Abstract
Adipocytes take up long chain FAs through diffusion and protein-mediated transport, whereas FA efflux is considered to occur by diffusion. To identify potential membrane proteins that are involved in regulating FA flux in adipocytes, the expression levels of 55 membrane transporters without known function were screened in subcutaneous adipose samples from obese patients before and after bariatric surgery using branched DNA methodology. Among the 33 solute carrier (SLC) transporter family members screened, the expression of 14 members showed significant changes before and after bariatric surgery. One of them, Slc43a3, increased about 2.5-fold after bariatric surgery. Further investigation demonstrated that Slc43a3 is highly expressed in murine adipose tissue and induced during adipocyte differentiation in primary preadipocytes and in OP9 cells. Knockdown of Slc43a3 with siRNA in differentiated OP9 adipocytes reduced both basal and forskolin-stimulated FA efflux, while also increasing FA uptake and lipid droplet accumulation. In contrast, overexpression of Slc43a3 decreased FA uptake in differentiated OP9 cells and resulted in decreased lipid droplet accumulation. Therefore, Slc43a3 seems to regulate FA flux in adipocytes, functioning as a positive regulator of FA efflux and as a negative regulator of FA uptake.
Keywords: lipid droplet, adipose metabolism, membrane transporters, fatty acid uptake, fatty acid efflux, solute carrier family 43 member 3
FAs play important roles in a variety of cellular functions, such as the production and storage of energy, synthesis of phospholipids, glycolipids, and cellular signaling messengers, modification of proteins for targeting to cellular membranes, and regulation of gene expression (1–3). Because most cells, with the exception of adipocytes, have a very limited capacity for storing FAs, in the form of triglycerides (or for de novo synthesis of FA), circulating plasma FAs are the most important source of FAs for most tissues (4). Short and medium chain FAs have high plasma membrane permeability and can diffuse freely and easily through the plasma membranes of cells. Long chain FAs (LCFAs), however, have much lower water solubility and otherwise bind to albumin to facilitate large volume fluxes. Different rates of FA uptake are observed among various cell types, with metabolically active cells having increased rates of FA uptake (5). The rates of cellular FA uptake are also regulated acutely and chronically by hormones (such as insulin) and metabolic status (in case of obesity) (6, 7).
In addition to the bidirectional flip-flop model for transporting FAs across the plasma membrane, studies have demonstrated the importance of specific LCFA transport systems in metabolically active tissues, such as intestine, heart, adipose tissue, and the liver (8). This transport appears to involve membrane proteins that can mediate FA uptake via a rapid, saturable, substrate-specific, and hormonally related mechanism. Further studies have established that LCFA uptake into heart and muscle is regulated acutely by contraction, insulin, and leptin and chronically in conditions like obesity and diabetes (7). In adipocytes, permeation of LCFAs across the plasma membrane relies on a high-affinity low-capacity protein-facilitated transport system. Studies have demonstrated the importance of several different proteins, such as FA transport proteins (FATPs), long chain fatty acyl-CoA synthetases (ACSLs), and FA translocase (also known as CD36), in the protein-facilitated process of FA transport. However, the mechanisms of how FAs traverse the plasma membrane to enter the soluble cytoplasm are not yet fully understood (9). Specifically, there is a debate on the rate-limiting step in the overall process of FA uptake and whether, and to what extent, one or more membrane-associated proteins could facilitate and/or regulate FA uptake. In contradistinction, FA efflux from adipocytes is attributed to diffusion without any proteins being associated with facilitating the process, though we previously reported evidence to suggest the possibility of a protein-facilitated process (10).
To find possible candidates for these mechanisms, we performed an in silico study of all transporter genes fulfilling the following criteria: unknown function, plasma membrane location, and significant expression in adipose tissue. One of the transporter gene families that comes into focus is the family of the ABC transporters, of which almost half are thought to facilitate the ATP-dependent translocation of lipids or lipid-related compounds (11–13). Another family of potentially relevant membrane transport proteins is the family of the solute carriers (SLCs), with over 400 members, most of which are located in the cell membrane (14–17).
In our present study, we investigated the mRNA expression levels of gene candidates from our in silico study in patients before and after bariatric surgery to test the hypothesis that the expression of proteins linked to FA efflux might be upregulated. Genes that showed a significant change in their expression level were then studied in OP9 murine adipocyte cells and primary adipocytes throughout differentiation. One of the interesting and promising genes identified was the gene for the Slc43a3 solute carrier. The expression levels of Slc43a3 in various mouse adipose depots and during adipocyte differentiation were explored in C57Bl6J mice and OP9 murine adipocyte cells. The functional significance of Slc43a3 in regulating FA transport was studied using overexpression or silencing of Slc43a3 in differentiated OP9 cells. Interestingly, knockdown of Slc43a3 reduced both basal and forskolin-stimulated FA efflux. Moreover, knockdown resulted in an increased uptake of FAs into cells, whereas overexpression of Slc43a resulted in a decreased uptake of FAs. Thus, we conclude that Slc43a3 regulates FA flux in adipocytes, functioning as a positive regulator of FA efflux and as a negative regulator of FA uptake.
MATERIALS AND METHODS
Chemicals and reagents
Reagents were obtained from the following sources: cAMP Complete ELISA kit from Enzo Life Sciences (Farmingdale, NY); NEFA HR series NEFA-HR(2) from Wako Diagnostics, Wako Life Sciences, Inc. (Mountain View, CA); QBT FA uptake assay kit from Molecular Devices Corporation (Sunnyvale, CA); ADIFAB2 FA indicator from FFA Sciences LLC (San Diego, CA). All other chemicals, including Roche cOmplete protease inhibitor cocktail, were from Sigma-Aldrich (St. Louis, MO).
Animals
Wild-type C57Bl6J and CD36 KO mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and were used for the collection of liver and fat tissues. Mice were housed in the animal facility at the Veterans Affairs Palo Alto Health Care System on a 12/12 h light/dark cycle. All procedures were in accordance with institution guidelines and approved by the Institutional Animal Care and Use Committee of the Veterans Affairs Palo Alto Health Care System.
Patient sample collection and RNA preparation
Patients undergoing an elective vertical sleeve gastrectomy (VSG) procedure were recruited and informed consent was obtained from each study participant. Inclusion criteria included men and women older than 21 years of age with a BMI ≥35.0 kg/m2, and meeting criteria by American Diabetes Association standards for prediabetes or T2DM. For laparoscopic VSG surgery, a 4.8 mm stapler load was used to divide the greater curvature of the stomach 5 cm from the pylorus and remaining 3 cm from the angularis incisura. Stapler loads (3.5 mm) were fired thereafter progressing up to the angle of His to complete the VSG. Details of the surgical procedures have been described by Jahansouz et al. (18). Demographic data on sex, age, and T2DM were collected for obese patients at the time of surgery and 7 days following bariatric surgery. Weight and height were measured immediately prior to surgery and during the postoperative visit. BMI was calculated as weight (kilograms) divided by height (square meters). Additional details as to the molecular changes occurring following bariatric surgery have been published by Jahansouz et al. (19). The University of Minnesota and St. Cloud Hospital Institutional Review Boards approved all investigations and informed consent was obtained from each participant. This work was conducted in accordance with the Declaration of Helsinki. Abdominal subcutaneous adipose tissue biopsies from patients were processed for tissue analysis. Approximately 2–3 g of fat were obtained from each subject and immediately frozen with liquid nitrogen and stored at −80°C until analyzed. For RNA preparation, approximately 0.3 g of adipose tissue were extracted using TRIzol® (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
QuantiGene 2.0 RNA assays for RNA level analysis
RNA levels were assayed using QuantiGene2.0 RNA assays from Affymetrix, which are hybridization-based assays that utilize a branched DNA (bDNA) technology for signal amplification for the direct quantitation of gene expression transcripts. The QuantiGene Plex assay combines bDNA signal amplification with Luminex multi-analyte profiling bead (xMAP®) technology to enable the capture and quantitation of multiple RNA targets simultaneously. First, samples are lysed to release the target RNA. Second, the lysates are incubated overnight with an oligonucleotide probe set and color-coded bead mix. During this incubation, the probes cooperatively hybridize to both the target RNA and the capture probes conjugated to the microspheres, capturing the target RNA onto designated beads. Third, signal amplification is achieved through sequential hybridizations of the bDNA preamplifier, amplifier, and biotinylated label probe molecules to the target. Finally, addition of streptavidin-phycoerythrin generates a fluorescent signal that is proportional to the amount of target RNA captured. Fluorescence intensity is measured using a Luminex instrument. This assay system provides a robust and accurate measurement utilizing xMAP Luminex beads for multiplexing RNA targets. We selected 22 members of the ABC transporter family and 33 members from the SLC family with unknown functions together with control genes (supplemental Table S1) for analysis simultaneously on the Luminex® platform. Total RNA (250 ng) for each sample was incubated overnight at 54°C with unique fluorescent beads that have target-specific capture extenders and label extenders. The beads are coated with capture probes specific to the capture extenders, thus cooperatively hybridizing each target gene to a unique bead. After overnight incubation, the bDNA signal amplification portion was initiated by first washing the beads, followed by a 1 h incubation with the preamplifier DNA mix at 50°C. The beads were next washed, followed by a 1 h incubation with the amplifier DNA solution mix at 50°C. Biotinylated DNA label probe was added to the wells and incubated at 50°C for 1 h. Finally, the beads were washed, followed by incubation at room temperature for 30 min with streptavidin phycoerythrin. The beads were washed and read on the Luminex® instrument.
