Abstract
Acne is one of the most common dermatological conditions, but the details of its pathology are unclear, and current management regimens often have adverse effects. Cutibacterium acnes is known as a major acne-associated bacterium that derives energy from lipase-mediated sebum lipid degradation. C. acnes is commensal, but lipase activity has been observed to differ among C. acnes types. For example, higher populations of the type IA strains are present in acne lesions with higher lipase activity. In the present study, we examined a conserved lipase in types IB and II that was truncated in type IA C. acnes strains. Closed, blocked, and open structures of C. acnes ATCC11828 lipases were elucidated by X-ray crystallography at 1.6–2.4 Å. The closed crystal structure, which is the most common form in aqueous solution, revealed that a hydrophobic lid domain shields the active site. By comparing closed, blocked, and open structures, we found that the lid domain-opening mechanisms of C. acnes lipases (CAlipases) involve the lid-opening residues, Phe-179 and Phe-211. To the best of our knowledge, this is the first structure-function study of CAlipases, which may help to shed light on the mechanisms involved in acne development and may aid in future drug design.
Keywords: phospholipases, lysophospholipid, skin lipid metabolism, protein structure, acne, lysophosphatidylcholine, lysophospholipase, X-ray crystallography
Acne is a common skin condition that is widespread across all cultures and ages. The prevalence rates are up to 85% in adolescents and 40% in adults (1–3). Acne leaves scarring, symptomatic pain, and emotional and psychological disturbances. While neither life threatening nor physically debilitating, acne can cause social and psychological discomfort. Even mild facial acne is more commonly associated with significant depression compared with other skin conditions, such as alopecia or atopic dermatitis (4). Current studies focus on the Cutibacterium acnes colonization of sebaceous follicles as a major cause of acne (5). The pathogenicity is thought to be due to substances generated by C. acnes, such as free fatty acids, extracellular enzymes, or virulence factors (6, 7). However, it is one of the most common bacteria on human skin that metabolizes sebum and releases fatty acids by secreting lipases in healthy skin. In this process, skin acidity is maintained and this functions as a natural barrier from harmful pathogens, providing innate skin immunity (8). Recently, studies have shown that the proliferation of C. acnes is not the trigger of acne but disturbances to a tight equilibrium between phylotypes might perform a key role (5, 9).
To date, three phylogenetic groups of C. acnes have been described, types I, II, and III, according to genome sequences or biological characteristics (lipase activity). Type I is further divided into IA1, IA2, IB, and IC (10–12). Recent evidence suggests that type IA is strongly associated with acne while IB, II, and III are less involved (11–13). Type IA was predominant in severe, moderate, and mild acne, but type II was preferentially present in healthy skin (14, 15). The notable characteristics of type IA strains are increased lipase activity and secretion. In contrast, the distinctive features of type II strains are decreased secretion of virulence factors, including lipase (16, 17). To understand different roles depending on C. acnes types, genetic studies based on sequence data are being conducted (18, 19).
Because lipolytic activity is an essential metabolic pathway for C. acnes, we focused on lipase from the type II C. acnes ATCC11828 (CAlipase hereafter). This gene is absolutely conserved across type II strains, while it shows 85–95% sequence homology with type IB, IC, or III strains. The known pathogenic type IA strains possess the corresponding gene but a 13 nucleotide deletion was detected. The deletion causes a premature stop codon, resulting in truncated fragments of CAlipase in type IA strains. Consequently, the catalytic triad is dissociated into two different fragments. This implies that CAlipase is originally conserved across all types of C. acnes but selectively mutated in genetic levels during evolution into new strains. In this regard, we could expect that conserved CAlipase performs well-controlled lipolysis in healthy skin, but the mutation causes deficiency of controlled lipase in acne lesions. By revealing how the conserved CAlipase regulates their activities, we could surmise different behavior of type I C. acnes as an opportunistic pathogen. In the present study, the crystal structures of closed, blocked, and open states of CAlipase were identified. We elucidated the protein shields and revealed an active site with bulky side chains. Our study will promote a better understanding of the CAlipase mechanism on a molecular basis.
MATERIALS AND METHODS
Cloning, expression, and purification
The predicted ORF of CAlipase was amplified from C. acnes ATCC 11828 genomic DNA by PCR. The NdeI and XhoI restriction sites were used for cloning into the pET-21a(+) vector (Novagen). The resulting construct has eight additional residues (LEHHHHHH) that encode a C-terminal hexa-histidine tag. The sequences of the cloned genes were confirmed by DNA sequencing (results not shown). To prepare mutants, the QuickChange site-directed mutagenesis kit (Agilent) was used to generate point mutations in the recombinant pET-21a(+) plasmid. The point mutations resulted in multiple recombinant plasmids, specifically F176F179W192F211A (active site shielding residues mutant), F179W192A (lipophilic path forming residues mutant), and E5D54D202K205D206A (dimer mutant). Each recombinant plasmid was transformed into Escherichia coli BL21 (DE3). Cells were grown at 37°C in LB medium supplemented with ampicillin (50 μg/ml). For the selenoproteins, cells were grown in minimal medium supplemented with selenomethionine. Recombinant protein expression was induced by the addition of IPTG to 0.5 mM when the OD600 reached 0.5. After an additional 4 h of growth, cells were harvested by centrifugation at 4,500 g at 4°C. Cell pellet was resuspended in 50 mM Tris (pH 7.5), 500 mM NaCl, and 20 mM imidazole buffer and disrupted using an ultrasonic processor (Cole-Parmer). The cell lysate was centrifuged at 20,000 g for 1 h at 4°C. The cleared supernatant was purified by binding to a Ni-NTA (Ni2+-nitrilotriacetate) affinity column (Qiagen, Germany; 3ml of resin per liter of cell culture) previously equilibrated with the same buffer. The Ni2+ bound protein was eluted with elution buffer [50 mM Tris (pH 7.5), 500 mM NaCl, and 200 mM imidazole] until there was no detectable absorbance at 280 nm. Further purification and buffer exchange were achieved by size-exclusion chromatography using a Superdex 75 (10/300 GL) column (GE Healthcare Life Sciences) that was previously equilibrated with 50 mM Tris (pH 7.5) and 150 mM NaCl. The purity of each recombinant protein was estimated to be over 95% by SDS-PAGE. The purified protein was concentrated to 10 mg/ml by ultrafiltration in 10,000 Da molecular mass cut-off spin columns (Millipore).
Crystallization, data collection, and structure determination
Crystallization was performed at 20°C using 24-well VDX plates (Hampton Research). Initial crystallization conditions were established using screening kits from Hampton Research (Crystal Screen I and II, Index, PEG/Ion, and MembFac) and from Emerald Biosystems (Wizard I, II, III, and IV). For the optimal growth of CAlipase crystals, each hanging drop was prepared on a siliconized cover slip by mixing 1 μl of precipitant solution [30% (w/v) PEG 8K and 0.1 M Tris (pH 8.0)] and 1 μl of protein solution (10 mg/ml). This drop was equilibrated against a 1 ml reservoir of precipitant solution. This condition yielded rod-shaped crystals in 3 days. Selenoprotein was crystallized in the same procedure. The cocrystal with LPC was prepared by addition of 20 mM LPC to the drop. The best crystals were grown from 2 M (NH4)2SO4, 0.2 M MgCl2, and 0.1 M Bis-Tris (pH 6.2) as a diamond shape.
