contribute to catalysis. Although a clear function for G177, G186, and L188 is not evident, these residues appear to lie in close proximity to the active site. Important goals for future studies include comparison of GGpp binding between purified WT and SCD-associated UBIAD1 and determining whether GGpp hydrolysis is required for ER-to-Golgi transport of UBIAD1.
Fig. 6.
SCD-associated mutations cluster around the substrate-binding and catalytic active site of human UBIAD1. A: Structure of a UbiA homolog from Archaeoglobus fulgidus (AfUbiA) bound to Gpp and Mg2+. Residues corresponding to SCD-associated mutations in human UBIAD1 are indicated by magenta dots. B: Amino acid sequence alignment of conserved motifs in human, mouse, Drosophila UBIAD1 (hUBIAD1, mUBIAD1, and dUBIAD1, respectively), and AfUbiA. Conserved residues are shaded in gray and SCD-associated mutations are indicated and highlighted in red. C: Zoomed view of human UBIAD1 catalytic active site model built by PyMOL using AfUbiA as a template. The side chains (stick) of residues mutated in SCD are as follows: coordination of Mg2+ ion (black), coordination of isoprenyl phosphate group (magenta), positioned in putative catalytic lid (magenta), and active site residues (white).
During the course of characterizing MK-4 synthetic activity in cells, we noticed that SCD-associated variants of UBIAD1 accumulated to levels 2- to 10-fold relative to the WT enzyme (Fig. 2C, D). Similar results have been observed in the liver and other tissues of mice harboring a knock-in mutation that changes N100 in UBIAD1 to a serine residue (N100S) (39), which corresponds to the SCD-associated N102S mutation in human UBIAD1. Several lines of evidence indicate this accumulation results from sequestration of SCD-associated UBIAD1 in the ER, which protects the protein from basal autophagic degradation in lysosomes. First, trapping UBIAD1 in the ER by chronically depleting cells of nonsterol isoprenoids with compactin or blocking lysosomal degradation with the vacuolar type H+-ATPase inhibitor, bafilomycin A1, stabilized both endogenous and overexpressed UBIAD1 (Fig. 3A, B). Second, UBIAD1 was markedly stabilized by RNAi-mediated knockdown of several components of the autophagy-lysosomal pathway (Fig. 3C and D). Finally, treatment of cells with bafilomycin A1 not only stabilized UBIAD1, but also caused the protein to accumulate in an intracellular compartment that housed the autophagosome marker LC3-II (Figs. 4, and supplemental Fig. S6). The finding that UBIAD1 failed to colocalize with LC3-II (Pearson correlation coefficient = 0.152, see supplemental Fig. S6) when cells were treated with both bafilomycin A1 and compactin is consistent with the conclusion that ER sequestration protects UBIAD1 from autophagic degradation. It is important to note that UBIAD1 (N102S), which is completely sequestered in the ER, failed to be stabilized by bafilomycin A1 (Fig. 3B) and did not redistribute to LC3-II-positive organelles in the presence of the drug (Fig. 5A).
The enhanced stability of ER-localized UBIAD1 may have important clinical implications. Statins are a group of drugs widely prescribed to lower plasma levels of low density lipoprotein cholesterol and reduce the incidence of atherosclerotic cardiovascular disease (40–44). However, statins block synthesis of sterol and nonsterol isoprenoids that mediate feedback regulation of reductase. This results in a marked accumulation of reductase (up to 200-fold) in the livers of animals and humans (13, 45, 46) that allows for continued synthesis of cholesterol and thereby blunts the cholesterol-lowering effects of statins (47–50). The statin-induced accumulation of hepatic reductase is blunted (5-fold) in knock-in mice harboring mutations that prevent sterol-induced ubiquitination of reductase compared with their WT counterparts (51). Thus, inhibition of ERAD significantly contributes to statin-induced accumulation of reductase in the liver. Our recent studies of mice consuming a statin-containing diet (39) are consistent with a scenario in which GGpp depletion stabilizes UBIAD1 by trapping the protein in ER membranes where it binds to and inhibits reductase ERAD. Considering this, we postulate that mimicking GGpp in stimulating ER-to-Golgi transport of UBIAD1 may alleviate accumulation of reductase associated with statin therapy.