Cell culture
OP9 cells (obtained from ATCC) were grown to confluence in Dulbecco’s modified Eagle’s medium containing 20% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Before RNA isolation, OP9 cells were treated with 0.5 mM isobutyl-methylxanthine and 1 μM dexamethasone on day 0 and with 1 μg/ml insulin on day 2. Cells were harvested on day 0, day 3 (72 h), and day 9 (216 h).
Preadipocytes were isolated by collection of white adipose tissue from wild-type C57Bl6/J mice, followed by collagenase II digestion and then cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Before RNA isolation, preadipocytes were differentiated as described above and harvested on day 0, day 7, and day 14.
Transfections
Slc43a3 cDNA was cloned into the KpnI and XhoI of pCDNA6/V5-HisB vector (from Invitrogen); the construct was confirmed via sequencing. Slc43a3 Silencer Predesigned siRNAs were purchased from Ambion Life Technologies, Inc. (Austin, TX). Scrambled siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used as a negative control. OP9 cells were used for transfection with Slc43a3 siRNA and scrambled siRNA. Cells were plated in 96-well culture plates and transfection of siRNA or overexpression plasmid was performed with PolyJet™ reagent (SignaGen Laboratories, Gaithersburg, MD) following a reverse transfection protocol. Cells were cultured at 37°C, 5% CO2 for 5 h; then the medium was replaced with fresh medium containing 0.5 mM isobutyl-methylxanthine and 1μM dexamethasone and subsequently cultured according to the differentiation protocol. After 120 h (5 days), the cells were used for FA efflux and uptake assays, as well as NEFA, glycerol, and cAMP assays.
RNA isolation and quantitative real-time PCR analysis
For RNA isolation OP9 cells and SVCs were cultured and then collected in 1ml TRIzol reagent (if cultured in a 6-well plate) or using the Direct-zol RNA isolation kit from Zymo Research, Irvine, CA (if cultured in a 96-well plate). After the ethanol precipitation step, total RNA was dissolved in 30 μl RNase-free water, reamplified to aRNA, and then converted to cDNA for real-time PCR analysis. Real-time PCR was performed with the cDNA prepared as above using an ABI Prism 8500 system using SYBR green master mix reagent. The relative mass of specific RNA was calculated by the comparative cycle of threshold detection method according to the manufacturer’s instructions. Genes examined included: Plin1, Hsl, Fabp4, Slc22a15. Slc43a3, Slc16a6, Slc45a3, Slc25a33, Slc35c2, and Abcd4. Supplemental Table S2 shows the primer sets used for each gene.
FA uptake assay
OP9 cells were seeded into a black-wall/clear-bottom 96-well plate (Costar), transfected with Slc43a3 Predesigned Silencer Select siRNA or scrambled siRNA following a reverse transfection protocol and then cultured for 120 h (5 days) according to the differentiation protocol (described above). On day 5, a FA uptake assay was performed according to the manufacturer’s protocol. Briefly, the medium was removed from the wells and replaced with 90 μl/well serum-free medium and 10 μl/well HBSS (1×) containing 0.2% FAacid-free BSA. The assay plate was then incubated for 1 h at 37°C, 5% CO2. Before the test compound, Bodipy-dodecanoic acid, was added, a time zero read was performed. One hundred microliters of the test compound, diluted in HBSS (1×) and 0.2% BSA, were added to each well and immediately read in a fluorescence microplate reader for a kinetic reading at 485 mm (every 30 s for 30 min) using a bottom-read mode.
Quencher-based free FA efflux assay
The efflux assay was conducted as previously described (10) with minor modifications. OP9 cells were seeded into a black-wall/clear-bottom 96-well plate (Costar) transfected with Slc43a3 Predesigned Silencer Select siRNA or scrambled siRNA following a reverse transfection protocol and then cultured for 120 h (5 days) according to the differentiation protocol for OP9 cells. The cells were then loaded with 2 μM of the fluorescent lipid BODIPY C1-C12 bound to 1% BSA in HBSS (1×) for 16 h. After washing the assay plates with PBS, 100 μl of fresh buffer containing HBSS (1×) with 1% BSA buffer and BackDrop Green background suppressor-ready probe reagent (2 drops/ml) (Molecular Probes by Life Technologies) were added and the wells were once read for an initial time zero reading. Half of the wells of each group were then stimulated with 20 μM forskolin and 1 mM isobutyl-methylxanthine. The assay plates were read for 30 min in a fluorescent plate reader utilizing a bottom-read setting (kinetic, 485 mm).
FA efflux assay using 14C-palmitic acid
OP9 cells were seeded into 12-well plates (Costar) and transfected with Slc43a3 Predesigned Silencer Select siRNA or scrambled siRNA control for knockdown, and Slc43a3 expression plasmid and pCDNA6/V5-HisB as blank control for overexpression. Following a reverse transfection protocol, the cells were cultured for 120 h (5 days) according to the differentiation protocol for OP9 cells (described above). The cells were then loaded with 20 μM of 14C-palmitic acid [1 μCi of 60 mCi/mmol from Perkin-Elmer (Waltham, MA)] bound to 1% BSA in growth medium for 3 h. After washing the assay plates with PBS, 750 μl of fresh buffer containing HBSS (1×) with 1% BSA buffer were added to the cells. Cells were then stimulated with either 20 μM forskolin and 1 mM isobutyl-methylxanthine or 100 nM isoproterenol for lipolysis. Medium was collected from each well 60 min following lipolytic stimulation and counted in a Beckman LS 6500 scintillation counter (Beckman-Coulter, Mountain View, CA).
Free glycerol assay
A glycerol assay kit (Sigma-Aldrich) was used to assay the glycerol released. Differentiated OP9 cells were incubated for 60 min with or without lipolytic stimuli (20 μM forskolin, 1 mM isobutyl-methylxanthine) or 100 nM isoproterenol in 1% BSA in HBSS (1×) at 37°C and 5% CO2. The incubation media were collected and assayed for glycerol content by adding glycerol kinase and glycerol phosphate oxidase in the assay system. The resulting colorimetric product was quantified at 570 nm using a plate reader (Molecular Devices, Sunnyvale, CA). A standard curve was generated for the calculation of the quantity of glycerol released.
Intracellular free FA measurement
Intracellular free FAs, both protein bound and unbound, were measured using the ADIFAB2 FA indicator. Differentiated OP9 cells were incubated for 60 min with or without 100 nM isoproterenol in 1% BSA in HBSS (1×) at 37°C and 5% CO2. Cells were homogenized in 10 mM Tris (pH 7.4) and 1 mM EDTA with a glass homogenizer for more than 10 times. Cell lysates were centrifuged at 14,000 g for 10 min. The protein bound and unbound free FAs were assayed in 50 mM HEPES with 140 mM NaCl, 5 mM KCl, and 1 mM Na2HPO4 (pH 7.4) with or without 1 μM ADIFAB2 indicator, and 5 nM oleic acid (OA):BSA were used as standard. The ratio of fluorescence emission at 550 nm and 457 nm was measured, and the amount of aqueous free FAs and the FAs that bound to ADIFAB were calculated according to the manufacturer’s instructions.
FA oxidation assay
Cellular FA oxidation (FAO) activity was determined by following the conversion of 3H-palmitic acid into 3H2O using a modification of a previously described procedure (20). OP9 cells were seeded into 12-well plates and transfected with Slc43a3 Predesigned Silencer Select siRNA or scrambled siRNA control for knockdown. After transfection and differentiation for 5 days following the protocol for OP9 cells (described above), cells were treated with or without 100 nM isoproterenol in the presence of 0.7% BSA bound palmitic acid (500 μM with 0.4 μCi 3H-palmitate). After 1 h incubation at 37°C, the reaction was terminated with addition of 20 vol of organic solvent (chloroform:methanol, 2:1) for Folch extraction. The aqueous phase, which contains the 3H2O, was collected and counted in a Beckman LS 6500 scintillation counter (Beckman-Coulter, Mountain View, CA). FAO activity was normalized with cellular protein content.
cAMP assay
A cAMP complete enzyme immunoassay kit (Enzo Life Sciences) was used to measure forskolin-induced cAMP formation intra- and extracellularly. Differentiated OP9 cells were incubated with or without lipolytic stimuli (20 μM forskolin, 1 mM isobutyl-methylxanthine, or 100 nM isoproterenol) in 1% BSA in HBSS (1×) at 37°C and 5% CO2. The incubation media were collected after 5 and 15 min incubation and the cells were extracted at 15 min using 0.1 M HCl to avoid degradation of cAMP, and cAMP levels were measured in whole cell lysates as well as the media by using the complete cAMP immunoassay kit (Enzo Life Sciences) according to the manufacturer’s instructions. The level of cAMP production was normalized to the amount of total protein.