CAlipase crystals were transferred to a cryoprotectant solution with 30% (v/v) glycerol in the crystallization condition for several minutes before being flash-frozen in a stream of nitrogen gas at 100 K. For the comparison, the same crystal was transferred to a different cryoprotectant solution, Al’s Oil (Hampton Research). CAlipase crystals and CAlipase:LPC cocrystals were transferred to Al’s Oil before freezing for several minutes. Diffraction data were collected on beamlines 5C and 7A at the Pohang Light Source, Korea. The raw data were processed and scaled using the HKL2000 program suite (20). Further data analysis was carried out using the CCP4 suite (21). The CAlipase crystals belong to space group P21212 and contained two molecules per asymmetric unit. The CAlipase:LPC cocrystals were determined as P31 with 26 molecules per asymmetric unit.
To determine the CAlipase structure, the MAD dataset was used for solving the structure using the Autosol and Autobuild Wizards in the Phenix package (22). The selenium sites were identified, and the initial phases calculated from these sites were further improved by density modification. The structures for Al’s Oil-soaked and CAlipase:LPC cocrystals were determined using molecular replacement with the program MolRep within the CCP4 suite (23). Iterative cycles of model building were performed using COOT followed by refinement with Refamc5 (24). A portion of data (5%) was set aside for the refinement calculations of Rfree data collection, and the final crystallographic statistics are summarized in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
| Closed CAlipase (Wild-Type) | Blocked CAlipase (BMK-Bound) | Open CAlipase (LPC-Bound) | |
| Data collection | |||
| Beamline | PAL-5C | PAL-7A | PAL-7A |
| Wavelength (Å) | 0.98 | 0.98 | 0.98 |
| Resolution range (Å)a | 38.36–1.75 | 31.89–1.60 | 35.88–2.40 |
| Space group | P21212 | P21212 | P31 |
| Unit cell parameters (Å) | a = 55.3 | a = 94.5 | a = 185.9 |
| b = 93.3 | b = 129.7 | b = 185.9 | |
| c = 129.7 | c = 55.5 | c = 205.1 | |
| Observations (total/unique) | 68,476/63,249 | 91,178/89,860 | 311,475/68,958 |
| Completeness (%) | 92.8 (87.6) | 98.7 (84.0) | 97.6 (83.3) |
| Rsymb | 7.7 (29.5) | 6.1 (23.3) | 10.7 (34.1) |
| CC1/2 | 0.95 (0.82) | 0.99 (0.96) | 0.99 (0.83) |
| Redundancy | 3.7 (3.7) | 12.4 (5.9) | 3.5 (2.5) |
| I/sigma | 21.0 (10.1) | 25.7 (7.8) | 19.0 (5.8) |
| Refinement | |||
| Rworkc/Rfreec (%) | 17.9/22.2 | 14.5/16.7 | 18.5/25.6 |
| Protein/ligand atoms | 4,712/60 | 4,712/114 | 61,283/204 |
| Water molecules | 521 | 705 | 2515 |
| Average B value (Å2) | 18.0 | 14.0 | 32.0 |
| r.m.s.d. bond (Å) | 0.013 | 0.014 | 0.010 |
| r.m.s.d. angle (°) | 1.728 | 1.638 | 1.686 |
| Ramachandran plot (%) | |||
| Favored | 93.97 | 94.95 | 93.05 |
| Allowed | 5.70 | 5.05 | 6.79 |
| Disallowed | 0.33 | 0.00 | 0.16 |
Numbers in parentheses indicate the statistics for the last resolution shell.
Rsym = ∑(|Ihkl−<Ihkl>|/∑<Ihkl>, where Ihkl = single value of measured intensity of hkl reflection, and <Ihkl> = mean of all measured value intensity of hkl reflection.
Rwork = ∑(|Fobs−Fcalc|/∑Fobs, where Fobs = observed structure factor amplitude, and Fcalc = structure factor calculated from model. Rfree is computed in the same manner as Rwork, but from a test set containing 5% of data excluded from the refinement calculation.
PAGE
SDS-PAGE was conducted according to the Laemmli method using a 12% (w/v) polyacrylamide gel (25). The samples were treated with 1% (w/v) SDS and 5% (v/v) 2-mercaptoethanol at 100°C for 5 min before electrophoresis in a vertical Mini Gel system (Bio-Rad Laboratories). The proteins were stained with Coomassie Brilliant Blue R250 (Thermo Scientific). Additionally, for the separation of CAlipase depending on the solvent condition, native PAGE was performed, and analysis was conducted using a 10% (w/v) polyacrylamide gel without either SDS or 2-mercaptoethanol. Native PAGE was performed in a buffer [25 mM Tris (pH 8.3) and 192 mM glycine]. The staining was performed as described above for SDS-PAGE.
Lipase/phospholipase activity assay
The colorimetric assay was performed to detect hydrolysis of the thioester bond (26, 27). To monitor the lipase activity, 3 mM of 2,3-dimercapto-1-propanol tributyrate was used (Sigma). For lysophospholipase activity, the same amount of palmitoyl thio-phosphocholine (Cayman) was used. The released free thiols are subsequently reacted with 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) (Sigma). Ten micrograms of CAlipase, F176F179W192F211A (active site shielding residues mutant), or F179W192A (lipophilic path forming residues mutant) were incubated for 30 min at 37°C with 10 μl of DTNB in a buffer containing 25 mM Tris (pH 7.5), 10 mM CaCl2, and 100 mM KCl. The free thiol groups reacted with DTNB to produce a yellow precipitate and absorbance was measured at 412 nm using TECAN Microplate Reader Spark (TECAN, Switzerland).
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was performed using a MicroCal PEAQ ITC (Malvern Instruments, UK). Three hundred microliters of 40 μM CAlipase in 50 mM Tris (pH 7.5) and 150 mM NaCl were injected into the sample cell at 25°C and buffer only into the control cell. Repeated 0.4–2 μl injections of 400 μM LPC in the same buffer were applied to both cells every 150 s with 500 rpm mixing. Thermograms were obtained and fitted via the nonlinear least squares minimization method to determine the KD and the change in the enthalpy of binding (ΔH). The Gibbs free energy of binding, ΔG, was calculated from Ka values, and the entropic term, TΔS, was derived from the Gibbs-Helmholtz equation using the recorded ΔH values. The data presented are the mean ± SEM of four separate experiments.
Data availability statement
Protein coordinates and structure factors have been deposited in the Research Collaboratory for Structural Informatics Protein Data Bank (RCSB-PDB) under code 6KHK for closed, 6KHL for blocked, and 6KHM for open state structures.
RESULTS
Conserved lipases in different types of C. acnes
C. acnes releases lipases to degrade sebum lipids, providing energy to the bacteria and skin with hydration. While the lipase serves in the nutrition of C. acnes and maintenance of healthy skin, excessive products (fatty acids) are known as major factors of inflammation. In inflammatory lesions, the quantities of C. acnes were similar to those in healthy skin, but preferential proliferation of type IA C. acnes over type II C. acnes is detected. Although there are only limited genetic studies to reveal the phenotypic and functional differences, virulent and lipase genes showed apparent differences between type IA and type II C. acnes. Indeed, the degree of lipase activity was elevated in type IA C. acnes (16, 28, 29). Each strain produces ∼15 lipases, and many of them are well conserved within types. However, these lipases are not conserved across types. The conserved lipases in type IA are not observed in types IB, IC, or II, and type II conserved lipases are not observed in type IA. The examples of conserved lipases in type IA are shown in supplemental Table S1.