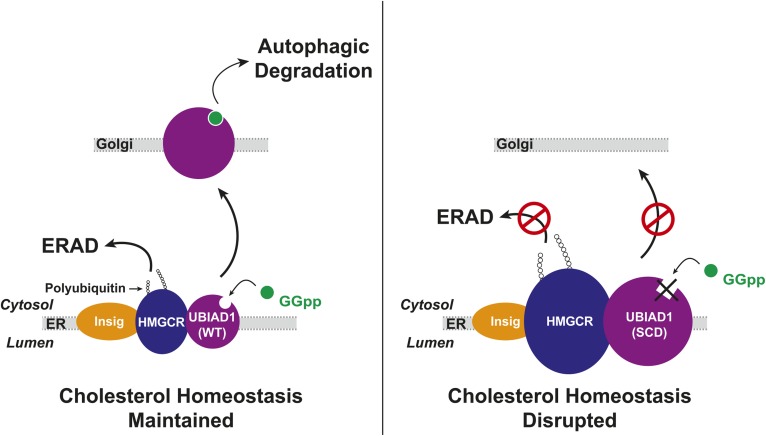
The accumulation of SCD-associated UBIAD1, which is defective in sensing membrane-embedded GGpp, has important implications for the pathology of the disease. SCD is autosomal-dominant, and our previous studies revealed that SCD-associated variants of UBIAD1 inhibit reductase ERAD in a dominant-negative fashion (25), explaining how cholesterol accumulation occurs in the corneas of SCD patients harboring heterozygous UBIAD1 mutations. We postulate that accumulation of SCD-associated UBIAD1 results from its sequestration in the ER, which protects the protein from basal autophagic degradation in lysosomes. The accumulation of ER-sequestered SCD-associated UBIAD1 magnifies its inhibitory effect on reductase ERAD, which augments synthesis and intracellular accumulation of cholesterol (Fig. 7). Agents that accelerate degradation of SCD-associated UBIAD1, trigger ER-to-Golgi transport of the variants, or prevent its interaction with reductase may prevent the cholesterol accumulation and corneal opacification associated with SCD.
Fig. 7.
SCD-associated UBIAD1 accumulates in ER membranes where it binds to HMG-CoA reductase and blocks its ERAD thereby disrupting cholesterol homeostasis.
Supplementary Material
Acknowledgments
The authors thank Kristina Garland-Brasher and Gennipher Young for excellent technical assistance, Lisa Beatty, Shomanike Head, Ijeoma Onweneme for help with tissue culture, and Dr. Xiaochun Li for helpful discussions.
Footnotes
Abbreviations:
- AfUbiA
- Archaeoglobus fulgidus UbiA prenyltransferase
- CHO
- Chinese hamster ovary
- ERAD
- ER-associated degradation
- GGpp
- geranylgeranyl pyrophosphate
- GGOH
- geranylgeraniol
- Gpp
- geranyl pyrophosphate
- MK-4
- menaquinone-4
- SCD
- Schnyder corneal dystrophy
- TLC
- thin layer chromatography
- UBIAD1
- UbiA prenyltransferase domain-containing protein-1
This work was supported by National Institutes of Health Grants HL020948 and GM134700 (to R.A.D-B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Klintworth G. K. 2009. Corneal dystrophies. Orphanet J. Rare Dis. 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orr A., Dube M. P., Marcadier J., Jiang H., Federico A., George S., Seamone C., Andrews D., Dubord P., Holland S., et al. 2007. Mutations in the UBIAD1 gene, encoding a potential prenyltransferase, are causal for Schnyder crystalline corneal dystrophy. PLoS One. 2: e685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss J. S., Kruth H. S., Kuivaniemi H., Tromp G., White P. S., Winters R. S., Lisch W., Henn W., Denninger E., Krause M., et al. 2007. Mutations in the UBIAD1 gene on chromosome short arm 1, region 36, cause Schnyder crystalline corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 48: 5007–5012. [DOI] [PubMed] [Google Scholar]
- 4.Weiss J. S. 2007. Visual morbidity in thirty-four families with Schnyder crystalline corneal dystrophy (an American Ophthalmological Society thesis). Trans. Am. Ophthalmol. Soc. 105: 616–648. [PMC free article] [PubMed] [Google Scholar]
- 5.Li W. 2016. Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem. Sci. 41: 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson M. L., Bosley A. D., Weiss J. S., Kostiha B. N., Hirota Y., Brandt W., Esposito D., Kinoshita S., Wessjohann L., Morham S. G., et al. 2013. The UBIAD1 prenyltransferase links menaquinone-4 [corrected] synthesis to cholesterol metabolic enzymes. Hum. Mutat. 34: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawa K., Hirota Y., Sawada N., Yuge N., Watanabe M., Uchino Y., Okuda N., Shimomura Y., Suhara Y., and Okano T.. 2010. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature. 468: 117–121. [DOI] [PubMed] [Google Scholar]