ATP assay
A luminescent ATP detection assay kit (Abcam, Cambridge, UK) was used to assay total cellular ATP levels. Differentiated OP9 cells were incubated for 60 min with or without lipolytic stimuli (20 μM forskolin, 1 mM isobutyl-methylxanthine, or 100 nM isoproterenol) in 1% BSA in HBSS (1×) at 37°C and 5% CO2. Cells were harvested and cellular ATP levels were assayed by following the protocol from the manufacturer; briefly, cells were lysed in detergent to inactivate ATPase and then substrate containing D-luciferin was added to the cell lysates. After incubation, luminescence was measured in a luminometer (Molecular Devices).
ACSL activity assay
Differentiated OP9 cells were treated with or without triacsin (5 nM) for 1 h prior to harvest. Cells were lysed in a buffer containing 250 mM sucrose, 10 mM Tris (pH 7.4), 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail (Roche), and phosphatase inhibitor cocktail (Sigma). Cell lysates were centrifuged at 16,000 g for 10 min at 4°C. Protein concentrations in supernatants were determined by bicinchoninic acid assay, and aliquots were stored at −80°C. Initial rates of total ACSL activity in cell lysates were measured using 10 μg of cell lysates at 37°C in the presence of 175 mM Tris (pH 7.4), 8 mM MgCl2, 5 mM DTT, 10 mM ATP, 250 μM CoA, 50 μM [3H]AA, [3H]palmitic acid, or [3H]OA in 0.5 mM Triton X-100 and 10 μM EDTA in a total volume of 0.1 ml. The reaction was initiated by the addition of the cell lysates and terminated by the addition of 1 ml Dole’s reagent, as previously described (21). Generated [3H]AA-CoA, [3H]palmitic acid-CoA, and [3H]OA-CoA were extracted, and the radioactivity was determined in a scintillation counter. The radioactivity in a reaction that contained all components but omitted cell lysate was included as a negative control.
Measurement of fatty acyl-CoA levels
Cellular fatty acyl-CoAs were measured using a previously reported method (22), with slight modifications. Differentiated OP9 cells were extracted in 0.8 ml 0.1 M KH2PO4, 0.8 ml isopropanol, 0.1 ml saturated aqueous ammonium sulfate, and 1.6 ml acetonitrile with 10 nmol C10-CoA internal standard. The sample was vortexed and centrifuged (1,400 g, 10 min, 4°C). The supernatant was diluted with 8 ml 0.1 M KH2PO4. Four milliliters of this sample were loaded onto an Oasis HLB 1 cc (30 mg) extraction cartridge (Waters) preconditioned with 3 ml acetonitrile and 2 ml 25 mM KH2PO4. The cartridge was washed once with water (4 ml) and eluted with 0.5 ml of 40:60 v/v acetonitrile:water containing 15 mM NH4OH. A portion of the eluant (50 μl) was injected onto an Agilent G6410B QQQ instrument for LC-MS analysis with unnatural standard.
Statistics
Data are expressed as mean ± SEM. Statistical analyses were performed by one-way ANOVA using Prism 6.02 for Mac OS X (GraphPad Software, Inc., La Jolla, CA). Differences between groups were considered statistically significant when P < 0.05.
RESULTS
mRNA expression levels of membrane transport proteins in patients before and after bariatric surgery
It is common for patients to show significant improvement in insulin sensitivity 1 week post bariatric surgery, before significant weight loss has been observed (23), but coincident with high rates of FAO. In search of genes that are potentially involved in FA transport, particularly any potentially involved in FA efflux, we examined the mRNA expression levels of 22 membrane transport proteins from the ABC transporter family and 33 from the SLC family (see list in supplemental Table S1), which at the time of study did not have a known ligand, in the adipose tissue of patients post bariatric surgery at this critical time. Table 1 shows the expression levels, determined using bDNA analysis, of genes that displayed statistically significant differences before and 1 week after bariatric surgery. Of the genes examined, eight genes from the SLC family showed significant increases after bariatric surgery and six showed significant decreases. The function of these SLC family members have not yet been determined.
TABLE 1.
mRNA expression levels of membrane transport proteins in patients before and after bariatric surgery showing statistically significant changes
| Pre-Surgery Average (/TBP) | Post-Surgery Average (/TBP) | Fold Post/Pre | t-Test | |
| ABC transporter family | ||||
| ABCC10 | 0.157 | 0.246 | 1.567 | 0.002 |
| ABCD4 | 0.789 | 1.181 | 1.497 | 0.009 |
| ABCB10 | 0.646 | 0.942 | 1.459 | 0.016 |
| ABCA6 | 4.636 | 2.273 | 0.490 | 0.017 |
| ABCA5 | 0.460 | 0.196 | 0.426 | 4.78E-04 |
| ABCA10 | 1.680 | 0.586 | 0.349 | 1.51E-04 |
| ABCA9 | 3.122 | 1.073 | 0.344 | 0.002 |
| ABCA8 | 3.878 | 1.229 | 0.317 | 0.002 |
| ABCC6 | 1.242 | 0.208 | 0.167 | 9.49E-05 |
| ABCD2 | 3.385 | 0.564 | 0.167 | 7.99E-06 |
| SLC transporter family | ||||
| SLC22A15 | 0.005 | 0.062 | 11.808 | 6.96E-06 |
| SLC16A6 | 0.060 | 0.379 | 6.271 | 0.014 |
| SLC45A3 | 0.066 | 0.244 | 3.680 | 0.006 |
| SLC35F2 | 0.065 | 0.226 | 3.460 | 0.044 |
| SLC43A3 | 1.151 | 2.791 | 2.424 | 0.012 |
| SLC16A5 | 0.136 | 0.280 | 2.058 | 0.001 |
| SLC35C2 | 0.922 | 1.734 | 1.880 | 0.001 |
| SLC35E3 | 0.372 | 0.531 | 1.428 | 0.054 |
| SLC25A46 | 1.740 | 1.197 | 0.688 | 4.48E-05 |
| SLC25A30 | 0.863 | 0.562 | 0.652 | 1.22E-04 |
| SLC25A36 | 0.975 | 0.531 | 0.545 | 0.001 |
| SLC25A42 | 0.441 | 0.191 | 0.432 | 0.001 |
| SLC25A33 | 1.780 | 0.627 | 0.352 | 2.19E-04 |
| SLC6A16 | 0.073 | 0.011 | 0.153 | 0.001 |
| P450 superfamily | ||||
| TBXAS1 | 0.942 | 2.882 | 3.059 | 0.001 |
| Leukotriene metabolism | ||||
| LTA4H | 2.191 | 3.617 | 1.651 | 0.028 |
| LTB4R | 0.258 | 0.422 | 1.636 | 0.026 |
| CYSLTR1 | 0.873 | 0.614 | 0.703 | 0.040 |
| Others | ||||
| SIRT3 | 0.770 | 0.619 | 0.804 | 0.012 |
| GSTA4 | 0.895 | 0.493 | 0.550 | 0.001 |
| PDE3B | 3.764 | 0.810 | 0.215 | 0.005 |
mRNA expression of membrane transporters in OP9 cells and mouse tissues
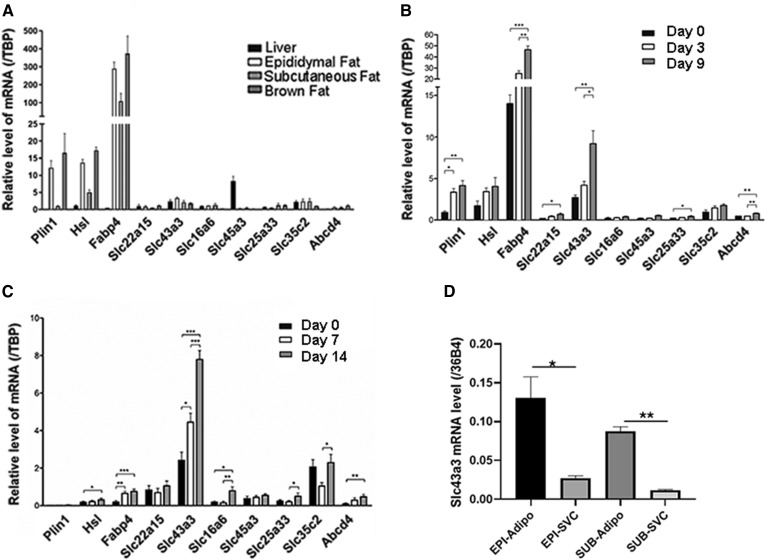
We then analyzed the mRNA expression of the six plasma membrane proteins in the SLC family that showed substantial basal expression levels and more than 1.5-fold increase after bariatric surgery in liver and various adipose depots in mice. For this analysis, 4-month-old male wild-type C57Bl6J mice were euthanized and their liver and fat tissues were collected for analysis of mRNA expression by RT-qPCR. From all analyzed genes, Slc43a3 showed the most significant expression in epididymal, subcutaneous, and brown adipose tissue (Fig. 1A). Its expression pattern is comparable to genes that are involved in control of lipid homeostasis in adipose tissue, i.e., Plin1, Hsl, and Fabp4. Further analysis during the differentiation of OP9 from preadipocytes to adipocytes showed that expression of Slc43a3 increased significantly between day 0 and day 9 (P < 0.01) and between day 3 and day 9 (P < 0.05) (Fig. 1B). We also analyzed the gene expression of these specific plasma membrane proteins in primary preadipocytes, which were isolated from wild-type mice and differentiated for 14 days. As shown in Fig. 1C, Slc43a3 showed the most significant changes in expression throughout differentiation: between day 0 and day 3 (P < 0.05), between day 3 and day 9 (P < 0.001), and between day 0 and day 9 (P < 0.001). Slc43a3 mRNA expression is ∼8-fold higher in adipocytes than in the stromal vascular cells isolated from the same fat pad (Fig. 1D), further evidence of its preferential expression in adipocytes.