Bioinformatics showed that two of conserved hydrolases from type IA C. acnes are relatively short in size. Most lipases consist of an enzymatically active core domain controlled by a lid domain. Therefore, lipases are typically >30 kDa in size, which makes these short hydrolases unique. Inasmuch as genes for two short hydrolases are adjacently positioned in type IA, nucleotides in the region are aligned together with types IB, IC, and II corresponding lipase genes. The alignment shows 97, 96, and 83% homology with types IB, IC, and II, respectively. Notably, the 13 nucleotide deletion in type IA genes is observed and this causes a frameshift, resulting in a premature stop codon (TGA). This mutation yields two short hydrolases, sequence identification numbers WP_002515192.1 and WP_002519208.1 (supplemental Fig. S1A). To gain further insight into truncated in type IA but intact lipase in other types (IB, IC, and II), we investigated the structure and function of type II CAlipase (WP_002514103.1).
CAlipase dimer shows core and lid domain
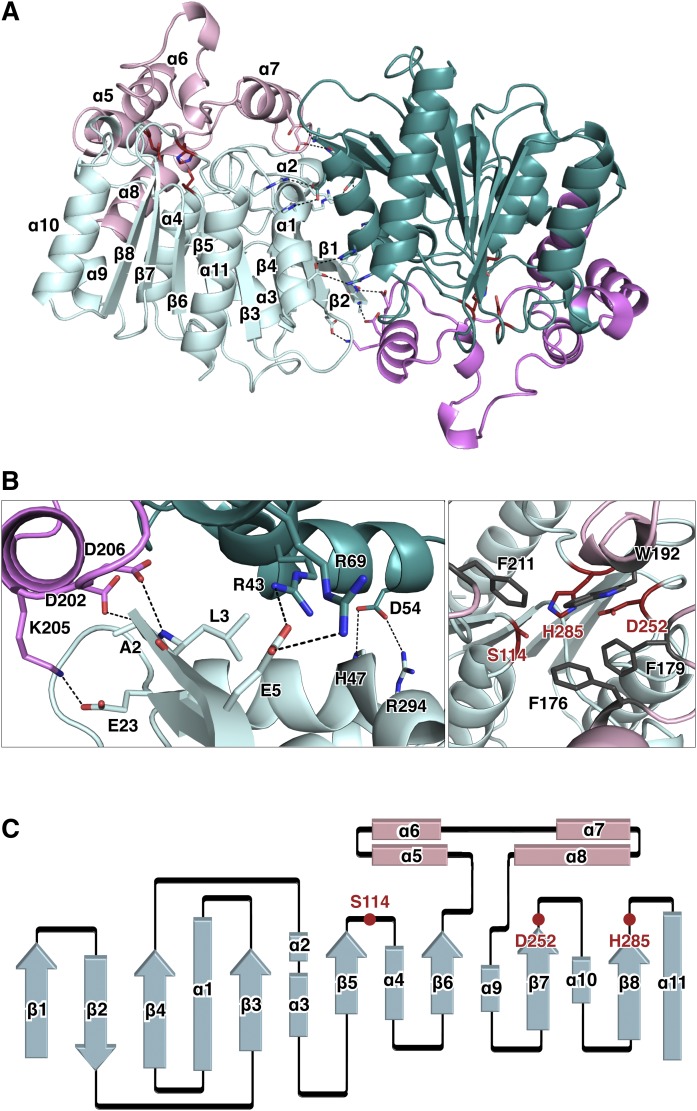
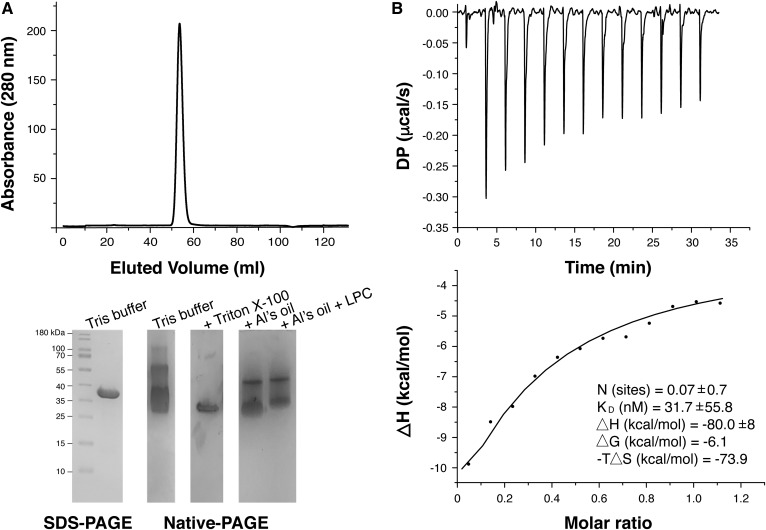
The crystal structure of CAlipase was determined to a 1.75 Å resolution. The asymmetric unit contains two subunits and this dimeric state was supported by size-exclusion chromatography (Fig. 1A). A single sharp peak eluted with an apparent molecular mass of 66 kDa, suggesting the formation of a dimer. Six salt bridges (Ala2-Asp202, Arg69-Glu5, Glu5-Arg43, Glu23-Lys205, His47-Asp54, and Asp54-Arg294) and a hydrogen bond at Leu3-Asp206 form a stable dimerization interface. This dimerization mode is unique, and these residues show poor conservation across similar structures. Most structurally known lipases are monomers and function as such. Moreover, when they form crystallographic dimers, these interfaces are highly different from the CAlipase dimer (Figs. 1B, 2) (30–33). The dimeric state of CAlipase contributes solubility and stability. The dimerization interface mutant, E5D54D202K206D206A, was expressed in inclusion bodies, although the wild-type is highly soluble (supplemental Fig. S2).
Fig. 1.
Crystal structure of CAlipase. A: The CAlipase dimer is shown in ribbon representation. The core domain is colored light cyan and teal while the lid domain is colored pink and magenta. B: Detailed view of the dimerization interface. Hydrogen bonds and salt bridges are indicated (left). The catalytic triad (colored red) is covered by bulky hydrophobic residues (colored gray) (right). C: Secondary structure diagram of the CAlipase monomer. The locations of the catalytic triad are indicated.