- 8.Lin B. R., Frausto R. F., Vo R. C., Chiu S. Y., Chen J. L., and Aldave A. J.. 2016. Identification of the first de novo UBIAD1 gene mutation associated with Schnyder corneal dystrophy. J. Ophthalmol. 2016: 1968493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng W., and Li W.. 2014. Structural insights into ubiquinone biosynthesis in membranes. Science. 343: 878–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., Levin E. J., Liu S., Bai Y., Lockless S. W., and Zhou M.. 2014. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 12: e1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirota Y., Nakagawa K., Sawada N., Okuda N., Suhara Y., Uchino Y., Kimoto T., Funahashi N., Kamao M., Tsugawa N., et al. 2015. Functional characterization of the vitamin K2 biosynthetic enzyme UBIAD1. PLoS One. 10: e0125737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards P. A., and Ericsson J.. 1999. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway 63. Annu. Rev. Biochem. 68: 157–185. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein J. L., and Brown M. S.. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 14.Wang M., and Casey P. J.. 2016. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17: 110–122. [DOI] [PubMed] [Google Scholar]
- 15.Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A.. 2003. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell. 11: 25–33. [DOI] [PubMed] [Google Scholar]
- 16.Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., and DeBose-Boyd R. A.. 2003. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278: 52479–52490. [DOI] [PubMed] [Google Scholar]
- 17.Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., and Goldstein J. L.. 1985. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 260: 522–530. [PubMed] [Google Scholar]
- 18.Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A. Jr., and Simoni R. D.. 1992. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 117: 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., and Debose-Boyd R. A.. 2010. Sterol-induced dislocation of HMG CoA reductase from ER membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 285: 19288–19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris L. L., Hartman I. Z., Jun D. J., Seemann J., and DeBose-Boyd R. A.. 2014. Sequential actions of the AAA-ATPase valosin-containing protein (VCP)/p97 and the proteasome 19 S regulatory particle in sterol-accelerated, endoplasmic reticulum (ER)-associated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 289: 19053–19066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song B. L., Sever N., and DeBose-Boyd R. A.. 2005. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 19: 829–840. [DOI] [PubMed] [Google Scholar]
- 22.Jo Y., Lee P. C., Sguigna P. V., and DeBose-Boyd R. A.. 2011. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. Proc. Natl. Acad. Sci. USA. 108: 20503–20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L. Y., Jiang W., Tian N., Xiong Y. N., Liu J., Wei J., Wu K. Y., Luo J., Shi X. J., and Song B. L.. 2018. Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase. J. Biol. Chem. 293: 4047–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elsabrouty R., Jo Y., Dinh T. T., and DeBose-Boyd R. A.. 2013. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from membranes of permeabilized cells. Mol. Biol. Cell. 24: 3300–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher M. M., Elsabrouty R., Seemann J., Jo Y., and DeBose-Boyd R. A.. 2015. The prenyltransferase UBIAD1 is the target of geranylgeraniol in degradation of HMG CoA reductase. eLife. 4: e05560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher M. M., Jun D. J., Johnson B. M., and DeBose-Boyd R. A.. 2018. UbiA prenyltransferase domain-containing protein-1 modulates HMG-CoA reductase degradation to coordinate synthesis of sterol and nonsterol isoprenoids. J. Biol. Chem. 293: 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher M. M., Jun D. J., Jo Y., Seemann J., and DeBose-Boyd R. A.. 2016. Geranylgeranyl-regulated transport of the prenyltransferase UBIAD1 between membranes of the ER and Golgi. J. Lipid Res. 57: 1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein J. L., Basu S. K., and Brown M. S.. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., and Russell D. W.. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 39: 27–38. [DOI] [PubMed] [Google Scholar]
- 30.Mosley S. T., Brown M. S., Anderson R. G., and Goldstein J. L.. 1983. Mutant clone of Chinese hamster ovary cells lacking 3-hydroxy-3 -methylglutaryl coenzyme A reductase. J. Biol. Chem. 258: 13875–13881. [PubMed] [Google Scholar]
- 31.Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., and Brown M. S.. 1983. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J. Biol. Chem. 258: 8450–8455. [PubMed] [Google Scholar]