Fig. 1.
mRNA expression of membrane transporters in OP9 cells and tissues. A: Levels of membrane transporter mRNA in wild-type mouse liver, epididymal fat, subcutaneous fat, and brown adipose tissue, each with n = 4. B: Levels of membrane transporter mRNA in OP9 cells. OP9 cells were treated with 0.5 mM isobutyl-methylxanthine, 1 μM dexamethasone, and 1 μg/ml insulin for differentiation for 5 days before mRNA analysis. Results are representative of three independent experiments, each with n = 6. C: Levels of membrane transporter mRNA in differentiated primary mouse preadipocytes. D: Slc43a3 mRNA expression in adipocytes versus stromal vascular cells. Results are representative of three independent experiments, each with n = 6. *P < 0.05, **P < 0.01, ***P < 0.001.
Effects of Slc43a3 knockdown and overexpression on free FA transport
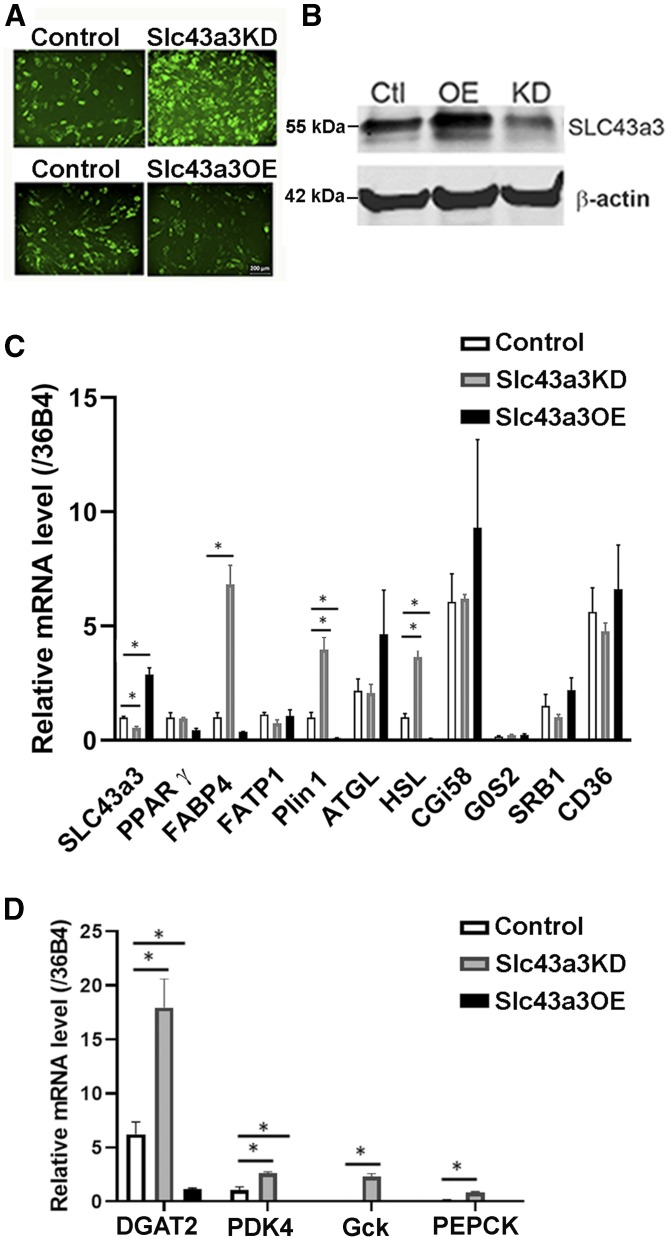
To characterize the function of Slc43a3, we manipulated levels of Slc43a3 in OP9 cells by knockdown using siRNA or overexpression (pEZ-M51-Slc43a3) and examined the cells after 5 days of differentiation. There was significantly more accumulation of lipid droplets in OP9 cells at day 5 after differentiation with knockdown of Slc43a3, whereas there was a significant decrease in lipid accumulation with overexpression of Slc43a3 (Fig. 2A). The expression levels of Slc43a3 in transfected and control (scrambled siRNA; pcDNA6) cells were detected by Western blot (Fig. 2B) and RT-qPCR (Fig. 2C). Slc43a3 siRNA knockdown significantly decreased Slc43a3 expression compared with control scrambled siRNA (P < 0.05), whereas transfection of cells with pEZ-M51-Slc43a3 significantly increased Slc43a3 expression compared with control (P < 0.001) (Fig. 2C). The mRNA expression of the master regulator for adipogenesis, PPARγ, did not show significant changes with either knockdown or overexpression of Slc43a3; however, there was a trend for its expression to be reduced with overexpression of Slc43a3. In contrast, several genes involved in lipid droplet metabolism, Plin1, FABP4, and HSL, were increased when Slc43a3 was knocked down and decreased when Slc43a3 was overexpressed (Fig. 2C), suggesting that overexpression of Slc43a3 might have interfered with normal adipocyte differentiation. However, no significant changes in mRNA levels were observed with either knockdown or overexpression of Slc43a3 in FATP1, ATGL, CGI58, G0S2, SR-B1, or CD36. In contrast, the expression levels of several genes involved in esterification, DGAT2, PDK4, Gck, and PEPCK, were increased when Slc43a3 was knocked down and decreased when Slc43a3 was overexpressed (Fig. 2D).
Fig. 2.
Effects of Slc43a3 knockdown and overexpression on genes involved in lipid metabolism. A: Lipid accumulation in differentiated OP9 cells transfected with scrambled siRNA (control), Slc43a3 siRNA, pcDNA6 (control), or pEZ-M51-Slc43a3-His clone. OP9 cells were transfected with the scrambled siRNA (control), Slc43a3 siRNA, pcDNA6 (control), or pEZ-M51-Slc43a3-His clone and differentiated for 5 days. Bodipy 493/503 (1 nM) was added in the media before fluorescent microscopy. Results are representative of three independent experiments. Scale bar 200 microns. B: Western blot of Slc43a3 in OP9 cells transfected with scrambled siRNA (control), Slc43a3 siRNA, pcDNA6 (control), or pEZ-M51-Slc43a3-His clone. C: Effects of knockdown and overexpression of Slc43a3 in OP9 cells on genes involved in lipid metabolism. Results are representative of two to three independent experiments. D: Effects of knockdown and overexpression of Slc43a3 in OP9 cells on genes involved in esterification. Results are representative of two independent experiments, each with n = 4–6. *P < 0.05.