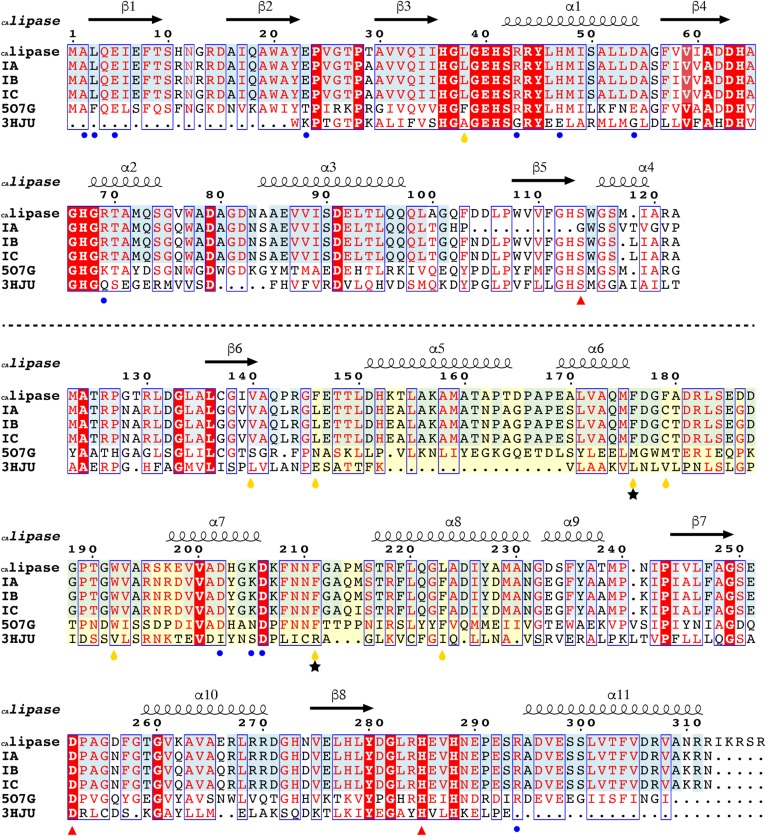
Fig. 2.
Sequence comparison of CAlipase with related proteins. The sequence alignment of CAlipase with type IB/IC lipases, type IA C. acnes short hydrolases (separated by dotted line), and the two highest structural matches from the DALI algorithm. Identical residues are colored white on a red background and similar residues are red on white background. Additionally, identical residues across type IA/IB/IC/II C. acnes have a light blue background. The CAlipase shows 83, 97, 96, 39, and 22% sequence identities with types IA, IB, and IC CAlipase, B. coagulans carboxyl esterase (5O7G), and human monoglyceride lipase (3HJU), respectively. Secondary structure elements (springs, α-helices; arrows, β-strands) are shown above. The residues responsible for dimeric formation are marked as blue circles. Catalytic triad residues are marked as red triangles. Two phenylalanine residues that open up closed structure are indicated as black stars. Hydrophobic residues around fatty acid are shown by yellow drops. The lid domain region including α5, α6, α7, and α8 is indicated with a pale yellow background.
Each monomer consists of two domains: a core domain that possesses a catalytic triad and a lid domain (residues 144-231). The core domain shares the characteristics of a classic α/β hydrolase fold: 1) mostly parallel eight-stranded β-sheet surrounded on both sides by α-helices; 2) a catalytic triad serine (Ser114) is coordinated after the β5-strand, aspartate (Asp252) after the β7-strand, and histidine (His285) after the β8-strand (Fig. 1C) (34). The CAlipase shows typical characteristics in the core domain but the distinctive features are in the lid domain. In our structure, four α-helices between the β6-strand and β7-strand form the lid domain and widely cover one side of the core domain. The lid domain shields the active site by bulky hydrophobic residues, preventing approaches from other substances. Especially (as shown in Fig. 1B), Phe176, Phe179, Trp192, and Phe211 coordinated around the active site pocket with side chains being headed toward pocket. The two short hydrolases in type IA C. acnes, WP_002515192.1 and WP_002519208.1, lack the β5-strand that includes active serine; therefore, hydrolysis is not expected. When the structures are modeled based on CAlipase crystal structure using SWISS-MODEL workspace, both are considered unstable (35) (supplemental Fig. S1B). The hydrophobic residues are exposed to solvent areas. While lid and core domains are tightly interacting in CAlipase, protein WP_002519208.1 loops are exposed freely to solvent areas. Protein WP_002515192.1 shows two folded domains separated by a single loop, which is physically unstable.
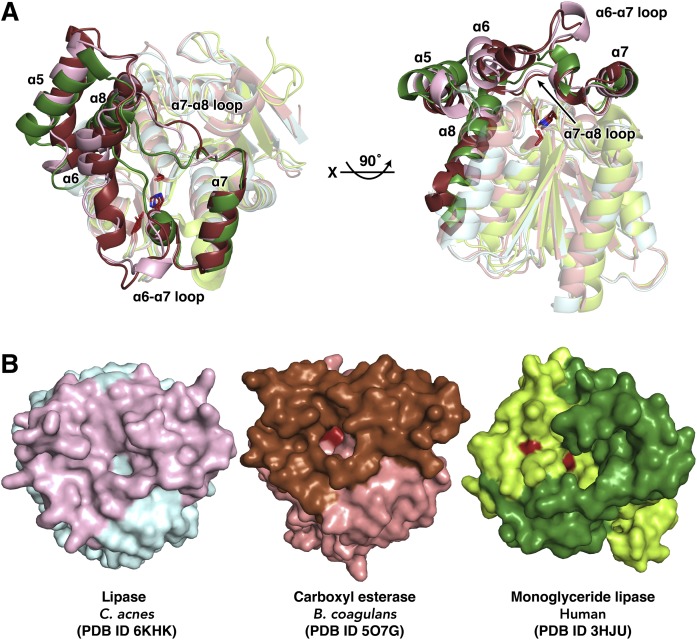
Comparison of CAlipase with related proteins
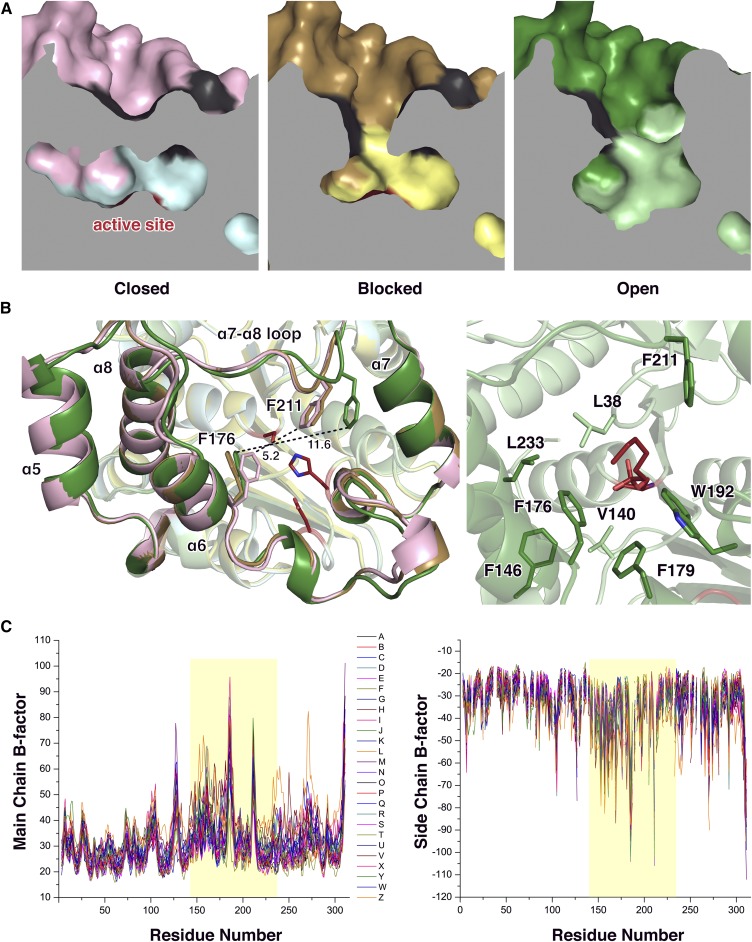
Using the DALI server, the two strongest matches (Z-scores over 30), Bacillus coagulans carboxyl esterase [Protein Data Bank identification number (PDB ID) 5O7G, 1.5 r.m.s.d., 39% sequence identity] and human monoglyceride lipase (PDB ID 3HJU, 2.2 r.m.s.d., 22% sequence identity) were found (30, 33) (Fig. 2). When these structures are aligned, clear differences are apparent in the lid domain. In the crystal structure of human monoglyceride lipase, one α-helix that corresponds to the α6-helix of CAlipase is absent and this causes a different arrangement in the loop between the α6-helix and α7-helix. Comparing CAlipase with the highest Z-scored B. coagulans carboxyl esterase, although both structures show four α-helices in the lid domain, a longer loop between the α6-helix and α7-helix and two 310 helical turns are distinct to CAlipase. Notably, the loop between the α7-helix and α8-helix shows major differences while the α7-helix coordination is well conserved (Fig. 3A). Another notable feature can be found in the surface structure. Although the lid domains cover active sites in B. coagulans carboxyl esterase and human monoglyceride lipase, the nucleophile serine residue is solvent accessible. Contrastingly, the CAlipase lid domain completely shields the catalytic triad (Fig. 3B). The structural differences in the lid domain are important because lipolysis activity is controlled by rearrangement of lid domain structure generally. However, the open-close mechanisms are not predictable due to the poor structural conservation (36). Judging from the crystal structure of CAlipase, the lid domain should be open farther and expose the hidden active site for the lipolysis activity.