- 32.Diao A., Rahman D., Pappin D. J., Lucocq J., and Lowe M.. 2003. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman E. J., Siebers A., and Altendorf K.. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA. 85: 7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., and Tashiro Y.. 1991. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266: 17707–17712. [PubMed] [Google Scholar]
- 35.Yu L., Chen Y., and Tooze S. A.. 2018. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y.. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23: 33–42. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27: 107–132. [DOI] [PubMed] [Google Scholar]
- 38.Ohsumi Y. 2014. Historical landmarks of autophagy research. Cell Res. 24: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo Y., Hamilton J. S., Hwang S., Garland K., Smith G. A., Su S., Fuentes I., Neelam S., Thompson B. M., McDonald J. G., et al. 2019. Schnyder corneal dystrophy-associated UBIAD1 inhibits ER-associated degradation of HMG CoA reductase in mice. eLife. 8: e44396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein J. L., and Brown M. S.. 2015. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 161: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heart Protection Study Collaborative Group. 2002. MRC/BHF Heart Protection Study of cholesterol lowering with simvastatin in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 360: 7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long-Term Intervention with Pravastatin in Ischaemic Disease (LIPID) Study Group. 1998. Prevention of cardiovascular events and death with pravastatin in patients with coronary heart disease and a broad range of initial cholesterol levels. N. Engl. J. Med. 339: 1349–1357. [DOI] [PubMed] [Google Scholar]
- 43.Sacks F. M., Pfeffer M. A., Moye L. A., Rouleau J. L., Rutherford J. D., Cole T. G., Brown L., Warnica J. W., Arnold J. M., Wun C. C., et al. 1996. The effect of pravastatin on coronary events after myocardial infarction in patients with average cholesterol levels. Cholesterol and Recurrent Events Trial investigators. N. Engl. J. Med. 335: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 44.1994. Randomised trial of cholesterol lowering in 4444 patients with coronary heart disease: the Scandinavian Simvastatin Survival Study (4S). Lancet. 344: 1383–1389. [PubMed] [Google Scholar]
- 45.Kita T., Brown M. S., and Goldstein J. L.. 1980. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. J. Clin. Invest. 66: 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reihnér E., Rudling M., Stahlberg D., Berglund L., Ewerth S., Bjorkhem I., Einarsson K., and Angelin B.. 1990. Influence of pravastatin, a specific inhibitor of HMG-CoA reductase, on hepatic metabolism of cholesterol. N. Engl. J. Med. 323: 224–228. [DOI] [PubMed] [Google Scholar]
- 47.Engelking L. J., Evers B. M., Richardson J. A., Goldstein J. L., Brown M. S., and Liang G.. 2006. Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116: 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldberg I. J., Holleran S., Ramakrishnan R., Adams M., Palmer R. H., Dell R. B., and Goodman D. S.. 1990. Lack of effect of lovastatin therapy on the parameters of whole-body cholesterol metabolism. J. Clin. Invest. 86: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grundy S. M., and Bilheimer D. W.. 1984. Inhibition of 3-hydroxy-3-methylglutaryl-CoA reductase by mevinolin in familial hypercholesterolemia heterozygotes: effects on cholesterol balance. Proc. Natl. Acad. Sci. USA. 81: 2538–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schonewille M., de Boer J. F., Mele L., Wolters H., Bloks V. W., Wolters J. C., Kuivenhoven J. A., Tietge U. J., Brufau G., and Groen A. K.. 2016. Statins increase hepatic cholesterol synthesis and stimulate fecal cholesterol elimination in mice. J. Lipid Res. 57: 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang S., Hartman I. Z., Calhoun L. N., Garland K., Young G. A., Mitsche M. A., McDonald J., Xu F., Engelking L., and DeBose-Boyd R. A.. 2016. Contribution of accelerated degradation to feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and cholesterol metabolism in the liver. J. Biol. Chem. 291: 13479–13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.