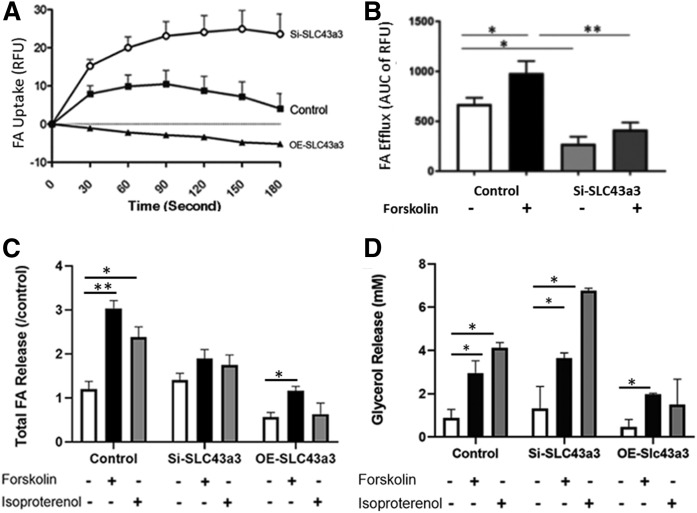
FA uptake and efflux were assayed in cells following Slc43a3 knockdown or overexpression. As shown in Fig. 3A, direct measurement of FA (Bodipy-dodecanoic acid, C1-C12 FA) uptake showed that knockdown of Slc43a3 significantly increased the rate of FA uptake (P < 0.001), whereas overexpression of Slc43a3 significantly decreased the rate of FA uptake (P < 0.001). The impact of Slc43a3 on FA efflux was also examined. Interestingly, knockdown of Slc43a3 significantly (P < 0.05) decreased the rates of FA efflux compared with control cells under basal conditions and substantially (P < 0.01) blunted FA efflux when cells were treated with forskolin to stimulate lipolysis and FA efflux (Fig. 3B). Whereas these FA flux experiments were performed with a fluorescently labeled C12 FA, we sought to confirm the efflux observations using cells loaded with radiolabeled 14C-palmitic acid (C16). As shown in Fig. 3C (FA efflux) and Fig. 3D (glycerol efflux), treatment of control OP9 cells with either forskolin or isoproterenol increased both FA and glycerol release, respectively. In contrast and similar to that observed using Bodipy-dodecanoic acid, treatment of OP9 cells in which Slc43a3 had been knocked down by siRNA with either forskolin or isoproterenol failed to stimulate FA efflux (Fig. 3C); however, both agents potently stimulated glycerol release (Fig. 3D), thus documenting activation of lipolysis. Overexpression of Slc43a3 resulted in low basal and, though forskolin stimulated FA and glycerol efflux, it remained low. This, however, might reflect the relatively low degree of differentiation observed with overexpression of Slc43a3.
Fig. 3.
Effects of Slc43a3 knockdown and overexpression on FA transport. A: Effects of knockdown and overexpression of Slc43a3 in OP9 cells on FA uptake. Results are representative of two to three independent experiments. Slc43a3 knockdown, n = 22; Slc43a3 overexpression, n = 12; Control combined, n = 36. *P < 0.05, **P < 0.01, ***P < 0.001. B: Effects of knockdown of Slc43a3 in OP9 cells on FA efflux. Transfected OP9 cells were loaded with fluorescent lipid BODIPY C1-C12 bound to 1% BSA, and FA efflux assayed following the quencher-based FA efflux assay. Results are representative of three to four independent experiments with replication of nine in each treatment. The cumulative FA efflux was calculated for the first 5 min of the reaction. *P < 0.05, **P < 0.01. C: Effects of knockdown of Slc43a3 in OP9 cells on FA efflux using 14C-palmitic acid. Transfected and differentiated OP9 cells were loaded with 20 μM 14C-palmitic acid. Cells were then stimulated either with 20 μM forskolin and 1 mM isobutyl-methylxanthine or 100 nM isoproterenol for lipolysis. Media were collected after 60 min for measurement of FA radioactivity and glycerol released (D). *P < 0.05, **P < 0.01.
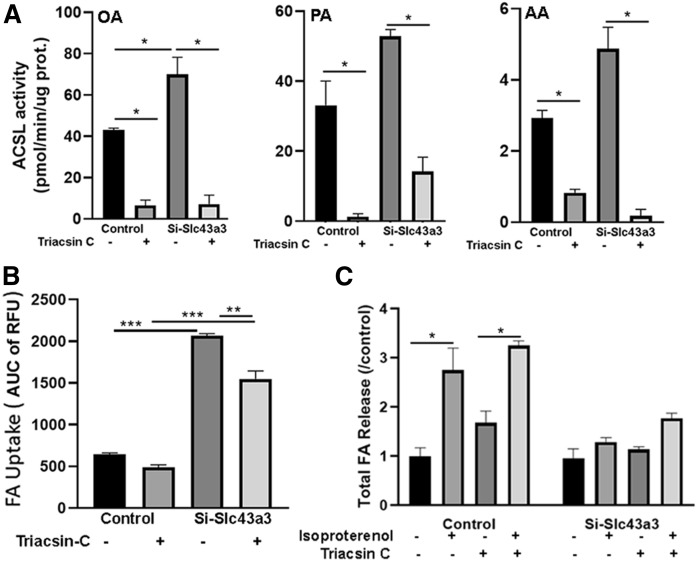
In view of the increased expression levels of several genes involved in esterification observed with knockdown of Slc43a3, we examined the contribution of esterification/re-esterification to the increased FA influx and the decreased FA efflux seen with knockdown of Slc43a3 by inhibiting acyl-CoA synthetase activity because esterification requires the activation of FAs to fatty acyl-CoA, mediated through the actions of the family of ACSLs (24). As shown in Fig. 4A, ACSL activity, as measured using oleate, palmitate, and arachidonate as substrates, was significantly increased in OP9 cells in which Slc43a3 had been knocked down (P < 0.05), consistent with the increased expression of genes involved in esterification. However, we were not able to detect any differences in cellular fatty acyl-CoAs in control and Slc43a3 siRNA-treated cells (data not shown). Incubation of OP9 cells with triacsin resulted in almost complete inhibition of ACSL activity in both control (P < 0.05) and Slc43a3 siRNA-treated (P < 0.05) cells, though inhibition was not as complete in Slc43a3 knockdown cells using palmitate as substrate. When FA influx was examined in control and Slc43a3 siRNA-treated cells that had been pretreated with triacsin (Fig. 4B), inhibition of ACSL activity had no effect on FA uptake in control cells. In contrast, Slc43a3 siRNA-treated cells displayed increased FA uptake in the absence of triacsin (P < 0.001), and inhibition of ACSL activity resulted in a small, but significant (P < 0.01), reduction in FA uptake. Thus, a portion of the increased FA influx observed with Slc43a3 knockdown appears to be due to a stimulation of esterification. In contradistinction to FA influx, inhibition of ACSL activity had no effects on FA efflux following lipolytic stimulation in either control or Slc43a3 siRNA-treated cells (Fig. 4C). Consequently, treatment with isoproterenol of OP9 cells in which Slc43a3 had been knocked down by siRNA failed to stimulate FA efflux even when FA reesterification had been inhibited by triacsin, suggesting that an increase in reesterification does not contribute to the block in FA efflux with Slc43a3 knockdown.
Fig. 4.
Effects of ACSL inhibition on FA transport. A: Effects of triacsin on ACSL activity in control and Slc43a3 knockdown OP9 cells. OP9 cells were transfected with scrambled siRNA (control) or Slc43a3 siRNA and differentiated for 5 days. Cells were incubated with triacsin (5 nM) for 1 h prior to measurement of ACSL activity. OA, palmitic acid (PA), and AA were assayed separately as substrates. B: Effects of triacsin on FA uptake in control and Slc43a3 knockdown OP9 cells. OP9 cells were transfected with scrambled siRNA (control) or Slc43a3 siRNA and differentiated for 5 days. Cells were preincubated with triacsin (5 nM) for 1 h prior to incubation with fluorescent lipid BODIPY C1-C12 for measurement of FA uptake. C: Effects of triacsin on FA efflux in control and Slc43a3 knockdown OP9 cells. OP9 cells were transfected with scrambled siRNA (control) or Slc43a3 siRNA and differentiated for 5 days. Differentiated cells were loaded with 20 μM 14C-palmitic acid, and then preincubated with triacsin (5 nM) for 1 h prior to incubation with 100 nM isoproterenol. Media were collected after 1 h for measurement of FA radioactivity released. Results are representative of two independent experiments, each with n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
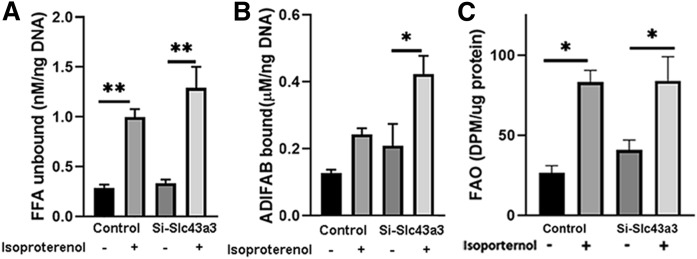
Because knockdown of Slc43a3 appeared to block lipolytically stimulated FA efflux, and reesterification did not appear to contribute to this, FAs would be expected to increase intracellularly following lipolysis in the setting of Slc43a3 knockdown. To assess this directly, we measured intracellular free FAs, both unbound (Fig. 5A) and protein bound (Fig. 5B), in control and Slc43a3 siRNA-treated OP9 cells under basal conditions and following stimulation of lipolysis with isoproterenol. Treatment of control cells with isoproterenol resulted in a significant increase in intracellular nonprotein bound free FAs (P < 0.05), without a change in protein bound free FAs, as has been previously reported (25), since most of the FAs released upon lipolysis efflux from the cell. It is important to note that the vast majority of intracellular FAs in adipocytes are protein bound by FA binding proteins (FABPs), such as FABP4 and FABP5 (26), and only a very small fraction is non-protein bound. Treatment of Slc43a3 knockdown cells with isoproterenol also resulted in a significant increase in intracellular non-protein bound free FAs (P < 0.05) similar to control; however, in contrast to control cells, there was a significant 2-fold increase in protein bound free FAs (P < 0.05), consistent with a block in FA efflux and the resultant buffering of the excess free FAs by FABPs. To examine whether the increase in intracellular free FAs had a metabolic effect, we assessed the rates of fatty oxidation (FAO) in control and Slc43a3 siRNA-treated OP9 cells under basal conditions and following stimulation of lipolysis with isoproterenol (Fig. 5C). Basal FAO was similar in control and Slc43a3 siRNA-treated OP9 cells, though there was a nonsignificant trend for an increase in Slc43a3 knockdown cells. Isoproterenol increased FAO to a similar extent in control and Slc43a3 knockdown cells.