Fig. 3.
Structural comparison of CAlipase with related proteins. A: The superposition of CAlipase with carboxyl esterase from B. coagulans (core and lid domains are colored in salmon and brown) and monoglyceride lipase from human (core and lid domains are colored in lime and green). The core domain is colored in light color. The conserved catalytic triads (serine, aspartate, and histidine) are indicated as sticks. B: Surface structures of superposed structures in A. The catalytic triad is colored in red and detected under the lid region, but is completely hidden in CAlipase.
Lipase and lysophospholipase activity
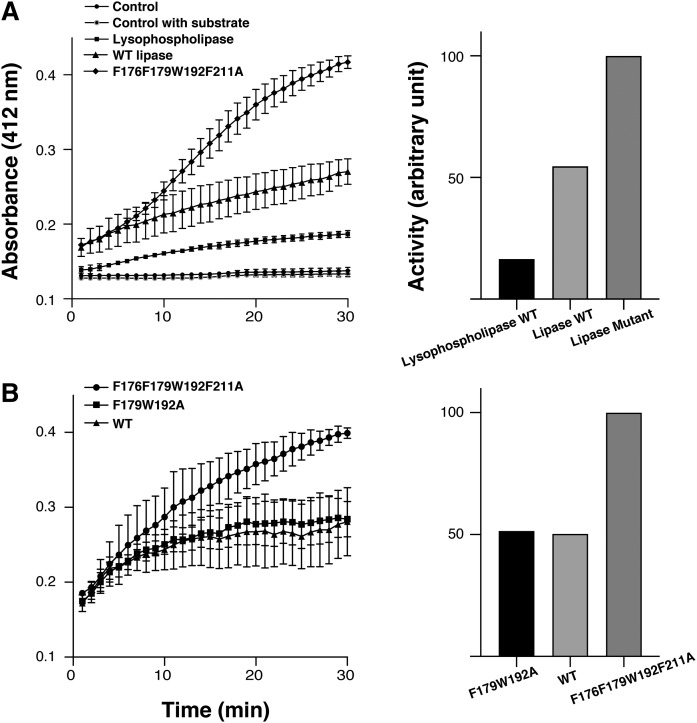
The lipolysis activity was tested using two different sebum lipids, triglyceride and lysophosphatidyl choline. Triglyceride and lysophosphatidyl choline are natural modulators that emulsify and moisturize skin or hair to prevent dryness. However, free fatty acids digested by CAlipases from triglyceride and lysophosphatidyl choline induce marked inflammation (37). In this regard, understanding the regulatory mechanisms underlying activation of CAlipases will provide new strategies for relieving inflammation in acne patients. To understand hydrolysis activities depending on structural characteristics, wild-type, active site shielding residue mutant (F176F179W192F211A), and lipophilic path forming residue mutant (F179W192A) were used. Both substrates were degraded actively by protein, showing higher affinity to triglyceride. Because triglyceride occupies the predominant proportion of sebum, the substrate selectivity toward triglyceride is convincing (38). Interestingly, the active site-shielding residue mutant showed higher activity than the wild-type or lipophilic path-forming residue mutant (F179W192A). This implies that CAlipase activity is closely related to bulky side chain residues, especially Phe176 and Phe211, which shield the active site in the wild-type (Fig. 4).
Fig. 4.
Lipase and lysophospholipase activity. A (Left): Activity of wild-type CAlipase (filled triangles ▲) and lysophospholipase (filled squares ■) was measured by colorimetry. The wild-type CAlipase hydrolyzes both substrates but preferred activity toward lipase substrate is observed. The active site shielding residues mutant F176F179W192F211A (filled diamonds ◆) indicates higher activity. A (Right): Relative initial rate of hydrolysis. Lipolytic activity is normalized to the active site shielding residue mutant. B: To compare the mutants’ activity, the lipase activity of active site shielding residue mutant F176F179W192F211A (filled circles ●) was compared with wild-type CAlipase (filled squares ■) and lipophilic path-forming residue mutant F179W192A (filled triangles ▲). The active site shielding residue mutant showed higher activity. Values are the mean of three separate determinations.
Interaction study with lysophosphatidyl choline by ITC
Lipase and lysophospholipase activities were identified, but our crystal structure showed a completely closed conformation. Lipolysis in lipases requires the catalytic triad to be accessible by solvents, but most lipases possess a lid domain adjacent to an active site. Accordingly, some lid opening mechanisms of lipases are known; these include temperature-dependent activation, oil-water interfacial activation, and polarity-dependent activation (36). These activation mechanisms are markedly different between species, lid conformations, and even specific residues (39). To open the lid domain of CAlipase, we changed buffer conditions by adding surfactant or oil in the presence of LPC and checked changes of the charge states (Fig. 5A). The final concentration of 0.1% Triton X-100 was used as surfactant or Al’s Oil (combination of paraffin and silicon oil) was suspended in buffers. A single intense peak in size-exclusion chromatography indicates homogenous size in aqueous solution, but the native PAGE result shows various surface charges. With the addition of 0.1% Triton X-100, bands are reduced as a single band. When excess oil was mixed with CAlipase protein, multiple bands were condensed. Native PAGE bands are further reduced by the concurrent addition of oil and LPC.
Fig. 5.
Charge states of CAlipase and binding assay. A: The size-exclusion chromatography of CAlipase and corresponding SDS-PAGE and native PAGE. The native PAGE shows broadened bands. Bands were reduced upon addition of Al’s Oil and Triton X-100. Addition of LPC in Al’s Oil induced further band reduction. B: ITC experiment of raw data and binding isotherms fitted to a one site binding model. The dissociation constant, ΔG, ΔH, and -TΔS are indicated in the figure.
To characterize the interaction between LPC and CAlipase, ITC experiments were performed. Under Tris buffer, the result clearly confirmed interaction and binding of LPC and CAlipase. ITC data also indicate a stoichiometry of 0.07, implying that most of the CAlipase remains in the closed state as determined from crystal structure (Fig. 5B). However, ITC data did not indicate any binding under 0.1% Triton X-100 (data not shown). For the Al’s Oil, ITC experiments were not able to be conducted due to the buffer phase separation. From this experiment, the closed form of CAlipase is expected as the major form in aqueous solution, and it is shown that changing the characteristics of the reaction buffer will cause structural differences in CAlipase.
Lid opening mechanism for substrate entry
Because the addition of Al’s Oil and LPC showed condensed bands in native PAGE, we tried cocrystallization of CAlipase with LPC. The cocrystals were soaked in Al’s Oil before freezing. CAlipase:LPC complexes have been found with a different space group (P31) from wild-type crystal structure (P21212). CAlipase:LPC has 13 dimers (26 monomers) in an asymmetric unit. For the comparison, crystals prepared in the absence of LPC were also soaked with Al’s Oil. Two different structures (blocked and open) were identified in addition to the previous closed conformation. In the absence of LPC, benzyl methyl ketone (BMK hereafter), one of the components of Al’ Oil, was covalently bound to Ser114 (blocked structure).
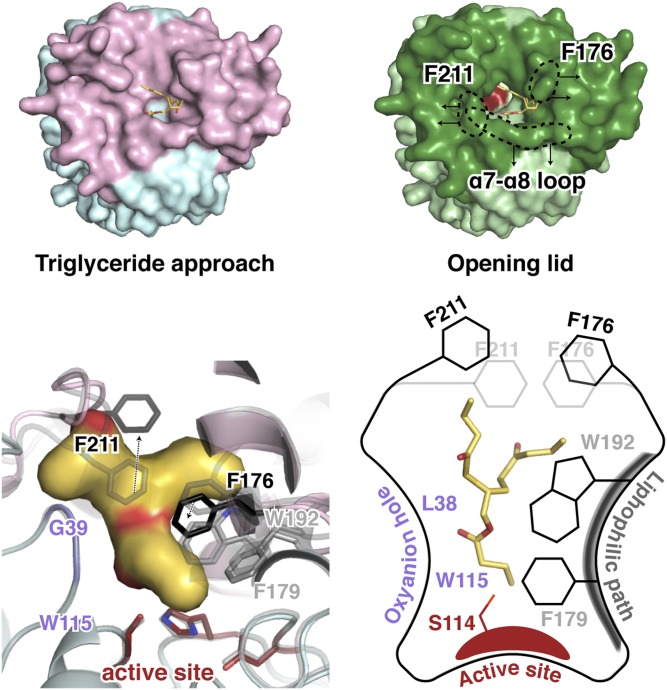
Structural differences of the closed, blocked, and open states of CAlipase are found in the lid domain (Fig. 6A). While the α7-helix (which is responsible for dimerization) is fixed, three other helices are shifted slightly toward the active site pocket. The notable movement is in the loop between the α7- and α8-helix that includes Phe211, which is located right above the active serine. In the closed state structure, Phe211 occludes the active site pocket with Phe176. The distance between Cζ atoms of both phenylalanines is 5.2 Å, not allowing ligands to pass through. In the blocked structure with BMK binding, this obstruction was relieved by a changed coordination of Phe176. Furthermore, after introduction of LPC in the open structure, Phe211 moves with the loops and the distance between the Cζ atoms of Phe211 and Phe176 becomes 11.6 Å, providing enough space for the ligand LPC. After the carbonyl carbon of LPC is attacked, products and fatty acids remain bound to serine. Many hydrophobic residues, such as Leu38, Val140, Phe146, Phe176, Phe179, Trp192, Phe211, and Leu223, form a stable hydrophobic path for fatty acids (Fig. 6B). These hydrophobic residues, including key lid-opening residues (Phe176 and Phe211), are not conserved across similar structures, indicating that this lid opening mechanism is unique in CAlipase (Fig. 2).
Fig. 6.
Structural comparison of closed, blocked, and open CAlipase. A: Cross-section of CAlipase structures. A path to the catalytic triad (colored ed) is closed, blocked, and open in each crystal structure. B (Left): Superposition of crystal structures of closed (core domain, cyan; lid domain, pink), blocked (core domain, yellow; lid domain, gold), and open (core domain, pale green; lid domain, green) state of CAlipase. Two phenylalanine residues that show rearrangement are represented as sticks. Distances are given in angstroms. B (Right): Hydrophobic residues around modified serine in open state crystal structure are indicated as sticks. C: B-factor difference plots of open state CAlipase crystal structure. The B-factors of all 26 chains are shown [main chain (left), side chain (right). The lid domain region is highlighted.
Although crystallography only provides a protein’s static structure, crystallographic temperature factors, known as B-factors, constitute a measurement of a structure’s flexibility. Moreover, the open structure of the CAlipase:LPC complex has 26 chains in an asymmetric unit, showing a slightly different structural state. Relatively large mean B-factors are monitored around the lid domain (residues 144-231). Considering B-factors, the closed structure is rigid showing constant values under 40 throughout the whole region, and we could check the flexibility of lid domain region after the introduction of substrate (Fig. 6C). From three different crystal structures, we determined that CAlipase moves its lid domain and that two phenylalanine residues are responsible for the exposure of the core domain active site to the solvent area.
The modified serine residue after hydrolysis
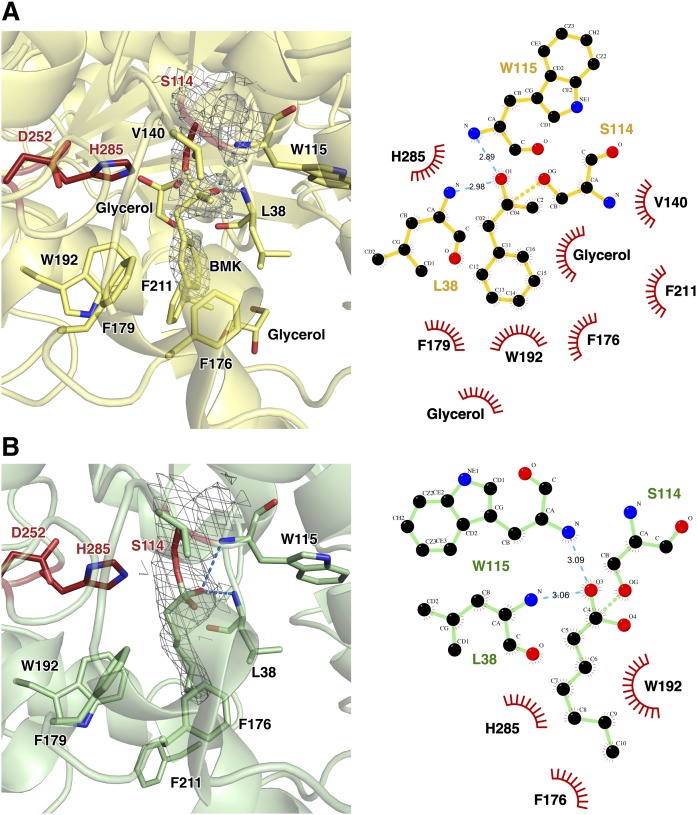
The structure of BMK is similar to PMSF, a well-known serine protease inhibitor that results in sulfonate ester after binding to the active serine residue (supplemental Fig. S3). There are lipase crystal structures with PMSF bound to active serine (PDB ID 5MII, 3RLI, and 3H17), but the PMSF molecule was not added in any step of purification or crystallization in our experiment (40–42). In the blocked crystal structure, Ser114-bound BMK forms stable tetrahedral intermediate coordination. When His285 acquires positive charge, Asp252 is stabilized. The BMK carbonyl oxygen forms hydrogen bonds with the main chain amides of Leu38 and Trp115 (oxyanion hole). The remaining moieties interact dominantly with hydrophobic residues: methyl group with Val140 and benzyl group with Phe176, Phe179, Trp192, and Phe211 (Fig. 7A).