Fig. 5.
Effects of Slc43a3 knockdown on intracellular free FA concentrations and FAO. OP9 cells were transfected with scrambled siRNA (control) or Slc43a3 siRNA and differentiated for 5 days. Differentiated cells were treated without or with 100 nM isoproterenol, and then intracellular protein bound (B) and unbound (A) free FA concentrations measured using an ADIFAB2 FA indicator. Results are representative of two independent experiments, each with n = 3. *P < 0.05, **P < 0.01. C: FAO. OP9 cells were transfected with scrambled siRNA (control) or Slc43a3 siRNA and differentiated for 5 days. Differentiated cells were treated without or with 100 nM isoproterenol in the presence of 3H-palmitate, and then FAO was measured as described in the Materials and Methods. Results are representative of three independent experiments, each with n = 5–6. *P < 0.05.
Expression of Slc43a3 in CD36 KO mice
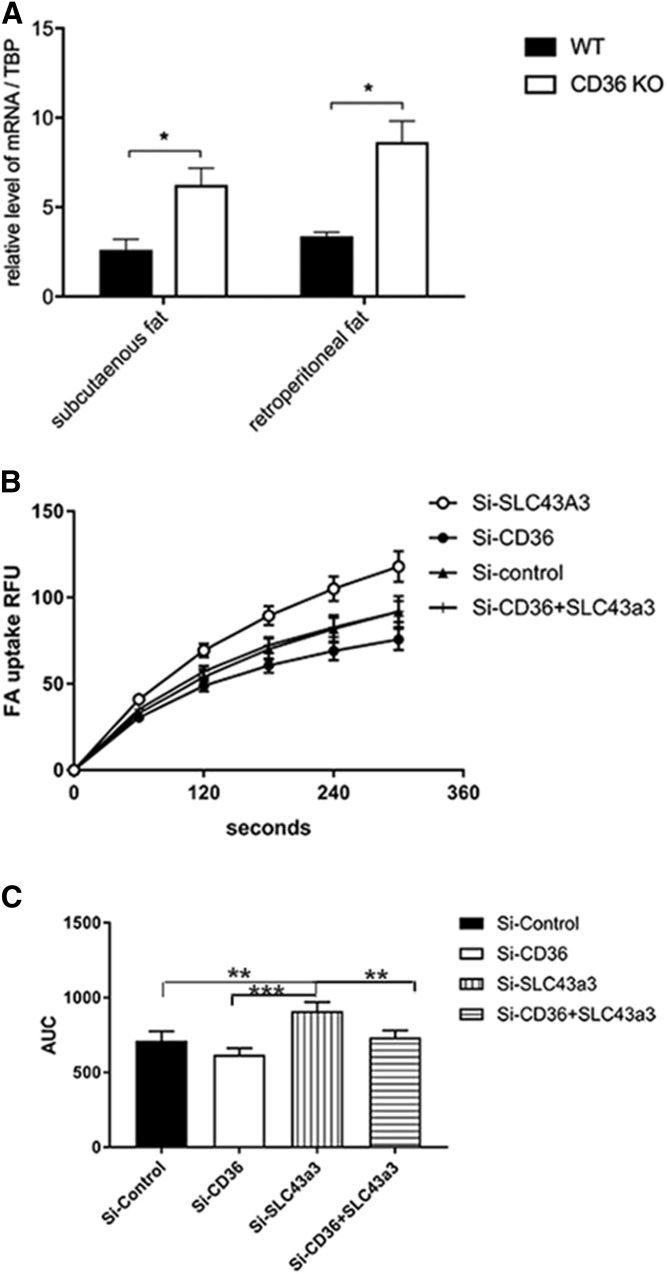
CD36 is known to be important in FA transport and regulation, such that there is decreased FA uptake in adipose tissue of CD36 KO mice. We, therefore, examined expression levels of Slc43a3 in adipose tissues of CD36 KO mice. As shown in Fig. 6A, there is a significant upregulation of expression of Slc43a3 (P < 0.05) compared with control (wild-type mice) in both the subcutaneous and retroperitoneal adipose tissue of CD36 KO mice. To further explore this possible relationship, we manipulated the expression of both Slc43a3 and CD36 in OP9 cells. OP9 cells were transfected with Slc43a3 siRNA, CD36 siRNA, both Slc43a3 and CD36 siRNA at the same time, or with control scrambled siRNA. Data presented in Fig. 6B show that knockdown of CD36 decreases the rate of FA uptake as compared with control, consistent with prior observations (27). Compatible with prior data (Fig. 2), knockdown of Slc43a3 again increased the rate of FA uptake compared with control. Interestingly, when both CD36 and Slc43a3 were knocked down, the rate of FA uptake was similar to control, suggesting that the effects of Slc43a3 are mediated independently from CD36.
Fig. 6.
Levels of Slc43a3 in adipose tissue of CD36 KO mice. A: Levels of Slc43a3 RNA in adipose tissue of wild-type and CD36 KO mice. Total RNA was prepared from subcutaneous and retroperitoneal adipose tissues from wild-type and CD36 KO mice. Levels of Slc43a3 were assayed using TaqMan RT-PCR as described in the Materials and Methods. Results are the summary of three independent assays with n = 3–4 in each group. B: Effects of Slc43a3, CD36, and double knockdown in OP9 cells on free FA uptake compared with control (scrambled siRNA). C: AUC of FA uptake for the first 6 min. Results are representative of three independent experiments, each with n = 8. *P < 0.05, **P < 0.01, ***P < 0.001.
Effects of Slc43a3 on cAMP and ATP levels
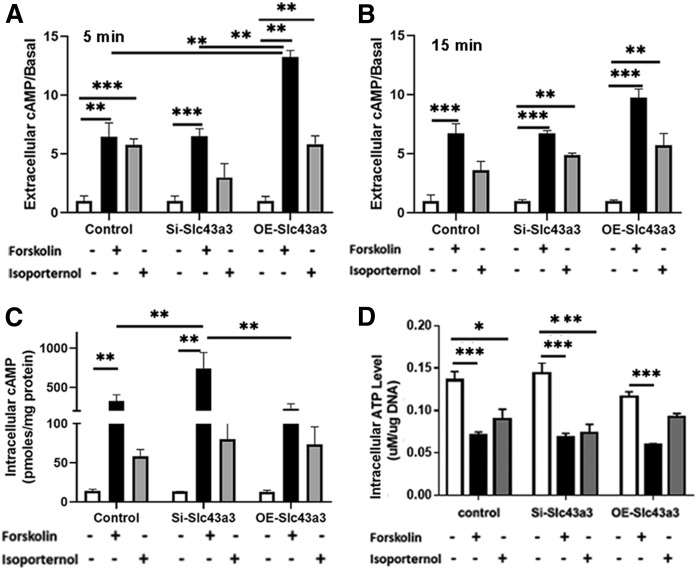
In exploring potential mechanisms through which Slc43a3 might influence cellular homeostasis, we examined changes in cAMP levels with either knockdown or with overexpression of Slc43a3 because Slc43a3 has been reported to be involved in the cellular uptake of extracellular purine nucleobases (28). In general, extracellular levels of cAMP are ∼10-fold higher than intracellular levels. As the basal level of extracellular cAMP varied among experiments from 86.3 pmoles/ml to 388 pmoles/ml, the changes in extracellular cAMP are presented as fold changes normalized to the basal level in each experiment (Fig. 7). Extracellular cAMP levels were significantly higher in cells overexpressing Slc43a3 than in control cells (P < 0.01) or in cells with knockdown of Slc43a3 (P < 0.01) following treatment with forskolin and isobutyl-methylxanthine for 5 min (Fig. 7A); however, there were no statistical differences among the cells at 15 min (Fig. 7B). Isoproterenol stimulated extracellular cAMP to similar levels in control, knockdown, and overexpressing cells at both 5 and 15 min. Intracellular cAMP levels were significantly increased by forskolin and isobutyl-methylxanthine in Slc43a3 knockdown cells compared with control (P < 0.01) and Slc43a3 overexpressing cells (P < 0.01) (Fig. 7C). Therefore, none of the small differences in cAMP appears to provide a mechanistic basis for alterations in FA flux associated with manipulation of Slc43a3 expression. Cellular ATP levels were similar in control, Slc43a3 knockdown, and Slc43a3 overexpressed cells (Fig. 7C). Forskolin and isoproterenol decreased ATP levels similarly in all cells.
Fig. 7.