Fig. 7.
Modified serine at residue 114. A: The crystal structure of blocked Ser114 with BMK (blocked structure). The catalytic triad is shown as red sticks. Ligplot showing residues involved in interactions between BMK and CAlipase. The covalently bound BMK forms hydrogen bonds with Leu38 and Trp115. The methyl and benzene moieties are stabilized predominately by hydrophobic interactions with Val140, Phe176, Phe179, Trp192, and Phe211. B: Ser114 is modified as the fatty acid bound form in open structure by introduction of LPC. Ligplot analysis for bound LPC and CAlipase is shown. The hydrophobic interactions between Val140, Phe179, and Phe211 are absent due to the lack of methyl and benzyl groups.
In the CAlipase:LPC crystal structure (open structure), the flexibility of 16 carbon chains led to poor electron density, and they could not be resolved. However, the 2Fo-Fc map indicates a covalently bound ester moiety to Ser114, and about five carbon chains are resolved in each domain. The carbon chains are extended through hydrophobic path of the lid domain. Compared with the blocked structure, although hydrogen bonds between one oxygen atom from LPC is maintained, missing hydrophobic interactions with Val140, Phe179, and Phe211 are detected. Because LPC does not have a methyl group and a sterically bulky benzyl group, Val140 and Phe179 lost their hydrophobic interaction. Phe211 is relocated by ∼5 Å away from the active site in the open structure (Fig. 7B).
DISCUSSION
Acne is the most common skin problem with a prevalence rate of nearly 100%. Most people are affected at some point during their lifetime. Although acne is not a fatal threat, severe acne can lead to disfigurement and permanent scarring. The psychosocial aspects of acne are of great importance for the patients in dealing with interpersonal relationships or study/work (43). Production of an excessively oily substance, known as sebum, can be caused by a number of factors, including hormones, foods, stresses, and hereditary conditions. When the excess sebum forms a plug in the follicle, C. acnes in the follicle digests sebum, creating inflammation and leading to papules, pustules, nodules, or cysts. C. acnes is a normally harmless bacteria, but recent studies have revealed that the proportion of C. acnes strains, not the population of bacteria, changes between healthy and acne patients (5, 9). Type IA strains are common in patients with inflammatory lesions, while types IB/IC and II are not. This is due to the different lipases expressed from diverse typed strains (11–13). In the present study, we focused a highly conserved lipase across types IA, IB, IC, and II but truncated only in virulence type IA C. acnes.
At present, the available remedies, such as benzoyl peroxide, retinoids, and antibiotics, focus on reducing symptoms. However, there is no direct drug treatment for acne. Because of limited studies about the pathology, the aim for the therapy is relieving symptoms or killing bacteria using antibiotics. To facilitate intelligent drug design, high-resolution structures provide a starting point for structure-based drug design and enable comparisons to find successful approaches in similar species. However, in the case of lipases, the activity is hard to predict because of their poor conservation in lid domains. The crystal structures of closed, blocked, and open CAlipases, which are the first structures of CAlipase, revealed the novel lid opening mechanism (Fig. 8, supplemental Movie S1). In healthy skin, lipase controls its activity depending on environmental lipid level. When the lipase is exposed to a certain lipid level, the loop between the α7- and α8-helix moves by ∼2.5 Å, providing enough space for Phe211 side chain flipping. Additionally, Phe176 changes its position to open up the blockage and make the active site ligand accessible. Hydrophobic residues, such as Phe179 and Trp192, form a lipophilic path for carbon chains of substrate and amides from Leu38 and Trp115 constitute oxyanion hole for the catalytic reaction. This pattern is unique compared with previously known lid-opening mechanisms. Candida antarctica lipase B (CALB) structure opens up by releasing salt bridges between aspartate and lysine residues. A whole α-helix movement makes a path for the fatty acids in Saccharomyces cerevisiae monoglyceride lipase Yju3p (PDB ID 4ZXF) (31, 44). To date, genome sequencing for various types of C. acnes strains are being studied and the biological behavior of type IA is now being revealed. From our study, we could gain insight into the properties of conserved CAlipase. Further research on various CAlipases, exposed to different environments, will aid novel inhibitor development, which has been neglected for over a decade.
Fig. 8.
Mode of action of CAlipase. In aqueous solution, the major form of lipase is the closed form, shielding the active site (colored red). However, when the lipase is exposed to a high level of lipid, the lid domain moves to reveal the active site. Two bulky side chain residues, Phe176 and Phe211, are responsible for the lid opening. The entry of the substrate is helped by the lipophilic path, followed by the catalytic reaction through the active site and oxyanion hole (colored red and purple, respectively).
Supplementary Material
Acknowledgments
The authors are grateful to the staff of Pohang Light Source (beamlines 5C and 7A) in Korea for their help with the X-ray experiments. The authors also thank to Dr. Weng Chan from the University of Nottingham and Dr. Sarah Kuehne from the University of Birmingham for informative discussions.
Footnotes
Abbreviations:
- BMK
- benzyl methyl ketone
- CAlipase
- Cutibacterium acnes lipase)
- DTNB
- 5,5-dithiobis(2-nitrobenzoic acid)
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- PDB ID
- Protein Data Bank identification number
This work was supported through the National Research Foundation of Korea (NRF) funded by the Ministry of Education of the Korean government (Grants 2017R1D1A1B03033857 and 2018R1A5A2024425). The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Poli F., Dreno B., and Verschoore M.. 2001. An epidemiological study of acne in female adults: results of a survey conducted in France. J. Eur. Acad. Dermatol. Venereol. 15: 541–545. [DOI] [PubMed] [Google Scholar]
- 2.Tasoula E., Gregoriou S., Chalikias J., Lazarou D., Danopoulou I., Katsambas A., and Rigopoulos D.. 2012. The impact of acne vulgaris on quality of life and psychic health in young adolescents in Greece. Results of a population survey. An. Bras. Dermatol. 87: 862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li D., Chen Q., Liu Y., Liu T., Tang W., and Li S.. 2017. The prevalence of acne in Mainland China: a systematic review and meta-analysis. BMJ Open. 7: e015354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta M. A., and Gupta A. K.. 1998. Depression and suicidal ideation in dermatology patients with acne, alopecia areata, atopic dermatitis and psoriasis. Br. J. Dermatol. 139: 846–850. [DOI] [PubMed] [Google Scholar]
- 5.Dréno B., Pécastaings S., Corvec S., Veraldi S., Khammari A., and Roques C.. 2018. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J. Eur. Acad. Dermatol. Venereol. 32 (Suppl. 2): 5–14. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y.b., Byun E. J., and Kim H. S.. 2019. Potential role of the microbiome in acne: a comprehensive review. J. Clin. Med. 8: e987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mollerup S., Friis-Nielsen J., Vinner L., Hansen T. A., Richter S. R., Fridholm H., Herrera J. A., Lund O., Brunak S., Izarzugaza J. M., et al. . 2016. Propionibacterium acnes: disease-causing agent or common contaminant? Detection in diverse patient samples by next-generation sequencing. J. Clin. Microbiol. 54: 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cogen A. L., Nizet V., and Gallo R. L.. 2008. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 158: 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland C., Mak T. N., Zimny-Arndt U., Schmid M., Meyer T. F., Jungblut P. R., and Bruggemann H.. 2010. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 10: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDowell A., Barnard E., Nagy I., Gao A., Tomida S., Li H., Eady A., Cove J., Nord C. E., and Patrick S.. 2012. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic’, ‘commensal’ and antibiotic resistant strains. PLoS One. 7: e41480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon H. H., Yoon J. Y., Park S. Y., and Suh D. H.. 2013. Analysis of distribution patterns of Propionibacterium acnes phylotypes and Peptostreptococcus species from acne lesions. Br. J. Dermatol. 169: 1152–1155. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y., Champer J., Agak G. W., Kao S., Modlin R. L., and Kim J.. 2016. Different Propionibacterium acnes phylotypes induce distinct immune responses and express unique surface and secreted proteomes. J. Invest. Dermatol. 136: 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDowell A., Perry A. L., Lambert P. A., and Patrick S.. 2008. A new phylogenetic group of Propionibacterium acnes. J. Med. Microbiol. 57: 218–224. [DOI] [PubMed] [Google Scholar]
- 14.Lomholt H. B., and Kilian M.. 2010. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 5: e12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakase K., Hayashi N., Akiyama Y., Aoki S., and Noguchi N.. 2017. Antimicrobial susceptibility and phylogenetic analysis of Propionibacterium acnes isolated from acne patients in Japan between 2013 and 2015. J. Dermatol. 44: 1248–1254. [DOI] [PubMed] [Google Scholar]
- 16.Tomida S., Nguyen L., Chiu B. H., Liu J., Sodergren E., Weinstock G. M., and Li H.. 2013. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 4: e00003–e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achermann Y., Goldstein E. J., Coenye T., and Shirtliff M. E.. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin. Microbiol. Rev. 27: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horváth B., Hunyadkurti J., Voros A., Fekete C., Urban E., Kemeny L., and Nagy I.. 2012. Genome sequence of Propionibacterium acnes type II strain ATCC 11828. J. Bacteriol. 194: 202–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasimatis G., Fitz-Gibbon S., Tomida S., Wong M., and Li H.. 2013. Analysis of complete genomes of Propionibacterium acnes reveals a novel plasmid and increased pseudogenes in an acne associated strain. BioMed Res. Int. 2013: 918320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z., and Minor W.. 1997. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- 21.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., et al. . 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W.. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65: 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin A., and Teplyakov A.. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66: 22–25. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Lohkamp B., Scott W. G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 26.Petrovic N., Grove C., Langton P. E., Misso N. L., and Thompson P. J.. 2001. A simple assay for a human serum phospholipase A2 that is associated with high-density lipoproteins. J. Lipid Res. 42: 1706–1713. [PubMed] [Google Scholar]
- 27.Santana C. C., Barbosa L. A., Junior I. D. B., Nascimento T. G. D., Dornelas C. B., and Grillo L. A. M.. 2017. Lipase activity in the larval midgut of Rhynchophorus palmarum: biochemical characterization and the effects of reducing agents. Insects. 8: E100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higaki S., Kitagawa T., Kagoura M., Morohashi M., and Yamagishi T.. 2000. Correlation between Propionibacterium acnes biotypes, lipase activity and rash degree in acne patients. J. Dermatol. 27: 519–522. [DOI] [PubMed] [Google Scholar]
- 29.Brüggemann H., Lomholt H. B., Tettelin H., and Kilian M.. 2012. CRISPR/cas loci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I P. acnes. PLoS One. 7: e34171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labar G., Bauvois C., Borel F., Ferrer J. L., Wouters J., and Lambert D. M.. 2010. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. ChemBioChem. 11: 218–227. [DOI] [PubMed] [Google Scholar]
- 31.Stauch B., Fisher S. J., and Cianci M.. 2015. Open and closed states of Candida antarctica lipase B: protonation and the mechanism of interfacial activation. J. Lipid Res. 56: 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaschner M., Schillinger O., Fettweiss T., Nutschel C., Krause F., Fulton A., Strodel B., Stadler A., Jaeger K. E., and Krauss U.. 2017. A combination of mutational and computational scanning guides the design of an artificial ligand-binding controlled lipase. Sci. Rep. 7: 42592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Vitis V., Nakhnoukh C., Pinto A., Contente M. L., Barbiroli A., Milani M., Bolognesi M., Molinari F., Gourlay L. J., and Romano D.. 2018. A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold. FEBS J. 285: 903–914. [DOI] [PubMed] [Google Scholar]
- 34.Nardini M., and Dijkstra B. W.. 1999. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9: 732–737. [DOI] [PubMed] [Google Scholar]
- 35.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., et al. . 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46: W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan F. I., Lan D., Durrani R., Huan W., Zhao Z., and Wang Y.. 2017. The lid domain in lipases: structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marples R. R., Downing D. T., and Kligman A. M.. 1971. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J. Invest. Dermatol. 56: 127–131. [DOI] [PubMed] [Google Scholar]
- 38.Greene R. S., Downing D. T., Pochi P. E., and Strauss J. S.. 1970. Anatomical variation in the amount and composition of human skin surface lipid. J. Invest. Dermatol. 54: 240–247. [DOI] [PubMed] [Google Scholar]
- 39.Skjold-Jørgensen J., Bhatia V. K., Vind J., Svendsen A., Bjerrum M. J., and Farrens D.. 2015. The enzymatic activity of lipases correlates with polarity-induced conformational changes: a Trp-induced quenching fluorescence study. Biochemistry. 54: 4186–4196. [DOI] [PubMed] [Google Scholar]
- 40.Nam K. H., Kim S. J., Priyadarshi A., Kim H. S., and Hwang K. Y.. 2009. The crystal structure of an HSL-homolog EstE5 complex with PMSF reveals a unique configuration that inhibits the nucleophile Ser144 in catalytic triads. Biochem. Biophys. Res. Commun. 389: 247–250. [DOI] [PubMed] [Google Scholar]
- 41.Rengachari S., Bezerra G. A., Riegler-Berket L., Gruber C. C., Sturm C., Taschler U., Boeszoermenyi A., Dreveny I., Zimmermann R., Gruber K., et al. . 2012. The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim. Biophys. Acta. 1821: 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavazzini D., Grossi G., Levati E., Vallese F., Montanini B., Bolchi A., Zanotti G., and Ottonello S.. 2017. A family of archaea-like carboxylesterases preferentially expressed in the symbiotic phase of the mychorrizal fungus Tuber melanosporum. Sci. Rep. 7: 7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazarika N., and Archana M.. 2016. The psychosocial impact of acne vulgaris. Indian J. Dermatol. 61: 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aschauer P., Rengachari S., Lichtenegger J., Schittmayer M., Das K. M., Mayer N., Breinbauer R., Birner-Gruenberger R., Gruber C. C., Zimmermann R., et al. . 2016. Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p. Biochim. Biophys. Acta. 1861: 462–470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein coordinates and structure factors have been deposited in the Research Collaboratory for Structural Informatics Protein Data Bank (RCSB-PDB) under code 6KHK for closed, 6KHL for blocked, and 6KHM for open state structures.