Effects of Slc43a3 on cAMP and ATP levels. cAMP and ATP levels were measured by ELISA. Changes of extracellular (A) and intracellular (B) cAMP levels, and cellular ATP levels (C) after transfection with Slc43a3 siRNA and pEZ-M51-Slc43a3-His clone in OP9 cells and treatment with forskolin with isobutyl-methylxanthine, and isoproterenol for 5 and 15 min. Intracellular values were obtained at 15 min. Results are representative of three independent experiments, each with n = 3–6. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
As a cell with endocrine function, adipocytes actively take up and release FAs under different physiological conditions, contribute to the regulation of lipid homeostasis, and, in turn, participate in whole-body metabolism. Dysregulation of FA metabolism is a major factor contributing to the development of disorders such as diabetes, cardiovascular disease, and nonalcoholic fatty liver disease (29–31).
Bariatric surgery has been shown to be the most effective treatment of obesity and obesity related type 2 diabetes (32–34). One of the interesting observations is that by 1 week postsurgery, most patients have improved insulin sensitivity without substantial weight loss (23). Studies have shown that in response to the major changes of bariatric surgery, several main regulators of adipose tissue metabolism, such as PPARγ, PPARδ, and UCP2, have altered expression during the first week postsurgery, attenuating lipid storage and promoting fatty oxidation (19). Indeed, FA efflux would be expected to be markedly increased at this time because caloric intake is substantially reduced.
In view of these dramatic metabolic changes, our search for potential regulators of FA transport in adipocytes showed that there are many alterations in adipocyte plasma membrane transporters, which currently have unknown functions, during this first week of metabolic transition. One of the transporters that showed increased expression after bariatric surgery was Slc43a3. The SLC43 family of transporters is composed of only three members: two amino acid system L transporters (LAT3 and LAT4) and the orphan transporter embryonic epithelia gene 1 (EEG1) (35–37). Human LAT3 and LAT4 share approximately 57% amino acid sequence identity, whereas human EEG1 is a distant member of the family with only approximately 27% amino acid sequence identity with the other two members. Mouse Eeg1 was identified as a gene expressed in a cellular model of renal tubulogenesis (38), and the human gene has been assigned as SLC43A3 (39). However, its specific function in adipose tissue and its involvement in lipid metabolism had not yet been investigated prior to the current studies. Slc43a3 was shown to be expressed during embryogenesis in liver and lung and has recently been reported to be involved in the cellular uptake of extracellular purine nucleobases (28).
Our analysis of mRNA expression in normal adult control mice showed that the expression of Slc43a3 in adipose tissue is similar to that in the liver and that it is expressed predominantly in adipocytes as opposed to stromal vascular cells within adipose depots. In vitro study in the murine OP9 adipocyte cell line and in primary mouse preadipocytes showed that Slc43a3 expression is induced during the differentiation of adipocytes. Interestingly, siRNA-mediated knockdown of Slc43a3 resulted in increased rates of FA uptake and increased lipid accumulation in cells. Surprisingly, knockdown of Slc43a3 significantly decreased the rates of FA efflux compared with control cells under basal conditions and substantially blunted FA efflux when cells were treated with forskolin or isoproterenol to stimulate lipolysis and FA efflux. Importantly, the efflux of glycerol upon lipolytic stimulation by forskolin or isoproterenol was not suppressed by knockdown of Slc43a3, thus the actions of Slc43a3 were specific for FA efflux and were observed whether C12 or C16 FAs were examined. This dissociation between FA and glycerol efflux is similar to prior observations where a chemical inhibitor was identified that blocked FA, but not glycerol, efflux (10), suggesting that Slc43a3 might have been the target of the chemical inhibitor. Overexpression of Slc43a3 in OP9 cells resulted in decreased rates of FA uptake and lipid accumulation in the cells along with blunted FA and glycerol efflux; however, Slc43a3 overexpression also resulted in reduced expression of PPARγ, Plin1, FABP4, and HSL, suggesting that its overexpression might have interfered with normal adipocyte differentiation. Thus, taken together, Slc43a3 appears to influence FA flux in adipocytes, functioning as a positive regulator of FA efflux and as a negative regulator of FA uptake.
Studies to attempt to explore potential mechanisms whereby Slc43a3 might be altering rates of FA flux showed that knockdown of Slc43a3 resulted in a small increase in forskolin-stimulated intracellular cAMP generation, but no changes in total or extracellular cAMP, changes which were unlikely to explain the blunted FA efflux, particularly in view of the robust glycerol efflux observed with either forskolin or isoproterenol. Overexpression of Slc43a3 resulted in an increase in forskolin-stimulated extracellular cAMP generation, but no changes in intracellular cAMP, again changes which were unlikely to explain the alterations in FA flux. Moreover, neither the knockdown nor overexpression of Slc43a3 had any significant effects on cellular ATP levels. In addition, knockdown of Slc43a3 did not significantly reduce the expression of a number of genes, such as SR-B1, CD36, HSL, ATGL, CGI58, G0S2, and FATP1, known to be involved in cellular FA and lipid metabolism. Furthermore, we showed that the actions of Slc43a3 on FA uptake are independent of CD36 expression. The mechanism(s) through which Slc43a3 mediates these effects on FA flux remains to be definitively elucidated and is the focus of ongoing studies. On the one hand, a primary function of Slc43a3 might be to directly facilitate FA efflux. Indeed, measurement of intracellular free FA concentrations showed that lipolytic stimulation of Slc43a3 knockdown cells resulted in a significant increase in intracellular nonprotein bound free FAs similar to control cells; however, in contrast to control cells, there was a significant increase in protein bound free FAs. It is noteworthy that adipocytes express very high levels of FABPs, such as FABP4 and FABP5 (26), which are capable of binding and buffering the vast majority of intracellular FAs. This finding of elevated intracellular free FAs following lipolytic stimulation can be considered to be consistent with a direct inhibition of FA efflux in the absence of Slc43a3 and with the resultant buffering of the excess free FAs by FABPs. Interestingly, the increase in intracellular free FAs was not associated with any significant changes in FAO. On the other hand, it is possible that Slc43a3 is not directly involved in FA flux, but somehow either influences the function of other proteins involved in facilitating FA flux or affects the diffusion or flip-flop of FAs across the plasma membrane. In regard to this latter possibility, we can speculate that Slc23a3 might be involved in maintaining the electroneutrality of the cell during FA flux, functioning either as a symporter or antiporter of protons, which would be required to offset the intracellular changes associated with the flux of FA anions. Alternatively, Slc43a3 might directly or indirectly inhibit (re)esterification, thus leading to increased FA trapping (esterification) in its absence. This latter possibility is consistent with the observation of the increase in expression of genes involved in (re)esterification and with the increase in ACSL activity when Slc43a3 was knocked down. However, experiments utilizing triacsin to inhibit ACSL activity, and thus (re)esterification, had no effects on the blockade of lipolytically stimulated FA efflux in the setting of Slc43a3 knockdown, suggesting that (re)esterification does not contribute to the effects of Slc43a3 on FA efflux. Nonetheless, triacsin did decrease some of the increased FA uptake observed with Slc43a3 knockdown, consistent with at least a portion of the elevated FA influx in the setting of Slc43a3 knockdown being related to cellular responses that increase esterification. Whatever its precise mechanism of action, we conclude that Slc43a3 regulates FA flux in adipocytes, functioning as a positive regulator of FA efflux and as a negative regulator of FA uptake. Moreover, the demonstration that manipulating a plasma membrane protein can dramatically affect the ability of a cell to export FA provides a new direction for gaining a deeper understanding of the processes mediating FA flux in mammalian cells.
Acknowledgments
The authors thank Jonathan Long and Stephanie Terrell for measurement of cellular fatty acyl CoAs.
Footnotes
Abbreviations:
- ACSL
- long chain fatty acyl-CoA synthetase
- bDNA
- branched DNA
- EEG1
- embryonic epithelia gene 1
- FABP
- FA binding protein
- FAO
- FA oxidation
- FATP
- FA transport protein
- LAT
- amino acid system L transporter
- LCFA
- long chain FA
- OA
- oleic acid
- SLC
- solute carrier
- VSG
- vertical sleeve gastrectomy
This work was supported by U.S. Department of Veterans Affairs (Biomedical Laboratory Research Development Program) Merit Review Awards I01BX001923 (S.A.) and I01BX000398 (F.B.K.) and Senior Research Career Scientist Award IK6B004200 (S.A.) and National Institutes of Health Grants P30DK116074 (F.B.K) and DK053189 (D.A.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Berg J. M., Tymoczko J. L., and Stryer L.. 2002. Biochemistry. 5th edition. W. H. Freeman, New York. [Google Scholar]
- 2.Glatz J. F. 2011. Challenges in fatty acid and lipid physiology. Front. Physiol. 2: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das U. N. 2006. Essential fatty acids: biochemistry, physiology and pathology. Biotechnol. J. 1: 420–439. [DOI] [PubMed] [Google Scholar]
- 4.Xu S., Jay A., Brunaldi K., Huang N., and Hamilton J. A.. 2013. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry. 52: 7254–7261. [DOI] [PubMed] [Google Scholar]
- 5.Knittle J. L., and Hirsch J.. 1965. Effect of chain length on rates of uptake of free fatty acids during in vitro incubations of rat adipose tissue. J. Lipid Res. 6: 565–571. [PubMed] [Google Scholar]
- 6.Berk P. D., Zhou S. L., Kiang C. L., Stump D., Bradbury M., and Isola L. M.. 1997. Uptake of long chain free fatty acids is selectively up-regulated in adipocytes of Zucker rats with genetic obesity and non-insulin-dependent diabetes mellitus. J. Biol. Chem. 272: 8830–8835. [DOI] [PubMed] [Google Scholar]
- 7.Bonen A., Benton C. R., Campbell S. E., Chabowski A., Clarke D. C., Han X. X., Glatz J. F., and Luiken J. J.. 2003. Plasmalemmal fatty acid transport is regulated in heart and skeletal muscle by contraction, insulin and leptin, and in obesity and diabetes. Acta Physiol. Scand. 178: 347–356. [DOI] [PubMed] [Google Scholar]
- 8.Stahl A. 2004. A current review of fatty acid transport proteins (SLC27). Pflugers Arch. 447: 722–727. [DOI] [PubMed] [Google Scholar]
- 9.Glatz J. F., and Luiken J. J.. 2017. From fat to FAT (CD36/SR-B2): understanding the regulation of cellular fatty acid uptake. Biochimie. 136: 21–26. [DOI] [PubMed] [Google Scholar]
- 10.Henkin A. H., Ortegon A. M., Cho S., Shen W. J., Falcon A., Kraemer F. B., Lee S. J., and Stahl A.. 2012. Evidence for protein-mediated fatty acid efflux by adipocytes. Acta Physiol. (Oxf.). 204: 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarling E. J., de Aguiar Vallim T. Q., and Edwards P. A.. 2013. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean M., Hamon Y., and Chimini G.. 2001. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 42: 1007–1017. [PubMed] [Google Scholar]
- 13.Rees D. C., Johnson E., and Lewinson O.. 2009. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10: 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colas C., Ung P. M., and Schlessinger A.. 2016. SLC transporters: structure, function, and drug discovery. MedChemComm. 7: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin L., Yee S. W., Kim R. B., and Giacomini K. M.. 2015. SLC transporters as therapeutic targets: emerging opportunities. Nat. Rev. Drug Discov. 14: 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hediger M. A., Clemencon B., Burrier R. E., and Bruford E. A.. 2013. The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol. Aspects Med. 34: 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.César-Razquin A., Snijder B., Frappier-Brinton T., Isserlin R., Gyimesi G., Bai X., Reithmeier R. A., Hepworth D., Hediger M. A., Edwards A. M., et al. 2015. A call for systematic research on solute carriers. Cell. 162: 478–487. [DOI] [PubMed] [Google Scholar]
- 18.Jahansouz C., Xu H., Hertzel A. V., Serrot F. J., Kvalheim N., Cole A., Abraham A., Luthra G., Ewing K., Leslie D. B., et al. 2016. Bile acids increase independently from hypocaloric restriction after bariatric surgery. Ann. Surg. 264: 1022–1028. [DOI] [PubMed] [Google Scholar]
- 19.Jahansouz C., Xu H., Hertzel A. V., Kizy S., Steen K. A., Foncea R., Serrot F. J., Kvalheim N., Luthra G., Ewing K., et al. 2018. Partitioning of adipose lipid metabolism by altered expression and function of PPAR isoforms after bariatric surgery. Int. J. Obes. (Lond.). 42: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H., Li Y., Hu J., Shen W. J., Singh M., Hou X., Bittner A., Bittner S., Cortez Y., Tabassum J., et al. 2015. Effect of creosote bush-derived NDGA on expression of genes involved in lipid metabolism in liver of high-fructose fed rats: relevance to NDGA amelioration of hypertriglyceridemia and hepatic steatosis. PLoS One. 10: e0138203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong B., Kan C. F., Singh A. B., and Liu J.. 2013. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J. Lipid Res. 54: 1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long J. Z., Li W., Booker L., Burston J. J., Kinsey S. G., Schlosburg J. E., Pavón F. J., Serrano A. M., Selley D. E., Parsons L. H., et al. 2009. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 5: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gastaldelli A., Iaconelli A., Gaggini M., Magnone M. C., Veneziani A., Rubino F., and Mingrone G.. 2016. Short-term effects of laparoscopic adjustable gastric banding versus Roux-en-Y gastric bypass. Diabetes Care. 39: 1925–1931. [DOI] [PubMed] [Google Scholar]
- 24.Coleman R. A. 2019. It takes a village: channeling fatty acid metabolism and triacylglycerol formation via protein interactomes. J. Lipid Res. 60: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kampf J. P., and Kleinfeld A. M.. 2004. Fatty acid transport in adipocytes monitored by imaging intracellular free fatty acid levels. J. Biol. Chem. 279: 35775–35780. [DOI] [PubMed] [Google Scholar]
- 26.Bernlohr D. A., Simpson M. A., Vogel Hertzel A., and Banaszak L. J.. 1997. Intracellular lipid-binding proteins and their genes. Annu. Rev. Nutr. 17: 277–303. [DOI] [PubMed] [Google Scholar]
- 27.Coburn C. T., Knapp F. F., Febbraio M., Beets A. L., Silverstein R. L., and Abumrad N. A.. 2000. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275: 32523–32529. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa J., Inoue K., Maeda J., Yasujima T., Ohta K., Kanai Y., Takada T., Matsuo H., and Yuasa H.. 2015. Functional identification of SLC43A3 as an equilibrative nucleobase transporter involved in purine salvage in mammals. Sci. Rep. 5: 15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazar M. A. 2005. How obesity causes diabetes: not a tall tale. Science. 307: 373–375. [DOI] [PubMed] [Google Scholar]
- 30.Frayn K. N. 2005. Obesity and metabolic disease: is adipose tissue the culprit? Proc. Nutr. Soc. 64: 7–13. [DOI] [PubMed] [Google Scholar]
- 31.Reddy J. K., and Rao M. S.. 2006. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am. J. Physiol. Gastrointest. Liver Physiol. 290: G852–G858. [DOI] [PubMed] [Google Scholar]
- 32.Ikramuddin S., Korner J., Lee W. J., Connett J. E., Inabnet W. B., Billington C. J., Thomas A. J., Leslie D. B., Chong K., Jeffery R. W., et al. 2013. Roux-en-Y gastric bypass vs intensive medical management for the control of type 2 diabetes, hypertension, and hyperlipidemia: the Diabetes Surgery Study randomized clinical trial. JAMA. 309: 2240–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chondronikola M., Harris L. L., and Klein S.. 2016. Bariatric surgery and type 2 diabetes: are there weight loss-independent therapeutic effects of upper gastrointestinal bypass? J. Intern. Med. 280: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Corsino L., Shantavasinkul P. C., Grant J., Portenier D., Ding L., and Torquati A.. 2016. Gastric bypass surgery leads to long-term remission or improvement of type 2 diabetes and significant decrease of microvascular and macrovascular complications. Ann. Surg. 263: 1138–1142. [DOI] [PubMed] [Google Scholar]
- 35.Bodoy S., Martin L., Zorzano A., Palacin M., Estevez R., and Bertran J.. 2005. Identification of LAT4, a novel amino acid transporter with system L activity. J. Biol. Chem. 280: 12002–12011. [DOI] [PubMed] [Google Scholar]
- 36.Babu E., Kanai Y., Chairoungdua A., Kim D. K., Iribe Y., Tangtrongsup S., Jutabha P., Li Y., Ahmed N., Sakamoto S., et al. 2003. Identification of a novel system L amino acid transporter structurally distinct from heterodimeric amino acid transporters. J. Biol. Chem. 278: 43838–43845. [DOI] [PubMed] [Google Scholar]
- 37.Cole K. A., Chuaqui R. F., Katz K., Pack S., Zhuang Z., Cole C. E., Lyne J. C., Linehan W. M., Liotta L. A., and Emmert-Buck M. R.. 1998. cDNA sequencing and analysis of POV1 (PB39): a novel gene up-regulated in prostate cancer. Genomics. 51: 282–287. [DOI] [PubMed] [Google Scholar]
- 38.Stuart R. O., Pavlova A., Beier D., Li Z., Krijanovski Y., and Nigam S. K.. 2001. EEG1, a putative transporter expressed during epithelial organogenesis: comparison with embryonic transporter expression during nephrogenesis. Am. J. Physiol. Renal Physiol. 281: F1148–F1156. [DOI] [PubMed] [Google Scholar]
- 39.Bodoy S., Fotiadis D., Stoeger C., Kanai Y., and Palacin M.. 2013. The small SLC43 family: facilitator system l amino acid transporters and the orphan EEG1. Mol. Aspects Med. 34: 638–645. [DOI] [PubMed] [Google Scholar]